Significance
Alternaria brassicicola infection causes dark spots on the leaves of most Brassica species, reducing the yield of economically important oilseed crops. In response to A. brassicicola infection, Arabidopsis thaliana and other Brassicaceae produce and secrete camalexin, a major phytoalexin imparting resistance to A. brassicicola. Because camalexin is toxic to the plant itself, specific transporters are needed for secretion. Here we show that the ABC transporter ABCG34 mediates the secretion of camalexin from the epidermal cells to the surface of leaves and thereby confers resistance to A. brassicicola infection. This work establishes a complete picture of a plant defense system, consisting of a toxic secondary metabolite, its transporter, and the disease phenotype caused by an economically important pathogen.
Keywords: AtABCG34, ABC transporters, camalexin, A. brassicicola, B. cinerea
Abstract
Plant pathogens cause huge yield losses. Plant defense often depends on toxic secondary metabolites that inhibit pathogen growth. Because most secondary metabolites are also toxic to the plant, specific transporters are needed to deliver them to the pathogens. To identify the transporters that function in plant defense, we screened Arabidopsis thaliana mutants of full-size ABCG transporters for hypersensitivity to sclareol, an antifungal compound. We found that atabcg34 mutants were hypersensitive to sclareol and to the necrotrophic fungi Alternaria brassicicola and Botrytis cinerea. AtABCG34 expression was induced by A. brassicicola inoculation as well as by methyl-jasmonate, a defense-related phytohormone, and AtABCG34 was polarly localized at the external face of the plasma membrane of epidermal cells of leaves and roots. atabcg34 mutants secreted less camalexin, a major phytoalexin in A. thaliana, whereas plants overexpressing AtABCG34 secreted more camalexin to the leaf surface and were more resistant to the pathogen. When treated with exogenous camalexin, atabcg34 mutants exhibited hypersensitivity, whereas BY2 cells expressing AtABCG34 exhibited improved resistance. Analyses of natural Arabidopsis accessions revealed that AtABCG34 contributes to the disease resistance in naturally occurring genetic variants, albeit to a small extent. Together, our data suggest that AtABCG34 mediates camalexin secretion to the leaf surface and thereby prevents A. brassicicola infection.
Plants are exposed to a multitude of pathogens, but they usually resist infection by using their unique defense systems and compounds, including secondary metabolites produced either constitutively (phytoanticipins) or in response to pathogen attack (phytoalexins). Plants produce tens of thousands of secondary metabolites, which are classified as phenolics, terpenoids, alkaloids, glucosinolates, cyanogenic glucosides, and betanins. Secondary metabolites are secreted from the infected cells and their surrounding cells after pathogen attack or are released, hydrolyzed, and become toxic when the cells are destroyed by pathogens (1), thus inhibiting pathogen growth on the plant. Many secondary metabolites are dangerous to the plants because of their toxicity to cellular metabolism. To avoid self-toxicity, plants either secrete these compounds to the leaf surface or sequester them in the vacuole, a compartment with low metabolic activity. In both cases, specific transporters are required either to deliver the secondary metabolites to the site where the pathogens are located or to store them as weapons against pathogen attack.
Several full-size ABCG transporters are involved in the transport of secondary metabolites. Nicotiana plumbaginifolia PDR1/ABC1 and Nicotiana tabacum PDR1 are induced by jasmonate, a defense-related hormone highly expressed in the leaf epidermal cells and trichomes, and transport sclareol, a diterpene alcohol secreted by Nicotiana species in response to pathogen attack (2–4). Two functionally redundant Nicotiana benthamiana full-size ABCG transporters, NbABCG1 and NbABCG2, are essential for resistance to Phytophthora infestans. Mutants with defects in these two ABCG transporters export much less capsidiol than do the corresponding wild-type plants, indicating that these transporters transport capsidiol (5). NtABCG5/NtPDR5 is induced by insect herbivory, and the absence of this transporter decreases resistance to insects, indicating that it is involved in the transport of a hitherto unidentified insecticidal compound (6). Absence of PhPDR2 increases insect herbivory in Petunia hybrida leaves and decreases concentrations of the insecticidal steroid petuniasterone (7). CrTPT2, expressed at the plasma membrane of Catharanthus roseus, secretes catharanthine to the leaf surface (8). Lr34, an ABCG transporter in Triticum aestivum (wheat), is one of the few durable resistance genes that protect the plant from multiple pathogens, namely Puccinia triticina and Puccinia striiformis, which cause leaf rust, and Blumeria graminis, which causes powdery mildew (9). Furthermore, when heterologously expressed in Oryza sativa (rice), Lr34 confers resistance to multiple isolates of Magnaporthe oryzae (10) and therefore is a promising candidate ABC transporter to improve crop plants’ resistance to a broad spectrum of pathogens. However, the substrate transported by Lr34 is still unknown.
Among the 129 ABC proteins present in Arabidopsis thaliana, only two have been implicated in the defense response. PDR8/ABCG36/PEN3, which is localized at the plasma membrane, recruited to infection sites (11), and involved in glucosinolate-dependent pathogen defense at the contact site (12), blocks the penetration of nonhost fungal pathogens (13). Recently, a report suggested that this protein transports 4-O-β-d-glucosyl-indol-3-yl formamide (4OGlcI3F) (14); however, this possibility remains to be proven. atabcg40/pdr12 mutants exhibit sensitivity to sclareol, and AtABCG40/PDR12 is induced by pathogen inoculation (15). However, sclareol is not likely to be the natural substrate of AtABCG40, because it is not synthesized in A. thaliana. Thus, AtABCG40 might transport chemicals similar to sclareol that are produced by A. thaliana in response to pathogen attack.
We hypothesized that additional ABCG transporters are involved in pathogen defense, because (i) the transcript levels of many additional full-size ABCG transporters have been reported to be up-regulated in plants exposed to pathogens (16, 17) and (ii) pathogen defense is often mediated by secondary metabolites, many of which are transported by ABC transporters (18). To identify such additional ABCG transporters involved in plant defense against pathogens, we screened A. thaliana transfer DNA (T-DNA) insertion mutants of full-size ABCG genes for altered sensitivity to the secondary metabolite sclareol, because sclareol hypersensitivity might provide a clue as to which ABCG proteins are involved in secondary metabolite transport. Having established that atabcg34 is hypersensitive to sclareol, we analyzed the function of AtABCG34 in relation to the transport of secondary metabolites implicated in pathogen defense.
Results
atabcg34 Mutants Exhibit Hypersensitivity to Sclareol.
To identify additional transporters involved in pathogen defense, we exposed 13 of the 15 full-length A. thaliana abcg transporter mutants (knockout mutants of atabcg42 and atabcg43 were not available at the time of screening) to a toxic concentration of sclareol (65 µM). Among the mutants, atabcg34-1 (ko-1; SAIL_5_G10) and atabcg34-2 (ko-2; SALK_036087) (Fig. S1) exhibited the strongest reduction in both fresh weight and root length in sclareol-containing medium (Fig. 1 A, C, and D). Expression of the genomic fragment of AtABCG34 (ABCG34pro:sGFP:ABCG34; genomic DNA) rescued the mutants from sensitivity to sclareol; both fresh weight and root length of the complementation lines recovered to levels similar to those of wild-type plants in half-strength Murashige–Skoog (1/2MS) medium supplemented with 65 µM sclareol (Fig. 1 B–D for ko-1 and Fig. S2 for ko-2). Interestingly, although AtABCG39 is the closest relative of AtABCG34 among the full-size ABCG members (Fig. S3A), and its amino acid sequence is 89% identical with that of AtABCG34, the atabcg39 mutant was not sensitive to sclareol (Fig. S3 B and C).
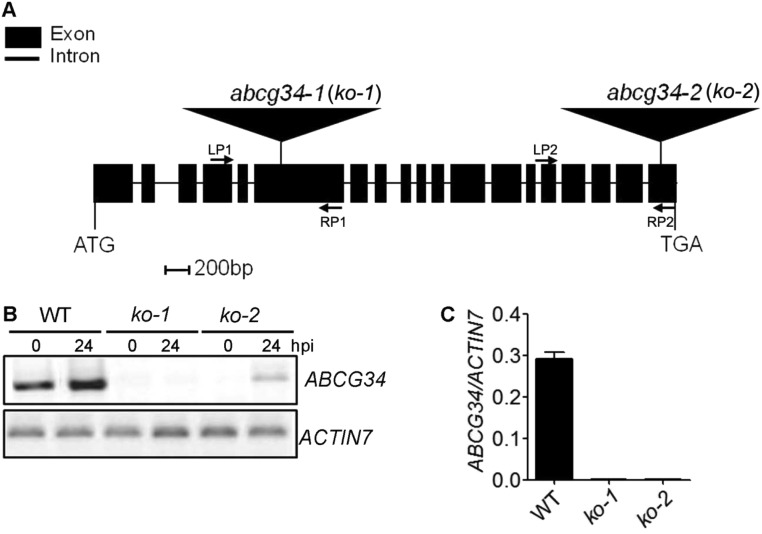
Fig. S1.
Isolation of atabcg34-knockout mutants. (A) Structure of AtABCG34 showing the positions of the T-DNA insertions. Exons and introns are indicated by black boxes and lines, respectively. The T-DNA insertion sites in the SAIL_5_G10 (atabcg34-1) and SALK_036087 (atabcg34-2) mutants are indicated by flags. (B) RT-PCR analysis of AtABCG34 expression 24 hpi in plants inoculated with A. brassicicola (5 × 105 spores/mL) and in untreated plants. Total RNA was extracted from 4-wk-old wild-type (WT), ko-1 (atabcg34-1), and ko-2 (atabcg34-2) leaves. AtACTIN7 was used as a loading control. A low level of AtABCG34 was induced in the ko-2 mutant at 24 hpi with A. brassicicola. (C) The graph represents mean values (±SE) of transcript levels of AtABCG34 in wild-type and abcg34 (ko-1 and ko-2) mutant seedlings, analyzed by qRT-PCR and normalized by AtACTIN7 transcripts from three technical replicates (n = 3).
Fig. 1.
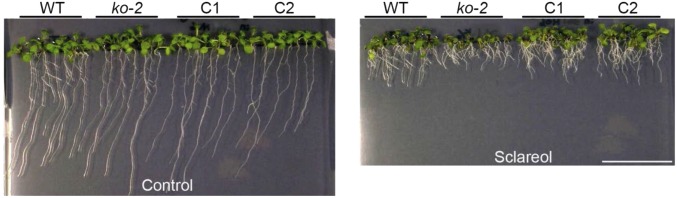
Sclareol-sensitive phenotype of atabcg34-knockout plants. (A) atabcg34 mutants (ko-1 and ko-2) exhibit enhanced sclareol sensitivity compared with the wild-type plants. Plants were grown on 1/2MS-agar plates in the absence (Control) or presence of 65 µM sclareol for 2 wk. (B) Sclareol tolerance was restored in complementation lines (C1–C3) expressing ABCG34pro:sGFP:ABCG34 in the ko-1 background. Plants were grown on 1/2MS-agar plates without (Control) or with supplementation of 65 µM sclareol for 2 wk. (Scale bar, 1 cm.) (C and D) Plant growth quantified by measuring fresh weight (C) and the longest root length (D) of 2-wk-old plants. Results are mean values (±SE) of three independent experiments with three replicates each. Different lowercase letters indicate that the means are significantly different between genotypes or treatments [Tukey’s honestly significant difference (HSD) test; P < 0.01; n = 3].
Fig. S2.
The sclareol sensitivity of abcg34-2 is restored by complementation of ABCG34pro:sGFP:ABCG34 in the atabcg34-2 (ko-2) background (C1 and C2). Plants were grown for 2 wk on 1/2MS-agar plates in the absence (Control) (Left) or presence (Right) of 65 μM sclareol (n = 3). (Scale bar, 1 cm.)
Fig. S3.
Phylogenic tree of A. thaliana full-size ABCG proteins and growth of atabcg39 mutants in sclareol-containing medium. (A) Phylogenic tree of A. thaliana full-size ABCG (PDR) proteins produced using MEGA4 software. NpPDR1 clusters with AtABCG34, AtABCG39, and AtABCG40. (B) qRT-PCR analysis of the AtABCG39 transcript. The graph represents mean values (±SE) of transcript levels of AtABCG39 in wild-type plants and atabcg39 mutants. AtACTIN7 was used as an internal control (n = 3). (C) Growth of the atabcg39 mutant compared with wild-type plants. Plants were grown on 1/2MS-agar plates without (Control) or with supplementation with 65 μM sclareol for 3 wk (n = 3). (Scale bar, 1 cm.)
AtABCG34 Contributes to Plant Defense Against Necrotrophic Fungi.
Because sclareol is known to restrict fungal growth (19, 20), we speculated that AtABCG34 might be involved in defense against fungal pathogens. We evaluated this possibility by inoculating the atabcg34 mutants (ko-1 and ko-2) with two different necrotrophic fungi, Botrytis cinerea and Alternaria brassicicola. Although the wild-type leaves inoculated with the two pathogens became necrotic only at the inoculation sites, the mutant leaves exhibited much more extensive necrotic areas upon infection with A. brassicicola (Fig. 2 A and B) or B. cinerea (Fig. S4A, Left), suggesting that the mutants were hypersensitive to both necrotrophic fungal pathogens. Quantification of the pathogen growth confirmed that the atabcg34 mutants were hypersensitive to the fungal pathogens. A. brassicicola growth, quantified using qPCR of A. brassicicola cutinase A genomic DNA relative to A. thaliana α-shaggy kinase genomic DNA, was higher in the mutant leaves than in leaves of the wild-type plants or the complementation lines (Fig. 2C). B. cinerea growth, measured by counting spore numbers (Fig. S4A, Right), was also higher in the mutant leaves than in the wild-type leaves. By contrast, the mutants’ response to a bacterial pathogen Pseudomonas syringae pv. tomato DC3000 (Pst DC3000) was similar to that of the wild-type plants (Fig. S4B). Furthermore, expression of the genomic fragment of AtABCG34 (ABCG34pro:sGFP:ABCG34; genomic DNA) rescued the mutants from sensitivity to A. brassicicola (Fig. 2).
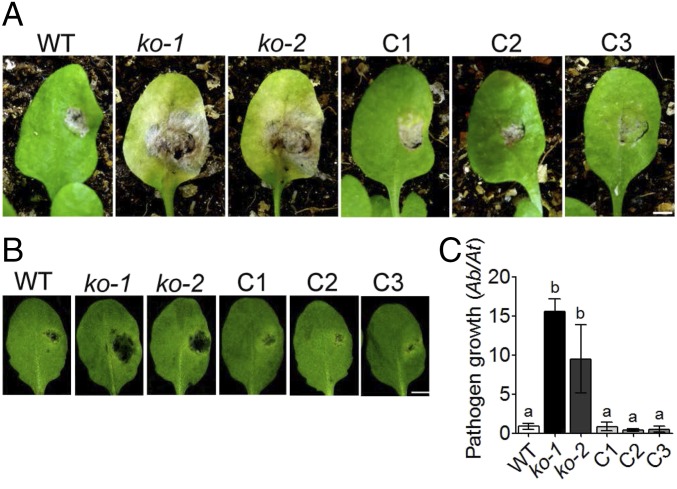
Fig. 2.
AtABCG34 is required for resistance to A. brassicicola. (A and B) Disease symptoms of atabcg34 mutants (ko-1 and ko-2) and complementation lines (C1–C3) on intact plants at 2 wk postinoculation (A) and on detached leaves 5 dpi (B). (Scale bar, 5 mm.) (C) Fungal growth, as determined by amplification of the A. brassicicola cutinase A (Ab) gene relative to the A. thaliana α shaggy kinase (At) gene by qPCR at 5 dpi with A. brassicicola (5 × 105 spores/mL). The graph represents mean values (±SE) of three biological replicates with at least 21 disease lesions. Different lowercase letters indicate that the means are significantly different between the genotypes (Tukey’s HSD test, P < 0.01; n = 3).
Fig. S4.
AtABCG34 is required for resistance to B. cinerea but not for Pst DC3000. (A) Disease symptoms on detached leaves of atabcg34 mutants (ko-1 and ko-2) and wild-type plants after B. cinerea treatment. (Left) Photographs of the detached leaves. (Scale bar, 1 cm.) (Right) The graph represents mean values (±SE) of fungal growth, as determined by counting the spores (number per unit area) at 5 dpi. n = 14 for spore population. (B, Left) Photographs showing that the atabcg34 mutants and wild-type plants exhibited no difference in sensitivity to Pst DC3000 (n = 10). (Right) The graph represents mean values (±SE) of bacterial growth at 4 dpi. cfu, colony-forming units.
AtABCG34 Expression Is Induced by Methyl Jasmonate and Necrotrophic Pathogens.
We then analyzed whether the expression level of AtABCG34 changes in response to treatment with either methyl jasmonate (MeJA) or salicylic acid (SA), hormones that are known to function in the pathogen-defense response. AtABCG34 expression was induced up to sevenfold within 1 h of MeJA treatment (Fig. 3A) but was only slightly induced by SA treatment (Fig. 3B). Next, we analyzed the transcript levels of AtABCG34 in plants exposed to necrotrophic fungi (A. brassicicola and B. cinerea). AtABCG34 transcript levels increased three- to sixfold in a time-dependent manner in response to treatment with necrotrophic fungal pathogens (Fig. 3C and Fig. S5A).
Fig. 3.
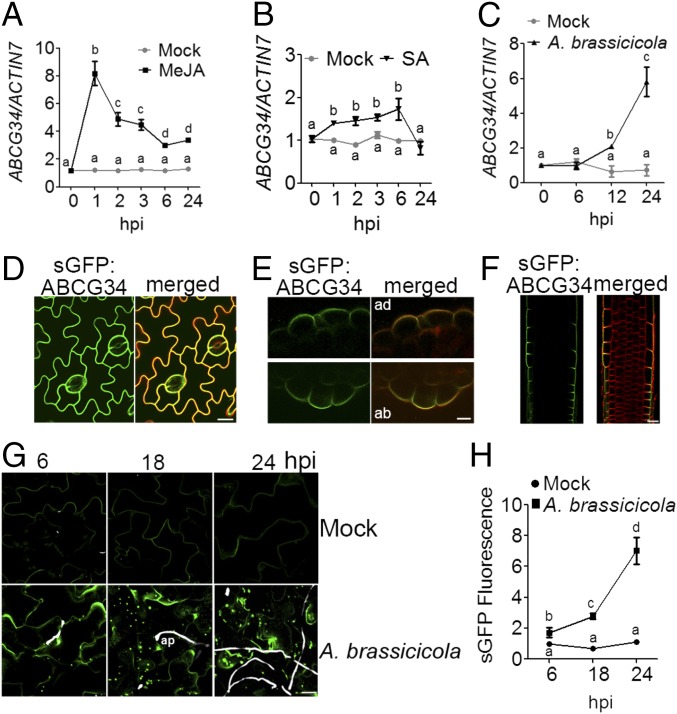
Expression profile after treatment with MeJA, SA, and A. brassicicola and polar localization of AtABCG34. (A–C) Time-dependent changes in the transcript level of AtABCG34 in mature leaves treated with 50 µM MeJA (A), 1 mM SA (B), or A. brassicicola (5 × 105 spores/mL) (C), quantified relative to that of AtACTIN7. Data represent mean values (±SE) of three independent experiments. Different lowercase letters indicate that the means are significantly different (Tukey’s HSD test, P < 0.01; n = 3). (D) Plasma membrane localization of ABCG34pro:sGFP:ABCG34 expressed in the ko-1 mutant at the epidermal cells of the mature rosette leaves. The images were taken at a higher gain than the images in G. (E and F) Polar localization of sGFP:ABCG34 in transverse sections of mature leaf at the adaxial (Upper) and abaxial (Lower) surface (E) and root (F). FM4-64 (10 µM) was used as a plasma membrane marker. (Scale bars in D–F, 10 µm.) (G and H) Increase in GFP fluorescence of ABCG34pro:sGFP:ABCG34-expression in the epidermal cells of leaves inoculated with A. brassicicola (5 × 105 spores/mL) observed at 6, 18, and 24 hpi (G). (Scale bar, 70 µm.) Fungal hyphae (white) were stained with Calcofluor white stain. ap, appressorium. In H, sGFP fluorescence intensity was quantified and normalized to that from mock-treated cells at 6 hpi, which was set to 1, using ImageJ, from photographs as shown in G. Different lowercase letters indicate that the means are significantly different (Tukey’s HSD test, P < 0.05; n = 10).
Fig. S5.
Expression profile of AtABCG34 after treatment with B. cinerea. (A) Transcript level of AtABCG34 after B. cinerea treatment. The transcript was analyzed by qRT-PCR after mock or B. cinerea treatment at different time points. Data represent mean values (±SE) of three independent experiments (n = 3). Different lowercase letters indicate that the means are significantly different (Tukey’s HSD test, P < 0.01). (B) Leaves of 3-wk-old ABCG34pro:GUS-expressing plants were inoculated with buffer or with B. cinerea. Asterisks indicate the site of inoculation. The leaves were punctured with needles before inoculation for efficient infection. A GUS assay was executed at 3 dpi.
β-Glucuronidase (GUS) staining of ABCG34pro:GUS lines at 3 d postinoculation (dpi) also revealed the high expression of AtABCG34 around the B. cinerea infection sites (Fig. S5B). Thus, AtABCG34 expression was strongly induced by necrotrophic fungal pathogens (A. brassicicola and B. cinerea) and by MeJA but not by SA.
AtABCG34 Displays Polar Localization at the Outer Surface of the Plasma Membrane of Epidermal Cells.
Next, we examined the subcellular localization of AtABCG34 in complementation lines expressing ABCG34pro:sGFP:ABCG34 in the atabcg34-1 background. The leaves of the transgenic plants were stained with FM4-64 for only 10 min on ice to prevent the dye from entering the cells. Under this condition, the red fluorescence of FM4-64 delineated only the plasma membrane, and the green fluorescence of sGFP-ABCG34 was colocalized with the red fluorescence (Fig. 3D), suggesting that AtABCG34 is localized to the plasma membrane. Interestingly, observation of the transverse sections of the leaves revealed that AtABCG34 was polarly localized at the outer half of the epidermal cells at both the adaxial and the abaxial surfaces (Fig. 3E). A similar pattern of polar localization at the outer surface of the epidermal cells was observed in the root (Fig. 3F).
To determine whether pathogens affect AtABCG34 localization, we observed synthetic GFP (sGFP) fluorescence in leaves expressing ABCG34pro:sGFP:ABCG34 in the atabcg34-1 background. When treated with A. brassicicola, the green fluorescence of sGFP:ABCG34 increased continuously until 24 h postinoculation (hpi) (Fig. 3 G and H), indicating induction of AtABCG34 by the pathogen, in agreement with the results of our qRT-PCR analysis (Fig. 3C and Fig. S5A) and GUS expression assay (Fig. S5B). Interestingly, in contrast to nontreated plants, the fluorescence was localized not only at the plasma membrane but also in dot-like structures in the cells (Fig. 3G).
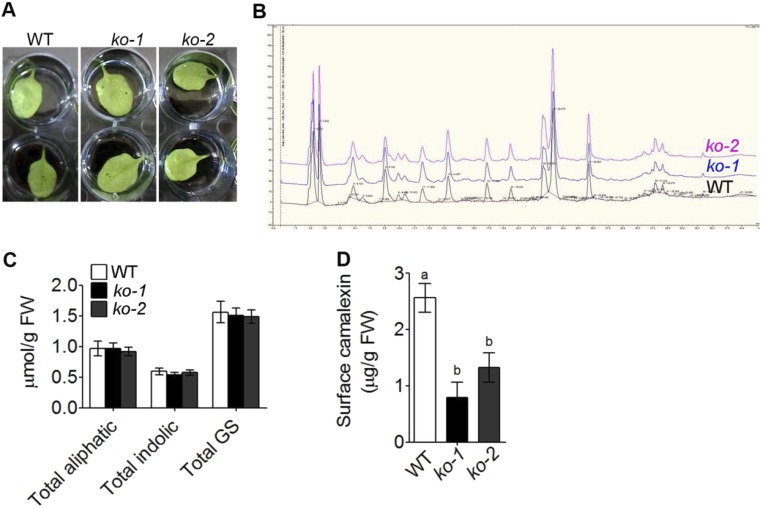
atabcg34 Exhibits Reduced Camalexin Abundance at the Leaf Surface.
The polar localization of AtABCG34 at the plasma membrane and hypersensitivity of atabcg34 to necrotrophic pathogens suggest that AtABCG34 may be required for the secretion of chemicals that protect plants from pathogen attack. Two different classes of such chemicals may be involved in this process: surface-coating materials, as shown for AtABCG32/PEC1 (21, 22), and secondary metabolites, as shown for AtABCG36/PDR8/PEN3 (14). We tested the first possibility by an ethanol penetration assay that detects permeability defects but could not observe any difference in ethanol penetration between the wild-type plants and atabcg34 mutants (Fig. S6A).
Fig. S6.
AtABCG34 is not required for cutin formation or for the transport of glucosinolates and phenolic compounds. (A) Ethanol penetration assay for cuticle permeability. The leaves of 3-wk-old wild-type plants and atabcg34 mutants (ko-1, ko-2) were immersed in 100% ethanol, and penetration was assessed after 5 min of immersion (n = 3). (B) Chromatogram of phenolics extracted from wild-type and atabcg34 (ko-1 and ko-2) mutant plants at 24 hpi with B. cinerea. The experiment was performed using four individuals per genotype (n = 2). (C) Glucosinolates measured from wild-type plants and atabcg34 (ko-1 and ko-2) mutants at 24 hpi with A. brassicicola. The graph represents mean values (±SE) of two independent experiments with three replicates each (n = 2). (D) Surface camalexin measured from whole rosettes of wild-type and atabcg34 (ko-1 and ko-2) mutant plants at 48 hpi of A. brassicicola. The graph represents mean values (±SE) from five different experiments with at least six rosettes per genotype in each experiment. Different lowercase letters indicate that the means are significantly different between genotypes (Tukey’s HSD test, P < 0.01; n = 5).
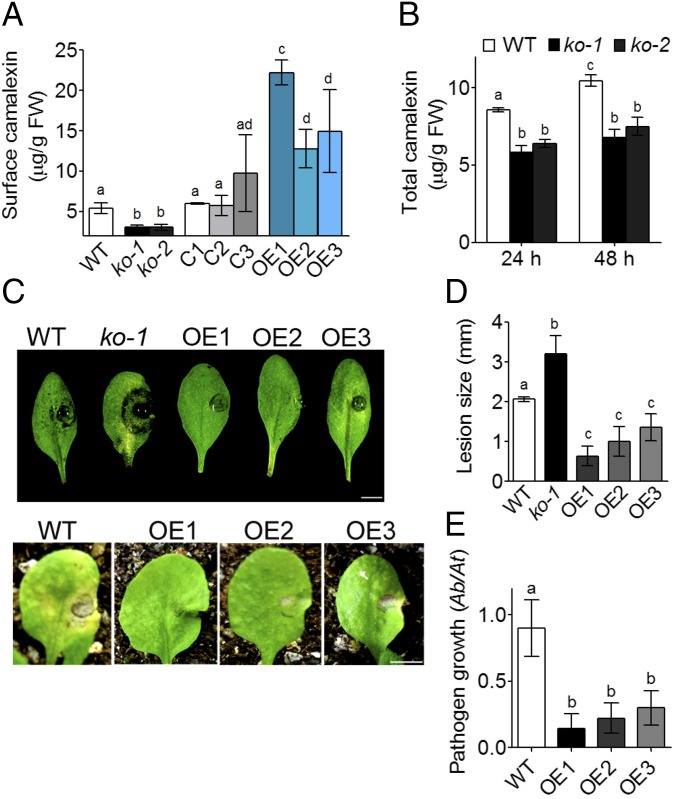
To test the second possibility, that AtABCG34 mediates the secretion of secondary metabolites, we compared the levels of some secondary metabolites in the wild-type plants and the atabcg34 mutants. The levels and the kinds of phenolic compounds in plants infected with B. cinerea did not differ between the atabcg34 mutants and wild-type plants (Fig. S6B). Furthermore, there was no distinct difference in glucosinolate content in plants infected by A. brassicicola (Fig. S6C). Camalexin is a major phytoalexin produced by A. thaliana and other Brassicaceae and is known to be induced by A. brassicicola infection and to be required for resistance against A. brassicicola (23). Consequently, we determined the camalexin contents at the surface of whole rosettes and observed that it was reduced to about half in atabcg34 mutants compared with the wild-type plants at both 24 (Fig. 4A) and 48 hpi (Fig. S6D) with A. brassicicola. At 24 hpi the camalexin levels at the rosette leaf surface in the complementation lines were similar to those in the wild-type plants (Fig. 4A). The total camalexin content of the whole rosette of atabcg34 mutants was also reduced compared with that of the wild-type plants at 24 and 48 hpi with A. brassicicola (Fig. 4B). These data suggest that AtABCG34 is necessary for camalexin secretion to the surface upon A. brassicicola infection.
Fig. 4.
AtABCG34 mediates the secretion of camalexin to the leaf surface. (A) Surface camalexin measured from whole rosettes of wild-type plants, atabcg34 lines (ko-1 and ko-2), complementation lines (C1–C3), and overexpression lines (OE1–OE3) expressing 35Spro:sGFP:ABCG34 in the wild-type background at 24 hpi of A. brassicicola (5 × 105 spores/mL). The graph represents mean values (±SE) from five different experiments with at least six rosettes per genotype per experiment. Different lowercase letters indicate that the means are significantly different between genotypes (Tukey’s HSD test, P < 0.01; n = 5). (B) Total camalexin measured from whole rosettes of wild-type A. thaliana and atabcg34 (ko-1 and ko-2) mutants at 24 hpi of A. brassicicola. Graphs represent mean values (±SE) from five different experiments with at least six rosettes per genotype for each time point in each experiment. Different lowercase letters indicate that the means are significantly different between genotypes or time points (Tukey’s HSD test, P < 0.01; n = 5). (C) Photographs of disease symptoms of wild-type plants, atabcg34-1 mutants, and AtABCG34-overexpressing lines (OE1–OE3) on detached leaves (Upper) and intact plants (Lower) taken at 3 and 14 dpi of A. brassicicola, respectively. (Scale bar, 5 mm.) (D and E) Lesion diameter (n = 3) (D) and A. brassicicola growth quantified by amplification of the A. brassicicola cutinase A (Ab) gene relative to the A. thaliana α shaggy kinase (At) gene by qPCR (n = 3) (E). Graphs represent mean values (±SE) from three biological experiments with at least 15 leaves per genotype and experiment. Different lowercase letters indicate that the means are significantly different between genotypes (Tukey’s HSD test, P < 0.01).
We further tested this possibility by generating AtABCG34 overexpression lines in the wild-type background. The rosette leaves of plants expressing 35Spro:sGFP:ABCG34 (OE1–OE3) exhibited higher levels of surface camalexin at 24 hpi than did the wild-type rosette leaves (Fig. 4A). The increased secretion of camalexin was accompanied by improved disease resistance to A. brassicicola (Fig. 4 C–E): Lesion diameter and pathogen growth were significantly reduced in all three overexpression lines treated with this pathogen compared with the wild-type plants (Fig. 4 D and E).
AtABCG34 Expression Improves Tolerance to Camalexin in A. thaliana and BY2 Cells.
Because camalexin is known to be toxic to A. thaliana cells in suspension cultures (24), we next compared the effect of exogenously applied camalexin on the mature leaf surface of wild-type plants and atabcg34 mutants. Cell death induced by camalexin treatment was analyzed by Evans blue staining. The leaves of both atabcg34 mutants exhibited significantly higher rates of cell death in response to camalexin treatment than did the wild-type plants and the complementation lines. However, the cell-death response in the leaves of the atpad3-1 mutant, which has a point mutation in the gene encoding a key enzyme in camalexin biosynthesis and exhibits susceptibility to A. brassicicola (25), did not differ from that of the wild-type plants or the complementation lines following camalexin treatment (Fig. 5A, Middle). By contrast, atabcg34 and atpad3-1 mutants were similar in their responses to A. brassicicola treatment, exhibiting significantly increased levels of cell death compared with the wild-type plants and the complementation lines (Fig. 5A, Bottom).
Fig. 5.
Camalexin toxicity assay in A. thaliana and BY2 cells. (A) Photographs of cell death in 3- to 4-wk-old plants using Evans blue staining 24 h after treatment with DMSO (1%) (Top), camalexin (1 mg/mL) (Middle), or A. brassicicola (5 × 105 spores/mL) (Bottom). The experiment was repeated four independent times with similar results (n = 4). (Scale bar, 5 mm.) (B) BY2 cells expressing 35Spro:sGFP empty vector (EV) (Top) and 35Spro:sGFP:ABCG34 (ABCG34-1 and ABCG34-2) (Middle and Bottom, respectively). Merged indicates colocalization with red fluorescence of FM4-64 (10 µM), a plasma membrane marker. (Scale bar, 20 µm.) (C) Camalexin-induced cell death in BY2 cells expressing EV or AtABCG34. A 6-d-old culture of BY2 cells was treated with either DMSO (0.05%) or camalexin (250 µM), and cell death was analyzed by Evans blue staining 24 h after treatment. (D) Camalexin-induced growth inhibition of BY2 cells. A 3-d-old culture of BY2 cells was treated with either DMSO (0.01%) or camalexin (50 µM), and 4 d after treatment the OD at 600 nm was measured. Graphs in C and D represent mean values (±SE) of three independent experiments. Different lowercase letters indicate that the means are significantly different (Tukey’s HSD test, P < 0.01; n = 3).
We then tested whether AtABCG34 can confer camalexin tolerance in a heterologous system. For this purpose, we expressed 35Spro:sGFP:ABCG34 [coding DNA sequence (CDS)] in tobacco Bright Yellow 2 (BY2) cells. AtABCG34 was localized at the plasma membrane in BY2 cells (Fig. 5B, Middle and Bottom), similarly as in A. thaliana cells (Fig. 3D), by contrast to the cytosolic localization of free sGFP [empty vector (EV)] (Fig. 5B, Top). When cultured in medium containing camalexin, AtABCG34-expressing BY2 cells (ABCG34-1 and ABCG34-2) exhibited reduced rates of cell death (Fig. 5C) and increased cell density (Fig. 5D) compared with the EV-expressing cells.
AtABCG34 Contributes to Preinvasion Resistance Against A. brassicicola.
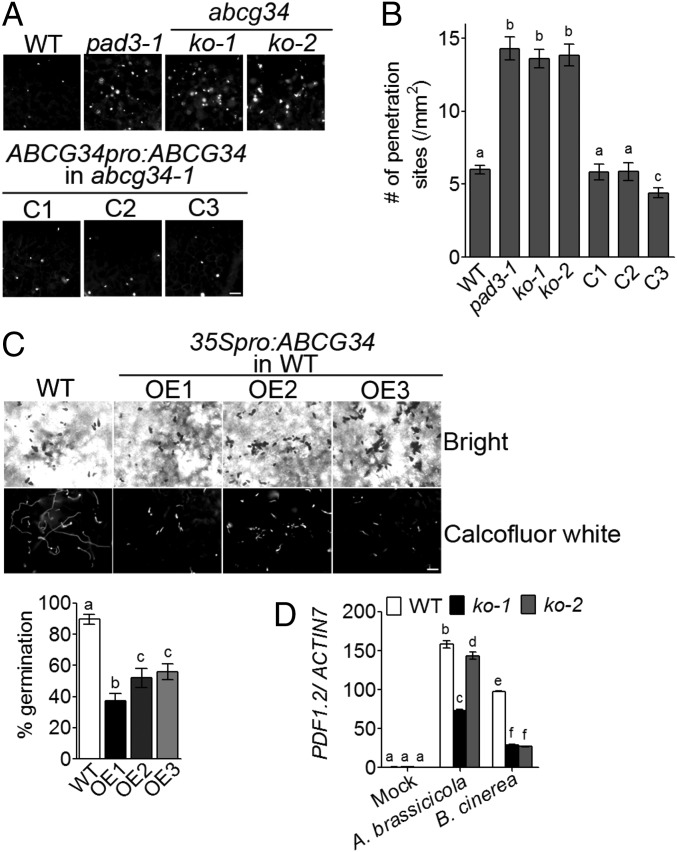
Plants exhibit a multilayered defense response to A. brassicicola that is composed of pre- and postinvasion stages. Formation of an appressorium, a swollen structure at the tip of growing hyphae that penetrates the plant cell wall (26), marks the preinvasion stage. Under our experimental conditions, appressoria were observed at 18 hpi with A. brassicicola (Fig. 3G), consistent with a previous report that appressoria formed at 12–24 hpi with A. brassicicola (27). Because AtABCG34 expression induced by A. brassicicola treatment was apparent at 12 hpi (Fig. 3 C and H), a relatively early time point during the pathogen-defense response (i.e., before appressorium formation), we suspected that AtABCG34 might contribute to resistance at the preinvasion stage. We tested this possibility using three different methods. First, we counted the pathogen penetration sites by callose staining in the wild-type plants, atabcg34 mutants, and complementation lines. Callose is deposited at the penetration site in response to A. brassicicola treatment (28), and a high number of callose depositions indicates a failure to overcome the pathogen at the preinvasion stage (5, 29). atabcg34 mutants exhibited a significantly higher number of penetration sites than did the wild-type and complementation lines (Fig. 6 A and B). We also analyzed the atpad3-1 mutant and found that it exhibited a high penetration frequency similar to that of the atabcg34 mutants (Fig. 6 A and B). These results support the notion that the reduction of camalexin content/secretion compromised defense against A. brassicicola at the preinvasion stage. Second, we examined the A. brassicicola conidia germination in the wild-type and AtABCG34 over-expressing lines. We found that overexpression lines exhibited significantly lower rates of conidia germination than did the wild-type plants (Fig. 6C), indicating that AtABCG34 plays an important role in preventing the conidia germination before invasion to the leaf. Third, we tested whether PDF1.2 induction by pathogens differs between the wild-type plants and the mutants. PDF1.2 is induced by A. brassicicola infection and plays an important role in the preinvasion stage of resistance (30). PDF1.2 expression increased in both the wild-type and the mutants inoculated with A. brassicicola or B. cinerea, but the extent of the increase was significantly less in atabcg34 mutants than in the wild-type plants (Fig. 6D).
Fig. 6.
AtABCG34 is required for preinvasion resistance to A. brassicicola. (A and B) Penetration sites in the leaves of various A. thaliana lines infected with A. brassicicola. (A) Callose deposited at the penetration sites was stained with aniline blue at 24 hpi. (Scale bar, 50 µm.) (B) The number of penetration sites was counted. Data represent mean values (±SE) of three independent experiments with at least 10 leaves per genotype in each experiment. Different lowercase letters indicate that the means are significantly different between genotypes (Tukey’s HSD test, P < 0.01; n = 3). (C, Upper) Bright-field (Upper Row) and Calcofluor white fluorescent (Lower Row) microscopy images of A. brassicicola conidia and hyphae. (Scale bars, 50 µm.) (Lower) Percent germinated A. brassicicola conidia on the leaves of wild-type and AtABCG34-overexpressing plants at 24 hpi (n = 15). (D) Transcript levels of AtPDF1.2 after A. brassicicola or B. cinerea infection in wild-type plants and atabcg34 mutants. The graphs in C, Lower and D represent mean values (±SE) of three replicates (n = 3). Different lowercase letters indicate that the means are significantly different between the genotypes (Tukey’s HSD test, P < 0.05).
Accessions Assay to Evaluate the Contribution of AtABCG34 to Resistance Against A. brassicicola.
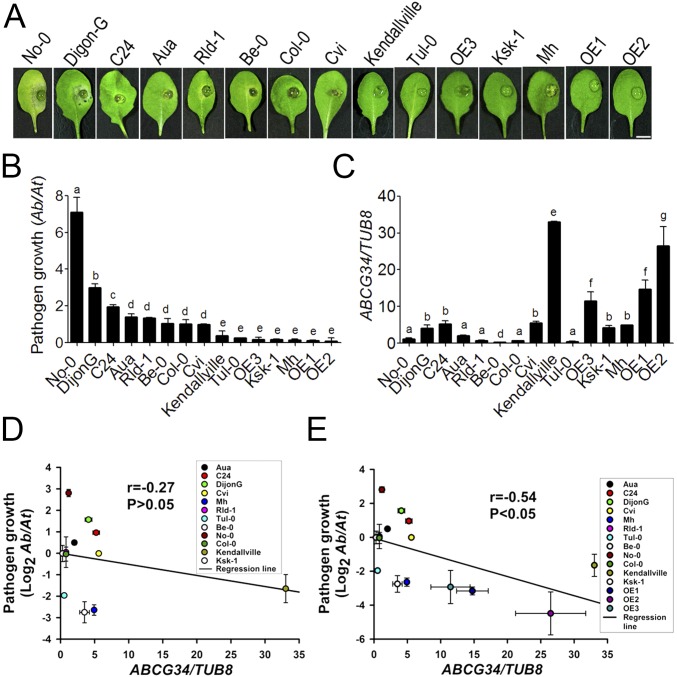
Next, we asked whether AtABCG34 is an important factor in determining the defense response to A. brassicicola among natural Arabidopsis accessions. To address this question, we used Arabidopsis accessions that were reported to exhibit different levels of resistance to A. brassicicola infection (31): Dijon-G and C24 as susceptible phenotypes; Aua/Rhon (Aa), Cape Verde (Cvi), RLD-1, and Columbia-0 (Col-0) as phenotypes with intermediate resistance; and Kendallville, Muehlen (Mh), Ksk-1, Turk Lake (Tul-0), Bensheim (Be-0), and Nossen (No-0) as resistant phenotypes. We evaluated the different accessions for their AtABCG34 transcript levels and their sensitivity to A. brassicicola inoculation. The responses of the accessions to A. brassicicola fell into two categories. Consistent with the results of a previous report (31), the susceptible group included Dijon-G, C24, Aua, RLD-1, Col-0, and Cvi (Fig. 7 A and B). Contrary to the results of the previous report (31), accessions No-0 and Be-0 were also susceptible to A. brassicicola under our experimental conditions (Fig. 7 A and B). The more resistant group included the remaining accessions.
Fig. 7.
A. brassicicola growth and AtABCG34 expression levels in different accessions and AtABCG34-overexpressing lines of Arabidopsis. (A) Disease symptoms in detached leaves. Photographs were taken at 5 dpi with A. brassicicola (5 × 105 spores/mL). (Scale bar, 5 mm.) (B) Growth of A. brassicicola, as determined by amplification of the A. brassicicola cutinase A (Ab) and the A. thaliana α shaggy kinase (At) genes by qPCR. The graph represents mean values (±SE) from three biological replicates with 27 lesions from 15 different plants (n = 3). (C) AtABCG34 transcript levels in leaves of 3- to 4-wk-old plants under control conditions. The graph represents mean values (±SE) from three biological replicates using qRT-PCR. Different lowercase letters in B and C indicate that the means are significantly different between the accessions (Tukey’s HSD test, P < 0.05; n = 3). (D and E) Regression analysis for the correlation between AtABCG34 expression (mean ± SE) and A. brassicicola growth (logarithmic transformed geometric mean ± SE) in 12 different accessions (D) and in 12 different accessions and three overexpression lines (OE1–OE3) (E).
Four of the seven susceptible accessions, No-0, Aua, RLD-1, and Be-0, expressed AtABCG34 at similar or lower levels than did Col-0 (Fig. 7C). The three remaining susceptible accessions, Dijon-G, C24, and Cvi, expressed AtABCG34 at five- to sixfold higher levels than did Col-0. By contrast, three of the four more resistant accessions, Kendallville, Ksk-1, and Mh, expressed AtABCG34 at four- to 41-fold higher levels than did Col-0. The AtABCG34 overexpression lines we generated fell into the more resistant group and expressed AtABCG34 at 14- to 33-fold higher levels than did Col-0. Among the more resistant accessions, Tul-0 was the only accession that expressed AtABCG34 at a level comparable to Col-0.
To quantify the relationship between AtABCG34 expression level and pathogen growth in different accessions and overexpression lines, we performed regression analysis. The coefficient of correlation was −0.27 when only the 12 accessions were included in the analysis (Fig. 7D), indicating a weak negative correlation between pathogen growth and AtABCG34 expression levels in the accessions tested. However, the coefficient of correlation changed to −0.54 when three overexpression lines were included together with the 12 accessions (Fig. 7E), indicating a moderate negative correlation between pathogen growth and expression levels of AtABCG34.
Discussion
AtABCG34 Is Important for Defense Against Necrotrophic Fungi.
In this study, we identified AtABCG34 as a strong factor conferring tolerance to the antifungal diterpene sclareol (Fig. 1 and Fig. S2) and hence as a potential player in pathogen defense. We report three lines of evidence supporting the role of AtABCG34 in defense against necrotrophic fungal pathogens. First, atabcg34 mutants exhibited hypersensitivity to the necrotrophic fungi A. brassicicola (Fig. 2) and B. cinerea (Fig. S4A). Second, AtABCG34 expression was induced by A. brassicicola (Fig. 3 C, G, and H) and B. cinerea (Fig. S5) infection in a time-dependent manner. Third, AtABCG34 expression was induced substantially by the application of MeJA (Fig. 3A) and only slightly by SA (Fig. 3B). The induction of AtABCG34 by MeJA and necrotrophic fungal pathogens is similar to that observed for the NpPDR1 and NtPDR1 transporters, which secrete antifungal molecules in response to pathogen infection (4). MeJA was reported to be important for resistance to necrotrophic pathogens rather than biotrophic pathogens (32). Moreover, jasmonic acid (JA), but not SA, is necessary for the resistance to A. brassicicola, as evidenced by the reduced tolerance of a JA-signaling mutant, coi1, to the pathogen and the similar tolerance levels for the pathogen of the SA-signaling mutant npr1, SA-deficient NahG-expressing plants, and the wild-type plants, respectively (32). The pattern of AtABCG34 induction (Fig. 3) suggests that AtABCG34 is involved in the defense against A. brassicicola through the JA-signaling pathway but not through the SA-signaling pathway. AtABCG34 does not seem to be involved in biotrophic pathogen resistance because (i) it was not induced significantly by SA, which is of major importance for biotrophic pathogen resistance (33), and (ii) the atabcg34 mutants did not exhibit sensitivity to the biotrophic bacterial pathogen Pst DC3000 (Fig. S4B).
Possible Importance of AtABCG34 in Natural Habitat.
In nature, multiple factors usually contribute to a phenotype. Thus, in an accession study, a very high correlation between the level of one factor and the phenotype is rare (34, 35). Consistently, in our results as well, there was only a weak negative correlation between A. brassicicola growth and AtABCG34 expression levels (Fig. 7D). However, there was a significant, moderate negative correlation when overexpression lines were included together with the 12 accessions in the statistical analysis (Fig. 7E). Moreover, three of the four ecotypes resistant to A. brassicicola expressed AtABCG34 at higher levels than did Col-0, whereas only three of the seven susceptible accessions did so (Fig. 7C). These results suggest that AtABCG34 contributes to the disease resistance in naturally occurring genetic variants. AtABCG34 may be one among many different factors contributing to A. brassicicola resistance, and only a subset of the accessions may have evolved this strategy. Other factors contributing to A. brassicicola resistance include other JA-response pathways (36), other secondary metabolites (such as phenolics), and a mechanism to tolerate the toxins secreted by the pathogen (31). Further studies, including analyses of additional accessions for resistance to fungal disease, are necessary to evaluate this hypothesis.
It is interesting that the expression level of AtABCG34 was about 41 times higher in the Kendallville accession than in Col-0. Thus, in wild populations of Arabidopsis, there may be a wide range of expression levels of this gene. However, we do not expect that there would be many other accessions with even higher levels of expression of AtABCG34 than the Kendallville ecotype, because the OE1 and OE3 lines expressed AtABCG34 at lower levels than found in this accession but exhibited similar levels of resistance to A. brassicicola (Fig. 7B).
AtABCG34 Localization Before and After A. brassicicola Infection.
AtABCG34 was localized at the outward-facing sides of the plasma membrane in epidermal cells of the leaves and roots (Fig. 3 E and F). Such polar localization of ABC transporters was reported previously for AtABCG32 (21), AtABCG36, and AtABCG37 (37). These transporters secrete compounds important for cuticle formation (21) or defense (13) or transport auxin precursor to the rhizosphere (38), respectively. The polar localization of AtABCG34 suggests that it secretes some compounds to the leaf surface. The mechanism of polar localization of AtABCG34 might resemble that of AtABCG36, another ABCG subfamily member involved in pathogen defense [i.e., polar secretion, constitutive endocytic recycling, and restricted lateral diffusion (39)].
AtABCG34 is highly induced by A. brassicicola infection (Fig. 3 C and H) and accumulates at the plasma membrane, as indicated by the increase in sGFP:ABCG34 fluorescence when plants are infected with the pathogen (Fig. 3G). Previous studies of AtABCG36/PEN3 and NbABCG1/2 showed that these transporters are preferentially localized at the pathogen penetration sites (5, 11). However, we did not observe such preferential accumulation of AtABCG34 at the penetration site. Rather, we observed its accumulation at broad areas surrounding the inoculation site of A. brassicicola (Fig. 3G) and B. cinerea (Fig. S5B). Interestingly, A. brassicicola infection also induced accumulation of sGFP:ABCG34 in dot-like structures that, to our knowledge, have not been reported for plant transporters during pathogen infection. Further studies are needed to decipher the formation and function of these dot-like structures.
AtABCG34 Most Likely Transports Camalexin.
Our data described above indicate that AtABCG34 is involved in transporting secondary metabolites to the leaf surface to inhibit the growth of necrotrophic pathogens. Initially, we selected AtABCG34 based on a screen for hypersensitivity to sclareol, a diterpene alcohol. However, based on several independent lines of evidence, we propose that the secondary metabolites that AtABCG34 transports include an indole alkaloid, camalexin (Fig. 8). First, surface camalexin in the rosette leaves was significantly reduced in atabcg34 mutants (Fig. 4A and Fig. S6D), whereas the surface camalexin levels were increased in the AtABCG34-overexpressing lines (Fig. 4A). Second, expression of sGFP:ABCG34 in BY2 cells improved resistance to a toxic concentration of camalexin (Fig. 5 B–D), suggesting that AtABCG34 mediates the removal of camalexin from the BY2 cells. Third, when treated with exogenous camalexin, atabcg34 mutants exhibited high levels of cell death, but the atpad3-1 mutant did not (Fig. 5A), most likely because the functional AtABCG34 in the atpad3-1 mutant plant cells was able to secrete camalexin from the cells. By contrast, there was no difference in the cell-death responses of the atpad3-1 and atabcg34 mutants to A. brassicicola treatment, indicating that both camalexin synthesis and transport are necessary for resistance against A. brassicicola infection. Fourth, the frequency of A. brassicicola penetration in the atpad3-1 mutant was similar to that in the atabcg34 mutants (Fig. 6 A and B). This result is consistent with our explanation that the two proteins function in the same pathway of defense against the pathogen: That PAD3 catalyzes the biosynthesis of camalexin, whereas AtABCG34 mediates transport of camalexin. Fifth, both AtABCG34 (Fig. 3G and Fig. S5B) and the camalexin biosynthesis genes (40) are induced around the A. brassicicola and B. cinerea infection site. By contrast, we did not obtain any clear evidence that the transporter transports surface-coating material (Fig. S6A), nor were there any clear differences in the levels of phenolics (in plants infected by B. cinerea) or glucosinolates (in plants infected by A. brassicicola) between the atabcg34 and wild-type plants (Fig. S6 B and C). AtABCG34 might transport multiple substrates, such as an indole alkaloid camalexin and some diterpene alcohols similar to sclareol produced by A. thaliana. Such transport of multiple substrates would not be unusual, because a single ABCG transporter can transport many chemically unrelated substrates (41, 42), and ABCG transporters have a broad range of substrates, including alkaloids, terpenoids, various hormones, and lipids (41).
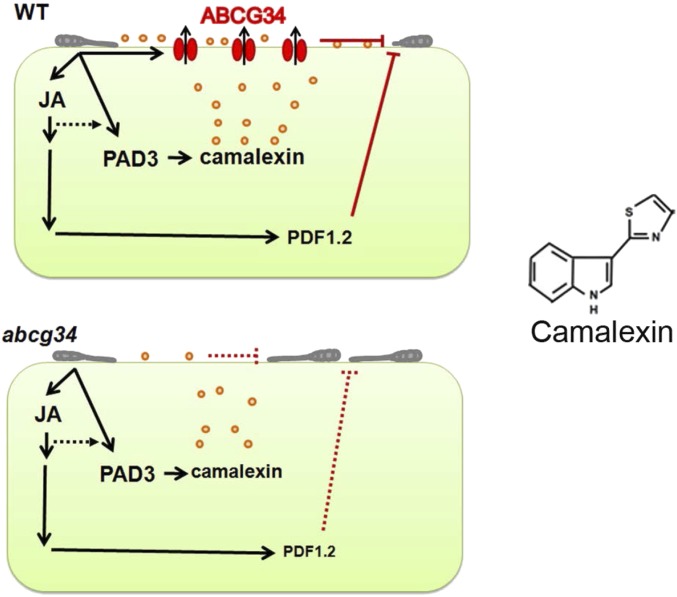
Fig. 8.
Working model for the function of AtABCG34. Inoculation of A. thaliana leaves with A. brassicicola induces the expression of AtABCG34, the camalexin biosynthesis gene PAD3, and JA-signaling genes. JA also induces the expression of AtABCG34 and AtPDF1.2. (Upper Left) AtABCG34-mediated secretion of camalexin to the surface inhibits A. brassicicola growth in wild-type plants (solid red lines). (Lower Left) Reduced camalexin secretion and AtPDF1.2 expression (small font) allow pathogen growth in the atabcg34 mutant (dotted red lines). Orange circles represent camalexin. (Right) The chemical structure of camalexin.
In addition, a comparison of our results with some previous reports supports the involvement of AtABCG34 in camalexin transport. Similar to AtABCG34, camalexin biosynthesis is induced by necrotrophic fungal pathogens (43). AtABCG34 is induced in response to JA (Fig. 3A), and JA-signaling components are involved in the biosynthesis of camalexin (25). Interestingly, a full-size ABCG transporter, BcatrB, in B. cinerea is necessary for the export of camalexin (44). This transporter is a critical virulence factor, as evidenced by the failure of a BcatrB-knockout mutant to infect A. thaliana. Thus, there may have been an evolutionary arms race between plants and the pathogen to develop camalexin transporters; plants developed ABC transporters to secrete camalexin, and fungal pathogens developed similar ABC transporters to export camalexin from their cytosol. Evolutionary studies on these transporters may lead to interesting findings on coevolution.
We attempted to conduct camalexin transport assays using BY2 cells. However, we did not find any conditions that allowed time-dependent loading of camalexin into cells. Similar difficulties in carrying out transport assays were reported for sclareol in BY2 cells (2). Direct transport assays of these secondary metabolites await further understanding of the chemical nature and permeability of the chemicals through lipid membranes. Nonetheless, our results strongly suggest that AtABCG34 has a critical role in secreting camalexin to the surface of the plant, where it inhibits invasion of A. brassicicola.
Because the level of surface camalexin was reduced to half that in the wild-type plants, we propose that other mechanisms, such as diffusion, or other transporters, such as ABC transporters, mediate the secretion of the remaining half of camalexin to the leaf surface. The only other ABCG member that has been reported to affect camalexin levels is AtABCG36; an increase in total camalexin content was observed in the pdr8/pen3/abcg36 mutant in response to powdery mildew fungal infection (45). However, it is unclear whether the pdr8 mutant has increased levels of surface camalexin, and the response of this mutant to A. brassicicola or B. cinerea has not been tested. Thus, whether there are additional ABCG transporters for camalexin remains to be determined.
AtABCG34 Functions at the Preinvasion Stage.
Some accessions of Arabidopsis exhibit an incompatible interaction with A. brassicicola and have evolved two layers of defense: pre- and postinvasion defense (46). Preinvasion defense culminates when the pathogen tries to penetrate the plant cell. Four of our findings suggest that AtABCG34 functions at the preinvasion step of defense. First, the level of camalexin was reduced in atabcg34 mutants compared with wild-type plants (Fig. 4 A and B and Fig. S6D). Camalexin, a major phytoalexin in A. thaliana, is known to be important for preinvasion resistance against powdery mildew, and atpad3-1, a camalexin-deficient mutant, is defective in the preinvasion resistance to both powdery mildew (47) and A. brassicicola (33). Second, A. brassicicola conidia germination was significantly reduced in the AtABCG34-overexpressing lines (Fig. 6C), which had higher levels of surface camalexin (Fig. 4A). Inhibition of conidia germination on the plant surface is most likely caused by the higher level of camalexin secreted by AtABCG34. Third, atabcg34 mutants exhibited higher penetration frequency than did the wild-type plants, and wild-type levels of penetration were restored in the complementation lines (Fig. 6 A and B). Furthermore, the atpad3-1 mutant also exhibited high penetration frequency similar to that of the atabcg34 mutants. In general, mutants compromised in preinvasion defense exhibit high penetration frequency (marked by callose deposition) at the inoculation site (29, 48). Fourth, the expression levels of AtPDF1.2, a JA- and A. brassicicola-responsive gene, were significantly reduced in atabcg34 mutants compared with the wild-type plants (Fig. 6D). AtPDF1.2 is induced in response to necrotrophic fungal pathogens and JA (49), similar to AtABCG34, and constitutive expression of PDF1.2 enhances preinvasion resistance (30). These results indicate that AtABCG34 has an important function in preinvasion resistance.
Other ABC transporters important for resistance at the preinvasion stage include NtPDR1 and NpPDR1, which transport sclareol (2, 3), an important factor for preinvasion resistance (5). Recently, NbABCG1 and NbABCG2 were reported to be important for both pre- and postinvasion resistance in N. benthamiana (5).
Application Potentials of AtABCG34.
Our data revealed that AtABCG34 is an important factor required for defense against the necrotrophic fungal pathogens A. brassicicola and B. cinerea and that AtABCG34 functions by secreting camalexin to the surface. A. brassicicola infection results in dark leaf spots on most Brassica species, including economically important oilseed crops (50). Infection by A. brassicicola considerably reduces the quality and quantity of harvested Brassica crops (51), resulting in annual yield losses of 15–70% (51). Interestingly, overexpression of AtABCG34 in A. thaliana enhanced the surface camalexin content (Fig. 4A) and resistance to A. brassicicola (Fig. 4C). We predict that high levels of AtABCG34 expression in economically important Brassica species, such as Brassica napus and Brassica oleracea, will be associated with reduced disease severity and yield loss following A. brassicicola infection because of the increased secretion of camalexin to the plant’s surface. Such a goal may be attained by genetic approaches or by screening and selecting for accessions of B. napus and B. oleracea that have naturally high levels of expression of the ABCG34 ortholog.
Materials and Methods
Plant Materials and Growth Conditions.
A. thaliana (L.) Heynh. ecotype Col-0 wild-type and transgenic plants were grown on 1/2MS agar plates with 1% sucrose in the presence or absence of sclareol (65 µM) for 2–3 wk at a 16/8 h light/dark, 22/18 °C cycle. For long-term experiments, plants were transferred to soil and grown in a controlled growth chamber (16/8 h light/dark; 22/18 °C). For pathogen infection and camalexin analysis, 3- to 4-wk-old plants were used. For accessions analysis, Kendallville (CS76344), Bensheim (Be-0- CS76345), Aua/Rhon (CS900), Cape Verde (Cvi-CS902), Muehlen (Mh-CS904), C24 (CS906), Dijon-G (CS910), Col-0 (CS1092), Ksk-1 (CS1634), Rld-1 (CS76588), Tul-0 (CS76618), and No-0 (CS28564) strains were used.
Fungal Culture and Disease Assays.
B. cinerea was grown on 2× V8 agar [36% V8 juice (Campbell’s), 0.2% CaCO3, and 2% Bacto-agar] at 25 °C for 1–2 wk, and spores were resuspended in 1% Sabouraud maltose broth buffer (SMBB; Difco). A. brassicicola strain KACC40036 was grown on potato dextrose agar and was resuspended in distilled water for plant inoculation (52). To examine gene-expression patterns, whole A. thaliana plants were inoculated with B. cinerea or A. brassicicola by spraying the plants with a spore suspension (5 × 105 spores/mL) in SMBB or distilled water, respectively. For the disease assay, 8 μL of the spore suspension (5 × 105 spores/mL) was dropped onto detached leaves. Inoculated leaves were kept under high humidity until 5 dpi. The B. cinerea spore population was counted as described previously (23). A. brassicicola growth was quantified as follows: leaf disks were collected from the infection area at 5 dpi, using a No. 2 cork borer. The genomic DNA was extracted, and qPCR was performed as described previously (53). To estimate the growth of A. brassicicola cutinase A (ABU03393), DNA was amplified using primers listed in Table S1. The value was normalized by amplifying genomic DNA of A. thaliana α-Shaggy kinase (At5g26751) using iASK1 and iASK2 primers (Table S1). Standard curves were prepared using genomic DNA extracted from the A. thaliana leaf disks and A. brassicicola grown on potato dextrose agar plates. For the intact-plant assay, 10 μL of spore suspension (5 × 105 spores/mL) in distilled water was dropped on the fully expanded leaves of 4-wk-old plants.
Table S1.
Primers used in this study
| Purpose | Name | Primer sequence (5′–3′) |
| Knockout mutant isolation | ||
| LP1(RT-F) | ATGTTAGGACGAGATGAAGATC | |
| RP1(RT-R) | TTGAGAAGTCATGCACTGATAC | |
| LP2(RT-F) | CATTCCCTATGCAATTTCTCA | |
| RP2(RT-R) | CCTTCTTTGGAAGTTGAGGA | |
| qRT-PCR | ||
| ABCG34-F | CCGGGATCGAATGATCTCTA | |
| ABCG34-R | CGAGGAACAATACAGCAGCA | |
| Actin7-F | AATGGTGAAGGCTGGTTTTG | |
| Actin7-R | TGCCTCTGTGAGTAGAACTG | |
| ABCG39-F | CCTTACCTGCTTGGCTTCTG | |
| ABCG39-R | AAGGGCCATCTGATTCACAC | |
| Tubulin 8-F | TCGATTCGACGGGAAGATAC | |
| Tubulin 8-R | CGGACCGGATCTGACACTAT | |
| Tubulin8-Ksk-1-F | GCCGAGCACGGGATCGAT | |
| qPCR | ||
| CG9 | GCATGTCCGCTCACCAATATC | |
| CG10 | GCCTGGGATCTTGGAATGC | |
| iASK1 | CTTATCGGATTTCTCTATGTTTGGC | |
| iASK2 | GAGCTCCTGTTTATTTAACTTGTACATACC |
Camalexin Level Assay.
Four-week-old plants were sprayed with spore suspension (5 × 105 spores/mL) of A. brassicicola, and whole rosettes were collected at the indicated time points (Fig. 4 A and B and Fig. S6D). Total camalexin was extracted with hot methanol and chloroform, and surface camalexin was extracted with dichloromethane, using established protocols (54, 55) with slight modifications. To extract surface camalexin, rosette leaves of A. thaliana were gently rotated in dichloromethane for 30 s. Extracted samples were loaded onto a TLC plate and were separated, and the camalexin band was visualized by its blue fluorescence under an UV lamp (365 nm). The silica containing camalexin was scraped off the plate, and camalexin was extracted into 500 μL of methanol. The emission at 385 nm after excitation at 315 nm was measured to quantify the camalexin in a 96-well black microplate (Greiner Bio-One) with a Tecan fluorometer (M200PRO). The camalexin concentration was calculated by comparison with a standard curve obtained using commercially available camalexin purchased from Glixx Laboratories.
Camalexin Toxicity Assay.
Three-week-old plants were treated with 1% DMSO (solvent control treatment), 1 mg/mL camalexin, or 5 × 105 spores/mL of A. brassicicola (positive control). Evans blue staining was performed 24 h after treatment as described previously (56). Briefly, 0.1% Evans blue solution (wt/vol) was vacuum infiltrated (25 mmHg for 5 min, followed by a 5-min hold and subsequent slow release), and the leaves were stained for 8 h at room temperature. After staining, samples were washed with a PBS solution containing 0.05% (vol/vol) Tween-20 until the washing solution became clear.
For BY2 cells, the 6-d-old culture was treated with 250 µM camalexin for 30 min at 25 °C in darkness (24). The cells were washed with water and harvested by centrifugation at 13,800 × g for 5 min at room temperature. To analyze cell death, cells were stained with 0.05% Evans blue for 15 min at 25 °C and then were washed three times with water (56). Evans blue stain was extracted from the cells with 50% methanol+1% SDS for 60 min at 60 °C, and the blue color was quantified by measuring absorbance at 595 nm and normalized by OD at 600 nm. Cell death caused by boiling, used as a positive control, was set to 100%, and relative cell death caused by either DMSO or camalexin treatment was analyzed. For cell growth analysis, a 3-d-old culture was treated with either 0.01% DMSO (solvent control) or camalexin (50 µM), and OD at 600 nm was measured 3 d after camalexin treatment using a spectrophotometer.
Callose Staining.
Callose staining was performed as described previously (12). Briefly, 3- to 4-wk-old plants were treated with A. brassicicola solution (5 × 105 spores/mL). Leaf disks from the inoculation sites were collected 24 hpi and fixed in ethanol:acetic acid (3:1) by brief vacuum infiltration (at 25 mmHg for 5 min, followed by a 5-min hold and subsequent slow release) and then were destained by gentle rotation until the disks became translucent, washed with 150 mM K2HPO4 for 30 min, and stained with aniline blue (1%) staining solution for 2 h on a shaker. Stained disks were observed with a Zeiss fluorescence microscope (Axioskop 2 MOT) using the DAPI channel after mounting with 50% glycerol.
Regression Analysis Between AtABCG34 Expression and Pathogen Growth in Natural Accessions.
The expression levels of AtABCG34 and pathogen growth in different ecotypes were analyzed as mentioned above using sets of primers listed in Table S1. The AtABCG34 sequences of all ecotypes tested contained fragments that precisely matched the sequences of the primers used. The tubulin 8 sequences differed from those of the primers (both forward and reverse) by 1 bp in two ecotypes, Kendallville and RLD-1. However, the transcript levels of tubulin 8 in these two ecotypes were similar to those of other the ecotypes evaluated.
The Ksk-1 and Bensheim ecotypes have not been sequenced. Thus, we sequenced parts of the coding regions of AtTubulin 8 and AtABCG34. No differences were observed in the regions corresponding to the AtABCG34 primer sequences in either of the two ecotypes. One base pair change was detected in the center of both the forward and reverse primers of AtTubulin 8 in Bensheim. The tubulin 8 sequence of Ksk-1 differed at 3 bp of the forward primer. Thus, we designed a different forward primer (listed in Table S1) to compare the transcript levels of tubulin 8 in Ksk-1 and Col-0.
Regression analysis between AtABCG34 expression and pathogen growth was performed by comparing the Pearson’s correlation coefficient between average data of AtABCG34 expression and pathogen growth with 10,000 randomly permuted datasets from all 12 accessions either including or excluding three overexpression (OE1–OE3) lines. Logarithmic values of relative copy numbers of A. brassicicola cutinase A were used for pathogen growth.
For other methods, please see SI Materials and Methods.
SI Materials and Methods
Isolation of Knockouts.
T-DNA insertion mutants of atabcg34, atabcg39, and a point mutant of atpad3-1 (25) were obtained from the Arabidopsis Biological Resource Center. T-DNA insertional mutants from the Syngenta Arabidopsis Insertion Library (SAIL) collection (abcg34-1) and the Salk Institute (abcg34-2) were isolated by genotyping using a gene-specific primer and a T-DNA left border-specific primer. Total RNA was extracted from wild-type, abcg34-1, and abcg34-2 SALK_067836 (abcg39) seedlings, and transcript levels were checked by RT-PCR and qRT-PCR using primers listed in Table S1.
Bacterial Disease Assay.
The leaves of 4-wk-old A. thaliana plants were inoculated with bacterial suspensions (OD600 = 0.0001 in 10 mM MgCl2) of Pst DC3000 by syringe infiltration. To quantify bacterial growth, leaf disks were collected from the leaves at 1, 2, and 3 dpi. The leaf disks were incubated for 2 d at 28 °C, and bacterial colonies were counted as described previously (57).
Measurement of Phenolics and Glucosinolates.
Leaves from 3- to 4-wk-old plants mock-treated or treated with B. cinerea were lyophilized and used for extraction. Samples were incubated overnight in darkness at 4 °C in 0.5 mL of 80% (vol/vol) MeOH and then were macerated with a pestle and centrifuged at 15,000 × g for 5 min. The supernatant was dried and resuspended in 0.1 mL of 80% MeOH. Aliquots (20 μL) were analyzed by HPLC (Dionex). Absorbance spectra were recorded with a PDA-100 photodiode array detector (Dionex). Data integration analysis was conducted using Chromeoleon software (version 6.8; Dionex). The peak height was quantified at 280 nm. Chromatographic conditions were an OD2 Hypersil 100-5 C18 column (5-μm particle size; 2.1-mm i.d. × 250-mm length; Thermo Scientific); flow rate: 1.00 mL/min; gradient (step, time, %A over B): 1, 25 min, 10–25%; 2, 10 min, 25–70%; 3, 5 min, 70–100%; and 4, 10 min, 100–10%). Solvent A was acetonitrile, and solvent B was H2O, 0.1% (vol/vol) H3PO4. For glucosinolate measurement, leaves from 3- to 4-wk-old mock-treated plants or plants treated with A. brassicicola were lyophilized, extracted with 70% MeOH (vol/vol), and analyzed by HPLC as previously described (58).
Acknowledgments
We thank Professor Jane Glazebrook for kindly providing camalexin and Dr. C. Douglas Grubb for measuring glucosinolates. This work was supported by National Research Foundation of Korea (NRF) Grant NRF-2015R1A2A1A01004294 (to Y.L.) and by NRF Grant R1A2A2A01002476 (to K.-H.P.). The NRF is funded by the Ministry of Science, ICT and Future Planning of the Korean Government.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1702259114/-/DCSupplemental.
References
- 1.Srikantaramas S, Yamazaki M, Saito K. Mechanisms of resistance to self-produced toxic secondary metabolites in plants. Phytochem Rev. 2008;7:467–477. [Google Scholar]
- 2.Crouzet J, et al. NtPDR1, a plasma membrane ABC transporter from Nicotiana tabacum, is involved in diterpene transport. Plant Mol Biol. 2013;82:181–192. doi: 10.1007/s11103-013-0053-0. [DOI] [PubMed] [Google Scholar]
- 3.Jasiński M, et al. A plant plasma membrane ATP binding cassette-type transporter is involved in antifungal terpenoid secretion. Plant Cell. 2001;13:1095–1107. [PMC free article] [PubMed] [Google Scholar]
- 4.Stukkens Y, et al. NpPDR1, a pleiotropic drug resistance-type ATP-binding cassette transporter from Nicotiana plumbaginifolia, plays a major role in plant pathogen defense. Plant Physiol. 2005;139:341–352. doi: 10.1104/pp.105.062372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shibata Y, et al. The Full-size ABCG transporters Nb-ABCG1 and Nb-ABCG2 function in pre- and post-invasion defense against Phytophthora infestans in Nicotiana benthamiana. Plant Cell. 2016;28:1163–1181. doi: 10.1105/tpc.15.00721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bienert MD, et al. A pleiotropic drug resistance transporter in Nicotiana tabacum is involved in defense against the herbivore Manduca sexta. Plant J. 2012;72:745–757. doi: 10.1111/j.1365-313X.2012.05108.x. [DOI] [PubMed] [Google Scholar]
- 7.Sasse J, et al. Petunia hybrida PDR2 is involved in herbivore defense by controlling steroidal contents in trichomes. Plant Cell Environ. 2016;39:2725–2739. doi: 10.1111/pce.12828. [DOI] [PubMed] [Google Scholar]
- 8.Yu F, De Luca V. ATP-binding cassette transporter controls leaf surface secretion of anticancer drug components in Catharanthus roseus. Proc Natl Acad Sci USA. 2013;110:15830–15835. doi: 10.1073/pnas.1307504110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Krattinger SG, et al. A putative ABC transporter confers durable resistance to multiple fungal pathogens in wheat. Science. 2009;323:1360–1363. doi: 10.1126/science.1166453. [DOI] [PubMed] [Google Scholar]
- 10.Krattinger SG, et al. The wheat durable, multipathogen resistance gene Lr34 confers partial blast resistance in rice. Plant Biotechnol J. 2016;14:1261–1268. doi: 10.1111/pbi.12491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Underwood W, Somerville SC. Perception of conserved pathogen elicitors at the plasma membrane leads to relocalization of the Arabidopsis PEN3 transporter. Proc Natl Acad Sci USA. 2013;110:12492–12497. doi: 10.1073/pnas.1218701110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clay NK, Adio AM, Denoux C, Jander G, Ausubel FM. Glucosinolate metabolites required for an Arabidopsis innate immune response. Science. 2009;323:95–101. doi: 10.1126/science.1164627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stein M, et al. Arabidopsis PEN3/PDR8, an ATP binding cassette transporter, contributes to nonhost resistance to inappropriate pathogens that enter by direct penetration. Plant Cell. 2006;18:731–746. doi: 10.1105/tpc.105.038372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lu X, et al. Mutant allele-specific uncoupling of PEN3 functions reveals engagement of the ABC25 transporter in distinct tryptophan metabolic pathways. Plant Physiol. 2015;168:814–827. doi: 10.1104/pp.15.00182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Campbell EJ, et al. Pathogen-responsive expression of a putative ATP-binding cassette transporter gene conferring resistance to the diterpenoid sclareol is regulated by multiple defense signaling pathways in Arabidopsis. Plant Physiol. 2003;133:1272–1284. doi: 10.1104/pp.103.024182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kang J, et al. Plant ABC transporters. Arabidopsis Book. 2011;9:e0153. doi: 10.1199/tab.0153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ji H, et al. ATP-dependent binding cassette transporter G family member 16 increases plant tolerance to abscisic acid and assists in basal resistance against Pseudomonas syringae DC3000. Plant Physiol. 2014;166:879–888. doi: 10.1104/pp.114.248153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yazaki K. ABC transporters involved in the transport of plant secondary metabolites. FEBS Lett. 2006;580:1183–1191. doi: 10.1016/j.febslet.2005.12.009. [DOI] [PubMed] [Google Scholar]
- 19.Bailey JA, Garter GA, Burden RS, Wain RL. Control of rust diseases by diterpenes from Nicotiana glutinosa. Nature. 1975;255:328–329. [Google Scholar]
- 20.Kennedy BS, et al. Leaf surface chemicals fromNicotiana affecting germination of Peronospora tabacina (adam) sporangia. J Chem Ecol. 1992;18:1467–1479. doi: 10.1007/BF00993221. [DOI] [PubMed] [Google Scholar]
- 21.Bessire M, et al. A member of the PLEIOTROPIC DRUG RESISTANCE family of ATP binding cassette transporters is required for the formation of a functional cuticle in Arabidopsis. Plant Cell. 2011;23:1958–1970. doi: 10.1105/tpc.111.083121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fabre G, et al. The ABCG transporter PEC1/ABCG32 is required for the formation of the developing leaf cuticle in Arabidopsis. New Phytol. 2016;209:192–201. doi: 10.1111/nph.13608. [DOI] [PubMed] [Google Scholar]
- 23.van Wees SC, Chang HS, Zhu T, Glazebrook J. Characterization of the early response of Arabidopsis to Alternaria brassicicola infection using expression profiling. Plant Physiol. 2003;132:606–617. doi: 10.1104/pp.103.022186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rogers EE, Glazebrook J, Ausubel FM. Mode of action of the Arabidopsis thaliana phytoalexin camalexin and its role in Arabidopsis-pathogen interactions. Mol Plant Microbe Interact. 1996;9:748–757. doi: 10.1094/mpmi-9-0748. [DOI] [PubMed] [Google Scholar]
- 25.Zhou N, Tootle TL, Glazebrook J. Arabidopsis PAD3, a gene required for camalexin biosynthesis, encodes a putative cytochrome P450 monooxygenase. Plant Cell. 1999;11:2419–2428. doi: 10.1105/tpc.11.12.2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lawrence CB, et al. At death’s door: Alternaria pathogenicity mechanisms. Plant Pathol J. 2008;24:101–111. [Google Scholar]
- 27.Cho Y, Ohm RA, Grigoriev IV, Srivastava A. Fungal-specific transcription factor AbPf2 activates pathogenicity in Alternaria brassicicola. Plant J. 2013;75:498–514. doi: 10.1111/tpj.12217. [DOI] [PubMed] [Google Scholar]
- 28.Flors V, et al. Interplay between JA, SA and ABA signalling during basal and induced resistance against Pseudomonas syringae and Alternaria brassicicola. Plant J. 2008;54:81–92. doi: 10.1111/j.1365-313X.2007.03397.x. [DOI] [PubMed] [Google Scholar]
- 29.Lipka V, et al. Pre- and postinvasion defenses both contribute to nonhost resistance in Arabidopsis. Science. 2005;310:1180–1183. doi: 10.1126/science.1119409. [DOI] [PubMed] [Google Scholar]
- 30.Hiruma K, et al. Arabidopsis ENHANCED DISEASE RESISTANCE 1 is required for pathogen-induced expression of plant defensins in nonhost resistance, and acts through interference of MYC2-mediated repressor function. Plant J. 2011;67:980–992. doi: 10.1111/j.1365-313X.2011.04651.x. [DOI] [PubMed] [Google Scholar]
- 31.Kagan IA, Hammerschmidt R. Arabidopsis ecotype variability in camalexin production and reaction to infection by Alternaria brassicicola. J Chem Ecol. 2002;28:2121–2140. doi: 10.1023/a:1021020512846. [DOI] [PubMed] [Google Scholar]
- 32.Thomma BPHJ, et al. Separate jasmonate-dependent and salicylate-dependent defense-response pathways in Arabidopsis are essential for resistance to distinct microbial pathogens. Proc Natl Acad Sci USA. 1998;95:15107–15111. doi: 10.1073/pnas.95.25.15107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Glazebrook J. Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu Rev Phytopathol. 2005;43:205–227. doi: 10.1146/annurev.phyto.43.040204.135923. [DOI] [PubMed] [Google Scholar]
- 34.Stein RJ, Waters BM. Use of natural variation reveals core genes in the transcriptome of iron-deficient Arabidopsis thaliana roots. J Exp Bot. 2012;63:1039–1055. doi: 10.1093/jxb/err343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Barah P, et al. Genome-scale cold stress response regulatory networks in ten Arabidopsis thaliana ecotypes. BMC Genomics. 2013;14:722. doi: 10.1186/1471-2164-14-722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cramer RA, Lawrence CB. Identification of Alternaria brassicicola genes expressed in planta during pathogenesis of Arabidopsis thaliana. Fungal Genet Biol. 2004;41:115–128. doi: 10.1016/j.fgb.2003.10.009. [DOI] [PubMed] [Google Scholar]
- 37.Langowski L, Růzicka K, Naramoto S, Kleine-Vehn J, Friml J. Trafficking to the outer polar domain defines the root-soil interface. Curr Biol. 2010;20:904–908. doi: 10.1016/j.cub.2010.03.059. [DOI] [PubMed] [Google Scholar]
- 38.Ružicka K, et al. Arabidopsis PIS1 encodes the ABCG37 transporter of auxinic compounds including the auxin precursor indole-3-butyric acid. Proc Natl Acad Sci USA. 2010;107:10749–10753. doi: 10.1073/pnas.1005878107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Łangowski Ł, et al. Cellular mechanisms for cargo delivery and polarity maintenance at different polar domains in plant cells. Cell Discov. 2016;2:16018. doi: 10.1038/celldisc.2016.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Glawischnig E, Hansen BG, Olsen CE, Halkier BA. Camalexin is synthesized from indole-3-acetaldoxime, a key branching point between primary and secondary metabolism in Arabidopsis. Proc Natl Acad Sci USA. 2004;101:8245–8250. doi: 10.1073/pnas.0305876101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Crouzet J, Trombik T, Fraysse AS, Boutry M. Organization and function of the plant pleiotropic drug resistance ABC transporter family. FEBS Lett. 2006;580:1123–1130. doi: 10.1016/j.febslet.2005.12.043. [DOI] [PubMed] [Google Scholar]
- 42.Jungwirth H, Kuchler K. Yeast ABC transporters– A tale of sex, stress, drugs and aging. FEBS Lett. 2006;580:1131–1138. doi: 10.1016/j.febslet.2005.12.050. [DOI] [PubMed] [Google Scholar]
- 43.Zhao J, Last RL. Coordinate regulation of the tryptophan biosynthetic pathway and indolic phytoalexin accumulation in Arabidopsis. Plant Cell. 1996;8:2235–2244. doi: 10.1105/tpc.8.12.2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stefanato FL, et al. The ABC transporter BcatrB from Botrytis cinerea exports camalexin and is a virulence factor on Arabidopsis thaliana. Plant J. 2009;58:499–510. doi: 10.1111/j.1365-313X.2009.03794.x. [DOI] [PubMed] [Google Scholar]
- 45.Bednarek P, et al. A glucosinolate metabolism pathway in living plant cells mediates broad-spectrum antifungal defense. Science. 2009;323:101–106. doi: 10.1126/science.1163732. [DOI] [PubMed] [Google Scholar]
- 46.Mysore KS, Ryu C-M. Nonhost resistance: How much do we know? Trends Plant Sci. 2004;9:97–104. doi: 10.1016/j.tplants.2003.12.005. [DOI] [PubMed] [Google Scholar]
- 47.Consonni C, et al. Tryptophan-derived metabolites are required for antifungal defense in the Arabidopsis mlo2 mutant. Plant Physiol. 2010;152:1544–1561. doi: 10.1104/pp.109.147660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Egusa M, Miwa T, Kaminaka H, Takano Y, Kodama M. Nonhost resistance of Arabidopsis thaliana against Alternaria alternata involves both pre- and postinvasive defenses but is collapsed by AAL-toxin in the absence of LOH2. Phytopathology. 2013;103:733–740. doi: 10.1094/PHYTO-08-12-0201-R. [DOI] [PubMed] [Google Scholar]
- 49.Lazniewska J, Macioszek VK, Lawrence CB, Kononowicz AK. Fight to the death: Arabidopsis thaliana defense response to fungal necrotrophic pathogens. Acta Physiol Plant. 2010;32:1–10. [Google Scholar]
- 50.Nowicki M, Nowakowska M, Niezgoda A, Kozik E. Vegetable Crops Research Bulletin. Versita; Warsaw: 2012. Alternaria black spot of crucifers: Symptoms, importance of disease, and perspectives of resistance breeding. [DOI] [Google Scholar]
- 51.Kumar D, et al. Alternaria blight of oilseed Brassicas: A comprehensive. Afr J Microbiol Res. 2014;8:2816–2829. [Google Scholar]
- 52.Choi J, et al. The cytokinin-activated transcription factor ARR2 promotes plant immunity via TGA3/NPR1-dependent salicylic acid signaling in Arabidopsis. Dev Cell. 2010;19:284–295. doi: 10.1016/j.devcel.2010.07.011. [DOI] [PubMed] [Google Scholar]
- 53.Gachon C, Saindrenan P. Real-time PCR monitoring of fungal development in Arabidopsis thaliana infected by Alternaria brassicicola and Botrytis cinerea. Plant Physiol Biochem. 2004;42:367–371. doi: 10.1016/j.plaphy.2004.04.001. [DOI] [PubMed] [Google Scholar]
- 54.Glazebrook J, Ausubel FM. Isolation of phytoalexin-deficient mutants of Arabidopsis thaliana and characterization of their interactions with bacterial pathogens. Proc Natl Acad Sci USA. 1994;91:8955–8959. doi: 10.1073/pnas.91.19.8955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Savatin DV, Bisceglia NG, Gravino M, Fabbri C, Pontiggia D, Mattei B. Camalexin quantification in leaves infected with B. cinerea. Bio Protoc. 2015;5:e1379. doi: 10.21769/BioProtoc.1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ohno R, et al. Cryptogein-induced cell cycle arrest at G2 phase is associated with inhibition of cyclin-dependent kinases, suppression of expression of cell cycle-related genes and protein degradation in synchronized tobacco BY-2 cells. Plant Cell Physiol. 2011;52:922–932. doi: 10.1093/pcp/pcr042. [DOI] [PubMed] [Google Scholar]
- 57.Mengiste T, Chen X, Salmeron JM, Dietrich RA. The BOTRYTIS SUSCEPTIBLE1 gene encodes an R2R3MYB transcription factor protein that is required for biotic and abiotic stress responses in Arabidopsis. Plant Cell. 2003;15:2551–2565. doi: 10.1105/tpc.014167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Grubb CD, Gross HB, Chen DL, Abel S. Identification of Arabidopsis mutants with altered glucosinolate profiles based on isothiocyanate bioactivity. Plant Sci. 2002;162:143–152. [Google Scholar]