Significance
Ceramide is a bioactive lipid involved in numerous cellular functions and disease states that are critically dependent on its site of generation. nSMase2 generates ceramide at the inner leaflet of the plasma membrane and is a therapeutic target for cancer and neurological disorders. Although much is known about the cellular functions of nSMase2, there is limited insight into the molecular mechanisms regulating its activity. Here we present the crystal structure of nSMase2 and identify the lipid-binding N-terminal domain as an allosteric activation domain. Key to activation is a catalytic motif termed the “DK switch,” whose conformation is allosterically gated. This study reveals one mechanism for nSMase2 regulation by lipids and will help guide structure-based development of nSMase2-targeted therapeutics.
Keywords: sphingomyelinase, ceramide, enzyme, lipid, crystallography
Abstract
Neutral sphingomyelinase 2 (nSMase2, product of the SMPD3 gene) is a key enzyme for ceramide generation that is involved in regulating cellular stress responses and exosome-mediated intercellular communication. nSMase2 is activated by diverse stimuli, including the anionic phospholipid phosphatidylserine. Phosphatidylserine binds to an integral-membrane N-terminal domain (NTD); however, how the NTD activates the C-terminal catalytic domain is unclear. Here, we identify the complete catalytic domain of nSMase2, which was misannotated because of a large insertion. We find the soluble catalytic domain interacts directly with the membrane-associated NTD, which serves as both a membrane anchor and an allosteric activator. The juxtamembrane region, which links the NTD and the catalytic domain, is necessary and sufficient for activation. Furthermore, we provide a mechanistic basis for this phenomenon using the crystal structure of the human nSMase2 catalytic domain determined at 1.85-Å resolution. The structure reveals a DNase-I–type fold with a hydrophobic track leading to the active site that is blocked by an evolutionarily conserved motif which we term the “DK switch.” Structural analysis of nSMase2 and the extended N-SMase family shows that the DK switch can adopt different conformations to reposition a universally conserved Asp (D) residue involved in catalysis. Mutation of this Asp residue in nSMase2 disrupts catalysis, allosteric activation, stimulation by phosphatidylserine, and pharmacological inhibition by the lipid-competitive inhibitor GW4869. Taken together, these results demonstrate that the DK switch regulates ceramide generation by nSMase2 and is governed by an allosteric interdomain interaction at the membrane interface.
Ceramide is a bioactive lipid that regulates multiple cellular responses such as growth arrest, senescence, and apoptosis (1, 2). Rapid generation of ceramide in response to extracellular stimuli is mediated by the activation of sphingomyelinases (SMases), which hydrolyze sphingomyelin (SM), a major component of cell membranes, into ceramide and phosphocholine (3, 4). Three families of SMases (acid, neutral, and alkaline) have been identified that are classified by their pH optima, subcellular localization, protein fold, and function (5–8). Neutral sphingomyelinase 2 (nSMase2, product of the SMPD3 gene) is the major neutral sphingomyelinase (N-SMase) in mammalian cells for the stress-induced generation of ceramide (4). TNF-α (9–15), cell confluence (16, 17), oxidative stress (18–20), and inflammation (20, 21) are all potent activators of nSMase2.
nSMase2 is a membrane-associated enzyme located in the inner leaflet of the plasma membrane (PM) that has been implicated in several cellular and physiological processes including apoptosis (22), cell-cycle and growth arrest (23, 24), inflammation and immune responses (21, 25, 26), and bone homeostasis (27–29). Notably, nSMase2 also has been identified as a critical regulator of exosomes (30, 31), which package and transfer pathogenic factors that include metastasis-promoting microRNAs (32, 33), tau protein (34), and amyloid-β (35). This activity has established nSMase2 as a therapeutic target for cancer and Alzheimer’s disease.
NSMase2 contains a hydrophobic N-terminal domain (NTD) and a C-terminal catalytic domain (CAT). The NTD is an integral-membrane domain that contains two hydrophobic segments, which are predicted to be helical but do not span the membrane (36). The NTD is a lipid-binding domain that binds phosphatidylserine (PS), which stimulates and is required for full nSMase2 activity (23, 24, 37). Lipid-dependent activation is necessary for nSMase2 function and distinguishes nSMase2 from other SMases (23, 38). However, how PS binding by the NTD activates the CAT of nSMase2 is unclear. The CAT is predicted to belong to the DNase I superfamily of enzymes (39), and the crystal structures of several bacterial homologs have been reported (40–42). However, all bacterial SMases derive from human pathogens, share limited sequence homology with nSMase2 (<10%), and contain a soluble unregulated CAT; these considerations have limited our understanding of nSMase2 regulation.
Here, we present extensive structural and biochemical data indicating that the membrane-associated NTD directly binds and activates the soluble nSMase2 CAT. Activation is mediated by a universally conserved motif, which we term the “DK switch.” We identify a function for the DK switch in catalysis and demonstrate a role for it in PS activation. These findings are supported by the crystal structure of the human nSMase2 CAT at 1.85-Å resolution that reveals the structural features distinguishing nSMase2 from its unregulated bacterial counterparts and will aid in the development of nSMase2 as a therapeutic target.
Results
Identification of the CAT.
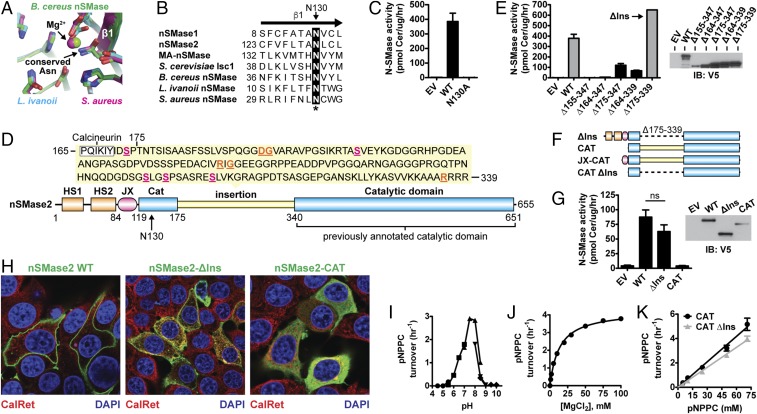
Sequence analysis found that the annotated CAT of nSMase2 (residues 341–655) lacked a strictly conserved active-site Asn residue. This conserved Asn residue is part of the first β-strand of the DNase I-type protein fold and is involved in metal coordination (Fig. 1A) (41). To identify this critical residue, we used a combination of secondary structure predictions and sequence alignments with bacterial and eukaryotic nSMases to search for any Asn residues present in predicted β-strands upstream of the annotated CAT. We identified Asn130 as the putative missing residue in human nSMase2, because it was present in the first and only predicted β-strand (residues 123–133) upstream of the annotated CAT (Fig. 1B and Fig. S1). Consistently, the point mutant N130A eliminated all nSMase2 activity (Fig. 1C). This finding suggested that the complete CAT of nSMase2 encompassed residues 119–655 and contained a large insertion that previously was referred to as a “collagen-like linker” (Fig. 1D) (23).
Fig. 1.
Domain architecture and characterization of nSMase2. (A) Superposition of bacterial SMase active sites highlights the Asn residue that is involved in Mg2+ coordination and is absent in the annotated nSMase2 CAT. (PDB ID codes: B. cereus, 2DDT, chain A; L. ivanovii, 1ZWX; S. aureus, 3I48 chain A.) (B) Sequence alignment of the β1-strand of N-SMases predicts N130 in human nSMase2 as the missing Asn residue involved in Mg2+ coordination. (C) The point mutant N130A eliminates SMase activity of nSMase2 from cell lysates of S. cerevisiae ΔIsc1p cells. Data represent mean ± SD (n = 2). (D) The newly identified domain architecture of nSMase2 consists of two N-terminal hydrophobic segments (HS1 and HS2), a JX region, and a CAT that contains a large insertion. Within the insertion are many regulatory elements including five phosphoserine sites (magenta, underlined), a calcineurin-binding site, and mutations associated with leukemia (orange, underlined). (E) SMase activity from cell lysates of S. cerevisiae ΔIsc1p cells expressing the indicated nSMase2 indel constructs. Data represent mean ± SD (n = 2). EV, empty vector; IB, immunoblot. (F) Domain architecture of nSMase2 constructs characterized in this article. (G) SMase activity from cell lysates of MCF7 cells transiently transfected with the indicated nSMase2 constructs. Data represent mean ± SD (n = 4). EV, empty vector. (H) Cellular localization of nSMase2 constructs (green) transiently transfected in MCF7 cells. ns, not significant. (I–K) Hydrolysis of pNPPC by purified nSMase2-CAT as a function of pH (each symbol corresponds to a different buffer system; see Materials and Methods) (I), magnesium ion concentration (J), and pNPPC concentration (K). Data represent mean ± SD (n = 3).
Fig. S1.
Domain architecture and structure-based sequence alignment of the N-SMase superfamily. (A) Domain architecture of N-SMases highlighting the hydrophobic domains that anchor eukaryotic N-SMases in the membrane. (B) Sequence alignment of the N-SMase superfamily. Secondary structure elements for human nSMase2 are shown above. Universally conserved residues are highlighted in black. Homologous residues are highlighted in gray. Residues involved in magnesium coordination and catalysis are denoted by a red asterisk. The position of the DK switch is labeled.
A Large CAT Insertion Is a Regulatory Region That Is Not Required for Full Enzyme Activity.
The majority of known regulatory elements in nSMase2 are found within the insertion. These include phosphoserine sites that affect protein stability (43), a docking site for the phosphatase calcineurin (44), and point mutations linked with leukemia (Fig. 1D) (45). Large CAT insertions in the phospholipase C isozymes play critical roles in regulating activity (46, 47). We therefore sought to define the boundaries of the insertion and to assess its role in nSMase2 regulation.
Sequence alignment of nSMase2 with distantly related bacterial SMases suggested that the insertion spanned residues 155–347 (Fig. S1). Deletion of this region (Δ155–347) eliminated all nSMase2 activity (Fig. 1E). We therefore shortened the deletion boundaries and thereby restored some activity. Ultimately, we deleted an internal stretch of 165 amino acids within the CAT (Δ175–339) (Fig. 1F) that slightly increased nSMase2 activity when expressed in Saccharomycetes cerevisiae cells (Fig. 1E) and did not significantly affect activity when expressed in MCF7 cells (Fig. 1G). We refer to this fully active construct (Δ175–339) as “nSMase2-ΔIns” or “ΔIns.”
Given its negligible effects on enzyme activity, we examined the role of the insertion in cellular localization, because translocation to the PM is critical for nSMase2 signaling (48). As previously reported (37), WT nSMase2 localized to the inner leaflet of the PM in confluent MCF7 cells (Fig. 1H). Deletion of the insertion altered nSMase2 trafficking to induce colocalization with the endoplasmic reticulum and the nuclear envelope marker calreticulin (Fig. 1H). nSMase2-ΔIns also colocalized with the PM, but with a more diffuse pattern (Fig. 1H). Taken together, these results showed that although the insertion regulates the cellular function of nSMase2 at multiple levels, it is dispensable for enzyme activity.
Characterization of the CAT.
Having identified the CAT (Fig. 1F), we set out to characterize it. Transfection of the nSMase2 CAT in MCF7 cells generated a cytoplasmic protein that no longer colocalized with cell membranes (Fig. 1H). This result suggested the CAT is soluble and that the hydrophobic NTD is the main determinant of nSMase2 membrane association. The activity of the CAT was first assessed in cell lysates of transfected MCF7 cells, but its activity was not significantly above the background activity of cells transfected with empty vector (Fig. 1G). Because the activity of the CAT was too low to facilitate biochemical characterization from cell lysates, we purified the recombinant CAT from Escherichia coli and assessed activity against the soluble substrate para-nitrophenyl-phosphocholine (pNPPC), which shares the phosphocholine headgroup with SM. The CAT hydrolyzed pNPPC with a neutral pH optimum (Fig. 1I) and Mg2+ dependence (Fig. 1J), two hallmark biochemical properties of nSMase2 (23). The purified CAT and CAT-ΔIns had comparable activities toward pNPPC (Fig. 1K), confirming that the insertion does not affect enzymatic function.
Structure Determination.
Structures of bacterial SMases have been determined (41). However, bacterial SMases are unregulated exotoxins from human pathogens that share limited sequence homology (∼10%) with nSMase2 and thus have provided little insight into the mechanisms that regulate nSMase2 function. All attempts to obtain diffraction-quality crystals of the nSMase2 CAT that retained the insertion were unsuccessful. However, using the aforementioned insight into the newly identified domain architecture of nSMase2, crystals of human CAT-ΔIns were obtained. The structure was determined using single-wavelength anomalous dispersion from a 2.9-Å dataset of selenomethione (SeMet)-derivatized protein. The final structure was refined to 1.85-Å resolution with an R factor of 16.25 and an Rfree of 19.60 (Table 1).
Table 1.
Data collection and refinement statistics
| Crystal | SeMet-nSMase2 | nSMase2 |
| Space group | P 21 21 2 | P 21 21 2 |
| Cell dimensions | ||
| a, b, c, Å | 72.56, 91.69, 49.82 | 73.59, 91.04, 50.03 |
| α, β, γ, ° | 90.0, 90.0, 90.0 | 90.0, 90.0, 90.0 |
| Data collection† | ||
| Resolution, Å | 43.94−2.89 (2.99−2.89) | 43.85−1.85 (1.855−1.849) |
| Wavelength, Å | 0.9793 | 1.075 |
| Reflections | ||
| Observed | 99,072 (6,830) | 191,989 (2,044) |
| Unique | 7,792 (542) | 29,448 (311) |
| Rmerge | 0.144 (0.788) | 0.076 (0.882) |
| Rmeas | 0.150 (0.822) | 0.095 (0.956) |
| Ranom | 0.142 (0.746) | N/A |
| Rpim | 0.042 (0.230) | 0.037 (0.365) |
| CC1/2 | 0.999 (0.961) | 0.999 (0.851) |
| CC* | 1.000 (0.944) | |
| I/σI | 17.1 (3.8) | 13.8 (2.2) |
| Completeness, % | 100 (100) | 100 (100) |
| Multiplicity | 12.7 (12.6) | 6.5 (6.6) |
| Wilson B | 66.9 | 23.9 |
| Refinement† | ||
| Resolution, Å | 43.85−1.85 (1.855−1.849) | |
| No. reflections | 29,448 (311) | |
| Rwork/Rfree | 0.1625/0.1960 | |
| No. atoms | ||
| Protein | 2,752 | |
| Solvent | 216 | |
| B-factors | ||
| Protein | 34.6 | |
| Solvent | 34.5 | |
| Rmsd | ||
| Bond lengths, Å | 0.01 | |
| Bond angles, ° | 1.06 | |
| Ramachandran | ||
| Favored, % | 97.3 | |
| Outliers, % | 0 | |
| PDB ID code | 5UVG |
Values in parenthesis are for the highest resolution shell.
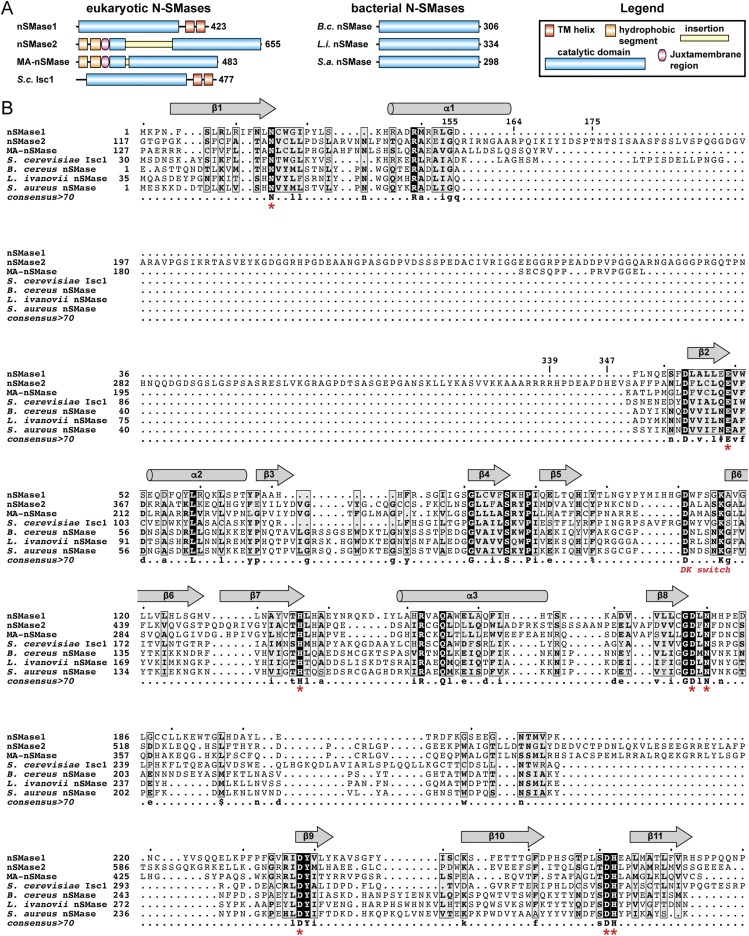
Overall Structure and Active Site.
The nSMase2 CAT adopts a DNase-I–type fold with a β-sandwich core and connecting loops and secondary structure elements forming the active site pocket (Fig. 2 A and B). As predicted, Asn130 is located in the first β-strand and helps coordinate a Ca2+ ion bound within the active site (Fig. 2C). This Ca2+ ion most likely derives from exogenous Ca2+ in the crystallization conditions and occupies the position of the primary Mg2+ ion that is required for the Mg2+-dependent activity of nSMase2 (23).
Fig. 2.
Crystal structure of the human nSMase2 CAT. (A) Cartoon representation of the human nSMase2 CAT structure (residues 117–651 Δ175–339) with the palmitoylation loop oriented to the membrane. The ΔIns junction that connects residues 174 and 340 is labeled. The Ca2+ ion in the active site is shown as a magenta sphere, β-strands in magenta, α-helices in cyan, and loops in salmon. (B) Structural comparison of human nSMase2 (Upper) and B. cereus bacterial SMase (PDB ID code 2UYR) (Lower) color coded to emphasize divergent functions and regulations of the key structural elements. The bacterial β-hairpin for membrane interaction, the insertion containing the calcineurin-binding site in human nSMase2, and distinct active site pockets are labeled. The β-sandwich core and active-site residues are largely conserved and are colored gray. (C) Active site of nSMase2 with conserved residues for SM recognition and hydrolysis. (D) All the conserved residues among bacterial and eukaryotic N-SMases (shown as sticks) cluster in the active site, suggesting a common mechanism for SM binding and hydrolysis. (PDB ID codes: nSMase2, 5UVG; B. cereus, 2UYR; L. ivanovii, 1ZWX; S. aureus, 3I48, chain B.)
Although the majority of the insertion was deleted to facilitate crystallization, the remaining portion formed two β-strands on the periphery of the CAT, as is consistent with their being required for stability and activity (Fig. 2B). One of these strands contains the binding motif for the calmodulin-activated phosphatase calcineurin (44), which is in close proximity to a predicted IQ calmodulin-binding motif in an adjacent helix.
The β-sandwich core of nSMase2 contains all the residues predicted to be involved in Mg2+ binding and catalysis. The select few residues that are absolutely conserved in the N-SMase superfamily (Fig. S1) all cluster within the active site of nSMase2 (Fig. 2D). The composition of these active-site residues aligns with and matches the hydrophobic and polar regions of SM (Fig. 2C). This conservation suggests these are the most critical residues for SM recognition and catalysis and suggests that eukaryotic and bacterial N-SMases use a similar mechanism both to bind and to hydrolyze SM.
This mechanism is magnesium dependent with one or two Mg2+ ions bound within the active site by the conserved set of His, Asp, Glu, and Asn residues (41) (Fig. 2D). The phosphodiester bond of SM is predicted to bind to and be stabilized by Mg2+, which activates a water molecule for nucleophilic attack to generate a negatively charged pentavalent phosphorous atom (41). The collapse of this intermediate results in cleavage of the bond to generate ceramide and phosphocholine.
Structural and Functional Divergence with Pathogenic Bacterial SMases.
Comparison of human nSMase2 and Bacillus cereus bacterial SMase (41), the member of the DNase I superfamily most closely related to human nSMase2, identified extensive structural differences that likely reflect their divergent functions and regulation (Fig. 2B). Notably, nSMase2 lacks the hydrophobic β-hairpin that soluble bacterial SMases use to interact with the outer leaflet of the PM (Fig. 2B) (41). This difference clearly establishes the β-hairpin as a selective feature that bacterial SMases have evolved to function as soluble secreted exotoxins that induce hemolysis (49).
In addition, the shape of the active site pocket is completely restructured in human nSMase2 because of the unique primary sequence, length, secondary structure, and spatial positioning of the α-helices and loops that form the active site pocket (Fig. 2B and Fig. S2). This restructuring creates a bowl-like cavity in nSMase2 that is in sharp contrast to the lipid-like contoured active site of bacterial SMase (Fig. 2B).
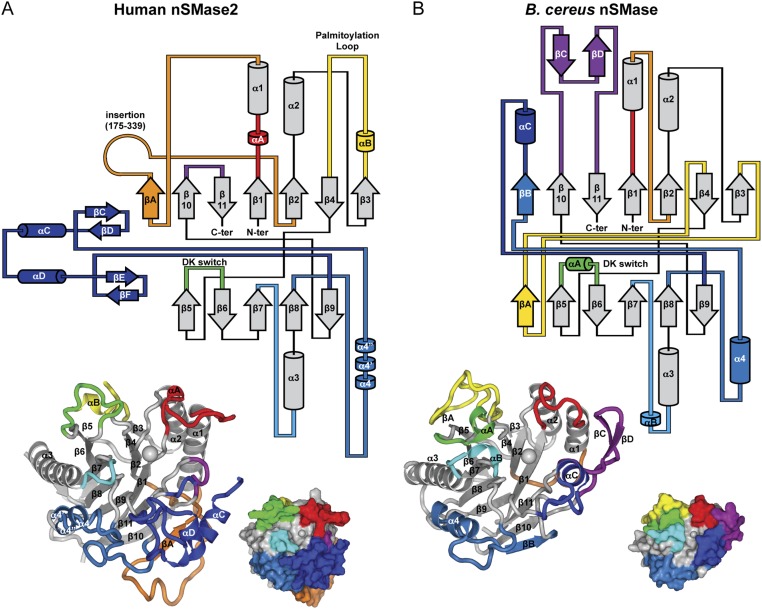
Fig. S2.
Topology diagram of human nSMase2 and B. cereus nSMase. (A and B) Topology diagrams of human nSMase2 (A) and B. cereus nSMase (B) highlighting the conserved β-sandwich core and the differing loops and helices that create the distinctive protein architectures. Elements that differ in sequence or structure are shown in matching colors. The 3D structures are shown below with labels. (B. cereus nSMase PDB ID code: 2UYR.)
Membrane Orientation.
To understand how SM would be extracted into the active site of nSMase2, we first sought to orient the CAT to the membrane interface. We took advantage of two adjacent Cys residues within the nSMase2 CAT that can be palmitoylated (50) and therefore must be in close proximity to the membrane (Fig. 2B). As a secondary guide, we used the β-hairpin of bacterial SMase that is known to interact with the membrane and is located on the opposite side of the CAT relative to the palmitoylation loop (Fig. 2B) (41). By aligning these two regions to the same 2D plane, we identified a hydrophobic groove at the membrane interface as the likely entryway for SM into the active site of both nSMase2 and bacterial SMase (Fig. 2B).
A Hydrophobic Groove for SM Binding Is Blocked in nSMase2.
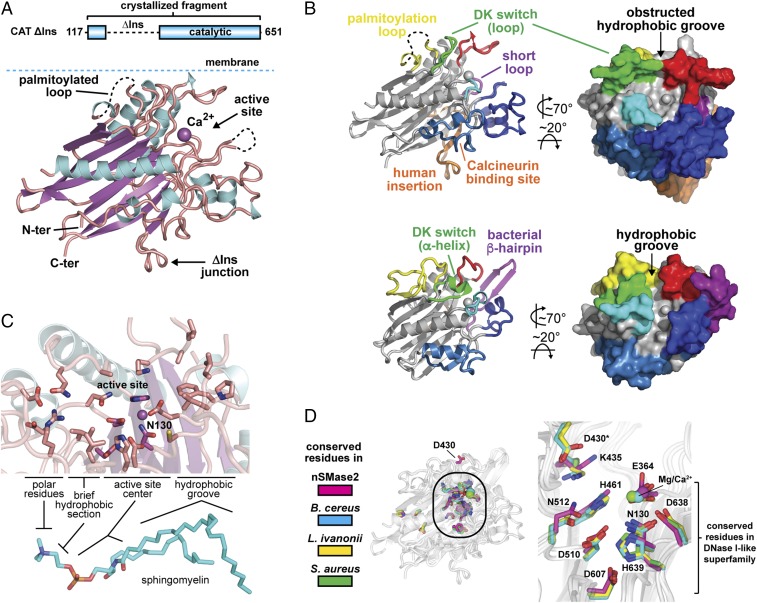
Next, we assessed whether SM could be accommodated within the different active sites of human nSMase2 and bacterial SMase. SM was easily placed manually in the lipid-contoured active site of bacterial SMase with the phospho moiety near the catalytic Mg2+ ion (Fig. 3A). The aliphatic chains of SM were not fully enclosed within the CAT, suggesting that SM is only partially extracted from the membrane by N-SMases during hydrolysis. Superposition of nSMase2 on the SM-bound bacterial structure identified significant clashes between nSMase2 and SM that all localized to the entrance of the hydrophobic groove (Fig. 3B). This observation suggests that conformational changes are required for nSMase2 to bind SM.
Fig. 3.
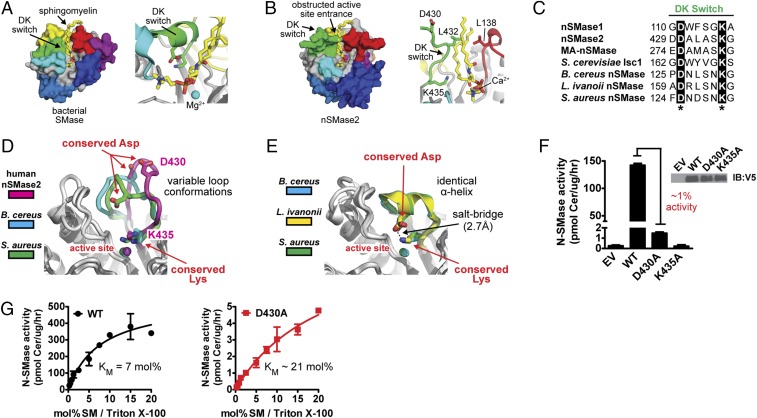
The DK switch is a universally conserved and flexible motif involved in SM catalysis. (A and B) SM is easily placed manually in the hydrophobic groove of bacterial SMase (PDB ID code 2UYR) (A) but clashes with the DK switch at the active site entrance in human nSMase2 (PDB ID code 5UVG) (B). (C) The Asp (D) and Lys (K) residues of the DK switch are universally conserved among mammalian, yeast, and bacterial N-SMases. (D and E) Structures of human nSMase2 and bacterial SMases highlighting the flexible nature of the DK switch in variable loop conformations that place the conserved Asp away from the active site (PDB ID codes: nSMase2, 5UVG; B. cereus, 2DDS, chain C; S. aureus, 3I48, chain A) (D) and a helical conformation that directs the conserved Asp into the active site to form a salt bridge with the conserved Lys residue of the DK switch. (PDB ID codes: B. cereus, 2UYR; L. ivanovii, 1ZWX; S. aureus, 3I48, chain B.) (E). (F) SMase activity of the nSMase2 DK switch point mutants from cell lysates of S. cerevisiae ΔIsc1p cells. Data represent mean ± SD (n = 3). (G) The D430A point mutant increases the Km of nSMase2, indicating a role in SM recognition. Data represent mean ± SD (n = 3).
A Universally Conserved DK Switch Motif Adopts Multiple Conformations.
A key factor in obstructing the hydrophobic groove is the element that we refer to as the DK switch and that previously was called the “P-loop–like domain” in the nSMase2 homolog of S. cerevisiae, Isc1p (51). We have renamed this element the DK switch because (i) it contains two key residues: an Asp (D) and Lys (K) that are absolutely conserved among N-SMases (Fig. 3C) but not in other DNase I type enzymes, and (ii) it folds into different conformations in nSMase2 and bacterial SMase (Fig. 3 D and E), indicating that this highly conserved element may “switch” conformations.
In nSMase2, the DK switch folds into an extended loop that juts into and obstructs the entrance to the hydrophobic groove (Fig. 3D). In this conformation, the conserved Asp residue (D430) is directed away from the active site (Fig. 3D). In the available structures of bacterial SMases, the DK switch adopts several different conformations in which it (i) forms different loop conformations that direct the Asp residue away from the active site in a similar manner to nSMase2 (Fig. 3D), (ii) forms a short α-helix that directs the conserved Asp residue into the active site to form a salt bridge with the conserved Lys residue (Fig. 3E), or (iii) is disordered with no observable electron density. Switching between these loop and helical conformations would reposition the universally conserved Asp residue from outside to inside the active site center while the position of the Lys residue remains constant. It is noteworthy that the DK switch in nSMase2 is involved in an extensive crystal contact in which the DK switch packs into the active site of another molecule; this packing most likely affects its conformation. In comparison, the DK switch in the different bacterial structures is sometimes involved in a crystal contact. However, the relevance for protein structure in these cases is unclear, because crystal contracts are found in both the loop and helical conformations. Taken together, these findings suggested that the DK switch is a flexible motif and that conformational changes in the DK switch may affect the SMase activity of nSMase2.
The DK Switch Is Involved in Catalysis.
Despite a previous study indicating that the conserved Asp and Lys residues of the DK switch were both essential for the activity of the nSMase2 homolog of S. cerevisiae Isc1p (51), the functional role of the DK switch in SM hydrolysis was unclear. We therefore generated the DK switch mutants D430A and K435A in full-length nSMase2 to assess their function. Similar to Isc1p, the K435A mutation completely eliminated nSMase2 activity (Fig. 3F). However, unlike Isc1p, the D430A mutant in full-length nSMase2 retained ∼1% activity (Fig. 3F), allowing us to probe the functional role of D430 in SM hydrolysis. The D430A point mutant dramatically reduced the Vmax by 100-fold in Triton X-100 mixed micelles (Fig. 3G). The Km of the D430A point mutant was also increased by approximately threefold, suggesting that D430 may be involved in SM recognition; however, this modest effect on Km could result from other causes such as loss of interaction between D430 and K435. Together, these results imply that the active state of the DK switch is the helical conformation that is observed in all the bacterial SMase structures of B. cereus, Listeria ivanovii, and Staphylococcus aureus. As in this conformation, the Asp side chain is directed into the active site where it forms a salt bridge with the conserved Lys side chain and could participate in the catalysis (Fig. 3E).
The Membrane-Associated NTD Stimulates SM Hydrolysis.
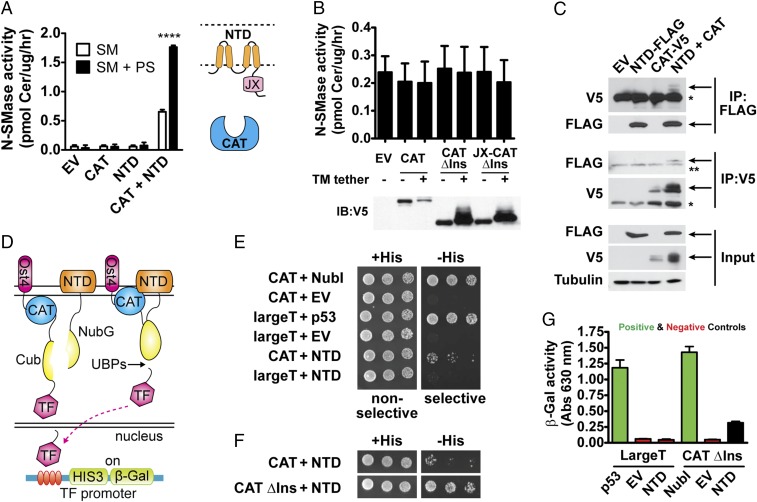
Before probing the mechanistic role of the DK switch, we first sought to delineate the activation mechanism of nSMase2. Based on our results, we hypothesized that the NTD plays a critical role in nSMase2 activation and that by deleting the NTD we trapped nSMase2-CAT in a low-activity state. We reasoned further that there would be a communication between the NTD and the CAT to impart this activation. Thus, we investigated whether coexpression of the NTD with the nSMase2 CAT would be sufficient to increase activity. Indeed, coexpression with the NTD stimulated the activity of the nSMase2 CAT (Fig. 4A). Furthermore, we speculated that the NTD, which contains two binding sites for PS (37), would mediate PS activation. Consistently, PS increased the activity of nSMase2 CAT when coexpressed with the NTD but not the activity of nSMase2 CAT alone (Fig. 4A).
Fig. 4.
The membrane-associated NTD interacts directly with and activates the soluble CAT of nSMase2. (A) Coexpression with the membrane-associated NTD stimulates the activity of the soluble CAT and promotes activation by PS. Data represent mean ± SD (n = 4). (B) Tethering the CAT of nSMase2 to the membrane with a transmembrane helix does not stimulate SMase activity. Immunoblots showing equivalent expression of the indicated proteins are shown below. Data represent mean ± SD (n = 3). (C) Interdomain activation is mediated through a direct interaction between the NTD and CAT as assessed by coimmunoprecipitation experiments. Arrows indicate specific bands. Single asterisks indicate the IgG-heavy chain band, and double asterisks indicate a nonspecific band. (D) Schematic of the split-ubiquitin membrane-yeast two-hybrid system with the CAT and NTD as bait and prey. (E and F) The NTD and the CAT interact directly as assessed by the membrane-yeast two-hybrid system. Serial dilutions of S. cerevisiae NMY51 cells coexpressing the indicated bait and prey constructs were spotted on nonselective (+His) and selective (−His) plates and were imaged after 3 d at 30 °C. Positive controls include NubI, a self-activating ubiquitin, and largeT and p53. Negative controls include CAT + empty vector (EV), largeT + EV, and largeT + NTD. (G) The relative strength of the NTD–CAT interaction is less than that with positive controls, indicating a putative transient interaction as assessed by β-galactosidase activity from cell lysates of S. cerevisiae NMY51 cells coexpressing the indicated bait and prey constructs. Data represent mean ± SD (n = 3).
This result suggested that the NTD functioned as an allosteric activator. However, we wanted to rule out the possibility that the NTD simply recruited nSMase2 CAT to the membrane and that this recruitment was sufficient for activation. To delineate these mechanisms, we assessed whether tethering nSMase2 CAT to the membrane with another transmembrane domain would stimulate activity. Membrane tethering of nSMase2 CAT was insufficient to increase activity (Fig. 4B). To ensure that this absence of effect was not caused by a linker of insufficient length, we included the entire region connecting the CAT and second hydrophobic region of the NTD. This extended linker was also insufficient to stimulate activity (Fig. 4B). Overall, these results suggest that the NTD serves as both a membrane-anchor and an allosteric activator of the CAT.
The NTD and the CAT Interact Directly.
Allosteric activation implies a direct interaction between the NTD and the CAT. To assess this interdomain interaction, we conducted coimmunoprecipitation experiments with the individual domains. We found that immunoprecipitation of the NTD pulled down the CAT, and vice versa, that immunoprecipitation of the CAT pulled down the NTD (Fig. 4C).
To confirm this interaction, we also applied a split-ubiquitin membrane yeast two-hybrid system (52) with the individual domains as bait and prey (Fig. 4D). An interaction between the NTD and the CAT would induce recombination of the split ubiquitins (the N-terminal half, Nub, and the C-terminal half, Cub) and recognition by ubiquitin proteases. This recognition would release a transcription factor to induce genes for growth on selective plates and β-galactosidase for quantitation (Fig. 4D). Using this system, we found that the NTD and the CAT of nSMase2 interacted directly, as assessed by growth on selective plates (Fig. 4E). Deletion of the insertion further promoted growth on selective plates (Fig. 4F). We attributed this further growth to reduced steric factors, given that nSMase2 and nSMase2-ΔIns had similar enzymatic activities.
To compare the relative strength of these interactions, we used β-galactosidase induction as a quantitative readout. The quantitated strength of the interaction between the NTD and the CAT was weaker than either a positive protein–protein interaction (p53–largeT) or an interaction with a self-activating prey (NubI) (Fig. 4G), suggesting that the interaction may be transient and might be potentiated by factors that activate nSMase2, such as the lipid activator PS.
The Juxtamembrane Region Is an Allosteric Activation Domain That Stimulates SM Hydrolysis and Increases the Affinity for SM.
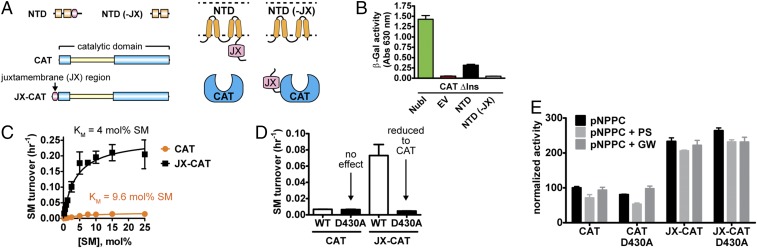
To determine which regions of the NTD were necessary for interdomain interactions, we generated a series of deletion constructs and found that deletion of the juxtamembrane (JX) region eliminated the interaction (Fig. 5 A and B). This finding suggested that the JX region, which does not contain any predicted secondary structural elements, interacts directly with the nSMase2 CAT.
Fig. 5.
The JX region is an allosteric activation domain that stimulates nSMase2 activity through the DK switch. (A) Domain architecture and membrane topology of nSMase2 constructs with and without the JX region. (B) Deletion of the JX region in the NTD [NTD (−JX)] eliminates the interdomain interaction between the NTD and CAT as assessed by β-galactosidase activity in the membrane-yeast two-hybrid system. (C) The addition of the JX region stimulates the SMase activity of the nSMase2 CAT and increases the affinity for SM. (D) The DK switch D430A point mutant eliminates the stimulatory effect of the JX region on basal SMase activity and has no effect on the activity of CAT. (E) Normalized activity of the nSMase2 CAT and the JX-CAT toward pNPPC, a soluble substrate that shares the phosphocholine headgroup of SM. PS at 20 mol% in Triton-X 100 mixed micelles and GW4869 (100 μM) had no effect on CAT and JX-CAT activity toward pNPPC. Data represent mean ± SD from three independent experiments.
Next, we assessed the role of the JX region on SMase activity. The JX region was appended to the CAT (hereafter referred to as the “JX-CAT”) and then was purified from E. coli. The purified JX-CAT had a 25-fold increase in enzyme efficiency (kcat/Km) in comparison with the nSMase-CAT (Fig. 5C). This increase was attributable to both an increase in the turnover rate (kcat) and, interestingly, an increased affinity for SM (a decreased Km). Although the JX region increased activity, the overall turnover rates of SM were slow for both the JX-CAT and the CAT. Notably, activation by the JX region was specific for the endogenous substrate SM, because the JX-CAT and the CAT had comparable activity against pNPPC (Fig. 5E).
The DK Switch Is Necessary for Activation by the JX Region.
Given the increase in SM affinity with the presence of the JX region and our finding that the point mutant D430A slightly increased the Km toward SM, we speculated that the JX region activated nSMase2 by modulating the DK switch. Thus, we generated the point mutant D430A in the nSMase2 CAT and JX-CAT and determined the effects on both SM and pNPPC hydrolysis. Consistent with a role in SM recognition, the D430A point mutant had no effect on pNPPC hydrolysis in both the CAT and JX-CAT (Fig. 5E). Strikingly, the D430A point mutant reduced the SMase activity of the JX-CAT back to the same level as in the CAT but notably did not affect the activity of the CAT toward SM (Fig. 5D). Taken together, these differential effects suggest that the JX region stimulates nSMase2 activity by promoting an active state of the DK switch.
PS Activation and the DK Switch.
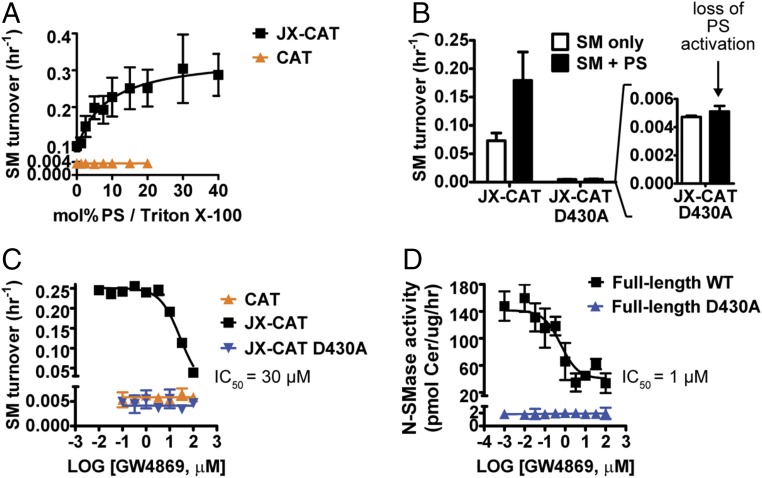
To probe the mechanism of nSMase2 further, we assessed whether the JX region and DK switch were necessary and sufficient for PS activation of nSMase2. Interestingly, PS stimulated the activity of the JX-CAT in a concentration-dependent manner but had no effect on the activity of the CAT (Fig. 6A). As a result, the SMase activity of the JX-CAT was increased approximately 100-fold over that of the CAT. PS did not stimulate the activity of JX-CAT D430A (Fig. 6B), thus providing an additional link between the gain of functions induced by the JX region and the DK switch. pNPPC hydrolysis by the CAT or JX-CAT was not stimulated by PS (Fig. 5E). Taken together, these findings identified the DK switch as a major element contributing to PS activation of nSMase2.
Fig. 6.
The JX region and DK switch are necessary for activation by PS and inhibition by the lipid-competitive inhibitor GW4869. (A) The JX region is necessary and sufficient for PS activation of the nSMase2 CAT. (B) Mutation of the DK switch eliminates PS activation of JX-CAT. (C) The JX region is necessary and sufficient for GW4869 inhibition of the nSMase2 CAT, and mutation of the DK switch eliminates GW4869 inhibition of JX-CAT. (D) The SMase activity of the full-length DK switch point mutant D430A is not inhibited by GW4869. All data in Fig. 6 represent the mean ± SD from three independent experiments.
GW4869 Inhibition and the DK Switch.
GW4869 is a noncompetitive (for the substrate SM) pharmacological inhibitor of nSMase2 that is competitive with PS binding to nSMase2 (22). We used GW4869 as an additional probe to assess the role of the JX region and the DK switch in nSMase2 activation. Consistent with the effects of PS activation, the CAT was not inhibited by GW4869, but the JX-CAT was inhibited by GW4869 with an IC50 of ∼30 μM (Fig. 6C). Again, the D430A point mutant eliminated the inhibition of the JX-CAT by GW4869 (Fig. 6C). These inhibitory effects were specific for SM, because GW4869 did not inhibit pNPPC hydrolysis for either the CAT or JX-CAT at concentrations up to 100 μM (Fig. 5E).
Next we tested the effects of GW4869 inhibition on full-length nSMase2. Consistent with our original report (22), GW4869 inhibited full-length nSMase2 with an IC50 of ∼1 μM (Fig. 6D). Interestingly, the residual activity of full-length D430A was no longer inhibited by GW4869 (Fig. 6D). These results are consistent with the original identification of GW4869 as a PS-competitive inhibitor of nSMase2.
Discussion
Almost 15 y ago, nSMase2 was cloned and identified as the PS-activated, Mg2+-dependent N-SMase (23, 24). Since then nSMase2 has been established as a key regulator of exosome secretion and cell-stress responses and has been implicated in cancer (32, 33), Alzheimer’s disease (34, 35), and cardiovascular disease (53). Based on our biochemical and structural experiments, we now are able to propose a model for the activation of nSMase2.
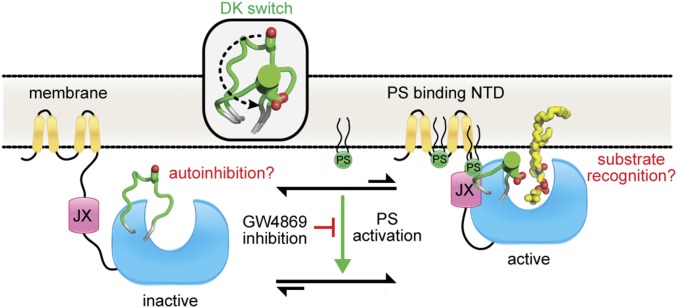
Our data suggest that nSMase2 is regulated by an interdomain interaction at the membrane interface between the membrane-associated NTD and the soluble CAT. This interaction modulates the DK switch to promote SM hydrolysis (Fig. 7). The DK switch may play multiple roles, because our crystal structure suggests that it prevents SM binding by occluding the active site entrance, but our biochemical results reveal that its role in catalysis is dependent on allosteric activation by the JX region of the NTD. Together, these findings have identified the lipid-binding NTD as an allosteric activation domain that both serves as a membrane anchor and actively participates in catalysis by interacting directly with and stimulating the activity of the CAT.
Fig. 7.
Model for nSMase2 activation. Interdomain interactions between the membrane-associated NTD and soluble CAT induce a conformational change in the DK switch (Inset) to promote SM hydrolysis. The requirement for both the JX region and the DK switch for both PS activation and GW4869 inhibition suggests a potential mechanism for lipid activation of nSMase2.
The membrane topology of this integral membrane domain suggests that a large portion of the NTD is exposed to solvent and could make extensive contacts with the CAT at multiple sites (36). The JX, which comprises only one of these regions, is sufficient to induce several gains of function, including increased SMase activity, activation by PS, and inhibition by the lipid-competitive inhibitor GW4869. All these gains of function that are induced by the JX region are dependent on the DK switch and are substrate specific, suggesting that at least one major component of PS activation is to promote the helical state of the DK switch, where the Asp and Lys residues form a salt bridge in the active site and can participate in SM catalysis. Notably, the DK switch is the only region of bacterial SMases that displays any conformational flexibility. Although the underlying reason for this conformational flexibility in the unregulated bacterial SMases is unknown, we speculate that its conformation may change upon SM binding or product release. However, in the nSMase2 CAT the D430A point mutant of the DK switch did not affect activity. This finding argues against an induced fit model for nSMase2 and suggests that the JX region allosterically regulates this universally conserved element.
Whether the NTD- and PS-binding induce more global conformational changes is a major remaining question and will require additional structural studies of full-length nSMase2. This question is important because the JX region harbors one previously identified PS-binding site, and the NTD harbors an additional PS-binding site (37). In addition, the SM turnover rate by JX-CAT was slow, possibly because of the loss of membrane association, suggesting that additional factors may contribute to nSMase2 activation. These studies of full-length nSMase2 will be aided by the ΔIns construct, which retains full enzyme activity but is more amenable to structural characterization.
To date, the majority of characterized lipid-dependent enzymes are either soluble targets that are activated by recruitment to the membrane or integral membrane enzymes whose activity and stability are dependent on specific lipids. NSMase2 is unique in that it contains both soluble and integral membrane domains. We speculate that regulation by interdomain interactions and PS provides a distinct advantage for the localized production of ceramide, whose signaling functions are critically dependent on the site of generation. The recent finding that PS colocalizes with detergent-resistant sphingolipid membrane microdomains (54) in the PM suggests that localized activation of nSMase2 may be a critical factor in regulating the formation and secretion of exosomes (30) as well as in cellular responses to chemotherapy (55).
Our crystal structure of the nSMase2 CAT revealed several differences with the structures of bacterial SMases. Strikingly, the differing contours of the active-site pockets suggest that nSMase2 may undergo global conformational changes upon activation. However, additional structures of nSMase2 are necessary to reveal these changes because, in general, nSMase2 contains a more elaborate structure built around the β-sandwich core with either extended linkers or unique subdomains that create this distinct pocket. One of these unique regions is the binding motif for the phosphatase calcineurin, which was present on the undeleted region of the insertion. This motif is located on the periphery of the CAT and provides a clear anchor point to recruit the phosphatase calcineurin to regulate nSMase2 stability (44). Although the exact function for the remaining elements is not clear, they may also be involved in protein–protein interactions. For example, the inflammatory cytokine TNF-α activates nSMase2 by recruiting it to the PM through interactions between the CAT and the multiprotein complex TNF receptor-FAN-RACK1-EED (11, 12, 14). Importantly, nSMase2 lacked the β-hairpin that bacterial SMases use to bind and recognize glycolipids on the outer leaflet of the PM. This element is functionally replaced in nSMase2 by the palmitoylation sites within the CAT (50) and by the membrane-associated NTD, whose PS-binding sites are necessary for proper cellular localization (37).
nSMase2 is emerging as a therapeutic target for cancer and neurological disorders. This identification is based on several studies that have used GW4869 to inhibit exosome secretion (30), to prevent cancer metastasis (32, 33), and to prevent tau protein and Aβ peptide secretion (35, 56). Our data suggest that the PS-competitive inhibitor GW4869 inhibits nSMase2 by targeting the activation of the DK switch. Thus, its success as a pharmacological tool may result from the inhibition of the endogenous activation mechanism of nSMase2. Last, our crystal structure provides avenues to develop nSMase2 as a therapeutic target.
Materials and Methods
Plasmid Constructs and Site-Directed Mutagenesis.
Various human nSMase2 constructs (full-length WT: residues 1–655; ΔIns: residues 1–655 Δ175–339; and CAT: residues 119–655) were cloned into the S. cerevisiae expression vector pYES3 using BamHI and NotI restriction sites in frame with the C-terminal V5-His tag. For mammalian expression, nSMase2 constructs were subcloned into the pEF6 plasmid using the flanking BamHI and PmeI restriction sites. Point mutants and internal deletions were generated using either a QuikChange Mutagenesis Kit (Agilent) or overlap extension methods. All plasmids were directly sequenced.
Protein Expression and Purification.
Fragments of human nSMase2 encoding for the CAT (residues 117–651) and the CAT-JX (residues 85–651) were cloned into the ppSUMO vector using BamHI and NotI restriction sites, which added a ULP-1 cleavable, N-terminal His-tagged SUMO fusion. For overexpression, plasmids were transformed into E. coli BL21 (DE3) RIPL cells. Cells were grown at 37 °C in Ultra-High Yield flasks (Thomson Instrument Company) to an OD600 ∼1.5–2.0 and then were cooled at 10 °C for 2 h. Protein expression was induced with isopropyl β-d-1-thiogalactopyranoside (IPTG) at 15 °C for 40 h. Harvested cells were resuspended in buffer A [50 mM sodium phosphate buffer (pH 7.5), 60 mM imidazole, 500 mM NaCl, 5% (vol/vol) glycerol, and 2 mM β-mercaptoethanol (βME)], lysed by sonication, centrifuged, and applied to a HisTrap FF column (GE Healthcare). After washing, proteins were eluted in buffer B [50 mM sodium phosphate buffer (pH 7.0), 300 mM imidazole, 500 mM NaCl, 5% glycerol]. Pooled fractions were supplemented with 10 mM βME and 10 mM DTT, incubated overnight at 4 °C with purified ULP-1 to cleave the His-SUMO tag, and applied to a Hi-Load 26/60 Superdex 200 column (GE Healthcare) equilibrated with 20 mM Bis-Tris (pH 6.5), 100 mM NaCl, 10 mM βME, and 2 mM DTT. Pooled fractions were supplemented with additional DTT to 10 mM. Concentrated proteins (10 mg/mL) were aliquoted, flash frozen, and stored at −80 °C. nSMase2 protein derivatized with selenomethionine (SeMet) was produced using B834 (DE3) cells. B834 cells were grown in M9 minimal medium supplemented with amino acids, SeMet, and Kao and Michayluk Vitamin Solution (Sigma).
Crystallization and Data Collection.
Crystals of native and SeMet nSMase2-CAT ΔIns (residues 117–651 Δ175–339) were grown using the hanging-drop method. Reservoir solution (2 μL) containing 20–30% PEG 400, 0.2 M CaCl2, 0–20% glycerol, and 0.1 M Hepes (pH 7.5) was mixed with 2 μL of protein solution (5–10 mg/mL) and incubated at 16 °C for 2–7 d. Crystals were cryoprotected by the addition of glycerol and were immediately frozen in liquid nitrogen before data collection. X-ray diffraction data for SeMet and native nSMase2 were collected on beamlines X25 and X29 of the National Synchrotron Light Source at Brookhaven National Laboratory using wavelengths of 0.9793 Å (the Se absorption edge) and 1.075 Å, respectively.
Data Processing, Structure Determination, and Refinement.
Diffraction data were integrated and scaled using XDS (57) and Aimless (58) as implemented in the autoPROC pipeline (59). All four selenium sites were located with SHELXD (60). Phases were calculated to 2.9 Å via single-wavelength anomalous dispersion (61) using SHARP (62). Several iterations of density modification and automated model building were carried out with Solomon (63), Buccaneer (64), and Parrot (65). The resulting map showed clear electron density for side chains, and the model was further improved by manual model building in Coot. This procedure yielded a nearly complete search model for molecular replacement with a 1.85-Å native dataset using Phaser (66). Subsequent model adjustments were carried out in Coot (67) followed by refinement with Phenix (68) and BUSTER (69). The geometric quality of the refined model was assessed with MolProbity (70) and the structure validation tools in the Phenix suite. Data collection and refinement statistics are shown in Table 1.
Analysis of Catalytic Activity Using pNPPC.
The catalytic activity of the purified nSMase2 CAT was determined using the generic phospholipase C substrate pNPPC or O-(4-Nitrophenylphosphoryl)choline (Sigma Aldrich). To determine pH optima (Fig. 1I), different overlapping buffers [sodium acetate (pH 4.5–5.5), Mes (pH 5.5–7.0), Hepes (pH 7.0–8.5), TAPS (N-Tris[hydroxymethyl]methyl-3-aminopropanesulfonic acid, pH 8.0–9.0), and CHES (N-Cyclohexyl-2-aminoethanesulfonic acid, pH 8.5–10.0)] were used to eliminate any buffer effects on enzyme activity. Before quantitation, the pH was adjusted with a 1-M NaOH solution to deprotonate the para-nitrophenol (pNP) product fully. The standard reaction contained 50 mM pNPPC, 100 mM MgCl2, and 50 mM of buffer in a 96-well plate. For Fig. 1J (Mg2+) and Fig. 1K (pNPPC) the amount of product, pNP, was followed in real time by monitoring the absorbance at 405 nm using a BioTek Synergy HT plate reader, and the kinetic curves were fit for accurate slope and rate determination. The pH was not adjusted with NaOH; such adjustment would increase the quantitated values slightly (∼15%). A molar extinction coefficient for pNP of 18,000 M/cm was used.
SMase Activity.
N-SMase activity was determined as previously described (71) using SM that was 14C-labeled in the methyl carbons of the choline headgroup. Briefly, SM from porcine brain (860062P; Avanti Polar Lipids), PS (840036P; Avanti Polar Lipids), and trace amounts of 14C-SM were dried under N2 gas in a glass vial and were resuspended in Triton X-100 (Research Products International) mixed micelles by sonication. The final reaction conditions contained 0.1% Triton X-100, 100 mM Tris (pH 7.5), 50 mM MgCl2, 5 mM βME, and 5 mM DTT. A standard reaction contained 5 mol% SM and 10 mol% PS, unless otherwise indicated. Cell lysates or purified protein were added to 100 μL of assay buffer and were incubated for 30–60 min at 37 °C (for cell lysates) or for 2–20 h at 25 °C (for purified protein). Reactions were quenched with the addition of CHCl3/MeOH (in a 2:1 ratio), and modified Bligh–Dyer extractions were performed. The aqueous phase, which contained the liberated 14C-labeled phosphocholine from SM hydrolysis, was transferred to scintillation vials, and counts per minute were quantitated using a Beckman LS 6500 scintillation counter. For determination of hydrolyzed ceramide, 20 μL of assay buffer was used to quantitate the ratio of picomoles of ceramide to the counts per minute.
Immunoprecipitation.
S. cerevisiae ΔIsc1 cells were transformed with constructs encoding for the C-terminally FLAG-tagged NTD and the C-terminally V5-tagged CAT. Cells were grown in Trp−Ura− synthetic dropout (Clontech) medium with glucose and were washed with water, and protein expression was induced by incubation in Trp−Ura− synthetic dropout medium with Gal/Raf for 18 h at 30 °C. Cells were washed and resuspended in lysis buffer [50 mM Tris⋅HCl (pH 7.4), 150 mM NaCl, 1 mM EDTA, and 1% Triton X-100] with protease and phosphatase inhibitors. Cell lysates were prepared using a bead beater and centrifugation at 0.6 × g. The supernatants were incubated with anti-FLAG M2 affinity resin or anti-V5 agarose affinity gel (Sigma) overnight at 4 °C as described by the manufacturer. The next day, the resins were washed three times (10 min each) with lysis buffer, and immunoprecipitated proteins were detected by Western blotting using anti-V5 (Life Sciences) and anti-FLAG (Sigma) antibodies.
Membrane Yeast Two-Hybrid System.
Interdomain interactions were assessed using the DUALHunter system (Dualsystems Biotech) (52). The bait proteins (nSMase2 CAT and CAT ΔIns) were cloned into the pDHB1 vector using SfiI restriction sites to generate a fusion at the N terminus with the small membrane protein Ost4p and a fusion at the C terminus with the C-terminal half of ubiquitin (Cub) followed by a transcription factor (LexA-VP16). Prey proteins of the NTD were cloned into the pPR3N vector using SfiI restriction sites to generate a fusion at the N terminus with the N-terminal half of ubiquitin (Nub). Bait and prey were transformed into the S. cerevisiae NMY51 reporter strain [MATa his3 200 trp1-901 leu2-3,112 ade2 LYS2::(lexAop)4-HIS3 ura3::(LexAop)8-lacZ ade2::(lexAop)8-ADE2 GAL4] using the standard lithium acetate method and were selected for using synthetic dropout-Leu-Trp plates. Qualitative assessment of interdomain interactions was determined using spot assays on nonselective (synthetic dropout-Leu-Trp plates) or selective (synthetic dropout-Leu-Trp-His plates). Quantitation was carried out using the β-galactosidase assay with cell lysis carried out using a Vortex Genie with TurboMix attachment.
Immunofluorescence and Confocal Microscopy.
The intracellular localization of nSMase2 proteins was determined using immunofluorescence confocal microscopy. MCF7 cells were plated onto poly-d-lysine–coated 35-mm confocal dishes (MatTek Corporation). The following day, cells were transfected using Lipofectamine (Invitrogen) with pEF6 plasmids containing nSMase2 constructs with C-terminal V5-His tags. After 24 h, cells were fixed with a 4% paraformaldehyde solution, washed, and permeabilized with 0.1% Triton X-100 in PBS. Cells were blocked in 2% human serum and incubated with primary antibodies (V5, 1:200) in 2% serum overnight at 4 °C. Secondary antibody incubation was carried out for 1 h before visualization. Cells were stained with DAPI 10 min before visualization. Images were captured with a confocal laser microscope (LSM510; Zeiss) and were analyzed with the Leica software package.
Acknowledgments
We thank the X25 and X29 beamlines at the National Synchrotron Light Source, Brookhaven National Laboratory for access to their facilities. This work was supported by NIH Grants R35GM118128 (to Y.A.H.), R01GM100021 (to M.G.-D.), F32GM100679 (to M.V.A.), and F30ES022930 (to K.E.G.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors reported in this paper have been deposited in the Protein Data Bank, www.wwpdb.org/ (PDB ID code 5UVG).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1705134114/-/DCSupplemental.
References
- 1.Obeid LM, Linardic CM, Karolak LA, Hannun YA. Programmed cell death induced by ceramide. Science. 1993;259:1769–1771. doi: 10.1126/science.8456305. [DOI] [PubMed] [Google Scholar]
- 2.Hannun YA, Obeid LM. Principles of bioactive lipid signalling: Lessons from sphingolipids. Nat Rev Mol Cell Biol. 2008;9:139–150. doi: 10.1038/nrm2329. [DOI] [PubMed] [Google Scholar]
- 3.Jenkins RW, Canals D, Hannun YA. Roles and regulation of secretory and lysosomal acid sphingomyelinase. Cell Signal. 2009;21:836–846. doi: 10.1016/j.cellsig.2009.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shamseddine AA, Airola MV, Hannun YA. Roles and regulation of neutral sphingomyelinase-2 in cellular and pathological processes. Adv Biol Regul. 2015;57:24–41. doi: 10.1016/j.jbior.2014.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xiong Z-J, Huang J, Poda G, Pomès R, Privé GG. Structure of human acid sphingomyelinase reveals the role of the saposin domain in activating substrate hydrolysis. J Mol Biol. 2016;428:3026–3042. doi: 10.1016/j.jmb.2016.06.012. [DOI] [PubMed] [Google Scholar]
- 6.Gorelik A, Illes K, Heinz LX, Superti-Furga G, Nagar B. Crystal structure of mammalian acid sphingomyelinase. Nat Commun. 2016;7:12196. doi: 10.1038/ncomms12196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou Y-F, et al. Human acid sphingomyelinase structures provide insight to molecular basis of Niemann-Pick disease. Nat Commun. 2016;7:13082. doi: 10.1038/ncomms13082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gorelik A, Liu F, Illes K, Nagar B. Crystal structure of the human alkaline sphingomyelinase provides insights into substrate recognition. J Biol Chem. 2017;292:7087–7094. doi: 10.1074/jbc.M116.769273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim M-Y, Linardic C, Obeid L, Hannun Y. Identification of sphingomyelin turnover as an effector mechanism for the action of tumor necrosis factor alpha and gamma-interferon. Specific role in cell differentiation. J Biol Chem. 1991;266:484–489. [PubMed] [Google Scholar]
- 10.Dressler KA, Mathias S, Kolesnick RN. Tumor necrosis factor-alpha activates the sphingomyelin signal transduction pathway in a cell-free system. Science. 1992;255:1715–1718. doi: 10.1126/science.1313189. [DOI] [PubMed] [Google Scholar]
- 11.Adam D, Wiegmann K, Adam-Klages S, Ruff A, Krönke M. A novel cytoplasmic domain of the p55 tumor necrosis factor receptor initiates the neutral sphingomyelinase pathway. J Biol Chem. 1996;271:14617–14622. doi: 10.1074/jbc.271.24.14617. [DOI] [PubMed] [Google Scholar]
- 12.Adam-Klages S, et al. FAN, a novel WD-repeat protein, couples the p55 TNF-receptor to neutral sphingomyelinase. Cell. 1996;86:937–947. doi: 10.1016/s0092-8674(00)80169-5. [DOI] [PubMed] [Google Scholar]
- 13.Belka C, et al. Tumor necrosis factor (TNF)-alpha activates c-raf-1 kinase via the p55 TNF receptor engaging neutral sphingomyelinase. EMBO J. 1995;14:1156–1165. doi: 10.1002/j.1460-2075.1995.tb07099.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Philipp S, et al. The Polycomb group protein EED couples TNF receptor 1 to neutral sphingomyelinase. Proc Natl Acad Sci USA. 2010;107:1112–1117. doi: 10.1073/pnas.0908486107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clarke CJ, Cloessner EA, Roddy PL, Hannun YA. Neutral sphingomyelinase 2 (nSMase2) is the primary neutral sphingomyelinase isoform activated by tumour necrosis factor-α in MCF-7 cells. Biochem J. 2011;435:381–390. doi: 10.1042/BJ20101752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marchesini N, et al. Role for mammalian neutral sphingomyelinase 2 in confluence-induced growth arrest of MCF7 cells. J Biol Chem. 2004;279:25101–25111. doi: 10.1074/jbc.M313662200. [DOI] [PubMed] [Google Scholar]
- 17.Hayashi Y, Kiyono T, Fujita M, Ishibashi M. cca1 is required for formation of growth-arrested confluent monolayer of rat 3Y1 cells. J Biol Chem. 1997;272:18082–18086. doi: 10.1074/jbc.272.29.18082. [DOI] [PubMed] [Google Scholar]
- 18.Liu B, et al. Glutathione regulation of neutral sphingomyelinase in tumor necrosis factor-α-induced cell death. J Biol Chem. 1998;273:11313–11320. doi: 10.1074/jbc.273.18.11313. [DOI] [PubMed] [Google Scholar]
- 19.Barth BM, Gustafson SJ, Kuhn TB. Neutral sphingomyelinase activation precedes NADPH oxidase-dependent damage in neurons exposed to the proinflammatory cytokine tumor necrosis factor-α. J Neurosci Res. 2012;90:229–242. doi: 10.1002/jnr.22748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rutkute K, Asmis RH, Nikolova-Karakashian MN. Regulation of neutral sphingomyelinase-2 by GSH: A new insight to the role of oxidative stress in aging-associated inflammation. J Lipid Res. 2007;48:2443–2452. doi: 10.1194/jlr.M700227-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Karakashian AA, Giltiay NV, Smith GM, Nikolova-Karakashian MN. Expression of neutral sphingomyelinase-2 (NSMase-2) in primary rat hepatocytes modulates IL-β-induced JNK activation. FASEB J. 2004;18:968–970. doi: 10.1096/fj.03-0875fje. [DOI] [PubMed] [Google Scholar]
- 22.Luberto C, et al. Inhibition of tumor necrosis factor-induced cell death in MCF7 by a novel inhibitor of neutral sphingomyelinase. J Biol Chem. 2002;277:41128–41139. doi: 10.1074/jbc.M206747200. [DOI] [PubMed] [Google Scholar]
- 23.Hofmann K, Tomiuk S, Wolff G, Stoffel W. Cloning and characterization of the mammalian brain-specific, Mg2+-dependent neutral sphingomyelinase. Proc Natl Acad Sci USA. 2000;97:5895–5900. doi: 10.1073/pnas.97.11.5895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marchesini N, Luberto C, Hannun YA. Biochemical properties of mammalian neutral sphingomyelinase 2 and its role in sphingolipid metabolism. J Biol Chem. 2003;278:13775–13783. doi: 10.1074/jbc.M212262200. [DOI] [PubMed] [Google Scholar]
- 25.Mittelbrunn M, et al. Unidirectional transfer of microRNA-loaded exosomes from T cells to antigen-presenting cells. Nat Commun. 2011;2:282. doi: 10.1038/ncomms1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Montecalvo A, et al. Mechanism of transfer of functional microRNAs between mouse dendritic cells via exosomes. Blood. 2012;119:756–766. doi: 10.1182/blood-2011-02-338004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aubin I, et al. A deletion in the gene encoding sphingomyelin phosphodiesterase 3 (Smpd3) results in osteogenesis and dentinogenesis imperfecta in the mouse. Nat Genet. 2005;37:803–805. doi: 10.1038/ng1603. [DOI] [PubMed] [Google Scholar]
- 28.Stoffel W, Jenke B, Blöck B, Zumbansen M, Koebke J. Neutral sphingomyelinase 2 (smpd3) in the control of postnatal growth and development. Proc Natl Acad Sci USA. 2005;102:4554–4559. doi: 10.1073/pnas.0406380102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Khavandgar Z, et al. A cell-autonomous requirement for neutral sphingomyelinase 2 in bone mineralization. J Cell Biol. 2011;194:277–289. doi: 10.1083/jcb.201102051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Trajkovic K, et al. Ceramide triggers budding of exosome vesicles into multivesicular endosomes. Science. 2008;319:1244–1247. doi: 10.1126/science.1153124. [DOI] [PubMed] [Google Scholar]
- 31.Kosaka N, et al. Secretory mechanisms and intercellular transfer of microRNAs in living cells. J Biol Chem. 2010;285:17442–17452. doi: 10.1074/jbc.M110.107821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tominaga N, et al. Brain metastatic cancer cells release microRNA-181c-containing extracellular vesicles capable of destructing blood-brain barrier. Nat Commun. 2015;6:6716. doi: 10.1038/ncomms7716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kosaka N, et al. Neutral sphingomyelinase 2 (nSMase2)-dependent exosomal transfer of angiogenic microRNAs regulate cancer cell metastasis. J Biol Chem. 2013;288:10849–10859. doi: 10.1074/jbc.M112.446831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Asai H, et al. Depletion of microglia and inhibition of exosome synthesis halt tau propagation. Nat Neurosci. 2015;18:1584–1593. doi: 10.1038/nn.4132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dinkins MB, Dasgupta S, Wang G, Zhu G, Bieberich E. Exosome reduction in vivo is associated with lower amyloid plaque load in the 5XFAD mouse model of Alzheimer’s disease. Neurobiol Aging. 2014;35:1792–1800. doi: 10.1016/j.neurobiolaging.2014.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tani M, Hannun YA. Analysis of membrane topology of neutral sphingomyelinase 2. FEBS Lett. 2007;581:1323–1328. doi: 10.1016/j.febslet.2007.02.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu BX, et al. Identification of novel anionic phospholipid binding domains in neutral sphingomyelinase 2 with selective binding preference. J Biol Chem. 2011;286:22362–22371. doi: 10.1074/jbc.M110.156471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tomiuk S, Hofmann K, Nix M, Zumbansen M, Stoffel W. Cloned mammalian neutral sphingomyelinase: Functions in sphingolipid signaling? Proc Natl Acad Sci USA. 1998;95:3638–3643. doi: 10.1073/pnas.95.7.3638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Airola MV, Hannun YA. Sphingolipids: Basic Science and Drug Development. Springer; Vienna: 2013. Sphingolipid metabolism and neutral sphingomyelinases; pp. 57–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Openshaw AE, Race PR, Monzó HJ, Vázquez-Boland J-A, Banfield MJ. Crystal structure of SmcL, a bacterial neutral sphingomyelinase C from Listeria. J Biol Chem. 2005;280:35011–35017. doi: 10.1074/jbc.M506800200. [DOI] [PubMed] [Google Scholar]
- 41.Ago H, et al. Structural basis of the sphingomyelin phosphodiesterase activity in neutral sphingomyelinase from Bacillus cereus. J Biol Chem. 2006;281:16157–16167. doi: 10.1074/jbc.M601089200. [DOI] [PubMed] [Google Scholar]
- 42.Huseby M, et al. Structure and biological activities of beta toxin from Staphylococcus aureus. J Bacteriol. 2007;189:8719–8726. doi: 10.1128/JB.00741-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Filosto S, Ashfaq M, Chung S, Fry W, Goldkorn T. Neutral sphingomyelinase 2 activity and protein stability are modulated by phosphorylation of five conserved serines. J Biol Chem. 2012;287:514–522. doi: 10.1074/jbc.M111.315481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Filosto S, Fry W, Knowlton AA, Goldkorn T. Neutral sphingomyelinase 2 (nSMase2) is a phosphoprotein regulated by calcineurin (PP2B) J Biol Chem. 2010;285:10213–10222. doi: 10.1074/jbc.M109.069963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim WJ, et al. Mutations in the neutral sphingomyelinase gene SMPD3 implicate the ceramide pathway in human leukemias. Blood. 2008;111:4716–4722. doi: 10.1182/blood-2007-10-113068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hicks SN, et al. General and versatile autoinhibition of PLC isozymes. Mol Cell. 2008;31:383–394. doi: 10.1016/j.molcel.2008.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lyon AM, Begley JA, Manett TD, Tesmer JJG. Molecular mechanisms of phospholipase C β3 autoinhibition. Structure. 2014;22:1844–1854. doi: 10.1016/j.str.2014.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Milhas D, Clarke CJ, Idkowiak-Baldys J, Canals D, Hannun YA. Anterograde and retrograde transport of neutral sphingomyelinase-2 between the Golgi and the plasma membrane. Biochim Biophys Acta. 2010;1801:1361–1374. doi: 10.1016/j.bbalip.2010.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bernheimer AW. Interactions between membranes and cytolytic bacterial toxins. Biochim Biophys Acta. 1974;344:27–50. doi: 10.1016/0304-4157(86)90018-3. [DOI] [PubMed] [Google Scholar]
- 50.Tani M, Hannun YA. Neutral sphingomyelinase 2 is palmitoylated on multiple cysteine residues. Role of palmitoylation in subcellular localization. J Biol Chem. 2007;282:10047–10056. doi: 10.1074/jbc.M611249200. [DOI] [PubMed] [Google Scholar]
- 51.Okamoto Y, Vaena de Avalos S, Hannun YA. Functional analysis of ISC1 by site-directed mutagenesis. Biochemistry. 2003;42:7855–7862. doi: 10.1021/bi0341354. [DOI] [PubMed] [Google Scholar]
- 52.Stagljar I, Korostensky C, Johnsson N, te Heesen S. A genetic system based on split-ubiquitin for the analysis of interactions between membrane proteins in vivo. Proc Natl Acad Sci USA. 1998;95:5187–5192. doi: 10.1073/pnas.95.9.5187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pavoine C, Pecker F. Sphingomyelinases: Their regulation and roles in cardiovascular pathophysiology. Cardiovasc Res. 2009;82:175–183. doi: 10.1093/cvr/cvp030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fairn GD, et al. High-resolution mapping reveals topologically distinct cellular pools of phosphatidylserine. J Cell Biol. 2011;194:257–275. doi: 10.1083/jcb.201012028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hannun YA. Functions of ceramide in coordinating cellular responses to stress. Science. 1996;274:1855–1859. doi: 10.1126/science.274.5294.1855. [DOI] [PubMed] [Google Scholar]
- 56.Guo BB, Bellingham SA, Hill AF. The neutral sphingomyelinase pathway regulates packaging of the prion protein into exosomes. J Biol Chem. 2015;290:3455–3467. doi: 10.1074/jbc.M114.605253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kabsch W. XDS. Acta Crystallogr D Biol Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Evans PR, Murshudov GN. How good are my data and what is the resolution? Acta Crystallogr D Biol Crystallogr. 2013;69:1204–1214. doi: 10.1107/S0907444913000061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vonrhein C, et al. Data processing and analysis with the autoPROC toolbox. Acta Crystallogr D Biol Crystallogr. 2011;67:293–302. doi: 10.1107/S0907444911007773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schneider TR, Sheldrick GM. Substructure solution with SHELXD. Acta Crystallogr D Biol Crystallogr. 2002;58:1772–1779. doi: 10.1107/s0907444902011678. [DOI] [PubMed] [Google Scholar]
- 61.Dauter Z, Dauter M, Dodson E. Jolly SAD. Acta Crystallogr D Biol Crystallogr. 2002;58:494–506. doi: 10.1107/s090744490200118x. [DOI] [PubMed] [Google Scholar]
- 62.Vonrhein C, Blanc E, Roversi P, Bricogne G. Automated structure solution with autoSHARP. Methods Mol Biol. 2007;364:215–230. doi: 10.1385/1-59745-266-1:215. [DOI] [PubMed] [Google Scholar]
- 63.Abrahams JP, Leslie AG. Methods used in the structure determination of bovine mitochondrial F1 ATPase. Acta Crystallogr D Biol Crystallogr. 1996;52:30–42. doi: 10.1107/S0907444995008754. [DOI] [PubMed] [Google Scholar]
- 64.Cowtan K. The Buccaneer software for automated model building. 1. Tracing protein chains. Acta Crystallogr D Biol Crystallogr. 2006;62:1002–1011. doi: 10.1107/S0907444906022116. [DOI] [PubMed] [Google Scholar]
- 65.Cowtan K. Recent developments in classical density modification. Acta Crystallogr D Biol Crystallogr. 2010;66:470–478. doi: 10.1107/S090744490903947X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.McCoy AJ, et al. Phaser crystallographic software. J Appl Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zwart P, et al. Automated structure solution with the PHENIX suite. In: Kobe B, Guss M, Huber T, editors. Structural Proteomics. Vol 426. Springer; New York: 2008. pp. 419–435. [DOI] [PubMed] [Google Scholar]
- 69.Smart OS, et al. Exploiting structure similarity in refinement: Automated NCS and target-structure restraints in BUSTER. Acta Crystallogr D Biol Crystallogr. 2012;68:368–380. doi: 10.1107/S0907444911056058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chen VB, et al. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shamseddine AA, et al. P53-dependent upregulation of neutral sphingomyelinase-2: Role in doxorubicin-induced growth arrest. Cell Death Dis. 2015;6:e1947. doi: 10.1038/cddis.2015.268. [DOI] [PMC free article] [PubMed] [Google Scholar]