Significance
Protein evolution in response to different environments has long been of interest to both evolutionary biologists and biochemists. High-altitude specialist catfishes in the Andes mountains offer an opportunity to examine the molecular adaptations accompanying adaptation to cold environments. Rhodopsins and other visual pigments form the first step in vision and have long been a model system for studying the molecular basis of sensory adaptations; however, many of these studies have focused solely on spectral shifts. Recent studies suggest that other aspects of function are as important for visual performance. We demonstrate that high-altitude amino acid variants significantly accelerate RH1 kinetics. These results suggest that the activity–stability trade-off characterized in cold-adapted enzymes also affects adaptation of signaling proteins through similar molecular mechanisms.
Keywords: visual pigment, protein evolution, G protein-coupled receptor, Andean catfishes, in vitro expression
Abstract
High-altitude environments present a range of biochemical and physiological challenges for organisms through decreases in oxygen, pressure, and temperature relative to lowland habitats. Protein-level adaptations to hypoxic high-altitude conditions have been identified in multiple terrestrial endotherms; however, comparable adaptations in aquatic ectotherms, such as fishes, have not been as extensively characterized. In enzyme proteins, cold adaptation is attained through functional trade-offs between stability and activity, often mediated by substitutions outside the active site. Little is known whether signaling proteins [e.g., G protein-coupled receptors (GPCRs)] exhibit natural variation in response to cold temperatures. Rhodopsin (RH1), the temperature-sensitive visual pigment mediating dim-light vision, offers an opportunity to enhance our understanding of thermal adaptation in a model GPCR. Here, we investigate the evolution of rhodopsin function in an Andean mountain catfish system spanning a range of elevations. Using molecular evolutionary analyses and site-directed mutagenesis experiments, we provide evidence for cold adaptation in RH1. We find that unique amino acid substitutions occur at sites under positive selection in high-altitude catfishes, located at opposite ends of the RH1 intramolecular hydrogen-bonding network. Natural high-altitude variants introduced into these sites via mutagenesis have limited effects on spectral tuning, yet decrease the stability of dark-state and light-activated rhodopsin, accelerating the decay of ligand-bound forms. As found in cold-adapted enzymes, this phenotype likely compensates for a cold-induced decrease in kinetic rates—properties of rhodopsin that mediate rod sensitivity and visual performance. Our results support a role for natural variation in enhancing the performance of GPCRs in response to cold temperatures.
High-altitude environments impose a suite of biochemical and physiological constraints on organisms, such as hypoxia, low atmospheric pressure, and decreasing temperatures (1–3). At the biochemical level, proteins adapted to such conditions are of particular interest to evolutionary biologists (1, 4). Studies of high-altitude–adapted organisms, especially endotherms, have focused primarily on adaptation to hypoxia, including modification of proteins involved in oxygen metabolism (2, 5), and hemoglobin structure and function (6). In contrast, our understanding of biochemical adaptations for high altitude in ectothermic organisms, for which cold temperatures impose unique constraints (7–9), remains limited. Studies in prokaryotes have highlighted how cold adaptation in enzymes is attained by functional trade-offs between protein stability and activity (10), a trade-off also characterized in enzymes from ectothermic vertebrates, such as teleost fishes, where single mutations altering intramolecular hydrogen bonding networks (HBNs) decrease protein stability and ligand binding affinity to optimize kinetic rates for cold environments (11). It is currently unknown if and how high-altitude conditions impose comparable temperature-related constraints on nonenzyme proteins, such as G protein-coupled receptors (GPCRs).
The dim-light visual pigment rhodopsin presents an ideal system in which to investigate adaptation in a GPCR. Rhodopsin (RH1) has been extensively characterized across vertebrates, with studies largely focused on how its spectral properties (i.e., wavelength of maximum absorbance, λMAX) can be linked to adaptation to various spectral environments (12, 13). Beyond spectral tuning, recent in vitro work has also highlighted the importance of the kinetics of activation and stability of rhodopsin (14–16), aspects of the proteins function that are considered evolutionary innovations for dim-light vision and are likely similarly critical for proper receptor functioning and organismal fitness (17–21). These important functional properties are modulated by amino acid sites distributed across the protein structure (22–25); however, several are located near conserved GPCR elements maintaining stability (26–28), such as the intramolecular HBN (29–32). Recent evidence suggests that visual sensitivity within different habitats is likely maximized through natural variation shifting the spontaneous thermal- and light-activated decay rates of ligand-bound rhodopsin (28, 33, 34), two distinct kinetic processes of direct relevance to rod photosensitivity (17, 20, 21). Transgenic animal models have demonstrated that amino acid substitutions destabilizing these inactive and active-forms of rhodopsin can directly affect the dim-light sensitivity of rods (35, 36), and are likely responsible for a reduction in rhodopsin-mediated dim-light visual performance in ectotherms exposed to small changes in temperature (20). These kinetic processes are therefore sensitive to temperature changes highly similar to those over which cold adaptation in ectotherm enzymes has repeatedly occurred (11), suggesting that there may be selection for cold adaptation in rhodopsin within species historically exposed to relatively small temperature changes. Furthermore, because some of the largest shifts in rhodopsin kinetics (26, 27) are mediated by substitutions to conserved GPCR HBNs (32), it is possible that cold adaptation in enzymes and GPCRs may also proceed through similar molecular mechanisms.
Natural variation in rhodopsin spectral tuning has substantially advanced our understanding of the evolutionary forces shaping protein function and adaptation (12, 37, 38). Comparatively, functional studies seldom examine rhodopsin stability through an evolutionary lens (28, 34). This is likely in part because of the caveat that investigating rhodopsin cold adaptation in natural systems may be confounded by differences in environment. Naturally occurring habitat transitions may also be accompanied by other, concurrent, selective pressures on rhodopsin function, such as changes in light spectra (12, 13, 28), pressure (39, 40), and temperature (34). To disentangle such effects from other aspects of rhodopsin function, we selected a system that maximized consistency across ambient spectral environments. Armored catfish native to high-altitude environments in the Andes inhabit single mountain drainages that can span over 4,000 vertical meters (41–43), introducing drainage-specific populations to wide temperature ranges (42, 44) but fast-flowing clear mountainous streams (45). Indeed, temperature is likely a primary determinant of the nocturnal high-altitude specialist’s (Astroblepus) geographical distribution (46), which—unlike the closely related generalist Ancistrus—is constrained to high altitudes, likely because of temperature-related physiological constraints (42, 43, 46). Little is known of the molecular adaptations these fishes may have evolved in response to the decreased temperatures associated with a high-altitude lifestyle. The range of temperatures inhabited by these two genera (Ancistrus and Astroblepus) of low- and high-altitude catfish (from <10 °C to >25 °C) (42) presents an ideal opportunity to investigate the relationship between rhodopsin function and temperature in an evolutionary framework.
We hypothesized that rhodopsin in high-altitude Andean specialists has adapted to cold temperatures by increasing kinetic rates, specifically the spontaneous thermal- and light-activated [Metarhodopsin II (MII)] decay rates of rhodopsin, two kinetic processes of direct relevance to organismal visual sensitivity (20, 35, 36). We sequenced the rhodopsin gene (rh1) from catfish inhabiting a range of altitudes, and combined molecular evolutionary analyses with site-directed mutagenesis experiments to test for cold adaptation in this protein. We found that high-altitude catfish rhodopsin had unique natural variants located in important structural domains known to mediate rhodopsin kinetics, and that these amino acid sites were under positive selection in high-altitude lineages. We generated rhodopsin mutants containing these variants, determining thermal- and light-activated decay rates in vitro. When mutated, these high-altitude variants produced a substantial acceleration of the spontaneous thermal- and light-activated decay rates of rhodopsin, supporting the role of these kinetic processes in the cold adaptation of rhodopsin.
Results
High-Altitude Variants Are Near Important Structural Motifs Controlling Rhodopsin Kinetics.
In a series of collection trips to high-altitude Andean drainages in Bolivia, Ecuador, and Peru, we collected fish specimens from altitudes ranging up to 2,800 m above sea level (masl) (SI Appendix, Table S9). Using genomic DNA extracted from muscle tissues, we generated an alignment of catfish rhodopsin coding-sequences (rh1) from two Andean clades representing Astroblepus, a high-altitude specialist catfish, and Ancistrus, a generalist constrained to lower altitudes. This rh1 dataset reflected a range of altitudes (40–2,810 masl) (SI Appendix, Table S9) and consequent temperatures (14–25 °C) (42) and was used together with a set of publicly available rh1 sequences representing subtropical/tropical and lowland catfish lineages to infer a maximum likelihood (ML; PhyML) (SI Appendix, Fig. S1) and Bayesian (MrBayes) (SI Appendix, Fig. S2) gene tree. The Bayesian topology and branch lengths were used for additional analyses.
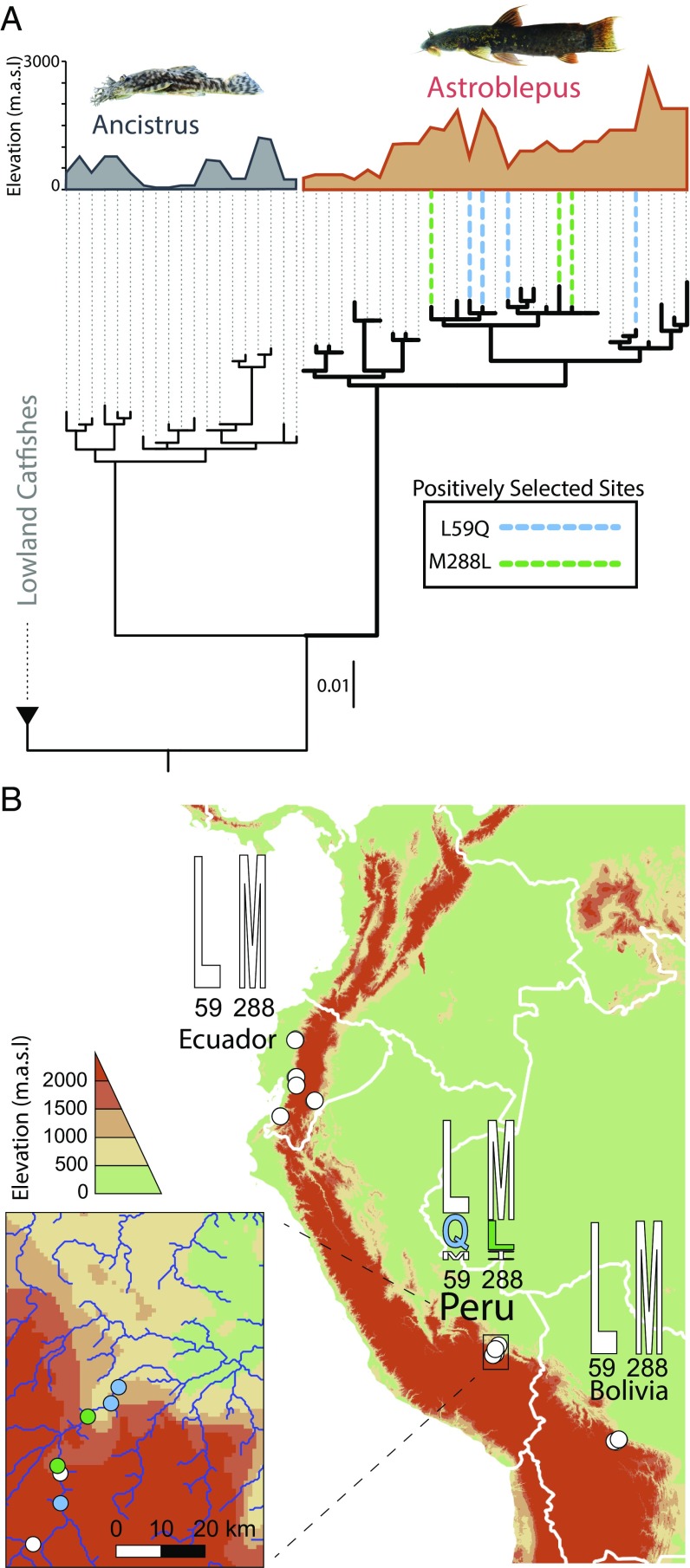
We predicted that high-altitude catfish rhodopsin may display unique natural variants near important structural domains mediating rhodopsin kinetics, similar to what has been found in many cold-adapted enzymes (10, 11). We identified two unusual single amino acid substitutions (L59Q, M288L), which repeatedly occur in different lineages of the high-altitude specialist Astroblepus (Fig. 1A). These positions are, to our knowledge, completely conserved in all known vertebrate rhodopsins, consistent with their proximity to the retinal binding pocket (RBP; site 288) and the NPxxY motif (site 59): two distinct hubs of the same intramolecular HBN stabilizing inactive and active rhodopsin (27, 47–49). To better understand what ecological factors may be driving substitutions at these highly conserved positions, we investigated the geographical distribution of natural variation at these positions across the Andean collection sites. We found that within our Astroblepus dataset, natural variation at sites 59 and 288 only occurred within populations from southern Peru (Fig. 1B), which represents both the highest altitudes from which specimens were collected (SI Appendix, Table S9), as well as the region where Astroblepus is believed to have been exposed to high altitudes for the longest period of time (50–52). These observations suggest that within high-altitude specialist rhodopsin, ecological conditions specific to high altitudes have changed the otherwise strongly maintained functional constraints at sites 59 and 288.
Fig. 1.
Positive and divergent selection in rh1 in high-altitude specialist catfish. (A) Rh1 coding sequences were obtained from high-altitude catfish collected from a range of elevations throughout the Andes Mountains in Bolivia, Ecuador, and Peru, with near sea level Ancistrus individuals collected from Guyana and Trinidad. The rh1 ML gene tree is shown. Collection site elevation is shown in meters above sea level. Bold lines indicate distinct evolutionary pressures, whereas dashed lines indicate lineages with amino acid substitutions to positively selected sites. (B) Overview of collection sites (white dots) in Ecuador, Peru, and Bolivia. Shown are rhodopsin sequence logos for the Astroblepus taxa collected and sequenced from each region. (Inset) Zoom-in of Peruvian collection sites where natural variant Astroblepus Q59 rhodopsin (blue dots) or L288 rhodopsin (green dots) originate. Elevations are shown as meters above sea level.
We investigated whether these functional constraints were shifting at high altitudes by searching for commensurate shifts in selective constraint (53). We searched for evidence of positive selection, which unlike relaxed selection, is consistent with adaptation in response to shifting functional constraints (53–56). We used multiple models of molecular evolution (57, 58) to estimate the selective pressures on high-altitude, as well as subtropical/tropical lowland catfish rh1, which revealed distinct selection pressures on all three catfish groups (SI Appendix, Tables S1 and S2). There was no evidence of positive selection in lowland rh1; however, there was robust support for a positively selected class of nonoverlapping sites in the rh1 of the high-altitude specialist (Astroblepus) and the altitude-generalist (Ancistrus) (SI Appendix, Tables S1 and S2). Further evolutionary model testing (59, 60) revealed significant evidence that only on the rh1 of the high-altitude specialist (Astroblepus) had the selective constraints significantly changed relative to lowland rh1 (SI Appendix, Tables S3–S5). Multiple Bayesian methods (57, 58) predicted with high confidence (SI Appendix, Tables S2 and S4) that these unique selection pressures were specifically targeting sites 59 and 288 in the rh1 of the high-altitude specialist (Fig. 1A), consistent with adaptation in response to shifting functional constraints.
No Evidence of Positive Selection in Nonvisual Control Genes.
Together, these analyses lend support for positive selection on high-altitude variants near structural motifs controlling rhodopsin kinetic rates. To ensure that these results were unique to rh1 rather than a consequence of genome-wide changes in selective constraint, or a reduction in the strength of selection because of small population size (61), we sequenced and analyzed two “control” genes, α- and β-actin (act) and cytochrome B (cytb) (SI Appendix, Tables S11 and S13). We found no evidence for positive selection in any lineage or clade, and overall both genes were under pervasive purifying selection (SI Appendix, Tables S6 and S7). This provided additional support for unique evolutionary constraints on high-altitude catfish RH1.
High-Altitude Variants Accelerate Rhodopsin Decay Rates Associated with Rod Photosensitivity.
To test whether the high-altitude sites under selection mediate kinetic properties of RH1, we examined the effects of rhodopsin mutants with natural high-altitude variants (L59Q and M288L) on the pigment’s dark-state and light-activated (MII) stability, two kinetic processes of direct relevance to organismal visual sensitivity (20, 35, 36). We tested this experimentally using the prototypical class A GPCR model, bovine (Bos taurus) rhodopsin, because it is well characterized and allows interpretation of the functional results within well-resolved crystal structures (62–64). We performed site-directed mutagenesis to alter WT residues to natural variants in Astroblepus, to explore the importance of these sites to rhodopsin function. WT rhodopsin, along with L59Q and M288L mutants, were expressed in HEK293T cells, regenerated with 11-cis-retinal, and immunoaffinity purified.
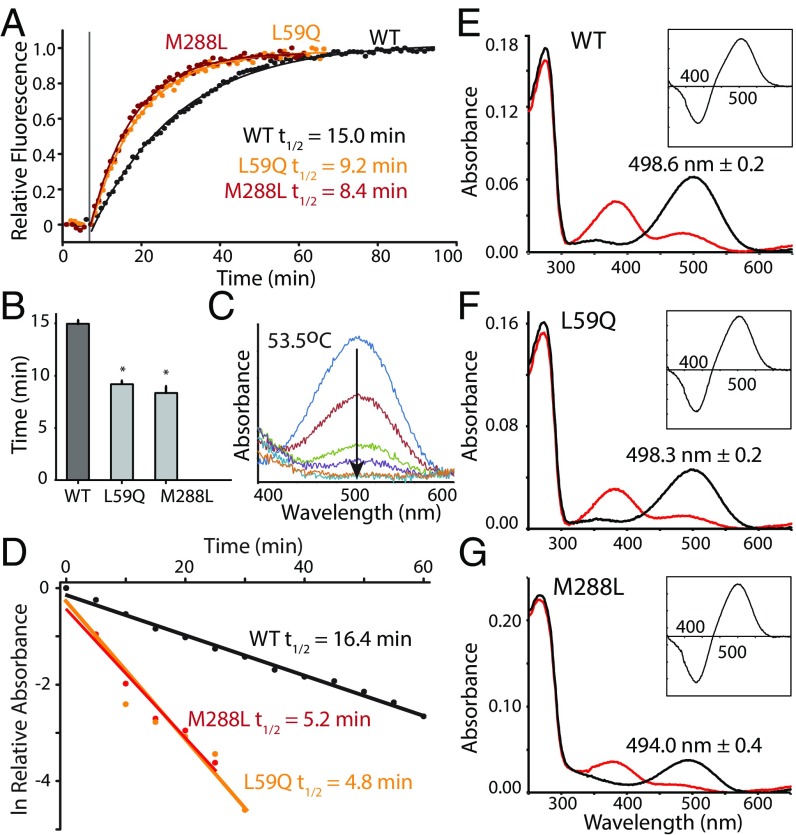
We monitored all–trans-retinal release in rhodopsin following photoactivation at 20 °C using a fluorescence assay (SI Appendix), a reliable method for tracking MII decay (16). The half-life value of retinal release for WT RH1 was 15.0 ± 0.2 min (Fig. 2 A and B), consistent with previously published results (16, 28). However, the half-life values of rhodopsin mutants with substitutions at positively and divergently selected sites, L59Q and M288L, were both significantly shorter at 9.2 ± 0.3 and 8.4 ± 0.6 min, respectively (Fig. 2 A and B). We also measured the thermal decay of dark-state WT rhodopsin and the mutant pigments to see if these mutations would affect the stability of the dark-state against spontaneous thermal activation. This was accomplished by monitoring the decrease of absorption at dark-state λMAX while incubating at 53.5 °C (Fig. 2C), a process caused by a combination of thermal isomerization and spontaneous hydrolysis of the Schiff base linkage (65). For WT RH1, the half-life of thermal decay was 16.4 min (Fig. 2D). For the rhodopsin mutants containing high-altitude amino acid variants, the results were similar to the retinal release experiment, with L59Q and M288L having shorter half-life values (4.8 and 5.2 min, respectively) (Fig. 2D). Taken together, these results suggest that natural variation at positively selected rhodopsin sites in the high-altitude specialist, Astroblepus, may promote increased kinetic rates as a response to the colder temperatures found at high altitudes.
Fig. 2.
Rhodopsin sites under positive selection in high-altitude catfish modulate kinetic rates of dark- and light-activated rhodopsin. (A) Decay of active-state (MII) WT and mutant rhodopsins, assayed by monitoring increasing fluorescence intensity upon release of all–trans-retinal after exposure to light (vertical line). Lines are data fits to first-order exponential equations, with half-life values calculated from the rate constant. (B) Differences in WT vs. mutant rhodopsin retinal release half-life values were statistically assessed. (C) Decay of dark-state WT and mutant rhodopsin at 53.5 °C, assayed by monitoring decreasing absorbance at λMAX in the dark over time (arrow). (D) Absorbance data converted to its natural logarithm and plotted against time with half-life values calculated from the rate constant. (E–G) UV-Vis absorbance spectra of dark-state and light-activated (E) WT, (F) L59Q, and (G) M288L rhodopsin, with dark-state λMAX values shown. All Insets show difference spectra between dark-state and MII, which indicates photoactivation as assessed by the well-established changes in spectral sensitivity between these rhodopsin species (SI Appendix).
We next explored whether sites containing high-altitude variants could be under positive selection as a result of putative spectral shifts at high altitude, especially because changes in λMAX tend to be linked with rhodopsin kinetic rates (14, 17, 34). We measured the dark-state λMAX of WT and mutant rhodopsins, finding that WT rhodopsin produced a λMAX of 498.6 nm ± 0.2, similar to previously published results (23) (Fig. 2E and SI Appendix, Table S8). Interestingly, L59Q produced no significant spectral shifts relative to WT, whereas M288L resulted in a blue-shift to 494.0 nm ± 0.4 (Fig. 2 F and G and SI Appendix, Table S8). Both natural variants therefore accelerate rhodopsin dark- and light-activated kinetic rates, whereas only one shifts spectral sensitivity. This finding is consistent with the structural locations of both substitutions (Fig. 3), where both are proximal to the same intramolecular HBN controlling rhodopsin kinetics, but only site 288 is involved in the section surrounding the RBP (23, 49, 66, 67) (Fig. 3A).
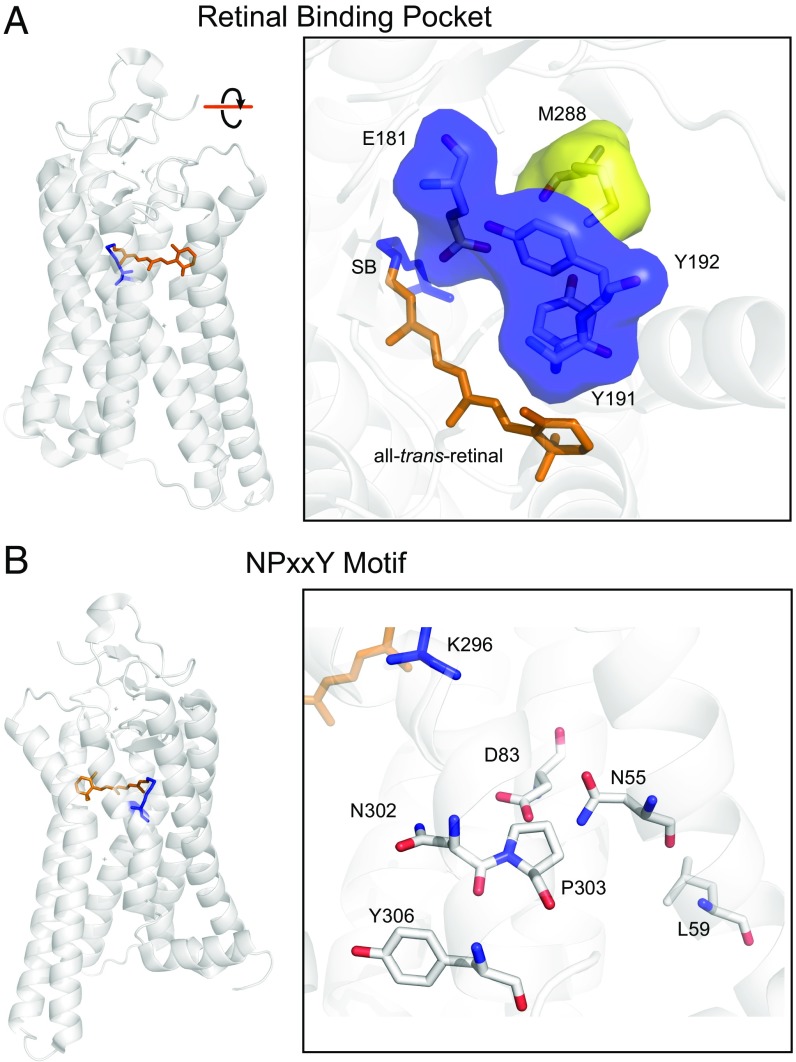
Fig. 3.
Sites accelerating kinetic rates of dark- and active-state rhodopsin are proximal to functionally key HBNs. Crystal structure of Meta II [PDB ID code 3PXO (62)] with the Schiff-base (SB; blue) and all–trans-retinal (orange). (A) Zoomed in view of the retinal binding pocket of MII, with residues proximal to M288 (yellow) and those involved in the local HBN shown as a Connolly surface (blue) (23, 66). (B) Stabilizing structural features proximal to L59: the NPxxY motif (N302-P303-Y306), N55, and D83.
Discussion
Using both computational and experimental methods, we tested for functional adaptation in high-altitude catfish (Astroblepus) rhodopsin. We found that unique high-altitude amino acid variants occurring at sites under positive selection are near important rhodopsin functional motifs maintaining the stability of ligand-bound RH1 in the dark- and light-activated states. Substituting natural high-altitude amino acid variants at these sites (L59Q or M288L) into rhodopsin produced pigments with accelerated dark-state and light-activated decay rates relative to a control pigment. Here, we discuss our results in the context of rhodopsin structure and function, and protein cold adaptation.
High-Altitude Variants Likely Modify Kinetic Properties of RH1 Through Alterations to Interconnected HBNs.
The ground-state inactivity of dark-state rhodopsin and the stability of light-activated rhodopsin are maintained in part by the HBN surrounding the chromophore (23, 66) and the tight conformation of the RBP (63) (Fig. 3A). Site 288 was under positive selection, and is located within 8 and 9 Å of the chromophore in the dark-state and MII crystal structures, respectively (62, 63). Given that the Astroblepus substitution, M288L, removes a sulfur atom while introducing a side chain with increased branching into the highly ordered configuration of the RBP, the resulting steric effects may severely alter the conformation of the RBP and the interactions of the surrounding HBN (63) (Fig. 3). Through interactions with water molecules, RBP HBN-participating residues stabilize both the dark-state and light-activated (MII) forms of rhodopsin, likely through interactions with the Schiff-base and counter ion (E113), where mutations to RBP HBN residues accelerate kinetic rates and tend also to blue-shift peak absorbance (23, 62, 63, 65, 66). Although previous evidence suggested the proximity of site 288 to functional RBP water molecules involved in photoactivation (68, 69), the exact functional role of site 288 in the RBP HBN and rhodopsin activation had not been directly investigated until recently (49). Our M288L rhodopsin functional results lend further support to the involvement of site 288 in the RBP HBN and steric organization.
Although they are located in opposite regions of the protein, the RBP HBN is connected to the HBNs of the NPxxY motif through intramolecular waters and interacting residues (27, 63, 67, 68, 70). Site 59 is directly adjacent to several of these interacting residues (Fig. 3B). L59 is within 6 Å of N55 in both dark-state and light-activated crystal structures (62, 63), a residue that forms a hydrogen bond with D83 (71, 72), which mediates the kinetics of rhodopsin light-activation (26, 33) and forms an HBN with N302 of the NPxxY (47). Although L59 is within 8 Å of Y306 of the NPxxY motif (63), it is directly adjacent to site 58, which was recently identified as a conserved GPCR element interacting with Y306 (48), forming part of a motif stabilizing the rhodopsin dark-state structure (47, 67). The Astroblepus L59Q substitution replaces a hydrophobic residue with a polar residue into a membrane-facing site, and thus may be indirectly perturbing the geometry of these nearby HBNs, resulting in our observed increases in dark-state and light-activated rhodopsin decay rates. Our findings that L59Q and M288L both significantly increase the decay-rates of light-activated rhodopsin, while also decreasing the stability of dark-state rhodopsin against thermal activation, are consistent with the emerging theory that distinct hubs of the rhodopsin HBN have interconnected roles in modulating the kinetic properties of rhodopsin and other GPCRs (47, 49, 65).
Evolution of Cold Adaptation in High-Altitude Rhodopsin.
Natural high-altitude variants occurring at sites under positive selection in Astroblepus dramatically accelerated the spontaneous thermal- and light-activated decay rates of rhodopsin, two distinct kinetic processes of direct relevance to rod photosensitivity, considered evolutionary innovations to dim light (17, 19–21, 35, 36). The spontaneous thermal activation of rhodopsin produces dark noise events, which decrease the sensitivity of rod photoreceptors, leading to worsening visual performance as temperature increases (20, 21, 73–75). The signal-to-noise ratio of rods relative to cones (20, 73, 74) is in part a result of the remarkable stability of rhodopsin against spontaneous thermal activation, an evolved scotopic adaptation (14, 23, 65, 76). Another molecular adaptation is the high signal amplification of light-activated rhodopsin (MII), which can activate hundreds of G transducin molecules per second (21, 77). Cone opsin MII decay rates are hundreds of times faster than rhodopsin MII, a consistent trend across species and cone opsin types (78), and functional evidence strongly suggests that the longer MII lifetime in rhodopsin relative to cone opsins (17, 78) promotes enhanced visual performance in dim-light environments (25, 33, 35). Our results suggest that both thermal- and light-activated decay rates are being modulated in response to temperature, in contrast to many cases where rhodopsin spectral sensitivity is shifted in response to photic environment (12, 13, 37).
Astroblepus is physiologically limited to high altitudes and cold temperatures, and is a predominantly nocturnal benthic feeder, strongly suggesting a dependence on rod-mediated scotopic vision (42, 46). Although the spectral composition of ambient light changes as altitude increases, this is mostly because of an increase in the UV at high altitudes (4), which falls outside the spectral sensitivity of rhodopsin, and absorption overlapping with rhodopsin sensitivity as a result of changes in atmospheric gases (i.e., ozone) appears to have minor effects on overall light attenuation between high and low altitudes (79, 80). This finding suggests that the changes in rhodopsin kinetics we describe here are not likely caused by spectral differences at high altitudes, because we examined drainage-specific Astroblepus populations found in clear mountain streams (41, 45). In contrast, temperature decreases (16–10 °C) well within the range of those in the Astroblepus/Ancistrus study system (<10 °C vs. >25 °C at sea level) (42) directly affect rhodopsin-mediated visual performance in ectotherms (20). This finding is consistent with our functional results, where the only unique high-altitude substitution shifting rhodopsin spectral sensitivity (M288L) occurs in the RBP HBN: a functional domain that has likely evolved to maintain rhodopsin dark- and light-state stability, but produces blue-shifts when mutated (23, 34, 66). To our knowledge, RBP HBN residues have not been otherwise targeted by natural selection as spectral tuning sites (12, 13, 37), likely because of their status as important regulators of rhodopsin kinetic rates.
The acceleration of rhodopsin kinetic rates in response to cold temperatures is highly consistent with many studies of cold-adapted psychrophilic enzymes, which—unlike thermophilic enzymes (81)—exhibit a trade-off between activity and stability, with decreasing stability improving activity relative to thermostable homologs at low temperatures (10, 82). Functional adaptations change the selective constraints acting on protein-coding genes (53), with site-specific selective pressures, such as positive selection capable of driving adaptive amino acid substitutions changing protein function in response to environmental variables (54, 55). This finding contrasts with a release of selective constraint, which indicates a relaxation of certain protein functional constraints (53). Unlike blind cave-dwelling fishes, where selective constraint on rh1 has been lost (56), rh1 in the high-altitude specialist Astroblepus is under strong positive selection, consistent with adaptation in response to changing functional constraints, such as temperature (53). Positive selection on sites containing rare natural variants, which accelerate the important kinetic properties of rhodopsin underlying rod photosensitivity, may therefore improve visual performance in cold high-altitude environments, where the reaction rates of rhodopsin functions are likely to be slowed relative to lowland environments (10, 82). Our results bear striking similarity to the molecular mechanisms of cold adaptation in teleost fish enzymes, such as A4-LDH, where protein stability and ligand-binding affinity are decreased by alterations to intramolecular hydrogen bonds through single amino acid substitutions in solvent-facing regions where conformational shifts occur (11). The molecular mechanisms of cold adaptation, combined with the importance and temperature sensitivity of rhodopsin kinetics, likely explains why natural variants on the periphery of HBNs controlling rhodopsin dark- and light-activated stability are under positive selection in an ectotherm specialized to high altitudes. This study provides evidence consistent with cold adaptation in a GPCR, and suggests that across different proteins, cold adaptation may proceed through similar mechanisms.
Materials and Methods
Also see SI Appendix for detailed descriptions.
Animals.
Fish specimens were collected from altitudes ranging from 40 masl to >2,800 masl, with consequent temperatures ranging from 14–25 °C, during expeditions to Bolivia (2011), Ecuador (2012), Guyana (2009), Peru (2010), and Trinidad (2010) (SI Appendix, Table S9). All fish specimens were killed after capture via overdose of clove oil, following approved Texas A&M University Institutional Animal Care and Use Committee guidelines. Subsamples of either fin or muscle tissue were dissected and preserved in 95% EtOH for genetic analysis, whereas whole voucher specimens were fixed in formalin, preserved in 70% EtOH.
Phylogenetic and Molecular Evolutionary Analyses.
Partial rh1 and act coding sequences were sequenced from genomic DNA extracted from the collected loricarioid catfish (including Astroblepus and Ancistrus genera) specimens using standard PCR- and Sanger-sequencing procedures (SI Appendix). Additional rh1 coding sequences and partial loricarioid cytb coding sequences were also obtained from GenBank, and some cytb coding sequences from a previous study (83). All coding sequences were aligned to produce rh1, act, and cytb phylogenetic datasets with gene trees estimated using ML and Bayesian methods (SI Appendix). In the rh1 trees, the topologies of the Astroblepus and Ancistrus clades were identical after clades with low support (<50% aLRT SH-like) were collapsed in the ML tree. Phylogenetic datasets were analyzed using PAML (58), the FUBAR model of HYPHY (57), and DIVERGE 3 (60) (SI Appendix).
Rhodopsin Expression and Spectroscopic Assays.
WT rhodopsin, along with L59Q and M288L mutants, were expressed in HEK293T cells, regenerated with 11-cis-retinal, and immunoaffinity purified essentially as previously described (28) (SI Appendix). MII decay was monitored using a fluorescence assay (Cary Eclipse; Agilent) tracking the release of all–trans-retinal in rhodopsin following photoactivation at 20 °C (16) (SI Appendix). Thermal decay of WT and mutant bovine rhodopsin was recorded at 53.5 °C as the decrease of absorbance at λMAX using a Cary 4000 double-beam spectrophotometer (Agilent), with data collected every 5 min for 2 h (SI Appendix).
Supplementary Material
Acknowledgments
The 11-cis-retinal was generously provided by Rosalie Crouch (Medical University of South Carolina). This work was supported by a National Sciences and Engineering Research Council Discovery grant (to B.S.W.C.); Vision Science Research Program Scholarships (to G.M.C., F.E.H., R.K.S., N.B., S.Z.D., and A.V.N.); Ontario Graduate Scholarship (to R.K.S., and F.E.H.); a grant from the Deutsche Forschungsgemeinschaft (to J.M.M.); the National Science Foundation Grant OISE-1064578 (N.K.L.); the Coypu Foundation (N.K.L.); National Geographic Committee for Research and Exploration Grant 8721-09 (N.K.L.); the estate of George and Carolyn Kelso via the International Sportfish Fund (N.K.L.); the Explorer’s Club of New York (N.K.L.); and the Royal Ontario Museum (N.K.L.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. KY945369–KY945484).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1705765114/-/DCSupplemental.
References
- 1.Storz JF, et al. The molecular basis of high-altitude adaptation in deer mice. PLoS Genet. 2007;3:e45. doi: 10.1371/journal.pgen.0030045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beall CM. Two routes to functional adaptation: Tibetan and Andean high-altitude natives. Proc Natl Acad Sci USA. 2007;104:8655–8660. doi: 10.1073/pnas.0701985104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hayes JP, Connor CSO, Hayes JP, Connor CSO. Natural selection on thermogenic capacity of high-altitude deer mice. Evolution. 1999;53:1280–1287. doi: 10.1111/j.1558-5646.1999.tb04540.x. [DOI] [PubMed] [Google Scholar]
- 4.Körner C. The use of ‘altitude’ in ecological research. Trends Ecol Evol. 2007;22:569–574. doi: 10.1016/j.tree.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 5.Song D, et al. Defective Tibetan PHD2 binding to p23 links high altitude adaption to altered oxygen sensing. J Biol Chem. 2014;289:14656–14665. doi: 10.1074/jbc.M113.541227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Projecto-Garcia J, et al. Repeated elevational transitions in hemoglobin function during the evolution of Andean hummingbirds. Proc Natl Acad Sci USA. 2013;110:20669–20674. doi: 10.1073/pnas.1315456110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dupré RK, Wood SC. Behavioral temperature regulation by aquatic ectotherms during hypoxia. Can J Zool. 1988;66:2649–2652. [Google Scholar]
- 8.Beitinger T, Bennett W, McCauley R. Temperature tolerances of North American freshwater fishes exposed to dynamic changes in temperature. Environ Biol Fishes. 2000;58:237–275. [Google Scholar]
- 9.Verberk WCEP, Bilton DT, Calosi P, Spicer JI. Oxygen supply in aquatic ectotherms: Partial pressure and solubility together explain biodiversity and size patterns. Ecology. 2011;92:1565–1572. doi: 10.1890/10-2369.1. [DOI] [PubMed] [Google Scholar]
- 10.Siddiqui KS, Cavicchioli R. Cold-adapted enzymes. Annu Rev Biochem. 2006;75:403–433. doi: 10.1146/annurev.biochem.75.103004.142723. [DOI] [PubMed] [Google Scholar]
- 11.Fields PA, Dong Y, Meng X, Somero GN. Adaptations of protein structure and function to temperature: There is more than one way to ‘skin a cat’. J Exp Biol. 2015;218:1801–1811. doi: 10.1242/jeb.114298. [DOI] [PubMed] [Google Scholar]
- 12.Hunt DM, Dulai KS, Partridge JC, Cottrill P, Bowmaker JK. The molecular basis for spectral tuning of rod visual pigments in deep-sea fish. J Exp Biol. 2001;204:3333–3344. doi: 10.1242/jeb.204.19.3333. [DOI] [PubMed] [Google Scholar]
- 13.Dungan SZ, Kosyakov A, Chang BS. Spectral tuning of killer whale (Orcinus orca) rhodopsin: Evidence for positive selection and functional adaptation in a cetacean visual pigment. Mol Biol Evol. 2016;33:323–336. doi: 10.1093/molbev/msv217. [DOI] [PubMed] [Google Scholar]
- 14.Gozem S, Schapiro I, Ferre N, Olivucci M. The molecular mechanism of thermal noise in rod photoreceptors. Science. 2012;337:1225–1228. doi: 10.1126/science.1220461. [DOI] [PubMed] [Google Scholar]
- 15.Johnson PJM, et al. Local vibrational coherences drive the primary photochemistry of vision. Nat Chem. 2015;7:980–986. doi: 10.1038/nchem.2398. [DOI] [PubMed] [Google Scholar]
- 16.Schafer CT, Fay JF, Janz JM, Farrens DL. Decay of an active GPCR: Conformational dynamics govern agonist rebinding and persistence of an active, yet empty, receptor state. Proc Natl Acad Sci USA. 2016;113:11961–11966. doi: 10.1073/pnas.1606347113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Imai H, et al. Single amino acid residue as a functional determinant of rod and cone visual pigments. Proc Natl Acad Sci USA. 1997;94:2322–2326. doi: 10.1073/pnas.94.6.2322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luo D-G, Yue WWS, Ala-Laurila P, Yau K-W. Activation of visual pigments by light and heat. Science. 2011;332:1307–1312. doi: 10.1126/science.1200172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lamb TD, et al. Evolution of vertebrate phototransduction: Cascade activation. Mol Biol Evol. 2016;33:2064–2087. doi: 10.1093/molbev/msw095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aho AC, Donner K, Hydén C, Larsen LO, Reuter T. Low retinal noise in animals with low body temperature allows high visual sensitivity. Nature. 1988;334:348–350. doi: 10.1038/334348a0. [DOI] [PubMed] [Google Scholar]
- 21.Baylor D. How photons start vision. Proc Natl Acad Sci USA. 1996;93:560–565. doi: 10.1073/pnas.93.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Janz JM, Fay JF, Farrens DL. Stability of dark state rhodopsin is mediated by a conserved ion pair in intradiscal loop E-2. J Biol Chem. 2003;278:16982–16991. doi: 10.1074/jbc.M210567200. [DOI] [PubMed] [Google Scholar]
- 23.Liu J, et al. Thermal properties of rhodopsin: Insight into the molecular mechanism of dim-light vision. J Biol Chem. 2011;286:27622–27629. doi: 10.1074/jbc.M111.233312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Piechnick R, et al. Effect of channel mutations on the uptake and release of the retinal ligand in opsin. Proc Natl Acad Sci USA. 2012;109:5247–5252. doi: 10.1073/pnas.1117268109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bickelmann C, et al. The molecular origin and evolution of dim-light vision in mammals. Evolution. 2015;69:2995–3003. doi: 10.1111/evo.12794. [DOI] [PubMed] [Google Scholar]
- 26.Weitz CJ, Nathans J. Rhodopsin activation: Effects on the metarhodopsin I-metarhodopsin II equilibrium of neutralization or introduction of charged amino acids within putative transmembrane segments. Biochemistry. 1993;32:14176–14182. doi: 10.1021/bi00214a016. [DOI] [PubMed] [Google Scholar]
- 27.Fritze O, et al. Role of the conserved NPxxY(x)5,6F motif in the rhodopsin ground state and during activation. Proc Natl Acad Sci USA. 2003;100:2290–2295. doi: 10.1073/pnas.0435715100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dungan SZ, Chang BSW. Epistatic interactions influence terrestrial—Marine functional shifts in cetacean rhodopsin. Proc Biol Sci. 2017;284:20162743. doi: 10.1098/rspb.2016.2743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nygaard R, Frimurer TM, Holst B, Rosenkilde MM, Schwartz TW. Ligand binding and micro-switches in 7TM receptor structures. Trends Pharmacol Sci. 2009;30:249–259. doi: 10.1016/j.tips.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 30.Weis WI, Kobilka BK. Structural insights into G-protein-coupled receptor activation. Curr Opin Struct Biol. 2008;18:734–740. doi: 10.1016/j.sbi.2008.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pardo L, Deupi X, Dölker N, López-Rodríguez ML, Campillo M. The role of internal water molecules in the structure and function of the rhodopsin family of G protein-coupled receptors. ChemBioChem. 2007;8:19–24. doi: 10.1002/cbic.200600429. [DOI] [PubMed] [Google Scholar]
- 32.Angel TE, Chance MR, Palczewski K. Conserved waters mediate structural and functional activation of family A (rhodopsin-like) G protein-coupled receptors. Proc Natl Acad Sci USA. 2009;106:8555–8560. doi: 10.1073/pnas.0903545106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sugawara T, Imai H, Nikaido M, Imamoto Y, Okada N. Vertebrate rhodopsin adaptation to dim light via rapid meta-II intermediate formation. Mol Biol Evol. 2010;27:506–519. doi: 10.1093/molbev/msp252. [DOI] [PubMed] [Google Scholar]
- 34.Luk HL, et al. Modulation of thermal noise and spectral sensitivity in Lake Baikal cottoid fish rhodopsins. Sci Rep. 2016;6:38425. doi: 10.1038/srep38425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Imai H, et al. Molecular properties of rhodopsin and rod function. J Biol Chem. 2007;282:6677–6684. doi: 10.1074/jbc.M610086200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yue WWS, et al. Spontaneous activation of visual pigments in relation to openness/closedness of chromophore-binding pocket. Elife. 2017;6:e18492. doi: 10.7554/eLife.18492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yokoyama S, Tada T, Zhang H, Britt L. Elucidation of phenotypic adaptations: Molecular analyses of dim-light vision proteins in vertebrates. Proc Natl Acad Sci USA. 2008;105:13480–13485. doi: 10.1073/pnas.0802426105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bowmaker JK. Evolution of vertebrate visual pigments. Vision Res. 2008;48:2022–2041. doi: 10.1016/j.visres.2008.03.025. [DOI] [PubMed] [Google Scholar]
- 39.Partridge JC, White EM, Douglas RH. The effect of elevated hydrostatic pressure on the spectral absorption of deep-sea fish visual pigments. J Exp Biol. 2006;209:314–319. doi: 10.1242/jeb.01984. [DOI] [PubMed] [Google Scholar]
- 40.Porter ML, Roberts NW, Partridge JC. Evolution under pressure and the adaptation of visual pigment compressibility in deep-sea environments. Mol Phylogenet Evol. 2016;105:160–165. doi: 10.1016/j.ympev.2016.08.007. [DOI] [PubMed] [Google Scholar]
- 41.Lujan NK, Conway KW. Life in the fast lane: A review of rheophily in freshwater fishes. In: Riesch R, Tobler M, Plath M, editors. Extremophile Fishes: Ecology Evolution, and Physiology of Teleosts in Extreme Environments. Springer International; Cham, Switzerland: 2015. pp. 107–136. [Google Scholar]
- 42.Lujan NK, et al. Aquatic community structure across an Andes-to-Amazon fluvial gradient. J Biogeogr. 2013;40:1715–1728. [Google Scholar]
- 43.Schaefer SA, Chakrabarty P, Geneva AJ, Sabaj Pérez MH. Nucleotide sequence data confirm diagnosis and local endemism of variable morphospecies of Andean astroblepid catfishes (Siluriformes: Astroblepidae) Zool J Linn Soc. 2011;162:90–102. [Google Scholar]
- 44.Velez-Espino LA. Population viability and perturbation analyses in remnant populations of the Andean catfish Astroblepus ubidiai. Ecol Freshw Fish. 2005;14:125–138. [Google Scholar]
- 45.Sommaruga R, Psenner R, Schafferer E, Koinig KA, Sommaruga-Wograth S. Dissolved organic carbon concentration and phytoplankton biomass in high-mountain lakes of the Austrian Alps: Potential effect of climatic warming an UV underwater attenuation. Arct Antarct Alp Res. 1999;31:247–253. [Google Scholar]
- 46.Schaefer SA, Arroyave J. Rivers as islands: Determinants of the distribution of Andean astroblepid catfishes. J Fish Biol. 2010;77:2373–2390. doi: 10.1111/j.1095-8649.2010.02822.x. [DOI] [PubMed] [Google Scholar]
- 47.Ernst OP, et al. Microbial and animal rhodopsins: Structures, functions, and molecular mechanisms. Chem Rev. 2014;114:126–163. doi: 10.1021/cr4003769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Venkatakrishnan AJ, et al. Diverse activation pathways in class A GPCRs converge near the G-protein-coupling region. Nature. 2016;536:484–487. doi: 10.1038/nature19107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kimata N, et al. Retinal orientation and interactions in rhodopsin reveal a two-stage trigger mechanism for activation. Nat Commun. 2016;7:12683. doi: 10.1038/ncomms12683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lujan NK, Meza-Vargas V, Astudillo-Clavijo V, Barriga-Salazar R, López-Fernández H. A multilocus molecular phylogeny for Chaetostoma clade genera and species with a review of Chaetostoma (Siluriformes: Loricariidae) from the Central Andes. Copeia. 2015;103:664–701. [Google Scholar]
- 51.Lundberg JG, Sullivan JP, Rodiles-Hernández R, Hendrickson DA. Discovery of African roots for the Mesoamerican Chiapas catfish, Lacantunia enigmatica, requires an ancient intercontinental passage. Proc Acad Nat Sci Phila. 2007;156:39–53. [Google Scholar]
- 52.Gregory-Wodzicki KM. Uplift history of the Central and Northern Andes: A review. Geol Soc Am Bull. 2000;112:1091–1105. [Google Scholar]
- 53.Echave J, Spielman SJ, Wilke CO. Causes of evolutionary rate variation among protein sites. Nat Rev Genet. 2016;17:109–121. doi: 10.1038/nrg.2015.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Elde NC, Child SJ, Geballe AP, Malik HS. Protein kinase R reveals an evolutionary model for defeating viral mimicry. Nature. 2009;457:485–489. doi: 10.1038/nature07529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Patel MR, Loo Y-M, Horner SM, Gale M, Jr, Malik HS. Convergent evolution of escape from hepaciviral antagonism in primates. PLoS Biol. 2012;10:e1001282. doi: 10.1371/journal.pbio.1001282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Niemiller ML, Fitzpatrick BM, Shah P, Schmitz L, Near TJ. Evidence for repeated loss of selective constraint in rhodopsin of amblyopsid cavefishes (Teleostei: Amblyopsidae) Evolution. 2013;67:732–748. doi: 10.1111/j.1558-5646.2012.01822.x. [DOI] [PubMed] [Google Scholar]
- 57.Murrell B, et al. FUBAR: A fast, unconstrained Bayesian approximation for inferring selection. Mol Biol Evol. 2013;30:1196–1205. doi: 10.1093/molbev/mst030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yang Z. PAML 4: Phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 59.Bielawski JP, Yang Z. A maximum likelihood method for detecting functional divergence at individual codon sites, with application to gene family evolution. J Mol Evol. 2004;59:121–132. doi: 10.1007/s00239-004-2597-8. [DOI] [PubMed] [Google Scholar]
- 60.Gu X. Statistical methods for testing functional divergence after gene duplication. Mol Biol Evol. 1999;16:1664–1674. doi: 10.1093/oxfordjournals.molbev.a026080. [DOI] [PubMed] [Google Scholar]
- 61.Lanfear R, Kokko H, Eyre-Walker A. Population size and the rate of evolution. Trends Ecol Evol. 2014;29:33–41. doi: 10.1016/j.tree.2013.09.009. [DOI] [PubMed] [Google Scholar]
- 62.Choe H-W, et al. Crystal structure of metarhodopsin II. Nature. 2011;471:651–655. doi: 10.1038/nature09789. [DOI] [PubMed] [Google Scholar]
- 63.Okada T, et al. The retinal conformation and its environment in rhodopsin in light of a new 2.2 A crystal structure. J Mol Biol. 2004;342:571–583. doi: 10.1016/j.jmb.2004.07.044. [DOI] [PubMed] [Google Scholar]
- 64.Hofmann KP, et al. A G protein-coupled receptor at work: The rhodopsin model. Trends Biochem Sci. 2009;34:540–552. doi: 10.1016/j.tibs.2009.07.005. [DOI] [PubMed] [Google Scholar]
- 65.Guo Y, et al. Unusual kinetics of thermal decay of dim-light photoreceptors in vertebrate vision. Proc Natl Acad Sci USA. 2014;111:10438–10443. doi: 10.1073/pnas.1410826111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Janz JM, Farrens DL. Role of the retinal hydrogen bond network in rhodopsin Schiff base stability and hydrolysis. J Biol Chem. 2004;279:55886–55894. doi: 10.1074/jbc.M408766200. [DOI] [PubMed] [Google Scholar]
- 67.Palczewski K. G protein-coupled receptor rhodopsin. Annu Rev Biochem. 2006;75:743–767. doi: 10.1146/annurev.biochem.75.103004.142743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Angel TE, Gupta S, Jastrzebska B, Palczewski K, Chance MR. Structural waters define a functional channel mediating activation of the GPCR, rhodopsin. Proc Natl Acad Sci USA. 2009;106:14367–14372. doi: 10.1073/pnas.0901074106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sun X, Ågren H, Tu Y. Functional water molecules in rhodopsin activation. J Phys Chem B. 2014;118:10863–10873. doi: 10.1021/jp505180t. [DOI] [PubMed] [Google Scholar]
- 70.Nygaard R, Valentin-Hansen L, Mokrosinski J, Frimurer TM, Schwartz TW. Conserved water-mediated hydrogen bond network between TM-I, -II, -VI, and -VII in 7TM receptor activation. J Biol Chem. 2010;285:19625–19636. doi: 10.1074/jbc.M110.106021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Okada T, et al. Functional role of internal water molecules in rhodopsin revealed by X-ray crystallography. Proc Natl Acad Sci USA. 2002;99:5982–5987. doi: 10.1073/pnas.082666399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Palczewski K, et al. Cyrstal structure of rhodopsin: A G protein-coupled receptor. Science. 2000;289:739–745. doi: 10.1126/science.289.5480.739. [DOI] [PubMed] [Google Scholar]
- 73.Baylor DA, Matthews G, Yau KW. Two components of electrical dark noise in toad retinal rod outer segments. J Physiol. 1980;309:591–621. doi: 10.1113/jphysiol.1980.sp013529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Angueyra JM, Rieke F. Origin and effect of phototransduction noise in primate cone photoreceptors. Nat Neurosci. 2013;16:1692–1700. doi: 10.1038/nn.3534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ala-Laurila P, Donner K, Koskelainen A. Thermal activation and photoactivation of visual pigments. Biophys J. 2004;86:3653–3662. doi: 10.1529/biophysj.103.035626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rieke F, Baylor DA. Origin and functional impact of dark noise in retinal cones. Neuron. 2000;26:181–186. doi: 10.1016/s0896-6273(00)81148-4. [DOI] [PubMed] [Google Scholar]
- 77.Heck M, Hofmann KP. Maximal rate and nucleotide dependence of rhodopsin-catalyzed transducin activation: Initial rate analysis based on a double displacement mechanism. J Biol Chem. 2001;276:10000–10009. doi: 10.1074/jbc.M009475200. [DOI] [PubMed] [Google Scholar]
- 78.Imai H, et al. Molecular properties of rod and cone visual pigments from purified chicken cone pigments to mouse rhodopsin in situ. Photochem Photobiol Sci. 2005;4:667–674. doi: 10.1039/b416731g. [DOI] [PubMed] [Google Scholar]
- 79.Horvath H. Atmospheric light absorption—A review. Atmos Environ Gen Top. 1993;27:293–317. [Google Scholar]
- 80.Chevalier A, et al. Influence of altitude on ozone levels and variability in the lower troposphere: A ground-based study for Western Europe over the period 2001–2004. Atmos Chem Phys. 2007;7:4311–4326. [Google Scholar]
- 81.Nguyen V, et al. Evolutionary drivers of thermoadaptation in enzyme catalysis. Science. 2016;355:289–294. doi: 10.1126/science.aah3717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Feller G. Protein stability and enzyme activity at extreme biological temperatures. J Phys Condens Matter. 2010;22:323101. doi: 10.1088/0953-8984/22/32/323101. [DOI] [PubMed] [Google Scholar]
- 83.Lujan NK, Armbruster JW, Lovejoy NR, López-Fernández H. Multilocus molecular phylogeny of the suckermouth armored catfishes (Siluriformes: Loricariidae) with a focus on subfamily Hypostominae. Mol Phylogenet Evol. 2015;82:269–288. doi: 10.1016/j.ympev.2014.08.020. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.