Abstract
We report the characterization of Cep170, a forkhead-associated (FHA) domain protein of previously unknown function. Cep170 was identified in a yeast two-hybrid screen for interactors of Polo-like kinase 1 (Plk1). In human cells, Cep170 is constantly expressed throughout the cell cycle but phosphorylated during mitosis. It interacts with Plk1 in vivo and can be phosphorylated by Plk1 in vitro, suggesting that it is a physiological substrate of this kinase. Both overexpression and small interfering RNA (siRNA)-mediated depletion studies suggest a role for Cep170 in microtuble organization and cell morphology. Cep170 associates with centrosomes during interphase and with spindle microtubules during mitosis. As shown by immunoelectron microscopy, Cep170 associates with subdistal appendages, typical of the mature mother centriole. Thus, anti-Cep170 antibodies stain only one centriole during G1, S, and early G2, but two centrioles during late G2 phase of the cell cycle. We show that Cep170 labeling can be used to discriminate bona fide centriole overduplication from centriole amplification that results from aborted cell division.
INTRODUCTION
The centrosome is the major microtubule (MT) organizing center of animal cells and exerts a profound influence on most MT-related cellular functions, including cell motility and polarity, the transport of organelles, and cell division (Nigg, 2004). It comprises a pair of barrel-shaped centrioles that are surrounded by pericentriolar material (PCM). The structure of centrioles in cultured mammalian cells has been analyzed in detail by electron microscopy (EM), and characteristic cell cycle-related changes are well described (Rieder and Borisy, 1982; Chretien et al., 1997). The proximal and distal ends of centrioles, as well as their degree of maturation, can be distinguished by the presence or absence of specific structures: only the older of the two centrioles (often referred to as the mother centriole) carries appendages at its distal end, and so these structures serve as markers for the age of centrioles. A second type of appendages, termed subdistal, also has been described. These are generally thought to be acquired during G1 by the older centriole but then disappear again at the onset of mitosis (Chretien et al., 1997; Bornens, 2002; Meraldi and Nigg, 2002). MTs are nucleated from the centriole-associated PCM regardless of the presence or absence of appendages, but they are preferentially anchored at the distal end of the mother centriole (Piel et al., 2000). Several proteins specifically associated with mother centrioles have recently been described. These include ODF2/cenexin (Lange and Gull, 1995; Nakagawa et al., 2001), ε-tubulin (Chang et al., 2003), ninein (Piel et al., 2000), ninein-like protein (Nlp; Casenghi et al., 2003), adenomatous polyposis coli and EB1 (Louie et al., 2004), and centriolin (Gromley et al., 2003). Ninein is thought to form part of a MT-anchoring complex (reviewed in Bornens, 2002; Mogensen, 2004) and important roles in MT organization have also been attributed to Nlp and EB1 (Casenghi et al., 2003; Louie et al., 2004). ε-Tubulin has been reported to be required for centrosome duplication in frog egg extracts (Chang et al., 2003), and centriolin has been implicated in cytokinesis (Gromley et al., 2003).
The precise function of centrioles remains mysterious, but several intriguing roles have been proposed. Injection of antibodies directed against posttranslationally modified centriolar tubulin induced not only a transient disassembly of centrioles, but also the dispersal of the PCM, indicating that centrioles are critical for centrosome stability (Bobinnec et al., 1998). Moreover, a specific movement of the mother centriole to the midbody has been linked to the terminal separation of dividing cells (abscission), indicating that centrioles contribute to the control of cytokinesis (Piel et al., 2001). The mechanical removal or laser-mediated ablation of centrioles, though not preventing entry into mitosis, caused frequent cytokinesis failure and cell cycle arrest in the subsequent G1 phase (Hinchcliffe et al., 2001; Khodjakov and Rieder, 2004).
Structure and activity of the centrosome are regulated throughout the cell cycle (Fry and Hames, 2004). One important change occurs at the onset of mitosis, when the centrosome undergoes a process called maturation. This is reflected by both the acquisition and loss of specific centrosome components and results in increased MT nucleation capacity. Concomitantly, subdistal appendages are lost from centrioles, leading to a more diffuse anchoring of MTs at spindle poles (Chretien et al., 1997; Bornens, 2002). Prominent among the enzymes implicated in the regulation of centrosome maturation is Polo-like kinase 1 (Plk1) (Sunkel and Glover, 1988; Lane and Nigg 1996; Barr et al., 2004). Candidate centrosomal substrates of this kinase have recently been identified (do Carmo Avides et al., 2001; Casenghi et al., 2003), but a complete understanding of the role of Plk1 at the centrosome remains elusive.
In the present study, we have used a yeast two-hybrid screen to search for novel interaction partners of Plk1. We report the characterization of a human protein of ∼170 kDa that contains a forkhead-associated (FHA) domain in its N terminus. We have localized this protein to the centrosome and hence refer to it as Cep170 (for centrosomal protein of ∼170 kDa). Our data indicate that Cep170 is involved in the control of cell morphology and interacts with Plk1 under physiological conditions. Finally, we show that Cep170 constitutes an excellent marker for centriole maturation during the cell cycle and that the acquisition of Cep170 can be used to distinguish distinct mechanisms leading to centrosome amplification.
MATERIALS AND METHODS
Antibody Production
Polyclonal rabbit antibodies were raised against N- (aa 15–754) and C-terminal (aa 755-1460) fragments of KIAA0470/Cep170γ, both His-tagged at their N-termini and expressed in Escherichia coli. Two rabbits were immunized for each antigen (BioGenes, Berlin, Germany). Antibodies against the N-terminal half of Cep170 were affinity purified on recombinant immunogen bound to an AffiGel 15 (Bio-Rad, Hercules, CA) column and used throughout this study. Importantly, both antibodies produced qualitatively similar results.
Cell Culture, Synchronization, and Transfections
All cells were grown at 37°C in a 5% CO2 atmosphere in DMEM, supplemented with 10% fetal calf serum (FCS) and penicillin-streptomycin (100 I.U./ml and 100 μg/ml, respectively). U2OS cells stably expressing centringreen fluorescent protein (GFP) (kindly provided by M. Bornens, Institut Curie, Paris, France) were maintained as described previously (Duensing et al., 2001). For synchronization, HeLa cells were treated with 2 mM thymidine and then released in medium for 14 h, arrested again by 14 h of aphidicolin treatment (1.6 μg/ml), and harvested at different times after release into fresh medium. For M phase arrest, cells were treated with 0.5 μg/ml nocodazole for 16 h. Alternatively, 0.1 μg/ml nocodazole was added 8 h after release from a G1/S arrest, and cells were collected by shake-off 6 h later. For in vivo labeling experiments, cells were synchronized and labeled as described previously (Mayor et al., 2002). NIH/3T3 were serum starved by culturing in DMEM supplemented with 0.5% FCS for 48 h. U2OS/centrin-GFP cells were treated with either 0.6 μg/ml cytochalasin D for 48 h to block cytokinesis or with 1 mM hydroxyurea (HU) for 72 h to induce centrosome overduplication.
U2OS cells were transfected using the calcium phosphate precipitation method as described previously (Seelos, 1997). When transfection was combined with RNA interference (RNAi), cells were transfected with the small interfering RNA (siRNA) oligos as described below (“siRNA experiments”); after 60 h cells were transfected with the DNA plasmid and, 12 h later, fixed and processed for immunofluorescence (IF) microscopy. U2OS/centrin-GFP cells were transfected with 2 μg of plasmid DNA encoding human papillomavirus type 16 (HPV-16) E7 or empty pCMVneo vector (Duensing et al., 2000) by using FuGENE 6 (Roche Diagnostics, Mannheim, Germany). Cells were cotransfected with 0.2 μg of vector encoding cyan fluorescent protein (CFP) with a mitochondrial target sequence (BD Biosciences Clontech, Palo Alto, CA). Cells were processed for IF microscopy at 48 h posttransfection.
Plasmid Preparation
The KIAA0470 cDNA was obtained from the Kazusa DNA Research Institute. It was subcloned into pT7-EGFP-C1 or pT7-EGFP-N1 (modified from pEGFP-C1 or -N1; BD Biosciences Clontech), after removing the ATG or the stop codon, respectively. The following KIAA0470 deletion clones were produced in pT7-EGFP-C1: N-term (aa 2–754, until XmnI site in the cDNA), C-term (aa 755-1460, from XmnI site to the end of cDNA), C-termI (aa 755-1014, XmnI-HindIII internal fragment), C-termII (aa 1015–1460, from HindIII in the C-term to the end of cDNA), and ΔFHA (aa 74–1460, from AccIII site to the end of cDNA). The following KIAA0470-encoded proteins were produced in pQE bacterial expression vectors (QIAGEN, Hilden, Germany): full length (pQE32), N-term (aa 15–754, BssSI-XmnI fragment), C-term, C-termI, and C-termII (pQE30) as described above for the enhanced green fluorescent protein (EGFP)-tagged clones.
Immuno-EM
For preembedding immuno-EM of whole cells, U2OS or HeLa cells were grown on coverslips, fixed with 3% paraformaldehyde (PFA)/2% sucrose for 10 min, and permeabilized with phosphate-buffered saline (PBS) + 0.5% Triton X-100 for 1 min. Blocking and primary antibody incubations were performed as described for IF microscopy, followed by incubation with goat anti-rabbit IgG-nanogold (1:75; Nanoprobes, Yaphank, NY). Cells were further processed as described (Fry et al., 1998). For ultrathin cryosection labeling, cells were fixed with 4% PFA in PBS for 60 min on ice, embedded in 10% gelatin, and infiltrated in 2.1 M sucrose in PBS. After freezing in liquid nitrogen, 100-nm cryosections were obtained using a Leica Ultracut UCT/EM FCS cryoultramicrotome at –110°C. Thawed cryosections were incubated with affinity-purified rabbit anti-Cep170 IgGs (1 μg/ml, diluted in PBS containing 1% milk powder, 0.5% bovine serum albumin) and goat anti-rabbit IgGs coupled to nanogold (Nanoprobes; diluted 1:50 as described above) or protein A-6-nm gold. Silver enhancement was performed as described previously (Stierhof et al., 1991). Final embedding was done in 2% methylcellulose (Sigma-Aldrich, St. Louis, MO) containing 0.3% uranyl acetate.
IF Microscopy
Cells grown on coverslips were fixed in –20°C methanol for 6 min and processed for IF as described in Meraldi et al., 2002. Purified centrosomes were spun onto coverslips, fixed, and processed as described for whole cells. Tween (0.05%) was included in all IF incubations. For competition experiments, mixtures containing the anti-Cep170 primary antibody were incubated for 1 h in the presence of a nitrocellulose filter preloaded with antigen. Primary antibodies were rabbit anti-Cep170 (affinity purified, 1 μg/ml), mouse anti α-tubulin (1:2000, B-5-1-2; Sigma-Aldrich), mouse anti-acetylated-tubulin (1:1000, 6-11B-1; Sigma-Aldrich), mouse anti-γ-tubulin (1:1000, GTU-88; Sigma-Aldrich), rabbit anti-centrin2 (affinity purified, 1 μg/ml), and mouse GT335 (1:1000 dilution from ascites; a kind gift of B. Eddé, Centre de Recherches de Biochimie Macromoleculaire, CNRS, Montpellier, France). Where indicated, anti-Cep170 was directly labeled with fluorescein by using the fluorescein labeling kit (Roche Diagnostics). Secondary antibodies were biotinylated donkey anti-rabbit IgGs (1:200; Amersham Biosciences, Piscataway, NJ), followed by Texas Red-conjugated streptavidin (1:100; Amersham Biosciences), Alexa Fluor-488–conjugated goat anti-mouse (1:1000; Molecular Probes, Eugene, OR), and 7-amino-4-methylcoumarin-3-acetic acid (AMCA)-conjugated goat anti-mouse IgGs (1:200; Jackson ImmunoResearch Laboratories, West Grove, PA). IF microscopy was performed using a Zeiss Axioplan II microscope and APOCHROMAT 40×/1.4, 63×/1.4, or 100 ×/1.4 oil immersion objectives. Images were captured using a Micromax charge-coupled device camera (Princeton Scientific Instruments, Monmouth Junction, NJ) and Metaview software (Universal Imaging, Downingtown, PA). For high-resolution images, a Deltavision microscope on a Nikon TE200 base (Applied Precision, Issaquah, WA), equipped with an APOPLAN 100×/1.4 oil immersion objective, was used for collecting 0.2-μm–distanced optical sections in the z-axis. Images at single focal planes were processed with a deconvolution algorithm and then projected into one picture by using Softworx (Applied Precision). Images were processed using Adobe Photoshop 6.0 and Adobe Illustrator 10 (Adobe Systems, Mountain View, CA)
U2OS/centrin-GFP–transfected cells grown on coverslips were fixed in 4% PFA in PBS for 15 min at room temperature (RT) and permeabilized using 1% Triton X-100 in PBS for 15 min at RT. After blocking with 10% normal donkey serum (Jackson ImmunoResearch Laboratories), cells were incubated with anti-Cep170 antibody at 0.5 μg/ml PBS overnight at 4°C followed by a rhodamine red-conjugated donkey anti-rabbit antibody (1:1000; Jackson ImmunoResearch Laboratories) for 2 h at 37°. Cells were counterstained with 4,6-diamidino-2-phenylindole (DAPI) and analyzed by epifluorescence microscopy.
Immunoblotting and Immunoprecipitation (IP) Experiments
Cells were directly lysed in gel sample buffer, resolved by SDS-PAGE, and electrophoretically transferred to nitrocellulose membranes (Schleicher & Schuell, Keene, NH) by using a semidry system. Approximately 30 μg of extract was loaded for each lane; for analysis of centrosomal fractions, 7.5 × 106 centrosomes and extracts from 7.5 × 104 KE37 cells were loaded. Blocking and antibody dilutions were made in 5% low-fat dry milk in PBS containing 0.1% Tween 20. Washing steps were in PBS containing 0.1% Tween 20. Membranes were blocked overnight at 4°C, primary antibodies were incubated for 1 h, secondary antibodies for 45 min, at RT. Bound horseradish peroxidase (HRP)-conjugated secondary antibodies were visualized by enhanced chemiluminescence detection. Primary antibodies were used at the following concentrations: rabbit anti-Cep170 (affinity purified 0.5 μg/ml), mouse anti-α-tubulin (1:2000), mouse anti-γ-tubulin (1:1000), mouse anti-Plk1 (PL6, 1:10 dilution from ascites; Golsteyn et al., 1994), mouse anti-His-tag (undiluted hybridoma supernatant; a kind gift from G. Gerisch, Max Planck Institute of Biochemistry, Munich, Germany). Secondary antibodies were goat anti-rabbit or goat anti-mouse HRP-conjugated IgGs (1:3000; Amersham Biosciences). In competition experiments, the diluted antibody solution was incubated overnight on nitrocellulose membranes preloaded with equal amounts of immunogen or a nonoverlapping protein fragment for control.
For IP experiments, cells were washed once in PBS + 1 mM phenylmethylsulfonyl fluoride (PMSF) and lysed on ice in extraction buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.4% NP-40, 1 mM EDTA, 1 mM PMSF, aprotinin, leupeptin, pepstatin at 1 μg/ml each, 1 mM Na3VO4) for 40 min. Extracts were clarified (14,000 rpm, 4°C, 10 min), preabsorbed on protein A beads (Affiprep; Bio-Rad) for 45 min at 4°C and incubated with beads bearing rabbit anti-Cep170 or goat anti-Plk1 antibodies, and rabbit IgGs or sheep anti-GFP IgGs for control, at 4°C for 2 h. For IP of in vivo 32P-labeled samples, prepared as described in Mayor et al., (2002), antibody-conjugated beads were preblocked with unlabeled HeLa cell extract and then used as described above. Immunoprecipitates were washed six times in extraction buffer and resuspended in gel sample buffer.
Laser Scanner Cytometry
DNA contents were determined using a laser scanning cytometer (CompuCyte, Cambridge, MA). Coverslips were fixed in cold methanol, DNA was stained with 1 μg/ml DAPI, and DNA profiles were obtained using the WinCyte software (CompuCyte). For each sample, 10,000 events were acquired.
Plk1 Kinase Assays
Recombinant His6-tagged Cep170γ and deletion mutants were expressed and purified from E. coli as described below. In vitro kinase assays were performed as described previously (Descombes and Nigg, 1998; Kelm et al., 2002), by using equal amounts of recombinant proteins with wild-type or catalytically inactive His6-Plk1 purified from baculovirus-infected Sf9 insect cells. Casein was used as control. Incubation was performed for 30 min at 30°C and then reactions were stopped by adding 5× gel sample buffer. After electrophoresis, phosphate incorporation was determined by autoradiography.
Purification of Recombinant Proteins
His6-tagged Cep170γ and deletion mutants were expressed in E. coli JM109 or JM109pRIL strains and purified under native conditions by using standard protocols (QIAexpressionist system; QIAGEN). After purification, buffer was exchanged to PBS + 1 mM PMSF, or PBS only for rabbit injections.
siRNA Experiments
The siRNA sequence targeting Cep170 corresponds to the coding region 2237–2255 (relative to the KIAA0470 cDNA sequence): GAA GGA AUC CUC CAA GUC A. For control, a duplex (GL-2) targeting luciferase was used (Elbashir et al., 2001). The 21-nt RNA duplexes were purchased from Dharmacon Research. Annealing of the siRNAs and transfections using Oligofectamine (Invitrogen, Carlsbad, CA) were performed as described previously (Elbashir et al., 2001).
Miscellaneous
The isolation of human centrosomes from KE37 cells was performed by spinning the cell lysates onto a 50% sucrose cushion followed by sedimentation of centrosomes in a discontinuous sucrose gradient (40, 50, and 70%) as described in Moudjou and Bornens, 1994. Yeast two hybrid screens have been described previously (Casenghi et al., 2003).
RESULTS
Identification of Cep170 (KIAA0470) as a Centrosomal Protein
To identify proteins interacting with Polo-like kinases, a yeast two-hybrid screen was performed with catalytically inactive Plx1N172A (Plk1 of Xenopus laevis) as bait. One cDNA isolated in this screen, PXP-6, showed homology to a human database entry (KIAA0470, also termed KAB [for KARP-1 binding protein], predicted to give rise to three splice variants). These cDNAs (accession nos. AB022657, AB022658, and AB022659) are predicted to code for proteins with molecular weights of ∼170 kDa, which display a region of 64–68% identity to the PXP-6–predicted protein product (depicted schematically in Figure 1A). By reverse transcription-polymerase chain reaction, all three splice forms are expressed in U2OS and HeLa cells (our unpublished data). All three proteins harbor a putative FHA-domain in the N terminus (aa 22–73, by using the numbering of splice form 1), as well as a serine-rich region (aa 968-1228) and a small predicted coiled-coil region (aa 1467–1495) in the C-terminus (Figure 1A). Partial cDNAs coding for homologues of these human proteins have been described in other vertebrates (Mus musculus, Rattus norvegicus and X. laevis; Do et al., 2003) but not in invertebrates or yeast.
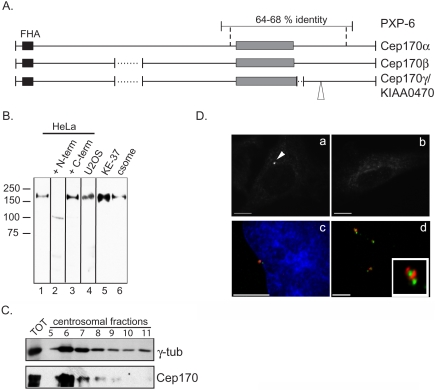
Figure 1.
Cep170 localizes to centrosomes. (A) Schematic representation of the predicted protein products of a X. laevis cDNA isolated in the yeast two-hybrid screen (PXP-6) and of three homologous human cDNA splice variants. The black and gray boxes indicate the FHA and the serine-rich domains, respectively. Dotted lines indicate gaps and the open triangle an insertion, presumably resulting from differential splicing. Vertical dashed lines delineate the region of homology between the X. laevis and human proteins, as calculated using the FASTA protein program. We believe that the start codon for the corresponding cDNAs is the ATG positioned at nucleotide 54 in the KIAA0470 clone (accession no. AB007939), because of 1) the presence of stop codons in the 5′ untranslated region, 2) the presence of an A at -3 respect to the ATG (consensus Kozak sequence), and 3) sequence conservation with a mouse cDNA sequence (accession no. BC050077). (B) Western blot using the Cep170 antibody on total extracts from HeLa (lanes 1–3), U2OS (lane 4), or KE37 (lane 5) cells or the isolated centrosomal fraction from KE37 cells (csome, lane 6). Primary antibody was preincubated with buffer (lane 1), the immunogen (+ N-term, lane 2) or a nonoverlapping fragment of Cep170 (+ C-term, lane 3). Molecular weight markers are indicated on the left. Vertical lines delineate separate nitrocellulose stripes. (C) Western blot using Cep170 and γ-tubulin antibodies on fractions (5–11) from a discontinuous sucrose gradient (see Materials and Methods) harboring partially purified centrosomes. Total KE37 extract (TOT) was loaded as control. (D) IF staining of HeLa cells (a–c), or isolated centrosomes (d). Cells were stained with Cep170 antibody (a and b; the arrowhead indicates the centrosomal staining) after preincubation with buffer (a) or the immunogen (b), or double stained (c and d) with anti-Cep170 (red) and anti-γ-tubulin (c) or GT335 (d) (green) antibodies. Bars, 10 μm (a–c), 5 μm (d).
When tagged with GFP, both the product of the partial PXP-6 cDNA and a full-length human KIAA0470 protein colocalized with Plx1/Plk1 at the centrosome (our unpublished data; but see Figure 4). This colocalization prompted us to further characterize this human protein and to investigate its possible interaction with Plk1. Polyclonal rabbit antibodies were raised against nonoverlapping N- and C-terminal halves of the human KIAA0470 protein expressed in E. coli. Both antibodies were expected to recognize all three splice products. By Western blotting on a total HeLa cell extract, the anti-N-terminal antibody reacted with a band of the size expected for KIAA0470 protein (Figure 1B), and this immunoreactivity was readily competed by preincubation with recombinant N-terminal but not C-terminal fragment of the antigen (compare lanes 2 and 3). A band of identical migration also was seen in U2OS and KE37 cell extracts and in purified centrosomes (Figure 1B). Moreover, in a sucrose gradient preparation of centrosomes, the KIAA0470 protein cofractionated with the γ-tubulin centrosomal marker (Figure 1C). Together with the immunolabeling data shown below, these results confirm the prediction of a recent proteomic survey of the human centrosome (Andersen et al., 2003) and identify KIAA0470 as a novel centrosome component. In line with the convention proposed by Andersen et al. (2003), we will henceforth refer to this protein as Cep170 (centrosomal protein of approximately 170 kDa) and to the three splice forms as Cep170 α, β, and γ, respectively (Figure 1A).
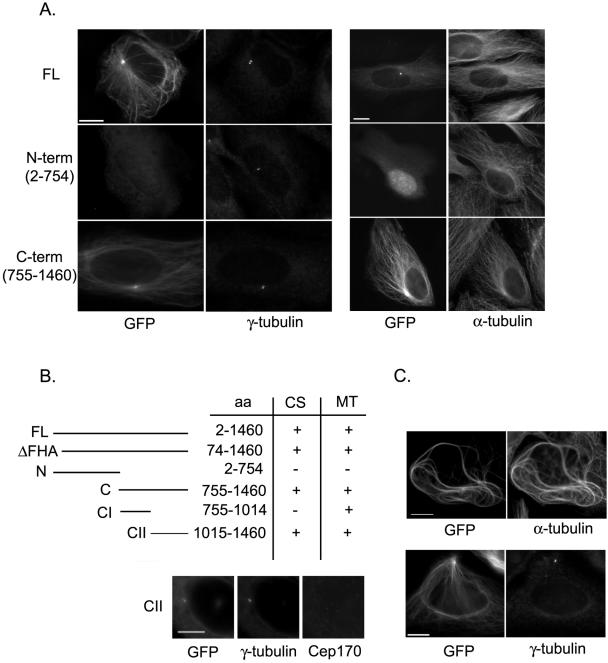
Figure 4.
Overexpression of Cep170 in mammalian cells. (A) U2OS cells were transfected with the indicated GFP-Cep170 constructs (FL, full-length; N-term; C-term). In the left-hand panel, the GFP signal of the Cep170 fusion proteins was compared with the distribution of γ-tubulin (to the right), whereas in the right-hand panel, the GPF signal was compared with α-tubulin staining (to the right). (B) Summary describing the ability of various GFP-Cep170 deletions to localize to centrosomes (CS) or MTs. The immunofluorescence panels at the bottom show the centrosomal localization of the GFP-CII construct after depletion of the endogenous Cep170 by siRNA; counterstaining with γ-tubulin is used to identify the centrosome. (C) Cells transfected with GFP-Cep170 (C-term) were stained for α-tubulin (top) or γ-tubulin (bottom), to illustrate MT bundling and centrosome displacement. Bars, 10 μm.
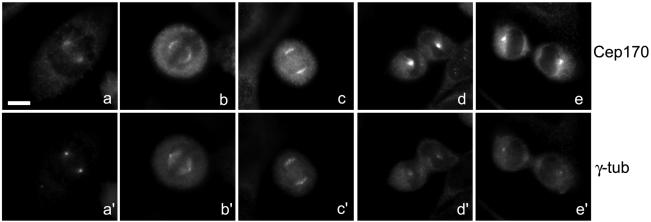
The localization of endogenous Cep170 in human cells was determined by IF analysis. In most cells, staining of a small, often circular or horseshoe-shaped structure was observed (Figure 1D, a). This staining was not seen with preimmune serum (our unpublished data) and was effectively competed by incubation with antigen (Figure 1D, b). As expected, the structures stained by anti-Cep170 antibodies colocalized with γ-tubulin (Figure 1D, c). The centrosome association of Cep170 was not abolished by nocodazoleinduced depolymerization of MTs (our unpublished data), and isolated centrosomes also stained strongly positive (Figure 1D, d). In many cases, Cep170 associated preferentially with one of two centrioles (Figure 1D, c and d), a finding to which we will return in more detail below. With the onset of mitosis, staining of the centrosome became progressively more diffuse, so that by metaphase, Cep170 was associated with spindle MTs in the region of the poles, but staining of the poles themselves was not pronounced (Figure 2, a and a′ and b and b′). The MT-associated staining persisted during anaphase (Figure 2, c and c′). In early telophase, strong staining of astral MTs could be seen (Figure 2, d and d′), and in late telophase Cep170 reassociated with the centrosomes (Figure 2, e and e′). These localization data demonstrate that Cep170 displays a dynamic association with elements of the MT cytoskeleton throughout the cell cycle.
Figure 2.
Cep170 localization during mitosis. HeLa cells were costained with Cep170 (a–e) and γ-tubulin (a′–e′) antibodies. Prometaphase (a and a′), metaphase (b and b′), anaphase (c and c′), early (d and d′) and late (e and e′) telophases are shown. Bar, 10 μm.
Cep170 Is Phosphorylated during M Phase and Interacts with Plk1
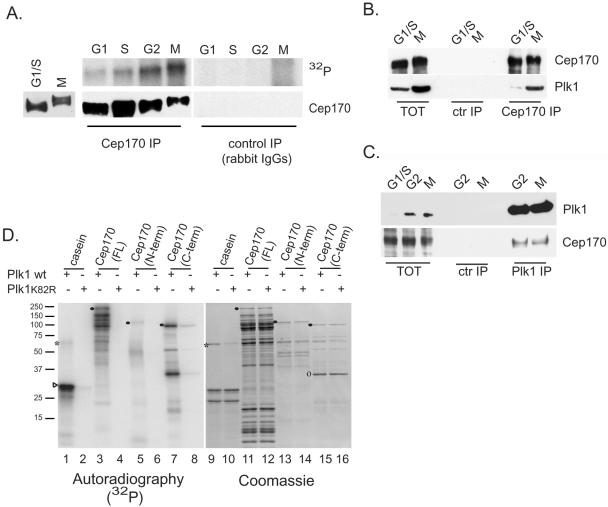
Western blots using extracts from G1/S or M phase-arrested HeLa cells revealed little difference in abundance of Cep170 during the cell cycle but a striking change in mobility, suggesting a mitosis-specific posttranslational modification (Figure 3A, left). IP experiments performed on 32P-labeled cells confirmed that Cep170 is a phosphoprotein, with a phosphorylation peak in G2/M (Figure 3A, top). As shown by Western blotting, comparable amounts of Cep170 were immunoprecipitated from all cell cycle stages (Figure 3A, bottom), and no significant signals were observed in control IP (Figure 3A). Considering that the phosphorylation peak of Cep170 coincides with the time of maximal Plk1 activity in the cell cycle (Golsteyn et al., 1995) and that Cep170 was isolated in a yeast two-hybrid screen with Plk1 as bait, we asked whether these two proteins could interact in human cells. Reciprocal coIPs were performed using extracts prepared from synchronized HeLa cells (Figure 3, B and C). Plk1 was readily detectable in Cep170 IPs from M phase samples but barely in G1/S samples (Figure 3B). This was expected because the amount of Plk1 was much higher in G2 and M than in G1/S phase samples (Figure 3, B and C), consistent with previous data (Golsteyn et al., 1995), whereas Cep170 levels were nearly constant. Cep170 also was detected in Plk1 IPs from G2 and M phase samples, whereas no precipitation of either Plk1 or Cep170 was detected in controls (Figure 3C). These results indicate that Plk1 and Cep170 interact in vivo at endogenous levels, either directly or indirectly.
Figure 3.
Cep170 is a mitotic phosphoprotein and interacts with Plk1. (A) Left, Western blot of total extracts from thymidine/aphidicolin-(G1/S) or nocodazole (M)-arrested HeLa cells with anti-Cep170 antibody. Right, in vivo labeling of Cep170. IP was performed using either anti-Cep170 (left) or control rabbit IgGs (right) and extracts from G1, S, G2, or nocodazole-arrested (M) HeLa cells, labeled with [32P]phosphoric acid. Top, 32P incorporation (film exposure); bottom, Western blot with anti-Cep170 antibody. To confirm the comigration of the 32P-labeled protein with Cep170, the Western membrane was exposed to autoradiography (our unpublished data). (B) IPs from G1/S or M phase-arrested cells using the anti-Cep170 antibody or rabbit IgG (ctr IP). Western blots using anti-Cep170 (top) or anti-Plk1 (bottom) antibody are shown. Total extracts (TOT) were loaded as control at 1/25th of the amount of IP samples. (C) IPs from G2 or M phase cells using anti-Plk1 or control IgG. Western blots using anti-Plk1 (top) or anti-Cep170 (bottom) antibody are shown. TOT were loaded as control at 1/15th of the amount of the IP samples. (D) In vitro kinase assay using wild-type (wt) or catalytically impaired (K82R) Plk1 on bacterially expressed His-Cep170 or deletion mutants. Both Coomassie-stained gels (right, lanes 9–16) and film exposure (left, lanes 1–8) are shown. Symbols indicate the migration of Plk1 (*), casein (▹), and full-length His-Cep170 (FL), His-Cep170 aa 15–754 (N-term), and His-Cep170 aa 755-1460 (C-term) (•). A degradation product of His-Cep170 (C-term) is also indicated (o). Molecular weight markers are indicated on the left.
To examine the possibility of a direct enzyme–substrate relationship between Plk1 and Cep170, His-tagged Cep170γ was incubated with recombinant Plk1 in the presence of [γ-32P]ATP. As shown in Figure 3D, Cep170γ was readily phosphorylated by wild-type Plk1 but not by a catalytically impaired Plk1K82R mutant (compare lanes 3 and 4), in line with results obtained with the control substrate casein (lanes 1 and 2). Two nonoverlapping fragments of Cep170γ (N-term, aa 15–754; C-term, aa 755-1460) also were assayed as substrates. The C-term was strongly phosphorylated, whereas little phosphate incorporation was seen with the N-term (compare lanes 5 and 7). Yet, both substrates were present at comparable levels (lanes 13 and 15).
These results demonstrate that Cep170 is a mitotic phosphoprotein able to interact with Plk1 in vivo and to serve as a Plk1 substrate in vitro. Domain analysis further suggests that phosphorylation site(s) for Plk1 are located primarily in the C-terminal part of Cep170.
Overexpression of Cep170 Affects the MT Cytoskeleton
When overexpressed in U2OS cells, GFP-tagged full-length Cep170 localized to the centrosome, regardless of whether the tag was located at the N or C terminus (Figure 4A; our unpublished data). In addition, GFP-Cep170 associated with filamentous structures emanating from the centrosome (Figure 4A), identified as MTs by costaining with α-tubulin (right-hand panels) and their sensitivity to cold or nocodazole treatment (our unpublished data). Staining of MTs also was seen occasionally for endogenous Cep170, and although this staining was faint during interphase (our unpublished data), it became prominent during mitosis (Figure 2).
To identify domains important for centrosome and/or MT association of Cep170, the subcellular localization of deletion constructs was analyzed. A Cep170 mutant lacking the FHA domain (ΔFHA construct) effectively localized to both centrosomes and MTs (Figure 4B), indicating that the FHA region does not influence Cep170 localization. In fact, the C-terminal half of Cep170 was sufficient to confer localization to both centrosomes and MTs, whereas the N-terminal half of Cep170 was diffusely distributed throughout the nucleus and cytoplasm (Figure 4A). Further mapping of the C-terminal part showed that a fragment encompassing aa 1015–1460 is sufficient for centrosome localization (Figure 4B). This does not reflect dimerization with endogenous centrosome-localized Cep170, because identical results were obtained after depletion of endogenous Cep170 by siRNA (Figure 4B, bottom). Moreover, expression of the C-terminal fragment CII displayed endogenous Cep170 from centrosomes (our unpublished data), suggesting that it acted as a dominant negative mutant, possibly by saturating centrosomal binding sites. Interestingly, both fragments 755-1014 and 1015–1460 showed MT association (Figure 4B), albeit to a different extent, suggesting that the C terminus of Cep170 contains at least two MT-binding domains.
Cells expressing GFP-Cep170 often displayed long and wavy MT bundles, similar in appearance to those observed in response to overexpression of EB1 or CLIP-115 (Hoogenraad et al., 2000; Ligon et al., 2003). Bundling was seen with all Cep170 constructs that displayed an ability to bind MTs, but it was particularly prominent with the C-terminal construct (755–1460) (Figure 4C, top). Interestingly, cells transfected with this construct often showed a striking change in centrosome positioning, characterized by a displacement of centrosomes toward the cell periphery (Figure 4C, bottom). Given that a very similar phenotype has been seen after treatment of cells with taxol or inhibition of the motor protein dynein (Burakov et al., 2003), this suggests that Cep170 overexpression affects centrosome positioning through a disturbance of MT dynamics.
RNAi-mediated Cep170 Depletion Perturbs Cytoskeletal Organization and Cell Shape
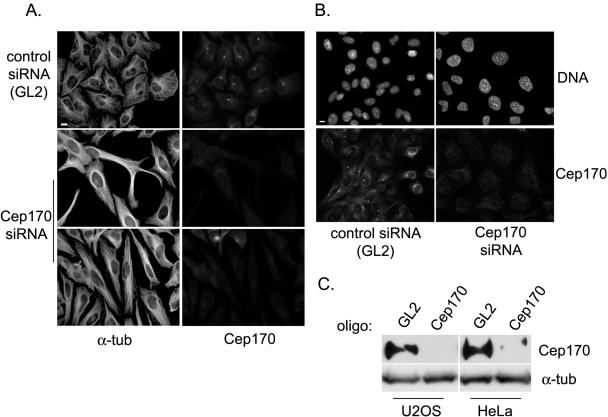
To explore the function of Cep170 in human cells, siRNA experiments were carried out. Transfection of both HeLa and U2OS cells with a siRNA duplex oligonucleotide specific for Cep170 resulted in substantial depletion of Cep170 after 60 h, as shown by both IF microscopy (Figure 5, A and B) and Western blotting (Figure 5C), whereas no depletion was seen in response to a GL2 control oligonucleotide. Cep170 depletion did not significantly affect the centrosomal recruitment of any protein tested (γ-tubulin, centrin-2, polyglutamylated tubulin [GT335], C-Nap1, Nek2, Plk1, and Aurora-A). Instead, striking changes were apparent in MT organization and in the shape of interphasic cells. Whereas the shape of HeLa cells changed from a typical epithelial (cobblestone) to a more fibroblastic (elongated) appearance (Figure 5A), U2OS cells depleted of Cep170 showed an apparent increase in size, affecting both the nucleus and the cytoplasm (Figure 5B). It is possible that Cep170-depleted U2OS cells were arrested or delayed during interphase, because mitotic cells were almost absent from such cultures (0.5% ± 0.1 compared with the 4.4% ± 0.6 in control cultures). However, neither FACS analysis nor centrosome counting provided evidence for an increase in ploidy (our unpublished data). Cep170-depleted HeLa cultures did not show a significant decrease in the percentage of mitotic cells (7.4% ± 1.4 compared with the 7% ± 0.6 of control cultures). We cannot presently explain why HeLa and U2OS cells displayed somewhat different responses to the depletion of Cep170, but the observed morphological changes clearly suggest a role for Cep170 in cytoskeletal organization.
Figure 5.
siRNA-mediated depletion of Cep170. HeLa (A) or U2OS (B) cells were transfected with control (GL2) or Cep170-specific oligos and costained for Cep170 and α-tubulin or Cep170 and DNA, as indicated. Bars, 10 μm. (C) Western blots on total extracts from U2OS or HeLa cells transfected with control (GL2) or specific (Cep170) oligos, using anti-Cep170 or anti-α-tubulin antibodies.
Changes in Cep170 Localization during the Cell Cycle
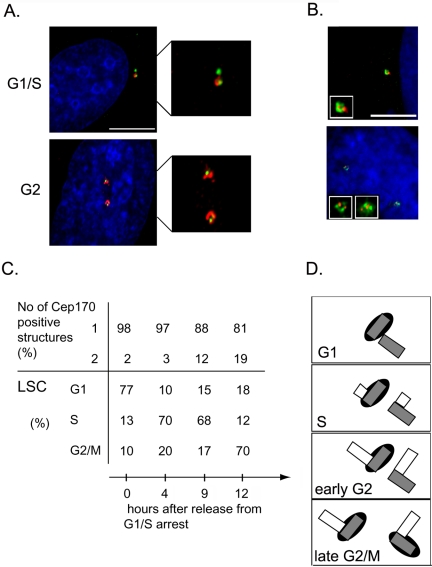
The localization studies described above had suggested that endogenous Cep170 preferentially associates with only one of two parental centrioles within the interphase centrosome (Figure 1D). This asymmetric staining was intriguing, because the two centrioles differ in both structure and function (Bornens, 2002). To determine whether the asymmetric localization of Cep170 varies during the cell cycle, HeLa cells were synchronized and costained for Cep170 and either γ-tubulin or centrin-2 (Figure 6, A and B). To obtain cultures enriched in G1, S, or G2 phases, cells were arrested at the G1/S transition by thymidine/aphidicoline treatment and then released and harvested at different time points. In parallel, the DNA content of the corresponding cell populations was determined by laser scanning cytometry (Figure 6C). In the majority of cells arrested at G1/S, Cep170 was clearly associated with only one of the two γ-tubulin–positive centrioles, and the same was true after a 4-h release from the block, when cells were passing through S phase (Figure 6, A and C). However, upon progression through G2 (9–12 h after release), an increasing number of cells showed Cep170 association with both γ-tubulin dots (Figure 6, A and C). In many of these latter cells, centrosomes were separated by >5 μm, suggesting that they were in late G2. Cells also were stained with an anti-centrin-2 antibody, to visualize single centrioles. In all cells that contained two centrioles (i.e., the G1 configuration), Cep170 associated with only one (Figure 6B, top), and even in cells in which four centrioles could clearly be discerned by centrin staining (i.e., the G2 configuration), two Cep170-positive structures were seen only in a subpopulation (our unpublished data). In contrast, in all cells in which the anti-Cep170 antibody labeled two structures, four centrin-positive centrioles could be seen (Figure 6B, bottom). Together, these data indicate that the recruitment of Cep170 to the second parental centriole begins late in G2 and probably continues into M phase (Figure 6D).
Figure 6.
Asymmetric Cep170 localization during the cell cycle. (A) Costaining of HeLa cells at the indicated cell cycle phases with antibodies against Cep170 (red) and γ-tubulin (green); DNA is shown in blue. Enlargements of centrosomes are shown on the right. (B) HeLa cells containing two (top) or four (bottom) centrioles, costained with antibodies against Cep170 (FLUOS-conjugated, green) and centrin-2 (red). DNA is shown in blue. Insets show single centrosomes. (C) Quantitative analysis of the experiment shown in A. The upper part of the table indicates the percentage of cells with Cep170 staining on one or two centrioles. The lower part shows the percentage of cells in G1, S, and G2/M phases, as determined by laser scanner cytometry. (D) Schematic representation of Cep170 association with centrioles during the cell cycle. Gray and empty rectangles represent parental centrioles and newly emerging (pro-) centrioles, respectively and the black ovals symbolize Cep170 localization. Bars, 10 μm.
Cep170 Localizes to Centrosome Appendages
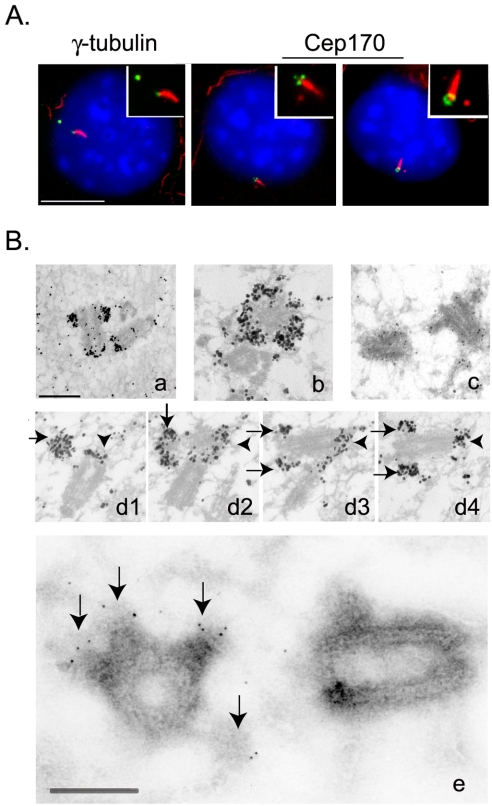
In response to serum starvation, NIH 3T3 fibroblasts grow a primary cilium specifically from the older, mature centriole (Albrecht-Buehler and Bushnell, 1980). This primary cilium can be visualized by antibodies against acetylated tubulin, affording a convenient assay for determining which of the two parental centrioles was labeled by the Cep170 antibody (Figure 7A). Whereas γ-tubulin was present on both centrioles in serum-starved NIH 3T3 cells (Figure 7A, left), Cep170 was invariably seen on the one centriole that grew the primary cilium (Figure 7A, middle and right). This unequivocally demonstrates that Cep170 localizes to the older of the two parental centrioles. As seen in human cells, Cep170 staining in these murine cells was often ring-shaped, but sometimes three dots could clearly be seen (Figure 7A). This suggested that Cep170 associated with structures that are specific to the mature centriole. To explore the localization of Cep170 at an ultrastructural level, HeLa and U2OS cells were analyzed by both preembedding immuno-EM (Figure 7B, a–d) and labeling of ultrathin cryosections (Figure 7B, e), performed on whole cells. Using the preembedding technique, anti-Cep170 antibodies produced strong labeling at the distal end of one of the two centrioles (Figure 7B, a and b, and d1–d4). In addition, a weaker staining was observed at the proximal end of both centrioles (Figure 7B, a and d1–d4). The distal staining was largely associated with the subdistal appendages, characteristic of the mother centriole (Bornens, 2002). Ultrathin cryosections of nonpermeabilized cells largely confirmed these results (Figure 7B, e). Again, subdistal appendages were clearly labeled, although staining at proximal ends was not observed. These results demonstrate that Cep170 preferentially associates with appendages that are typical of the fully mature centriole.
Figure 7.
Localization of Cep170 to appendages on the mature centriole. (A) Costaining of serum-starved NIH 3T3 cells with antibodies against acetylated tubulin (red) and either γ-tubulin or Cep170 (green), as indicated. Insets show enlargements of centrosomes and cilia. Bar, 10 μm. (B) Immuno-EM localization of Cep170 in U2OS cells. Preembedding (a–d): longitudinal section of one centrosome (a); transversal section of one centriole with the typical “crown” labeling produced by the anti-Cep170 antibody (b); control staining with secondary antibody only (c); and serial longitudinal sections through one centrosome (arrows indicate labeling of appendages, arrowheads mark the proximal staining at both centrioles) (d1–d4). Ultrathin cryosection (e), enlarged image of one centrosome; arrows indicate labeling of appendages. Bars, 250 nm.
Use of Cep170 for Studying Centriole Amplification Mechanisms
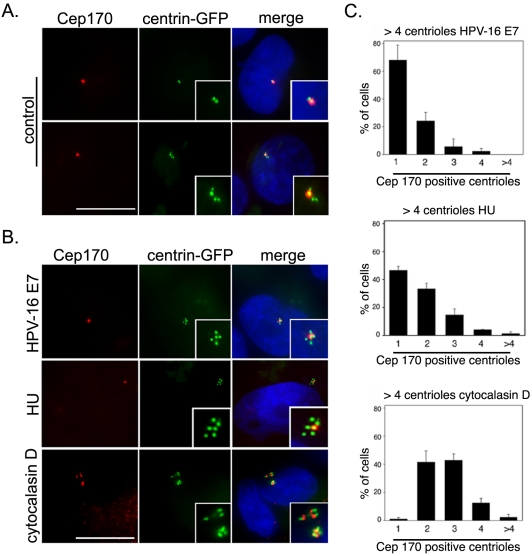
It occurred to us that markers for mature centrioles, such as Cep170, could be used to investigate the molecular mechanisms that underlie numerical centrosome aberrations. Such aberrations are frequently seen in tumor cells, but their molecular origins are presently unclear. As one important example of cells displaying a centrosome amplification phenotype, we examined U2OS cells in which supernumerary centrioles were induced by expression of HPV-16 oncoprotein E7 (Duensing et al., 2000; Munger et al., 2001). In parallel, we analyzed U2OS cells in which supernumerary centrosomes were deliberately induced through treatment with either HU, known to cause repeated rounds of centrosome duplication (Balczon et al., 1995; Meraldi et al., 1999), or cytochalasin D, known to cause division failure (Meraldi et al., 2002). U2OS cells stably expressing centrin-GFP (Duensing et al., 2000) were transiently transfected with HPV-16 E7 or empty vector, together with a transfection marker, and 48 h later they were stained with anti-Cep170 antibodies (Figure 8). As expected, the expression of HPV-16 E7 significantly prolonged S phase and increased the percentage of cells with abnormal numbers of centrioles (>4/cell) compared to controls (10.5 ± 1.3 and 3.1 ± 1.9%; p ≤ 0.005), consistent with previous results (Duensing et al., 2000, 2001). When analyzed for Cep170 staining, the majority of HPV-16 E7-transfected cells (68.1%) with abnormal centriole numbers contained only one Cep170-positive centriole (Figures 8B, top, and C). About 24.2% of cells showed 2 Cep170-positive centrioles, suggesting that these cells had progressed sufficiently far in the cell cycle (late G2/M) to allow maturation of the second parental centriole. A very similar result was obtained when analyzing the frequency of Cep170-positive centrioles in HU-treated U2OS cells (Figures 8B, middle, and C). These results strongly indicate that E7 causes centrosome amplification by an overduplication mechanism similar to the one operating during prolonged S phase arrest in HU-treated U2OS cells. In striking contrast, virtually all of the cytochalasin D-treated U2OS/centrin-GFP cells displayed two or more (up to 5) Cep170-positive centrioles (Figures 8B, bottom, and C). This result is in line with expectation, because supernumerary centrosomes arising through division failure are predicted to harbor mature centrioles. Together, these results illustrate that Cep170 staining can distinguish a centrosome amplification phenotype arising through deregulation of centriole duplication from the consequences of aborted divisions. From a more general perspective, they suggest that Cep170 (or other markers for mature centrioles) may constitute invaluable diagnostic tools for exploring the mechanisms underlying the genesis of supernumerary centrioles both in vitro and in vivo.
Figure 8.
Cep170 localization in cells with centrosome amplification. (A) U2OS/centrin-GFP (green) cells were stained with anti-Cep170 antibodies (red) and DAPI (blue): examples of Cep170 staining of only one centriole (see insets with centrosome enlargement) both before (top) or after (bottom) centriole duplication. Merge is shown on the right. Bar, 10 μm. (B) Cep170 staining (red) of U2OS/centrin-GFP (green) cells with centrosome amplification produced by HPV-16 E7 transfection (top), HU treatment (middle), or cytochalasin D treatment (bottom). Merge is shown in the right-hand panels. Insets show enlarged images of centrioles. DAPI is shown in blue. Bar, 10 μm. (C) Quantification of the number of Cep170-positive centrioles in cells showing abnormal centriole numbers after HPV-16 E7 transfection (top), HU treatment (middle), or cytochalasin D treatment (bottom). A total of at least 150 cells were counted in three independent experiments. Mean and SEs are shown.
DISCUSSION
We have characterized a centrosome protein, Cep170, which we isolated as a novel interaction partner of Plk1. The centrosome association of this protein confirms a recent prediction based on mass spectrometry and protein correlation profiling (Andersen et al., 2003). We show that Cep170 is expressed throughout the cell cycle. It associates with the centrosome in interphase and with the spindle apparatus during mitosis. In the G2 and M phases, Cep170 is phosphorylated extensively, and several lines of evidence point to Plk1 as a possible upstream kinase: 1) Plk1 is activated at the time in the cell cycle when Cep170 is phosphorylated in vivo; 2) Cep170 interacts with Plk1 in both a yeast two-hybrid assay and, at endogenous levels, within human cells; and 3) Cep170 is an in vitro substrate of recombinant Plk1. A detailed analysis of the function and physiological relevance of Cep170 phosphorylation will have to await the identification and subsequent mutation of physiological phosphorylation sites. However, it is intriguing that the site/s phosphorylated in vitro by Plk1, the Plk1–Cep170 interaction domain, and the centrosomal targeting domain of Cep170 all map to the C-terminal half.
Overexpression of Cep170 in U2OS cells induced strong MT bundling, suggesting that Cep170 is able to bind MTs with a high affinity. Deletion mapping studies implicated the C-terminal half of Cep170 in both MT binding and bundling and suggested the existence of more than one MT-binding domain. Although no previously described MT-binding motif could be recognized in the Cep170 sequence, the C terminus harbors a serine-rich region, a domain that is implicated in MT binding in the case of the CLIP-115 protein (Hoogenraad et al., 2000). Interestingly, cells overexpressing the C terminus of Cep170 frequently showed displacement of the centrosome from its typical perinuclear location to the cell periphery, implying a major rearrangement and polarization of the cytoskeleton. Because this phenotype resembles that caused by inhibition of dynein (Burakov et al., 2003), it is possible that Cep170 and dynein cooperate in centrosome–MT organization. In support of this possibility, preliminary results indicate that dynactin and Cep170 can interact in human cells (Guarguaglini and Nigg, unpublished data).
siRNA could be successfully used to deplete Cep170 from human cells. Depletion of Cep170 did not cause any obvious defects in centrosome structure. All centrosome proteins examined localized correctly, and microtubule nucleation was not significantly impaired (Guarguaglini and Nigg, unpublished data). Instead, drastic changes in cell shape and overall organization of the interphase cytoskeleton were observed, suggesting that Cep170 is involved in the regulation of microtubule dynamics. How Cep170 contributes to control cell shape is not yet clear, but a priori, it could act not only through the microtubule but also the actomyosin cytoskeleton. In support of the latter possibility, it is intriguing that, similar to Cep170 (see below), p160ROCK localizes to mother centrioles (Chevrier et al., 2002). This kinase coordinates actin organization with the MT network, and its inhibition was reported to cause the formation of elongated MTs, similar to the phenotype observed here upon depletion of Cep170. Moreover, inhibition of p160ROCK caused a relocalization of the mother centriole toward the cell periphery, reminiscent of the phenotype seen upon overexpression of the Cep170 C terminus. In view of these similarities, it will be interesting to investigate a possible functional link between p160ROCK and Cep170.
A striking feature of Cep170 is its asymmetric distribution within the centrosome. As we demonstrated by both IF and EM microscopy, Cep170 belongs to a growing group of proteins that preferentially localize to the mother centriole, usually by virtue of an association with subdistal appendages (Lange and Gull, 1995; Piel et al., 2000; Nakagawa et al., 2001; Casenghi et al., 2003; Chang et al., 2003; Gromley et al., 2003; Louie et al., 2004). Although these proteins have been implicated in different processes, subdistal appendages seem to be important sites for MT anchoring (Bornens, 2002; Mogensen, 2004). It is plausible, therefore, that several proteins work in concert to control MT anchoring to centriolar appendages, which in turn may contribute to regulate different aspects of centrosome functions during the cell cycle.
Cep170 clearly constitutes a specific marker for centriole maturation. Here, we have explored the possibility of using Cep170 for the selective identification of mature parental centrioles in cells displaying a centriole amplification phenotype. It is well established that several oncoproteins, notably the human papillomavirus E7 protein, induce the appearance of supernumerary centrosomes in both cultured cells and tumors. By staining centrioles with the anti-Cep170 antibody, we were able to show that HPV16/E7-transfected cells contain a normal number of mature (Cep170-positive) centrioles (1 or 2) and an increased number of immature (Cep170-negative) ones. Very similar results were obtained when centrosome overduplication was induced by culturing cells in HU, indicating that the overproduction of centrioles in the absence of passage through mitosis is not accompanied by the acquisition of maturation markers on newly generated centrioles. Conversely, cells in which centrioles accumulated due to failed cytokinesis generally displayed at least two, and often more than two, Cep170-positive centrioles, reflecting the fact that centrosome amplification in these cells had occurred through passage through at least one aborted mitosis, with the concomitant acquisition of Cep170 on the maturing centriole(s). These results illustrate that centriole maturity markers such as Cep170 can be used as powerful tools to discriminate between distinct mechanisms leading to centriole accumulation. In principle, such markers may prove useful to study any process in which the degree of centriole maturation can provide mechanistic information on centriole biogenesis.
The majority of known centrosomal proteins contain large coiled-coil regions. In contrast, Cep170 contains only a very short coiled coil. Instead, it harbors a putative FHA domain in its N terminus. Such domains are generally implicated in phosphopeptide recognition (Hofmann and Bucher, 1995; Li et al., 2000; Durocher and Jackson, 2002) and frequently observed in proteins that function in DNA damage response pathways. This is intriguing for two reasons: first, growing evidence links DNA damage responses to centrosomes and the G2/M transition (Sibon and Theurkauf, 2004), and second, Plk1 is inactivated after DNA damage and may interact with the FHA-containing protein Chk2, a central regulator of the DNA damage response (Smits et al., 2000; van Vugt et al., 2001; Tsvetkov et al., 2003). The identification of an FHA domain in the Plk1 interactor Cep170 strongly suggests that this protein is involved in a signaling pathway. The identification of this pathway constitutes one of the major challenges for future enquiry into the function of Cep170.
Acknowledgments
We thank R. Kopajtich, F. Barr, G. Maridor, B. Eddé, and G. Gerisch for antibodies and the laboratories of F. Barr, E. Nigg, and P. Lavia for sharing materials and helpful discussions. G. G. was supported by fellowships from Marie Curie EU, the Max-Planck Society, and Italian Foundation for Cancer Research.
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E04-10-0939) on December 22, 2004.
Abbreviations used: Cep170, centrosomal protein 170; FHA, forkhead-associated; HPV-16, human papillomavirus-16; HU, hydroxyurea; IF, immunofluorescence; IP, immunoprecipitation; MT, microtubule; PCM, pericentriolar material; Plk1, polo-like kinase 1; siRNA, small interfering RNA.
References
- Albrecht-Buehler, G., and Bushnell, A. (1980). The ultrastructure of primary cilia in quiescent 3T3 cells. Exp. Cell. Res. 126, 427-437. [DOI] [PubMed] [Google Scholar]
- Andersen, J. S., Wilkinson, C. J., Mayor, T., Mortensen, P., Nigg, E. A., and Mann, M. (2003). Proteomic characterization of the human centrosome by protein correlation profiling. Nature 426, 570-574. [DOI] [PubMed] [Google Scholar]
- Balczon, R., Bao, L., Zimmer, W. E., Brown, K., Zinkowski, R. P., and Brinkley, B. R. (1995). Dissociation of centrosome replication events from cycles of DNA synthesis and mitotic division in hydroxyurea-arrested Chinese hamster ovary cells. J. Cell. Biol. 130, 105-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr, F. A., Silljé, H.H.W., and E. A. Nigg. (2004). Polo-like kinases and the orchestration of cell division. Nat. Rev. Mol. Cell. Biol. 5, 429-440. [DOI] [PubMed] [Google Scholar]
- Bobinnec, Y., Khodjakov, A., Mir, L. M., Rieder, C. L., Edde, B., and Bornens, M. (1998). Centriole disassembly in vivo and its effect on centrosome structure and function in vertebrate cells. J. Cell. Biol. 143, 1575-1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bornens, M. (2002). Centrosome composition and microtubule anchoring mechanisms. Curr. Opin. Cell. Biol. 14, 25-34. [DOI] [PubMed] [Google Scholar]
- Burakov, A., Nadezhdina, E., Slepchenko, B., and Rodionov, V. (2003). Centrosome positioning in interphase cells. J. Cell. Biol. 162, 963-969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casenghi, M., Meraldi, P., Weinhart, U., Duncan, P. I., Korner, R., and Nigg, E. A. (2003). Polo-like kinase 1 regulates Nlp, a centrosome protein involved in microtubule nucleation. Dev. Cell 5, 113-125. [DOI] [PubMed] [Google Scholar]
- Chang, P., Giddings, T. H., Jr., Winey, M., and Stearns, T. (2003). Epsilontubulin is required for centriole duplication and microtubule organization. Nat. Cell. Biol. 5, 71-76. [DOI] [PubMed] [Google Scholar]
- Chevrier, V., Piel, M., Collomb, N., Saoudi, Y., Frank, R., Paintrand, M., Narumiya, S., Bornens, M., and Job, D. (2002). The Rho-associated protein kinase p160ROCK is required for centrosome positioning. J. Cell. Biol. 157, 807-817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chretien, D., Buendia, B., Fuller, S. D., and Karsenti, E. (1997). Reconstruction of the centrosome cycle from cryoelectron micrographs. J. Struct. Biol. 120, 117-133. [DOI] [PubMed] [Google Scholar]
- Descombes, P., and Nigg, E. A. (1998). The polo-like kinase Plx1 is required for M phase exit and destruction of mitotic regulators in Xenopus egg extracts. EMBO J. 17, 1328-1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- do Carmo, A. M., Tavares, A., and Glover, D. M. (2001). Polo kinase and Asp are needed to promote the mitotic organizing activity of centrosomes. Nat. Cell. Biol. 3, 421-424. [DOI] [PubMed] [Google Scholar]
- Do, E., Taira, E., Irie, Y., Gan, Y., Tanaka, H., Kuo, C. H., and Miki, N. (2003). Molecular cloning and characterization of rKAB1, which interacts with KARP-1,localizes in the nucleus and protects cells against oxidative death. Mol. Cell. Biochem. 248, 77-83. [DOI] [PubMed] [Google Scholar]
- Duensing, S., Lee, L. Y., Duensing, A., Basile, J., Piboonniyom, S., Gonzalez, S., Crum, C. P., and Munger, K. (2000). The human papillomavirus type 16 E6 and E7 oncoproteins cooperate to induce mitotic defects and genomic instability by uncoupling centrosome duplication from the cell division cycle. Proc. Natl. Acad. Sci. USA 97, 10002-10007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duensing, S., Duensing, A., Crum, C. P., and Munger, K. (2001). Human papillomavirus type 16 E7 oncoprotein-induced abnormal centrosome synthesis is an early event in the evolving malignant phenotype. Cancer Res. 61, 2356-2360. [PubMed] [Google Scholar]
- Durocher, D., and Jackson, S. P. (2002). The FHA domain. FEBS Lett. 513, 58-66. [DOI] [PubMed] [Google Scholar]
- Elbashir, S. M., Harborth, J., Lendeckel, W., Yalcin, A., Weber, K., and Tuschl, T. (2001). Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411, 494-498. [DOI] [PubMed] [Google Scholar]
- Fry, A. M., Mayor, T., Meraldi, P., Stierhof, Y. D., Tanaka, K., and Nigg, E. A. (1998). C-Nap1, a novel centrosomal coiled-coil protein and candidate substrate of the cell cycle-regulated protein kinase Nek2. J. Cell Biol. 141, 1563-1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry, A. M., and Hames, R. S. (2004). The role of the centrosome in cell cycle progression. In: Centrosomes in Development and Disease, ed. E. A. Nigg, Weinheim: Wiley-VCH, 143-166.
- Golsteyn, R. M., Schultz, S. J., Bartek, J., Ziemiecki, A., Ried, T., and Nigg, E. A. (1994). Cell cycle analysis and chromosomal localization of human Plk1, a putative homologue of the mitotic kinases Drosophila polo and Saccharomyces cerevisiae Cdc5. J. Cell Sci. 107, 1509-1517. [DOI] [PubMed] [Google Scholar]
- Golsteyn, R. M., Mundt, K. E., Fry, A. M., and Nigg, E. A. (1995). Cell cycle regulation of the activity and subcellular localization of Plk1, a human protein kinase implicated in mitotic spindle function. J. Cell. Biol. 129, 1617-1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gromley, A., Jurczyk, A., Sillibourne, J., Halilovic, E., Mogensen, M., Groisman, I., Blomberg, M., and Doxsey, S. (2003). A novel human protein of the maternal centriole is required for the final stages of cytokinesis and entry into S phase. J. Cell. Biol. 161, 535-545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinchcliffe, E. H., Miller, F. J., Cham, M., Khodjakov, A., and Sluder, G. (2001). Requirement of a centrosomal activity for cell cycle progression through G1 into S phase. Science 291, 1547-1550. [DOI] [PubMed] [Google Scholar]
- Hofmann, K., and Bucher, P. (1995). The FHA domain: a putative nuclear signalling domain found in protein kinases and transcription factors. Trends Biochem. Sci. 20, 347-349. [DOI] [PubMed] [Google Scholar]
- Hoogenraad, C. C., Akhmanova, A., Grosveld, F., De Zeeuw, C. I., and Galjart, N. (2000). Functional analysis of CLIP-115 and its binding to microtubules. J. Cell. Sci. 113, 2285-2297. [DOI] [PubMed] [Google Scholar]
- Kelm, O., Wind, M., Lehmann, W. D., and Nigg, E. A. (2002). Cell cycle-regulated phosphorylation of the Xenopus polo-like kinase Plx1. J. Biol. Chem. 277, 25247-25256. [DOI] [PubMed] [Google Scholar]
- Khodjakov, A., and Rieder, C. L. (2004). A synergy of technologies: using green fluorescent protein tagging and laser microsurgery to study centrosome function and duplication in vertebrates. In: Centrosomes in Development and Disease, ed. E. A. Nigg, Weinheim, Wiley-VCH, 191-210.
- Lane, H. A., and Nigg, E. A. (1996). Antibody microinjection reveals an essential role for human polo-like kinase 1 (Plk1) in the functional maturation of mitotic centrosomes. J. Cell. Biol. 135, 1701-1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange, B. M., and Gull, K. (1995). A molecular marker for centriole maturation in the mammalian cell cycle. J. Cell. Biol. 130, 919-927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J., Lee, G. I., Van Doren, S. R., and Walker, J. C. (2000). The FHA domain mediates phosphoprotein interactions. J. Cell. Sci. 113, 4143-4149. [DOI] [PubMed] [Google Scholar]
- Ligon, L. A., Shelly, S. S., Tokito, M., and Holzbaur, E. L. (2003). The microtubule plus-end proteins EB1 and dynactin have differential effects on microtubule polymerization. Mol. Biol. Cell. 14, 1405-1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louie, R. K., Bahmanyar, S., Siemers, K. A., Votin, V., Chang, P., Stearns, T., Nelson, W. J., and Barth, A. I. (2004). Adenomatous polyposis coli and EB1 localize in close proximity of the mother centriole and EB1 is a functional component of centrosomes. J. Cell. Sci. 117, 1117-1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayor, T., Hacker, U., Stierhof, Y. D., and Nigg, E. A. (2002). The mechanism regulating the dissociation of the centrosomal protein C-Nap1 from mitotic spindle poles. J. Cell. Sci. 115, 3275-3284. [DOI] [PubMed] [Google Scholar]
- Meraldi, P., Lukas, J., Fry, A. M., Bartek, J., and Nigg, E. A. (1999). Centrosome duplication in mammalian somatic cells requires E2F and Cdk2-cyclinA. Nat. Cell. Biol. 1, 88-93. [DOI] [PubMed] [Google Scholar]
- Meraldi, P., and Nigg, E. A. (2002). The centrosome cycle. FEBS Lett. 521, 9-13. [DOI] [PubMed] [Google Scholar]
- Meraldi, P., Honda, R., and Nigg, E. A. (2002). Aurora-A overexpression reveals tetraploidization as a major route to centrosome amplification in p53-/- cells. EMBO J. 21, 483-492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogensen, M. M. (2004). Microtubule organizing centers in polarized epithelial cells. In: Centrosomes in Development and Disease, ed. E. A. Nigg, Weinheim, Wiley-VCH, 299-320.
- Moudjou, M., and Bornens, M. (1994) Isolation of centrosomes from cultured animal cells. In: Cell Biology: A Laboratory Handbook, ed. J. E. Celis, London, Academic Press, 595-604.
- Munger, K., Basile, J. R., Duensing, S., Eichten, A., Gonzalez, S. L., Grace, M., and Zacny, V. L. (2001). Biological activities and molecular targets of the human papillomavirus E7 oncoprotein. Oncogene 20, 7888-7898. [DOI] [PubMed] [Google Scholar]
- Nakagawa, Y., Yamane, Y., Okanoue, T., Tsukita, S., and Tsukita, S. (2001). Outer dense fiber 2 is a widespread centrosome scaffold component preferentially associated with mother centrioles: its identification from isolated centrosomes. Mol. Biol. Cell 12, 1687-1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigg, E. A. (2004). Centrosomes in Development and Disease, Weinheim, Wiley-VCH.
- Piel, M., Meyer, P., Khodjakov, A., Rieder, C. L., and Bornens, M. (2000). The respective contributions of the mother and daughter centrioles to centrosome activity and behavior in vertebrate cells. J. Cell. Biol. 149, 317-330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piel, M., Nordberg, J., Euteneuer, U., and Bornens, M. (2001). Centrosomedependent exit of cytokinesis in animal cells. Science 291, 1550-1553. [DOI] [PubMed] [Google Scholar]
- Rieder, C. L., and Borisy, G. G. (1982). The centrosome cycle in PTK2 cells - asymmetric distribution and structural changes in the pericentriolar material. Biology of the Cell 44, 117-132. [Google Scholar]
- Seelos, C. (1997). A critical parameter determining the aging of DNA-calciumphosphate precipitates. Anal. Biochem. 245, 109-111. [DOI] [PubMed] [Google Scholar]
- Sibon, O.C.M., and Theurkauf, W. E. (2004). Centrosome regulation in response to environmental and genotoxic stress. In: Centrosomes in Development and Disease, ed. E. A. Nigg, Weinheim, Wiley-VCH, 211-224.
- Smits, V. A., Klompmaker, R., Arnaud, L., Rijksen, G., Nigg, E. A., and Medema, R. H. (2000). Polo-like kinase-1 is a target of the DNA damage checkpoint. Nat. Cell. Biol. 2, 672-676. [DOI] [PubMed] [Google Scholar]
- Stierhof, Y. D., Humbel, B. M., and Schwarz, H. (1991). Suitability of different silver enhancement methods applied to 1 nm colloidal gold particles: an immunoelectron microscopic study. J. Electron Microsc. Tech. 17, 336-343. [DOI] [PubMed] [Google Scholar]
- Sunkel, C. E., and Glover, D. M. (1988). polo, a mitotic mutant of Drosophila displaying abnormal spindle poles. J. Cell Sci. 89, 25-38. [DOI] [PubMed] [Google Scholar]
- Tsvetkov, L., Xu, X., Li, J., and Stern, D. F. (2003). Polo-like kinase 1 and Chk2 interact and co-localize to centrosomes and the midbody. J. Biol. Chem. 278, 8468-8475. [DOI] [PubMed] [Google Scholar]
- van Vugt, M. A., Smits, V. A., Klompmaker, R., and Medema, R. H. (2001). Inhibition of Polo-like kinase-1 by DNA damage occurs in an ATM- or ATR-dependent fashion. J. Biol. Chem. 276, 41656-41660. [DOI] [PubMed] [Google Scholar]