Significance
Many activating immunoreceptors associate, via interactions between transmembrane domains, with adaptor molecules that mediate signaling for leukocyte activation. To date, the best characterized form of receptor complex assembly depends on a single basic transmembrane (TM) domain residue. We now describe a second, completely different solution for TM-mediated receptor assembly, found in three different Fc receptor TM domains, and involving a more complex polar/aromatic interface. Residues in the core of this interaction motif can also regulate receptor protein turnover. Thus, multiple solutions for TM-mediated receptor assembly with signaling modules have evolved. These findings may provide more broadly useful insights into how other immune receptors that do not contain charged residues in their TM domains assemble into complexes with signaling adaptor molecules.
Keywords: activating immunoreceptors, signaling adaptor molecules, Fc receptors, transmembrane interactions
Abstract
Many activating immunoreceptors associate with signaling adaptor molecules like FcεR1γ or CD247. FcεR1γ and CD247 share high sequence homology and form disulphide-linked homodimers that contain a pair of acidic aspartic acid residues in their transmembrane (TM) domains that mediate assembly, via interaction with an arginine residue at a similar register to these aspartic acids, with the activating immunoreceptors. However, this model cannot hold true for receptors like CD16A, whose TM domains do not contain basic residues. We have carried out an extensive site-directed mutagenesis analysis of the CD16A receptor complex and now report that the association of receptor with the signaling adaptor depends on a network of polar and aromatic residues along the length of the TM domain. Molecular modeling indicates that CD16A TM residues F202, D205, and T206 form the core of the membrane-embedded trimeric interface by establishing highly favorable contacts to the signaling modules through rearrangement of a hydrogen bond network previously identified in the CD247 TM dimer solution NMR structure. Strikingly, the amino acid D205 also regulates the turnover and surface expression of CD16A in the absence of FcεR1γ or CD247. Modeling studies indicate that similar features underlie the association of other activating immune receptors, including CD64 and FcεR1α, with signaling adaptor molecules, and we confirm experimentally that equivalent F, D, and T residues in the TM domain of FcεR1α markedly influence the biology of this receptor and its association with FcεR1γ.
Human FcγRIIIA (CD16A) is a low-affinity receptor for the Fc portion of IgG expressed by human CD56dim natural killer (NK) cells, subsets of monocytes, dendritic cells, and rare T cells. FcγRIIIB (CD16B) is encoded by a distinct gene and is preferentially expressed by neutrophils (1). CD16B is a GPI-anchored glycoprotein whereas CD16A is a type 1 membrane glycoprotein with a single transmembrane (TM) domain and a short cytoplasmic tail whose expression at the cell surface depends on association with the signaling adaptor molecules CD247 (TCRζ) and/or FcεR1γ (1–4). First discovered as components of the TCR:CD3 complex and the high affinity receptor for IgE, respectively (5–7), CD247 and FcεR1γ are integral membrane proteins that have subsequently been found to be obligate signaling adaptors for many immunoreceptors in different cell types. Both adaptors have very short extracellular domains and cytoplasmic tails that contain immunoreceptor tyrosine-based activating motifs (ITAMs) for signal transduction. Moreover, both CD247 and FcεR1γ form dimers in the endoplasmic reticulum (ER) that are stabilized by a disulphide bond at the junction between the extracellular and TM domains so that, at the cell surface, CD247 and FcεR1γ in complex with their client receptors are dimeric. In general, the receptors known to associate with CD247 and FcεR1γ have short intracellular tails and thus are completely dependent on these adaptors to signal, although phosphorylation of a protein kinase C (PKC) motif in the cytoplasmic tail of CD16A can modulate the outcome of receptor ligation (8).
The incorporation of CD247 into the TCR/CD3 complex is the rate-limiting step in the process of assembly of the receptor complex for cell surface expression because incomplete TCR/CD3 complex is otherwise retained in the cis-Golgi and targeted for degradation (9). Similarly, association with the FcεR1γ or CD247 molecules is also required for the correct maturation and expression at the cell surface of the CD16A protein (2–4). The interaction of CD247 with the TCR, like many immunoreceptor complex interactions, occurs via a precise interface involving the TM domains of the different subunits in the complex (10). Detailed analysis of the interactions underlying the associations between the different subunits of the T-cell receptor complex have shown that interactions between basic and acidic residues localized at precise positions of the TM domains of the different subunits are necessary, and often sufficient, for complex assembly (11). Subsequent studies on the assembly of NK cell receptor complexes, such as NKG2D–DAP10 (12) or NKG2C–DAP12 (13), have shown that the association of the subunits in these complexes is also mediated by similar interactions, where pairs of aspartic acids in the adaptor molecules interact with either an arginine or lysine residue in the receptor TM domain. Indeed, for KIR2DS2 or NKG2D, all residues of the receptor TM domain can be mutated to valine or leucine, and the interactions of the lysine of KIR2DS2, or the arginine of NKG2D, with aspartic acids in DAP12 and DAP10, respectively, are sufficient to maintain the receptor/adaptor complex (14). Surprisingly, however, the TM domain sequence of CD16A, which is devoid of basic residues and contains an aspartic acid, pairs with FcεR1γ and CD247 which also contain aspartic acids in their TM. Thus, a mechanism of assembly between CD16A and FcεR1γ or CD247, distinct from those known for other immunoreceptor complexes, must exist.
We examined in detail the TM interactions that mediate association of CD16A with FcεR1γ and CD247 signaling molecules and found that multiple polar and aromatic residues, distributed along an extended helical face of the CD16A TM domain, contribute to the receptor/adaptor interaction. Our analysis revealed a particularly important role for the CD16A TM sequence F202D205T206 in both assembly with signaling adaptors and intracellular retention/degradation of unassembled CD16A protein. An independent molecular dynamics (MD) simulation approach identified a highly favored TM assembly mode in which nearly all of the CD16A TM residues identified in mutagenesis experiments directly contact the surfaces of FcεR1γ and CD247 signaling dimers. The model that emerged provides a structural rationale for the role of the F202D205T206 trio in establishing favorable contacts with signaling dimers and sequestering D205, which constitutes the key CD16A retention/degradation signal, within the interface. Unbiased MD simulations revealed that the related Fc receptors FcεR1α and FcγR1 (CD64) likely form similar interfaces with the FcεR1γ dimer that include nearly identical interactions mediated by conserved FxxDT (FcεR1α) and FxxNT (CD64) TM sequences. We confirmed the importance of this sequence by examining the assembly and surface expression of FcεR1α in cells. These results indicate that the concepts identified here may serve as a useful guide to understand the assembly and cell biology of multiple activating immunoreceptor complexes where interactions between amino acids of complementary charge potential are not obviously relevant.
Results
CD16A Associates with Either FcεR1γ or CD247.
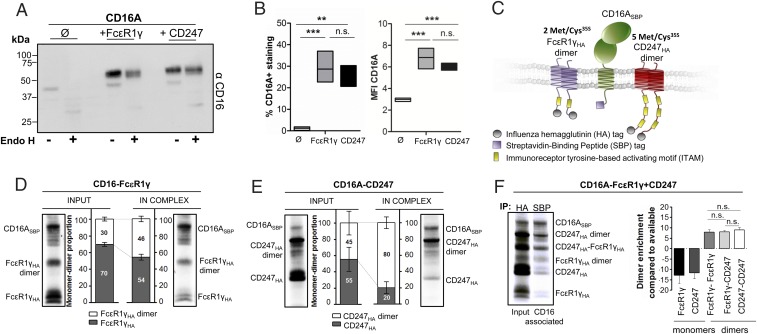
It has previously been reported that cell surface expression of the CD16A receptor depends on a noncovalent association with the signaling dimers FcεR1γ and/or CD247 (2–4). Ex vivo analysis of CD16A cell surface expression on primary human NK cells deficient in either FcεR1γ or CD247 has suggested that CD16A does not discriminate between these adaptor molecules (15), and we confirmed these data in vitro when fibroblasts were transfected with CD16A alone or in combination with either FcεR1γ or CD247 adaptor molecules. Only small amounts of CD16A protein, which correspond to an immature (EndoH-sensitive) species, were detected in cells transfected with the receptor construct alone (Fig. 1A), and almost no surface expression of CD16A was observed (Fig. 1B). In contrast, when the CD16A construct was cotransfected with plasmids driving expression of the FcεR1γ or CD247 adaptor molecules, abundant glycosylated and EndoH-resistant CD16A protein was observed (Fig. 1A). These experiments showed that CD16A was retained in the ER and probably degraded, in the absence of adaptor (Fig. 1A). Cell-surface expression of CD16A seemed equivalent when either FcεR1γ or CD247 was present in the system (Fig. 1B). Therefore, these data confirm prior work demonstrating that association of the adaptor modules FcεR1γ or CD247 with CD16A not only is required for progression of this receptor through the secretory pathway and cell surface expression but also protects the CD16A protein from degradation (16).
Fig. 1.
CD16A requires association with adaptor modules for cell surface expression but shows no preference for assembly with either CD247 or FcεR1γ. An expression vector encoding CD16A was transfected, either alone or in combination with plasmids for CD247 or FcεR1γ, into 293T cells, and cell lysates were analyzed by Western blot (A). Aliquots of the same lysates were treated with EndoH endoglycosidase for determination of glycosylation state as a measure of maturation state (A). Representative image of three independent experiments is shown. Surface expression of CD16A was also analyzed by flow cytometry of transfected cells. (B) Percentage of cells expressing CD16 at the cell surface and mean fluorescence intensity (MFI) of CD16 staining was determined using Kaluza Flow Cytometry Analysis Software. Data represent mean of three experiments, and statistical significance was determined using the one-way ANOVA-Tukey´s multiple comparison test (**P < 0.01; ***P < 0.001; n.s., not statistically significant P > 0.05). (C) Schematic showing CD16A-SBP and HA-tagged signaling dimers used for IVT assay. The different numbers of 35S labeling positions are indicated. (D–F) CD16A mRNA was cotranslated with FcεR1γ (D), CD247 (E), or both FcεR1γ and CD247 (F) adaptor molecule mRNAs (in the presence of radioactively labeled 35S methionine and cysteine). These reactions were done in ER-like conditions to allow assembly of the receptor complex. Each completed assembly reaction was then divided in two aliquots: one for HA immunoprecipitation, to quantify total full-length transcribed FcεR1γHA and CD247HA protein and labeled “INPUT” control in our analysis and the other labeled “IN COMPLEX,” where CD16-SBP was immunoprecipitated and associated HA-tagged adaptor molecules could be analyzed. The immunoprecipitates were analyzed in 12% SDS/PAGE gels in nonreducing conditions and transferred to PVDF membranes for phospho-imaging. Dimer and monomer species were quantified, normalized to the number of labeling positions, and plotted as mean ± SD. Statistical significance was calculated using one-way ANOVA. Images are representative of at least three independent experiments.
Adaptor Modules FcεR1γ and CD247 Are Recruited Equally by CD16A.
To address in detail how the CD16A receptor associates with the FcεR1γ or CD247 adaptors, we used an in vitro translation (IVT)-based experimental system that had previously been used to define the assembly of the TCRαβ/CD3εγδ/CD247 complex (11) and several NK receptor complexes (10). Streptavidin-binding peptide (SBP)-tagged CD16A was cotranslated with either HA-tagged FcεR1γ or HA-CD247 in the presence of ER-derived microsomes and 35S-labeled methionine/cysteine, and then complex formation was quantitated in coimmunoprecipitation experiments (Fig. 1C and Fig. S1).
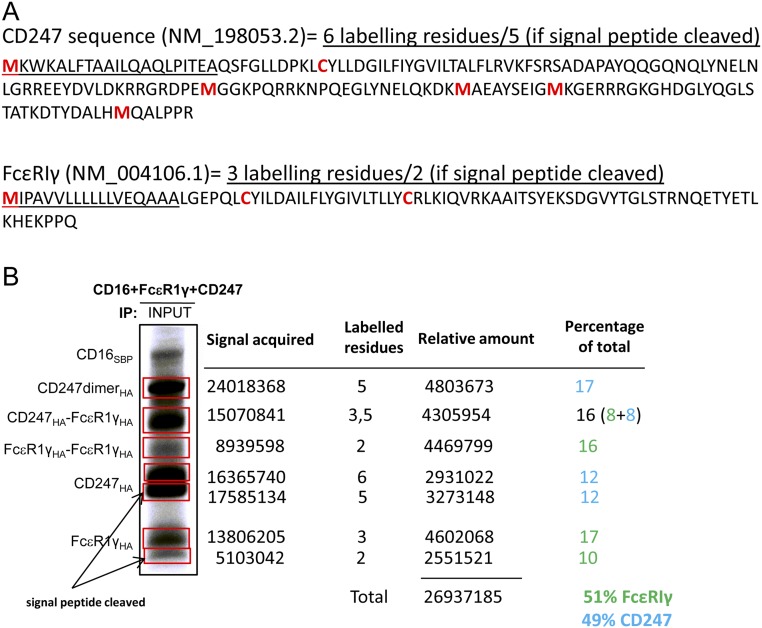
Fig. S1.
Quantitation of in vitro translations. In vitro-transcribed mRNAs were translated using a methionine- and cysteine-free amino acid mixture supplemented with 35S-labeled amino acids. (A) Each protein is labeled to a different level, depending on the protein sequence. In some cases, the signal peptide (underlined) is only partially cleaved, and then an extra Met residue is present in some bands. (B) For quantitation, the signal from each band (shown boxed in red) is normalized by dividing the signal acquired by the number of labeled residues (shown in red in A) from each protein to obtain the relative amounts of protein. Because the HA and SBP epitope tags are located at the C termini of the proteins, HA immunoprecipitation was used as “INPUT” control to only pull down and quantify full-length translated molecules of FcεR1γ and CD247.
Inspection of the lanes corresponding to “INPUT” controls (Fig. 1 D and E) showed that HA-tagged FcεR1γ and CD247 species ran on nonreducing SDS/PAGE gels as dimers and monomers, but that, after immunoprecipitation of CD16A, the fraction of dimer species was clearly enriched (Fig. 1 D and E, “IN COMPLEX”). Quantitation of CD16A immunoprecipitates from these reactions confirmed that CD16A preferentially recruited adaptor molecule dimers rather than monomers (Fig. 1 D and E). Indeed, because dimerization of CD247 and FcεR1γ is driven by polar residues in the TM domain, we cannot exclude the possibility that a proportion of the monomeric species detected after SDS/PAGE were actually noncovalent dimeric species in which the disulphide bond had not yet formed. Experiments in which CD16A was cotranslated with both FcεR1γ and CD247 in the same reaction showed once again that dimeric species were highly enriched in CD16A immunoprecipitates, compared with monomeric adaptor molecules (Fig. 1F). Moreover, after integrating the radioactive signal in each band and correcting for the number of methionine/cysteine labeling positions in each dimeric species, no preference for FcεR1γ-FcεR1γ, FcεR1γ-CD247, or CD247-CD247 dimers was observed, suggesting that CD16A binds FcεR1γ and CD247 with similar affinity. This observation suggested that the association of CD16A with these adaptor molecules is mediated by similar protein interactions; this hypothesis was tested in the next set of experiments.
A Polar and Aromatic Interface Mediates the Association of CD16A with Adaptor Modules.
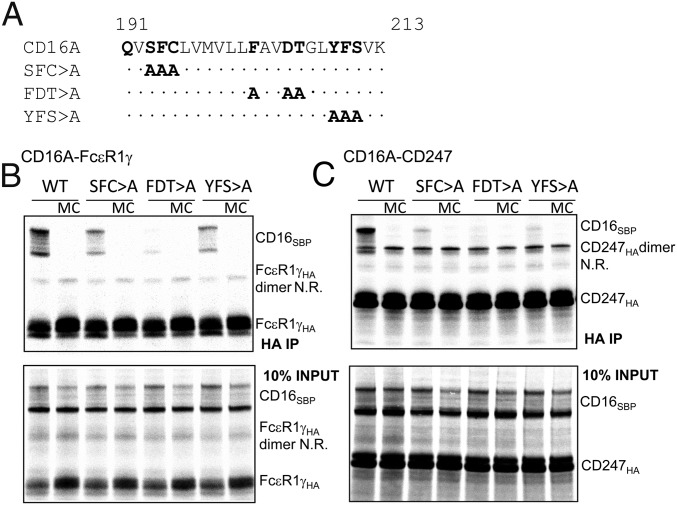
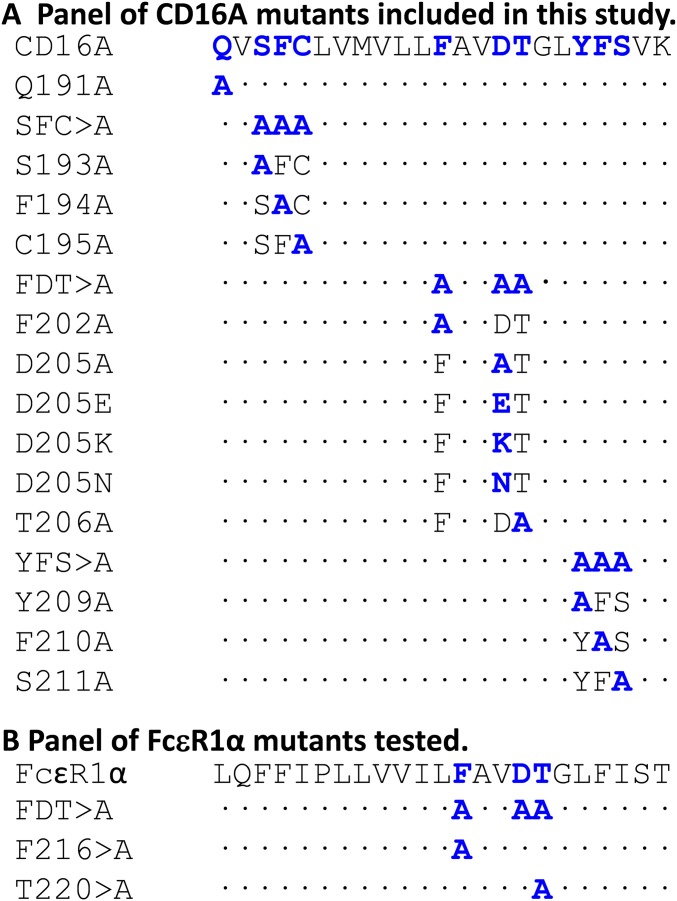
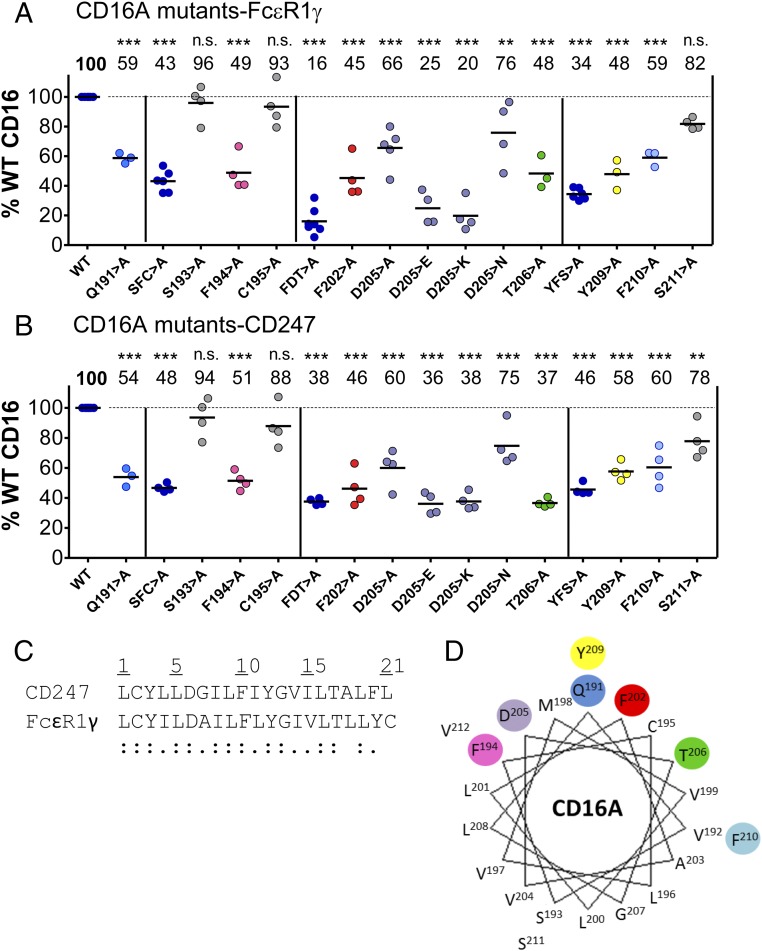
To identify the TM residues involved in CD16A association with adaptor modules, a panel of mutants was prepared in three blocks of three mutations. Based on the documented roles of polar and aromatic residues in driving TM helix associations (17, 18), we mutated these amino acid types in the CD16A TM sequence (Fig. 2A) to alanine and evaluated the association of mutants with each signaling molecule using the IVT system described above. All three of the “triple” mutants (S193F194C195 > A, F202D205T206 > A, and Y209F210S211 > A) showed marked defects in association with both FcεR1γ and CD247 (Fig. 2 B and C). To determine the contribution of specific residues to the defective association of the triple mutants with FcεR1γ, a panel of single mutants (Fig. S2A) was prepared and tested in quantitative analyses (Fig. 3A). This panel included a Q191 > A mutant that had not been tested in the block analysis but caused ∼40% reduction in CD16A association, identifying an additional polar amino acid involved in the assembly. The mutation of S193F194C195 > A decreased CD16A association by more than 50%, and analysis of the individual substitutions showed that the single F194 > A mutation was sufficient to cause this decrease (Fig. 3A) whereas mutations at S193 and C195 had no significant effects. Dissection of the F202D205T206 > A triple mutant revealed that single substitutions at either F202 or T206 provoked significant reductions in the association with FcεR1γ whereas the substitution of alanine at D205 of the CD16A TM domain had a lesser effect (Fig. 3A). This result was surprising because of the prior observations on the crucial roles played by acidic and basic amino acids in the formation of other immunoreceptor complexes (10). To further explore the role of this acidic residue, substitutions of D205 with E, K, or N were tested for complex formation with FcεR1γ. Substitution with either E or K essentially abolished complex formation, suggesting that the size and charge at this position have an important impact on the association with FcεR1γ. Replacement with N (polar, nonionizable) also led to only a mild reduction in formation of the receptor complex, suggesting that the ionization of D205 is not critical for assembly. When single mutants of the Y209F210S211 > Ala triple mutant were tested, alteration of the aromatic residues Y209 and F210 had the greatest effect on the CD16A–FcεR1γ complex formation whereas replacement of S211 with alanine produced only a mild defect (Fig. 3A). We tested the same CD16A TM mutants for their effects on association with CD247, and very similar results were obtained (Fig. 3B), consistent with the high sequence similarity between the TM domains of FcεR1γ and CD247 (Fig. 3C).
Fig. 2.
The effects of CD16A triple block mutants on association with CD247 or FcεR1γ. Three triple mutants targeting groups of polar and aromatic residues along the length of the CD16A transmembrane domain were prepared (A), and the ability of these mutants to assemble with adaptor modules, FcεR1γ or CD247, was evaluated (B and C). After immunoprecipitation using anti-HA antibody, complex formation with FcεR1γ (B) and CD247 (C) was analyzed in 12% SDS/PAGE gels in reducing conditions. Then 10% of assembly reactions without immunoprecipitation were analyzed in parallel as loading controls. As can be observed in the gels, dimers of FcεR1γ, and especially CD247, were not completely reduced in these conditions. MC, mixing control in which CD16A and FcεR1γ/CD247 were translated in separate reactions and mixed just before detergent extraction; N.R., nonreduced.
Fig. S2.
Panels of CD16A and FcεR1α mutants used in these studies. Shown are alignments of the transmembrane domains of (A) CD16A and (B) FcεR1α indicating the mutants studied in these experiments. Mutated residues are shown in blue.
Fig. 3.
The CD16A-FcεR1γ TM interface is dominated by polar and aromatic residues. Triple- and single-residue CD16A mutants were tested for association with FcεR1γ (A) or CD247 (B). Each dot represents an independent experiment where complex formation for WT or the indicated CD16A mutant was quantified. Statistical significance was calculated using one-way ANOVA-Dunnett posttest for multiple comparison (**P < 0.01; ***P < 0.001; n.s., not statistically significant P > 0.05). (C) Alignments of the predicted TM domains of FcεR1γ and CD247 showing the similarity between these adaptors. For ease of comparison between the two sequences, which have different extracellular lengths, the sequences have been numbered 1 to 21, beginning with the first TM residue. Double dot (:), fully conserved residue; single dot (.), weak similarity. (D) TM residues important for complex association (Q191, F194, F202, D205, T206, Y209, and F210) were highlighted in a helical wheel representation of the predicted CD16A TM domain, showing their localization to the same aspect of CD16A-TM (residues are color-coded to their corresponding data points in A and B).
Mapping the amino acids that contributed most to the interactions of CD16A with FcεR1γ and CD247 onto a helical wheel projection (Fig. 3D), we observed that these residues are colocated on one face of the receptor TM domain whereas the only two polar residues located on the opposite side of the TM helix (S193 and S211) had no effect on complex formation. The replacement of C195 with alanine on the interacting face also had no significant effect, indicating that disruption of the association with adaptor molecules is likely a specific effect of particular mutations rather than a generally disruptive effect of alanine substitution on the TM helix structure. This interpretation is consistent with the view that the helical structure of TM domains is driven primarily by backbone hydrogen bonds and the observation that alanine is, after leucine, the second most common α-helical TM residue (19). We therefore concluded that interactions between CD16A and its signaling adaptors FcεR1γ and CD247 are mediated by a specific helical face of the CD16A TM domain that is composed of mostly polar and aromatic amino acids.
Analysis of the Interactions Between CD16A and Signaling Adaptors in Live Cells.
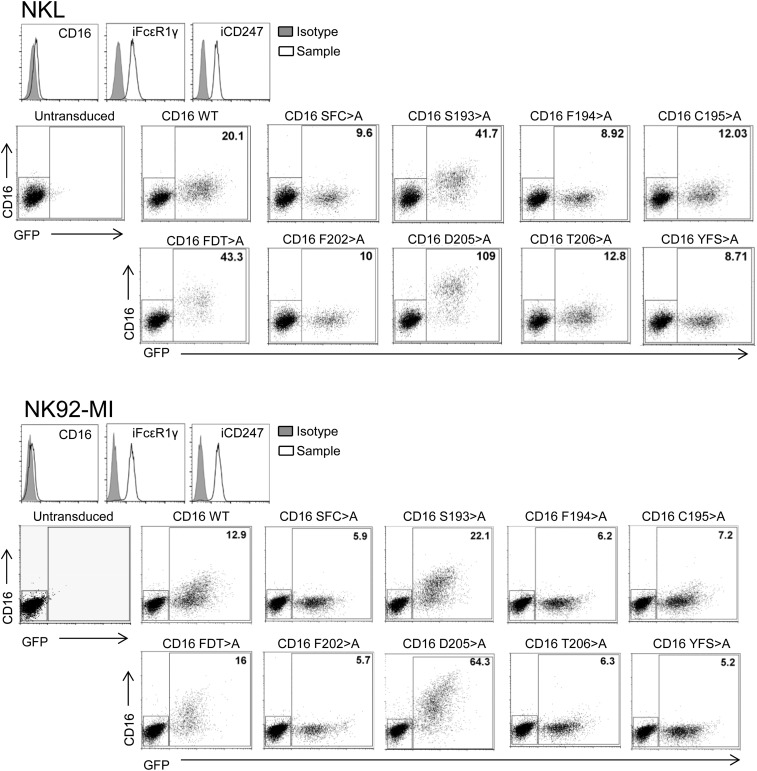
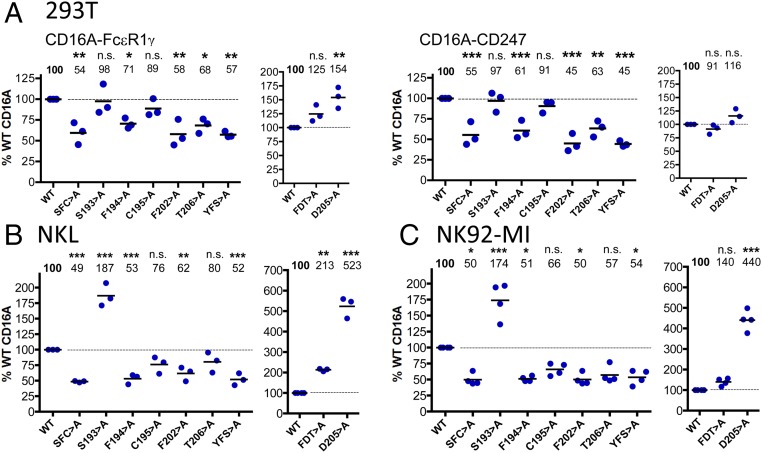
Although the IVT technique facilitates highly quantitative comparisons of mutants in receptor assembly, it has the drawback that the use of isolated ER microsomes means that only a fraction of the secretory pathway that membrane proteins must traverse to reach the cell surface is present in these experiments. Thus, it was important to analyze the CD16A-signaling adaptor association in live cells. To this end, 293T cells and two NK cell lines, NKL (20) and NK92-MI (21) (that express endogenous FcεR1γ and CD247) (Fig. S3), were transfected with selected TM mutants of CD16A. Flow cytometry analysis of 293T cells transfected with CD16A WT or selected mutants, in the presence of either FcεR1γ or CD247, confirmed that surface expression of the triple mutants S193F194C195 > A and Y209F210S211 > A was significantly reduced compared with WT receptor (Fig. 4A), consistent with the reduced ability of these mutants to associate with either FcεR1γ or CD247 in the IVT experiments (Fig. 3). Surprisingly, however, the F202D205T206 > A mutant receptor reached the cell surface, despite being essentially unable to associate with the adaptor molecules in IVT experiments. Subsequent analysis of point mutants showed that substitution of any of the TM amino acids F194, F202, and T206 with alanine significantly impaired CD16A expression at the cell surface, supporting the conclusion that these residues contribute to the association with FcεR1γ and CD247. In contrast, the single change D205 > A sufficed to permit normal, or increased, levels of CD16A expression at the plasma membrane. Analysis of the NK cell lines NKL and NK92-MI transduced with the same panel of CD16A mutants confirmed these findings (Fig. 4 B and C) (representative FACS plots are shown in Fig. S3). These data strongly support the conclusion from IVT experiments that the CD16A TM residues F194, F202, and T206 mediate important contacts with the adaptor molecules and further indicate that D205 markedly influences intracellular retention of CD16A. In these experiments, the CD16A S193 > A mutant was also expressed at increased levels on the surface of the NKL and NK92-MI cell lines, but not on 293T cells. The IVT data showed that mutation of this amino acid had no effect on the interaction with signaling adaptor molecules, consistent with its predicted location on the opposite side of the TM helix from the residues that do participate directly in the interaction. It is possible that this increased expression simply reflects an increased ability to reach the cell surface due to the substitution of a nonpolar for a polar amino acid in the TM domain, but this hypothesis does not explain why this effect occurs in NK cell lines, but not 293T cells. Thus, we favor the idea that the S193 > A mutation leads to increased expression of CD16A at the cell surface by modulating the association of this receptor with some other molecule expressed in lymphoid cells (e.g., CD2) (22), but further experiments will be required to resolve this issue definitively.
Fig. S3.
Representative flow cytometry analyses of the NK cell lines NKL and NK92-MI. Histograms show that FcεR1γ and CD247 are expressed in unmanipulated NK92-MI and NKL cell lines, but CD16A is only present at very low levels on the cell surface. Dot plots show CD16A expression at the cell surface of the NK92-MI and NKL cell lines transduced with either WT CD16A or the indicated mutants. Transduced cells were gated using GFP as marker of the lentiviral-based vector used. The number indicates the MFI of CD16A surface staining in the GFP-positive gate for each transduced cell line.
Fig. 4.
Analysis of the surface expression of CD16A mutants in live cells. (A) The 293T cells were transfected with CD16A combined with either FcεR1γ or CD247. Surface expression of the different mutants was analyzed by flow cytometry, and the data are shown after normalization to WT CD16A. NK cell lines NKL (B) and NK92-MI (C) were transduced with lentivirus to express CD16A WT or selected mutants, and cell surface expression of these receptors was analyzed by flow cytometry. Representative dot plots of NK cell staining are shown (Fig. S3). Statistical significance was calculated using one-way ANOVA-Dunnett posttest for multiple comparison (*P < 0.05; **P < 0.01; ***P < 0.001; n.s., not statistically significant P > 0.05).
The TM Amino Acid D205 Regulates Intracellular Trafficking of CD16A.
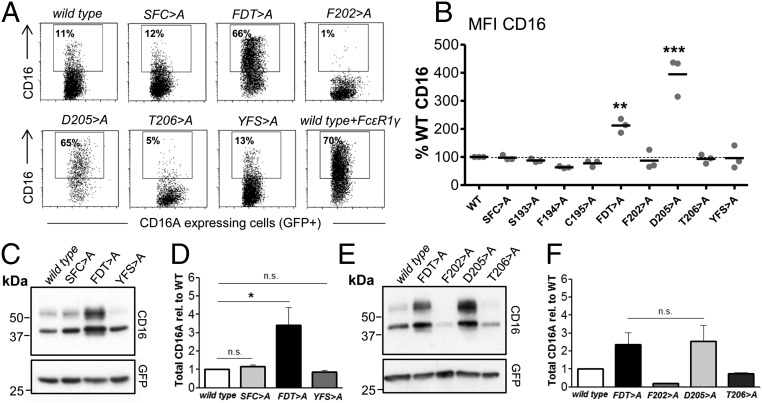
Because CD16A is unable to reach the cell surface in the absence of adaptor modules and is instead degraded, it has been suggested that the association of CD16A with either FcεR1γ or CD247 masks a motif in the CD16A TM domain that might direct protein degradation (16) or retention (23). To test this hypothesis, 293T fibroblasts were transfected with different CD16A TM mutant proteins in the absence of adaptor proteins and analyzed by flow cytometry and Western blot. As previously shown (Fig. 1A), in the absence of signaling adaptors, WT CD16A accumulates in the ER, and little to no mature glycosylated species or cell surface expression can be detected (Fig. 5). In these experiments, the S193F194C195 > A and Y209F210S211 > A mutants behaved comparably with WT CD16A, showing no significant differences in cell surface expression (Fig. 5 A and B) or total cellular protein levels (Fig. 5 C and D). In contrast, cells transfected with the F202D205T206 > A mutant in the absence of signaling adaptors showed a level of surface expression comparable with WT CD16A coexpressed with FcεR1γ (Fig. 5A), and a 3.5-fold increase in levels of both immature and fully glycosylated CD16A protein was detected by Western blot (Fig. 5 C and D). The CD16A D205 > A mutant was also expressed at very high levels on the cell surface, and, again, marked increases in the levels of total CD16A protein were obvious on Western blot analysis (Fig. 5 E and F). Together with the low surface expression and total cellular protein levels of the F202 > A and T206 > A mutants, these data confirm that D205 is the major TM residue controlling the intracellular retention before eventual degradation of CD16A and support the hypothesis that association with either FcεR1γ or CD247 masks this aspartic acid, facilitating export from the ER and expression at the plasma membrane.
Fig. 5.
Analysis of cell surface expression and stability of CD16A mutants in the absence of adaptor protein. The indicated CD16A TM mutants were transfected into 293T fibroblasts in the absence of adaptor protein. Representative dot plots of transfected cells are shown (A). Cells expressing CD16A at the cell surface are marked by a square. The mean fluorescence intensity (MFI) of cells expressing CD16A at the cell surface staining (B) was analyzed using Kaluza Flow Cytometry Analysis Software. Western blots analyzing total CD16A protein expression (C and E) was quantified using ImageJ and plotted after normalization to GFP as a control for transfection efficiency (D and F). Statistical significance was calculated using one-way ANOVA-Dunnett post test for multiple comparison (*P < 0.05; **P < 0.01; ***P < 0.001; n.s., not statistically significant P > 0.05).
The Influence of TM Residues on CD16A Function.
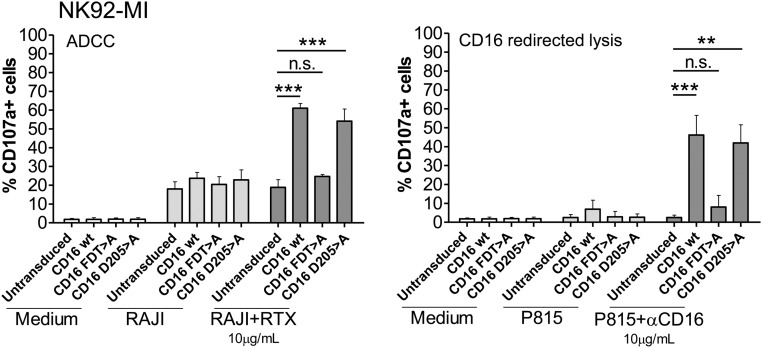
The above data show clearly that the FxxDT sequence element not only critically influenced the association of CD16A with FcεR1γ, but also the intracellular traffic and expression of the receptor at the cell surface. Thus, it was important to address the ability of the F202D205T206 > A and D205 > A mutants to trigger NK cell activation (degranulation measured by CD107a exposure) after stimulation via CD16A (Fig. 6). These experiments clearly showed that the F202D205T206 > A mutant, which is expressed at higher than WT levels at the cell surface, was nonetheless unable to signal for activation, consistent with the impaired association of this receptor with the FcεR1γ and CD247 adaptor molecules observed in IVT experiments (Figs. 2 and 3). In contrast, the D205 > A mutant, which retained some ability to interact with adaptors in IVT experiments (Fig. 3), was able to trigger NK cell cytotoxicity. However, despite the roughly fivefold higher surface expression of this mutant compared with WT, NK cell activation via CD16A D205 > A was not increased, suggesting that, although the D205 > A mutant reaches the surface very efficiently, the association with FcεR1γ and/or CD247 is only partially intact. Overall, these observations strongly support our prior conclusions that the CD16A TM residues F202 and T206 mediate important contacts with the signaling adaptor molecules. Although D205 also contributes to these interactions, the major role of this amino acid seems to be to influence the intracellular trafficking of the receptor.
Fig. 6.
Functional analysis of FDT > A and D205 > A NK92-MI transduced cell lines. NK92-MI cell lines transduced with CD16A WT, FDT > A, or D205 > A mutants were functionally tested in ADCC and redirected lysis experiments. NK cells were cultivated alone or in combination with Raji or P815 cell lines loaded, or not, with αCD20 (Rituximab) and CD16-specific mAb (3G8), respectively. After 2-h coculture, CD107a staining was measured by flow cytometry. Data represent mean of three experiments, and statistical significance was calculated using one-way ANOVA-Dunnett posttest for multiple comparison (**P < 0.01; ***P < 0.001; n.s., not statistically significant P > 0.05).
Structural Model of the Interface Between CD16A and Signaling Adaptors.
To gain insight into how the residues identified in our mutagenesis experiments contribute to assembly with CD247 and FcεR1γ, we performed replica exchange molecular dynamics (REMD) simulations in an implicit bilayer model examining possible modes of association of the CD16A TM domain with dimeric signaling adaptors. A 30-aa CD16A fragment encompassing the predicted α-helical TM domain was tested in unrestrained assembly simulations with the previously reported CD247 TM dimer structure (24) (Fig. S4) and with an FcεR1γ dimer modeled on this structure (Fig. 7 A and D) (simulation parameters are described in Materials and Methods). Cluster analysis of the resulting trimeric models revealed a clearly favored conformation for each simulation in which the polar/aromatic face identified in our mutagenesis screen mediates extensive contacts to the composite surfaces formed by the CD247 (Fig. S4) and FcεR1γ dimers (Fig. 7 A and D). Of 1,000 structures randomly chosen for analysis from the CD16A-CD247 simulation (Fig. S4), 931 clustered together with only 1.21 Å average root mean square deviation (rmsd) compared with the centroid structure. For CD16A-FcεR1γ (Fig. 7 A and D), 800/1,000 structures clustered together with an average rmsd of 1.58 Å compared with the centroid structure. The two representative models from these independent simulations were essentially identical, with only 0.61 Å rmsd between the centroid structures.
Fig. S4.
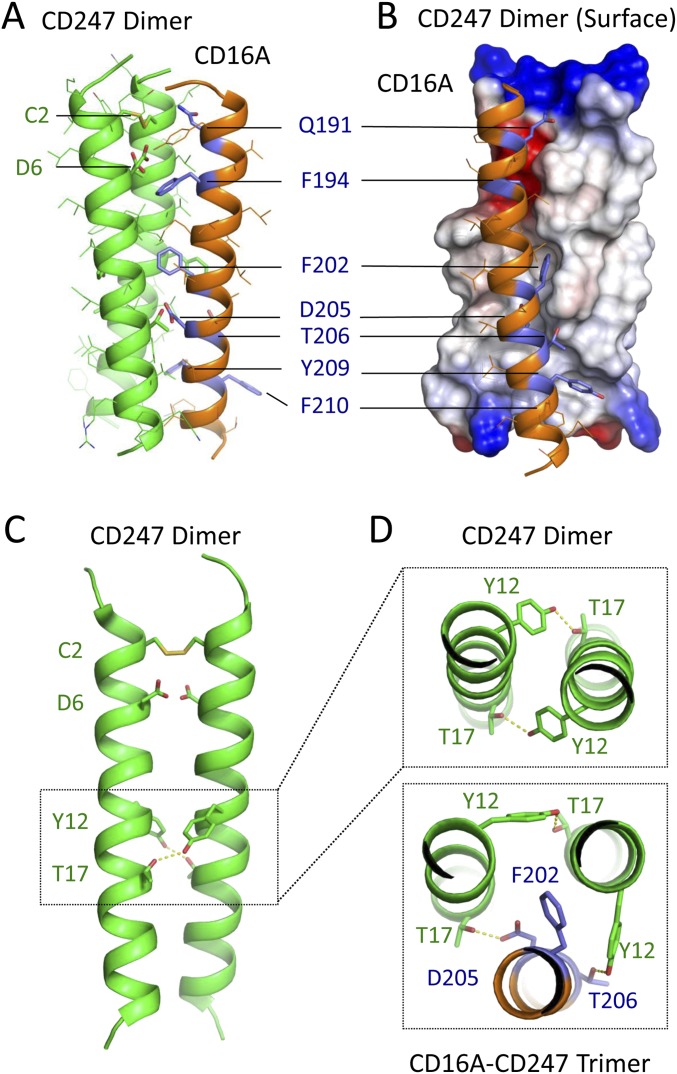
Model of CD16A assembled with CD247 dimer. The previously reported solution NMR structure of human CD247 dimer (PDB ID code 2HAC) was assembled with a helical model of the human CD16A TM sequence in a 60-ns REMD simulation in a model lipid bilayer as described in Materials and Methods. The major cluster contained 800/1,000 randomly chosen models with an average rmsd of 1.58 Å compared with the centroid structure shown here. In the ribbon diagram shown in A, CD247 is colored green, and CD16A is colored orange. Key residues are shown as sticks, with residue type and number labeled. Interface residues also identified in the CD16A mutagenesis are colored purple. (B) Rotated view of the structure in A, with CD247 shown as an electrostatic surface. (C) “Front” view of the solution NMR structure of CD247 dimer alone for comparison. Key residues are shown as sticks. (D) A view down the long axis of the CD247 dimer (Top) and CD247-CD16A trimer (Bottom) highlighting the rearrangement of the Y12-T17 hydrogen-bond network to accommodate new contact to CD16A. Colored as in A.
Fig. 7.
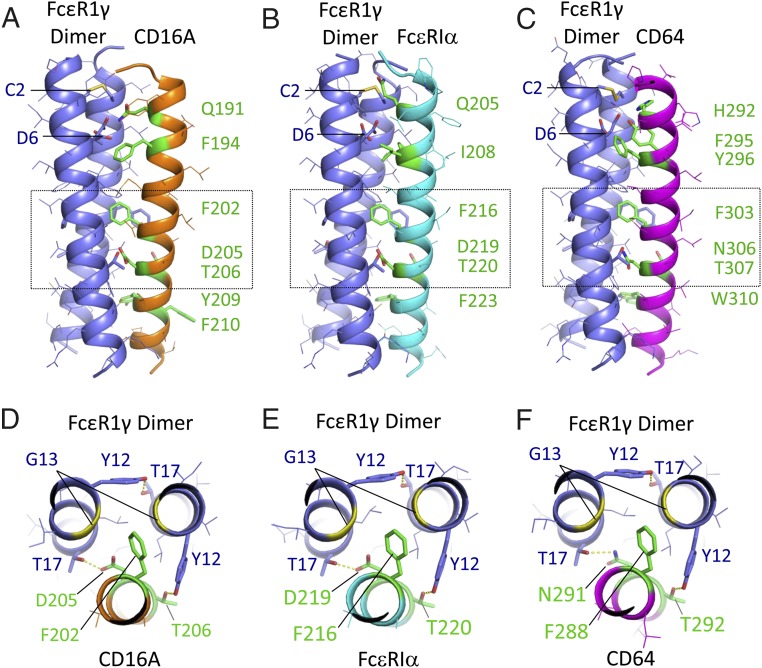
Structural models of CD16A and related Fc receptors assembled with the FcεR1γ dimeric signaling adaptor. Centroid structures from the dominant cluster are shown for REMD assembly simulations of CD16A (A and D), FcεR1α (B and E), and CD64 (C and F) with FcεR1γ disulphide-linked dimers in model membranes (Materials and Methods for simulation and cluster analysis procedures). FcεR1γ is shown in purple, and receptor TM domains are shown in orange (CD16A), cyan (FcεR1α), or magenta (CD64). Side views (A–C) are shown with key interface residues in stick representation and colored green in receptor TM domains. The FcεR1γ intermolecular disulphide bond (at C2) and aspartic acid pair (D6) are also indicated. For each model, the boxed region is also shown in a view down the long axis of the trimeric complex (D–F), highlighting the key features of the proposed core TM packing region. Hydrogen-bonding interactions discussed in the main text are represented by yellow dashed lines. A pair of glycine (G13) residues in FcεR1γ (yellow ribbons) create a cavity that accommodates the phenylalanine side-chain in the core packing motif. All figures were prepared in MacPyMol.
In the trimeric complex structures, the F202D205T206 trio that accounted for the most severe assembly defects in our screen of CD16A mutants forms the central core of the trimeric interface through highly favorable polar and van der Waals contacts. Comparison of the trimeric structures with the previously reported solution NMR structure of the CD247 dimer alone (Fig. S4) showed that one side of a symmetrical tyrosine-threonine hydrogen-bond pair that is essential for CD247 dimer formation (24) rearranges to accommodate favorable contacts to CD16A. The receptor-facing side of the signaling dimer opens to establish two new hydrogen bonds to CD16A (CD247-FcεR1γ T17 with CD16A D205 and CD247-FcεR1γ Y12 with CD16A T206) (Fig. S4 and Fig. 7D). CD16A F202 contributes to the surface complementarity in this region by occupying a cavity at G13 that is present in the interface of both signaling dimers (see surface representations in Fig. S4). Because the “rear” Y12-T17 hydrogen bond in the signaling dimer is maintained, these new contacts to CD16A create a “belt” of hydrogen bonds in the lipid bilayer interior that constitutes a major stabilizing feature of the trimeric assembly. Although the relatively mild effect of alanine substitution at D205 suggests that loss of this hydrogen bond alone is not sufficient to abrogate assembly, its interior position in the closely packed interface explains why larger side-chains were not tolerated (Fig. 3).
The interactions near the outer (at the top of Fig. 7) and inner (at the bottom of Fig. 7) limits of the membrane also show interfacial contacts involving residues identified in our mutagenesis experiments. CD16A Q191 is positioned to make contacts to one of the aspartic acids in the signaling dimer whereas F194 also packs closely against the aspartic acid pair, where it may contribute favorable π-electronic interactions (25). Of the two aromatic residues at the bottom of the CD16A TM helix, only Y209 contacts the signaling dimer, fitting neatly into a groove between two aliphatic residues one helical turn apart in the signaling dimer (see surface representations in Fig. S4). The detrimental effects of mutations at F210, which faces away from the interface, may be due to a role in stabilizing CD16A membrane insertion, a role commonly ascribed to aromatic residues near the inner leaflet of the lipid bilayer.
CD16A, FcεR1α, and FcγR1 (CD64) Form Similar Interfaces with FcεR1γ.
Of the Fc receptors that require assembly with dimeric FcεR1γ (CD16A, FcεR1, FcγR1/CD64, and FcαR1/CD89), only CD89 contains a basic amino acid in its TM domain for assembly. Like CD16A, FcεR1α and CD64 both contain FxxDT-like sequences in their TM domains (FxxDT in FcεR1α and FxxNT in CD64). Given this sequence similarity and others (Fig. 8 A and B), we sought to determine whether these receptors could form comparable TM interfaces with the signaling adaptor. In independent and unrestrained REMD simulations of assembly with FcεR1γ dimers in a model membrane, both receptor TM domains indeed converged on very similar interfaces to that observed in the CD16A simulations. The models shown in Fig. 7 B and C represent the majority clusters in each assembly: 726/1,000 FcεR1α-FcεR1γ structures analyzed clustered together with an average rmsd of 1.40 Å compared with the centroid (Fig. 7B), and 839/1,000 CD64-FcεR1γ structures clustered together with average rmsd of only 1.10 Å compared with the centroid (Fig. 7C). Consistent with sequence differences in the N-terminal regions of the three TM domains, the major contacts to FcεR1γ at the outer membrane limit are variable. FcεR1α has a glutamine in the same position as CD16A Q191, and, although it does not contact the aspartic acid pair in the centroid model shown (Fig. 7B), hydrogen bonds to the acidic side-chains are sampled in ∼27% of structures in the majority cluster, suggesting that this polar residue may indeed play a similar role to Q191 in CD16A. In FcεR1α, an isoleucine (I) takes the place of CD16A F194. In the CD64 model (Fig. 7C), the position of CD16A F194 is maintained, but neighboring histidine (H) and tyrosine (Y) residues provide the polar contacts to FcεR1γ aspartic acids. At the inner membrane limit (bottom), both receptors contain an aromatic residue in the position of CD16A Y209 (F in FcεR1α; W in CD64).
Fig. 8.
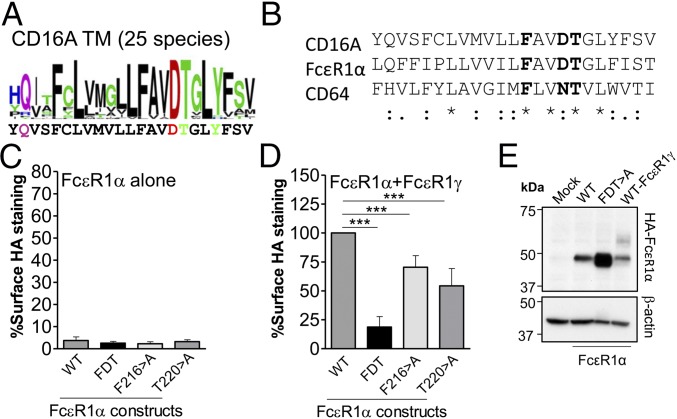
The FxxDT element in FcεR1α significantly influences association with FcεR1γ and receptor degradation. (A) Sequence logo illustrating the degree of amino acid conservation within TM sequences of CD16A and FcεR1α from multiple mammalian species. The height of each letter stack indicates relative conservation at that position whereas the height of each individual letter indicates its relative prevalence at that position. The sequences of human CD16A and FcεR1α are shown below sequence logos for reference. Logo graphics were generated using the WEBLOGO application (weblogo.berkeley.edu). (B) Alignment of CD16A, FcεR1α, and CD64 TM sequences showing the conservation of the FxxD/NT sequence. *, conserved residue; double dot (:), strong similarity; single dot (.), weak similarity. WT FcεR1α or selected mutants were transfected into 293T cells alone (C) or in combination with FcεR1γ (D), and their expression at the cell surface was analyzed by flow cytometry. Data represent mean of three experiments, and statistical significance was calculated using one-way ANOVA-Dunnett posttest for multiple comparison (***P < 0.001; n.s., not statistically significant P > 0.05). The expression and maturation of FcεR1α WT or the FDT > A mutant in the absence of adaptor molecule was studied using lysates of transfected 293T cells and analyzed by Western blot (E).
Remarkably, the FxxDT trio in FcεR1α and the FxxNT trio in CD64 established contacts to FcεR1γ dimers essentially identical to those we observed in the CD16A assembly models (Fig. 7 D–F). The similar role of aspartic acid and asparagine in these assembly models is fully consistent with the outcome of the D205 > N mutation in CD16A IVT experiments (Fig. 3), which we found was well-tolerated. These results indicate that Fc receptors without basic TM residues may all assemble based on core TM interactions similar to those we described for CD16A above.
We tested this hypothesis for one of the two additional receptors, FcεR1α, by examining the role of the FxxDT sequence in assembly and surface expression in cells (Fig. 8). Consistent with previous data (26), WT FcεR1α chain was not expressed at the cell surface unless cotransfected with FcεR1γ (Fig. 8 C and D). The FcεR1α F216D219T220 > A triple mutant was unable to reach the cell surface regardless of whether FcεR1γ was coexpressed or not (compare Fig. 8 C and E). This observation confirms a role for this sequence in assembly but suggests that, unlike CD16A D205, FcεR1α D219 does not play a dominant role in intracellular retention. This idea is consistent with previous data (27, 28). A Western blot analysis (Fig. 8E) shows that the F216D219T220 > A mutant does accumulate intracellular protein to a much greater level than WT FcεR1α although it does not progress through the endocytic pathway and acquire mature glycosylation, suggesting that the D219 residue does play some role in regulating the turnover of FcεR1α protein. As in CD16A, substitution of either the F216 or T220 residues with alanine significantly reduced the ability of cotransfected FcεR1γ to promote receptor expression at the cell surface (Fig. 8E), and together these two mutations account for most of the expression defect observed in the triple mutant. These data show that FcεR1α assembles with its signaling adaptor through a similar core TM arrangement as CD16A.
Discussion
A key feature underlying the formation of activating immune receptor complexes is the formation of an interface between the TM helix of the ligand-binding receptor subunit and the paired TM helices of the dimeric adaptor module that is crucial for intracellular signaling. In this paper, we have characterized how TM helices can associate to form an activating immunoreceptor complex in the absence of complementary charge potential and have gone on to show that a subset of the residues in the receptor TM domain that mediate association with the adaptor molecules also have a marked influence on the cell biology of the receptor.
Many activating immunoreceptors associate with signaling adaptor molecules via the interaction of a basic amino acid in the TM domain of the receptor with a pair of acidic residues in the TM region of the dimeric signaling module (10). However, our studies of the complex formed between CD16A and the signaling adaptor molecules FcεR1γ and CD247 have revealed that this three-helix interface can also form via interactions between polar and aromatic residues extending over the whole length of the TM domain. In this mode of association, multiple residues located on a single aspect of the receptor TM helix contribute to the interaction with the signaling dimer. These data contrast markedly with receptor complexes such as KIR2DS2–DAP12 and NKG2D–DAP10, wherein a single, properly placed lysine or arginine residue within a polyvaline or polyleucine TM sequence is sufficient for interaction with a pair of aspartic acid residues in the signaling adaptor module and cell surface expression (14). It has been argued that focusing the interaction of signaling adaptor molecules on a single charged residue facilitates the interaction of these adaptors with multiple receptor TM domains with widely varying sequences (14), but our data demonstrate that CD247 and FcεR1γ can also enlarge the repertoire of receptors with which they interact by associating with TM domains in more than one way. Indeed, it could be argued that the mode of interaction of FcεR1γ or CD247 with CD16A might, in evolutionary terms, be more robust because there is no one pair of interacting residues that is absolutely critical and so multiple mutations would have to occur to ablate this association.
Our independent and unbiased REMD simulations are in excellent agreement with the mutagenesis analysis, identifying the polar/aromatic face of the CD16A TM domain as the key assembly surface and reaching nearly identical conformations for assembled CD16A–CD247 and CD16A–FcεR1γ complexes. Similar to the previously published solution NMR structure of the NKG2C–DAP12 trimeric TM complex (29), the receptor TM helix is predicted to bind along an extended groove formed by the interface of the two signaling module α-helices. However, where NKG2C assembly is governed by a single, centrally located lysine residue binding to the paired DxxxT motifs in DAP12, CD16A establishes a more extensive interface, dependent on polar and aromatic residues, with CD247/FcεR1γ that is anchored around the central F202D205T206 trio but also has significant contributions at both ends of the helices. Thus, the modeling data strongly support the suggestion from the experimental data that interactions involving specific polar and aromatic residues of CD16A are critical for the association between the receptor and adaptor molecules.
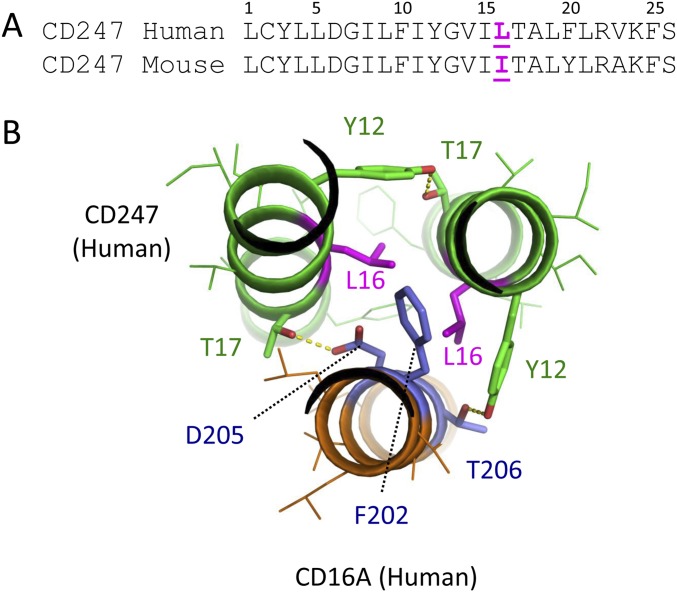
The rearrangement of the previously described CD247 interface (24) to accommodate the new contacts to CD16A is striking in view of the recent report suggesting that CD247 also undergoes a significant structural alteration upon incorporation into the TCR–CD3 complex (30), and this flexibility may be particularly important for the demonstrated ability of both CD247 and FcεR1γ to form stable assemblies with many different receptor TM sequences. Interestingly, despite the apparent interchangeability of human FcεR1γ and CD247, murine CD247 is essentially unable to function in the assembly of either human or mouse CD16A so that, in murine NK cells, only FcεR1γ participates in the assembly of this receptor complex (4). This phenotype has been mapped to an isoleucine for leucine change in murine CD247 (16) at a position that is buried within the trimeric interface formed with the CD16A F202D205T206 sequence in our models (Fig. S5). This positioning suggests that the β-branched structure of isoleucine is disruptive to the central intermolecular packing motif, providing a molecular explanation for the inability of murine CD247 to associate with CD16A and additional support for our assembly model.
Fig. S5.
A single isoleucine residue in the mouse CD247 TM sequence that prevents assembly with CD16A is located in the core TM packing interface. (A) Alignment of human and mouse CD247 TM sequence highlighting the position of the variation identified in ref. 16 as blocking association with CD16A (magenta). (B) A view down the long axis of the human CD247-CD16A trimer model showing the internal position of L16 in the F202D205T206 interface. We hypothesize that substitution of leucine with the β-branched side-chain of isoleucine is not well accommodated in the tightly packed interface, leading to defective assembly.
The observation that CD16A associates equally well with human CD247 and FcεR1γ homodimers, as well as the heterodimer (Fig. 1), is consistent with the convergence of the modeling results. It therefore seems likely that the major factors influencing whether CD16A associates with CD247 or FcεR1γ are related to the relative levels of expression of the different adaptors and possible competition from other receptors that also bind CD247 and FcεR1γ. For example, a population of terminally differentiated human NK cells lose expression of FcεR1γ (31), and, in these cells, CD16A associates exclusively with CD247 (15). In contrast, the majority of circulating human NK cells express both FcεR1γ and CD247, but they also express multiple NK receptors that associate with these adaptor molecules. These receptors show little sequence similarity between their TM domains, and it is possible that they recruit CD247 and FcεR1γ unequally from the pool of available adaptors because an interaction between amino acids of complementary charge in the hydrophobic core of the lipid bilayer could differ in “affinity” from one that depends on interactions involving polar and aromatic residues. It is also possible that the specificity of the interaction could be modulated by steric hindrance between domains of the receptors flanking the TM region (13, 14).
Apart from the mechanism of association of CD16A with FcεR1γ and CD247, the other striking observation emerging from these experiments is that D205 within the CD16A TM, unless sequestered within the three-helix interface upon assembly with either FcεR1γ or CD247, promotes intracellular retention of the receptor. It has long been appreciated that the formation of specific polar contacts between TM domain amino acids is a key mechanism to ensure the accurate assembly of multicomponent protein complexes to be expressed on the cell surface (9, 10) and that, in the absence of these contacts, unassembled subunits and partially formed complexes are retained intracellularly and eventually degraded (32). In the case of the TCR, it is the same charged residues providing the core assembly contacts that mediate this quality-control function. In contrast, we found that CD16A D205 makes only a limited contribution to the receptor/adaptor interaction, but its sequestration within the assembly interface seems to act as a quality control mechanism to ensure that only complete receptor complexes can traffic to the cell surface. Thus, in CD16A, the quality-control function has been separated, at least partially, from the assembly function although both are still located in the TM domain. This FxxDT sequence is highly conserved among the predicted TM sequences of CD16A from multiple mammalian species (Fig. 8A) and is also present in FcεR1α and CD64 (as FxxNT) (Fig. 8B), both of which formed trimeric complexes with FcεR1γ in our assembly simulations that used CD16A-like interfaces (Fig. 7). Indeed, mutation of these residues in FcεR1α caused loss of surface expression and intracellular retention of the receptor, indicating that this sequence also regulates cell-surface expression of FcεR1α, most likely through assembly with FcεR1γ. In contrast to our observations with CD16A, the FDT triple mutant of FcεR1α was not expressed at the cell surface in the absence of adaptor molecules, but the large accumulation of intracellular protein detected by Western blot suggests that mutation of the aspartic acid did indeed protect unassembled FcεR1α protein from degradation. The lack of cell surface expression of this mutant likely reflects the action of ER retention motifs present in the cytoplasmic tail of this receptor that have previously been identified as key elements regulating the trafficking of FcεR1α to the plasma membrane (27, 28).
Overall, these data strongly suggest that the Fxx(D/N)T sequences found in three different Fc receptor TM domains form the core membrane-embedded interaction motifs that drive assembly with signaling adaptor molecules and, at least in a subset of these receptor complexes, regulate receptor protein turnover. Thus, at least two completely different solutions for TM-mediated receptor assembly with CD247/FcεR1γ signaling modules have arisen during evolution, one driven by a single basic TM residue and another based on the more complex polar/aromatic interface described here. These findings may provide more broadly useful insights into the biology of other immune receptors that do not contain charged residues in their TM domains and how they assemble into complexes with signaling adaptor molecules.
Materials and Methods
Transmembrane Domain Mutagenesis.
A WT human FcγRIIIA (CD16A) sequence was amplified by PCR and cloned into a modified pSP64 vector (11) for coupling to a streptavidin binding protein (SBP) tag. FcεR1γ was amplified and cloned into a vector bearing an HA tag. The CD247 construct has been described previously (11). Multiple and single amino acid substitutions in CD16A were generated by QuikChange PCR-based mutagenesis. The sequences of the oligonucleotides used for cloning and mutagenesis are shown in Table S1. The integrity of all WT and mutant CD16A, FcεR1γ, and CD247 constructs was verified by sequencing (GATC-Biotech). In some cases, CD16A WT and mutant cDNAs were then subcloned into the lentiviral vector pHRSIN-C56W-UbEM (a gift of Paul Lehner, Cambridge Institute for Medical Research, Cambridge, United Kingdom) to simultaneously express CD16A under the SFFV promoter and the GFP derivative protein, Emerald, under a ubiquitin promoter. A cDNA clone encoding the mature FcεR1α cDNA was amplified by PCR and cloned into the pMX-HA puro vector (a gift of Chiwen Chang, Department of Pathology, University of Cambridge, Cambridge, UK) for expression as an N-terminally HA-tagged protein.
Table S1.
Primers used in the preparation of WT and mutant constructs of CD16A, FcεR1γ, and FcεR1α
| Primer name | Sequence 5′–3′ |
| CD16A WT cloning For | GCCAAGCTTGCCACCATGTGGCAGCTGCTCCTCCC |
| CD16A WT cloning Rev | GCCGGATCCTTTGTCTTGAGGGTCCTTTCTC |
| CD16ATM Q191 > A For | CCTGGGTACGCAGTCTCTTTC |
| CD16ATM Q191 > A Rev | GAAAGAGACTGCGTACCCAGG |
| CD16ATM S193 > A For | GTACCAAGTCGCTTTCTGCTT |
| CD16ATM S193 > A Rev | AAGCAGAAAGCGACTTGGTAC |
| CD16ATM F194 > A For | CCAAGTCTCTGCCTGCTTGGTG |
| CD16ATM F194 > A Rev | CACCAAGCAGGCAGAGACTTGG |
| CD16ATM C195 > A For | AGTCTCTTTCGCCTTGGTGATG |
| CD16ATM C195 > A Rev | CATCACCAAGGCGAAAGAGACT |
| CD16ATM F202 > A For | GGTACTCCTTGCTGCAGTGGAC |
| CD16ATM F202 > A Rev | GTCCACTGCAGCAAGGAGTACC |
| CD16ATM D205 > A For | TTTGCAGTGGCCACAGGACTA |
| CD16ATM D205 > A Rev | TAGTCCTGTGGCCACTGCAAA |
| CD16ATM D205 > E For | TTGCAGTGGAAACAGGACTAT |
| CD16ATM D205 > E Rev | ATAGTCCTGTTTCCACTGCAA |
| CD16ATM D205 > K For | TTTTGCAGTGAAGACAGGACTAT |
| CD16ATM D205 > K Rev | ATAGTCCTGTCTTCACTGCAAAA |
| CD16ATM D205 > N For | TTTTGCAGTGAACACAGGACT |
| CD16ATM D205 > N Rev | AGTCCTGTGTTCACTGCAAAA |
| CD16ATM T206 > A For | CCTTTTTGCAGTGGACGCAGGACTATATTTC |
| CD16ATM T206 > A Rev | GAAATATAGTCCTGCGTCCACTGCAAAAAGG |
| CD16ATM Y209 > A For | CACAGGACTAGCTTTCTCTGTG |
| CD16ATM Y209 > A Rev | CACAGAGAAAGCTAGTCCTGTG |
| CD16ATM F210 > A For | AGGACTATATGCCTCTGTGAAG |
| CD16ATM F210 > A Rev | CTTCACAGAGGCATATAGTCCT |
| CD16ATM S211 > A For | ACTATATTTCGCTGTGAAGAC |
| CD16ATM S211 > A Rev | GTCTTCACAGCGAAATATAGT |
| CD16ATM S193F194C195 > A For | CCTGGGTACCAAGTCGCTGCCGCCTTGGTGATGGTACT |
| CD16ATM S193F194C195 > A Rev | AGTACCATCACCAAGGCGGCAGCGACTTGGTACCCAGG |
| CD16ATM Y209F210S211 > A For | GTGGACACAGGACTAGCTGCCGCTGTGAAGACAAACA |
| CD16ATM Y209F210S211 > A Rev | TGTTTGTCTTCACAGCGGCAGCTAGTCCTGTGTCCAC |
| CD16ATM D205T206 > A For | CTCCTTGCTGCAGTGAAGGCAGGACTATATTTCT |
| CD16ATM D205T206 > A Rev | AGAAATATAGTCCTGCCTTCACTGCAGCAAGGAG |
| FcεR1γ WT cloning For | GCCAAFCTTGCCACCATGATTCCAGCAGTGGTCTTGC |
| FcεR1γ WT cloning Rev | GCCGGATCCCTGTGGTGGTTTCTCATGCTTC |
| FcεR1α WT cloning For | GCGAATTCGTCCCTCAGAAACCTAAGGTC |
| FcεR1α WT cloning Rev | GCTGCGGCCGCTAGTTGTTTTTGGGGTTTGGCTTAG |
| FcεR1αTM F216 > A For | GTGATTCTGGCTGCTGTGGACACAG |
| FcεR1αTM F216 > A Rev | CTGTGTCCACAGCAGCCAGAATCAC |
| FcεR1αTM T220 > A For | GCTGTGGACGCAGGATTATTTATC |
| FcεR1αTM T220 > A Rev | GATAAATAATCCTGCGTCCACAGC |
| FcεR1αTM D219T220 > A For | CTGCTGTGGCCGCAGGATTATTTATC |
| FcεR1αTM D219T220 > A Rev | GATAAATAATCCTGCGGCCACAGCAG |
For, forward; Rev, reverse.
In Vitro Translation Assay.
In vitro-transcribed mRNAs encoding all full-length receptor subunits and mutants were pretested for matched translation and ER microsome import. CD16A, FcεR1γ, and CD247 were then cotranslated in the presence of ER microsomes for 30 min at 30 °C before addition of oxidized glutathione (4 mM final concentration) to initiate oxidative folding and assembly. After a further 2-h incubation at 30 °C, the completed assembly reactions were stopped by dilution in 0.5 mL of ice-cold Tris-buffered saline (TBS) (pH 8) containing 10 mM iodoacetamide. The membrane fraction was collected by centrifugation and washed with cold TBS before extraction with 1% digitonin in TBS containing 10 mM iodoacetamide to block disulphide bond formation after extraction and during handling. Lysates were cleared by centrifugation and immunoprecipitated with antibody-coupled agarose beads (4 °C for 2 h). Final products were eluted in SDS, separated on 12% NuPAGE gels (Life Technologies) under nonreducing conditions and transferred to PVDF membranes for phospho-imaging.
Cell Lines.
Cells were maintained at 37 °C and 5% CO2 in a humidified incubator and split as necessary. The 293T cell line was cultivated in Dulbecco´s modified Eagle’s medium (DMEM) with 10% FCS. NKL and NK92-MI cells were grown in RPMI with 5% FCS, 5% human serum, and 50 U/mL IL-2 (Peprotech). Media were supplemented with 2 mM l-glutamine, 0.1 mM sodium pyruvate, 100 U/mL penicillin, 100 U/mL streptomycin, and 50 μM β-mercaptoethanol. The 293T cells were transfected with plasmids bearing CD16A WT or mutant constructs using the jetPEI transfection reagent. Twenty-four hours after transfection, cells were either lysed or recovered for flow cytometry analysis. Lentiviral transduction was used to express CD16A and mutants in NKL and NK92-MI cell lines.
Lentiviral Gene Transduction.
Lentiviruses were generated by transfection of 293T cells with the indicated lentivector together with the plasmids pCMVR8.91 and pMD2G. Two days after transfection, the culture media containing the lentiviruses were harvested, filtered, and stored at −80 °C. For each lentiviral transduction, 0.1 × 106 cells were mixed with 0.75 mL of virus supernatant in the presence of either 1 µM TBK1 inhibitor BX795 (InvivoGen), 8 μg/mL protamine sulfate (Sigma-Aldrich), and 50 U/mL IL-2 (NKL and NK92-MI cells) and seeded in five wells of a 96-well plate (BD Biosciences). The plates were then centrifuged at 120 × g for 1 h at 33 °C. After centrifugation, without removing viral supernatants, the plates were incubated at 37 °C, 5% CO2 for 4 to 6 h and then spun again at 120 × g for 1 h at 33 °C. The supernatants were then removed from the wells, and fresh growth medium was added.
Flow Cytometry.
Cells were washed and incubated in PBS/0.5% (wt/vol) BSA/1% (vol/vol) FBS/0.1% sodium azide buffer (PBA buffer) and stained using purified CD16A-specific mAb (3G8) or the HA-specific mAb 12CA5, followed by anti-mouse–Ig coupled to PE (phyco-erythrin) (Dako Cytomation) for 30 min at 4 °C. Cells were analyzed using FACSCalibur (BD Biosciences) and Gallios (Beckman Coulter) cytometers. Data were analyzed with FlowJo and Kaluza Flow Cytometry Analysis programs.
Degranulation Assay.
For degranulation assays quantifying cell surface CD107a expression, 1 × 105 resting NK92MI cells were washed twice in PBS and added to 2 × 105 target cells in 200 μL of complete medium. Two target cells were used in these antibody-dependent cell-mediated cytotoxicity (ADCC) and redirected cytotoxicity experiments; Raji cells and P815 cells sensitized, or not, with Rituximab or the CD16-specific mAb 3G8, respectively (33). Cells were spun down for 3 min at 100 × g and incubated for 2 h at 37 °C in 5% CO2. The cells were then washed once in ice-cold PBS/2% BSA/2mM EDTA/0.05% sodium azide (PBA) and stained with PE-conjugated anti-CD94 for NK cell gating and APC-conjugated anti-CD107a mAbs for 30 min at 4 °C. The cells were washed, resuspended in PBA, and analyzed by flow cytometry.
Western Blot.
Cells were collected, washed once with ice-cold PBS, and lysed in radioimmunoprecipitation assay (RIPA) lysis buffer [50 mM Tris, pH 7.4, 150 mM NaCl, 1% Triton X-100, 1% sodium deoxycholate, and 0.1% SDS with protease inhibitors (1 μM pepstatin A, 1 μM leupeptin)] and 0.5 mM iodoacetamide for at least 30 min on ice. Lysates were centrifuged to pellet insoluble material for 15 min at 4 °C and then quantified; 25 to 30 μg of protein were loaded onto 8 to 12% SDS reducing polyacrylamide gels and electrophoresed, followed by transfer to PVDF (Immobilon-P; Millipore). Membranes were blocked in 5% dried skimmed milk (1 h, 25 °C), washed three times in 0.05% Tween/TBS, and incubated (overnight, 4 °C) in primary antibodies: mouse monoclonal anti-CD16A (clone DJ130c), rabbit polyclonal anti-GFP (both from Santa Cruz Biotechnology), mouse monoclonal anti-β actin (Sigma Aldrich), or 3F10 (HA-specific; Roche) in 0.05% Tween/TBS. Membranes were then washed in 0.05% Tween/TBS (5 min, 2×), and bound antibodies were visualized using goat anti-mouse or goat anti-rabbit secondary reagents (Dako Cytomation) and the ECL-Western Blot Detection Reagent (GE Healthcare). For EndoH treatment, a 25-μg sample was denatured for 10 min at 100 °C and treated with 0.25 units of EndoH enzyme (New England Biolabs) for 1 h at 37 °C and visualized as mentioned before. Western quantification was carried out using ImageJ and normalized against GFP as reporter of transfection efficiency.
TM Assembly Modeling.
To model complex structures of CD247-CD16A, FcεR1γ-CD16A, FcεR1γ-FcεR1α, and FcεR1γ-CD64 TM domains, we used the replica exchange molecular dynamics (REMD) simulation method (34). The TM sequences used in this study were DPKLCYLLDGILFIYGVILTALFLRVKFS (CD247), EPQLCYILDAILFLYGIVLTLLYCRLKIQ (FcεR1γ), SPPGYQVSFCLVMVLLFAVDTGLYFSVKTN (CD16A), EKYWLQFFIPLLVVILFAVDTGLFISTQQQ (FcεR1α), and TPVWFHVLFYLAVGIMFLVNTVLWVTIRKE (CD64). We assigned the aspartic acid residues (bold) in CD16A and FcεR1α to the protonated state based on our observation that mutation to asparagine was tolerated and the results of test simulations indicating that the deprotonated state destabilizes CD16A TM in the membrane. The histidine residue in CD64 was protonated at both N atoms based on preliminary simulations where conformations of the complex based on neutral histidine in CD64 did not converge. The CD247 and FcεR1γ homodimers were modeled with one protonated and one deprotonated aspartic acid based on previous simulations indicating that a mixed ionization state is likely in signaling adaptor dimers (35, 36). The solution NMR structure of CD247 TM homodimer (PDB ID code 2HAC) (24) was used to model the initial structure of the FcεR1γ TM homodimer, and an α-helical secondary structure was assumed for the initial state of CD16A TM. The initial configurations were generated by first placing the adaptor TM dimer’s xy-center of mass (xy-COM) at x = y = 0 on the xy-plane (perpendicular to the membrane normal) and then positioning the CD16A xy-COM along a circle of radius 25 Å (every 11.25° for 32 complex systems, each with random rotation along the membrane normal). The initial configurations of FcεR1γ-FcεR1α and FcεR1γ-CD64 TM helices were generated in the same manner.
Using these initial configurations, 60-ns (CD247-CD16A and FcεR1γ-CD16A), 70-ns (FcεR1γ-CD64), and 110-ns (FcεR1γ-FcεR1α) REMD simulations with 32 replicas each in a temperature range of 300 to 750 K were carried out using CHARMM (37). The membrane environment was mimicked by a generalized Born with a simple switching (GBSW) implicit membrane model (38), where we used the default options provided in Implicit Solvent Modeler in CHARMM-GUI (39), except with an empirical surface tension coefficient (0.03 kcal⋅mol−1⋅Å−2) for the nonpolar solvation contribution. A time-step of 2 fs was used with the SHAKE algorithm (40), and the collision frequency was set as γ = 5 ps−1 for the Langevin dynamics simulation. Weak dihedral restraints (k = 50 kcal⋅mol−1⋅rad−2) were applied to each TM domain to hold α-helical structure at high-T replicas. Replica exchanges were controlled by the REPDSTR module (41) in CHARMM, with exchange attempts at every 1 ps. Conformations during the last 20-ns trajectory at 300 K were sampled every 20 ps (1,000 conformations) and clustered based on (pairwise) rmsd of Cα atoms with a cutoff value of 3.0 Å, where the time interval 20 ps was chosen to balance the number of samples and computational cost.
Acknowledgments
We thank Dr. Cesar Santiago for helpful advice on design of the mutagenesis strategy; Dr. Mar Valés Gómez for critical reading of the manuscript; Ruth López-Caro, Dr. Mario Mellado, Pilar Lucas, Dr. Laura Martínez-Muñoz, and Dr. Blanca Soler-Palacios for constructive advice and help; and Dr. Gloria Esteso for daily encouragement and support. This work was supported by Ministerio de Economia y Competitividad (MINECO) Grant SAF2014-58752-R (to H.T.R.), MINECO PhD Studentship SVP-2014-068263 (to A.B.-M.), Australian Research Council Future Fellowship FT120100145 (to M.J.C.), Extreme Science and Engineering Discovery Environment Grant MCB070009 (to W.I.), National Science Foundation Grant MCB-1727508 (to W.I.), and National Institutes of Health Grant R01-GM092950 (to W.I.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1706483114/-/DCSupplemental.
References
- 1.Ravetch JV, Perussia B. Alternative membrane forms of Fc gamma RIII(CD16) on human natural killer cells and neutrophils: Cell type-specific expression of two genes that differ in single nucleotide substitutions. J Exp Med. 1989;170:481–497. doi: 10.1084/jem.170.2.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hibbs ML, et al. Mechanisms for regulating expression of membrane isoforms of Fc gamma RIII (CD16) Science. 1989;246:1608–1611. doi: 10.1126/science.2531918. [DOI] [PubMed] [Google Scholar]
- 3.Lanier LL, Yu G, Phillips JH. Co-association of CD3 zeta with a receptor (CD16) for IgG Fc on human natural killer cells. Nature. 1989;342:803–805. doi: 10.1038/342803a0. [DOI] [PubMed] [Google Scholar]
- 4.Kurosaki T, Ravetch JV. A single amino acid in the glycosyl phosphatidylinositol attachment domain determines the membrane topology of Fc gamma RIII. Nature. 1989;342:805–807. doi: 10.1038/342805a0. [DOI] [PubMed] [Google Scholar]
- 5.Samelson LE, Patel MD, Weissman AM, Harford JB, Klausner RD. Antigen activation of murine T cells induces tyrosine phosphorylation of a polypeptide associated with the T cell antigen receptor. Cell. 1986;46:1083–1090. doi: 10.1016/0092-8674(86)90708-7. [DOI] [PubMed] [Google Scholar]
- 6.Weissman AM, Samelson LE, Klausner RD. A new subunit of the human T-cell antigen receptor complex. Nature. 1986;324:480–482. doi: 10.1038/324480a0. [DOI] [PubMed] [Google Scholar]
- 7.Perez-Montfort R, Kinet JP, Metzger H. A previously unrecognized subunit of the receptor for immunoglobulin E. Biochemistry. 1983;22:5722–5728. doi: 10.1021/bi00294a007. [DOI] [PubMed] [Google Scholar]
- 8.Li X, et al. The unique cytoplasmic domain of human FcγRIIIA regulates receptor-mediated function. J Immunol. 2012;189:4284–4294. doi: 10.4049/jimmunol.1200704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ashwell JD, Klusner RD. Genetic and mutational analysis of the T-cell antigen receptor. Annu Rev Immunol. 1990;8:139–167. doi: 10.1146/annurev.iy.08.040190.001035. [DOI] [PubMed] [Google Scholar]
- 10.Call ME, Wucherpfennig KW. Common themes in the assembly and architecture of activating immune receptors. Nat Rev Immunol. 2007;7:841–850. doi: 10.1038/nri2186. [DOI] [PubMed] [Google Scholar]
- 11.Call ME, Pyrdol J, Wiedmann M, Wucherpfennig KW. The organizing principle in the formation of the T cell receptor-CD3 complex. Cell. 2002;111:967–979. doi: 10.1016/s0092-8674(02)01194-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garrity D, Call ME, Feng J, Wucherpfennig KW. The activating NKG2D receptor assembles in the membrane with two signaling dimers into a hexameric structure. Proc Natl Acad Sci USA. 2005;102:7641–7646. doi: 10.1073/pnas.0502439102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feng J, Garrity D, Call ME, Moffett H, Wucherpfennig KW. Convergence on a distinctive assembly mechanism by unrelated families of activating immune receptors. Immunity. 2005;22:427–438. doi: 10.1016/j.immuni.2005.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feng J, Call ME, Wucherpfennig KW. The assembly of diverse immune receptors is focused on a polar membrane-embedded interaction site. PLoS Biol. 2006;4:e142. doi: 10.1371/journal.pbio.0040142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vales-Gomez M, et al. Natural killer cell hyporesponsiveness and impaired development in a CD247-deficient patient. J Allergy Clin Immunol. 2016;137:942–945 e944. doi: 10.1016/j.jaci.2015.07.051. [DOI] [PubMed] [Google Scholar]
- 16.Kurosaki T, Gander I, Ravetch JV. A subunit common to an IgG Fc receptor and the T-cell receptor mediates assembly through different interactions. Proc Natl Acad Sci USA. 1991;88:3837–3841. doi: 10.1073/pnas.88.9.3837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johnson RM, Hecht K, Deber CM. Aromatic and cation-pi interactions enhance helix-helix association in a membrane environment. Biochemistry. 2007;46:9208–9214. doi: 10.1021/bi7008773. [DOI] [PubMed] [Google Scholar]
- 18.Moore DT, Berger BW, DeGrado WF. Protein-protein interactions in the membrane: Sequence, structural, and biological motifs. Structure. 2008;16:991–1001. doi: 10.1016/j.str.2008.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ulmschneider MB, Sansom MS. Amino acid distributions in integral membrane protein structures. Biochim Biophys Acta. 2001;1512:1–14. doi: 10.1016/s0005-2736(01)00299-1. [DOI] [PubMed] [Google Scholar]
- 20.Robertson MJ, et al. Characterization of a cell line, NKL, derived from an aggressive human natural killer cell leukemia. Exp Hematol. 1996;24:406–415. [PubMed] [Google Scholar]
- 21.Tam YK, et al. Characterization of genetically altered, interleukin 2-independent natural killer cell lines suitable for adoptive cellular immunotherapy. Hum Gene Ther. 1999;10:1359–1373. doi: 10.1089/10430349950018030. [DOI] [PubMed] [Google Scholar]
- 22.Grier JT, et al. Human immunodeficiency-causing mutation defines CD16 in spontaneous NK cell cytotoxicity. J Clin Invest. 2012;122:3769–3780. doi: 10.1172/JCI64837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim MK, et al. Fcgamma receptor transmembrane domains: Role in cell surface expression, gamma chain interaction, and phagocytosis. Blood. 2003;101:4479–4484. doi: 10.1182/blood.V101.11.4479. [DOI] [PubMed] [Google Scholar]
- 24.Call ME, et al. The structure of the zetazeta transmembrane dimer reveals features essential for its assembly with the T cell receptor. Cell. 2006;127:355–368. doi: 10.1016/j.cell.2006.08.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Philip V, et al. A survey of aspartate-phenylalanine and glutamate-phenylalanine interactions in the protein data bank: Searching for anion-π pairs. Biochemistry. 2011;50:2939–2950. doi: 10.1021/bi200066k. [DOI] [PubMed] [Google Scholar]
- 26.Albrecht B, Woisetschläger M, Robertson MW. Export of the high affinity IgE receptor from the endoplasmic reticulum depends on a glycosylation-mediated quality control mechanism. J Immunol. 2000;165:5686–5694. doi: 10.4049/jimmunol.165.10.5686. [DOI] [PubMed] [Google Scholar]
- 27.Letourneur F, Hennecke S, Démollière C, Cosson P. Steric masking of a dilysine endoplasmic reticulum retention motif during assembly of the human high affinity receptor for immunoglobulin E. J Cell Biol. 1995;129:971–978. doi: 10.1083/jcb.129.4.971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cauvi DM, Tian X, von Loehneysen K, Robertson MW. Transport of the IgE receptor alpha-chain is controlled by a multicomponent intracellular retention signal. J Biol Chem. 2006;281:10448–10460. doi: 10.1074/jbc.M510751200. [DOI] [PubMed] [Google Scholar]
- 29.Call ME, Wucherpfennig KW, Chou JJ. The structural basis for intramembrane assembly of an activating immunoreceptor complex. Nat Immunol. 2010;11:1023–1029. doi: 10.1038/ni.1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee MS, et al. A mechanical switch couples t cell receptor triggering to the cytoplasmic juxtamembrane regions of CD3ζζ. Immunity. 2015;43:227–239. doi: 10.1016/j.immuni.2015.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hwang I, et al. Identification of human NK cells that are deficient for signaling adaptor FcRγ and specialized for antibody-dependent immune functions. Int Immunol. 2012;24:793–802. doi: 10.1093/intimm/dxs080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bonifacino JS, Cosson P, Klausner RD. Colocalized transmembrane determinants for ER degradation and subunit assembly explain the intracellular fate of TCR chains. Cell. 1990;63:503–513. doi: 10.1016/0092-8674(90)90447-m. [DOI] [PubMed] [Google Scholar]
- 33.Perussia B, Loza MJ. Assays for antibody-dependent cell-mediated cytotoxicity (ADCC) and reverse ADCC (redirected cytotoxicity) in human natural killer cells. Methods Mol Biol. 2000;121:179–192. doi: 10.1385/1-59259-044-6:179. [DOI] [PubMed] [Google Scholar]
- 34.Sugita Y, Okamoto Y. Replica-exchange molecular dynamics method for protein folding. Chem Phys Lett. 2000;314:141–151. [Google Scholar]
- 35.Petruk AA, et al. The structure of the CD3 ζζ transmembrane dimer in POPC and raft-like lipid bilayer: A molecular dynamics study. Biochim Biophys Acta. 2013;1828:2637–2645. doi: 10.1016/j.bbamem.2013.07.019. [DOI] [PubMed] [Google Scholar]
- 36.Knoblich K, et al. Transmembrane complexes of DAP12 crystallized in lipid membranes provide insights into control of oligomerization in immunoreceptor assembly. Cell Reports. 2015;11:1184–1192. doi: 10.1016/j.celrep.2015.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brooks BR, et al. CHARMM: The biomolecular simulation program. J Comput Chem. 2009;30:1545–1614. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Im W, Feig M, Brooks CL., 3rd An implicit membrane generalized born theory for the study of structure, stability, and interactions of membrane proteins. Biophys J. 2003;85:2900–2918. doi: 10.1016/S0006-3495(03)74712-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jo S, Kim T, Iyer VG, Im W. CHARMM-GUI: A web-based graphical user interface for CHARMM. J Comput Chem. 2008;29:1859–1865. doi: 10.1002/jcc.20945. [DOI] [PubMed] [Google Scholar]
- 40.Ryckaert J-P, Ciccotti G, Berendsen JC. Numerical integration of the cartesian equations of motion of a system with constraints: Molecular dynamics of n-alkanes. J Comput Phys. 1977;23:327–341. [Google Scholar]
- 41.Woodcock HL, 3rd, et al. Interfacing Q-Chem and CHARMM to perform QM/MM reaction path calculations. J Comput Chem. 2007;28:1485–1502. doi: 10.1002/jcc.20587. [DOI] [PubMed] [Google Scholar]