Significance
Although the potent antiproliferative effects of the Arf tumor suppressor depend on p53, Arf also exhibits less well understood p53-independent activities. Arf-null mice are blind and males exhibit defective spermatogenesis, two focal anomalies not observed in Trp53-null animals. An N-terminally truncated and unstable Arf protein (smArf), translated from a second internal AUG codon within the full-length Arf mRNA, lacks amino acid residues required for p53 activation and has been reported to localize to mitochondria and trigger mitophagy. Missing potent p53-dependent tumor suppressor activity, p15smArf can surprisingly act independently of p19Arf to correct the focal developmental defects of Arf-null mice. The unusual features of the Ink4a-Arf (Cdkn2a) locus, which include alternative reading frames and internal translational initiation, are unprecedented in mammals.
Keywords: Arf tumor suppressor, p53, BCR-ABL acute lymphoblastic leukemia, spermatogenesis, hyaloid vasculature
Abstract
The mouse p19Arf (human p14ARF) tumor suppressor protein, encoded in part from an alternative reading frame of the Ink4a (Cdkn2a) gene, inhibits the Mdm2 E3 ubiquitin ligase to activate p53. Arf is not expressed in most normal tissues of young mice but is induced by high thresholds of aberrant hyperproliferative signals, thereby activating p53 in incipient tumor cells that have experienced oncogene activation. The single Arf mRNA encodes two distinct polypeptides, including full-length p19Arf and N-terminally truncated and unstable p15smArf (“small mitochondrial Arf”) initiated from an internal in-frame AUG codon specifying methionine-45. Interactions of p19Arf with Mdm2, or separately with nucleophosmin (NPM, B23) that localizes and stabilizes p19Arf within the nucleolus, require p19Arf N-terminal amino acids that are not present within p15smArf. We have generated mice that produce either smARF alone or M45A-mutated (smArf-deficient) full-length p19Arf proteins. BCR-ABL–expressing pro/pre-B cells producing smArf alone are as oncogenic as their Arf-null counterparts in generating acute lymphoblastic leukemia when infused into unconditioned syngeneic mice. In contrast, smArf-deficient cells from mice of the ArfM45A strain are as resistant as wild-type Arf+/+ cells to comparable oncogenic challenge and do not produce tumors. Apart from being prone to tumor development, Arf-null mice are blind, and their male germ cells exhibit defects in meiotic maturation and sperm production. Although ArfM45A mice manifest the latter defects, smArf alone remarkably rescues both of these p53-independent developmental phenotypes.
Three exons (1α, 2, and 3) of the Cdkn2a (Ink4a) gene encode spliced mRNA segments that specify the Cdk4/6 inhibitor p16Ink4a, whereas mRNA initiated from a distinct upstream 5′ exon (1β) opens an alternative reading in Ink4a exons 2 and 3 to yield p19Arf (1), an inhibitor of the p53 ubiquitin ligase Mdm2 (2–5). As such, the unprecedented structure of the compact Ink4a-Arf locus, conserved in mammals, specifies two distinct tumor suppressors that interface with both the retinoblastoma protein (RB) and p53. The separate promoters upstream of exon-1α (encoding Ink4a) and exon-1β (encoding Arf) are induced by hyperproliferative signals generated by activated oncoproteins, triggering RB- and p53-dependent programs, respectively, that counter cellular self-renewal in response to oncogene stress (6, 7). Deletion or epigenetic silencing of the Ink4a-Arf locus coordinately diminishes the tumor suppressing effects of both RB and p53 and, as would be expected, inactivation of this gene cluster is one of the most frequently observed events in human cancer.
The stability and nucleolar sequestration of p19Arf depend on its interaction with nucleophosmin (NPM or B23), and Arf’s ability to bind to either Mdm2 or NPM is determined by N-terminal amino acid residues encoded solely by exon-1β (8–11). Indeed, a small polypeptide specified by exon-1β alone is capable of interacting with NPM and Mdm2 and inducing acute p53-dependent cell cycle arrest (12, 13). The single ORFs of the mouse (Arf) and human (ARF) mRNAs contain two, and only two, AUG codons (encoded by exon-1β) that specify methionines (M)-1 and -45 of p19Arf (M48 in human p14ARF), each of which can initiate translation (14). The N-terminally truncated protein, p15smArf, initiated at M45 is highly unstable (14), lacks amino acid residues required for p53 activation and NPM binding (15), localizes to mitochondria (14), and has been reported to trigger autophagy and/or mitophagy when overexpressed (16–18). By generating mice that produce either p15smArf or full-length p19Arf alone, we have now elucidated distinct physiological roles played by the two Arf proteins.
Results and Discussion
Generation of Mouse Strains Expressing Either p19Arf or p15smArf Alone.
RNAs encoding forward and reverse TALEN nucleases directed to spacer sequences flanking the first Arf ATG codon were injected into C57BL/6J × FvN/N mouse zygotes and yielded mosaic pups in which a 25-bp deletion, including the 5′ ATG, generated a frame-shift mutation in which translation was initiated at M45 (19). A second strain with a single base-pair deletion immediately 3′ to the first ATG similarly initiated translation at M45 and was indistinguishable from the first. Following germ-line transmission of mutant Arf alleles, heterozygous progeny producing both p19Arf and p15smArf proteins were bred to homozygosity and backcrossed through multiple generations to yield a pure (−25 bp) C57BL/6 smArf strain (>99.6% C57BL/6 at the time the following experiments were first undertaken).
To generate p15smArf-deficient mice encoding p19Arf alone, we initially undertook pilot experiments in which we transfected C57BL/6 embryonic stem (ES) cells with an all-in-one construct expressing Cas9 and a 5′-CTTTCGTGAACATGTTGTTG-3′ guide RNA targeting the internal Arf initiation codon (bold italic) encoding M45. Cotransfection of a 127-base single-strand oligodeoxynucleotide (ssODN) containing 5′-CTTTCGTGAACGCCCTGTTGCGC-3′ plus 31-base 5′ and 72-base 3′ homology arms was used to mutate the ATG (M) to GCC (A), to simultaneously disrupt the guide RNA-targeting site by an additional C to T substitution, and to convert the 3′ AGG PAM sequence to CGC (bold italic). The latter base substitutions were included to prevent repeated CRISPR/Cas9 nucleolytic activity at the correctly edited and recombined target site. Apart from the M45A mutation, alteration of the four additional bases within the targeted region did not change their coded amino acids. These experiments indicated that of 47 expanded ES clones subjected to Sanger sequencing, the overall indel frequency was 73%; 8 ES clones contained a correct M45A knock-in of one Arf allele and an indel in the other. Encouraged by the respectable mutational frequencies and correct gene editing achieved in ES cells, microinjections into C57BL/6 zygotes of the in vitro transcribed guide RNA and Cas9 mRNA together with the same ssODN were carried forward. Individual surrogate females each received 20 injected zygotes; when brought to term, these animals yielded 65 pups, 9 of which contained single ArfM45A alleles, again confirmed by Sanger sequencing. M45A founders were bred to homozygosity, and the colonies were expanded. Subsequent experiments were performed with syngeneic C57BL/6 wild-type Arf+/+, smArf (p15smArf only), M45A mutant (p19Arf-M45A only), and Arf−/− (double-null) homozygotes.
Mitochondrial Localization of the Endogenously Produced smArf Protein.
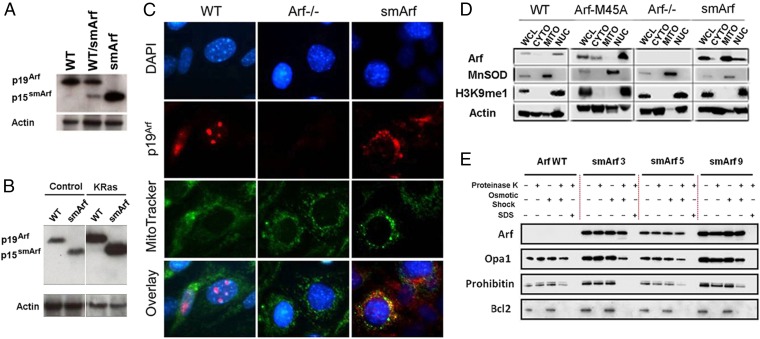
The same Arf mRNA is responsible for cotranslation of both p19Arf and p15smArf, as demonstrated by the almost equally efficient translation of both proteins in vitro (14). However, the relative instability of p15smArf (t1/2 < 60 min) in wild-type (WT) Arf+/+ cells results from its rapid proteasomal degradation, normally leading to its steady-state accumulation at less than 10% of the levels achieved by the full-length p19Arf protein (11, 14). This finding is consistent with previous metabolic [3H]leucine “pulse–chase” kinetic analyses, which demonstrated that p19Arf mutants lacking amino acids 2–14 and 26–37 failed to bind both NPM and Mdm2 and were highly unstable (11). By contrast, in passaged primary mouse embryo fibroblasts (MEFs) from both Arf/smArf heterozygous and smArf homozygous strains, the p15smArf protein was readily observed (Fig. 1A), confirming that removal of the first initiation codon significantly increases the efficiency of translation from the second (14). Notably, the two Arf isoforms are equally detected by use of a monoclonal antibody that recognizes a conserved epitope in both polypeptides (amino acid residues 54–62 in p19Arf) (20). Because the Arf promoter responds to hyperproliferative signals triggered by numerous oncoproteins (6), we reasoned that both p19Arf and p15smArf would be induced following infection of cells with a retroviral vector encoding mutant KRas (G12V). Indeed, the levels of each Arf protein were significantly increased by oncogenic KRas, but even under these conditions, synthesis of p15smArf in WT Arf+/+ cells was barely detectable (Fig. 1B). MEFs expressing smArf alone, like Arf-null MEFs (2), adapted a spindle-shaped morphology, lost contact inhibition, and overgrew one another following KRas transduction, implying that they were defective in tumor suppression (see below).
Fig. 1.
p15smArf localizes within the mitochondria of primary MEFs. (A) p15smArf is not normally detected by immunoblotting in wild-type MEFs because of its rapid turnover, but it accumulates when translation is efficiently initiated at M45. (B) Infection of MEFs with a retroviral vector encoding a KRas (G12V) oncoprotein induces the Arf promoter to yield increased levels of Arf proteins; even under this condition, p15smArf is only barely detected in WT Arf+/+ cells. The four lanes illustrated were cropped, two by two, from a single immunoblot developed after a single exposure. Actin was used as a protein loading control. (C) For immunofluorescence, cells were treated with membrane-permeable MitoTracker (green) (Invitrogen), fixed, stained with 5C3-1 monoclonal antibody to p19Arf followed by goat anti-rat Ig (red), and counterstained with DAPI (blue) to visualize nuclei. p19Arf expressed in WT cells localizes to nucleoli, whereas p15smArf colocalizes with Mitotracker (green) in mitochondria. (D) Subcellular fractionation confirms mitochondrial (MITO) enrichment of p15smArf together with superoxide dismutase (MnSOD), whereas p19Arf colocalizes in the nuclear (NUC) fraction with H3K9me1-marked histones. Fractionation experiments were performed with three independently derived MEF clones of each indicated genotype; representative data are illustrated. Fractionated organelles from whole cell lysates (WCL) were denatured in loading buffer, and equal amounts of protein loaded in each lane were separated on SDS/PAGE gels and immunoblotted (43). Antibodies to MnSOD (BD Biosciences), histone H3K9me1 (Invitrogen), and actin (Millipore) were used to chart enrichment of organelles in different fractions. Signals for p19Arf and p15smArf, simply indicated at the left as Arf, were aligned for convenient illustration despite their different electrophoretic mobilities on denaturing gels. (E) p15smArf localizes within the mitochondrial matrix. Purified mitochondria (42) were treated as described (23) with proteinase K to remove exposed proteins. Osmotic shock disrupted the OMM, and SDS solubilized the OMM and IMM to release matrix components. Lysates were subjected to immunoblotting by using submitochondrial markers including BCL2 (OMM), Opa1 (IMM and intramembrane space), and Prohibitin (inner matrix). Three primary MEF strains designated −3, −5, and −9 expressing smArf were analyzed in parallel on a single gel together WT Arf+/+ cells.
Enforced vector-mediated overexpression of smArf has been reported to reduce mitochondrial membrane potential and to trigger selective autophagy (mitophagy) (16, 18) through mechanisms that do not trigger cytochrome c release or loss of ATP production, and that depend on direct binding to the matrix protein p32 (gC1qR, C1QBP) (21) and on the autophagy regulators ATG5 and Beclin-1 (14), PINK1, and Parkin (22). Unlike p19Arf which is directed to the nucleolus and stabilized by NPM, the endogenously produced smArf protein is imported into mitochondria (Figs. 1C and 1D), where it accumulates in the matrix and is protected from proteolysis by the outer and inner mitochondrial membranes (Fig. 1E). Using previously applied methods to analyze mitochondrial functions (23), studies with three independently derived primary smArf MEFs revealed that levels of oxygen consumption and ATP production were similar to those of WT MEFs and reversed defects observed in cultured Arf-null MEFs (Fig. S1). In contrast to the reported perturbations of mitochondrial membrane potential observed in response to enforced smArf expression following transient transfection or in stably transfected cells expressing inducible smArf (14, 18, 22), endogenous smArf expression did not depolarize mitochondria in cultured MEFs or affect production of reactive oxygen species (Fig. S1). Given that ectopically expressed smArf has to exceed some threshold level to damage mitochondria (14), it is conceivable that potentially deleterious effects of smArf expression in mitochondria were adaptively counterselected during development of the smArf mouse strain.
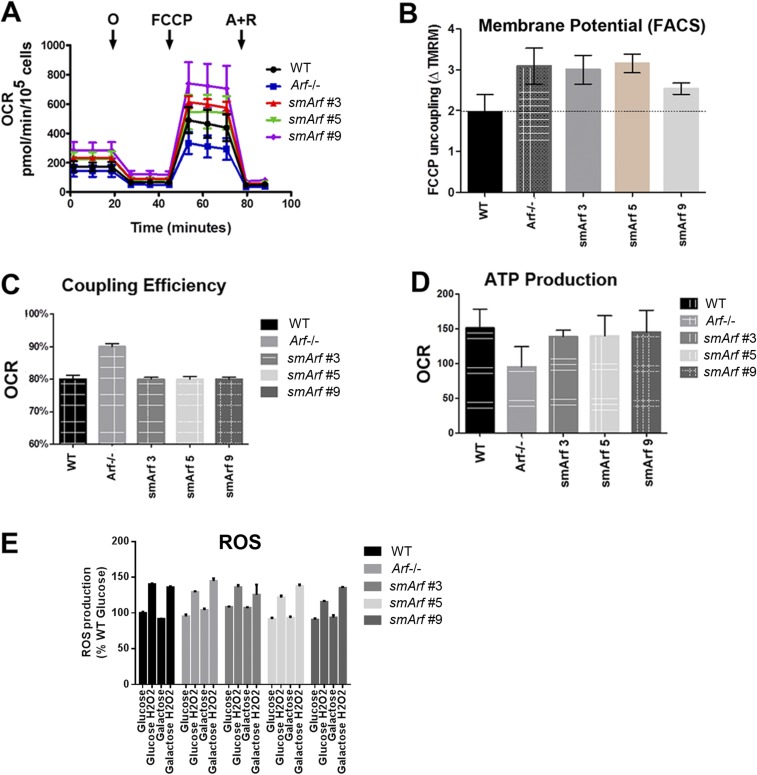
Fig. S1.
Mitochondrial function in MEFs of various Arf genotypes. (A) Measurements of oxygen consumption rate (OCR) were made with a Seahorse Xfe24 Bioanalyzer (Agilent) as per manufacturer’s instructions. Oligomycin (O) was used to poison complex V of the electron transfer chain (ETC); FCCP (carbonyl cyanide-p-trifluoromethoxyphenyl-hydrazone), a protonophore added to collapse the inner membrane gradient, was used to allow the ETC to function at its maximal rate; and antimycin A plus rotenone (A+R) were added to shut down ETC function to reveal residual nonmitochondrial respiration. ATP-linked respiration is calculated by subtracting the oligomycin rate from baseline cellular OCR, whereas proton leak respiration is calculated by subtracting nonmitochondrial respiration from the oligomycin rate. Mitochondrial reserve capacity is calculated by subtracting basal respiration from maximal respiratory capacity. These studies indicated that Arf inactivation decreased overall oxygen consumption, whereas three independently derived smArf MEF strains exhibited slightly higher average oxygen consumption than WT cells, rescuing the deficiencies in ATP production and maximal respiratory capacity of Arf-null MEFs. (B) When MEFs were stained with 2.5 µM tetramethylrhodamine methyl ester (TMRM, detected by FACS at 574 nm) and normalized to TMRM-stained cells treated with FCCP, Arf-deficient and smArf cells displayed subtly increased mitochondrial membrane potential compared with the WT strain, differing from the decreased membrane potential reported for cells acutely transfected with smArf cDNA (14). (C) Seahorse analysis of Arf-null MEFs revealed an increased coupling efficiency compared with smArf and WT strains. The coupling efficiency is defined as the proportion of the oxygen consumed to drive ATP synthesis compared with that driving proton leak respiration and is calculated as the fraction of basal mitochondrial OCR used for ATP synthesis (ATP-linked OCR/basal OCR). (D) WT and smArf MEFs were indistinguishable with regard to ATP-related oxygen consumption. Taken together, smArf MEFs manifest similar respiratory capacity to WT MEFs. (E) WT, Arf-null, and smArf MEFs plated in 96-well dishes were treated with either glucose- or galactose-containing medium for 4 h. ROS was induced with 1 mM H2O2 and measured by using ROS red stain solution (Abcam) on a Beckman Coulter microplate reader. Upon oxidation, the staining solution emits fluorescence at Ex/Em 520/605 nm. Results were normalized to WT ROS levels in glucose. No significant differences were detected between genotypes.
SmArf Lacks Potent Antiproliferative, Tumor Suppressor Activity.
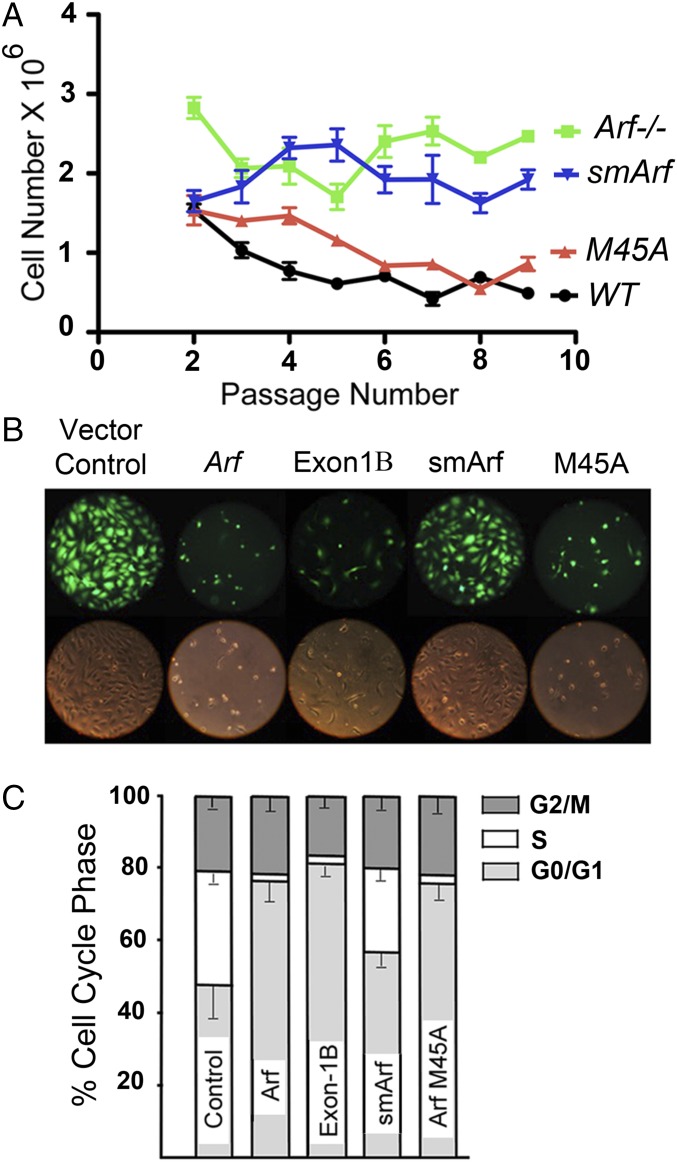
Although the exon-1β–coded segment of p19Arf is both necessary and sufficient for interaction with Mdm2 and for p53 activation (12, 15), we reasoned that smArf might still retain p53-independent tumor suppressive activity (24–26). Wild-type MEFs accumulate p19Arf during continuous passage and undergo p53-dependent senescence, as did MEFs of the M45A strain (Fig. 2A). In contrast, Arf-null MEFs do not senesce (2), a feature mimicked by MEFs of the smArf strain (Fig. 2A), implying that chronic smArf expression does not confer significant antiproliferative activity. Transducing immortalized Arf-null, but Trp53 wild-type, NIH 3T3 cells with retroviral vectors encoding green fluorescent protein (GFP) plus either p19Arf, the smArf-deficient p19M45A mutant, or p15smArf again revealed that smArf alone was markedly deficient in inducing cell cycle arrest. Again, introduction of ArfM45A closely mimicked wild-type Arf in acutely arresting colony formation (Fig. 2 B and C). As confirmed in this experiment, Arf exon-1β alone had potent inhibitory activity, causing rapid G1 phase arrest and stasis without cell death.
Fig. 2.
Arf-dependent antiproliferative activities of cultured cells. (A) 3T3 senescence assay (44). Newly isolated MEFs derived from nine embryos of each of four Arf genotypes were cultured in six-well 35-mm-diameter dishes (3 × 105 cells per well). At 3-d passage intervals, cells recovered by trypsinization were counted and recorded, and 3 × 105 cells were serially replated. WT Arf+/+ cells undergoing senescence experience fewer net population doublings and yield fewer total cells at each passage as indicated in the graphs. Arf-null cells do not senesce. MEFs expressing p15smArf-only phenocopied Arf-null MEFs, whereas MEFs of the M45A strain senesced. [WT or M45A vs. smArf or Arf−/−, P < 0.0001; WT vs. M45A P = 0.12; Arf−/− vs. smArf, P > 0.05]. (B) Subconfluent, immortal NIH 3T3 cells (Arf-null, Trp53+/+) were infected with GFP-expressing retroviral vectors encoding p19Arf, p19Arf-M45A, p15smArf, or a 64-aa Arf exon-1β polypeptide. Cells were photographed for GFP fluorescence 48 h after infection. (C) Infected cells from four independent experiments were trypsinized, stained with propidium iodide, and analyzed in triplicate for DNA content; phases of the cell cycle were determined by flow cytometric measurement to record G0/G1 (2N), G2/M (4N), and S-phase fractions (between 2N to 4N). Error bars indicate ± SEM.
We next used a well-characterized in vivo model of BCR-ABL oncogene-induced acute lymphoblastic leukemia (ALL) to test the ability of p15smArf and the p19M45A mutants to act as tumor suppressors in living mice. In this system, pro/pre-B cells derived from the bone marrow of Arf-null C57BL/6 mice are transduced with retroviruses coexpressing the p185BCR-ABL oncoprotein together with marker proteins [either GFP or luciferase (Luc)], and infected cells are infused into the tail veins of normal, unconditioned syngeneic mice monitored for subsequent tumor development. This model exhibits several key characteristics: (i) recipient mice receiving syngeneic, transduced pro/pre-BCR-ABL–positive, Arf-null B cells need not be preconditioned with drugs or radiation to succumb to ALL; (ii) wild-type donor cells do not induce ALL, consistent with Arf’s oncogene-induced tumor suppressive activity; (iii) Trp53-null BCR-ABL–positive donor cells induce ALL with the same kinetics as Arf-null donor cells, implying that loss of Arf mimics p53 tumor suppression; (iv) ALLs arising in recipient mice are polyclonal, and administration of only 20 BCR-ABL–positive, Arf-null donor cells kill recipient mice within 30 d, revealing that BCR-ABL and Arf loss of function are themselves sufficient to induce ALL; and (v) inhibition of the BCR-ABL kinase with targeted drugs induces ALL remission, implying that persistent BCR-ABL signaling in the absence of Arf-dependent tumor suppression is required for tumor maintenance (27–30).
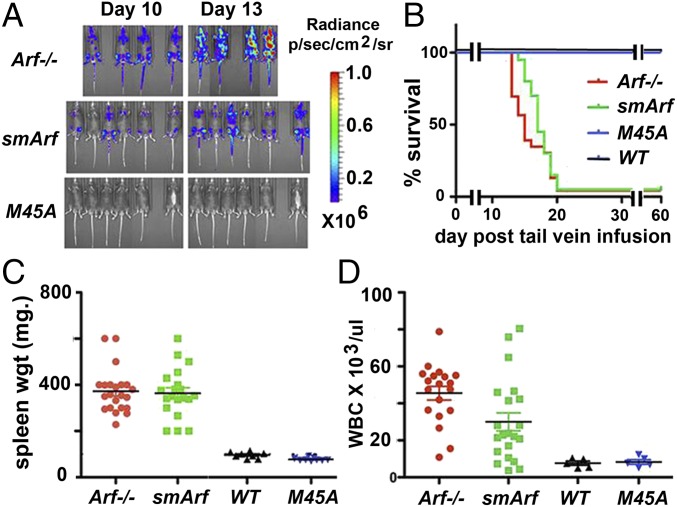
We generated p185BCR-ABL-positive pro/pre-B cells from cultured bone marrow progenitors of wild-type, Arf-null, smArf, and M45A mutant mice (27, 31), confirmed uniform expression of marker proteins (B-cell B220, vector-driven GFP or Luc) after vector transduction, and infused 2 × 105 donor cells per animal into cohorts of normal, unconditioned syngeneic mice. Notably, Arf-null, BCR-ABL–positive donor cells expand rapidly in vivo, increasing almost 10-fold in number every 3 d and killing all recipient mice within only 20 d (28–30). Living mice were screened for luciferase activity (Fig. 3A) and monitored for typical signs of tumor development. Moribund mice were necropsied to confirm ALL involvement manifested by increased blood leukocyte counts, lymph node and splenic infiltration, and central nervous system meningeal invasion as described (28, 29) (Fig. 3B). Donor BCR-ABL–positive, Arf-null donor cells rapidly induced ALL, whereas wild-type and M45A donor cells did not generate tumors during a 60-d observation period, indicating that p19Arf alone was fully protective in this model (Fig. 3 A and B). In direct contrast, mice receiving smArf-only donor cells expired rapidly with ALL. By comparison with mice that received Arf-null donor cells, the temporal accumulation of luciferase-positive smArf-expressing cells in vivo was slightly retarded (Fig. 3A), but accompanying differences in survival were not significant (Fig. 3B). Spleen weights in both cohorts were comparable (Fig. 3C), but white blood cell counts were lower in mice that received smArf versus Arf−/− donor cells (Fig. 3D). These findings do not preclude that smArf may retain some relatively modest tumor suppressive activity, albeit insufficient to prevent rapid ALL development.
Fig. 3.
Arf tumor suppression in a mouse B-cell ALL model. (A) Cultured B220-positive, pro/pre-B cells derived from the bone marrows of C57BL/6 mice of the indicated Arf strains were infected with ecotropic retroviral vectors coexpressing p185BCR-ABL and either luciferase or GFP. Infected, cultured cells scored for GFP expression (>80% positive, to monitor infection) or luciferase were infused into the tail veins of naïve syngeneic C57BL/6 mice (2 × 105 cells per mouse) that were periodically imaged for accumulation of luciferase-positive cells detected by whole body fluorescence. (B) Animals were killed when moribund and necropsied to confirm leukemic involvement. The rapid time course of ALL development documented by Kaplan–Meier survival curves (22 mice per group) closely mimicked previous independent studies performed in separate laboratories (28–30). Differences in survival between mice receiving Arf-null and smArf donor cells were not significant. Spleen weights (C) and circulating leukocyte counts (D) in blood obtained from the hearts of moribund animals were recorded for individual mice that received p185BCR-ABL-positive donor cells of the indicated Arf genotypes. Differences in leukocyte counts between animals receiving Arf-null and smArf donor cells were significant (P < 0.01). Statistical comparisons of results obtained with different cohorts were determined by two-tailed t test.
Mice of the smArf Strain Do Not Manifest Focal Developmental Anomalies Seen in Arf-Null Mice.
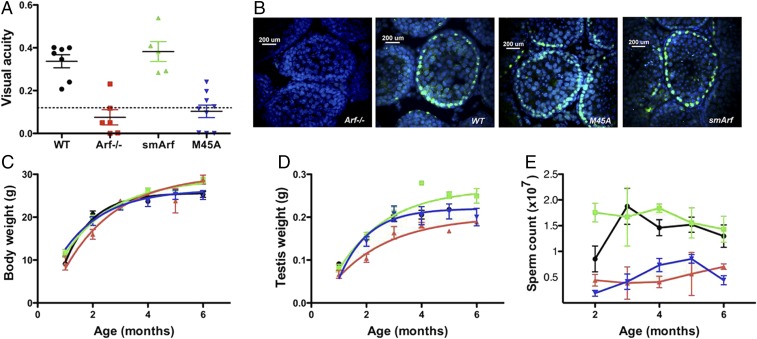
Arf-null mice exhibit late embryonal and early postnatal persistence of the hyaloid vasculature in the vitreous of their eyes because of an abnormal proliferation of mural mesenchymal cells that envelop the hyaloid capillary vascular endothelium (32). In wild-type mice, the hyaloid vasculature normally undergoes regression in the first weeks after birth to leave a clear avascular vitreous. However, in the Arf-null setting, aberrant accumulation of mural pericytes and development of a retrolental mass ultimately detaches the retina, which collapses into the lens and causes early postnatal blindness. Although Trp53-null mice on a C57BL/6 background exhibit certain strain-specific eye defects reminiscent of those observed in Arf-null animals (33), the extreme pathological manifestations that lead to blindness appear to be p53-independent (32). As noted by others, the resulting ocular phenotype closely mimics persistent hyperplastic primary vitreous, a congenital disease of unknown etiology in children that can similarly erode the posterior lens capsule and lead to retinal detachment and blindness (34, 35). The appearance of the accumulating mass within the vitreous of the developing eye depends on disruption of p19Arf-dependent suppression of platelet-derived growth factor-beta (PDGFβ) signaling within the hyaloid vasculature (35, 36), so that targeted disruption of PDGF receptor-β signaling in the eye rescues blindness (37). By use of an optokinetic system that depends on reflexive head movement in the direction of a rotating visual field (38), functional and quantitative measurements of visual acuity allow any underlying histopathological defects to be prospectively identified (37). Arf-null mice lacking both p19Arf and p15smArf and M45A mutant mice lacking only p15smArf were blind, whereas mice expressing smArf alone exhibited no detectable visual defects (Fig. 4A). Unexpectedly, then, p15smArf, rather than full-length p19Arf, rescues the Arf-null eye phenotype.
Fig. 4.
Focal developmental phenotypes of Arf−/−, smArf, and M45A mutant mice. (A) Tests of visual acuity reveal that wild-type (WT) Arf+/+ and smArf-only mice can see, whereas vision is defective in animals of the Arf−/− and M45A strains. Visual acuity of mice was performed as described (37, 38). The maximal spatial frequency capable of driving reflexive head tracking is numerically scored as cycles per degree. The dotted line indicates the cutoff below which animals are completely blind. (B) Immunofluorescence analysis of seminiferous tubules from 3- to 4-wk-old male mice reveals the presence of Arf proteins (green) in spermatogonia lining the basement membranes. Intratubular meiotic cells (spermatocytes), detached from the basement membrane, that have extinguished Arf expression (37, 39) are visualized with DAPI. Spermiogenesis has yet to occur in male mice of this age. The overall weights of mice of the different indicated genotypes (C), the weights of testes (D), and the numbers of sperm recovered from the epididymides (E) [WT vs. M45A, P < 0.005; WT vs. Arf−/−, P < 0.005; WT vs. smArf, nonsignificant by two-tailed t test] were recorded from birth to age 6 mo. Error bars indicate ± SEM; color code in C–E as in A.
Young Arf-null male mice are fertile, but they experience premature loss of sperm as they age (37, 39). In the testes of young males, expression of the p19Arf protein is restricted to mitotically dividing spermatogonia that line the inner seminiferous tubules, but once these cells detach from the basement membrane and enter meiotic prophase as intratubular spermatocytes, Arf expression is completely extinguished (39); as expected, smArf and M45A mice also express Arf proteins in spermatogonia but not in intratubular meiotic cells (Fig. 4B). Notably, spermatocytes that arise from p19Arf-deficient spermatogonia fail to properly correct synaptonemal DNA damage during prophase I, leading to a p53-dependent DNA damage response that eliminates meiotic progenitors during pachytene (39). Although the general paradigm for Arf-induced tumor suppression depends on the ability of p19Arf to antagonize Mdm2 and induce a p53 response, in seminiferous tubules it is instead the absence of Arf expression in mitotic progenitor cells that leads to DNA damage and p53-dependent apoptosis of their meiotic progeny. SmArf-only mice gain weight normally (Fig. 4C), but the weights of their testes are significantly increased, in direct contrast to Arf-null mice in which testicular mass is reduced relative to that of wild-type animals (Fig. 4D). Importantly, male smArf mice produce abundant sperm and maintain fertility as they age, in contradistinction to Arf-null and M45A mutant mice (Fig. 4E).
It has long been appreciated that expression of p19Arf (p14ARF in humans) and p16Ink4a, controlled by distinct promoters upstream of unique first exons, enlist strong tumor suppressive activities exerted by p53 and RB, respectively. Whereas the Arf exon-1β–coded N-terminal segment is sufficient to inhibit Mdm2 and activate p53, internally initiated p15smArf also lacks sequences necessary for its NPM-dependent nucleolar sequestration and accumulates instead in mitochondria. Shared exon-2 of human ARF and INK4a, translated in alternative reading frames and encoding smARF amino acid residues as well, sustains frequent mutations in cancer. More than a decade ago, Itahana and Zhang (40) reported that 560 of 661 already documented cancer-derived alterations in exon-2 included 118 mutations that truncated the C terminus of both p14ARF and p16INK4a, as well as 260-point mutations affecting both proteins, and 39 that targeted ARF alone. Some of these mutations are predicted to disrupt the binding of smARF to p32/C1QBP, required for smARF mitochondrial localization (21, 40), raising the possibility that some such mutations target smARF function in tumors. However, mouse p15smArf appears devoid of overt antiproliferative activity and was unable to limit oncogene-induced cell transformation, implying that cancer-associated exon-2 mutations likely target p16INK4a or p14ARF. Nonetheless, it remains surprising that expression of smArf in living mice corrects the focal phenotypes of Arf-null mice that lead to aberrant proliferation of mesenchymal cells in the hyaloid vasculature of the developing eye and that result in death of primary spermatocytes arising from Arf-null spermatogonial progenitors. Remarkably, these two p53-independent developmental phenotypes of Arf-null mice depend on p15smArf alone. These unexpected findings further underscore the unusual structure and unprecedented complexities of the Ink4a-Arf (Cdkn2a) locus.
Materials and Methods
Subcellular Localization of Arf Proteins.
Fractionation experiments were performed with three independently derived MEF clones of each indicated genotype, and subcellular components were recovered by differential centrifugation (41, 42). All steps were performed at 4 °C. Trypsinized MEFs suspended in 220 mM mannitol, 70 mM sucrose, 10 mM Hepes-KOH (pH 7.4) and containing a mixture of protease inhibitors (Sigma) were lysed by passage through a 30 gauge needle. Fractions enriched for various subcellular components were recovered by differential centrifugation and included discarded nuclei (600 × g, 10 min), crude mitochondria and heavy membranes (5,500 × g, 15 min), and cytosol (soluble supernatant cleared at 100,000 × g, 30 min). The crude mitochondria-enriched fraction was suspended in 250 mM mannitol, 5 mM Hepes (pH 7.4), and 0.5 mM EGTA, layered on 8 mL of Percoll, and ultracentrifuged in an SW41 rotor at 95,000 × g for 30 min. The pellet was suspended in 10 volumes of the same buffer and spun at 7,000 × g for 10 min to recover the mitochondrial fraction (MITO). Nuclear fractions were prepared by lysing cells for 30 min in 40 mM Hepes, pH 7.4, 120 mM KCl, 2 mM EGTA, 0.4% glycerol, 10 mM β-glycerophosphate, and 0.2% Nonidet-P40 (Nonidet P-40) containing a protease inhibitor mixture (Sigma). Nuclei were pelleted at 1,000 × g for 5 min, washed in the same buffer lacking Nonidet P-40, and resedimented. The pellet was suspended for 1 h in 10 mM Tris⋅HCl, pH 7.4, 1.5 mM KCl, 0.5% Triton X-100, 0.5% Na deoxycholate, 2.5 mM MgCl2, containing freshly prepared 0.2 mM LiCl and protease inhibitors. Nuclei were pelleted at 2,000 × g for 5 min, washed in the same buffer, and resedimented to yield the nuclear fraction (NUC). Fractionated organelles were lysed in SDS-containing polyacrylamide gel electrophoresis (PAGE) loading buffer, and equal amounts of protein loaded in each lane were separated on SDS/PAGE gels and immunoblotted by using a mouse monoclonal antibody (5C3-1) that detects an Arf exon-2-encoded epitope present in both full-length p19Arf and truncated p15smArf (20). Antibodies to superoxide dismutase (mitochondria) (BD Biosciences), histone H3K9me1 (nuclei) (Invitrogen), and actin (loading control) (Millipore) were used to chart enrichment of organelles in different fractions. Following described procedures (23), isolated mitochondria were treated with proteinase K to digest exposed proteins; osmotic shock was used to disrupt the outer mitochondrial membrane (OMM); and SDS was then used to disrupt both the OMM and inner mitochondrial membranes (IMM). Fractions were subjected to immunoblotting by using marker proteins indicated in Fig. 1. For immunofluorescence, cells were treated with membrane-permeable MitoTracker (green) (Invitrogen), fixed, stained with 5C3-1 monoclonal antibody to p19Arf followed by goat anti-rat Ig (red), and counterstained with DAPI (blue) to visualize nuclei.
Cell Proliferation Assays.
MEFs harvested from midgestation embryos of different mouse strains and cultured as described (43) were expanded on a 3T3 protocol (44) as indicated in Fig. 2. Subconfluent Arf-null NIH 3T3 cells were infected with GFP-expressing mouse stem cell ecotropic retroviral vectors (MSCV) (43) encoding either p19Arf, p19Arf-M45A, p15smArf, or a 64-aa Arf exon-1β polypeptide and scored 48 h after infection.
Analysis of Arf-Dependent Developmental Phenotypes.
The overall weights of mice of the different indicated genotypes, the weights of testes, and numbers of sperm recovered from the epididymides were recorded from birth to age 6 mo. Immunofluorescence of testes sections from 3- to 4-wk-old mice was performed (39) to detect Arf-positive spermatogonia rimming the inner seminiferous tubules; detached intratubular Arf-negative meiotic cells were revealed with DAPI.
Visual acuity of mice was performed as described (37) by using an OptoMotry virtual task system (Cerebral Mechanics) (38). In brief, mice placed on an elevated stage are exposed to a rotating drum that reveals a computer-generated pattern of interspersed white and black bars of defined periodicity. The visual capability of each eye is measured by the unrestrained mouse’s reflexive head turning response as the direction of the surrounding drum rotation is randomly changed. Increasing the spatial periodicity of the grated white and black bars limits the subject’s ability to respond to the stimulus, allowing quantitative measurement (38). Each alternating white and black bar is defined as a cycle, and the maximal spatial frequency capable of driving reflexive head tracking is numerically scored as cycles per degree.
BCR-ABL ALL Model.
All animal experiments were performed according to NIH guidelines and Institutional Animal Care and Use Committee-approved protocols. Pro/pre-B donor cells cultured from the bone marrow cells of C57BL/6 mice (27, 31) of the indicated Arf strains were infected with MSCV vectors encoding the human p185BCR-ABL oncoprotein together with either luciferase or GFP expressed from an internal ribosomal entry site and packaged with ecotropic gp70 envelope proteins into virions. GFP was used to document retroviral vector-mediated infection of cultured pro/pre-B cells and luciferase for imaging of live mice that received donor cells. Infected cells (2 × 105 per recipient animal) were infused into the tail veins of cohorts of syngeneic C57BL/6 recipient mice that were followed for signs of leukemia development (increased luciferase signals, ruffled fur, hunched posture, hind limb paralysis, seizures, respiratory distress) and confirmed at necropsy (elevated peripheral blood lymphocyte counts, enlarged lymph nodes and spleen, tumor dissemination to liver, spine, brain meninges) as described in detail (27–29). Statistical comparisons of results obtained with different cohorts were determined by two-tailed t test.
Acknowledgments
We thank the following colleagues at St. Jude Children’s Research Hospital (SJCRH): Martine F. Roussel for critical oversight of B-cell culture, retroviral vector design, preparation and usage, and overall counsel; Elizabeth Stewart for assistance with measurements of mouse visual acuity; Michelle Churchman for guiding immunofluorescence studies of Arf protein expression in seminiferous tubules; Frederique Zindy for supervision of animal husbandry; the SJCRH Transgenic Core Facility for recovery and microinjections of zygotes and their implantation into surrogate mothers; the SJCRH Imaging Core for visualization of seminiferous tubules; and Sarah Robinson, Dana Farmer, Shaela Wright, Judith Hyle, Jose Grenet, and Haiyan Xu for contributing expert technical assistance. This work was supported by NIH Comprehensive Cancer Center Core Grant CA-21765 and by the American Lebanese Syrian Associated Charities of SJCRH. C.J.S. is an Investigator of Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1707292114/-/DCSupplemental.
References
- 1.Quelle DE, Zindy F, Ashmun RA, Sherr CJ. Alternative reading frames of the INK4a tumor suppressor gene encode two unrelated proteins capable of inducing cell cycle arrest. Cell. 1995;83:993–1000. doi: 10.1016/0092-8674(95)90214-7. [DOI] [PubMed] [Google Scholar]
- 2.Kamijo T, et al. Tumor suppression at the mouse INK4a locus mediated by the alternative reading frame product p19ARF. Cell. 1997;91:649–659. doi: 10.1016/s0092-8674(00)80452-3. [DOI] [PubMed] [Google Scholar]
- 3.Pomerantz J, et al. The Ink4a tumor suppressor gene product, p19Arf, interacts with MDM2 and neutralizes MDM2's inhibition of p53. Cell. 1998;92:713–723. doi: 10.1016/s0092-8674(00)81400-2. [DOI] [PubMed] [Google Scholar]
- 4.Zhang Y, Xiong Y, Yarbrough WG. ARF promotes MDM2 degradation and stabilizes p53: ARF-INK4a locus deletion impairs both the Rb and p53 tumor suppression pathways. Cell. 1998;92:725–734. doi: 10.1016/s0092-8674(00)81401-4. [DOI] [PubMed] [Google Scholar]
- 5.Kamijo T, et al. Functional and physical interactions of the ARF tumor suppressor with p53 and Mdm2. Proc Natl Acad Sci USA. 1998;95:8292–8297. doi: 10.1073/pnas.95.14.8292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sherr CJ. Tumor surveillance via the ARF-p53 pathway. Genes Dev. 1998;12:2984–2991. doi: 10.1101/gad.12.19.2984. [DOI] [PubMed] [Google Scholar]
- 7.Zindy F, et al. Arf tumor suppressor promoter monitors latent oncogenic signals in vivo. Proc Natl Acad Sci USA. 2003;100:15930–15935. doi: 10.1073/pnas.2536808100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Itahana K, et al. Tumor suppressor ARF degrades B23, a nucleolar protein involved in ribosome biogenesis and cell proliferation. Mol Cell. 2003;12:1151–1164. doi: 10.1016/s1097-2765(03)00431-3. [DOI] [PubMed] [Google Scholar]
- 9.Bertwistle D, Sugimoto M, Sherr CJ. Physical and functional interactions of the Arf tumor suppressor protein with nucleophosmin/B23. Mol Cell Biol. 2004;24:985–996. doi: 10.1128/MCB.24.3.985-996.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Colombo E, et al. Delocalization and destabilization of the Arf tumor suppressor by the leukemia-associated NPM mutant. Cancer Res. 2006;66:3044–3050. doi: 10.1158/0008-5472.CAN-05-2378. [DOI] [PubMed] [Google Scholar]
- 11.Kuo ML, den Besten W, Bertwistle D, Roussel MF, Sherr CJ. N-terminal polyubiquitination and degradation of the Arf tumor suppressor. Genes Dev. 2004;18:1862–1874. doi: 10.1101/gad.1213904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Quelle DE, Cheng M, Ashmun RA, Sherr CJ. Cancer-associated mutations at the INK4a locus cancel cell cycle arrest by p16INK4a but not by the alternative reading frame protein p19ARF. Proc Natl Acad Sci USA. 1997;94:669–673. doi: 10.1073/pnas.94.2.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weber JD, Taylor LJ, Roussel MF, Sherr CJ, Bar-Sagi D. Nucleolar Arf sequesters Mdm2 and activates p53. Nat Cell Biol. 1999;1:20–26. doi: 10.1038/8991. [DOI] [PubMed] [Google Scholar]
- 14.Reef S, et al. A short mitochondrial form of p19ARF induces autophagy and caspase-independent cell death. Mol Cell. 2006;22:463–475. doi: 10.1016/j.molcel.2006.04.014. [DOI] [PubMed] [Google Scholar]
- 15.Weber JD, et al. Cooperative signals governing ARF-mdm2 interaction and nucleolar localization of the complex. Mol Cell Biol. 2000;20:2517–2528. doi: 10.1128/mcb.20.7.2517-2528.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reef S, Kimchi A. A smARF way to die: A novel short isoform of p19ARF is linked to autophagic cell death. Autophagy. 2006;2:328–330. doi: 10.4161/auto.3107. [DOI] [PubMed] [Google Scholar]
- 17.Reef S, Kimchi A. Nucleolar p19ARF, unlike mitochondrial smARF, is incapable of inducing p53-independent autophagy. Autophagy. 2008;4:866–869. doi: 10.4161/auto.6691. [DOI] [PubMed] [Google Scholar]
- 18.Budina-Kolomets A, Hontz RD, Pimkina J, Murphy ME. A conserved domain in exon 2 coding for the human and murine ARF tumor suppressor protein is required for autophagy induction. Autophagy. 2013;9:1553–1565. doi: 10.4161/auto.25831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li C, et al. Simultaneous gene editing by injection of mRNAs encoding transcription activator-like effector nucleases into mouse zygotes. Mol Cell Biol. 2014;34:1649–1658. doi: 10.1128/MCB.00023-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bertwistle D, Zindy F, Sherr CJ, Roussel MF. Monoclonal antibodies to the mouse p19(Arf) tumor suppressor protein. Hybrid Hybridomics. 2004;23:293–300. doi: 10.1089/hyb.2004.23.293. [DOI] [PubMed] [Google Scholar]
- 21.Reef S, Shifman O, Oren M, Kimchi A. The autophagic inducer smARF interacts with and is stabilized by the mitochondrial p32 protein. Oncogene. 2007;26:6677–6683. doi: 10.1038/sj.onc.1210485. [DOI] [PubMed] [Google Scholar]
- 22.Grenier K, Kontogiannea M, Fon EA. Short mitochondrial ARF triggers Parkin/PINK1-dependent mitophagy. J Biol Chem. 2014;289:29519–29530. doi: 10.1074/jbc.M114.607150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Perciavalle RM, et al. Anti-apoptotic MCL-1 localizes to the mitochondrial matrix and couples mitochondrial fusion to respiration. Nat Cell Biol. 2012;14:575–583. doi: 10.1038/ncb2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weber JD, et al. p53-independent functions of the p19(ARF) tumor suppressor. Genes Dev. 2000;14:2358–2365. doi: 10.1101/gad.827300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Modestou M, Puig-Antich V, Korgaonkar C, Eapen A, Quelle DE. The alternative reading frame tumor suppressor inhibits growth through p21-dependent and p21-independent pathways. Cancer Res. 2001;61:3145–3150. [PubMed] [Google Scholar]
- 26.Kuo ML, et al. Arf induces p53-dependent and -independent antiproliferative genes. Cancer Res. 2003;63:1046–1053. [PubMed] [Google Scholar]
- 27.Williams RT, Roussel MF, Sherr CJ. Arf gene loss enhances oncogenicity and limits imatinib response in mouse models of Bcr-Abl-induced acute lymphoblastic leukemia. Proc Natl Acad Sci USA. 2006;103:6688–6693. doi: 10.1073/pnas.0602030103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Williams RT, den Besten W, Sherr CJ. Cytokine-dependent imatinib resistance in mouse BCR-ABL+, Arf-null lymphoblastic leukemia. Genes Dev. 2007;21:2283–2287. doi: 10.1101/gad.1588607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boulos N, et al. Chemotherapeutic agents circumvent emergence of dasatinib-resistant BCR-ABL kinase mutations in a precise mouse model of Philadelphia chromosome-positive acute lymphoblastic leukemia. Blood. 2011;117:3585–3595. doi: 10.1182/blood-2010-08-301267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Appelmann I, et al. Janus kinase inhibition by ruxolitinib extends dasatinib- and dexamethasone-induced remissions in a mouse model of Ph+ ALL. Blood. 2015;125:1444–1451. doi: 10.1182/blood-2014-09-601062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Whitlock CA, Witte ON. Long-term culture of murine bone marrow precursors of B lymphocytes. Methods Enzymol. 1987;150:275–286. doi: 10.1016/0076-6879(87)50085-4. [DOI] [PubMed] [Google Scholar]
- 32.McKeller RN, et al. The Arf tumor suppressor gene promotes hyaloid vascular regression during mouse eye development. Proc Natl Acad Sci USA. 2002;99:3848–3853. doi: 10.1073/pnas.052484199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ikeda S, et al. Severe ocular abnormalities in C57BL/6 but not in 129/Sv p53-deficient mice. Invest Ophthalmol Vis Sci. 1999;40:1874–1878. [PubMed] [Google Scholar]
- 34.Goldberg MF. Persistent fetal vasculature (PFV): An integrated interpretation of signs and symptoms associated with persistent hyperplastic primary vitreous (PHPV). LIV Edward Jackson Memorial Lecture. Am J Ophthalmol. 1997;124:587–626. doi: 10.1016/s0002-9394(14)70899-2. [DOI] [PubMed] [Google Scholar]
- 35.Iqbal NS, Devitt CC, Sung CY, Skapek SX. p19(Arf) limits primary vitreous cell proliferation driven by PDGF-B. Exp Eye Res. 2016;145:224–229. doi: 10.1016/j.exer.2016.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Widau RC, et al. p19Arf represses platelet-derived growth factor receptor β by transcriptional and posttranscriptional mechanisms. Mol Cell Biol. 2012;32:4270–4282. doi: 10.1128/MCB.06424-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gromley A, Churchman ML, Zindy F, Sherr CJ. Transient expression of the Arf tumor suppressor during male germ cell and eye development in Arf-Cre reporter mice. Proc Natl Acad Sci USA. 2009;106:6285–6290. doi: 10.1073/pnas.0902310106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Douglas RM, et al. Independent visual threshold measurements in the two eyes of freely moving rats and mice using a virtual-reality optokinetic system. Vis Neurosci. 2005;22:677–684. doi: 10.1017/S0952523805225166. [DOI] [PubMed] [Google Scholar]
- 39.Churchman ML, Roig I, Jasin M, Keeney S, Sherr CJ. Expression of arf tumor suppressor in spermatogonia facilitates meiotic progression in male germ cells. PLoS Genet. 2011;7:e1002157. doi: 10.1371/journal.pgen.1002157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Itahana K, Zhang Y. Mitochondrial p32 is a critical mediator of ARF-induced apoptosis. Cancer Cell. 2008;13:542–553. doi: 10.1016/j.ccr.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shaiken TE, Opekun AR. Dissecting the cell to nucleus, perinucleus and cytosol. Sci Rep. 2014;4:4923. doi: 10.1038/srep04923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wieckowski MR, Giorgi C, Lebiedzinska M, Duszynski J, Pinton P. Isolation of mitochondria-associated membranes and mitochondria from animal tissues and cells. Nat Protoc. 2009;4:1582–1590. doi: 10.1038/nprot.2009.151. [DOI] [PubMed] [Google Scholar]
- 43.Zindy F, et al. Myc signaling via the ARF tumor suppressor regulates p53-dependent apoptosis and immortalization. Genes Dev. 1998;12:2424–2433. doi: 10.1101/gad.12.15.2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Todaro GJ, Green H. Quantitative studies of the growth of mouse embryo cells in culture and their development into established lines. J Cell Biol. 1963;17:299–313. doi: 10.1083/jcb.17.2.299. [DOI] [PMC free article] [PubMed] [Google Scholar]