Nearly half of the mammalian genome is occupied by repetitive transposon elements, including long-interspersed nuclear elements (LINEs), short-interspersed nuclear elements, and long terminal repeat retrotransposons. The ability of these transposable elements to propagate and insert randomly throughout the genome plays an important role in genome evolution (1). However, as transposon-mediated insertional mutagenesis has been shown to play a causative role in >65 human genetic diseases (2), it is critical that this process be tightly controlled, particularly in the germline. Remarkably, LINEs account for about one-fifth of the human or mouse genome (3), the majority of which are LINE1 (∼600,000 copies in the murine genome). LINE1 is an active and autonomous transposable element that propagates in the genome through retrotransposition, whereby a LINE1 transcript is reverse-transcribed and inserted into the genome at a different location. The full-length LINE1 encodes two proteins: ORF1p and ORF2p, both of which are needed for its mobilization. Because of its repetitive and highly abundant nature, it is nearly impossible to track the activity of individual endogenous LINE1 elements in animals. As reported in PNAS, Newkirk et al. have overcome this hurdle by generating a new LINE1 reporter transgene in mouse (4).
Although a number of LINE1 transgenic mouse models have been reported, these transgenes used either human LINE1 elements or non-LINE1 promoters (5–8), and therefore may not fully recapitulate the expression and activity of endogenous mouse LINE1 elements. Newkirk et al. (4) generated new LINE1 transgenes in which expression was driven under the control of the endogenous mouse LINE1 promoter encoded within its own 5′UTR. This new transgene consists of codon-optimized mouse ORF1 and ORF2 for improved translation and an intron-containing split EGFP for assaying retrotransposition.
Newkirk et al. (4) next asked whether regulation of these novel LINE1 transgenes mirrors that of endogenous LINE1 elements undergoing epigenetic reprogramming in embryonic gonocytes. Specifically, primordial germ cells (PGCs) undergo genome-wide demethylation. Soon after, PGCs differentiate into embryonic gonocytes, the genomes of which undergo genome-wide de novo DNA remethylation. At this point, LINE1s become methylated at their promoters and thus are silenced (Fig. 1). Newkirk et al. observed that a single-copy LINE1 transgene, referred to as SN1, exhibited the typical hypomethylation and remethylation characteristics of endogenous LINE1 in embryonic gonocytes and could thus be used to study mechanisms regulating LINE1 retrotransposition in vivo.
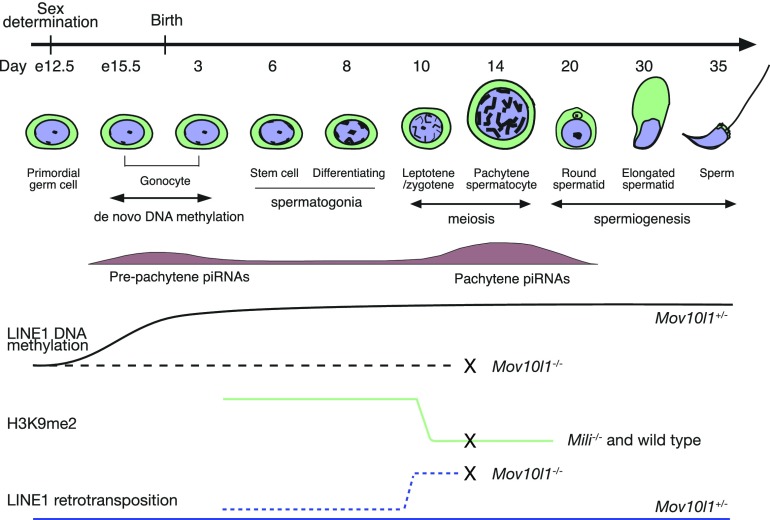
Fig. 1.
Dynamics of piRNAs, DNA methylation, histone modification, and LINE1 activity during mouse male germ-cell development. Two populations of piRNAs (prepachytene and pachytene) are shown. Prepachytene piRNAs are mostly derived from transposable elements, whereas the majority of pachytene piRNAs are derived from nonrepetitive regions. Crosses on the lines mark the time point of meiotic arrest in Mov10l1−/− and Mili−/− males (13, 20). Data from Newkirk et al. (4) support that H3K9me2 expression pattern in Mov10l1−/− testis is similar to that in Mili−/− testis (depicted here) (9). The figure is modified from figure 1 in ref. 15.
Because maintenance of genome integrity in the germline is particularly crucial, LINE1 retrotransposons are silenced by multiple mechanisms: DNA methylation, histone modification, and piRNAs (piwi-interacting RNAs) (Fig. 1) (9, 10). The piRNA pathway is a highly conserved process in metazoans required for silencing of transposable elements in gonads. piRNAs, an abundant class of small-noncoding RNAs in gonads, are mostly derived from transposable elements. piRNAs are used as guides to target transposon transcripts through antisense base pairing for degradation by Piwi proteins in the cytoplasm, and also target transposable elements for transcriptional silencing in the nucleus. Therefore, the piRNA pathway constitutes an adaptive “immune” system to protect the genome integrity of the germline (11). In mouse, piRNA pathway mutants exhibit de-silencing of retrotransposons and male sterility. Most of the piRNA mouse mutant strains display meiotic arrest, characterized by increased DNA damage and apoptosis in spermatocytes (Fig. 1). It has been widely assumed but yet unproven in the field that increased insertional mutagenesis caused by LINE1 overexpression leads to the demise of germ cells in piRNA pathway mutants. This has not been possible to test because of a lack of a suitable LINE1 reporter.
In PNAS, Newkirk et al. (4) address this issue by crossing the SN1 LINE1 transgenic mice to Mov10l1 mutant mice. MOV10L1 is a germ-cell–specific RNA helicase that initiates piRNA biogenesis by binding to piRNA precursors (12). Mov10l1−/− testes lack piRNAs (13–15). As expected, the SN1 LINE1 transgene is hypomethylated in Mov10l1−/− spermatogonia and spermatocytes, and highly expressed in Mov10l1−/− spermatocytes, showing that silencing of the LINE1 transgene, like that of endogenous LINE1s, depends on the intact piRNA pathway (4). Newkirk et al. (4) next quantify the SN1 LINE1 retrotransposition frequency based on the intron loss in new insertions as a result of splicing during retrotransposition (Fig. 1). New SN1 LINE1 insertion increased by 70-fold in Mov10l1−/− testes at postnatal day 14, when most germ cells are meiotic spermatocytes. When assayed in sorted germ cells, insertion increased by 144-fold in Mov10l1-deficient spermatocytes. These results demonstrate that the intact piRNA pathway is required to block LINE1 retrotransposition. By extrapolation of the SN1 LINE1 insertion frequency, Newkirk et al. estimated the frequency of endogenous LINE1 retrotransposition at ∼2.3 insertions per cell in Mov10l1−/− testes and concluded that insertional mutagenesis at this frequency is unlikely to responsible for the death of Mov10l1-deficient spermatocytes. This conclusion is further supported by the lack of rescue of the meiotic defects in Mov10l1−/− mice by inhibition of retrotransposition with a nucleoside reverse-transcriptase inhibitor. However, this result does not exclude the role of LINE1 de-silencing in the death of spermatocytes. The massive DNA damage in Mov10l1-deficient spermatocytes could be partially attributed to overexpression of the cytotoxic LINE1 ORF2 protein, which is an endonuclease. Indeed, LINE1 overexpression causes DNA damage in testes lacking MAEL, one of the protein factors in the piRNA pathway (16). Alternatively, meiotic defects in Mov10l1-deficient testes could also be related to altered epigenetic state in meiotic chromatin, as reported in testes lacking DNMT3L, a protein required for de novo DNA methylation (17).
LINE1 de-silencing exhibits a binary expression pattern in Mov10l1−/− testes: up-regulation in spermatocytes but not in spermatogonia (13). It turns out that LINE1 silencing in spermatogonia requires both piRNAs and H3K9me2 (dimethylation of histone H3 at lysine 9) (9). The silencing H3K9me2 modification is present in spermatogonia through zygotene spermatocytes and disappears
Newkirk et al. generated new LINE1 transgenes in which expression was driven under the control of the endogenous mouse LINE1 promoter encoded within its own 5′UTR.
in pachytene spermatocytes (Fig. 1) (9, 18). In piRNA-deficient spermatogonia, LINE1 is hypomethylated but remains silenced because of H3K9me2 (19). In PNAS, Newkirk et al. (4) show that SN1 LINE1 retrotransposition did not change in Mov10l1-deficient spermatogonia, providing additional support for the multiple epigenetic mechanisms in silencing of LINE1 in spermatogonia.
The germline is the most important battle front in the arms race between transposons and the host genome, as new insertions that occur in the germline will be passed onto the next generation. Newkirk et al. (4) show dramatically increased new insertions of LINE1 in spermatocytes in the absence of piRNAs, highlighting the sterility of piRNA-deficient males as an important mechanism for controlling the rate of insertional mutagenesis. The SN1 LINE1 transgene generated by Newkirk et al. can be used to study piRNA biogenesis. Why are some transcripts selected as piRNA precursors to be processed into mature piRNAs but others are not? Another question to be addressed is whether piRNAs are derived from the SN1 LINE1 transgene in the testes. In particular, do the codon-optimized ORF1/ORF2 and EGFP portion of the SN1 LINE1 transcript give rise to piRNAs? In addition, The SN1 LINE1 transgene will be a useful tool to track in vivo LINE1 mobilization in other mouse mutants and in other tissues, such as brain. Finally, this LINE1 transgene can be used for insertional mutagenesis if infertility is circumvented.
Acknowledgments
The author’s research is supported by the National Institute of Child Health and Human Development of the National Institutes of Health Grants R01HD069592 and U01HD084007, and by National Institute of General Medical Sciences of the National Institutes of Health Grant R35GM118052.
Footnotes
The author declares no conflict of interest.
See companion article on page E5635.
References
- 1.Huang CR, Burns KH, Boeke JD. Active transposition in genomes. Annu Rev Genet. 2012;46:651–675. doi: 10.1146/annurev-genet-110711-155616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Goodier JL, Kazazian HH., Jr Retrotransposons revisited: The restraint and rehabilitation of parasites. Cell. 2008;135:23–35. doi: 10.1016/j.cell.2008.09.022. [DOI] [PubMed] [Google Scholar]
- 3.Mandal PK, Kazazian HH., Jr SnapShot: Vertebrate transposons. Cell. 2008;135:192–192.e1. doi: 10.1016/j.cell.2008.09.028. [DOI] [PubMed] [Google Scholar]
- 4.Newkirk SJ, et al. Intact piRNA pathway prevents L1 mobilization in male meiosis. Proc Natl Acad Sci USA. 2017;114:E5635–E5644. doi: 10.1073/pnas.1701069114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ostertag EM, et al. A mouse model of human L1 retrotransposition. Nat Genet. 2002;32:655–660. doi: 10.1038/ng1022. [DOI] [PubMed] [Google Scholar]
- 6.An W, et al. Active retrotransposition by a synthetic L1 element in mice. Proc Natl Acad Sci USA. 2006;103:18662–18667. doi: 10.1073/pnas.0605300103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Muotri AR, et al. Somatic mosaicism in neuronal precursor cells mediated by L1 retrotransposition. Nature. 2005;435:903–910. doi: 10.1038/nature03663. [DOI] [PubMed] [Google Scholar]
- 8.Kano H, et al. L1 retrotransposition occurs mainly in embryogenesis and creates somatic mosaicism. Genes Dev. 2009;23:1303–1312. doi: 10.1101/gad.1803909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Di Giacomo M, et al. Multiple epigenetic mechanisms and the piRNA pathway enforce LINE1 silencing during adult spermatogenesis. Mol Cell. 2013;50:601–608. doi: 10.1016/j.molcel.2013.04.026. [DOI] [PubMed] [Google Scholar]
- 10.Yang F, Wang PJ. Multiple LINEs of retrotransposon silencing mechanisms in the mammalian germline. Semin Cell Dev Biol. 2016;59:118–125. doi: 10.1016/j.semcdb.2016.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Castañeda J, Genzor P, Bortvin A. piRNAs, transposon silencing, and germline genome integrity. Mutat Res. 2011;714:95–104. doi: 10.1016/j.mrfmmm.2011.05.002. [DOI] [PubMed] [Google Scholar]
- 12.Vourekas A, et al. The RNA helicase MOV10L1 binds piRNA precursors to initiate piRNA processing. Genes Dev. 2015;29:617–629. doi: 10.1101/gad.254631.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zheng K, et al. Mouse MOV10L1 associates with Piwi proteins and is an essential component of the Piwi-interacting RNA (piRNA) pathway. Proc Natl Acad Sci USA. 2010;107:11841–11846. doi: 10.1073/pnas.1003953107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frost RJ, et al. MOV10L1 is necessary for protection of spermatocytes against retrotransposons by Piwi-interacting RNAs. Proc Natl Acad Sci USA. 2010;107:11847–11852. doi: 10.1073/pnas.1007158107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zheng K, Wang PJ. Blockade of pachytene piRNA biogenesis reveals a novel requirement for maintaining post-meiotic germline genome integrity. PLoS Genet. 2012;8:e1003038. doi: 10.1371/journal.pgen.1003038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Soper SF, et al. Mouse maelstrom, a component of nuage, is essential for spermatogenesis and transposon repression in meiosis. Dev Cell. 2008;15:285–297. doi: 10.1016/j.devcel.2008.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zamudio N, et al. DNA methylation restrains transposons from adopting a chromatin signature permissive for meiotic recombination. Genes Dev. 2015;29:1256–1270. doi: 10.1101/gad.257840.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tachibana M, Nozaki M, Takeda N, Shinkai Y. Functional dynamics of H3K9 methylation during meiotic prophase progression. EMBO J. 2007;26:3346–3359. doi: 10.1038/sj.emboj.7601767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Di Giacomo M, Comazzetto S, Sampath SC, O’Carroll D. G9a co-suppresses LINE1 elements in spermatogonia. Epigenetics Chromatin. 2014;7:24. doi: 10.1186/1756-8935-7-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuramochi-Miyagawa S, et al. Mili, a mammalian member of piwi family gene, is essential for spermatogenesis. Development. 2004;131:839–849. doi: 10.1242/dev.00973. [DOI] [PubMed] [Google Scholar]