Abstract
Stromal stem cells proliferate in vitro and may be differentiated along several lineages. Freshly isolated, these cells have been too few or insufficiently pure to be thoroughly characterized. Here, we have isolated two populations of CD45-CD34+CD105+ cells from human adipose tissue which could be separated based on expression of CD31. Compared with CD31+ cells, CD31- cells overexpressed transcripts associated with cell cycle quiescence and stemness, and transcripts involved in the biology of cartilage, bone, fat, muscle, and neural tissues. In contrast, CD31+ cells overexpressed transcripts associated with endothelium and the major histocompatibility complex class II complex. Clones of CD31- cells could be expanded in vitro and differentiated into cells with characteristics of bone, fat, and neural-like tissue. On culture, transcripts associated with cell cycle quiescence, stemness, certain cytokines and organ specific genes were down-regulated, whereas transcripts associated with signal transduction, cell adhesion, and cytoskeletal components were up-regulated. CD31+ cells did not proliferate in vitro. CD45-CD34+CD105+CD31- cells from human adipose tissue have stromal stem cell properties which may make them useful for tissue engineering.
INTRODUCTION
A distinctive feature of adipose tissue is its ability to grow and regress throughout adult life. It is therefore not surprising that cells with the functional capability of stem cells have recently been cultured from adipose tissue. Adipose tissue–derived adult stem cells (ADASC) proliferate extensively and differentiate toward several mesodermal lineages in vitro (Zuk et al., 2001, 2002; Safford et al., 2002; Woodbury et al., 2002; Gronthos et al., 2003). In vivo, these cells have recently been used to heal critical-size skeletal defects in mice (Cowan et al., 2004). Therefore, ADASC offer an appealing cell type for the future use in tissue regeneration.
Stem cells have been isolated from other mesodermal tissues such as bone marrow (Pittenger et al., 1999), muscle (Howell et al., 2003), and perichondrium (Arai et al., 2002). Generally called stromal stem cells or mesenchymal stem cells (MSC), they share key characteristic features. These include the ability to self-renew in vitro by forming adherent fibroblastic-like colonies, termed colony-forming units-fibroblasts (CFU-F), while retaining the ability to differentiate into mesodermal tissues including bone, cartilage, and fat. Recent evidence also suggests that subpopulations of stromal stem cells possess the ability to differentiate to cells of nonmesodermal lineages in vitro such as neuronal cells (Safford et al., 2002) and hepatocyte-like cells (Jiang et al., 2002) and to contribute to most tissues after injection into a blastocyst (Jiang et al., 2002). These observations suggest that stromal stem cells may be less committed than previously thought.
Although stromal stem cells have been the focus of intense research over recent years, little is known about their biology in vivo. Such work has largely been hampered by the scarcity of stromal stem cells in tissues and the lack of reagents required to obtain freshly isolated cells of high purity and in quantities required for extensive analysis. In human bone marrow, for example, only 0.001–0.01% of nucleated cells form CFU-Fs (Pittenger et al., 1999). Thus, most of our current knowledge on stromal stem cells is based on cells expanded in culture.
Adipose tissue is easy to obtain in large quantities and harbors a large number of cells with CFU-F ability (Zuk et al., 2002); therefore, it should provide a readily available source of stromal stem cells in numbers sufficient to study their biology without having to resort to cell culture. Here, we have purified uncultured ADASC using a combination of monoclonal antibodies (MAbs) to cell surface antigens. The cells were isolated in sufficient quantities to perform extensive phenotype and global gene expression analyses. Polyclonal and monoclonal cell culture experiments demonstrate that these cells have stem cell properties.
MATERIALS AND METHODS
Chemicals
All chemicals were from Sigma (St. Louis, MO) unless otherwise stated.
Isolation of Stromal Vascular Cells from Human Adipose Tissue
Adipose tissue was obtained by liposuction from abdominal, hip, and thigh regions of healthy female donors aged 18–39. The stromal vascular fraction (SVF) was separated from adipose tissue using a procedure modified from Zuk and colleagues (Zuk et al., 2001). Briefly, lipoaspirate (300–400 ml) was washed with Hanks' balanced salt solution (HBSS; Life Technologies-BRL, Paisley, UK) containing antibiotics (100 IU/ml penicillin and 100 IU/ml streptomycin, Life Technologies-BRL) and 2.5 μg/ml amphotericin B (Life Technologies-BRL). Washed adipose tissue was digested for 2 h on a shaker at 37°C in HBSS containing 0.2% collagenase. Floating adipocytes were aspirated from pelleted SVF cells after centrifugation at 400 × g for 10 min. Pellets were resuspended in red blood cell lysis buffer (2.06 g/l Tris base, 7.49 g/l NH4Cl, pH 7.2) for 10 min at room temperature. After resuspending SVF cells in HBSS containing 2% fetal bovine serum (FBS), tissue clumps were allowed to settle for 1 min. Suspended cells were passed through 100-μm and then 40-μm cell sieves (Becton Dickinson, San Jose, CA). Cell suspensions (15 ml) were applied to Histopaque-1077 gradients (15 ml) in 50-ml tubes. After centrifugation (400 × g, 30 min), cells at the gradient interface were collected, washed in HBSS, and passed through a 30-μm mesh. Cell counts and viability assessment were performed.
Isolation of ADASC from SVF
CD45+ cells were removed using magnetic beads coupled to mouse antihuman CD45 MAb (Miltenyl Biotech, Bergish Gladbach, Germany) and a superMACS magnet according to instructions provided by the manufacturer. Magnetic beads were also used to split the remaining CD45- cells into CD31+ and CD31- cells. CD45- cells were incubated with FITC-conjugated mouse anti-human CD31 MAb (Serotec, Oxford, United Kingdom) at 1:40 dilution for 15 min at 4°C. After washing, antibody-coated cells were incubated with anti-FITC Microbeads (Milteny Biotech) for 15 min at 4°C. Microbead-coated CD31+ cells were retained in LS columns, allowing the initial isolation of CD31- cells, and later flushed out separately according to the manufacturer's protocol. Flow cytometry was used to confirm separation. We never observed more than 8% contaminating cells in either population (unpublished data).
Culture of Freshly Isolated ADASC
Immediately after separation, CD31+ and CD31- cell subsets were washed and resuspended in DMEM/F12 (Life Technologies-BRL) containing 20% FBS, 100 U/ml penicillin, 100 μg/ml streptomycin, and 2.5 μg/ml amphotericin B. Cells were added to 25-cm2 culture flasks (105 cells/flask) and cultured at 37°C in an atmosphere of 5% CO2 in humid air. After 7 d, attached cells were passaged by trypsinization and cultured in DMEM/F12 containing 10% FBS without amphotericin B. Medium was replaced every third day.
Generation of clonal cell lines from CD31- cells was achieved by culturing single fresh cells in separate wells. Using a micropipette, a single cell was placed into each well of 48-well plates containing DMEM/F12 with 20% FBS, antibiotics, and 2.5 μg/ml amphotericin B. Colony-forming ability was assessed after 21 d. At that time, colonies were passaged and cultured further in DMEM/F12 containing 10% FBS without amphotericin B.
Flow Cytometry
Freshly isolated CD31+ and CD31- cells and cultured CD31- cells (passage 4) were examined for surface and intracellular molecule expression using flow cytometry. The following MAbs conjugated to fluorochromes were used: anti-CD1a-FITC, CD3-APC, CD4-PE, CD8-PerCP, CD10-FITC, CD28-CY, CD33-APC, CD34-PerCP, CD36-PerCP, CD38-APC, CD49a-PE, CD62L-FITC, CD64-CY, CD71-FITC, CD117-PerCP-CY, CD152-CY, HLA ABC-APC, HLA DR-PerCP (BD Biosciences, San Diego, CA), CD13-PE, CD14-FITC, CD19-APC, CD41-PE, CD45-APC (Diatec, Oslo, Norway), CD44-PE, CD90-PE, CD106-FITC (Serotec), CD133-APC (Milteny Biotec), NGFR-FITC or PE (Chromaprobe, Aptos, CA), VEGFR2-APC (R&D Systems, Abingdon, United Kingdom). The CD105 (SH2, Endoglin) cell line hybridoma was purchased from ATCC (Cat. no. HB-10743, Manassas, VA), and the SH2 antibody was purified and conjugated with APC. Irrelevant control MAbs were included for all fluorochromes. Cells were coated with directly conjugated MAbs for 15 min, washed, and fixed in 1% paraformaldehyde. Cells were also stained with unconjugated MAbs directed against the following proteins: CD11a, CD11b, CD11c, CD18, CD29, CD43, CD47, CD49b, CD49c, CD49d, CD49e, CD49f, CD51, CD61, CD62E, CD62P, CD63, CD100, CD103, CD104, CD109, CD114, CD116, CD120, CD121, CD122, CD124, CD126, CD127, CDw119, CD146, membrane SCF, and integrin β7 (gift from Dr. F. L. Johansen) and von Willebrand factor (Immunotech, Marseille, France). For immunolabeling, cells were incubated with primary MAbs for 15 min, washed, and incubated with PE-conjugated goat anti-mouse antibodies (Southern Biotechnology Association, Birmingham, AL) for 15 min. After washing, cells were incubated with 20% mouse serum for 10 min and then with labeled secondary antibodies for 15 min, washed, and fixed.
For detection of intracellular proteins, cells were fixed in 1% paraformaldehyde for 4 h and incubated overnight with 1% Tween-20. Immunolabeling was as described above except that antibody incubations were for 30 min. Cells were analyzed using a FACSCalibur flow cytometer (Becton Dickinson). Gates were set based on staining with combinations of relevant and irrelevant MAbs so that no more than 1% of the cells were positive using irrelevant antibodies.
Real-time RT-PCR
Total RNA was extracted from cells using Trizol (Invitrogen, Gaithersburg, MD), treated with DNase I (Qiagen, Vento, Netherlands) and purified on RNeasy MinElute columns (Qiagen). Reverse transcription (RT) was performed according to manufacturer's protocol (Applied Biosystems, Foster City, CA) with 100 ng total RNA per 50 μl RT reaction. Real-time Quantitative RT-PCR assays for osteocalcin (forward primer: 5′-GGTGCAGCCTTTGTGTCCA-3′, reverse primer: 5′-ACAGTCCGGATTGAGCTCACA-3′, TaqMan probe: FAM (5′)-TCCCAGCCATTGATACAGGTAGCG-Darquencher (3′)) and PPAR-γ2 (forward primer: 5′-TCCATGCTGTTATGGGTGAAACT-3′, reverse primer: 5′-GTGTCAACCATGGTCATTTCTTGT-3′, TaqMan probe: FAM (5′)-AAGCGATTCCTTCACTGATACACTGTCTG-Darquencher (3′)) were designed using Primer Express v.1.5 (Applied Biosystems). Assays for collagen II and aggrecan were as described (Martin et al., 2001). The assay for osteomodulin was from Applied Biosystems (Cat No. 4331182 Assay ID Hs00192325_m1). All assays were designed to overlay a junction between two exons to avoid hybridization to genomic DNA. 18S RNA (Applied Biosystems) was included as an endogenous normalization control to adjust for unequal amounts of RNA. Quantification of mRNA was performed using the ABI Prism 7700 (Applied Biosystems). Each sample (each reaction: 2.5 μl cDNA, total volume 25 μl) was run in triplicate. Cycling parameters were 95°C for 10 min followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. Gene expression was calculated using the relative standard curve method (ABI Prism 7700 Sequence Detection System, User Bulletin 2, PE Applied Systems, Foster City, CA).
Microarray Analysis
RNA sample preparation and microarray assay were performed according to the Affymetrix GeneChip Expression Analysis Technical Manual (Affymetrix, Santa Clara, CA). Briefly, freshly isolated cells of CD31+ and CD31- subsets and cultured CD31- (passage 4) cells from three donors were snap-frozen in liquid nitrogen. Total RNA was extracted as described above. Because of small amounts of RNA in freshly isolated cells, cRNA was obtained according to the GeneChip Eukaryotic Small Sample Target Labeling Assay Version II (Affymetrix). For all samples, 10 μg of cRNA was hybridized to the HG-U133A array (Affymetrix) representing 22,284 probes and ∼14,500 genes. Arrays were scanned at 3 μm using the Agilent Gene Array Scanner (Affymetrix). Gene expression data were analyzed using the Affymetrix Microarray Suite (MAS) 5.0, Affymetrix MicroDB 3.0 and Affymetrix Data Mining Tool (DMT) 3.0 programs. Briefly, a target value of 100 was set for scaling signal intensities for all probe sets. For each comparison, differentially expressed genes were obtained as follows: genes with a present (P) or marginal (M) call in one or both populations were selected. Only genes that showed increased (I) or decreased (D) calls were kept for further analysis. Within these genes, only those with a log2 ratio >1.6 or < -1.6 were selected and published using MicroDB 3.0 into DMT 3.0 to obtain gene names and descriptions. Raw data of the microarray analyses are available at: http://www.ebi.ac.uk/arrayexpress/, under the accession nr. E-MEXP-167 and E-MEXP-168.
Mesodermal Lineage Differentiation of Cultured CD31- Cells
Both clonal (12 lines from 3 donors) and polyclonal (4 donors) cell lines generated from CD31- cells at passage 4 were cultured to confluency before induction toward mesodermal lineages. For adipogenic differentiation, confluent cultures were incubated with DMEM/F12 containing 10% FBS, 0.5 μM 1-methyl-3 isobutylxanthine, 1 μM dexamethasone, 10 μg/ml insulin (Novo Nordisk, Copenhagen, Denmark) and 100 μM indomethacin (Dumex-Alpharma, Copenhagen, Denmark) for 3 wk. Fixed cells (4% formalin) were washed in 50% isopropanol and subsequently incubated for 10 min with Oil-Red O to visualize lipid droplets. Cells were then washed in isopropanol and contrastained with hematoxylin (Fisher Scientific, Hampton, NH). For osteogenic differentiation, cells were incubated in DMEM/F12 containing 10% FBS, 100 nM dexamethasone, 10 mM β-glycerophosphate, and 0.05 mM l-ascorbic acid-2-phosphate for 3 wk. Mineralization of the extracellular matrix (ECM) was visualized by staining with Alizarin Red. For chondrogenic differentiation, trypsinized cells (0.2 × 106) were pelleted and resuspended in 500 μl chondrogenic induction media containing high-glucose DMEM (4.5 g/l; Life Technologies-BRL) supplemented with 500 ng/ml bone morphogenic protein-6 (R&D Systems), 10 ng/ml recombinant human transforming growth factor-β1 (R&D Systems), 1 mM sodium pyruvate, 0.1 mM ascorbic acid-2-phosphate, 100 nM dexamethasone, 1% ITS (25 mg insulin, 25 mg transferrin, and 25 μg sodium selenite), and 1.25 mg/ml bovine serum albumin. After 4 wk of culture in 15 ml tubes, sections were made from micromasses and stained with Toluidine blue. Real-time PCR analyses were performed on cells differentiated from the polyclonal cultures.
Neurogenic Differentiation and Immunocytochemistry
Trypsinized cells from polyclonal and clonal cell lines at passage 4 were added to wells on 48-well plates at a density of 1000 cells per cm2 in DMEM (with 4.5 g/l glucose) containing 10% FBS and antibiotics. To initiate differentiation, the medium was replaced the following day to also contain B27 (Invitrogen), 10 ng/ml epidermal growth factor and 20 ng/ml bFGF. After 5 d, cells were washed and incubated with induction media containing DMEM with 5 μg/ml insulin, 200 μM indomethacin and 0.5 mM 1-methyl-3-isobutylxanthine in the absence of FBS for 5 h. Cells were fixed with methanol at 20°C for 10 min and washed, and immunocytochemistry performed using standard methods. In brief, cells were blocked with 10% FBS and 5% milk and incubated with a polyclonal antibody against Neurofilament 200 (NF200; 1:200 dilution, Sigma) or a MAb against glial acidic fibrillary protein (GFAP; 1:200, Chemicon, Chandlers Ford, United Kingdom) overnight at 4°C. After washing, cells were incubated with goat anti-rabbit antibody conjugated to Cy2 (1:400, Amersham Biosciences, Uppsala, Sweden) for 2 h. An Olympus CKX41 inverted phase contrast microscope (Tokyo, Japan) with a DP camera and controller software was used to capture images.
RESULTS
Cell Surface Expression of Stem Cell Markers
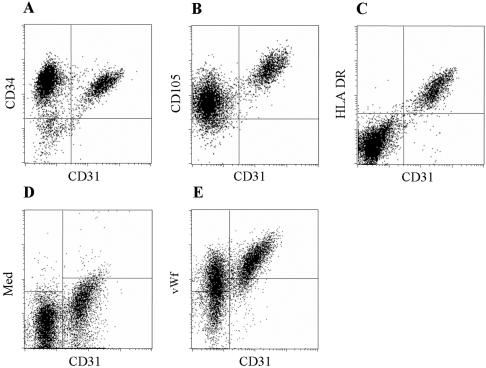
In bone marrow, MSC are known to bind the CD105-specific MAb SH2, whereas hematopoietic stem cells (HSC) express CD34 (Pittenger et al., 1999; Okuno et al., 2002). We used these markers in an initial screening for stem cells in the SVF obtained from liposuction material. The SVF contained a variable proportion of cells expressing the common leukocyte marker CD45. Most CD45+ cells expressed markers found on peripheral T-cells and monocytes (unpublished data). When CD45+ cells were removed from the SVF, flow cytometry revealed that the remaining cells could be separated into two distinct populations based on the expression of CD31, a marker for endothelial cells. All CD31+ cells expressed both CD34 and CD105 with high intensity, and most of CD31- cells expressed these markers (Figure 1, A and B).
Figure 1.
Phenotypic characterization of CD45- cells from human adipose tissue. Expression is shown for CD34 versus CD31 (A), CD105 (endoglin, using the SH2 MAb) versus CD31 (B), and HLA DR versus CD31 (C). Using intracellular staining, the expression of von Willebrand factor versus CD31 is shown in E. The result using medium in stead of vWf MAb is shown in D.
Using a panel of >70 MAbs, we undertook a cell surface phenotypic characterization of the CD31- and CD31+ cell populations (Table 1 and Supplementary Table S1). We found little difference between donors. The most striking difference between the subsets was found for expression of HLA DR. CD31- cells were uniformly negative for HLA DR, whereas all CD31+ cells expressed HLA DR (Figure 1C). Another difference was detected for CD49f (integrin α6). This marker, reported to be the only common putative stemness marker between various stem cell populations in several studies (Fortunel et al., 2003), was expressed on most CD31+ cells but was absent from CD31- cells. For all other markers examined, surface expression was either similar or marginally different for both subsets. Expression of integrins and endothelium-related molecules was found more frequently and with higher intensity on CD31+ cells than on CD31- cells (Table 1). Of note, von Willebrand factor was detected in all CD31+ cells but also in a proportion of CD31- cells (Figure 1, D and E). CD133, which in the bone marrow is coexpressed on practically all CD34+ cells and used to identify HSCs and hemangioblasts (Loges et al., 2004), was not detected on cells from adipose tissue. Similarly CD117 (stem cell factor receptor, c-KIT) was not detected. Nerve growth factor receptor (NGFR), which has been used to isolate MSCs from bone marrow (Jones et al., 2002), was found on a minor proportion of CD31- cells (Table 1).
Table 1.
Cell surface molecule expression by freshly isolated CD31+ and CD31- ADASC and by cultured CD31- cells at passage 4
| CD | Name or function | CD31+ uncultured | CD31- uncultured | Passage 4 |
|---|---|---|---|---|
| Endothelium related molecules | ||||
| CD62E | E-selectin | — | — | — |
| CD62P | P-selectin | +/- | — | — |
| CD63 | LAMP-3 | + | +/- | +/- |
| CD105 | SH-2, endoglin | + | (+) | +hi |
| CD146 | ENDO 3 | + | (+)/- | +/- |
| VEGFR2 | +/- | — | ||
| von Willebrand factor | + | (+) | (+) | |
| Integrins, adhesion molecules | ||||
| CD29 | β1 integrin, VLA-β | +hi | +lo | +hi |
| CD43 | Sialophorin | (+) | — | +/- |
| CD44 | H-CAM | +/- | + | + |
| CD47 | Neurophilin | (+) | (+) | + |
| CD49a | α1 integrin, VLA-1 | + | +/- | + |
| CD49b | α2 integrin, VLA-2 | — | — | + |
| CD49c | α3 integrin, VLA-3 | + | +/- | + |
| CD49d | α4 integrin, VLA-4 | — | — | + |
| CD49e | α5 integrin, VLA-5 | (+) | +/- | + |
| CD49f | α6 integrin, VLA-6 | (+) | — | (+) |
| CD51 | αV integrin, VNR-α | (+) | (+) | + |
| CD61 | β3 integrin | — | — | +/- |
| CD104 | β4 integrin | +/- | — | +/- |
| Miscellaneous | ||||
| CD13 | Aminopeptidase N | + | + | + |
| CD34 | + | (+) | —a | |
| CD36 | GPIIIb | +hi | +/- | +/- |
| CD38 | +lo | +/- | — | |
| CD45 | Leucocyte common R | — | — | — |
| CD90 | Thy-1 | + | + | + |
| CD117 | c-KIT, SCFR | — | — | — |
| CD133 | AC133 | — | — | — |
| HLA ABC | + | + | + | |
| HLA DR | + | — | — | |
| NGFR | — | +/- | — |
+, >90% of the cells positive for the marker; (+), <90% but >10% of the cells positive for the marker; +/-, <10% but >2% of the cells positive for the marker; —, <2% of the cells positive for the marker.
In one donor, expression of CD34 in cultured cells was (+)
Comparative Transcriptional Profiling of CD31- and CD31+ Cells
To determine the relationship between CD31- and CD31+ cells at the transcription level, we compared the global gene expression profile in these subsets for three different donors. Transcripts that were up-regulated more than threefold in either subset were included in the analysis. Supplementary Figure 1, A–C, shows that only a few hundred out of ∼22,000 probed sequences were differentially expressed at the >3-fold level. Of the differentially expressed transcripts, a very large proportion was shared between the three donors (Supplementary Figure 1, D and E). Subsequent analyses were performed only on these shared transcripts. Table 2 and Supplementary Table S2 show that a number of transcripts involved in cell cycle arrest, stem cell biology and development were up-regulated in CD31- cells. Transcripts involved in the biology of adipose tissue, bone, cartilage, muscle, and neuronal tissue were also enriched in this subset. The relative overexpression of the CD44 transcript in CD31- cells confirmed our flow cytometry results (see Table 1).
Table 2.
Selected genes overexpressed in freshly isolated CD31- cells compared with freshly isolated CD31+ cells
| Gene name | Fold difference |
|---|---|
| Cell cycle-related genes | |
| Inhibin, beta A (activin A, activin AB alpha polypeptide) | 18 |
| Growth arrest-specific 1 | 11 |
| Growth arrest-specific 7 | 10 |
| Stem cell- and development-related genes | |
| Zinc finger DAZ interacting protein 1 | 40 |
| Slit homolog | 27 |
| Sema domain, immunoglobulain domain, short basic domain, secreted (semaphorin) 3C | 26 |
| Fibroblast activation protein alpha | 21 |
| Mesencymal stem cell protein DSC54 | 12 |
| Wingless-type MMTV integration site family, member 5A | 9 |
| Dickkopf homolog 2 | 6 |
| Frizzled homolog 7 | 5 |
| Cytokine- and cytokine receptor-related genes | |
| Insulin-like growth factor1 (somatomedin C) | 25 |
| Platelet-derived growth factor receptor-like | 25 |
| Chemokine (C-X-C motif) ligand 14 | 23 |
| Insulin-like growth factor binding protein 6 | 13 |
| Insulin receptor substrate 1 | 8 |
| Chemokine (C-X-C motif) ligand 12 (stromal cell-derived factor 1) | 8 |
| Complement system | |
| Complement component 3 | 29 |
| Complement component 1s | 18 |
| Complement component D (adipsin) | 10 |
| Complement component 7 | 10 |
| Adipose tissue-related | |
| Phospholipase A2, group IIA | 21 |
| LR8 protein (VLDL receptor) | 17 |
| Apolipoprotein D | 12 |
| Phospholipase A2, group IVA | 12 |
| Bone-related | |
| Osteoglycin (osteoinducive factor, mimecan) | 22 |
| Osteomodulin | 21 |
| Cathepsin K | 15 |
| Osteonectin/sparc | 14 |
| Decorin | 13 |
| Cartilage- and extracellular matrix-related | |
| Microfibrillar-associated protein 4 | 83 |
| Fibulin 1 | 50 |
| Dermatopontin | 43 |
| EGF-containing fibulin-like extracellular matrix protein 1 | 31 |
| Glypican 3 | 26 |
| Fibulin 5 | 25 |
| Microfibril-associated glycoprotein-2 | 24 |
| Hyaluronan synthase 1 | 22 |
| Matrix metalloproteinase 2 (gelatinase A, 72 kDa) | 14 |
| Cartilage intermediate layer protein | 14 |
| Collagen, type I, alpha 1 | 14 |
| Collagen, type XIV, alpha 1 (undulin) | 13 |
| Collagen, type VI, alpha 2 | 12 |
| Matrix Gla protein | 11 |
| Nervous system-related | |
| Neuro-oncological ventral antigen1 | 28 |
| Doublecortin and CaM kinase like 1 | 17 |
| Glutamate receptor, ionotrophic, AMPA 3 | 15 |
| Myelin basic protein | 10 |
| Muscle-related | |
| Myosin heavy chain polypeptide 11, smooth muscle | 33 |
| Sarcospan | 15 |
| Laminin, alpha 2 | 14 |
| Phospholamban | 13 |
| Other | |
| KIAA0527 protein | 80 |
| Solute carrier family 22 (extraneuronal monoamine transporter), member 3 | 69 |
| Prostaglandin F receptor | 42 |
| WNT1 inducible signaling pathway protein 2 | 39 |
| Integrin, beta-like 1 (with EGF-like repeat domains) | 37 |
| CD44 antigen (H-CAM) | 11 |
The table presents the 30 most highly overexpressed transcripts plus selected transcripts with presumed biological relationship with some of the most highly overexpressed transcripts. In addition, some transcripts with relevance to biology discussed in the text are presented. If a gene was represented with more than one probe in the list, the probe with the highest degree of differential expression was selected. The values represent the median from three donors.
Table 3 and Supplementary Table S3 show some of the transcripts that were overexpressed in CD31+ cells compared with CD31- cells. Some transcripts associated with stemness and development were overexpressed in CD31+ cells. Surface expression of the putative stemness marker integrin α6 in CD31+, but not in CD31- cells, was confirmed by overexpression of integrin α6 mRNA. Four large groups of transcripts dominated the list of overexpressed transcripts in CD31+ cells, namely transcripts associated with endothelium, transcripts related to the MHC class II complex and antigen presentation, transcripts for some cytokines and cytokine receptors, and transcripts for proteins involved in signal transduction and transcription. Confirming our flow cytometric results, transcripts coding for vascular endothelium cadherin (CD31) and von Willebrand factor were highly enriched in CD31+ cells. The endoglin (CD105) transcript was also moderately overexpressed, in agreement with the moderately higher expression noted for CD105 using the SH2 MAb (Figure 1; Table 1). Complete lists of differentially expressed genes are presented in Supplementary Tables S2 and S3 online.
Table 3.
Selected genes overexpressed in freshly isolated CD31+ cells compared with freshly isolated CD31- cells
| Gene name | Fold difference |
|---|---|
| Stem cell- and development-related genes | |
| SRY (sex determining region Y)-box (Sox) 17 | 43 |
| Mesenchyme homeo box 1 | 27 |
| LIM homeobox 6 | 22 |
| Endothelial-related genes | |
| Vascular epithelium-cadherin (CD144) | 168 |
| Platelet endothelial cell adhesion molecule (PECAM, CD31) | 134 |
| Claudin 5 (endothelial specific component of tight junctions) | 109 |
| Von Willebrand factor | 52 |
| Selectin E | 40 |
| Kinase insert domain receptor (KDR, VEGFR-2) | 28 |
| TEK tyrosine kinase (endothelial) | 24 |
| Ephrin-B2 | 15 |
| Fms-related tyrosine kinase 1 (Flt-1, VEGFR-1) | 13 |
| Angiotensin II receptor-like 1 | 13 |
| Thrombomodulin (CD141) | 9 |
| Endoglin (CD105) | 6 |
| Selectin P | 6 |
| Notch homolog 4 | 6 |
| Eph B4 | 4 |
| MHC and antigen presentation related genes | |
| Major histocompatibility complex, class II DR alpha | 186 |
| Major histocompatibility complex, class II DQ beta 1 | 159 |
| Major histocompatibility complex, class II DQ beta 2 | 74 |
| HLA complex p5 | 48 |
| Major histocompatibility complex, class II DR beta 3 | 28 |
| Major histocompatibility complex, class II DQ alpha 1 | 21 |
| Major histocompatibility complex, class II DP alpha 1 | 19 |
| Invariant chain (CD74 antigen) | 19 |
| Major histocompatibility complex, class II DM alpha | 10 |
| Cytokines and -receptors, growth factors and -receptors | |
| Duffy antigen receptor for chemokines (CD234) (expressed on erythrocytes and endothelium) | 362 |
| Hepatocyte growth factor receptor | 140 |
| G protein-coupled receptor 56 (EGF-TM7-like) | 131 |
| Chemokine (C-C motif) receptor-like 2 | 131 |
| Colony stimulating factor 3 (granulocyte) | 130 |
| Epidermal growth factor-seven transmembrane (EGF-TM7) 1 | 92 |
| Tyrosine kinase with immunoglobulin and epidermal growth factor homology domains | 78 |
| Chemokine (C-C motif) ligand 15 | 52 |
| Interleukin 3 receptor alpha | 35 |
| Signal transduction and transcription related genes | |
| Ectopic viral integration site 1 | 91 |
| RAS guanyl releasing protein 3 (calcium- and DAG-regulated) | 61 |
| Liver receptor homolog 1 (nuclear receptor subfamily 5, group A, member 2) | 59 |
| Cdc42 effector protein 3 | 35 |
| Tumor-associated calcium signal transducer 2 | 30 |
| Others | |
| Alpha 6 integrin (CD49f) | 437 |
| Human immune associated nucleotide 2 | 254 |
| Matrix metalloproteinase 1 (interstitial collagenase) | 74 |
| Glia maturation factor gamma | 57 |
| v-ets erythroblastosis virus E26 oncogene-like (avian) | 51 |
| Myosin VIIA and Rab interacting protein | 48 |
| Carbonic anhydrase IV | 43 |
The table presents the 30 most highly overexpressed transcripts plus selected transcripts with presumed biological relationship with some of the most highly overexpressed transcripts. In addition, some transcripts with relevance to biology discussed in the text are presented. If a gene was represented with more than one probe in the list, the probe with the highest degree of differential expression was selected. The values represent the median from three donors.
Cultured CD31- Cells Display Proliferative and Multilineage Differentiation Capacity In Vitro
By definition, a stem cell needs to meet two criteria, namely the ability to self-renew and capacity to produce at least one mature cell type. To determine whether cells freshly isolated from adipose tissue met the first criterion, we cultured CD31- and CD31+ cells separately under standard ADASC culture conditions (Zuk et al., 2002). The CD31+ cells (Figure 2A) did not plate even after 7 d and remained in suspended aggregates. Under these conditions, these cells did not proliferate. In contrast, freshly isolated CD31- cells appeared as single, small, round cells (Figure 2B). After 1–2 d, ∼50% of the CD31- cells attached to the plastic surface and displayed a fibroblastic-like morphology. The remaining CD31- cells either attached without changing their phenotype or remained in suspension. These cells did not proliferate. After an initial period of apparent quiescence, a proportion of the fibroblastic CD31- cells began to divide with an average population doubling time of 65 h (Figure 2C). Lines of these cells have been kept in continuous culture without any noticeable loss in proliferative capacity for more than 8 mo.
Figure 2.
Photomicrograph of CD31+ (A) and CD31- (B) cells ∼1 h after establishment in cell culture (×400). (C) Typical ADASC at passage 4 (×100).
To assess the multilineage differentiation potential of cultured CD31- cells, polyclonal cell lines generated from four donors were cultured under conditions favorable for adipogenic, osteogenic, chondrogenic, and neurogenic differentiation (Safford et al., 2002; Pochampally et al., 2004). Differentiation to adipogenic (Figure 3A), osteogenic (Figure 3B), and chondrogenic (Figure 3C) phenotypes and to cells expressing Neurofilament 200 (Figure 3D; anti-NF200 labeling) was confirmed by staining assays. However, chondrogenic differentiation based on morphology and toluidine blue staining (Figure 3C) was only clearly positive for one of the four donors. Adipogenic and osteogenic phenotypes were not obtained for all cells in all cultures. In undifferentiated cultured ADASC, real-time RT-PCR analysis detected transcripts coding for osteocalcin, osteomodulin (weakly) and aggrecan, but not for collagen type II or PPARγ2. These results were similar to those obtained for these transcripts in microarray analyses (unpublished data). After differentiation, increased levels of osteomodulin and osteocalcin mRNAs were observed in cells driven toward the osteogenic direction, increased levels of mRNA coding for collagen type II, aggrecan, and osteomodulin were found in chondrogenic cells, and increased level of PPARγ2 mRNA was found in cells differentiated toward adipocytes (Table 4). Collectively, these results indicate that ADASCs can be induced to differentiate into multiple lineages in vitro.
Figure 3.
Staining of polyclonal cultures of ADASC after differentiation in adipogenic direction (Oil-Red O, A), osteogenic direction (Alzarin Red, B), chondrogenic direction (Toluidine blue, C) and neurogenic direction (NF-200, D). Differentiated cells are shown in the left-hand panels and undifferentiated cells in the righthand panels. In D, images obtained in fluorescent light is shown in the top panels, and images obtained in white light are shown in the bottom panels. All images (A–D): magnification ×200.
Table 4.
Relative mRNA expression of molecules characteristic for different lineages.
| Osteomodulin | Osteocalcin | Collagen II | Aggrecan | PPAR γ2 | |
|---|---|---|---|---|---|
| Control | 2 | 1.5 | nd | 5 | nd |
| Osteogenic | 29.7 | 2.4 | nd | 4 | 1 |
| Chondrogenic | 114.9 | 1.3 | 23.2 | 340 | 1.8 |
| Adipogenic | 1 | 1 | 1 | 1 | 111.6 |
mRNA levels were scaled according to the sample with the lowest detectable expression. This value was set to 1. nd, not detected. Adjusted for the concentration of expression of 18S. Values are median of four experiments.
Definitive experiments to determine multipotential differentiation capability were performed on clonal cell cultures. To this end, several monoclonal colonies were generated from single CD31- cells. Notably, only 16% (63/384) of cells cultured in solitude adhered to plastic and displayed the fibroblastic-like morphology observed in polyclonal cultures (unpublished data). Many of these cells failed to produce a CFU-F (>10 cells) after 3 wk, and only 15 clones could be expanded to more than 106 cells (∼20 population doublings). Experiments performed on 12 clones demonstrated that most of them could be differentiated in adipogenic, osteogenic, and neurogenic directions (Figure 4). However, within each clone, the proportion of cells that labeled for the organ specific markers varied considerably. Differentiation in the chondrogenic direction could not be evidenced on cells from these clones. Attempts were also made to assess HSC functionality using CFU assays for several hematopoietic lineages. We found no evidence that these cells possessed HSC properties (unpublished data).
Figure 4.
Staining of differentiated clonal cells. Four different clones obtained from two donors are shown to illustrate differences observed in differentiation between different clones. Staining is for adipogenic differentiation (Oil-Red O, left panels), osteogenic differentiation (Alzarin Red, middle panels), and neurogenic differentiation (anti-NF-200, right panels). Magnification, ×200.
Changes in CD31- Cells during Culture
After expansion in vitro, polyclonal CD31- cells were examined by flow cytometry and microarray analyses and compared with their uncultured predecessors. Expression of surface markers did not differ greatly between uncultured and cultured CD31- cells (Table 1; Supplementary Table S1). The most obvious differences were found within the group of integrins and adhesion molecules, where expression of several molecules was up-regulated in culture. Other key observations were increased expression of the MSC-related marker CD105 and loss of expression of the HSC marker CD34. Comparison of the transcriptomes of the cultured cells with those of the uncultured predecessors showed that the gene expression profile was largely similar (Supplementary Figure 2, A–C). Where genes were differentially expressed, the differences were predominantly the same among the three donors (Supplementary Figure 2). Down-regulated transcripts included molecules involved in cell cycle arrest, some stem cell-related transcripts and, surprisingly, organ- and function-associated transcripts belonging to the cytokine and complement system, adipose tissue, cartilage, bone, and nervous system (Table 5, Supplementary Table S4). Up-regulated transcripts belonged to a variety of functional groups, dominated by transcripts involved in the biology of the cytoskeleton and ECM and transcripts for a number of enzymes (Table 6, Supplementary Table S5).
Table 5.
Selected genes down-regulated in cultured ADASC compared with freshly isolated CD31- cells
| Gene name | Fold change |
|---|---|
| Cell cycle- and growth arrest-related genes | |
| Cyclin-dependent kinase inhibitor 1 C (p57, Kip2) | 32 |
| Growth arrest-specific 7 | 18 |
| Growth arrest and DNA-damage-inducible gamma | 18 |
| Growth arrest and DNA-damage-inducible beta | 14 |
| Stem cell- and development-related genes | |
| Wingless-type MMTV integration site family, member 11 | 75 |
| Frizzled-related protein | 20 |
| Dickkopf homolog 2 | 10 |
| Cytokine- and cytokine receptor—related genes | |
| Chemokine (C-X-C motif) ligand 14 | 1136 |
| Insulin-like growth factor 1 (somatomedin C) | 96 |
| Platelet-derived growth factor receptor-like | 86 |
| Suppressor of cytokine signaling 3 | 52 |
| Chemokine (C-X-C motif) ligand 3 | 31 |
| Interleukin 8 | 27 |
| Complement system | |
| Complement component 7 | 278 |
| Complement component D (adipsin) | 99 |
| Complement component 3 | 69 |
| Complement component H | 31 |
| Adipose tissue—related | |
| Apolipoprotein E | 146 |
| Apolipoprotein D | 128 |
| LR8 protein (VLDL receptor) | 111 |
| Fatty acid binding protein 4 | 52 |
| Apolipoprotein C1 | 43 |
| Peroxisome proliferative activated receptor, gamma (PPARγ) | 19 |
| Bone-related | |
| Osteoglycin (osteoinducive factor, mimecan) | 63 |
| Osteomodulin | 54 |
| Bone morphogenetic protein 2 | 38 |
| Cathepsin K | 9 |
| Cartilage- and extracellular matrix—related | |
| Dermatopontin | 191 |
| SPARC-like 1 | 182 |
| Tenomodulin protein | 50 |
| Cartilage intermediate layer protein | 41 |
| Glypican 3 | 39 |
| Matrix Gla protein | 34 |
| Collagen, type XIV, alpha 1 (undulin) | 15 |
| Nervous system—related | |
| Brain-specific protein | 131 |
| Neuralin 1 | 43 |
| EphA3 | 36 |
| Neuro-oncological ventral antigen 1 | 34 |
| Other genes | |
| FBJ murine osteosarcoma viral oncogene homolog B (Fos-B) | 315 |
| Cell adhesion molecule with homology to L1CAM (close homolog of L1) | 284 |
| H. sapiens mRNA full length insert cDNA clone EUROIMAGE 1630957 | 224 |
| CD14 antigen | 86 |
| WNT1 inducible signaling pathway protein 2 | 84 |
| Alcohol dehydrogenase 1B (class I), beta polypeptide | 84 |
| Periplakin | 70 |
| Myocilin (trabecular meshwork inducible glucocorticoid response) | 79 |
| Carboxypeptidase, vitellogenic-like | 64 |
| N-acetylated alpha-linked acidic dipeptidase 2 | 60 |
| Serine (or cysteine) proteinase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 3 | 56 |
| Inter-alpha trypsin inhibitor heavy chain precursor 5 | 53 |
| RGC32 protein | 52 |
| Bone marrow stromal cell antigen 2 | 40 |
| CD44 antigen | 7 |
| CD34 antigen | 6 |
The table presents the 30 most highly overexpressed transcripts plus selected transcripts with presumed biological relationship with some of the most highly overexpressed transcripts. In addition, some transcripts with relevance to biology discussed in the text are presented. If a gene was represented with more than one probe in the list, the probe with the highest degree of differential expression was selected. The values represent the median from three donors.
Table 6.
Selected genes up-regulated in cultured ADASC compared with freshly isolated CD31- cells
| Gene name | Fold difference |
|---|---|
| Development-related | |
| Forkhead box (FOX) D1 | 24 |
| Odd Oz/Ten-m homolog 3 | 19 |
| Maternal embryonic leucine zipper kinase | 14 |
| Cysteine knot superfamily 1, BMP antagonist 1 | 13 |
| Dickkopf homolog 1 | 10 |
| Wingless-type MMTV integration site family, member 5A | 4 |
| Signal transduction-, growth- and growth regulatory—related genes | |
| Stanniocalcin 2 | 48 |
| Hepatocyte growth factor receptor | 30 |
| Inhibin, beta A (activin A, activin AB alpha polypeptide) | 21 |
| CDC42 effector protein (Rho GTPase binding) 3 | 15 |
| v-myb myeloblastosis viral oncogene homolog (avian)-like 1 | 13 |
| Cell adhesion—related genes N-cadherin (neuronal) | |
| Activated leukocyte cell adhesion molecule (CD166, ALCAM) | 17 |
| Integrin alpha 4 (CD49d) | 8 |
| Bone-related | |
| Osteoprotegerin | 25 |
| Cartilage- and matrix-related | |
| Cartilage oligomeric matrix protein | 56 |
| Collagen, type XI, alpha 1 | 20 |
| Cartilage-linking protein | 18 |
| Proteoglycan 1, secretory granule | 15 |
| Aggrecan 1 | 11 |
| Fibronectin 1 | 10 |
| Cytoskeleton related—genes | |
| Keratin 7 | 288 |
| Keratin 18 | 22 |
| Desmoplakin | 17 |
| Tropomyosin 2 (beta) | 17 |
| Other | |
| NADPH dehydrogenase, quinone 1 | 75 |
| Solute carrier family 7, member 11 | 31 |
| KIAA1199 protein | 23 |
| Potassium channel, subfamily K, member 2 | 19 |
| A desintegrin and metalloproteinase (ADAM) domain 10 | 19 |
| Asp (abnormal spindle)-like, microcephaly-associated | 17 |
| Tumor protein D52-like 1 | 16 |
| Endoglin (CD105) | 4 |
The table presents the 30 most highly overexpressed transcripts plus selected transcripts with presumed biological relationship with some of the most highly overexpressed transcripts. In addition, some transcripts with relevance to biology discussed in the text are presented. If a gene was represented with more than one probe in the list, the probe with the highest degree of differential expression was selected. The values represent the median from three donors.
DISCUSSION
Isolation of Uncultured Stromal Stem Cells from Adipose Tissue
Conventional methods for isolation of multilineage cells from mesodermal tissues have relied on adhesion of stromal cells to plastic and their ensuing expansion in vitro. Their characterization has, therefore, been limited to cultured cells rather than on their uncultured progenitors. We show here that freshly isolated cells with CD45-CD34+CD105+CD31- phenotype are likely to be the precursors from which multilineage cells are derived by expansion of cells from the SVF of adipose tissue (Safford et al., 2002; Woodbury et al., 2002; Zuk et al., 2002; Cowan et al., 2004). Supporting evidence is provided by 1) the fact that cells of the CD31- phenotype adhered to plastic, changed to a fibroblastoid morphology and proliferated extensively under standard ADASC culture conditions in vitro. On culture, CD31- cells performed as previously reported for cultures of ADASC (Safford et al., 2002; Zuk et al., 2002). Cells of the CD31+ phenotype did not proliferate in vitro under these conditions. 2) Cell surface phenotypic characterization of CD31- cells before culture showed similarities with not only cultured ADASC (Zuk et al., 2002; Gronthos et al., 2003), but also with cultured bone marrow–derived stromal stem cells (Pittenger et al., 1999; unpublished observations in our lab). 3) Under appropriate conducive conditions, they change to cells with characteristics of bone, cartilage, and adipose tissue and cells expressing Neurofilament 200 in vitro. 4) Clonal analyses eliminated the possibility that CD31- cells were merely comprised of polyclonal populations of lineage committed progenitor cells, and support our conclusion that these cells represent stromal stem cells in adipose tissue. Nonetheless, the phenotype and gene expression profile of CD31- and CD31+ cells are sufficiently similar to suggest ontogenetic linkage.
Despite the morphologically homogenous appearance of the CD31- cells, an element of heterogeneity of the subset was evidenced in 1) expression patterns for particular cell surface markers, 2) ability of only some of the CD31- cells to adhere to plastic and subsequently proliferate, and 3) the differential capacity of clonal cell lines to differentiate. This heterogeneity in growth and differentiation potential, including the failure of several clones to differentiate toward a chondrogenic lineage, is similar to that described for bone marrow stromal stem cells (Pittenger et al., 1999; Gronthos et al., 2003). Suboptimal culture conditions (Gronthos et al., 2003), contamination with lineage restricted precursor cells and loss of multilineage potential after a given number of population doublings (Pittenger et al., 1999) may explain the differences observed between clones.
How Do ADASCs Compare with Uncultured Stem Cells from Other Sources?
Phenotypic comparison of uncultured ADASCs with previously published uncultured stem cell populations was only possible for some markers owing to difficulties observed in the previously published studies in obtaining sufficient numbers of cells of the other uncultured stem cell populations for extensive analysis. Morphologically, uncultured ADASCs resemble the RS-1 population of uncultured bone marrow–derived MSCs (Colter et al., 2000) more than the STRO-1BRIGHTVCAM+ from bone marrow (Gronthos et al., 2003). Furthermore, human bone marrow–derived MSCs express CD106 (VCAM-1; Gronthos et al., 2003), a molecule not detected on the cells described here. Expression of NGFR has been used to select cells with clonogenic properties among MSCs (Quirici et al., 2002). In the present study, a small proportion of CD31- cells expressed NGFR. Interestingly, CD34, a marker of human HSCs also expressed on uncultured murine hair follicle stem cells (Morris et al., 2004), was expressed on most CD31- cells in the current study. Moreover, murine hair follicle stem cells express α-6 integrin (CD49f), a marker apparently common to several stem cell populations (Fortunel et al., 2003), and β-1 integrin (CD29; Morris et al., 2004). Both of these markers were expressed at high levels in CD31+ cells. CD31- cells, however, expressed low levels of CD29 and no CD49f. Therefore, even though uncultured ADASCs share some characteristics with other uncultured adult stem cell populations, extensive characterization reveal that they display a unique phenotype.
Comparison between CD31- and CD31+ Adipose Tissue–derived Cells
The most prominent difference between uncultured CD31+ and CD31- cells was the cell surface and/or transcript expression by CD31+ cells of molecules associated with endothelial cells. These included VEGFR2 (albeit with very low expression on the cell surface) and VEGFR1, for E and P selectin (McGill et al., 1998), and transcripts for arterial (EphrinB2, Notch 4) and venous endothelium (EphB4) (Chi et al., 2003). Moreover, CD31+ cells overexpressed transcripts for several HLA class II molecules, and uniquely expressed HLA DR on the cell surface, a feature previously described for renal microvascular endothelial cells (Muczynski et al., 2003). Thus, compared with previously published gene expression profiles, the CD31+ cells in this study most closely resemble microvascular cells, with transcripts typical of both the arterial and the venous side (Chi et al., 2003). Surprisingly von Willebrand factor, a molecule with a very restricted expression profile including predominantly endothelial cells, was expressed by all CD31+ cells, but also by most of the CD31- cells.
By comparison, CD31- cells overexpressed transcripts associated with different mesodermal organs. As could be expected, we observed overexpression of transcripts related to fat metabolism. The high level of expression of several complement components is also likely to be related to the function of these cells as precursors of adipocytes (Kershaw and Flier, 2004). However, we generally failed to find transcripts associated with the endocrine function of adipose tissue (Kershaw and Flier, 2004). We also observed a high level of expression of several transcripts associated with ECM, suggesting a role for these immature cells in matrix production in adipose tissue. CD31- cells also overexpressed transcripts related to bone, cartilage and muscle biology. Several of these transcripts have been shown to be expressed early in osteoblast differentiation (osteoglycin, osteomodulin, decorin, collagens type I and VI; Balint et al., 2003), whereas cathepsin K is the most important osteoabsorbtive enzyme produced by osteoclasts (Vaaraniemi et al., 2004). Osteoblasts originate from stromal lineages, whereas osteoclasts derive from bone marrow hematopoietic cells. Finally, we also observed overexpression of transcripts typical of neuronal tissue, which developmentally derives from ectoderm. The differentiation plasticity of CD31- cells, extending across germ layer boundaries, therefore appears to be reflected in their transcriptional signature.
Changes in CD31- Cells Induced by Cell Culture
Comparative profiling of fresh ADASC with their cultured counterparts provided a unique insight into the changes that stromal stem cells undergo during culture. Culture was found to only marginally affect the cell surface phenotype, the most prominent changes being down-regulation of CD34 and up-regulation of certain integrins and adhesion molecules. Also, cultured ADASCs expressed higher levels of CD105 on the cell surface. Comparison with the published phenotype of bone marrow–derived MSC demonstrate that the two cultured stromal stem cell populations are predominantly similar, but that some notable differences exist (Pittenger et al., 1999). There is a general tendency for MSC to express some cytokine receptors not found on ADASC (receptors for tumor necrosis factor and interleukins 4, 6, and 7). Differences are also found among the adhesion molecules, where ADASC were found to express CD49a and CD49d, which were not expressed by MSC, whereas MSC express CD58 and CD106, which were not expressed by ADASC. Pittenger et al. (1999) did not observe von Willebrand factor on MSC. Using intracellular staining analysis, we found von Willebrand factor in ADASC. However, this difference may only reflect different staining strategies. These results confirm and extend observations made by Zuk et al. (2002).
At the gene transcript level, the most prominently up-regulated transcript in cultured ADASC was keratin 7, and some other transcripts associated with the cytoskeleton were also up-regulated. This strongly suggests that adherence to plastic introduces considerable changes in the composition of the cytoskeleton. Up-regulation of some of the transcripts coding for adhesion molecules confirmed our observation of increased cell surface expression of adhesion molecules. An indication of the degree of correlation between transcript and cell surface expression was noted for CD105 (endoglin): a fourfold up-regulation of this transcript corresponded to an increase from moderate expression on the surface of uncultured CD31- cells to high expression on the cultured cells. Overall, however, transcripts up-regulated in cultured CD31- cells did not suggest differentiation toward a specific lineage.
Conversely, a number of transcripts were down-regulated following culture. As expected, these included transcripts involved in cell cycle and growth arrest. Additionally, many tissue-specific transcripts expressed in uncultured ADASCs were down-regulated, such as transcripts related to properties of adipose tissue, osteoblastogenesis, ECM formation, neuronal function and cytokine signaling. Interestingly, transcripts found to be down-regulated upon culture were also found to be down-regulated in CD31+ cells compared with uncultured CD31- cells. This observation further supports the notion of an ontogenetic linkage between CD31- and CD31+ cells, with the CD31- ADASCs the less committed cell population.
In summary, we show that cells with CD45-CD34+CD105+ CD31- phenotype derived from the SVF of adipose tissue harbor stem cell properties. Under appropriate conducive conditions, polyclonal populations of these cells change to cells with characteristics of bone, cartilage, adipose, and neuronal-like tissues in vitro. Clonal cell lines displayed both proliferative and differentiation capacity and thus support our conclusion that these cells represent the adipose tissue–derived stem cells. Nevertheless, we found that the cultured cells differ from their uncultured counterparts. Whether these changes interfere with the stemness of these cells, or just represent the consequence of in vitro culture adherent to plastic surfaces, remains to be investigated. In either case these are cells which can be easily obtained in vivo, expanded in vitro to large numbers and probably be differentiated to several different functional end organ specific cells. Consequently, these are cells with great potential for therapeutic tissue regeneration.
Supplementary Material
Acknowledgments
We are grateful to Dr. Eric Dillerud (Fornebuklinikken, Oslo) for generously providing liposuction material, to professor Finn P. Reinholt (Institute of Pathology, Rikshospitalet) for histochemistry and immunohistochemistry, and to Marianne Enger for cell sorting. This work was supported through the Norwegian Center for Stem Cell Research by the Research Council of Norway, the Norwegian Cancer Society, the University of Oslo, and Gidske og Peter Jacob Sørensens Foundation for the Promotion of Science.
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E04-10-0949) on January 5, 2005.
Abbreviations used: ADASC, adipose tissue-derived adult stem cells; MSC, mesenchymal stem cells; CFU-F, colony-forming units-fibroblasts; SVF, stromal vascular fraction; ECM, extracellular matrix; HSC, hematopoietic stem cells; NGFR, nerve growth factor receptor.
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
References
- Arai, F., Ohneda, O., Miyamoto, T., Zhang, X. Q., and Suda, T. (2002). Mesenchymal stem cells in perichondrium express activated leukocyte cell adhesion molecule and participate in bone marrow formation. J. Exp. Med. 195, 1549-1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balint, E., Lapointe, D., Drissi, H., van der Meijden, C., Young, D. W., Van Wijnen, A. J., Stein, J. L., Stein, G. S., and Lian, J. B. (2003). Phenotype discovery by gene expression profiling: mapping of biological processes linked to BMP-2-mediated osteoblast differentiation. J. Cell. Biochem. 89, 401-426. [DOI] [PubMed] [Google Scholar]
- Chi, J. T. et al. (2003). Endothelial cell diversity revealed by global expression profiling. Proc. Natl. Acad. Sci. USA 100, 10623-10628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colter, D. C., Class, R., DiGirolamo, C. M., and Prockop, D. J. (2000). Rapid expansion of recycling stem cells in cultures of plastic-adherent cells from human bone marrow. Proc. Natl. Acad. Sci. USA 97, 3213-3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowan, C. M., Shi, Y. Y., Aalami, O. O., Chou, Y.F., Mari, C., Thomas, R., Quarto, N., Contag, C. H., Wu, B., and Longaker, M. T. (2004). Adipose-derived adult stromal cells heal critical-size mouse calvarial defects. Nat. Biotechnol. 22, 560-567. [DOI] [PubMed] [Google Scholar]
- Fortunel, N. O. et al. (2003). Comment on “`Stemness': transcriptional profiling of embryonic and adult stem cells” and “a stem cell molecular signature.” Science 302, 393. [DOI] [PubMed] [Google Scholar]
- Gronthos, S., Zannettino, A. C., Hay, S. J., Shi, S., Graves, S. E., Kortesidis, A., and Simmons, P. J. (2003). Molecular and cellular characterisation of highly purified stromal stem cells derived from human bone marrow. J. Cell Sci. 116, 1827-1835. [DOI] [PubMed] [Google Scholar]
- Howell, J. C., Lee, W. H., Morrison, P., Zhong, J., Yoder, M. C., and Srour, E. F. (2003). Pluripotent stem cells identified in multiple murine tissues. Ann. NY Acad. Sci. 996, 158-173. [DOI] [PubMed] [Google Scholar]
- Jiang, Y. et al. (2002). Pluripotency of mesenchymal stem cells derived from adult marrow. Nature 418, 41-49. [DOI] [PubMed] [Google Scholar]
- Jones, E. A., Kinsey, S. E., English, A., Jones, R. A., Straszynski, L., Meredith, D. M., Markham, A. F., Jack, A., Emery, P., and McGonagle, D. (2002). Isolation and characterization of bone marrow multipotential mesenchymal progenitor cells. Arthritis Rheum. 46, 3349-3360. [DOI] [PubMed] [Google Scholar]
- Kershaw, E. E. and Flier, J. S. (2004). Adipose tissue as an endocrine organ. J. Clin. Endocrinol. Metab. 89, 2548-2556. [DOI] [PubMed] [Google Scholar]
- Loges, S. et al. (2004). Identification of the adult human hemangioblast. Stem Cells Dev. 13, 229-242. [DOI] [PubMed] [Google Scholar]
- Martin, I., Jakob, M., Schafer, D., Dick, W., Spagnoli, G., and Heberer, M. (2001). Quantitative analysis of gene expression in human articular cartilage from normal and osteoarthritic joints. Osteoarthritis Cartilage 9, 112-118. [DOI] [PubMed] [Google Scholar]
- McGill, S. N., Ahmed, N. A., and Christou, N. V. (1998). Endothelial cells: role in infection and inflammation. World J. Surg. 22, 171-178. [DOI] [PubMed] [Google Scholar]
- Morris, R. J., Liu, Y., Marles, L., Yang, Z., Trempus, C., Li, S., Lin, J. S., Sawicki, J. A., and Cotsarelis, G. (2004). Capturing and profiling adult hair follicle stem cells. Nat. Biotechnol. 22, 411-417. [DOI] [PubMed] [Google Scholar]
- Muczynski, K. A., Ekle, D. M., Coder, D. M., and Anderson, S. K. (2003). Normal human kidney HLA-DR-expressing renal microvascular endothelial cells: characterization, isolation, and regulation of MHC class II expression. J. Am. Soc. Nephrol. 14, 1336-1348. [DOI] [PubMed] [Google Scholar]
- Okuno, Y., Iwasaki, H., Huettner, C. S., Radomska, H. S., Gonzalez, D. A., Tenen, D. G., and Akashi, K. (2002). Differential regulation of the human and murine CD34 genes in hematopoietic stem cells. Proc. Natl. Acad. Sci. USA 99, 6246-6251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittenger, M. F., Mackay, A. M., Beck, S. C., Jaiswal, R. K., Douglas, R., Mosca, J.D., Moorman, M. A., Simonetti, D. W., Craig, S., and Marshak, D. R. (1999). Multilineage potential of adult human mesenchymal stem cells. Science 284, 143-147. [DOI] [PubMed] [Google Scholar]
- Pochampally, R. R., Smith, J. R., Ylostalo, J., and Prockop, D. J. (2004). Serum deprivation of human marrow stromal cells (hMSCs) selects for a subpopulation of early progenitor cells with enhanced expression of OCT-4 and other embryonic genes. Blood 103, 1647-1652. [DOI] [PubMed] [Google Scholar]
- Quirici, N., Soligo, D., Bossolasco, P., Servida, F., Lumini, C., and Deliliers, G. L. (2002). Isolation of bone marrow mesenchymal stem cells by anti-nerve growth factor receptor antibodies. Exp. Hematol. 30, 783-791. [DOI] [PubMed] [Google Scholar]
- Safford, K. M., Hicok, K. C., Safford, S. D., Halvorsen, Y. D., Wilkison, W. O., Gimble, J. M., and Rice, H. E. (2002). Neurogenic differentiation of murine and human adipose-derived stromal cells. Biochem. Biophys. Res. Commun. 294, 371-379. [DOI] [PubMed] [Google Scholar]
- Vaaraniemi, J., Halleen, J. M., Kaarlonen, K., Ylipahkala, H., Alatalo, S. L., Andersson, G., Kaija, H., Vihko, P., and Vaananen, H. K. (2004). Intracellular machinery for matrix degradation in bone-resorbing osteoclasts. J. Bone Miner. Res. 19, 1432-1440. [DOI] [PubMed] [Google Scholar]
- Woodbury, D., Reynolds, K., and Black, I. B. (2002). Adult bone marrow stromal stem cells express germline, ectodermal, endodermal, and mesodermal genes prior to neurogenesis. J. Neurosci. Res. 69, 908-917. [DOI] [PubMed] [Google Scholar]
- Zuk, P. A., Zhu, M., Ashjian, P., De Ugarte, D. A., Huang, J. I., Mizuno, H., Alfonso, Z. C., Fraser, J. K., Benhaim, P., and Hedrick, M. H. (2002). Human adipose tissue is a source of multipotent stem cells. Mol. Biol. Cell 13, 4279-4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuk, P. A., Zhu, M., Mizuno, H., Huang, J., Futrell, J. W., Katz, A. J., Benhaim, P., Lorenz, H. P., and Hedrick, M. H. (2001). Multilineage cells from human adipose tissue: implications for cell-based therapies. Tissue Eng. 7, 211-228. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.