Abstract
J-domain cochaperones confer functional specificity to their heat shock protein (HSP)70 partner by recruiting it to specific substrate proteins. To gain insight into the functions of plastidic HSP70s, we searched in Chlamydomonas databases for expressed sequence tags that potentially encode chloroplast-targeted J-domain cochaperones. Two such cDNAs were found: the encoded J-domain proteins were named chloroplast DnaJ homolog 1 and 2 (CDJ1 and CDJ2). CDJ2 was shown to interact with a ∼28-kDa protein that by mass spectrometry was identified as the vesicle-inducing protein in plastids 1 (VIPP1). In fractionation experiments, CDJ2 was detected almost exclusively in the stroma, whereas VIPP1 was found in low-density membranes, thylakoids, and in the stroma. Coimmunoprecipitation and mass spectrometry analyses identified stromal HSP70B as the major protein interacting with soluble VIPP1, and, as confirmed by cross-linking data, as chaperone partner of CDJ2. In blue native-PAGE of soluble cell extracts, CDJ2 and VIPP1 comigrated in complexes of >>669, ∼150, and perhaps ∼300 kDa. Our data suggest that CDJ2, presumably via coiled-coil interactions, binds to VIPP1 and presents it to HSP70B in the ATP state. Our findings and the previously reported requirement of VIPP1 for the biogenesis of thylakoid membranes point to a role for the HSP70B/CDJ2 chaperone pair in this process.
INTRODUCTION
Chaperones of the heat shock protein (Hsp)70 family belong to the most conserved proteins known. Except for some Archaea, Hsp70s are found in all known organisms and are present in every compartment of the eukaryotic cell (Bukau and Horwich, 1998). Principally, Hsp70s consist of an N-terminal ATPase domain and a C-terminal substrate-binding domain. ATP hydrolysis at the ATPase domain regulates substrate binding and release. Substrate proteins recognized by Hsp70 expose hydrophobic regions, a characteristic feature not only of nonnative proteins, but also of native Hsp70 substrates. Binding of Hsp70 to hydrophobic regions prevents the formation of aggregates. In addition, the intrinsic secondary amide peptide bond cis-trans isomerase activity recently detected for DnaK (the Hsp70 of Escherichia coli) may introduce conformational changes to bound substrates that eventually allow nonnative proteins to reconvert to the native state (Schiene-Fischer et al., 2002). Thus, Hsp70s play a major role in the folding of nascent chains and in the renaturation of nonnative proteins that have accumulated during stress situations such as heat shock (Frydman, 2001). However, they also are involved in many highly specialized functions such as the regulation of the general stress response (Tomoyasu et al., 1998), the uncoating of clathrincoated vesicles (Ungewickell et al., 1995), or the translocation of proteins across membranes (Kang et al., 1990).
Specificity of Hsp70 function is mediated largely by its cochaperones, of which the J-domain cochaperones represent an important class. J-domain cochaperones contain a highly conserved J-domain that is responsible for the interaction with Hsp70. In addition, these cochaperones contain domains typical for protein–protein interactions, such as zinc finger or coiled-coil domains, by which specific substrates are bound (Cyr et al., 1994; Szabo et al., 1996; Miernyk, 2001). These substrates are then presented to the substrate binding domain of a specific Hsp70 chaperone. One Hsp70 may be recruited by several different J-domain cochaperones to fulfill a variety of specific tasks. In the Arabidopsis thaliana genome, for example, at least 89 genes encoding J-domain proteins (Miernyk, 2001) cooperate with only 12 Hsp70s (Sung et al., 2001).
Three Hsp70 systems have been identified in the chloroplast (Schroda, 2004). Two of these, Com70 and Hsp70 IAP, seem to be involved in the import of precursor proteins into the chloroplast (Schnell et al., 1994; Kourtz and Ko, 1997). The third plastidic Hsp70 system is located to the stroma. Stromal Hsp70 has been shown to cooperate with Cpn60 in the folding of proteins newly imported into the chloroplast (Madueño et al., 1993; Tsugeki and Nishimura, 1993; Bonk et al., 1996) and indirect evidence suggested that it also is involved in the refolding of denatured proteins (Schroda et al., 2001b).
Also chloroplast-specific functions can be attributed to stromal Hsp70. In Chlamydomonas, stromal HSP70B was shown to increase the cell's ability to cope with photoinhibition, possibly by either protecting photosystem II from irreversible photodamage or by stabilizing photodamaged photosystem II for a coordinated exchange of damaged D1 protein for de novo-synthesized D1 (Schroda et al., 1999, 2001a). Other specialized functions of stromal Hsp70 were deduced from functions attributed to plastidic DnaJ-like proteins. The maize bsd2 mutant impaired in Rubisco assembly was shown to lack a protein exhibiting high sequence similarity to the zinc finger domain of DnaJ proteins (Brutnell et al., 1999). Bsd2 was hypothesized to act together with ATJ11, which consists essentially of a J-domain to recruit a stromal Hsp70 for the folding of Rubisco subunits (Orme et al., 2001). An Arabidopsis mutant impaired in chloroplast division was shown to be defective in the ARC6 J-domain–like protein. ARC6 spans the inner envelope membrane and exposes its J-domain to the stroma, where it may recruit a stromal Hsp70 for a function in chloroplast division (Vitha et al., 2003).
Evidently, insight into the functions of chloroplast Hsp70s may be gained by the functional characterization of chloroplast-localized J-domain proteins. For this purpose, we have identified expressed sequence tags (ESTs) from Chlamydomonas databases that encode proteins harboring both a putative chloroplast transit peptide and a J-domain. In this study, we show that the chloroplast-targeted J-domain protein CDJ2 interacts with the vesicle-inducing protein in plastids 1 (VIPP1) and that both CDJ2 and VIPP1 interact with stromal HSP70B. VIPP1 (or IM30) was first described as a 30-kDa protein associated with the inner envelope and the thylakoid membranes of pea chloroplasts (Li et al., 1994). From this unusual localization, the authors proposed that VIPP1 might be involved in lipid transfer from the inner envelope to the thylakoids, perhaps by vesicle traffic (Li et al., 1994). Consistent with this hypothesis is the phenotype of the Arabidopsis hcf155 mutant, which expresses the VIPP1 gene at significantly lower levels, and, as a result, exhibits dramatic defects in the structure of its thylakoid membranes (Kroll et al., 2001). In addition, vesicle budding from the inner envelope was found to be abolished in hcf155. Also, in Synechocystis cells that expressed VIPP1 at low levels, distorted thylakoids were observed (Westphal et al., 2001). The data presented here suggest that the stromal HSP70B/CDJ2 chaperone pair might play a role in the functional cycle of VIPP1 required for the biogenesis/maintenance of thylakoid membranes.
MATERIALS AND METHODS
Strains and Culture Conditions
Chlamydomonas reinhardtii strains were grown mixotrophically in TAP medium (Harris, 1989) on a rotatory shaker at 25°C at ∼30 μE m-2 s-1. For chloroplast isolation, cells were grown in TAP medium supplemented with 0.5% peptone.
Heat Shock and Dark-to-Light Shift Kinetics, RNA and Protein Extractions, RNA Gels, and Hybridizations
For heat shock kinetics, Chlamydomonas cc124 cells were grown in 160 ml of TAP medium to a density of 6 × 106 cells/ml, harvested by centrifugation, and resuspended in 100 ml of TAP medium prewarmed to 40°C. For dark-to-light shift, Chlamydomonas cc124 cells were grown to a density of 6 × 106 cells/ml, incubated in the dark for 16 h, and transferred to dim light (∼30 μE m-2 s-1). Samples were taken at different time points and cooled by adding ice. Cells were harvested by centrifugation and resuspended in 600 μl of TAP medium, of which 100 μl for protein analyses were centrifuged; resuspended in 40 μl of 0.1 M NaCO3, 0.1 M dithiothreitol; and solubilized by adding 55 μl of 5%SDS, 30% sucrose. For RNA extraction, 500 μl of lysis buffer (100 mM Tris-HCl, pH 8.0, 600 mM NaCl, 4% SDS, and 10 mM EDTA) was added to the remaining 500 μl of cell suspension, followed by incubation at 65°C for 10 min. SDS was precipitated by adding 132 μl of 2 M KCl, incubation on ice for 15 min, and centrifugation. Remaining proteins were extracted once with phenol/chloroform and once with chloroform. After addition of 0.3 volumes of 8 M LiCl and incubation at 4°C overnight, RNA was pelleted by centrifugation. RNA was resuspended in diethylpyrocarbonate (DEPC)-treated water, precipitated with ethanol, and resuspended in DEPC-treated water. RNA (10 μg) per lane was separated on 1.2% formaldehyde agarose gels (Sambrook et al., 1989), incubated for 30 min with 50 mM NaOH, 10 mM NaCl and for 10 min with 100 mM Tris-HCl, pH 7.5, and blotted to Hybond-N membranes (Amersham Biosciences, Freiburg, Germany) by capillary transfer with 25 mM phosphate buffer, pH 6.5. After baking for 2 h at 80°C, membranes were hybridized with DNA probes prepared by the random priming technique (Feinberg and Vogelstein, 1983) by using [α-32P]dCTP (Amersham Biosciences). Hybridization and quantitation was done as described previously (Schroda et al., 1999). Probes used were a 2-kb NheI-AatII fragment containing the HSP70B coding region, a 1.9-kb SpeI-XhoI fragment containing the CDJ1 cDNA (AV626034), a 1.35-kb polymerase chain reaction (PCR) product containing the VIPP1 coding region and 3′ untranslated region, and a 1-kb cDNA of cβlp2 (Von Kampen et al., 1994).
Polyacrylamide Electrophoreses and Gel Blot Analyses
Blue native (BN)-PAGE for soluble proteins was carried out according to a published protocol (Schägger et al., 1994). The native high-molecular-weight marker (66–669 kDa) was purchased from Amersham Biosciences. SDS-PAGE was performed as described previously (Laemmli, 1970). For heat shock kinetics, proteins were loaded on the basis of equal chlorophyll concentrations. For fractionation experiments, 1 volume of 2× Laemmli sample buffer (125 mM Tris-HCl, pH 6.8, 20% glycerol, 4% SDS, 10% β-mercapto-ethanol, and 0.005% bromphenol blue) was added to the samples, and protein concentrations were determined by amido black (Popov et al., 1975).
Proteins in gels were stained with silver nitrate or transferred to nitrocellulose membranes (Hybond-ECL; Amersham Biosciences) by semidry blotting by using a discontinuous transfer system. Blocking and immunodecorations were performed in phosphate-buffered saline (Sambrook et al., 1989) containing 3% nonfat dry milk, immunodetection was done by enhanced chemiluminescence (Amersham Biosciences). Antisera described previously were against HSP70B (Schroda et al., 1999), CGE1 (Schroda et al., 2001b), mitochondrial carboanhydrase (Eriksson et al., 1996), and Cytf (Pierre and Popot, 1993).
Immunoprecipitations
Chlamydomonas cc124 was grown to a density of ∼5 × 106 cells/ml, harvested by centrifugation, and resuspended in lysis buffer (20 mM HEPES, pH 7.2, 10 mM KCl, 1 mM MgCl2, 154 mM NaCl, 10% glycerol, and 0.25× protease inhibitor cocktail; Roche Diagnostics, Mannheim, Germany). For the experiment in Figure 6, cells were split in two parts, to one-half carbonyl cyanide p-trifluoromethoxyphenylhydrazone (FCCP) at a final concentration of 20 μM was added, and then both halves were incubated on a shaker for 10 min at 25°C. To the FCCP-treated cells apyrase was added at a concentration of 0.3 U/ml; to nontreated cells Mg-ATP was added to a final concentration of 2.5 mM, and both halves were incubated on ice for 10 min before sonication. For all experiments, cells were sonicated on ice for 90 s. Lysates were loaded onto sucrose cushions (20 mM HEPES-KOH, pH 7.2, 0.6 M sucrose) and centrifuged in a TI50 rotor for 30 min at 152,000 × g and 4°C. For the experiments in Figures 5 and 6, supernatants were supplied with Triton X-100 to a final concentration of 0.5%. Protein A-Sepharose beads with coupled antibodies were equilibrated in lysis buffer and incubated with the cell lysates under agitation for 1 h at 4°C. Beads were washed four times with lysis buffer (containing 0.1% Triton X-100) and once with 10 mM Tris-HCl, pH 7.5, and proteins were eluted by boiling 45 s in 1× Laemmli sample buffer (Figure 3) or by shaking 30 min at 25°C with 1× Laemmli sample buffer lacking β-mercaptoethanol (Figures 5 and 6).
Figure 6.
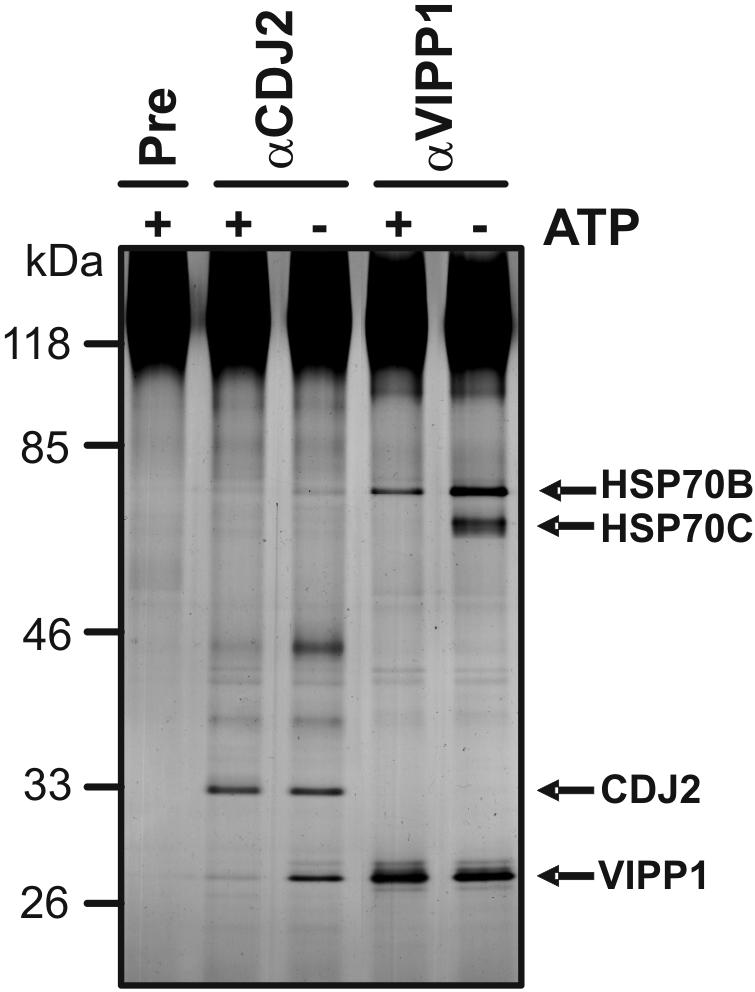
ATP dependence of the VIPP1–CDJ2–HSP70B interaction. Soluble extract from Chlamydomonas cells that were depleted from ATP or repleted with 2.5 mM Mg-ATP was incubated with protein A-Sepharose coupled to antibodies of either preimmune (Pre), anti-CDJ2, or anti-VIPP1 serum. Precipitated proteins were separated on a 7.5–15% SDS-polyacrylamide gel under nonreducing conditions and visualized by silver staining.
Figure 5.
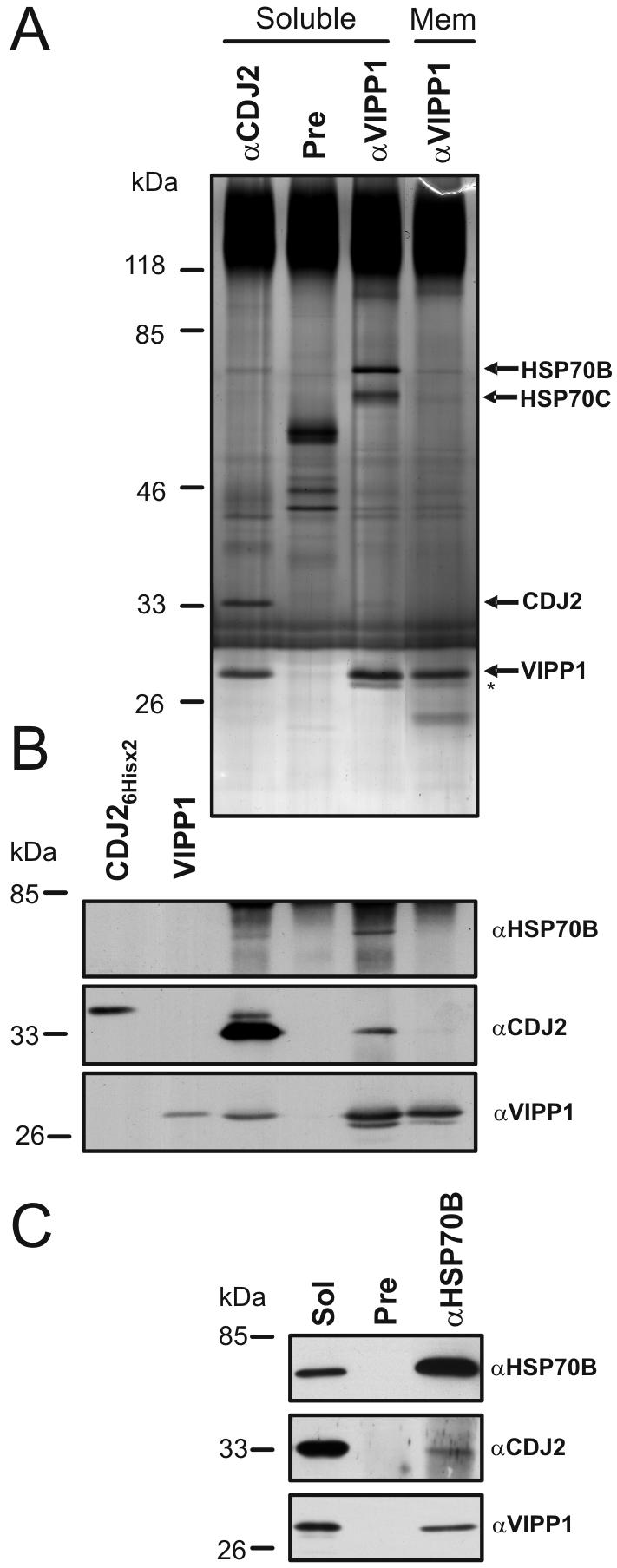
Immunoprecipitation of CDJ2, VIPP1, and HSP70B. (A) Chlamydomonas soluble extract from ∼1.5 × 1010 cells was incubated with protein A-Sepharose coupled to antibodies of either preimmune (Pre), anti-CDJ2, or anti-VIPP1 serum, and membranes solubilized with Triton X-100 (Mem) were incubated with anti-Vipp1 antibodies coupled to protein A-Sepharose. Precipitated proteins were separated on a 7.5–15% SDS-polyacrylamide gel under nonreducing conditions and visualized by silver staining. The asterisk indicates a slightly faster migrating VIPP1 form. (B) Proteins from the immunoprecipitation shown in A in addition to heterologously expressed CDJ2 (containing N- and C-terminal hexahistidine tags) and VIPP1 were separated on a 7.5–15% SDS-polyacrylamide gel, transferred to nitrocellulose, and immunodecorated with antibodies against HSP70B, CDJ2, and VIPP1. Note that the CDJ2 detection was exposed much longer than the VIPP1 detection. (C) Chlamydomonas total soluble protein (Sol) was incubated with protein A-Sepharose coupled to antibodies of Pre or anti-HSP70B serum. Total soluble and precipitated proteins were separated on a 7.5–15% SDS-poly-acrylamide gel, transferred to nitrocellulose, and immunodecorated with antibodies against HSP70B, CDJ2, and VIPP1.
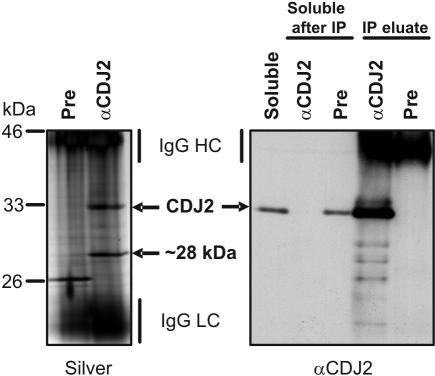
Figure 3.
Immunoprecipitation of CDJ2. Six milliliters of a soluble extract from ∼1010 Chlamydomonas cells were incubated with protein A-Sepharose coupled to antibodies of either preimmune serum (Pre) or anti-CDJ2 serum. Precipitated proteins were eluted under reducing conditions. Proteins were separated on a 7.5–15% SDS-poly-acrylamide gel and visualized by silver staining (left gel). Ten microliters of each of the soluble proteins before (Soluble) and after incubation with preimmune or anti-CDJ2 sera (Soluble after IP) and the proteins eluted from the immunoprecipitations (IP eluate) was separated on a 7.5–15% SDS-polyacrylamide gel, transferred to nitrocellulose, and immunodecorated with anti-CDJ2 antibodies (right gel). The positions of IgG heavy chains (HC) and light chains (LC) are indicated.
Cell Fractionations
All fractionations were carried out with cw15 strain CF185 (Schroda et al., 1999). Cells were grown to ∼5 × 106 cells/ml, harvested by centrifugation, and resuspended in 10 mM Tris-HCl, pH 7.6, 0.25× protease inhibitor cocktail. Cells were split into two parts and ruptured by either sonication on ice or four cycles of freeze-thawing. Broken cells were centrifuged in a TLA 100.2 rotor for 1 h at 355,000 × g and 4°C. The supernatant was regarded as soluble proteins and the pellet after resuspension in the initial volume of Tris-HCl buffer as membranes. Isolation of chloroplasts and fractionation into stroma, thylakoids, and low-density membranes was done as described previously (Zerges and Rochaix, 1998). Mitochondria were isolated following a published protocol (Eriksson et al., 1995).
Mass Spectrometry Analyses
Proteins in gels were visualized with Coomassie or silver staining and bands were excised and treated with trypsin before extraction of peptides for analysis by microliquid chromatography tandem mass spectrometry (μLC-MSMS) (Shevchenko et al., 1996). Samples were analyzed by μLC-MSMS with data-dependent acquisition (LCQ-DECA; Thermo Finnigan, San Jose, CA) after dissolution in 5 μl of 70% acetic acid (vol/vol). A reverse-phase column (200 μm × 10 cm; PLRP/S 5 μm, 300 Å; Michrom Biosciences, San Jose, CA) was equilibrated for 10 min at 1.5 μl/min with 95% A, 5% B (A, 0.1% formic acid in water; B, 0.1% formic acid in acetonitrile) before sample injection. A linear gradient was initiated 10 min after sample injection ramping to 60% A, 40% B after 50 min and 20% A, 80% B after 65 min. Column eluent was directed to a coated glass electrospray emitter (TaperTip, TT150-50-50-CE-5; New Objective, Woburn, MA) at 3.3 kV for ionization without nebulizer gas. The mass spectrometer was operated in “triple-play” mode with a survey scan (400–1500 m/z), data-dependent zoom scan, and MSMS with exclusion of singly charged ions (Whitelegge, 2003). Individual sequencing experiments were matched to a custom Chlamydomonas sequence database downloaded from JGI (http://www.jgi.doe.gov/) by using Sequest software (Thermo Finnigan). The search was run under the “no enzyme” mode to identify nontryptic peptides. The results of Sequest searches were carefully scrutinized. MSMS spectra of doubly charged ions with cross-correlation scores (Xcorr) >2.8, and triply charged ions with scores >3.2 were examined manually. Some noisy spectra were discarded despite high Xcorr scores. Nontryptic peptide returns were retained only if the data looked of especially high signal to noise.
Cloning, Expression, and Purification of CDJ2, CDΔ2, and VIPP1
The coding region of CDJ2 was amplified by PCR from cDNA clone AV387908 with primers 5′-AAGGATCCATGGCAGCGAAGAACTTCTACGAC-3′ and 5′-CGTGGGTAACCTAGCTTATTGAGCTTCTTCTTGAGC-3′. The ∼1-kb PCR product was digested with BamHI and BstEII and cloned into BamHI-BstEII–digested pCB785 (a pQE-9 derivative containing the HSP70B coding region cloned into BamHI and BstEII restriction sites), giving pMS254. The coding region of CDJ2 excluding the J-domain was amplified by PCR with primers 5′-GCTGCCCATGGGCTACGCCGGCGGGAGGA-3′ and 5′-CGTGGGTAACCTAGCTTATTGAGCTTCTTCTTGAGC-3′; digested with NcoI and BstEII; and after a subcloning step, ligated into pMS254, yielding pMS269. pMS254 and 269 were expressed in E. coli M15 (QIAGEN, Hilden, Germany) and purified by nickel-nitrilotriacetic acid agarose (Ni-NTA) according to the manufacturer's instructions (QIAGEN) with the following alterations. For the immunization of rabbits, cells expressing pMS254 were lysed in lysis buffer (6 M guanidine-HCl, 0.5 M NaCl, 20 mM Tris-HCl, pH 8.0, and 5 mM imidazole), and lysates were applied to a column containing 2 ml of Ni-NTA agarose. The column was washed with lysis buffer containing 6 M urea instead of guanidine-HCl and with urea buffer containing 30 mM imidazole instead of 5 mM. Proteins were eluted with 200 mM imidazole in urea buffer. For native proteins used for cross-linking studies, lysis, washing, and elution was done with imidazole concentrations as described above, but in 50 mM sodium-phosphate, 300 mM NaCl, pH 8.0.
The coding region of VIPP1 was amplified by PCR from cDNA clone AV632440 with primers 5′-GGACTAGTGCTCTTCGAACGCGAACCTGTTCTCTCGC-3′ and T7. The ∼1.35-kb PCR product was digested with SapI and XhoI and ligated into SapI-XhoI–digested pTYB11 (NEB, Frankfurt, Germany), giving pMS319. pMS319 was expressed in E. coli ER2566 and purified by chitin affinity chromatography according to the manufacturer's instructions (NEB). Pure VIPP1 was dialyzed extensively at 4°C against KMH buffer (20 mM HEPES-KOH, pH 7.2, 80 mM KCl, and 2.5 mM MgCl2). The calculated masses matched the masses determined for the purified proteins by mass spectrometry with a deviation of ± 0.01%.
Purification of HSP70B from Chlamydomonas
One liter of HSP70B-overexpressing strain CF184 (Schroda et al., 1999) was grown to a density of ∼8 × 106 cells/ml, harvested by centrifugation, and resuspended in a final volume of 10 ml of KMH buffer. Cells were supplemented with FCCP to a final concentration of 10 μM and incubated on a shaker for 15 min at 25°C. After addition of 0.25× protease inhibitor cocktail, cells were sonicated on ice for 90 s. The lysate was precleared by a 20-min centrifugation at 38,000 × g and 4°C, and the supernatant was loaded onto a sucrose cushion (20 mM HEPES-KOH, pH 7.2, 0.4 M sucrose) and centrifuged in a TI50 rotor for 30 min at 152,000 × g and 4°C. The supernatant was supplied with Triton-X 100 to a final concentration of 0.1% and stored at -80°C.
About 1 mg of CGE1 containing N- and C-terminal hexahistidine tags (Schroda et al., 2001b) was purified from an overexpressing E. coli strain by nickel-NTA chromatography. Purified CGE1 was incubated in KMH with a 10-× 5-cm nitrocellulose membrane (Amersham Biosciences) overnight at 4°C, washed once with KMHT (KMH, 0.1% Triton X-100), blocked for 2 h with 5% nonfat dry milk in KMHT at 25°C, and washed 3 × 10 min with KMHT at 25°C. The membrane was incubated with the CF184 lysate for 1 h at 25°C and washed 3 × 10 min with KMHT and 2 × with KMH. HSP70B was eluted by incubating the membrane with 10 ml of KMH containing 5 mM ATP for 20 min, concentrated by centrifugation in Amicon Ultra-4 tubes (Millipore, Molsheim, France), and dialyzed against 2 liters of KMH at 4°C overnight. The yield was ∼20 μg of HSP70B.
Glutaraldehyde Cross-linking
CDJ2, CDΔ2, and HSP70B (2.6 μM each) were mixed in a buffer containing 14 mM sodium phosphate, pH 8.0, 85 mM NaCl, 28 mM imidazole, 65 mM KCl, 16 mM HEPES, pH 7.2, and 2.2 mM MgCl2 and were incubated at 30°C for 10 min. After addition of glutaraldehyde to a final concentration of 0.1%, incubation at 30°C was continued for another 20 min. After that, 1 volume of 2× Laemmli sample buffer containing 400 mM glycine was added. The mixtures were incubated at room temperature for 40 min and separated on 4–18% SDS-PAGs.
RESULTS
To obtain cDNAs that encode chloroplast-targeted J-domain proteins, we searched the Chlamydomonas EST libraries generated recently (Asamizu et al., 1999, 2000; Shrager et al., 2003) with the amino acid sequence of the J-domain of E. coli DnaJ. Two partial cDNA contigs were assembled that potentially encoded J-domain proteins with N-terminal extensions which by the ChloroP program (Emanuelsson et al., 1999) were predicted to be chloroplast transit peptides. The corresponding cDNAs AV626034 and AV387908 were sequenced to completion (GenBank accession nos. AY387908 and AY696657, respectively). The mature protein potentially encoded by cDNA AY696656 has a calculated molecular mass of 40.26 kDa and exhibits 52% identity and 68% similarity to the chloroplast DnaJ homolog PCJ1 identified in pea (Schlicher and Soll, 1997). In addition to the J-domain, it contains a glycin/phenylalanin-rich region and a conserved cysteine cluster predicted to form a zinc finger-like motif involved in the binding of denatured polypeptides (Szabo et al., 1996). The gene product was named chloroplast DnaJ protein 1 (CDJ1), and it is encoded by a gene located on scaffold 49 (nt 2174–6823) of the 2.0-version of the Chlamydomonas genome sequence (http://genome.jgi-psf.org/chlre2/chlre2.home.html). As judged by its similarity to DnaJ, CDJ1 is likely to deliver denatured polypeptides to chloroplast Hsp70(s) for their refolding to the native state (Szabo et al., 1996). In support for this view, we found CDJ1 mRNA strongly induced by heat shock (Figure 1A). CDJ1 mRNA accumulation peaked in cells exposed to 40°C between 15 and 30 min; thereafter, CDJ1 mRNA levels decreased again. CDJ1 also was induced by a shift of dark-adapted cells to dim light; here, mRNA levels peaked 45 min after transfer to light and then decreased again (Figure 1C). Both heat shock and light induction of CDJ1 closely resembled that of the Chlamydomonas HSP70A-C genes (von Gromoff et al., 1989).
Figure 1.
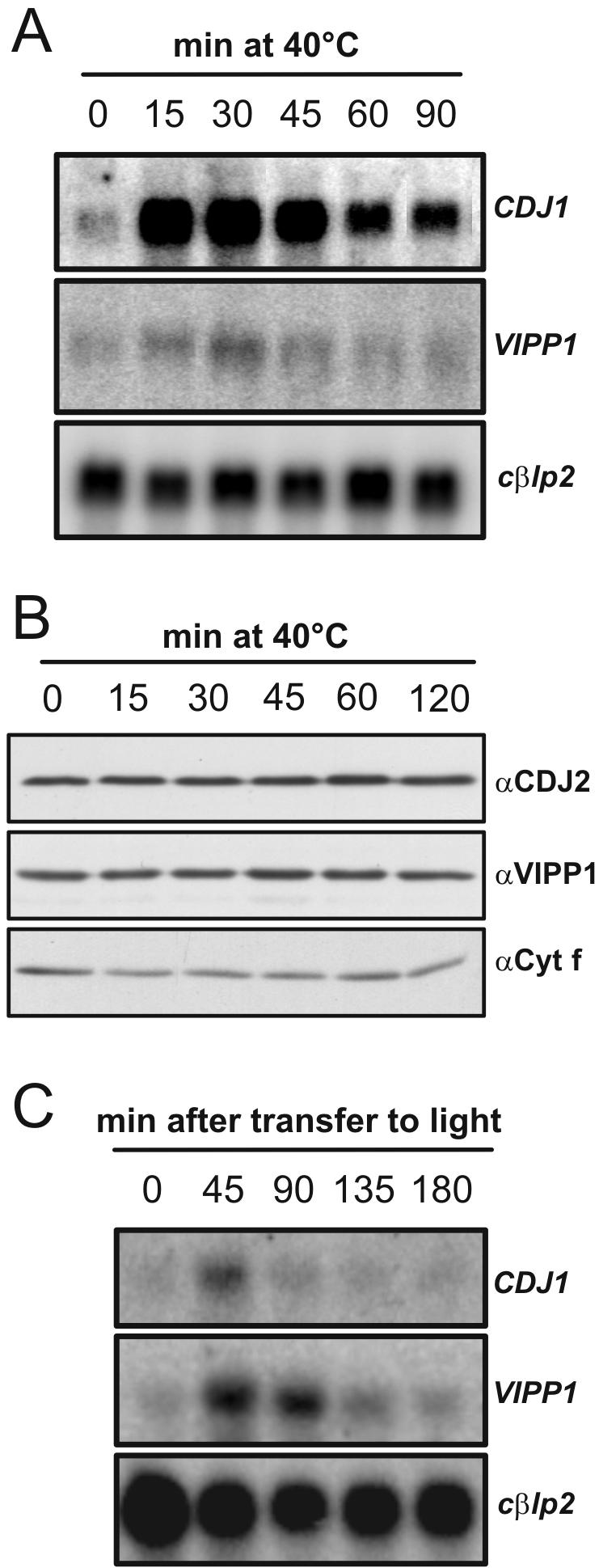
Induction of CDJ1, CDJ2, and VIPP1 after heat shock and dark-to-light shift. (A) CDJ1 and VIPP1 mRNA accumulation after shift of cells from 25 to 40°C. RNA gel blots with 10 μg of total RNA per lane were hybridized with probes for CDJ1, VIPP1, and the Chlamydomonas β-like protein 2 (cβlp2). The constitutively expressed cβlp2 served as loading control. (B) CDJ2 and VIPP1 protein accumulation after shift of cells from 25 to 40°C. Protein samples were from the same experiment depicted in A. Gels were loaded on the basis of equal chlorophyll concentrations (2 μg/lane). Constitutively expressed Cytf served as loading control. (C) CDJ1 and VIPP1 mRNA accumulation after shift of cells from 16-h dark to dim light. RNA gel blots with 10 μg of total RNA per lane were hybridized with the same probes used in A.
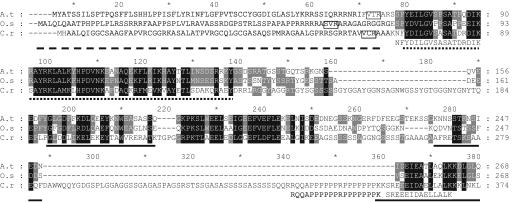
cDNA AY696657 encodes a protein of 38.4 kDa, which according to the TargetP program (Emanuelsson et al., 2000), is targeted to the chloroplast and processed to a 31.8-kDa mature form (Figure 2). The J-domain protein encoded by this cDNA contains neither a glycine/phenylalanine-rich region nor a zinc finger domain. The gene encoding this protein is located on scaffold 44 (nt 188990–198382) of the 2.0-version of the Chlamydomonas genome sequence and was named CDJ2. The 3′ untranslated region of CDJ2 transcript is remarkably short (87 nucleotides) compared with the average in Chlamydomonas (several hundred nucleotides; Silflow, 1998). CDJ2 mRNA was barely detectable by RNA gel blot analyses but seemed not to be induced by heat shock (unpublished data).
Figure 2.
Alignment of CDJ2 homologues. Aligned are amino acid sequences deduced from CDJ2 genes from Arabidopsis (A.t), rice (O.s), and Chlamydomonas (C.r). Residues highlighted in black are conserved in all three CDJ2 homologues, those highlighted in gray are conserved only in two of the three. Conserved amino acids are N/Q, D/E, R/K, S/T, F/Y, A/G, and V/I/L/M. Chloroplast transit peptides are indicated by an interrupted line, and cleavage sites as predicted by the TargetP program are boxed. The conserved J-domains are underlined with a dotted line, and putative coiled-coil regions as predicted by the COILS program are underlined with solid lines. Sequences of peptides identified by mass spectrometry from immunoprecipitated Chlamydomonas CDJ2 are given below the alignment. Alignments were made with the ClustalW program, refined manually and piled up with the GeneDoc program. Accessions for sequences used for the alignment of CDJ2 homologues are NP_200769 (Arabidopsis; atDjB42; Miernyk, 2001), AAO18454 (rice), and AY696657 (Chlamydomonas).
Database searches using the amino acid sequence located C-terminally to the CDJ2 J-domain revealed potentially chloroplast-targeted CDJ2 homologues in Arabidopsis and rice (Figure 2) and identified several higher plant ESTs that potentially encode CDJ2 homologues. In contrast, no CDJ2 homologues were identified in Cyanobacteria or other bacteria nor in nonphotosynthetic eukaryotes. Except for the J-domain, no conserved motifs were located within the CDJ2 proteins, and no function was yet attributed to them. Within the C termini of the CDJ2 proteins the COILS program (Lupas et al., 1991) located two regions that may form coiled-coil structures known to mediate protein–protein interactions (Figure 2). One of the predicted coiled-coil regions in Chlamydomonas CDJ2 is interrupted by a 74-amino acid sequence stretch rich in serine (27%), proline (20%), and glycine (13.5%). Adjacent to its J-domain, Chlamydomonas CDJ2 contains another 27-amino acid glycine-rich sequence (44% glycine) that is absent in the Arabidopsis and rice CDJ2 homologues (Figure 2). Thus, the predicted mature Arabidopsis and rice proteins are significantly smaller than those of Chlamydomonas CDJ2 (22–23 vs. 31.8 kDa). The high content of charged residues and the lack of putative transmembrane regions suggest that all three CDJ2 homologues are soluble proteins.
We conclude that CDJ2 is conserved from algae to higher plants. It lacks the glycine/phenylalanine-rich region and the zinc finger domain typical for DnaJ homologues involved in protein folding, but it contains domains able to mediate protein–protein interactions.
Coimmunoprecipitation of Chlamydomonas VIPP1 with CDJ2 Antibodies
To gain insight into the biological function of CDJ2, we used coimmunoprecipitation to identify proteins that interact with CDJ2. For this, a polyclonal antibody was raised against mature CDJ2 expressed in E. coli. In protein gel blot analyses of whole cell and soluble Chlamydomonas proteins, the CDJ2 antibody recognized a single band at ∼32 kDa (Figure 1B), which correlated well with the 31.8 kDa calculated for the mature protein. With the CDJ2 antibody, we could verify our notion from RNA gel blots that CDJ2 is not a heat shock-induced protein (Figure 1B).
One hundred microliters of CDJ2 antiserum was sufficient to precipitate quantitatively all CDJ2 protein from soluble extracts from ∼1010 Chlamydomonas cells (Figure 3). Silver staining of the proteins immunoprecipitated with the CDJ2 antibody revealed a protein of ∼28 kDa that coprecipitated with CDJ2 at about equal quantities. Subsequent immuno-detection revealed that this ∼28-kDa protein was not recognized by the CDJ2 antibody (Figure 3). The CDJ2 preimmune serum precipitated an unknown ∼26-kDa protein, but no CDJ2. Bands corresponding to CDJ2 and the ∼28-kDa CDJ2 coprecipitate were excised, digested with trypsin, and analyzed by mass spectrometry. Three peptides were identified for CDJ2 (Figure 2). For the ∼28-kDa coprecipitate, we found one peptide encoded by EST AV632440, which codes for a Chlamydomonas homolog of the VIPP1.
The Chlamydomonas VIPP1 Gene and Gene Product
We sequenced the VIPP1 cDNA corresponding to EST AV632440 (GenBank accession no. AY696658) and found that the VIPP1 gene is located on scaffold 23 (nt 582575–587082) of the 2.0-version of the Chlamydomonas genome sequence. In RNA gel blot analyses, we observed an approximately threefold induction of VIPP1 transcript after heat shock (Figure 1A) and an approximately fivefold induction after dark-to-light shift (Figure 1C). Chlamydomonas VIPP1 shares ∼50% identical and ∼73% similar residues with VIPP1 homologues from pea, Arabidopsis, and rice, which in turn share ∼78% identical and ∼90% similar residues with each other. Chlamydomonas and higher plant VIPP1 proteins all contain N-terminal extensions that according to ChloroP represent chloroplast transit peptides. From the cleavage site predicted by TargetP, mature Chlamydomonas VIPP1 has a size of 28.4 kDa. Accordingly, a polyclonal antibody raised against mature Chlamydomonas VIPP1 expressed in E. coli detected a protein of ∼28 kDa (Figure 1B). Despite the increase of VIPP1 mRNA after heat shock and dark-to-light shift, we observed no significant changes on VIPP1 protein level after these treatments (Figure 1B; unpublished data).
Intraplastidal Localization of Chlamydomonas VIPP1 in Stroma, Thylakoids, and Low-density Membranes
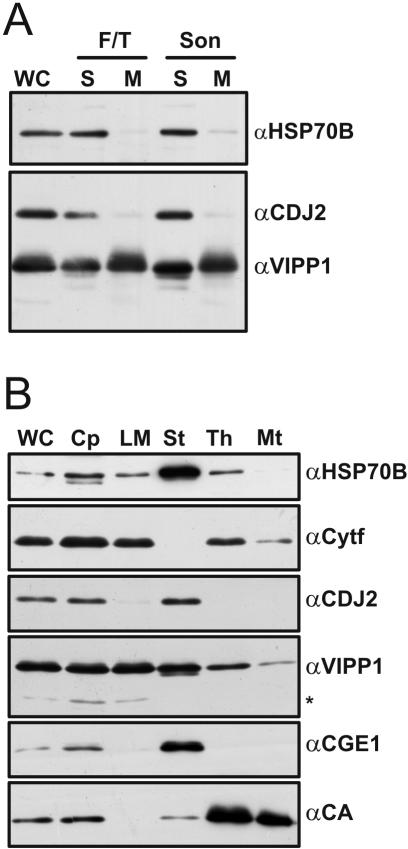
In previous studies, VIPP1 in Cyanobacteria (Westphal et al., 2001), pea (Li et al., 1994), and Arabidopsis (Kroll et al., 2001) was found exclusively in membrane fractions. Because we used soluble extracts for our immunoprecipitations, the identification of VIPP1 from these was unexpected. To address this discrepancy, we performed a crude fractionation of Chlamydomonas cells into membranes and soluble proteins. For this, a cell wall-deficient strain was ruptured by freeze-thawing or sonication, and soluble proteins were separated from membranes by a 355,000 × g centrifugation. The VIPP1 antibody detected a ∼28-kDa protein present in approximately equal amounts in soluble and membrane fractions of freeze-thawed and sonicated cells (Figure 4A). Because fractions were loaded on a volume basis, the data suggest that about one-half of the Chlamydomonas VIPP1 protein is soluble and the other half is membrane-associated. In contrast, only little HSP70B and CDJ2 were detected in the membrane fractions (Figure 4A).
Figure 4.
Intracellular localization of CDJ2 and VIPP1. (A) Chlamydomonas cell wall-deficient cells were sonicated (Son) or ruptured by freeze-thawing (F/T) and separated into soluble (S) or membrane-enriched (M) fractions on a volume basis. Proteins from whole cells (WC, 14 μg) and fractions (S, 4 μg; M, 10 μg) were separated on a 7.5–15% SDS-polyacrylamide gel, transferred to nitrocellulose, and immunodecorated with antibodies against HSP70B, CDJ2, and VIPP1. (B) Chlamydomonas chloroplasts (Cp) were isolated, lysed by hypoosmotic shock and separated into stroma (St), low-density membranes (LM) and thylakoid membranes (Th). Mitochondria (Mt) were isolated from the same strain. Proteins from whole cells (WC) and fractions (7 μg each) were separated on a 7.5–15% SDS-polyacrylamide gel, transferred to nitrocellulose, and immunodecorated with antibodies against HSP70B, Cytf, CDJ2, VIPP1, CGE1, and mitochondrial carbonic anhydrase (CA).
Next, we set out to verify the chloroplast localization of CDJ2 and VIPP1 and to determine which membranes Chlamydomonas VIPP1 is associated with. Chlamydomonas chloroplasts were isolated, lysed by hypoosmotic shock, and separated into stroma, thylakoid, and low-density membrane fractions. Low-density membranes are considered to consist of inner envelopes and of transitory membranes between inner envelope and thylakoids (Zerges and Rochaix, 1998). In addition, mitochondria were isolated. The purity of the fractions was tested with antibodies against mitochondrial carboanhydrase, stromal HSP70B, and CGE1 (the nucleotide exchange factor of HSP70B; Schroda et al., 2001b), and the integral thylakoid membrane protein Cytf. Chloroplasts were significantly contaminated by mitochondria and mitochondria slightly by thylakoids, as judged by the detection of carbonic anhydrase in the chloroplast fraction, and Cytf in the mitochondrial fraction, respectively (Figure 4B). The low-density membranes contained Cytf, but no mitochondrial carboanhydrase, whereas the thylakoids were heavily contaminated with carboanhydrase. The stroma fraction contained no Cytf and therefore was free of thylakoidal contaminations but contained some mitochondrial carboanhydrase (Figure 4B).
CDJ2 was detected in chloroplasts, stroma, and very weakly in low-density membranes, but it was absent in thylakoids and mitochondria (Figure 4B). Thus, CDJ2 exhibited exactly the same fractionation pattern as stromal CGE1. HSP70B also showed the same fractionation pattern as CGE1 but also was detected in low-density membranes and thylakoids, corroborating previous reports (Schroda et al., 2001b; Friso et al., 2004). VIPP1 was detected in chloroplasts, low-density membranes, stroma, thylakoids, and weakly in mitochondria (Figure 4B). The presence of some VIPP1 in mitochondria most likely is due to their slight contamination by thylakoids. Therefore, VIPP1 exhibited a combination of the fractionation patterns of Cytf and CGE1. Note that in independent fractionation experiments, the low-density membrane preparation was devoid of Cytf, but it still contained VIPP1 and some HSP70B. In addition to the major ∼28-kDa protein, the VIPP1 antibody also detected a minor protein at ∼26 kDa in whole cells, chloroplasts, and low-density membranes, which by mass spectrometry was revealed to be a truncated form of VIPP1 (Figure 4B, asterisk). Also in pea and Arabidopsis, two VIPP1 forms with identical N termini, but a size difference of ∼2 kDa, were detected (Kroll et al., 2001). The functional significance of these two VIPP1 forms is unclear.
In summary, our fractionation experiments indicate that CDJ2 and VIPP1 indeed are chloroplast proteins. Both seem to be localized in the stroma. VIPP1 also was located to low-density membranes and thylakoids.
Coimmunoprecipitation of CDJ2 with Anti-VIPP1 Antibodies
The size of the ∼28-kDa protein that coprecipitated with CDJ2 matched well with that predicted for VIPP1. However, the identification of only a single VIPP1 peptide is not sufficient to conclude that the ∼28-kDa CDJ2 coprecipitate is indeed VIPP1. To verify VIPP1 as a CDJ2 interaction partner, we used the VIPP1 antibody for the immunodetection of CDJ2 immunoprecipitations. As shown in Figure 5B, the VIPP1 antibody clearly recognized the major ∼28-kDa protein that coprecipitated with CDJ2. Moreover, when the VIPP1 antibody was used for immunoprecipitation of VIPP1 from soluble extracts, among others a minor coprecipitating protein of ∼32 kDa was detected by silver staining (Figure 5A). Immunodetection with CDJ2 antibodies identified this ∼32-kDa protein as CDJ2 (Figure 5B). Several proteins in the ∼40- to 60-kDa range, which were precipitated by the VIPP1 preimmune serum were not detected by either CDJ2 or VIPP1 antibodies. Interestingly, whereas anti-CDJ2 antibodies precipitated CDJ2 and VIPP1 in about equal amounts (Figures 3 and 5A), the anti-VIPP1 antibody coprecipitated only little CDJ2 (Figure 5A). Immunoprecipitation of VIPP1 from membranes solubilized with 2% Triton X-100 coprecipitated hardly any CDJ2 (Figure 5, A and B).
VIPP1 protein that was precipitated with anti-VIPP1 antibodies migrated as a double band at ∼28 kDa in SDS-gels (Figure 5A, asterisk). The double band consisted of a major and a minor band, the minor band migrating slightly faster than the major one. In contrast, VIPP1 that was coprecipitated with anti-CDJ2 antibodies seemed to consist only of the major, slow-migrating VIPP1 (Figure 5, A and B). Apparently, part of the VIPP1 pool is modified such that its migration is altered. To test, whether modified VIPP1 corresponds to the upper or to the lower band, we separated the VIPP1 protein that we had overexpressed and purified from E. coli for the generation of antibodies on the same gel next to the VIPP1 immunoprecipitations (Figure 5B). VIPP1 was expressed as a fusion protein that after cleavage had alanine 38 as N-terminal amino acid. Alanine 38 aligns with methionine 61, which has been identified as the N-terminal amino acid of mature pea VIPP1 (Westphal et al., 2001). VIPP1 that was purified from E. coli and verified by mass spectrometry to be nonmodified comigrated with the major ∼28-kDa VIPP1 band (Figure 5B). Therefore, the major ∼28-kDa VIPP1 band is likely to correspond to the unmodified protein and the faster migrating form seems to contain a modification that increases its migration properties in SDS-polyacrylamide gels, or simply is a VIPP1 degradation product.
In summary we show that the ∼28-kDa protein coprecipitating with CDJ2 is indeed VIPP1 and vice versa that CDJ2 coprecipitated with VIPP1.
Coimmunoprecipitation of HSP70B with CDJ2 and VIPP1 Antibodies
Because J-domain proteins are known to present bound proteins to a specific Hsp70 chaperone (Cyr et al., 1994), we wondered which Hsp70 may take over VIPP1 apparently presented by CDJ2. In the silver-stained gel shown in Figure 5A, one protein in the 70-kDa range was observed that specifically coprecipitated with CDJ2. A protein of the same size coprecipitated with VIPP1 but in much larger amounts. With VIPP1 a second ∼66-kDa protein of more diffuse migration pattern also was coprecipitated, which seemed to be absent in the CDJ2 precipitate. The two ∼66- and ∼70-kDa bands were excised, digested with trypsin, and analyzed by mass spectrometry. The protein in the ∼70-kDa band was clearly identified as HSP70B: four HSP70B peptides were detected from the ∼70-kDa protein that coprecipitated with CDJ2, and 18 HSP70B peptides were identified from the ∼70-kDa protein that coprecipitated with VIPP1. In addition, a specific HSP70B antibody detected this band in both precipitates (Figure 5B). Surprisingly, eight HSP70C peptides (the mitochondrial Hsp70 homolog; Schroda, 2004) were detected from the ∼66-kDa protein that coprecipitated with VIPP1. Because the immunoprecipitations were performed from whole cell extracts, we believe that native VIPP1 may expose Hsp70-binding motifs that also are recognized by other DnaK-type chaperones, like mitochondrial HSP70C.
When the HSP70B antibody was used in a reciprocal experiment to immunoprecipitate HSP70B from soluble proteins, VIPP1, and at low concentrations also CDJ2 coprecipitated with HSP70B (Figure 5C). Whereas the major stable interaction partner of CDJ2 seems to be VIPP1, VIPP1 seems to be mostly interacting with HSP70B. Note that membrane-associated VIPP1 only interacted with negligible amounts of both, CDJ2 and HSP70B (Figure 5, A and B).
HSP70s are known to bind substrate proteins with high affinity in the ADP state and with low affinity in the ATP state (Bukau and Horwich, 1998). To test whether the HSP70B–VIPP1 interaction is influenced by the ATP concentration, we immunoprecipitated VIPP1 from ATP-supplemented extracts or from extracts prepared from ATP-depleted cells. As shown in Figure 6, in cell extracts depleted from ATP, the interaction of VIPP1 with HSP70C and HSP70B was comparable with that observed in extracts from nontreated cells (Figure 5), indicating that the ATP concentrations in our cell extracts were low. In contrast, in ATP-supplemented extracts, the VIPP1 interaction with HSP70C was abolished and that with HSP70B was significantly reduced (Figure 6). This suggests that VIPP1 has a higher affinity for HSP70B than for HSP70C. Interestingly, CDJ2 that was immunoprecipitated from ATP-supplemented cell extracts coprecipitated significantly less VIPP1 and HSP70B. The identity of the ∼45-kDa protein that also coprecipitated with CDJ2 in an ATP-dependent manner (Figure 6) could not yet be revealed.
Together, the data suggest that the HSP70 partner of CDJ2 is stromal HSP70B and that native VIPP1 behaves like a substrate for HSP70B. The interaction of VIPP1 with CDJ2 and HSP70B is significantly weaker in the presence of ATP but apparently not abolished.
Verification of the Interaction between CDJ2 and HSP70B by Glutaraldehyde Cross-linking
Han and Christen (2003) provided evidence that DnaJ and DnaK may bind to different sites of the same substrate molecule. Thus, if this was the case also for CDJ2 and HSP70B binding to VIPP1, HSP70B may have coprecipitated with CDJ2 via VIPP1 without necessarily being involved in a common chaperoning process. To rule out this possibility, we had to confirm that CDJ2 and HSP70B also interacted physically in the absence of VIPP1. To test this, we performed in vitro glutaraldehyde cross-linking studies with purified HSP70B and CDJ2. To assay the role of the J-domain in this interaction, we also included a CDJ2 derivative lacking the N-terminal J-domain (CDΔ2). Glutaraldehyde cross-linking has been used successfully to monitor complex formation of Hsp70 and its cochaperones (Wu et al., 1996; Azem et al., 1997).
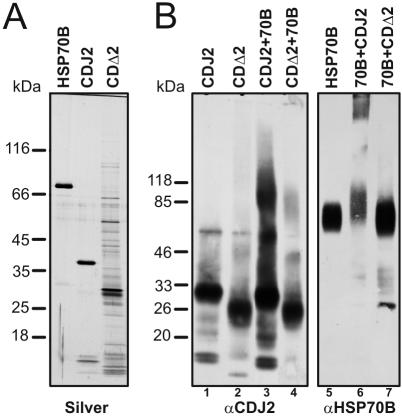
As judged by its inability to interact with the CGE1 co-chaperone in glutaraldehyde cross-linking experiments, HSP70B heterologously expressed in E. coli seemed to be nonfunctional (Willmund and Schroda, unpublished results). Thus, we purified the HSP70B protein from Chlamydomonas-soluble extracts by using the CGE1 cochaperone as an affinity matrix. Silver staining of the purified proteins after separation on an SDS-polyacrylamide gel revealed that HSP70B and CDJ2 contained few impurities, whereas the preparation of CDΔ2, for which the expression level in E. coli was very low, contained several impurities (Figure 7A).
Figure 7.
In vitro analysis of the interaction between HSP70B and CDJ2 by glutaraldehyde cross-linking. (A) HSP70B purified from Chlamydomonas and heterologously expressed CDJ2 and CDJ2 lacking its J-domain (CDΔ2) (both containing N- and C-terminal hexahistidine tags) were separated on a 7.5–15% SDS-polyacrylamide gel and visualized by silver staining. (B) CDJ2, CDΔ2, and HSP70B (2.6 μM each) were cross-linked, separated on 4–18% SDS-polyacrylamide gels, transferred to nitrocellulose, and immunodecorated with antibodies against CDJ2 or HSP70B.
The purified proteins were incubated alone or in equimolar amounts with their partner, cross-linked with glutaraldehyde, separated on SDS-polyacrylamide gels, transferred to nitrocellulose, and immunodecorated with antibodies against CDJ2 or HSP70B. CDJ2, CDΔ2, and HSP70B alone were present mainly as monomers (Figure 7B, lanes 1, 2, and 5). When incubated with HSP70B, CDJ2 formed a major complex of ∼95 kDa and CDΔ2 a minor one of ∼85 kDa with HSP70B (Figure 7B, lanes 3 and 4). HSP70B incubated with CDJ2 shifted to a complex of ∼95 kDa but also formed high-molecular-weight polymers that hardly entered the gel (Figure 7B, lane 6). When incubated with CDΔ2, HSP70B smeared weakly into a ∼85-kDa complex but did not form polymers (Figure 7B, lane 7).
In summary, our data suggest that CDJ2 and CDΔ2 both interact physically as monomers with monomeric HSP70B. The weak ∼85-kDa HSP70B–CDΔ2 complex may arise from a specific interaction but also from the recognition of CDΔ2 by HSP70B as a substrate due to misfolding of CDΔ2 induced by the absence of its J-domain. Compared with the weak HSP70B–CDΔ2 complex, the strong ∼95-kDa HSP70B–CDJ2 complex points to an important role of the CDJ2 J-domain for mediating the interaction of the two proteins. CDJ2's J-domain also is required to induce polymerization of HSP70B.
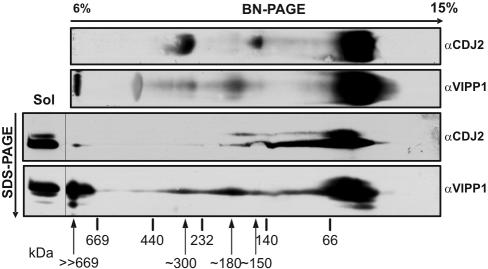
Analysis of Soluble CDJ2- and VIPP1-containing Complexes by BN-PAGE
We intended to analyze the complexes formed by soluble VIPP1 and CDJ2. For this, we separated native protein complexes from soluble Chlamydomonas extracts by size by using BN-PAGE and transferred the complexes directly to nitro-cellulose membranes. Immunodetection with the CDJ2 antibody revealed a strong signal <66 kDa from monomeric or dimeric CDJ2 and weaker signals from complexes of ∼150 and ∼300 kDa (Figure 8). The VIPP1 antibody gave a strong signal at <66 kDa from monomeric or dimeric VIPP1 and weak ones from complexes at ∼150, ∼180, ∼300, and >>669 kDa. A signal in the 550-kDa region seems to be a cross-reaction of the VIPP1 antibody with native Rubisco, which migrates in large quantities at this position.
Figure 8.
Analysis of CDJ2 and VIPP1 complexes by BN-PAGE. Chlamydomonas total soluble proteins (Sol) were separated on a 6–15% native gel (BN-PAGE) and transferred directly to nitrocellulose (top two gels) or separated in the second dimension on a 10% SDS-PAG and then transferred to nitrocellulose (bottom two gels). Membranes were immunodecorated with antibodies against CDJ2 and VIPP1. Arrows indicate the positions of complexes that contain CDJ2, VIPP1, or both.
CDJ2 in the second dimension was found mostly below 66 kDa and in a complex of ∼150 kDa (Figure 8). A minor complex was also observed in a region >>669 kDa, which was not detected in the first dimension. In contrast, the ∼300-kDa complex detected in the first dimension was absent in the second dimension, a finding we have reproduced many times. Either the CDJ2 antibody cross-reacts only with the native form of a protein in this ∼300-kDa complex or CDJ2 in this complex resists SDS-denaturation and does not enter the second dimension gel. In SDS-gels, some CDJ2 protein reproducibly migrated slightly slower than bulk CDJ2 (Figures 3, 5B, and 8). It is not clear whether this is due to a modification of CDJ2 or due to incomplete denaturation.
All VIPP1 complexes detected in the first dimension also were detected at similar intensities in the second dimension (Figure 8). With the HSP70B antibody, no distinct complexes but a smear into the higher molecular weight region was detected (our unpublished data; Schroda et al., 2001b). Exactly the same pattern of CDJ2- and VIPP1-containing complexes was detected when cells were lysed more gently by vortexing with glass beads instead of sonication (our unpublished data). This argues against the possibility of a nonspecific disassembly of the >>669-kDa VIPP1/CDJ2 complex by the harsh sonication procedure. When Chlamydomonas VIPP1 purified from E. coli was separated by BN-PAGE, VIPP1 was detected only at positions <66 and >>669 kDa (our unpublished data). This indicates that VIPP1 alone has the capability to form large oligomers and thus that the VIPP1 signal at >>669 kDa is not just low-molecular-weight VIPP1 associated with membrane vesicles, which may still be present in the soluble cell extracts and would hardly enter the native gel.
In summary, we can conclude that 1) most of soluble VIPP1 and CDJ2 exist as monomers or dimers; 2) VIPP1 alone can assemble into oligomers of >>669 kDa; and 3) CDJ2 and VIPP1 may form heterodimers and/or complexes of ∼150, >>669, and perhaps ∼300 kDa.
DISCUSSION
We report on the identification of two cDNAs that encode the CDJ1 and CDJ2 proteins. We show that the CDJ1 gene is strongly induced by heat shock and slightly by light (Figure 1) and potentially encodes a plastidic member of the DnaJ family involved in protein folding (Szabo et al., 1996). A plastidic DnaJ homologue, named PCJ1, has previously been reported in pea chloroplasts (Schlicher and Soll, 1997).
CDJ2 and HSP70B Form a Plastidic Chaperone Pair
For CDJ2, the following findings suggest that it forms a plastidic chaperone pair with HSP70B. 1) From soluble cell extracts, HSP70B coimmunoprecipitated with CDJ2, and vice versa; CDJ2 coprecipitated with HSP70B (Figure 5). 2) Both proteins colocalize to the chloroplast stroma (Figure 4B), to which at most two HSP70 chaperones are located, with HSP70B being the most prominent (Schroda, 2004). 3) In vitro cross-linking studies revealed that CDJ2 forms a complex with HSP70B in a 1:1 stoichiometry and that CDJ2 can catalytically induce polymerization of HSP70B into high-molecular-weight complexes without being part of these (Figure 7B). Stable interactions between J-domain proteins and Hsp70s already have been reported for some chaperone pairs, e.g., mammalian hsp40/Hsp70 (Sugito et al., 1995), ER Sec63p/BiP (Brodsky and Schekman, 1993), and bovine brain auxilin/Hsc70 (Jiang et al., 1997), but not for the E. coli DnaJ/DnaK couple (King et al., 1995). Even catalytic polymerization by J-domain proteins has been demonstrated for HSP70s of the eukaryotic cytosol (King et al., 1995; Jiang et al., 1997), but again not for bacterial DnaK (King et al., 1999). Therefore, our results show that polymerization also of a prokaryotic-type Hsp70 (stromal HSP70B) may be induced by the J-domain protein CDJ2.
According to a model developed previously to explain Hsp70 polymerization by J-domain proteins (King et al., 1999), the J-domain protein in the absence of its natural substrate instead binds an Hsp70 and presents it to the substrate-binding domain of another Hsp70. The latter undergoes ATP hydrolysis and binds the presented Hsp70 like a substrate. Bound Hsp70 may in turn bind another Hsp70 newly presented by a J-domain protein, eventually producing a chain of polymerized Hsp70s. This model implies that the respective J-domain protein is capable of binding Hsp70 in a region distinct from the J-domain. In support of this model, in our cross-linking studies we have indeed observed an interaction of CDΔ2 (CDJ2 with deleted J-domain) with HSP70B (Figure 7B). Whether Hsp70 polymerization occurs in vivo and what its role may be are unclear (King et al., 1999).
VIPP1 Is a Major Target Protein of the HSP70B/CDJ2 Chaperone Pair
We found that soluble VIPP1 is a major target protein of the HSP70B/CDJ2 chaperone pair. Antibodies against both CDJ2 and HSP70B coimmunoprecipitated VIPP1, and vice versa; antibodies against VIPP1 coprecipitated CDJ2 and HSP70B (Figure 5). Several lines of evidence indicate that the interactions between these three proteins are specific and not due to, e.g., a nonspecific recognition of nonnative VIPP1 by the HSP70B/CDJ2 chaperone pair. 1) In contrast to CDJ1, CDJ2 is induced neither by heat stress nor does it contain domains typical for DnaJ proteins involved in protein folding (a glycine/phenylalanine-rich region or a zinc finger-forming cysteine cluster). Thus, CDJ2 is more likely a J-domain protein specialized for a specific cellular function other than protein folding. 2) VIPP1, an unknown ∼45-kDa protein, and HSP70B in this order are the most prominent interaction partners of CDJ2 (Figure 6). If CDJ2 had a tendency to nonspecifically interact with other proteins, more of them would have been expected. 3) Recent studies have provided evidence that E. coli DnaK and DnaJ bind to different sites on the same substrate protein (Han and Christen, 2003). If this holds for J-domain protein/HSP70 chaperone pairs in general, the binding of a specialized J-domain protein and of its HSP70 partner to the same polypeptide suggests specificity. 4) CDJ2 and VIPP1 both contain regions predicted to form coiled-coils (Figure 2). 5) CDJ2, VIPP1, and HSP70B all are localized in the same compartment, i.e., the chloroplast stroma (Figure 4). 6) The interaction of CDJ2 with VIPP1 and with an ∼45-kDa protein is ATP dependent, i.e., less of them interact with CDJ2 in the presence of Mg-ATP (Figure 6). This observation is consistent with the model that J-domain proteins deliver bound substrates to their HSP70 partner in the ATP state, trigger ATP hydrolysis and dissociate once the substrate is processed by HSP70 (e.g., folded to the native state or translocated across a membrane) (Bukau and Horwich, 1998). In the absence of ATP, the J-domain protein keeps binding its substrate, because HSP70 cannot process it further. Thus, a protein non-specifically bound to a J-domain protein should remain bound also in the ATP-state, because it is unlikely that the HSP70 partner is competent for further processing. 7) Antibodies against chloroplast-targeted HSP90, bacterial GroEL, and bacterial ClpB detected proteins of the expected molecular weights in soluble extracts, but neither chaperone was detected in CDJ2 or VIPP1 immunoprecipitates (our unpublished data), suggesting that CDJ2 and VIPP1 specifically interact only with HSP70 chaperones. 8) Plastidic HSP70B has a higher affinity for VIPP1 than obviously nonspecifically binding mitochondrial HSP70C: even in the presence of ATP HSP70B exhibited some affinity for VIPP1, whereas HSP70C released VIPP1 entirely (Figure 6). 9) Immunoprecipitates prepared with antibodies against stromal CF1α or CF1β did not contain HSP70B or CDJ2 (our unpublished data), suggesting that the chaperone pair does not simply interact with any stromal protein.
VIPP1 is likely to have evolved from duplication of the pspA gene in Cyanobacteria (Westphal et al., 2001). Because we found CDJ2 homologues only in algae and higher plants but not in Cyanobacteria, CDJ2 as mediator for the VIPP1–HSP70 interaction may have evolved after the endosymbiotic event. Perhaps CDJ2 has evolved to link VIPP1 to DnaK2, the ancestor of today's stromal Hsp70s, so that a specific cyanobacterial DnaK system (DnaK1 or DnaK3) may have become redundant and for this reason got extinct?
Chlamydomonas VIPP1 Is Located to the Stroma and to Chloroplast Membranes
The coprecipitation of VIPP1 from soluble cell extracts was unexpected, because localization studies in pea (Li et al., 1994) and Arabidopsis (Kroll et al., 2001) detected VIPP1 in thylakoids and inner envelopes, and in Synechocystis VIPP1 was found exclusively in plasma membranes (Westphal et al., 2001). However, some pea VIPP1 that cofractionated with outer envelopes was interpreted as soluble, particulate VIPP1 with a density similar to that of outer envelopes (Li et al., 1994). In addition, the probable VIPP1 ancestor PspA was detected in the cytosol and the inner membrane of E. coli in about equal quantities (Brissette et al., 1990; Kleerebezem and Tommassen, 1993). The same localization pattern as for PspA also was observed for cyanobacterial VIPP1 expressed in E. coli (DeLisa et al., 2004). Careful fractionation of Chlamydomonas cells verified that about one-half of the VIPP1 protein is indeed in the soluble stroma fraction, whereas the other half is located to thylakoids and to low-density membranes, which to a large part consist of inner envelopes (Zerges and Rochaix, 1998) (Figure 4). It is not clear whether these rather diverse localization patterns of VIPP1 proteins in different organisms are due to the preparation techniques, differences in cell physiology, or rather to organism-specific differences in the affinity of VIPP1 for thylakoids and inner envelope/plasma membranes.
VIPP1 and CDJ2 Form Distinct Complexes
PspA in E. coli has been demonstrated to form dimers and perhaps higher order oligomers (Dworkin et al., 2000) and recently, PspA in vitro has been shown to form oligomeric rings of 1023 kDa that probably consist of 36 PspA subunits organized in ninefold symmetry (Hankamer et al., 2004). By BN-PAGE, we found VIPP1 in soluble cell extracts as monomers or dimers <66 kDa and in complexes of ∼150, ∼180, ∼300, and >>669 kDa. We hypothesize that the >>669-kDa VIPP1 complex found in soluble cell extracts and in purified VIPP1 samples also may represent large oligomeric rings. This view is consistent with immunogold studies with VIPP1 antibodies, in which gold particles were found in clusters (Li et al., 1994). CDJ2, too, was found in monomers or dimers <66 kDa and in complexes of ∼150, ∼300, and >>669 kDa (Figure 8). Thus, CDJ2 and VIPP1 may interact as heterodimers and/or in the ∼150-, ∼300-, and >>669-kDa complexes. Immunoprecipitation of CDJ2 led to the coprecipitation of approximately equal amounts of VIPP1 (Figures 3, 5, and 6), whereas immunoprecipitation of VIPP1 coprecipitated only small amounts of CDJ2 (Figure 5). Thus, in the stroma, either CDJ2 is much less abundant than VIPP1 and is quantitatively organized into complexes with VIPP1, or few CDJ2 proteins interact with larger VIPP1 oligomers. The finding that very little CDJ2 and almost stoichiometric amounts of HSP70B coprecipitated with soluble VIPP1 (Figures 5 and 6) suggests that CDJ2 may prime VIPP1 for the binding of HSP70B and in the presence of ATP seems to leave the HSP70B–VIPP1 complex once it has been established (Figure 6).
VIPP1 May Play Roles in Maintaining Membrane Integrity and/or in Vesicle Traffic
To discuss which cellular function the interaction of HSP70B and CDJ2 with VIPP1 may have, we first need to summarize briefly what is known about VIPP1. VIPP1 underexpression resulted in distorted thylakoids in Arabidopsis and Synechocystis, and vesicle budding from the inner envelope of Arabidopsis chloroplasts was abolished (Kroll et al., 2001; Westphal et al., 2001). Unfortunately, nothing is known about the mechanisms involved. Expression of the likely VIPP1 ancestor pspA is induced by stresses that affect membrane integrity, such as heat, ethanol, hyperosmolarity, pore-forming phage proteins, or the accumulation of aberrant secretion pathway intermediates within secretion pores (Brissette et al., 1990; Kleerebezem and Tommassen, 1993; Jones et al., 2003). All these stresses are believed to eventually result in a decreased transmembrane proton motif force (pmf), which is likely to trigger induction of the psp operon (Adams et al., 2003). PspA by an unknown mechanism seems to sustain membrane integrity, thereby maintaining the pmf (Kleerebezem et al., 1996) and ensuring a proper performance of the Sec (Kleerebezem and Tommassen, 1993) and of the Tat pathways (DeLisa et al., 2004). Importantly, VIPP1 can substitute PspA to improve the performance of an overloaded Tat pathway in E. coli, and vice versa; PspA can improve the performance of the thylakoidal Tat pathway in vitro (DeLisa et al., 2004). Still, some specificity for PspA and VIPP1 function must exist, because both are present in Cyanobacteria, but cyanobacterial PspA cannot substitute for cyanobacterial VIPP1 function in vivo (Westphal et al., 2001).
Thus, two possibilities may explain the VIPP1 mutant phenotype. First, VIPP1, like PspA, may function exclusively in sustaining membrane integrity but became specialized to fulfill this task on chloroplast membranes, e.g., as essential auxiliary factor for lumenal import pathways. The distorted thylakoid phenotype in VIPP1 mutants may be a consequence of the malfunctioning of lumenal import. Note that mutants defective in components of the Sec or Tat pathways display phenotypes that resemble that of VIPP1 mutants (Settles et al., 1997). The absence of vesicles may be due to a secondary effect caused by a lack of a thylakoid-derived signal required to trigger vesicle budding.
Alternatively, VIPP1 may have adopted a function in addition to that required for sustaining membrane integrity, i.e., a role in vesicle traffic. As suggested previously (Westphal et al., 2001), this additional function may be related to the C-terminal extension of ∼30 amino acids present in VIPP1 but absent in PspA. Perhaps the mechanism by which PspA sustains membrane integrity became essential for vesicle transport?
Hypothetical Roles of the HSP70B/CDJ2 Chaperone Pair in the Functional Cycle of VIPP1
The mechanism underlying Hsp70 function is the ability of this chaperone to bind hydrophobic motifs exposed by virtually all unfolded and some native proteins and to induce conformational changes in an ATP-dependent reaction (Rüdiger et al., 1997; Schiene-Fischer et al., 2002). The latter may result in the refolding of denatured proteins, and in the case of native protein complexes, in complex disassembly. In E. coli, for example, DnaK monomerizes RepA dimers and dissociates DnaB helicase-Lambda P complexes (Alfano and McMacken, 1989; Wickner et al., 1991); in the eukaryotic cytosol Hsc70 disassembles clathrin cages (Ungewickell et al., 1995) and primes disassembled clathrin for reassembly (Jiang et al., 2000).
We can envision that both a folding and a disassembling/assembling activity of HSP70B may support VIPP1 function. It is likely that in a first step CDJ2 binds to VIPP1 through coiled-coil interactions to recruit HSP70B via its J-domain and to lock HSP70B onto VIPP1. HSP70B may then target VIPP1 to sites where unfolded membrane proteins jeopardize membrane integrity. Such a targeting function may account for the finding that HSP70B and CDJ2 interact mainly with soluble VIPP1 and barely with membrane-associated VIPP1 (Figure 5). For a more specialized function in thylakoidal import, HSP70B associated with VIPP1 may verify the correct folding of lumenal precursors (unfolded proteins for the Sec pathway; correctly folded, monomeric and cofactor-bound proteins for the Tat pathway), thereby preventing proton leakage potentially caused by improperly translocating precursors. Accordingly, the induction of the VIPP1 and HSP70B genes by heat shock and light (Figure 1; von Gromoff et al., 1989) may account for a need in the chloroplast to cope with the accumulation of heat-denatured membrane proteins and to improve lumenal import at translocation pores that become crowded after the onset of light, respectively.
Alternatively, the HSP70B/CDJ2 chaperone pair may carry out disassembly and/or assembly of VIPP1 oligomers. Such a function would be reminiscent of the Hsc70–auxilin–clathrin triad, where the J-domain protein auxilin specifically interacts with assembled clathrin on clathrin-coated vesicles and recruits Hsc70. Hsc70 then drives clathrin disassembly, eventually resulting in the uncoating of clathrin-coated vesicles (Ungewickell et al., 1995). Like CDJ2, auxilin interacts rather stably with Hsc70 (Jiang et al., 1997). Hsc70 binds stoichiometrically to clathrin (Ungewickell et al., 1995), which also seems to hold for HSP70B binding VIPP1 (Figure 5A). Finally, after the uncoating reaction, Hsc70 in the ADP state forms a stable presteady-state complex with adaptor proteins and clathrin. After replacement of ADP by ATP, this complex turns into a structurally different steady-state complex, which is more labile but persists. In this complex, clathrin may be primed to interact again with vesicle membranes (Jiang et al., 2000). Apparently, HSP70B also interacts with VIPP1 more strongly in the ADP state, but affinity is not lost completely in the ATP state (Figure 6). In analogy to the auxilin/Hsc70 chaperone pair, CDJ2/HSP70B might disassemble and/or assemble VIPP1 oligomers to recycle the system for another turn of vesicle formation/transport. Alternatively, disassembly/assembly of VIPP1 might be required for its activation/inactivation in the process of sustaining membrane integrity.
Although we currently cannot distinguish between these possibilities, our finding that HSP70B and CDJ2 specifically interact with VIPP1 will pave the path for further studies to elucidate the roles that stromal chaperones play in the biogenesis/maintenance of chloroplast membranes in algae and higher plants.
Acknowledgments
We thank the Kazusa DNA Research Institute for providing cDNA clones AV626034, AV387908, and AV632440. We also thank Michael Hippler for help with mass spectrometry measurements; Francis-André Wollman for the antibodies against Cytf, CF1α, and CF1β; Bernd Bukau for the antibodies against GroEL and ClpB; and Mats Eriksson for the antibody against mitochondrial carbonic anhydrase. We are grateful to Olivier Vallon and Christoph Beck for critical reading of the manuscript and acknowledge Christoph Beck for the support with laboratory equipment. This work was supported by the Deutsche Forschungsgemeinschaft (Schr 617/2-1; 4-1; Be 903/12-1; 12-2).
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E04-08-0736) on January 5, 2005.
References
- Adams, H., Teertstra, W., Demmers, J., Boesten, R., and Tommassen, J. (2003). Interactions between phage-shock proteins in Escherichia coli. J. Bacteriol. 185, 1174-1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfano, C., and McMacken, R. (1989). Heat shock protein-mediated disassembly of nucleoprotein structures is required for the initiation of bacteriophage lambda DNA replication. J. Biol. Chem. 264, 10709-10718. [PubMed] [Google Scholar]
- Asamizu, E., Miura, K., Kucho, K., Inoue, Y., Fukuzawa, H., Ohyama, K., Nakamura, Y., and Tabata, S. (2000). Generation of expressed sequence tags from low-CO2 and high-CO2 adapted cells of Chlamydomonas reinhardtii. DNA Res. 7, 305-307. [DOI] [PubMed] [Google Scholar]
- Asamizu, E., Nakamura, Y., Sato, S., Fukuzawa, H., and Tabata, S. (1999). A large scale structural analysis of cDNAs in a unicellular green alga, Chlamydomonas reinhardtii. Generation of 3433 non-redundant expressed sequence tags. DNA Res. 6, 369-373. [DOI] [PubMed] [Google Scholar]
- Azem, A., Oppliger, W., Lustig, A., Jenö, P., Feifel, B., Schatz, G., and Horst, M. (1997). The mitochondrial hsp70 chaperone system. Effect of adenine nucleotides, peptide substrate, and mGrpE on the oligomeric state of mhsp70. J. Biol. Chem. 272, 20901-20906. [DOI] [PubMed] [Google Scholar]
- Bonk, M., Tadros, M., Vandekerckhove, J., Al-Babili, S., and Beyer, P. (1996). Purification and characterization of chaperonin 60 and heat-shock protein 70 from chromoplasts of Narcissus pseudonarcissus. Plant Physiol. 111, 931-939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brissette, J. L., Russel, M., Weiner, L., and Model, P. (1990). Phage shock protein, a stress protein of Escherichia coli. Proc. Natl. Acad. Sci. USA 87, 862-866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodsky, J. L., and Schekman, R. (1993). A Sec63p-BiP complex from yeast is required for protein translocation in a reconstituted proteoliposome. J. Cell. Biol. 123, 1355-1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brutnell, T. P., Sawers, R.J.H., Mant, A., and Langdale, J. A. (1999). BUNDLE SHEATH DEFECTIVE2, a novel protein required for post-translational regulation of the rbcL gene of maize. Plant Cell 11, 849-864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukau, B., and Horwich, A. L. (1998). The Hsp70 and Hsp60 chaperone machines. Cell 92, 351-366. [DOI] [PubMed] [Google Scholar]
- Cyr, D. M., Langer, T., and Douglas, M. G. (1994). DnaJ-like proteins: molecular chaperones and specific regulators of Hsp70. Trends Biochem. Sci. 19, 176-181. [DOI] [PubMed] [Google Scholar]
- DeLisa, M. P., Lee, P., Palmer, T., and Georgiou, G. (2004). Phage shock protein PspA of Escherichia coli relieves saturation of protein export via the Tat pathway. J. Bacteriol. 186, 366-373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dworkin, J., Jovanovic, G., and Model, P. (2000). The PspA protein of Escherichia coli is a negative regulator of σ54-δεπενδεντ transcription. J. Bacteriol. 182, 311-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson, O., Nielsen, H., Brunak, S., and von Heijne, G. (2000). Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 300, 1005-1016. [DOI] [PubMed] [Google Scholar]
- Emanuelsson, O., Nielsen, H., and von Heijne, G. (1999). ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci. 8, 978-984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson, M., Gardeström, P., and Samuelsson, G. (1995). Isolation, purification, and characterization of mitochondria from Chlamydomonas reinhardtii. Plant Physiol. 107, 479-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson, M., Karlsson, J., Ramazanov, Z., Gardestrom, P., and Samuelsson, G. (1996). Discovery of an algal mitochondrial carbonic anhydrase: molecular cloning and characterization of a low-CO2-induced polypeptide in Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. USA 93, 12031-12034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg, A. P., and Vogelstein, B. (1983). A technique for radiolabelling DNA restriction endonuclease fragments to high activity. Anal. Biochem. 132, 6-13. [DOI] [PubMed] [Google Scholar]
- Friso, G., Giacomelli, L., Ytterberg, A. J., Peltier, J. B., Rudella, A., Sun, Q., and Wijk, K. J. (2004). In-depth analysis of the thylakoid membrane proteome of Arabidopsis thaliana chloroplasts: new proteins, new functions, and a plastid proteome database. Plant Cell 16, 478-499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frydman, J. (2001). Folding of newly translated proteins in vivo: the role of molecular chaperones. Annu. Rev. Biochem. 70, 603-649. [DOI] [PubMed] [Google Scholar]
- Han, W., and Christen, P. (2003). Mechanism of the targeting action of DnaJ in the DnaK molecular chaperone system. J. Biol. Chem. 278, 19038-19043. [DOI] [PubMed] [Google Scholar]
- Hankamer, B. D., Elderkin, S., Buck, M., and Nield, J. (2004). Organization of the AAA+ adaptor protein PspA is an oligomeric ring. J. Biol. Chem. 279, 8862-8866. [DOI] [PubMed] [Google Scholar]
- Harris, E. H. (1989). The Chlamydomonas Source Book: A Comprehensive Guide to Biology and Laboratory Use, San Diego: Academic Press. [DOI] [PubMed]
- Jiang, R., Gao, B., Prasad, K., Greene, L. E., and Eisenberg, E. (2000). Hsc70 chaperones clathrin and primes it to interact with vesicle membranes. J. Biol. Chem. 275, 8439-8447. [DOI] [PubMed] [Google Scholar]
- Jiang, R.-F., Greener, T., Barouch, W., Greene, L., and Eisenberg, E. (1997). Interaction of auxilin with the molecular chaperone Hsc70. J. Biol. Chem. 272, 6141-6145. [DOI] [PubMed] [Google Scholar]
- Jones, S. E., Lloyd, L. J., Tan, K. K., and Buck, M. (2003). Secretion defects that activate the phage shock response of Escherichia coli. J. Bacteriol. 185, 6707-6711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang, P. J., Ostermann, J., Shilling, J., Neupert, W., Craig, E. A., and Pfanner, N. (1990). Requirement for hsp70 in the mitochondrial matrix for translocation and folding of precursor proteins. Nature 348, 137-143. [DOI] [PubMed] [Google Scholar]
- King, C., Eisenberg, E., and Greene, L. E. (1995). Polymerization of 70-kDa heat shock protein by yeast DnaJ in ATP. J. Biol. Chem. 270, 22535-22540. [DOI] [PubMed] [Google Scholar]
- King, C., Eisenberg, E., and Greene, L. E. (1999). Interaction between Hsc70 and DnaJ homologues: relationship between Hsc70 polymerization and ATPase activity. Biochemistry 38, 12452-12459. [DOI] [PubMed] [Google Scholar]
- Kleerebezem, M., Crielaard, W., and Tommassen, J. (1996). Involvement of stress protein PspA (phage shock protein A) in Escherichia coli in maintenance of the protonmotive force under stress conditions. EMBO J. 15, 162-171. [PMC free article] [PubMed] [Google Scholar]
- Kleerebezem, M., and Tommassen, J. (1993). Expression of the pspA gene stimulates efficient protein export in Escherichia coli. Mol. Microbiol. 7, 947-956. [DOI] [PubMed] [Google Scholar]
- Kourtz, L., and Ko, K. (1997). The early stage of chloroplast protein import involves Com70. J. Biol. Chem. 272, 2808-2813. [DOI] [PubMed] [Google Scholar]
- Kroll, D., Meierhoff, K., Bechtold, N., Kinoshita, M., Westphal, S., Vothknecht, U. C., Soll, J., and Westhoff, P. (2001). VIPP1, a nuclear gene of Arabidopsis thaliana essential for thylakoid membrane formation. Proc. Natl. Acad. Sci. USA 98, 4238-4242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli, U. K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680-685. [DOI] [PubMed] [Google Scholar]
- Li, H. M., Kaneko, Y., and Keegstra, K. (1994). Molecular cloning of a chloroplastic protein associated with both the envelope and thylakoid membranes. Plant Mol. Biol. 25, 619-632. [DOI] [PubMed] [Google Scholar]
- Lupas, A., Van Dyke, M., and Stock, J. (1991). Predicting coiled coils from protein sequences. Science 252, 1162-1164. [DOI] [PubMed] [Google Scholar]
- Madueño, F., Napier, J. A., and Gray, J. C. (1993). Newly imported iron-sulfur protein associates with both Cpn60 and Hsp70 in the chloroplast stroma. Plant Cell 5, 1865-1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miernyk, J. A. (2001). The J-domain proteins of Arabidopsis thaliana: an unexpectedly large and diverse family of chaperones. Cell Stress Chaperones 6, 209-218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orme, W., Walker, A. R., Gupta, R., and Gray, J. C. (2001). A novel plastid-targeted J-domain protein in Arabidopsis thaliana. Plant Mol. Biol. 46, 615-626. [DOI] [PubMed] [Google Scholar]
- Pierre, Y., and Popot, J.-L. (1993). Identification of two 4 kDa mini-proteins in the cytochrome b6f complex from Chlamydomonas reinhardtii. C R Acad. Sci. Ser. III 316, 1404-1409. [PubMed] [Google Scholar]
- Popov, N., Schmitt, S., and Matthies, H. (1975). Eine störungsfreie Mikromethode zur Bestimmung des Proteingehalts in Gewebshomogenaten. Acta Biol. Med. Ger. 34, 1441-1446. [PubMed] [Google Scholar]
- Rüdiger, S., Germeroth, L., Schneider-Mergener, J., and Bukau, B. (1997). Substrate specificity of the DnaK chaperone determined by screening of cellulose-bound peptide libraries. EMBO J. 16, 1501-1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E. F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual, Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
- Schägger, H., Cramer, W. A., and von Jagow, G. (1994). Analysis of molecular masses and oligomeric states of protein complexes by blue native electrophoresis and isolation of membrane protein complexes by two-dimensional native electrophoresis. Anal. Biochem. 217, 220-230. [DOI] [PubMed] [Google Scholar]
- Schiene-Fischer, C., Habazettl, J., Schmid, F. X., and Fischer, G. (2002). The hsp70 chaperone DnaK is a secondary amide peptide bond cis-trans isomerase. Nat. Struct. Biol. 9, 419-424. [DOI] [PubMed] [Google Scholar]
- Schlicher, T., and Soll, J. (1997). Chloroplastic isoforms of DnaJ and GrpE in pea. Plant Mol. Biol. 33, 181-185. [DOI] [PubMed] [Google Scholar]
- Schnell, D. J., Kessler, F., and Blobel, G. (1994). Isolation of components of the chloroplast protein import machinery. Science 266, 1007-1012. [DOI] [PubMed] [Google Scholar]
- Schroda, M. (2004). The Chlamydomonas genome reveals its secrets: chaperone genes and the potential roles of their gene products in the chloroplast. Photosynth. Res. 82, 221-240. [DOI] [PubMed] [Google Scholar]
- Schroda, M., Kropat, J., Oster, U., Rüdiger, W., Vallon, O., Wollman, F.-A., and Beck, C. F. (2001a). A role for molecular chaperones in assembly and repair of photosystem II?. Biochem. Soc. Trans. 29, 413-418. [DOI] [PubMed] [Google Scholar]
- Schroda, M., Vallon, O., Whitelegge, J. P., Beck, C. F., and Wollman, F.-A. (2001b). The chloroplastic GrpE homolog of Chlamydomonas: two isoforms generated by differential splicing. Plant Cell 13, 2823-2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroda, M., Vallon, O., Wollman, F.-A., and Beck, C. F. (1999). A chloroplast-targeted heat shock protein 70 (HSP70) contributes to the photoprotection and repair of photosystem II during and after photoinhibition. Plant Cell 11, 1165-1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Settles, A. M., Yonetani, A., Baron, A., Bush, D. R., Cline, K., and Martienssen, R. (1997). Sec-independent protein translocation by the maize Hcf106 protein. Science 278, 1467-1469. [DOI] [PubMed] [Google Scholar]
- Shevchenko, A., Wilm, M., Vorm, O., and Mann, M. (1996). Mass spectrometric sequencing of proteins from silver-stained polyacrylamide gels. Anal. Chem. 68, 850-858. [DOI] [PubMed] [Google Scholar]
- Shrager, J., Hauser, C., Chang, C. W., Harris, E. H., Davies, J., McDermott, J., Tamse, R., Zhang, Z., and Grossman, A. R. (2003). The Chlamydomonas rein-hardtii genome project. A guide to the generation and use of the cDNA information. Plant Physiol. 131, 401-408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silflow, C. D. (1998). Organization of the nuclear genome. In: Molecular Biology of Chlamydomonas: Chloroplasts and Mitochondria, ed. J.-D. Rochaix, M. Goldschmidt-Clermont, and S. Merchant S, Dordrecht, The Netherlands: Kluwer, 25-40.
- Sugito, K., Yamane, M., Hattori, H., Hayashi, Y., Tohnai, I., Ueda, M., Tsuchida, N., and Ohtsuka, K. (1995). Interaction between hsp70 and hsp40, eukaryotic homologues of DnaK and DnaJ, in human cells expressing mutant-type p53. FEBS Lett. 358, 161-164. [DOI] [PubMed] [Google Scholar]
- Sung, D. Y., Vierling, E., and Guy, C. L. (2001). Comprehensive expression profile analysis of the Arabidopsis Hsp70 gene family. Plant Physiol. 126, 789-800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo, A., Korszun, R., Hartl, F.-U., and Flanagan, J. (1996). A zinc finger-like domain of the molecular chaperone DnaJ is involved in binding to denatured protein substrates. EMBO J 15, 408-417. [PMC free article] [PubMed] [Google Scholar]
- Tomoyasu, T., Ogura, T., Tatsuta, T., and Bukau, B. (1998). Levels of DnaK and DnaJ provide tight control of heat shock gene expression and protein repair in Escherichia coli. Mol. Microbiol. 30, 567-581. [DOI] [PubMed] [Google Scholar]
- Tsugeki, R., and Nishimura, M. (1993). Interaction of homologues of Hsp70 and Cpn60 with ferredoxin-NADP+ reductase upon its import into chloroplasts. FEBS Lett. 320, 198-202. [DOI] [PubMed] [Google Scholar]
- Ungewickell, E., Ungewickell, H., Holstein, S.E.H., Lindner, R., Prasad, K., Barouch, W., Martin, B., Greene, L. E., and Eisenberg, E. (1995). Role of auxilin in uncoating clathrin-coated vesicles. Nature 378, 632-635. [DOI] [PubMed] [Google Scholar]
- Vitha, S., Froehlich, J. E., Koksharova, O., Pyke, K. A., van Erp, H., and Osteryoung, K. W. (2003). ARC6 is a J-domain plastid division protein and an evolutionary descendent of the cyanobacterial cell division protein Ftn2. Plant Cell 15, 1918-1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Gromoff, E. D., Treier, U., and Beck, C. F. (1989). Three light-inducible heat shock genes of Chlamydomonas reinhardtii. Mol. Cell. Biol. 9, 3911-3918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Kampen, J., Nieländer, U., and von Wettern, M. (1994). Stress-dependent transcription of a gene encoding a Gb-like polypeptide from Chlamydomonas reinhardtii. J. Plant Physiol. 143, 756-758. [Google Scholar]
- Westphal, S., Heins, L., Soll, J., and Vothknecht, U. C. (2001). Vipp1 deletion mutant of Synechocystis: a connection between bacterial phage shock and thylakoid biogenesis?. Proc. Natl. Acad. Sci. USA 98, 4243-4248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitelegge, J. P. (2003). Thylakoid membrane proteomics. Photosynth. Res. 78, 265-277. [DOI] [PubMed] [Google Scholar]
- Wickner, S., Hoskins, J., and McKenney, K. (1991). Monomerization of RepA dimers by heat shock proteins activates binding to DNA replication origin. Proc. Natl. Acad. Sci. USA 88, 7903-7907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, B., Wawrzynow, A., Zylicz, M., and Georgopoulos, C. (1996). Structure-function analysis of the Escherichia coli GrpE heat shock protein. EMBO J. 15, 4806-4816. [PMC free article] [PubMed] [Google Scholar]
- Zerges, W., and Rochaix, J.-D. (1998). Low density membranes are associated with RNA-binding proteins and thylakoids in the chloroplast of Chlamydomonas reinhardtii. J. Cell. Biol. 140, 101-110. [DOI] [PMC free article] [PubMed] [Google Scholar]