Abstract
Trafficking through the Golgi apparatus requires members of the Arf family of GTPases, whose activation is regulated by guanine nucleotide exchange factors (GEFs). Once activated, Arf-GTP recruits effectors such as coat complexes and lipid-modifying enzymes to specific membrane sites, creating a domain competent for cargo concentration and transport. GBF1 is a peripherally associated Arf GEF involved in both endoplasmic reticulum–Golgi and intra-Golgi transport. The mechanism of GBF1 binding to membranes is unknown. As a first step to understanding the mechanism of membrane association, we constructed a yellow fluorescent protein-tagged version of GBF1 and performed fluorescence recovery after photobleaching analysis to determine its residence time on Golgi membranes. We find that GBF1 molecules are not stably associated with the Golgi but rather cycle rapidly on and off membranes. The drug brefeldin A (BFA), an uncompetitive inhibitor of the exchange reaction that binds to an Arf–GDP–Arf GEF complex, stabilizes GBF1 on Golgi membranes. Using an in vivo assay to monitor Arf1-GTP levels, we show that GBF1 exchange activity on Arf1 is inhibited by BFA in mammalian cells. These results suggest that an Arf1–GBF1–BFA complex is formed and has a longer residence time on Golgi membranes than GBF1 or Arf1 alone.
INTRODUCTION
Arf GTPases play a central role in regulating membrane dynamics and protein transport in eukaryotic cells. Like all GTPases, Arf exists in inactive GDP-bound and active GTP-bound states. Arf activation leads to recruitment of coat proteins and lipid-modifying enzymes that in turn regulate the protein sorting and membrane deformation events associated with a given trafficking step (Nie et al., 2003). Arf is activated, that is, converted to its GTP-bound form, by guanine nucleotide exchange factors (GEFs) and returned to its GDP-bound form by GTPase activating proteins (GAPs) (Donaldson and Jackson, 2000). The Arf GEFs are responsible for the spatial and temporal regulation of Arf activation and all contain a centrally located Sec7 domain that is the catalytic domain for nucleotide exchange (Jackson and Casanova, 2000). There are six subfamilies of Arf GEFs in eukaryotic cells, one of which is the GEA/GBF subfamily, with homologues in all eukaryotes, including yeast, plants, and mammals (Cox et al., 2004). The Arf GEFs are all peripherally associated membrane proteins (Jackson and Casanova, 2000). The lower molecular weight GEFs such as ARNO and the cytohesins possess a pleckstrin homology (PH) domain that mediates membrane binding through direct interaction with specific phosphoinositides (Cullen and Chardin, 2000). The high-molecular-weight Arf GEFs associated with the Golgi apparatus, GBF1, BIG1, and BIG2, do not have recognizable PH domains, and little is known about the mechanism of their interaction with membranes (Cox et al., 2004). Because the Arf GEFs determine when and where Arf proteins will be activated, it is critical that they be localized to the correct membrane site. The Arf-related GTPase Sar1p acts in export of proteins from the ER and is activated by the Sec12p GEF, which is a transmembrane protein (Antonny and Schekman, 2001). Although there are parallels in the molecular events regulated by Sar1p and Arfs, the fact that Arf GEFs are recruited to membranes, whereas Sar1 GEFs are constitutively membrane-bound, is a clear difference.
GBF1 is a high-molecular-weight Arf GEF that at steady state is localized to early Golgi compartments in mammalian cells (Kawamoto et al., 2002; Zhao et al., 2002; Garcia-Mata et al., 2003). We constructed a yellow fluorescent protein (YFP)-tagged version of GBF1 to assess whether GBF1 associates stably or transiently with Golgi membranes. The drug brefeldin A (BFA) inhibits the activity of a subset of Sec7 domain Arf GEFs by binding to an Arf-GDP–Arf GEF complex in vitro (Peyroche et al., 1999). A yeast homologue of GBF1, Gea1p, is a target of BFA both in vitro and in vivo (Peyroche et al., 1999). It has been reported that GBF1 has in vitro GEF activity on both class I and class II Arf proteins and that this activity is resistant to BFA (Claude et al., 1999). GBF1 acts on both class I and II Arf proteins in cells, because it has been demonstrated that overexpression of GBF1 recruits Arf1, Arf3, and Arf5 to Golgi membranes (Kawamoto et al., 2002). These authors also demonstrate that overexpression of GBF1 prevents Arf1 from dissociating from Golgi membranes in response to BFA and that Arf3 and Arf5 levels at the Golgi are increased and resistant to BFA with GBF1 overexpression (Kawamoto et al., 2002). We find that GBF1 activity on the class I Arf protein Arf1 is inhibited by BFA in cells and that GBF1 is stabilized on Golgi membranes in the presence of BFA. Hence, GBF1 association with the Golgi is highly dynamic and its rapid cycling on and off membranes is significantly slowed by BFA treatment of cells. Our data support the conclusion that GBF1 is a direct target of BFA in cells.
MATERIALS AND METHODS
Antibodies and Reagents
All reagents were obtained from Sigma-Aldrich (St. Louis, MO) unless stated otherwise. Oligonucleotides were provided by Invitrogen (Carlsbad, CA) or QIAGEN (Valencia, CA).
Rhodamine-conjugated secondary antibodies were purchased from Southern Biotechnology (Birmingham, AL). Rabbit polyclonal anti-GBF1 antibodies were generated by immunization by using a conjugate of the 14 most C-terminal residues of human GBF1 (TPRPTDPIPTSEVN) with KLH (Zymed Laboratories, South San Francisco, CA). Mouse anti-hemagglutinin (HA) antibodies were purchased from Covance (Berkeley, CA), and horseradish peroxidase-conjugated secondary antibodies were purchased from Amersham Biosciences (Piscataway, NJ).
Cell Lines and Cell Culture
COS7 (African green monkey kidney) or normal rat kidney (NRK), both from American Type Tissue Culture Collection, Manassas, VA) cells were used in all experiments. Cells were maintained in Dulbecco's minimal essential medium (Biofluids, Rockville, MD) supplemented with 10% fetal bovine serum (FBS), 2 mM glutamine, 100 U/ml penicillin, and 100 mg/ml streptomycin at 37°C in a 5% CO2 incubator.
Transient transfections were performed using FuGENE 6 transfection reagent (Roche Diagnostics, Indianapolis, IN) according to the manufacturer's instructions. For live cell imaging, cells were grown in Lab-Tek chambered coverglasses (Nalge Nunc International, Rochester, NY) or 35-mm glass-bottom culture dishes (MatTek, Ashland, MA).
DNA Constructs
To construct YFP-GBF1, we used cDNA clone FLJ21677 encoding a GBF1 protein that is a different splice variant than has been reported previously (Claude et al., 1999; Kawamoto et al., 2002; Garcia-Mata et al., 2003). There is a three-nucleotide (one amino acid) insertion in the N-terminal portion that leads to an insertion of an additional Q at amino acid 338 of the Claude et al. (2002) GBF1 sequence, and a 12-nucleotide (four-amino acid) deletion in the C-terminal portion of the protein with respect to the originally reported GBF1 protein, amino acids 1494–1497 of the previously published sequence. To maintain consistency of numbering for Sec7 domain residues, we have started numbering our version of GBF1 at the amino acid directly after the initiating methionine. The different splice variants of GBF1, including the one we have used, have no effect on their ability to confer resistance to BFA when overexpressed in cells (Claude et al., 2003). This result is perfectly consistent with the fact that the changes lie well outside of the Sec7 domain (amino acids 698–893), which is the target of BFA. M832L and E794D were generated by QuikChangeXL mutagenesis kit according to manufacturer's instructions (Stratagene, La Jolla, CA).
Arf1, Arf3, Arf4, and Arf5 cDNAs (gifts from Julie Donaldson, National Institutes of Health, Bethesda, MD) were amplified by polymerase chain reaction and subcloned into pBiFC-YN155 (a gift from Tom Kerppola, University of Michigan Medical Center, Ann Arbor, MI) to generate Arf1-YN, Arf3-YN, Arf4-YN, and Arf5-YN, respectively. Full-length and the Sec7 domain deletion mutant of GBF1 (deletion of aa 710–894) were subcloned into pBiFC-YC155 (a gift from Tom Kerppola) to generate HA-GBF1-YC and ΔSEC7-GBF1-YC, respectively. All constructs were verified by sequencing. The pBiFC-YN155 and pBiFC-YC155 vectors contain FLAG- and HA-tag sequences, respectively, that result in fusion of these epitopes to the proteins expressed from the constructs. We tested expression and localization of the individual FLAG-Arf-YN and HA-GBF1-YC fusion proteins by immunofluorescene by using anti-FLAG and anti-HA antibodies, respectively. p58-CFP, p58-GFP, and Arf1-CFP were described previously (Ward et al., 2001; Presley et al., 2002). Elizabeth Sztul kindly provided p115-CFP.
Fixation and Staining of Cells
Cells grown on round coverslips (Daigger & Co., Wheeling, IL) were prepared for immunostaining by washing them three times with phosphate-buffered saline (PBS) (10 mM KH2PO4, 150 mM NaCl, pH 7.4) with 5% FBS. After fixation with 4% formaldehyde in PBS for 10 min at room temperature, excess formaldehyde was washed off with PBS, and cells were incubated with primary antibodies diluted in PBS with 0.2% saponin for 1 h at room temperature. After 1 h the antibody was aspirated, and cells were washed with PBS with 10% FBS. Cells were then incubated with secondary antibodies for 1 h at room temperature. After 1 h, the antibody was aspirated, and cells were washed with PBS with 10% FBS. Cells were mounted on slides in Fluoromount G and sealed with nail polish at the edge of the coverslip. Mounted cells were stored at 4°C protected from light up to several months. Cells were imaged using a Zeiss LSM 510 confocal microscope system (Carl Zeiss, Thornwood, NY).
Live Cell Imaging
All live cell imaging was done using a Zeiss LSM 510 confocal microscope system (Carl Zeiss). The stage was held at 37°C with a Nevtek airstream stage incubator (Burnsville, VA). Images not intended for quantitation were captured with a 63× 1.4 numerical aperture (NA) Zeiss PlanApochromat oil immersion objective. For quantitative imaging and photobleaching experiments, a 40× 1.3 NA Fluar oil immersion objective was used with the pinhole fully opened to collect fluorescence from the entire depth of the cell. For nonquantitative image several line scans were averaged to achieve higher image quality. For quantitative experiments only single line scans were acquired to increase the temporal resolution. Images were then analyzed using the Zeiss LSM software and converted to Quicktime movies with NIH Image 1.63. Single frames were processed with Adobe Photoshop 7 for presentation.
Quantitative Analysis and Photobleaching
For fluorescence recovery after photobleaching (FRAP) analysis, usually two prebleach images were taken at low laser intensity. Then, the region of interest (ROI) was scanned 5–10 times with the appropriate laser line at full laser power and zero attenuation to photobleach the fluorescence in this area as completely as possible. Fluorescence recovery into the ROI was monitored immediately after the bleach by time-lapse imaging (∼10 s/frame unless stated otherwise) at low-intensity illumination. As described above, average fluorescence intensities were measured using the Zeiss LSM software. The values were plotted using the following equation: F(t) = (FROI/FpreROI)/(Fcell/Fprecell) × 100, where Fprecell is the whole cell prebleach intensity, Fcell is the whole cell intensity at time t, FpreROI is the bleach ROI intensity before the bleach, and FROI is the intensity in the bleach ROI at time t. All values were background subtracted. For each quantitation, the number of cells imaged is indicated by “n.” These numbers refer to individual cells issuing from at least three independent transfections.
Arf-GTP Pull-Down Assay
To assess the amount of activated Arf-GTP in cells, we performed a pull-down assay by using a GST-GGA3-GAT domain construct (kindly provided by Julie Donaldson) as described previously (Santy and Casanova, 2001; Shinotsuka et al., 2002). Briefly, COS7 cells expressing Arf1-HA and YFP-GBF1 or Arf1-HA and vector alone were left untreated or treated with different concentrations of BFA. Cell lysates were prepared and incubated with glutathione-Sepharose beads containing a GST-GGA3-GAT fusion protein. Bound proteins were eluted from beads and loaded onto a gel. Western blotting analysis was performed using an enhanced chemiluminescence kit (Amersham Biosciences).
RESULTS
YFP-GBF1 Localizes to the Golgi
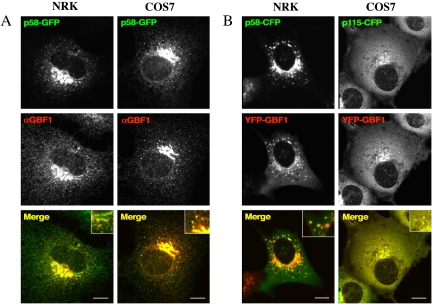
We constructed a fusion protein in which YFP was fused to the N terminus of GBF1. YFP-GBF1 was found to be present in cytoplasmic and Golgi membrane-associated pools in both NRK and COS7 cells (Figure 1). Fractionation experiments also have shown GBF1 to be distributed in both membrane and cytosolic pools (Claude et al., 1999). GBF1 has been described previously to localize to endoplasmic reticulum (ER) exit sites, VTC/intermediate compartment structures, and early Golgi compartments (Claude et al., 1999; Kawamoto et al., 2002; Zhao et al., 2002; Garcia-Mata and Sztul, 2003). YFP-GBF1, like endogenous GBF1, colocalizes with p58 at the Golgi and at peripheral sites (Figure 1). YFP-GBF1 also colocalizes very well with p115 (Figure 1B, right), as has been described previously for endogenous GBF1 (Garcia-Mata and Sztul, 2003; Garcia-Mata et al., 2003). Hence, YFP-GBF1 colocalizes with early Golgi markers and peripheral sites in a manner indistinguishable from endogenous GBF1. This result confirms that the YFP-tagged version of GBF1 retains the ability to localize correctly. Another function shared with untagged GBF1 is the ability of YFP-GBF1 to confer resistance to BFA when overexpressed in mammalian cells (Claude et al., 1999; see below). We also have confirmed that YFP-GBF1 is capable of stimulating production of Arf-GTP when overexpressed in mammalian cells (see below).
Figure 1.
Colocalization of GBF1 with p58 and p115 in NRK and COS7 cells. (A) NRK and COS7 cells were transfected with p58-GFP (green). Twenty-four hours after transfection, cells were fixed and stained with antibodies to GBF1 (red). Insets in merge images show colocalization of p58-GFP and endogenous GBF1. (B) NRK cells were cotransfected with p58-CFP (green) and YFP-GBF1 (red) and COS7 cells with p115-CFP (green) and YFP-GBF1 (red). Cells were visualized by 24 h after transfection. Insets in merge images show colocalization of p58-CFP or p115-CFP with overexpressed YFP-GBF1. Bar, 10 μm.
Dynamics of YFP–GBF1 Association with Golgi
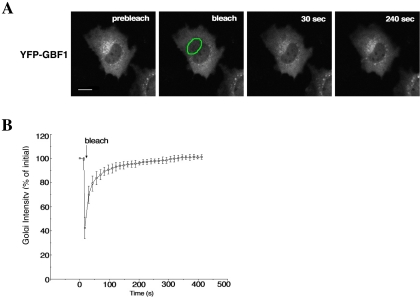
Previous studies have shown that the Arf1 GTPase and one of its effectors, COPI, cycle rapidly between Golgi membranes and cytoplasm in a manner independent of vesicle budding (Presley et al., 2002). To determine whether GBF1 cycles on and off membranes, or is more stably associated, we performed FRAP analysis. NRK cells were transiently transfected with YFP-GBF1, and the Golgi region was selected for photobleaching. Fluorescence recovered rapidly to the bleached region, indicating that GBF1 cycles rapidly on and off Golgi membranes (Figure 2A). The half-time of recovery of fluorescence was ∼30 s (Figure 2B). Hence, GBF1, like COPI and Arf, constantly binds to and is released from Golgi membranes.
Figure 2.
Kinetics of GBF1 binding to and dissociation from Golgi membranes. (A) NRK cells expressing YFP-GBF1 were photographed before photobleaching to obtain a prebleach image. Photobleaching of the Golgi region (outlined in green) was carried out by illumination with high-intensity laser light. After bleaching, images were taken at 13-s intervals to monitor the recovery of fluorescence to the bleached area. Images at representative time points of postbleach are shown. Bar, 10 μm. (B) Quantification of Golgi FRAP in NRK cells expressing YFP-GBF1 (n = 5). Error bars indicate the SD.
Overexpression of YFP-GBF1 Confers Resistance to BFA
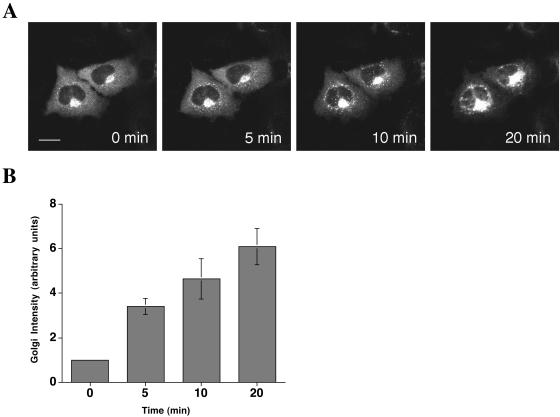
The drug BFA has profound effects on the secretory pathway in mammalian cells, causing rapid disassembly of the Golgi apparatus at high concentrations (Klausner et al., 1992; Sciaky et al., 1997). Overexpression of GBF1 confers resistance to the growth inhibition and Golgi breakdown caused by BFA treatment of Chinese hamster ovary and 293 cells (Claude et al., 1999). We obtained a similar result with COS7 cells overexpressing YFP-GBF1. When these cells were treated with a high concentration of BFA (5 μg/ml), ∼90% failed to undergo Golgi disassembly after 20 min of treatment, whereas in control cells not overexpressing YFP-GBF1, almost all cells had lost their Golgi structure under these conditions (our unpublished data). In NRK cells expressing YFP-GBF1, treatment with 5 μg/ml BFA led to Golgi disassembly in >80% of cells, probably due to the fact that NRK cells do not overexpress proteins to as high a level as do COS7 cells (Movie 1, supplemental materials; our unpublished data). Indeed, we found that at lower concentrations of BFA, overexpression of YFP-GBF1 protected cells from Golgi breakdown. At 0.5–1.0 μg/ml BFA, only a small fraction (10–30%) of NRK cells expressing YFP-GBF1 exhibited Golgi tubulation or redistribution into the ER (Figure 3A), even after 1 h of BFA treatment, whereas control cells did undergo these changes (our unpublished data). These results indicate that overexpression of YFP-GBF1 confers resistance to BFA in Golgi disassembly, as has been shown previously for untagged GBF1.
Figure 3.
GBF1 is stabilized on Golgi membranes by BFA. (A) NRK cells expressing YFP-GBF1 were treated with 1 μg/ml BFA for 20 min. Images were taken every 30 s after the addition of BFA (see movie in supplemental data). (B) Relative Golgi intensity was measured at representative time points (n = 4). Error bars indicate the SD. Bar, 10 μm.
GBF1 Is Stabilized on Golgi Membranes by BFA
When NRK cells expressing YFP-GBF1 were treated with 1 μg/ml BFA, and imaged over a period of 20 min, a dramatic increase in the amount of Golgi-localized GBF1 was observed (Figure 3A and Movie 2, supplemental materials). The Golgi/cytoplasm ratio of YFP-GBF1 increased at least sixfold after treatment with BFA (Figure 3B). The increase in signal was observed both in the Golgi region and at peripheral sites, indicating that recruitment of GBF1 to membranes by BFA is not restricted to a particular site, but it occurs at all membranes that GBF1 localizes to. When NRK cells were treated with 5 μg/ml BFA, >80% of cells eventually underwent Golgi disassembly, but even under these conditions, there was a rapid increase in the amount of YFP-GBF1 associated with both peripheral sites and the Golgi region (Movie 1, supplemental materials). After 10–15 min of BFA treatment, Golgi membranes fused with the ER in most cells, and at this point, at least a portion of YFP-GBF1 remained associated with ER membranes (Movie 1, supplemental materials). Hence, YFP-GBF1 is recruited to or stabilized on Golgi membranes by BFA.
Mechanism of BFA-induced Stabilization of GBF1 on Membranes
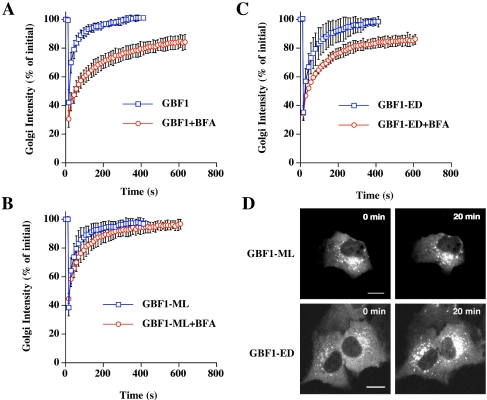
There are two possible explanations for the dramatic increase in GBF1 association with membranes in the presence of BFA. It could be that GBF1 is released more slowly from membranes in the presence of BFA or that GBF1 binding sites on membranes are limiting, and that BFA treatment leads to a dramatic increase in the number of these sites. To distinguish these possibilities, we performed FRAP experiments on YFP-GBF1–expressing cells treated with BFA. The kinetics of recovery of YFP-GBF1 to Golgi membranes in the presence of BFA is significantly slower than in its absence (Figure 4A). This result supports the first hypothesis, that in the presence of BFA, YFP-GBF1 dissociates from membranes much more slowly than in the absence of BFA. Over the time course tested, there is a slow, gradual recovery of YFP-GBF1 fluorescence to the Golgi region, indicating that there is a slow exchange of YFP-GBF1 from membrane to cytoplasm in BFA-treated cells.
Figure 4.
Golgi membrane binding and dissociation kinetics of GBF1 and Sec7 domain mutants in the presence and absence of BFA. Quantification of Golgi FRAP in NRK cells expressing YFP-GBF1 (n = 7) (A), YFP-GBF1-ML (n = 6) (B), or YFP-GBF1-ED (n = 7) (C) with or without addition of 1 μg/ml BFA. In the presence of BFA, FRAP was performed after 20 min of treatment. Error bars indicate the SD. (D) Images of cells expressing YFP-GBF1-ED and YFP-GBF1-ML with and without 1 μg/ml BFA. Bar, 10 μm.
One explanation for the results given above is that GBF1 is a direct target of BFA. However, it is formally possible that another direct target of BFA at the Golgi leads to stabilization of GBF1 on membranes. We previously identified residues within the Sec7 domain of Arf GEFs that are responsible for conferring resistance or sensitivity to BFA. One such mutation in yeast Gea1p, M699L, was able to confer resistance to BFA both in vivo and in vitro (Peyroche et al., 1999). The crystal structures of the Sec7 domain–Arf1–GDP–BFA complex have shown that M699 in Gea1 (corresponding to M832L in GBF1) makes a direct van der Waals contact with the BFA molecule in the complex (Mossessova et al., 2003; Renault et al., 2003). We introduced the corresponding mutation, M832L, into YFP-GBF1. If GBF1 is a direct target of BFA, we predict that GBF1-M832L will have a lower affinity for BFA and hence that cells expressing this mutant will be resistant to the effects of BFA. This model predicts the same rate of recovery from photobleaching for GBF1-M832L and wild-type GBF1. If a distinct Arf GEF at the Golgi is the target of BFA, then we predict that YFP-GBF1-M832L will have the same characteristics as wild-type YFP-GBF1, that is, slower recovery kinetics from FRAP of the Golgi region in cells treated with BFA compared with untreated cells. We performed FRAP analysis on cells expressing YFP-GBF1-M832L in the presence and absence of BFA. The half-time of recovery of fluorescence to the Golgi region in the absence of BFA was essentially identical to that of the wild-type protein (Figure 4B). Strikingly, in the presence of BFA, recovery was much faster than for wild-type GBF1 and was very similar to that of untreated cells (Figure 4, B and D). This result strongly supports the conclusion that GBF1 is a direct target of BFA.
We tested another Sec7 domain mutant of GBF1, GBF1-E794D, that carries a mutation in the catalytic glutamate residue and is predicted to be defective in exchange activity. In the ARNO protein, the corresponding E→D mutation decreases exchange activity by 400-fold in vitro (Beraud-Dufour et al., 1998). In contrast to the YFP-GBF1-E794K mutant, which causes fragmentation of the Golgi apparatus, when YFP-GBF1-E794D is expressed in cells, the Golgi apparatus remains intact (Figure 4D) and hence FRAP analysis of the Golgi region can be carried out. The recovery kinetics of YFP-GBF1-E794D after photobleaching of the Golgi region was very similar to those of wild-type YFP-GBF1, both in the presence and absence of BFA (Figure 4, C and D). Hence, not all mutations in the Sec7 domain of GBF1 confer resistance to BFA in the Golgi FRAP assay.
As a more direct measure of YFP-GBF1 dissociation from Golgi membranes in the presence and absence of BFA, we performed fluorescence loss in photobleaching experiments. In the absence of BFA, YFP-GBF1 fluorescence was lost rapidly from the Golgi region upon repeated bleaching of a region of the cytoplasm (Movie 3, supplemental materials). Fluorescence intensity of the Golgi region after 50 bleaches was <10 ± 2% of the level before photobleaching of a region of the cytoplasm, and it was <3 ± 1% of the level before photobleaching after 100 bleaches. These results show that the entire Golgi pool of GBF1 is exchanging with the cytoplasmic pool and rules out the possibility that photobleaching the Golgi in the FRAP experiments described above induces dissociation of an otherwise stable pool of YFP-GBF1 from these membranes. When cells expressing YFP-GBF1 were treated with BFA before photobleaching, the loss of GBF1 from Golgi membranes was significantly slowed (Movie 4, supplemental materials). Fluorescence intensity of the Golgi region after 50 bleaches was still 50 ± 10% of the original level. This result confirms the conclusion that GBF1 binding to membranes is stabilized upon BFA treatment.
In Vivo Physical Interaction between GBF1 and Arf1 by Using Bimolecular Fluorescence Complementation
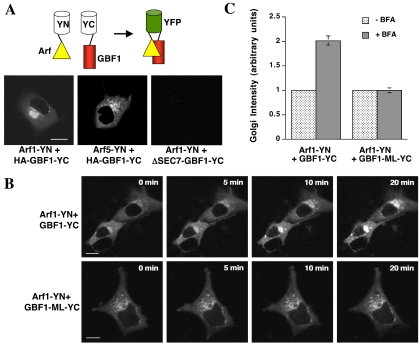
It has been reported that GBF1 acts specifically on class II Arf proteins such as Arf5 (Claude et al., 1999), although recent in vivo data support the idea that GBF1 interacts also with class I Arf proteins (Kawamoto et al., 2002). To test this directly in vivo, we made use of the bimolecular fluorescence complementation assay developed by Tom Kerppola and colleagues (Hu et al., 2002). In this assay, one-half of the YFP molecule is fused to a protein of interest and the other YFP half is fused to a potential partner. If the two proteins interact, the two halves of YFP are brought together and allow formation of the YFP fluorophore to produce a fluorescent signal. We expressed class I human Arf proteins (Arf1 and Arf3) fused at their C termini to the N terminal half of YFP and GBF1 fused to the C-terminal half of YFP. Each fusion protein expressed alone in COS7 cells produced no detectable fluorescence (our unpublished data). We saw a robust YFP fluorescence signal at the Golgi when either Arf1 or Arf3 was coexpressed with GBF1, indicating that these proteins interact directly in cells (Figure 5A; our unpublished data). GBF1 also interacts directly with class II Arf proteins in cells, because both Arf4 and Arf5 give a positive signal in the bimolecular fluorescence complementation assay (Figure 5A; our unpublished data). To confirm the specificity of these interactions, we tested a GBF1 construct in which the Sec7 domain was deleted (ΔSEC7-GBF1-YC) and saw no interaction with Arf1 (Figure 5A). We confirmed that both Arf1-YN and ΔSEC7-GBF1-YC were expressed in cells by immunofluorescence analysis (our unpublished data; see Materials and Methods). These results show that the GBF1 Sec7 domain physically interacts with class I and class II Arf proteins in vivo.
Figure 5.
Bimolecular fluorescence complementation analysis to monitor GBF1-Arf1 interaction in cells. (A) Schematic diagram showing principle of bimolecular fluorescence complementation assay (top). COS7 cells were transfected with Arf1-YN and either HA-GBF1-YC or ΔSEC7-GBF1-YC and were imaged 24 h after transfection (bottom). Bar, 10 μm. (B) COS7 cells were transfected with Arf1-YN and either HA-GBF1-YC or HA-GBF1-M794L-YC and were treated for the indicated times with 5 μg/ml BFA. Cells were imaged 24 h after transfection. Bar, 10 μm. (C) Quantitation of the results shown in B (n = 5). Error bars indicate the SD.
Having visualized a physical Arf1–GBF1 complex, we next tested whether this complex could be recruited to membranes by BFA. We found that this was indeed the case (Figure 5B, top). In contrast, BFA had no effect on the level of Arf1-YN–GBF1-M832L-YC complex present at the Golgi (Figure 5B, bottom). Quantitation of these results is shown in Figure 5C. These results support the conclusion that BFA stabilizes a GBF1–Arf1 complex at the Golgi in mammalian cells.
Evidence for Arf1 Stabilization on Golgi Membranes by BFA
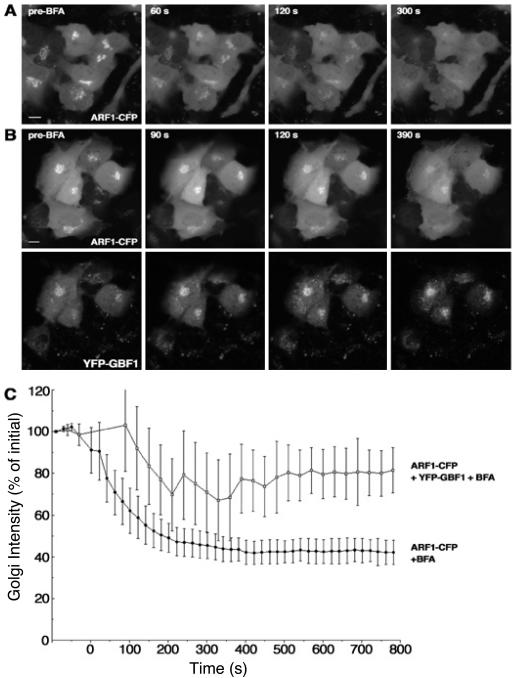
Arf1 is rapidly released from Golgi membranes when cells are treated with BFA, due to the fact that GEF activity is inhibited but GAP activity is not and continues to return Arf to its soluble GDP-bound form (Presley et al., 2002). This result would seem to contradict the conclusion that BFA is trapping Arf1 in a GBF1–Arf1 complex on membranes. However, Arf1 is a very abundant protein and likely is present in excess over GBF1, even at endogenous levels of expression. Hence, only a small fraction of total overexpressed Arf1-CFP would be in complex with endogenous GBF1 at the Golgi in the presence of BFA, likely below the level of detection compared with the large background of cytoplasmic Arf1. Increasing the amount of YFP-GBF1 should increase the amount of Arf1–GBF1–BFA complex to a level detectable over the background of cytoplasmic Arf1. To test this hypothesis, we treated cells expressing Arf1–CFP alone or coexpressing Arf1-CFP and YFP-GBF1 with BFA and monitored Arf1-CFP fluorescence in the Golgi region. Arf1-CFP was rapidly lost from Golgi membranes after treatment with BFA, as described previously (Figure 6). In cells overexpressing YFP-GBF1 and Arf1-CFP, a higher level of Arf1 was retained on Golgi membranes in the presence of BFA (Figure 6). These results show that both GBF1 and Arf1 are stabilized on Golgi membranes by BFA and support the idea that BFA is stabilizing a GBF–Arf1 complex on Golgi membranes.
Figure 6.
More Arf remains associated with Golgi membranes after BFA treatment in cells expressing GBF1. (A) NRK cells expressing Arf1-CFP were treated with 1 μg/ml BFA. Images at representative time points are shown. (B) NRK cells expressing Arf1-CFP and YFP-GBF1 were treated with 1 μg/ml BFA. Images of Arf1 (top) and GBF1 (bottom) taken at the same time point are shown. (C) Quantification of Arf1 release from membranes with (n = 4) or without (n = 6) YFP-GBF1 after BFA treatment. Error bars indicate the SD. Bar, 10 μm.
GBF1 Stimulates Production of Arf-GTP In Vivo
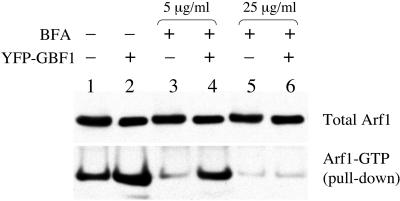
The results presented above strongly support the idea that BFA can inhibit the activity of GBF1 by binding directly to its Sec7 domain, as has been shown previously for the yeast homologues of GBF1, Gea1p, and Gea2p (Peyroche et al., 1999; Robineau et al., 2000; Mossessova et al., 2003). However, in vitro exchange assays have shown that the activity of GBF1 on class I Arf proteins is insensitive to BFA (Claude et al., 1999). It should be noted that the activity of GBF1 on class I Arf proteins in this study was only seen under very low magnesium concentration and that its activity on Arf was not detectable at physiological magnesium concentration (Claude et al., 1999). Many factors influence the exchange activity of a given Sec7 domain protein in vitro, and it may not fully reflect the in vivo situation. Hence, the possibility that GBF1 does catalyze nucleotide exchange on class I Arf proteins in cells cannot be ruled out. To test this possibility directly, we carried out in vivo assays to determine whether GBF1 can activate Arf1 in cells. We used a GST-GGA3-GAT domain fusion protein to monitor the amount of Arf1-GTP protein in lysates of cells expressing Arf1-HA (Santy and Casanova, 2001; Shinotsuka et al., 2002). GGA3 is an effector of Arf1, and the GAT domain binds with high affinity specifically to Arf1-GTP (Boman, 2001). We found a two- to threefold increase in the amount of Arf1-GTP present in cells overexpressing YFP-GBF1 compared with cells carrying vector alone (Figure 7, lanes 1 and 2). Hence, GBF1 is capable of catalyzing nucleotide exchange on Arf1 in vivo, and the YFP tag does not abolish this exchange activity. When cells were treated with BFA, a dramatic reduction in the amount of Arf1-GTP present in cells was observed (Figure 7, lanes 3 and 5). If GBF1 is a direct target of BFA, we predict that BFA should inhibit the increase in Arf-GTP that accompanies its overexpression in untreated cells. This prediction was borne out when COS7 cells expressing YFP-GBF1 or vector alone were treated with 25 μg/ml BFA (Figure 7, lanes 5 and 6). When cell were treated with 5 μg/ml BFA, GBF1 overexpression led to a significantly higher level of Arf-GTP in cells, as would be expected given the fact that GBF1 overexpression confers resistance to this concentration of BFA in the Golgi disassembly assay. These results also show that there is a correlation between the level of GBF1 expressed in cells and the amount of BFA that is required to have an effect, and they are consistent with the conclusion that GBF1 is a direct target of the drug. Our interpretation of the results is that in COS7 cells, at low concentrations of BFA (e.g., 5 μg/ml), the level of overexpression of GBF1 is high enough that not all molecules are inhibited by the drug and hence in vivo exchange activity is observed. At a high enough concentration of BFA (25 μg/ml), most or all molecules of GBF1 are bound by the drug and in vivo exchange activity is inhibited.
Figure 7.
Arf-GTP pull-down assay. COS7 cells were cotransfected with Arf1-HA and empty vector (lanes 1, 3, and 5) or Arf1-HA and YFP-GBF1 (lanes 2, 4, and 6). Sixteen hours after transfection, cells were either lysed directly (lanes 1 and 2) or treated with 5 μg/ml BFA (lanes 3 and 4) or 25 μg/ml BFA (lanes 5 and 6) for 20 min before lysis. Arf-GTP pull-downs by using GST-GGA3-GAT bound to glutathione-Sepharose was performed as described in Materials and Methods.
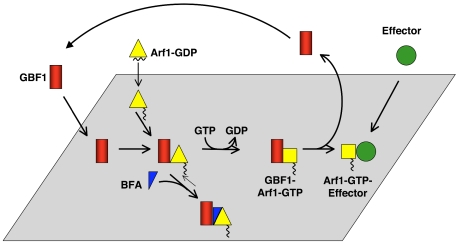
Our results are summarized in Figure 8. GBF1 and Arf1 are targeted independently to membranes. When the two proteins encounter each other, the exchange reaction occurs rapidly and GBF1 is released, leaving Arf1-GTP associated with membranes to recruit effectors. We propose that BFA binds to a GBF1–Arf1–GDP reaction intermediate and stabilizes this complex on Golgi membranes.
Figure 8.
Model for Arf activation at the Golgi. The Arf GEF GBF1 (red) and Arf1-GDP (yellow triangle) are recruited independently to Golgi membranes. The exchange reaction proceeds through several short-lived intermediates, and GBF1 is released from membranes upon GTP binding to Arf1. Arf1-GTP (yellow square) remains associated with membranes where it recruits effectors. BFA (blue triangle) binds to an early reaction intermediate, a GBF1–Arf–GDP complex, which is stabilized on Golgi membranes.
DISCUSSION
Activation of class I Arf proteins at the Golgi is essential to maintaining Golgi structure and function. The first step in Arf activation is recruitment of the Arf GEFs to Golgi membranes. We have demonstrated that GBF1 cycles rapidly on and off Golgi membranes and that BFA dramatically inhibits release of GBF1 from these membranes. We provide three lines of evidence that GBF1 is a BFA-sensitive Arf GEF that acts upon Arf1 in mammalian cells. First, introducing a mutation into the GBF1 Sec7 domain in an amino acid that has been shown to make a direct contact with BFA in the crystal structure of the Sec7 domain–Arf–GDP–BFA complex (Mossessova et al., 2003) renders GBF1 insensitive to BFA-induced stabilization on Golgi membranes. Second, direct visualization of an Arf1–GBF1 complex at the Golgi by using the bimolecular fluorescence complementation assay shows that this complex is recruited to Golgi membranes. Finally, we have assayed GBF1 activity on Arf1 in vivo by using the GGA3-GAT pull-down assay and show that GBF1 stimulates Arf1-GTP production in cells when it is overexpressed and that this activity is abolished by BFA treatment.
BFA forms an abortive Arf1–GDP–BFA–Sec7 domain protein complex (Peyroche et al., 1999). Our data support the idea that an Arf1–GDP–BFA–GBF1 complex binds membranes more stably than GBF1 alone in cells. Arf1-GDP is in a conformation that does not bind tightly to membranes on its own. Interestingly, even though Arf1 is in its GDP-bound conformation in the BFA complex, the Arf1–GDP–GBF1–BFA complex is tightly associated with Golgi membranes. This result suggests that in cells, the Arf1–GDP–GBF1 reaction intermediate that is the target of BFA is associated with membranes, perhaps through interaction of GBF1 and/or Arf with a membrane receptor. It is likely that Arf1 and GBF1 are targeted separately to membranes. Arf1-GDP interacts with p23, a member of the p24 family of transmembrane proteins that localize to the ER and early Golgi, and it could act as a membrane receptor for Arf1-GDP (Gommel et al., 2001). Rein et al. (2002) have shown that in yeast, Arf1 binds to SNAREs, which could potentially be involved in membrane recruitment of Arf1. We have identified hGmh1 as a potential membrane receptor for Gea2p and GBF1, although it clearly is not the only such receptor in cells (Chantalat et al., 2003; Niu and Jackson, unpublished data). p115 is a Golgi scaffold protein that has been shown to interact with GBF1 and could also be involved in recruiting GBF1 to membranes (Garcia-Mata and Sztul, 2003). Future experiments will be directed toward determining to which membrane proteins or lipids GBF1 and the Arf1–GDP–GBF1–BFA complex interact.
It was reported previously that GBF1 acts on the class II Arf protein, Arf5, and that this activity is not inhibited by BFA (Claude et al., 1999). We have demonstrated that in cells, GBF1 acts directly on a class I Arf protein, Arf1, in a manner that is sensitive to BFA. Our results do not conflict with the conclusions of Claude et al. (1999), and in fact support them, because we have confirmed by using the bimolecular fluorescence complementation assay that GBF1 physically interacts with Arf5 in cells. Although Claude et al. (1999) show that GBF1 has BFA-resistant exchange activity in vitro on Arf1, this activity was only seen at a nonphysiological magnesium concentration. Hence, these data do not conflict with our results, because we have tested activity of GBF1 in vivo under physiological conditions, where Claude et al. (1999) were unable to detect in vitro activity. Our results explain in simple terms why GBF1 was originally identified as a BFA-resistance factor. Overexpression of GBF1 confers resistance to BFA at the Golgi because it is a direct target of the drug in cells, and increasing its level for a given concentration of BFA overcomes the titration. BFA acts in the same way as a dominant negative mutant, freezing GBF1 in an inactive complex with Arf. Because GBF1 is less abundant than Arf, it is the rate-limiting component of the complex and hence its overexpression leads to resistance against BFA. This mechanism has been demonstrated previously for the Saccharomyces cerevisiae homologues of GBF1, the redundant Gea1p and Gea2p proteins (Peyroche et al., 1999), and hence is evolutionarily conserved. There are two other Arf GEFs in mammalian cells that have exchange activity sensitive to BFA, BIG1, and BIG2 (Togawa et al., 1999). Overexpression of BIG1 or BIG2 confers resistance to BFA at the trans-Golgi network, but not earlier Golgi cisternae (Shinotsuka et al., 2002; Smirnova and Jackson, unpublished observations). Hence, it is likely that GBF1 is a target of BFA at the early Golgi in mammalian cells.
Supplementary Material
Acknowledgments
We thank Elizabeth Sztul, Julie Donaldson, and Tom Kerppola for plasmids, and George Patterson for expert assistance in microscopy. We thank Julie Donaldson and Wei Liu for critical reading of the manuscript. A.C.P. was supported by a Ph.D. scholarship from Boehringer Ingelheim Fonds (Heidesheim, Germany).
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E04-07-0599) on December 22, 2004.
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
References
- Antonny, B., and Schekman, R. (2001). ER export: public transportation by the COPII coach. Curr. Opin. Cell Biol. 13, 438-443. [DOI] [PubMed] [Google Scholar]
- Beraud-Dufour, S., Robineau, S., Chardin, P., Paris, S., Chabre, M., Cherfils, J., and Antonny, B. (1998). A glutamic finger in the guanine nucleotide exchange factor ARNO displaces Mg2+ and the beta-phosphate to destabilize GDP on ARF1. EMBO J. 17, 3651-3659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boman, A. L. (2001). GGA proteins: new players in the sorting game. J. Cell. Sci. 114, 3413-3418. [DOI] [PubMed] [Google Scholar]
- Chantalat, S., Courbeyrette, R., Senic-Matuglia, F., Jackson, C. L., Goud, B., and Peyroche, A. (2003). A novel Golgi membrane protein is a partner of the ARF exchange factors Gea1p and Gea2p. Mol. Biol. Cell 14, 2357-2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claude, A., Zhao, B. P., Kuziemsky, C. E., Dahan, S., Berger, S. J., Yan, J. P., Armold, A. D., Sullivan, E. M., and Melancon, P. (1999). GBF 1, A novel Golgi-associated BFA-resistant guanine nucleotide exchange factor that displays specificity for ADP-ribosylation factor 5. J. Cell Biol. 146, 71-84. [PMC free article] [PubMed] [Google Scholar]
- Claude, A., Zhao, B. P., and Melancon, P. (2003). Characterization of alternatively spliced and truncated forms of the Arf guanine nucleotide exchange factor GBF1 defines regions important for activity. Biochem. Biophys. Res. Commun. 303, 160-169. [DOI] [PubMed] [Google Scholar]
- Cox, R., Mason-Gamer, R. J., Jackson, C. L., and Segev, N. (2004). Phylogenetic analysis of Sec7-domain-containing Arf nucleotide exchangers. Mol. Biol. Cell 15, 1487-1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen, P. J., and Chardin, P. (2000). Membrane targeting: what a difference a G makes. Curr. Biol. 10, R876-R878. [DOI] [PubMed] [Google Scholar]
- Donaldson, J. G., and Jackson, C. L. (2000). Regulators and effectors of the ARF GTPases. Curr. Opin. Cell Biol. 12, 475-482. [DOI] [PubMed] [Google Scholar]
- Garcia-Mata, R., and Sztul, E. (2003). The membrane-tethering protein p115 interacts with GBF1, an ARF guanine-nucleotide-exchange factor. EMBO Rep. 4, 320-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Mata, R., Szul, T., Alvarez, C., and Sztul, E. (2003). ADP-ribosylation factor/COPI-dependent events at the endoplasmic reticulum-Golgi interface are regulated by the guanine nucleotide exchange factor GBF1. Mol. Biol. Cell 14, 2250-2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gommel, D. U., Memon, A. R., Heiss, A., Lottspeich, F., Pfannstiel, J., Lechner, J., Reinhard, C., Helms, J. B., Nickel, W., and Wieland, F. T. (2001). Recruitment to Golgi membranes of ADP-ribosylation factor 1 is mediated by the cytoplasmic domain of p23. EMBO J. 20, 6751-6760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, C. D., Chinenov, Y., and Kerppola, T. K. (2002). Visualization of interactions among bZIP and Rel family proteins in living cells using bimolecular fluorescence complementation. Mol. Cell 9, 789-798. [DOI] [PubMed] [Google Scholar]
- Jackson, C. L., and Casanova, J. E. (2000). Turning on ARF: the Sec7 family of guanine-nucleotide-exchange factors. Trends Cell Biol. 10, 60-67. [DOI] [PubMed] [Google Scholar]
- Kawamoto, K., Yoshida, Y., Tamaki, H., Torii, S., Shinotsuka, C., Yamashina, S., and Nakayama, K. (2002). GBF1, a guanine nucleotide exchange factor for ADP-ribosylation factors, is localized to the cis-Golgi and involved in membrane association of the COPI coat. Traffic 3, 483-495. [DOI] [PubMed] [Google Scholar]
- Klausner, R. D., Donaldson, J. G., and Lippincott-Schwartz, J. (1992). Brefeldin A: insights into the control of membrane traffic and organelle structure. J. Cell. Biol. 116, 1071-1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mossessova, E., Corpina, R. A., and Goldberg, J. (2003). Crystal structure of ARF1*Sec7 complexed with brefeldin A and its implications for the guanine nucleotide exchange mechanism. Mol. Cell 12, 1403-1411. [DOI] [PubMed] [Google Scholar]
- Nie, Z., Hirsch, D. S., and Randazzo, P. A. (2003). Arf and its many interactors. Curr. Opin. Cell. Biol. 15, 396-404. [DOI] [PubMed] [Google Scholar]
- Peyroche, A., Antonny, B., Robineau, S., Acker, J., Cherfils, J., and Jackson, C. L. (1999). Brefeldin A acts to stabilize an abortive ARF-GDP-Sec7 domain protein complex: involvement of specific residues of the Sec7 domain. Mol. Cell. 3, 275-285. [DOI] [PubMed] [Google Scholar]
- Presley, J. F., Ward, T. H., Pfeifer, A. C., Siggia, E. D., Phair, R. D., and Lippincott-Schwartz, J. (2002). Dissection of COPI and Arf1 dynamics in vivo and role in Golgi membrane transport. Nature 417, 187-193. [DOI] [PubMed] [Google Scholar]
- Rein, U., Andag, U., Duden, R., Schmitt, H. D., and Spang, A. (2002). ARF-GAP-mediated interaction between the ER-Golgi v-SNAREs and the COPI coat. J. Cell. Biol. 157, 395-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renault, L., Guibert, B., and Cherfils, J. (2003). Structural snapshots of the mechanism and inhibition of a guanine nucleotide exchange factor. Nature 426, 525-530. [DOI] [PubMed] [Google Scholar]
- Robineau, S., Chabre, M., and Antonny, B. (2000). Binding site of brefeldin A at the interface between the small G protein ADP-ribosylation factor 1 (ARF1) and the nucleotide-exchange factor Sec7 domain. Proc. Natl. Acad. Sci. USA 97, 9913-9918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santy, L. C., and Casanova, J. E. (2001). Activation of ARF6 by ARNO stimulates epithelial cell migration through downstream activation of both Rac1 and phospholipase D. J. Cell Biol. 154, 599-610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sciaky, N., Presley, J., Smith, C., Zaal, K. J., Cole, N., Moreira, J. E., Terasaki, M., Siggia, E., and Lippincott-Schwartz, J. (1997). Golgi tubule traffic and the effects of brefeldin A visualized in living cells. J. Cell. Biol. 139, 1137-1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinotsuka, C., Yoshida, Y., Kawamoto, K., Takatsu, H., and Nakayama, K. (2002). Overexpression of an ADP-ribosylation factor-guanine nucleotide exchange factor, BIG2, uncouples brefeldin A-induced adaptor protein-1 coat dissociation and membrane tubulation. J. Biol. Chem. 277, 9468-9473. [DOI] [PubMed] [Google Scholar]
- Togawa, A., Morinaga, N., Ogasawara, M., Moss, J., and Vaughan, M. (1999). Purification and cloning of a brefeldin A-inhibited guanine nucleotide-exchange protein for ADP-ribosylation factors. J. Biol. Chem. 274, 12308-12315. [DOI] [PubMed] [Google Scholar]
- Zhao, X., Lasell, T. K., and Melancon, P. (2002). Localization of large ADP-ribosylation factor-guanine nucleotide exchange factors to different Golgi compartments: evidence for distinct functions in protein traffic. Mol. Biol. Cell 13, 119-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.