Abstract
Single nucleotide polymorphisms (SNPs) in MicroRNAs (miRNAs) are involved in the mechanism of carcinogenesis. Several studies have evaluated the association of rs4919510 SNP in miR-608 with cancer susceptibility in different types of cancer, with inconclusive outcomes. To obtain a more precise estimation, we carried out this meta-analysis through systematic retrieval from the PubMed and Embase database. A total of 10 case-control studies were analyzed with 6,000 cases and 7,664 controls. The results showed that 4919510 SNP in miR-608 was significantly associated with decreased cancer risk only in recessive model (CC vs. GG+GC: OR=0.89, 95% CI: 0.82-0.97, P=0.009). By further stratified analysis, we found that rs4919510 SNP had some relationship with decreased cancer risk in both homozygote model (CC vs. GG: OR=0.59, 95% CI: 0.36-0.96, P=0.034) and dominant model (CG+ CC vs. GG: OR=0.60, 95% CI: 0.37-0.98, P=0.042) in Caucasians but no relationship in any genetic model in Asians. These results indicated that miR-608 rs4919510 polymorphism may contribute to the decreased cancer susceptibility and could be a promising target to forecast cancer risk for clinical practice. However, to further confirm these results, well-designed large scale case–control studies are needed in the future.
Keywords: miR-608, rs4919510, polymorphism, cancer risk, meta-analysis
INTRODUCTION
MicroRNAs (miRNAs) are small (usually 21–23 nucleotides in length), evolutionarily conserved, noncoding RNA molecules which participate in post-transcriptional gene regulation by binding to the complementary sequences in 3’ untranslated region (3’ UTR) of target messenger RNAs (mRNAs) [1, 2]. These miRNAs function as negative regulators through degradation of mRNAs and translational repression [3]. Accumulated evidence has suggested that the aberrations of these miRNAs are involved in cell proliferation, differentiation, migration and apoptosis in the process of carcinogenesis [4, 5]. Studies have shown that approximately 50% miRNA genes are located in cancer-related chromosomal regions [6]. The loss or gain of specific function of several miRNAs are thought to be significant events in diverse types of cancer [7]. These evidences indicated that microRNAs could be a kind of biomarkers to evaluate cancer risk.
Single nucleotide polymorphisms (SNPs), the most common genetic variation, have been demonstrated to influence the expression or target site selection of miRNAs and thus are involved in a series of biological processes by interfering interaction between miRNAs and target mRNAs [8, 9]. SNPs present in the miRNA genes have been illustrated to be a potentially important mechanism in the development and progression of cancer [10, 11].
Recently, rs4919510 SNP in miR-608 has been reported to be a predictor of clinical outcomes for patients with renal cell carcinoma [12], colorectal adenocarcinoma [13, 14], esophageal squamous cell carcinoma [15] and nasopharyngeal carcinoma [16]. However, the relationship between cancer risk and rs4919510polymorphism in miR-608 is now inconclusive and controversial. Qiu and colleagues had found that rs4919510 (C > G) polymorphism showed a consistent association with nasopharyngeal carcinoma susceptibility in south area of China [17]. Other study reported that there existed no significant association between miRNA-608 rs4919510 and the risk of colorectal cancer [18]. By systematically summarizing the existing data, we performed a meta-analysis to further determine whether there is an association of rs4919510 polymorphism in miR-608 with cancer susceptibility.
RESULTS
Characteristics of the studies
A total of 51 literatures based on our searching strategy were selected out from PubMed and EMBASE database. After screening the title and abstract, 25 studies uncorrelated with cancer risk and SNPs were excluded and 26 literatures were then evaluated in detail. Finally ten case-control studies [17–26] meeting our inclusion criteria were included into our meta-analysis with 6,000 cases and 7,664 controls (Figure 1) (Table 1). One article composed by Ryan and colleagues contains two case-control studies with detailed genotype information of Caucasian and African Americans. To perform subgroup analysis more conveniently, we regarded this one as two independent studies according to ethnicity. In all included studies, genotype distributions of rs4919510(C > G) in the controls were in agreement with HWE. A variety of genotyping methods were applied including Taqman, RT-PCR, MassArray, SNaPshot, and SNPstream. Genomic DNA was isolated from blood samples in all included studies except one (using both blood and tissue) [18].
Figure 1. Study flow chart for the process of selecting the eligible publications.
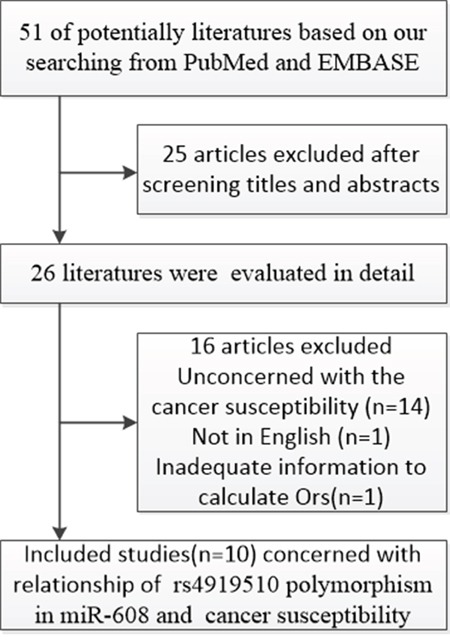
Table 1. Characteristics of studies in the meta-analysis.
| Author | Year | country | Ethnicity | Cancer type | Genotyping | Source of controls | Cases(n) | Controls(n) | P value for HWE h | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Total | GG | GC | CC | Total | GG | GC | CC | ||||||||
| Dong | 2015 | China | Asian | Thyroid tumor | MassArray | HB f | 369 | 136 | 186 | 47 | 751 | 279 | 370 | 102 | 0.494 |
| Zhang | 2015 | China | Asian | ESCC b | SNaPshot | PB g | 738 | 217 | 384 | 137 | 882 | 291 | 440 | 151 | 0.784 |
| Yin | 2015 | China | Asian | Lung cancer | Taqman | HB | 258 | 65 | 140 | 53 | 310 | 96 | 152 | 62 | 0.992 |
| Wei | 2015 | China | Asian | Thyroid tumor | MassArray | PB | 824 | 266 | 428 | 130 | 1031 | 326 | 503 | 202 | 0.950 |
| Qiu | 2015 | China | Asian | SCCHN c | TaqMan | PB | 906 | 255 | 460 | 191 | 1072 | 254 | 532 | 286 | 0.977 |
| Wang | 2014 | China | Asian | HCC d | MassArray | HB | 993 | 304 | 500 | 189 | 992 | 318 | 497 | 177 | 0.775 |
| Huang | 2012 | China | Asian | Breast cancer | SNPstream | PB | 1118 | 381 | 545 | 192 | 1417 | 456 | 684 | 277 | 0.776 |
| Kupcinskas | 2014 | Lithuania | Caucasian | Gastric cancer | RT-PCR | HB | 363 | 25 | 88 | 250 | 350 | 13 | 86 | 251 | 0.275 |
| Kupcinskas | 2014 | Lithuania | Caucasian | CRC e | RT-PCR | HB | 192 | 7 | 47 | 138 | 426 | 12 | 96 | 318 | 0.364 |
| Ryan | 2012 | USA | Mixed races a | CRC | Taqman | PB/HB | 239 | 19 | 96 | 124 | 433 | 36 | 166 | 231 | 0.729 |
a Caucasian and African Americans; b ESCC esophageal squamous cell carcinoma; c SCCHN squamous cell carcinoma of the head and neck; d HCC hepatocellular carcinoma; e CRC colorectal cancer; f HB hospital-based; g PB population-based; h HWE Hardy-Weinberg equilibrium.
Meta-analysis result
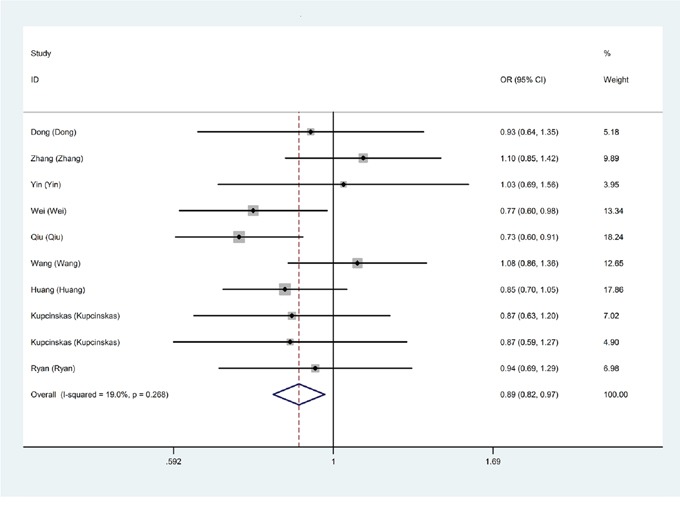
The main results of this meta-analysis were shown in Table 2. For overall studies, there existed a significant association of miR-608 rs4919510 polymorphism with decreased cancer risk only in recessive model (CC vs. GG+GC: OR=0.89, 95% CI: 0.82-0.97, P=0.009) (Figure 2). In other genotype model, the relationship still remain controversial.
Table 2. The result of meta-analysis for various genotype models.
| Test of Association | P Value for heterogeneity | I2 (%) | ||||
|---|---|---|---|---|---|---|
| OR (95%CI)b | Z | P Value | ||||
| Totala | CC vs. GG | 0.90 (0.77,1.06) | 1.23 | 0.219c | 0.031 | 51.1% |
| CG vs. GG | 1.01 (0.93,1.10) | 0.31 | 0.754d | 0.361 | 8.8% | |
| CG+CC vs.GG | 0.98 (0.88,1.10) | 0.28 | 0.776c | 0.093 | 39.7% | |
| GG+GC vs.CC | 0.89 (0.82,0.97) | 2.61 | 0.009d | 0.268 | 19.0% | |
| Asian | CC vs. GG | 0.93 (0.77,1.11) | 0.81 | 0.418c | 0.016 | 61.6% |
| CG vs. GG | 1.02 (0.94,1.11) | 0.51 | 0.610d | 0.353 | 9.9% | |
| CG+CC vs.GG | 1.00 (0.89,1.12) | 0.01 | 0.988c | 0.079 | 47.0% | |
| GG+GC vs.CC | 0.90 (0.79,1.03) | 1.51 | 0.130c | 0.090 | 45.3% | |
| Caucasian | CC vs. GG | 0.59 (0.36,0.96) | 2.12 | 0.034d | 0.832 | 0.0% |
| CG vs. GG | 0.65 (0.39,1.09) | 1.63 | 0.103d | 0.727 | 0.0% | |
| CG+CC vs.GG | 0.60 (0.37,0.98) | 2.03 | 0.042d | 0.802 | 0.0% | |
| GG+GC vs.CC | 0.84 (0.68,1.04) | 1.56 | 0.118d | 0.876 | 0.0% | |
aContrasts including homozygote model (CC vs. GG), heterozygote model (CG vs. GG), dominant model (CG+ CC vs. GG) and recessive model (CC vs. GG+GC) respectively.
bR odds ratio, CI confidence interval.
cP-value for significance under random-effects model.
dP-value for significance under fixed-effects model.
Figure 2. Overall meta-analysis of the relationship between miR-608 rs4919510 polymorphism and cancer risk in recessive model (CC vs. GG+GC).
For subgroup analysis of races, we found that in Caucasians miR-608 rs4919510 polymorphism had some relationship with decreased cancer risk in both homozygote model (CC vs. GG: OR=0.59, 95% CI: 0.36-0.96, P=0.034) and dominant model (CG+ CC vs. GG: OR=0.60, 95% CI: 0.37-0.98, P=0.042) (Figure 3). In Asians, Our results did not show any association of miR-608 rs4919510 polymorphism with cancer risk in any genotype model (Figure 4).
Figure 3. Subgroup analysis of the relationship between miR-608 rs4919510 polymorphism and cancer risk in Caucasians.
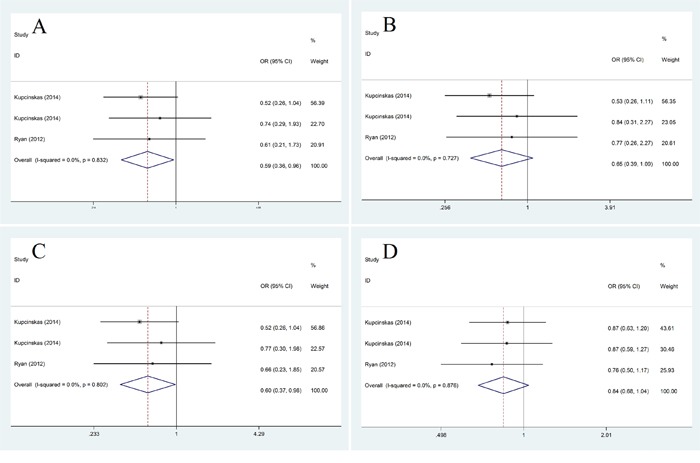
A. homozygote model (CC vs. GG); B. heterozygote model (CG vs. GG); C. dominant model (CG+ CC vs. GG); D. recessive model (CC vs. GG+GC).
Figure 4. Subgroup analysis of the relationship between miR-608 rs4919510 polymorphism and cancer risk in Asians.
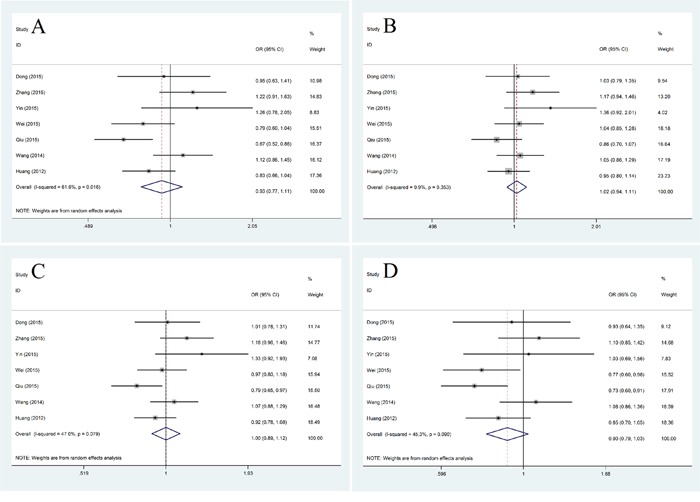
A. homozygote model (CC vs. GG); B. heterozygote model (CG vs. GG); C. dominant model (CG+ CC vs. GG); D. recessive model (CC vs. GG+GC).
Publication bias and sensitivity analysis
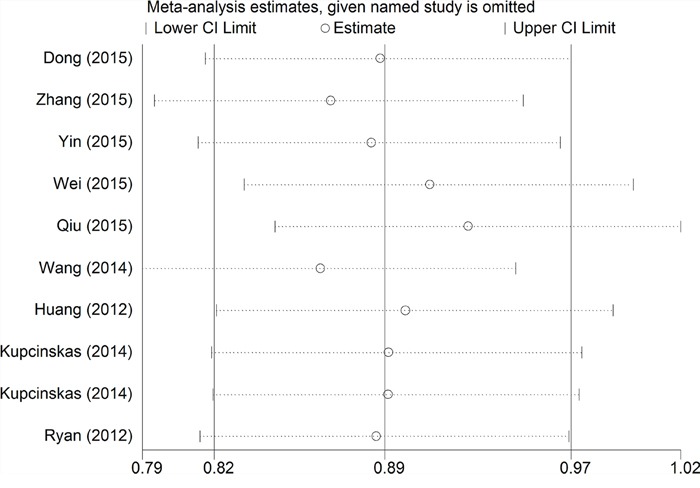
We utilized Funnel Plot, Begg's funnel plot and Egger's test to evaluate publication bias. The almost symmetrical shape of the funnel plots for four genetic models did not reveal any significant publication bias (Figure 5). There was also no evidence of publication bias in Begg's funnel for all genetic models (P > 0.05)(Figure 6). We also did not find out publication bias in Egger's test in homozygote model (CC vs. GG, P= 0.977), heterozygote model (CG vs. GG, P=0.744), dominant model (CG+ CC vs. GG, P=0.772) and recessive model (CC vs. GG+GC, P=0.440). Sensitivity analysis through evaluating the influence of each study on overall ORs showed that the omission of any study made no significant difference (Figure 7).
Figure 5. Funnel plot for publication bias test.
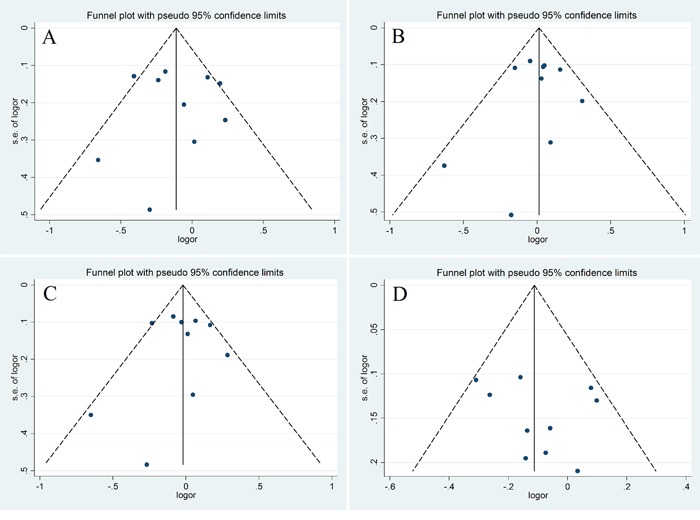
A. homozygote model (CC vs. GG); B. heterozygote model (CG vs. GG); C. dominant model (CG+ CC vs. GG); D. recessive model (CC vs. GG+GC).
Figure 6. Begg's funnel plot for publication bias test.
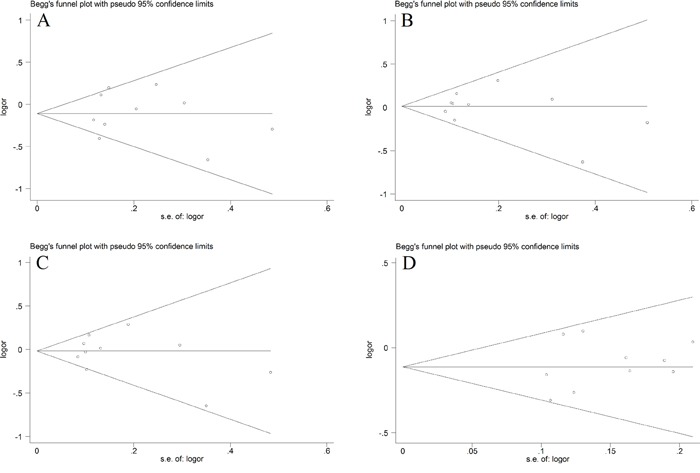
A. homozygote model (CC vs. GG); B. heterozygote model (CG vs. GG); C. dominant model (CG+ CC vs. GG); D. recessive model (CC vs. GG+GC).
Figure 7. The influence of individual studies on the overall OR in recessive model (CC vs. GG+GC).
DISCUSSION
Genetic mutations are responsible for cancer occurrence [27]. SNPs as the most common genetic sequence variation, could affect the function of a series of microRNAs by altering the formation of the primary transcript, pre-miRNA maturation, or miRNA-mRNA interactions [10, 28]. Recently, rs4919510 polymorphism in miR-608 has been reported to predict clinical outcomes for cancer patients in different cancer types [13, 14, 16]. In addition, Zhang and colleagues found that miR-608 expressions were reduced in chordoma cell lines and restoration of miR-608 inhibited chordoma cell proliferation and invasion [29]. All these studies provoked us to think about the association of miR-608 polymorphism and cancer risk.
In this meta-analysis, we demonstrated that individuals carrying CC genotype in rs4919510 have a decreased cancer risk. Moreover, Yang and colleagues reported that miR-608 suppressed the carcinogenesis of colon cancer cells in vitro and in vivo by eliminating NAA10 mRNA which participate as a key molecule in the process of carcinogenesis [30]. Taken together, these results strongly indicated that genetic variation of rs4919510 in miR-608 played an important role in cancer development.
By subgroup analysis, we found that in Caucasians miR-608 rs4919510 polymorphism had some relationship with decreased cancer risk in homozygote model and dominant model while in Asians there exist no relationship in any genotype model. These differences may be a result of various genetic backgrounds in different races and various mechanisms of carcinogenesis in different areas.
Nevertheless, some limitations in this meta-analysis should be paid attention to. First, included studies are still so limited that we cannot perform subgroup analysis for different cancer types although it is well known that one microRNA may play different functions in different types of cancer. Second, there exists a certain degree of heterogeneity between studies. After subgroup analysis stratified by races, it could be found that heterogeneity of Caucasians reduced significantly. Thus, it could be presumed that the heterogeneity partly resulted from differences in races. Simultaneously, the selection of subjects may become another source of heterogeneity. Third, only three studies were included in subgroup analysis for Caucasians and the results could be inaccurate and dubious. Fourth, only published articles were included, the unpublished and ongoing studies could convert our result. Last, we did not take sex, age, sex, family history and environmental factors into consideration and further detailed meta-analysis remain needed.
In conclusion, the results of meta-analysis indicated that rs4919510(C > G) polymorphism in miR-608 was significantly associated with decreased cancer risk and could become a promising target to forecast cancer risk for clinical practice. However, these results should be treated with caution due to the limitations above. For further verifying the results, well-designed large scale case–control studies are needed in the future.
MATERIALS AND METHODS
Publication search and data extraction
To identify all published studies concerning the relationship between miRNA polymorphisms and cancer risk, PubMed and Embase database (updated to Jan 1, 2016) were searched without language, publication, or date restrictions using the following search terms: (“microRNA 608” OR “microRNA-608” OR “miR-608” OR “rs4919510”) AND (“polymorphism” OR “SNP” OR “variation” OR “locus” OR “mutation”) AND (“cancer” OR “tumor” OR “malignance” OR “carcinoma” OR “neoplasm”). The included papers should meet criteria listed below: (1) Assessment of the relationship between miR-608 rs4919510 polymorphism and cancer risk; (2) a case-control design; (3) histologically confirmed for malignant tumors; (4) sufficient published data for further calculating odds ratios (ORs) and their 95% confidence intervals (95%CIs); (5) Meeting Hardy-Weinberg equilibrium (HWE) in the control group (P>0.05). Two reviewers (HQ Liu and BY Wang) in our group screened out the data independently and had reached a consensus on each term. All extracted data consisted of author (year), country, ethnicity, cancer type, genotyping method, source of controls, characteristics of cases and controls and P value for Hardy–Weinberg equilibrium (HWE) was exhibited in Table 1.
Statistical analysis
We calculated P value of HWE in control group by X2 test and considered P value>0.05 as fulfilling Hardy-Weinberg equilibrium [31]. The association between the miR-608 rs4919510 (C > G) SNP and the risk of cancer was measured by odds ratios (ORs) with 95% confidence intervals (CIs) based on different genetic models such as homozygote model (CC vs. GG), heterozygote model (CG vs. GG), dominant model (CG+ CC vs. GG) and recessive model (CC vs. GG+GC) respectively. Hierarchical analysis was conducted by Ethnicity (Asian and Caucasian). The statistical significance of the pooled OR was evaluated by Z test, and P value of <0.05 was regarded as significant. Heterogeneity assumption was tested among studies using a Chi-square-based Q-test. We considered P value of > 0.10 for Q-test as a lack of heterogeneity which indicate fixed-effects model should be used to perform the meta-analysis [32]. If significant heterogeneity was existing (P<0.10 for Q-test), the random effects model should be chosen as a more appropriate one [33]. To evaluate whether there existed publication bias, Funnel plots, Begg's test and Egger's test were applied [34]. The influence of each study on overall OR was also evaluated using metaninf order. Statistical analysis was all conducted using Stata12.0 Software.
Acknowledgments
Huiquan Liu and Dawei Ye designed and wrote this article. Huiquan Liu and Bangyan Wang screened out relevant articles and extracted data. Qingquan Liu, Yaqun Zhou, and Guangqin Xiao analyzed data. Weijuan Li and Shiying Yu revised this article.
Footnotes
CONFLICTS OF INTEREST
All authors declare no conflicts of interest.
GRANT SUPPORT
This work was supported by National Natural Science Foundation of P.R. China 81400917 and National Natural Science Foundation of Hubei Province 2014CFB449.
REFERENCES
- 1.Kong YW, Ferland-McCollough D, Jackson TJ, Bushell M. microRNAs in cancer management. Lancet Oncol. 2012;13:249–58. doi: 10.1016/S1470-2045(12)70073-6. [DOI] [PubMed] [Google Scholar]
- 2.Zhang B, Pan X, Cobb GP, Anderson TA. microRNAs as oncogenes and tumor suppressors. Dev Biol. 2007;302:1–12. doi: 10.1016/j.ydbio.2006.08.028. [DOI] [PubMed] [Google Scholar]
- 3.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–33. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–66. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 5.Anglicheau D, Muthukumar T, Suthanthiran M. MicroRNAs: small RNAs with big effects. Transplantation. 2010;90:105–12. doi: 10.1097/TP.0b013e3181e913c2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Garzon R, Marcucci G, Croce CM. Targeting microRNAs in cancer: rationale, strategies and challenges. Nat Rev Drug Discov. 2010;9:775–89. doi: 10.1038/nrd3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang F, Ma YL, Zhang P, Yang JJ, Chen HQ, Liu ZH, Peng JY, Zhou YK, Qin HL. A genetic variant in microRNA-196a2 is associated with increased cancer risk: a meta-analysis. Mol Biol Rep. 2012;39:269–75. doi: 10.1007/s11033-011-0735-0. [DOI] [PubMed] [Google Scholar]
- 8.Mishra PJ, Bertino JR. MicroRNA polymorphisms: the future of pharmacogenomics, molecular epidemiology and individualized medicine. Pharmacogenomics. 2009;10:399–416. doi: 10.2217/14622416.10.3.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gao Y, Liu Y, Liu GL, Ran LK, Zeng F, Wu JY, Song FZ. Association between the pre-mir-218 polymorphism and cancer risk in the Chinese population: a meta-analysis. Asian Pac J Cancer Prev. 2014;15:2517–22. doi: 10.7314/apjcp.2014.15.6.2517. [DOI] [PubMed] [Google Scholar]
- 10.Ryan BM, Robles AI, Harris CC. Genetic variation in microRNA networks: the implications for cancer research. Nat Rev Cancer. 2010;10:389–402. doi: 10.1038/nrc2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schetter AJ, Harris CC. Alterations of microRNAs contribute to colon carcinogenesis. Semin Oncol. 2011;38:734–42. doi: 10.1053/j.seminoncol.2011.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lin J, Horikawa Y, Tamboli P, Clague J, Wood CG, Wu X. Genetic variations in microRNA-related genes are associated with survival and recurrence in patients with renal cell carcinoma. Carcinogenesis. 2010;31:1805–12. doi: 10.1093/carcin/bgq168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lin M, Gu J, Eng C, Ellis LM, Hildebrandt MA, Lin J, Huang M, Calin GA, Wang D, Dubois RN, Hawk ET, Wu X. Genetic polymorphisms in MicroRNA-related genes as predictors of clinical outcomes in colorectal adenocarcinoma patients. Clin Cancer Res. 2012;18:3982–91. doi: 10.1158/1078-0432.CCR-11-2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pardini B, Rosa F, Naccarati A, et al. Ye Y, Wu X, di Gaetano C, Buchler T, Novotny J, Matullo G, Vodicka P. Polymorphisms in microRNA genes as predictors of clinical outcomes in colorectal cancer patients. Carcinogenesis. 2015;36:82–6. doi: 10.1093/carcin/bgu224. [DOI] [PubMed] [Google Scholar]
- 15.Yang PW, Huang YC, Hsieh CY, Hua KT, Huang YT, Chiang TH, Chen JS, Huang PM, Hsu HH, Kuo SW, Kuo ML, Lee JM. Association of miRNA-related genetic polymorphisms and prognosis in patients with esophageal squamous cell carcinoma. Ann Surg Oncol. 2014;21:601–9. doi: 10.1245/s10434-014-3709-3. [DOI] [PubMed] [Google Scholar]
- 16.Zheng J, Deng J, Xiao M, Yang L, Zhang L, You Y, Hu M, Li N, Wu H, Li W, Lu J, Zhou Y. A sequence polymorphism in miR-608 predicts recurrence after radiotherapy for nasopharyngeal carcinoma. Cancer Res. 2013;73:5151–62. doi: 10.1158/0008-5472.CAN-13-0395. [DOI] [PubMed] [Google Scholar]
- 17.Qiu F, Yang L, Zhang L, Yang X, Yang R, Fang W, Wu D, Chen J, Xie C, Huang D, Zhou Y, Lu J. Polymorphism in mature microRNA-608 sequence is associated with an increased risk of nasopharyngeal carcinoma. Gene. 2015;565:180–6. doi: 10.1016/j.gene.2015.04.008. [DOI] [PubMed] [Google Scholar]
- 18.Ryan BM, McClary AC, Valeri N, Robinson D, Paone A, Bowman ED, Robles AI, Croce C, Harris CC. rs4919510 in hsa-mir-608 is associated with outcome but not risk of colorectal cancer. PLoS One. 2012;7:e36306. doi: 10.1371/journal.pone.0036306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang P, Wang J, Lu T, Wang X, Zheng Y, Guo S, Yang Y, Wang M, Kolluri VK, Qiu L, Shen F, Fan L, Li J, et al. miR-449b rs10061133 and miR-4293 rs12220909 polymorphisms are associated with decreased esophageal squamous cell carcinoma in a Chinese population. Tumour Biol. 2015;36:8789–95. doi: 10.1007/s13277-015-3422-2. [DOI] [PubMed] [Google Scholar]
- 20.Yin Z, Cui Z, Guan P, Li X, Wu W, Ren Y, He Q, Zhou B. Interaction between Polymorphisms in Pre-MiRNA Genes and Cooking Oil Fume Exposure on the Risk of Lung Cancer in Chinese Non-Smoking Female Population. PLoS One. 2015;10:e0128572. doi: 10.1371/journal.pone.0128572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wei WJ, Wang YL, Li DS, Wang Y, Wang XF, Zhu YX, Pan XD, Wang ZY, Wu Y, Jin L, Wang JC, Ji QH. Association study of single nucleotide polymorphisms in mature microRNAs and the risk of thyroid tumor in a Chinese population. Endocrine. 2015;49:436–44. doi: 10.1007/s12020-014-0467-8. [DOI] [PubMed] [Google Scholar]
- 22.Wang R, Zhang J, Ma Y, Chen L, Guo S, Zhang X, Ma Y, Wu L, Pei X, Liu S, Wang J, Hu H, Liu J. Association study of miR149 rs2292832 and miR608 rs4919510 and the risk of hepatocellular carcinoma in a largescale population. Mol Med Rep. 2014;10:2736–44. doi: 10.3892/mmr.2014.2536. [DOI] [PubMed] [Google Scholar]
- 23.Kupcinskas J, Wex T, Link A, Leja M, Bruzaite I, Steponaitiene R, Juzenas S, Gyvyte U, Ivanauskas A, Ancans G, Petrenkiene V, Skieceviciene J, Kupcinskas L, Malfertheiner P. Gene polymorphisms of micrornas in Helicobacter pylori-induced high risk atrophic gastritis and gastric cancer. PLoS One. 2014;9:e87467. doi: 10.1371/journal.pone.0087467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kupcinskas J, Bruzaite I, Juzenas S, Gyvyte U, Jonaitis L, Kiudelis G, Skieceviciene J, Leja M, Pauzas H, Tamelis A, Pavalkis D, Kupcinskas L. Lack of association between miR-27a, miR-146a, miR-196a-2, miR-492 and miR-608 gene polymorphisms and colorectal cancer. Sci Rep. 2014;4:5993. doi: 10.1038/srep05993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang AJ, Yu KD, Li J, Fan L, Shao ZM. Polymorphism rs4919510: C>G in mature sequence of human microRNA-608 contributes to the risk of HER2-positive breast cancer but not other subtypes. PLoSOne. 2012;7:e35252. doi: 10.1371/journal.pone.0035252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dong G, Zhang R, Xu J, Guo Y. Association between microRNA polymorphisms and papillary thyroid cancer susceptibility. Int J Clin Exp Pathol. 2015;8:13450–7. [PMC free article] [PubMed] [Google Scholar]
- 27.Frixa T, Donzelli S, Blandino G. Oncogenic MicroRNAs: Key Players in Malignant Transformation. Cancers (Basel) 2015;7:2466–85. doi: 10.3390/cancers7040904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kang Z, Li Y, He X, Jiu T, Wei J, Tian F, Gu C. Quantitative assessment of the association between miR-196a2 rs11614913 polymorphism and cancer risk: evidence based on 45,816 subjects. Tumor Biol. 2014;35:6271–82. doi: 10.1007/s13277-014-1822-3. [DOI] [PubMed] [Google Scholar]
- 29.Zhang Y, Schiff D, Park D, Abounader R. MicroRNA-608 and microRNA-34a regulate chordoma malignancy by targeting EGFR, Bcl-xL and MET. PLoS One. 2014;9:e91546. doi: 10.1371/journal.pone.0091546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang H, Li Q, Niu J, Li B, Jiang D, Wan Z, Yang Q, Jiang F, Wei P, Bai S. microRNA-342-5p and miR-608 inhibit colon cancer tumorigenesis by targeting NAA10. Oncotarget. 2016;7:2709–20. doi: 10.18632/oncotarget.6458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.He B, Pan Y, Cho WC, Xu Y, Gu L, Nie Z, Chen L, Song G, Gao T, Li R, Wang S. The association between four genetic variants in microRNAs (rs11614913, rs2910164, rs3746444, rs2292832) and cancer risk: evidence from published studies. PLoS One. 2012;7:e49032. doi: 10.1371/journal.pone.0049032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst. 1959;22:719–48. [PubMed] [Google Scholar]
- 33.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7:177–88. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 34.Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:629–34. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]


