Abstract
The PH domains of OSBP and FAPP1 fused to GFP were used to monitor PI(4)P distribution in COS-7 cells during manipulations of PI 4-kinase (PI4K) activities. Both domains were associated with the Golgi and small cytoplasmic vesicles, and a small fraction of OSBP-PH was found at the plasma membrane (PM). Inhibition of type-III PI4Ks with 10 μM wortmannin (Wm) significantly reduced but did not abolish Golgi localization of either PH domains. Downregulation of PI4KIIα or PI4KIIIβ by siRNA reduced the localization of the PH domains to the Golgi and in the former case any remaining Golgi localization was eliminated by Wm treatment. PLC activation by Ca2+ ionophores dissociated the domains from all membranes, but after Ca2+ chelation, they rapidly reassociated with the Golgi, the intracellular vesicles and with the PM. PM association of the domains was significantly higher after the Ca2+ transient and was abolished by Wm pretreatment. PM relocalization was not affected by down-regulation of PI4KIIIβ or -IIα, but was inhibited by down-regulation of PI4KIIIα, or by 10 μM PAO, which also inhibits PI4KIIIα. Our data suggest that these PH domains detect PI(4)P formation in extra-Golgi compartments under dynamic conditions and that various PI4Ks regulate PI(4)P synthesis in distinct cellular compartments.
INTRODUCTION
Phosphoinositides have been known for some time for their signaling roles in mediating the actions of calcium-mobilizing hormones and neurotransmitters (Michell, 1975; Berridge, 1984). Phosphoinositides are also emerging as important regulators of molecular interactions critical for proper trafficking and function of cellular proteins (Martin, 1997; Odorizzi et al., 2000). There are a number of inositide kinase and phosphatase enzymes that have been shown to play a central role in the sorting of molecules to specific organelles (Fruman et al., 1998; Odorizzi et al., 2000). Phosphatidylinositol (PI) 4-kinases (PI4Ks), on the other hand, have long been considered only in the context of synthesis of polyphosphoinositides, the precursors of the PLC-generated second messengers, Ins(1,4,5)P3 and diacylglycerol in agonist-stimulated cells. This picture has begun to change only when the first two PI4Ks, Pik1 and Stt4, were cloned in yeast (Flanagan et al., 1993; Garcia-Bustos et al., 1994; Yoshida et al., 1994), and it was shown that the two proteins assumed nonredundant functions, related to Golgi to membrane trafficking and to cell-wall biogenesis, respectively (Hama et al., 1999; Walch-Solimena and Novick, 1999; Audhya et al., 2000). The mammalian homologues of the yeast enzymes, the type-III PI4ks have been identified as the wortmannin (Wm)-sensitive enzymes responsible for the synthesis of the hormone-sensitive pools of PI(4)P and PI(4,5)P2 (Nakanishi et al., 1995; Downing et al., 1996). However, localizations of the two type-III enzymes in mammalian cells are not particularly consistent with their function at the plasma membrane, the type-IIIβ enzymes being localized primarily to the Golgi (Wong et al., 1997), whereas the type-IIIα is found at the ER (Wong et al., 1997) and in the pericentriolar area that possibly also contains the Golgi compartment (Nakagawa et al., 1996). In addition to being produced by type-III PI4Ks, PI(4)P is also generated by the type-II PI4Ks, a separate family of Wm-insensitive enzymes that have been cloned recently and that also exist as an α and β form (Barylko et al., 2001; Minogue et al., 2001; Balla et al., 2002; Wei et al., 2002). The localization of these latter enzymes is more complex in that the endogenous forms are found in localization to trans-Golgi (Wei et al., 2002; Wang et al., 2003), whereas the expressed forms are also localized to endosomes, especially in the case of the type-IIα enzyme (Balla et al., 2002). This latter enzyme has also been found in noncaveolar Rafts (Minogue et al., 2001), and in a subcompartment of the ER (Waugh et al., 2003) and has been purified from the secretory granules of adrenal chromaffin cells (Barylko et al., 2001). The only enzyme that has been shown to translocate to the plasma membrane in stimulated cells was the type-IIβ enzyme using a Rac-dependent mechanism (Wei et al., 2002). Nonetheless, type-II PI4K activity has been shown to associate with epidermal growth factor (EGF) receptors (Kauffmann-Zeh et al., 1994) and with another group of membrane proteins, the tetraspanins (Berditchevski et al., 1997). All of these data suggest that localization alone cannot unveil the multiple roles of the PI4Ks and there is a need for additional methods that can assess the activity of these enzymes in the various cellular compartments preferably in living cells.
Recent advances in understanding the nature of phosphoinositide-protein interactions and the detailed characterization of protein domains that specifically recognize inositol lipids (Hurley and Meyer, 2001; Lemmon, 2003) allowed dynamic imaging of several phosphoinositide species in living cells (Balla et al., 2000; Balla and Varnai, 2002). Among these, the pleckstrin homology (PH) domains have been particularly popular because of their often (but not always) specific and high-affinity binding to specific phosphoinositides (Lemmon and Ferguson, 2000; Yu et al., 2004). The PH domains of the OSBP and FAPP1 proteins have been shown to recognize PI(4)P very specifically in vitro (Dowler et al., 2000). A detailed characterization of the cellular localization of these PH domains in yeast, on the other hand, led to the conclusion that in addition to PI(4)P, their localization is also determined by protein-protein interactions (Levine and Munro, 2002).
In the present study, we used the PH domains of the OSBP and FAPP1 proteins together with various manipulations of PI4K activities to obtain further spatial information about PI(4)P production by the distinct enzymes and about the usefulness of these PH domains for such analyses. In agreement with previous reports, our studies show that in steady state, the two PH domains detect PI(4)P formation in the Golgi by a mechanism that is also Arf1 dependent. However, our data also suggest that during de novo synthesis, both PH domains can reveal active PI(4)P formation in nonGolgi membranes and that distinct PI4Ks are responsible for the generation of this lipid in the various membrane compartments.
MATERIALS AND METHODS
Reagents
Ionomycin, Wm, and BAPTA were purchased from Calbiochem (La Jolla, CA), and brefeldin A (BFA) from Epicenter Technologies (Madison, WI). The primary antibody of gm130 was obtained from BD Transduction Laboratories (Franklin Lakes, NJ) and the monoclonal anti HA antibody (HA1.1) was form Covance (Berkley, CA). The polyclonal antibody against the type-IIα PI4K was kindly provided by Drs. Jun Guo and Pietro DeCamilli. The antibody against the PI4KIIIα was obtained by immunization of New Zeeland Rabbits with the peptide, KYLTASQLVPPDNQDTRS conjugated to KLH (Covance), and affinity purification of the immune-sera using the same peptide conjugated to Sulfolinc gel (New England Peptide, Gardner, MA) The secondary antibodies, Alexa-595 and Alexa-488 were from Molecular Probes (Eugene, OR). The lipofectamine 2000 reagent was purchased from Invitrogen (Carlsbad, CA).
DNA Constructs and Transfections
The PH domain of OSBP (residues 87–189) has been amplified from human brain cDNA (Quick-clone, Clontech, Palo Alto, CA) using the primer pairs of: fw: 5′-tttagatctggctcggctcgagagggctggctc-3′, rev: 5′-aaagaattctgccagcatcttcacagctttggc-3′. The PH domain of FAPP1 (residues 1–101) was amplified from a partial human EST clone: (IMAGE id: 287618) with the following primers: fw: 5′-aaagaattcaccatggagggggtgttgtacaag-3′, rev: 5′-aaaggatccttagtccttgtatcagtcaaacatgc-3′. Both PH domains were subcloned into the pEGFP plasmid, pEGFP-C1 (BglII/EcoRI) and pEGFP-N1 (EcoRI/BamHI) for the OSBP and FAPP1 PH-domains, respectively, in frame with the GFP protein. Mutations were generated with the QuikChange mutagenesis kit of Stratagene (La Jolla, CA). The construct containing the transmembrane targeting sequence of the β1,4-galactosyl transferase enzyme fused to GFP (N-GT-GFP) was created from the commercially available CFP-fused construct (Clontech) by exchanging the CFP to GFP. For siRNA studies, the double-stranded RNAs corresponding to nucleotides 888–908 of human PI4KIIα (NM_018425; Wang et al., 2003), 2684–2704 of PI4KIIIβ (NM_002651), and 1072–1092 of PI4KIIIα (NM_058004) were used. In some experiments, a pool of five different siRNAs (against sequences: 1072–1092, 1244–1264, 4516–4536, 6175–6195, and 6325–6345) were mixed and used to knock down the PI4KIIIα enzyme. Silencing RNAs were obtained from Qiagen (Valencia, CA). COS-7 cells (105 cells in 2 ml) were plated in 35-mm culture dishes 1 d before transfection with 20 μl of 20 μM siRNA using Oligofectamine (Invitrogen). After 6 h, the medium was changed to DMEM containing fetal bovine serum (FBS). Transfection with the siRNAs was repeated 24 h later, and cells were transfected with plasmid DNAs on the third day using Lipofectamine 2000. Cells were then studied on the following day either live or after fixation and immunostaining. The effect of siRNA treatment on the PI4K expression levels was also determined by Western blot analysis.
Immunocytochemistry and Confocal Microscopy
For immunostaining, COS-7 cells were grown on coverslips and fixed in 2% formaldehyde in phosphate-buffered saline (PBS; pH 7.4) for 10 min at room temperature. After three washes with PBS (5 min each), fixed cells were incubated in blocking solution (10% FBS and 0.2% Saponin in PBS) for 1 h to decrease the nonspecific binding of the antibodies and to improve the penetration of the antibodies through membranes. This blocking solution was also used for diluting the primary antibody (gm130 [1:500] or anti HA [1: 500]), and cells were incubated for 1 h. After three washes, cells were incubated in the same buffer with a fluorescent secondary antibody (1:1000) for 1 h at RT. This was followed by a last washing step (3 times for 5 min, in PBS), and then the cells were rinsed with distilled water, air-dried, and mounted on glass slides using Cytoseal 60 mounting medium (Stephens Scientific, Riverdale, NJ). Cells were then analyzed by using an inverted Zeiss LSM-410 or LSM-510 scanning laser confocal microscope (Thornwood, NY) or an Olympus IX70 inverted microscope (Melville, NY) equipped with a CCD camera (Hamamatsu, ORCA ER, Bridgewater, NJ) and a lambda DG-4 illuminator (Sutter, Novato, CA). Live cells were studied at 35°C using a temperature-controlled chamber (Harvard Instruments, Boston, MA) and an objective heater (Bioptech, Butler, PA).
Analysis of FRET in Cell Suspension
To have a quantitative measure of membrane localization of the various constructs, the CFP and YFP variants of all fusion proteins were created. These were cotransfected into COS-7 cells that were cultured in 10-cm culture dishes. One day after transfection, cells were removed from the dishes by mild trypsinization, washed, and centrifuged. Cells (∼3–5 million) were then resuspended in 2 ml of the Krebs-Ringer solution described above and placed in the thermostated cuvette holder of a fluorescence spectrophotometer used for ratiometric Ca2+ measurements. Recordings were made using an excitation of 425 nm and calculating a ratio from the emissions detected at 525 and 475 nm (20 nm bandwidth, each). Ionomycin (10 μM) was used to activate endogenous phospholipase C to hydrolyze the phospholipids and the decrease in the 525/475 ratio was taken as an index of translocation of the domains from the membrane to the cytosol (see van Der Wal et al., 2001 and Várnai and Balla, 1998 for details concerning the FRET approach, and ionomycin manipulation, respectively). Digitonin (15 μg/ml) was added at the end of the incubations so that the domains dissociate away from FRET distance. The slight baseline drift (which was increased by high Wm concentrations) was fitted for the control period and was subtracted from the traces. Traces were reproduced at least three times in independent experiments.
RESULTS
Cellular Distribution of the OSBP- and FAPP1-PH-GFP Proteins
Both PH domains were expressed as GFP fusion proteins and their positions relative to GFP were designed to follow those in their native proteins, i.e., FAPP1-PH was N-terminally and OSBP-PH C-terminally placed relative to GFP. Fusion proteins were expressed in COS-7 cells and were examined in a scanning laser confocal microscope either in live cells or after fixation with 2% paraformaldehyde. Occasionally, fixed cells were also examined in a wide-field fluorescence microscope equipped with a CCD camera. As observed earlier by the Munro group (Levine and Munro, 1998, 2001, 2002), both PH domains showed primarily Golgi localization (Figure 1). There was a significant heterogeneity in the morphology of cells in that some cells showed a very pronounced small vesicular distribution of OSBP-PH-GFP, especially at high expression levels, whereas in some cases a clear filamentous structure extending from the Golgi was observed (e.g., Figure 1). Such filaments were even more pronounced with the FAPP1-PH-GFP, but at higher expression levels, this construct caused the accumulation and aggregation of large cytoplasmic vesicles with FAPP1-PH-GFP in their walls and a parallel loss of Golgi localization. This was never observed with the OSBP-PH domain. A further difference between the distributions of the two proteins was the prominent nuclear accumulation of the OSBP-PH domain, which was not characteristic of the FAPP1-PH domain (Figure 1). When the distribution of the proteins was studied in fixed, rather than live cells, the localization of the OSBP-PH was very similar to that observed in live-cells, but the FAPP1-PH domain also appeared more in filamentous structures and the larger vesicular structures were not obvious in the fixed and immunostained samples. Mutant forms of the proteins (R18L-FAPP1-PH) or (R107E,R108E-OSBP-PH), which prevents their PI(4)P binding showed no localization to the Golgi (unpublished data), consistent with earlier findings (Levine and Munro, 2002).
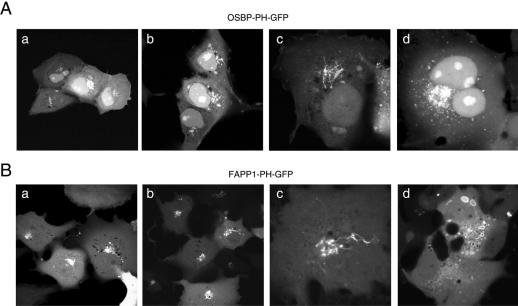
Figure 1.
Cellular distribution of OSBP-PH-GFP (A) and FAPP1-PH-GFP (B) expressed in COS-7 cells. Cells were transiently transfected with the indicated PH domain-GFP chimera for 24 h, and live cells were studied by confocal microscopy at 35°C. In most cells showing low-to-moderate expression levels both constructs show localization to the Golgi compartment (a and b panels in both A and B). In many cells expressing low levels of either construct dynamic tubular structures emanate from the Golgi, reminiscent of the effects of early BFA treatment (panels c). At higher expression levels, OSBP-PH showed several small vesicles around the Golgi area and also in more peripheral locations, whereas the FAPP1-PH domain generated multiple larger vesicles strongly positive for the domain in their limiting membranes (panels d on A and B, respectively). Notable difference is the nuclear accumulation of OSBP-PH but not FAPP1-PH, especially in cells expressing the protein at higher levels.
Effects of PH Domains on Golgi Morphology and Function
Next, we analyzed whether expression of the constructs altered the Golgi morphology especially because many cells expressing either of the constructs showed a filamentous tubular structure that was reminiscent of the acute effect of BFA on the Golgi. To assess Golgi morphology, we used either immunostaining for the endogenous cis-Golgi marker gm130 or the transmembrane domain of the β1,4-galactosyl transferase enzyme fused to GFP (N-GT-GFP) in live cells. As shown in Figure 2A, expression of either the OSBP- or FAPP1-PH-GFP caused tubulation and dispersion of the cis-Golgi marker. Importantly, the colocalization of the domains with the cis-Golgi marker was progressively lost in cells expressing higher levels of the fusion proteins (compare the structures shown by the arrows on Figure 2A). For a simultaneous detection of a Golgi marker with the PH domains in live cells, the latter were also created as RFP fusion proteins using the mRFP protein, which does not oligomerize (Campbell et al., 2002) in place of GFP. In these studies a GFP construct containing the N-terminal segment including the transmembrane domain of the β1,4-galactosyl transferase enzyme (N-GT-GFP) was used as a Golgi marker (Yamaguchi and Fukuda, 1995). In this construct, the GFP is located in the lumen of the Golgi. This marker also revealed the tubulation caused by moderate expression of both PH domains (Figure 2B, top and middle rows) and a small extent of Golgi dispersion at higher expression levels of the FAPP1, but less so with the OSBP-PH domain (Figure 2B, bottom row).
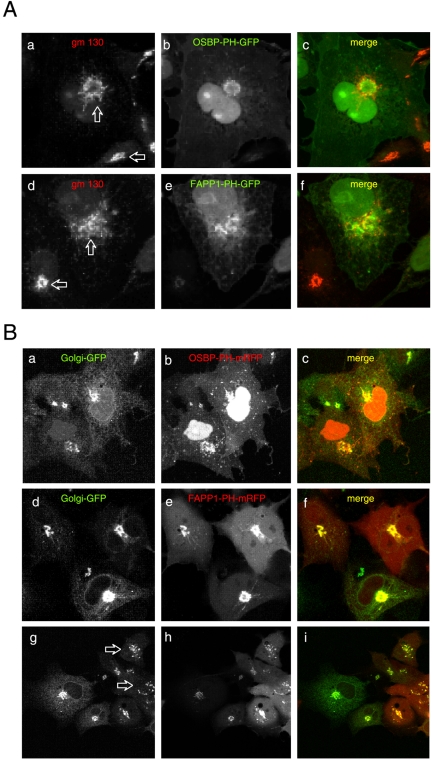
Figure 2.
Effects of overexpressed OSBP- and FAPP1-PH-GFP on the distribution of Golgi markers. COS-7 cells were transfected for 24 h with either the GFP-fused (A) or the mRFP-fused (B) respective PH domains and studied either as fixed cells immunostained for the cis-Golgi marker, gm130 (A) or cotransfected with a GFP construct containing the N-terminal transmembrane domain of the β1,4-galactosyl transferase enzyme (Golgi-GFP) and observed live (B). Note the fragmentation and tubulation of the cis-Golgi marker in the cells expressing higher levels of either PH domains (compare the structures indicated by the two arrows on A) and the loss of tight colocalization observed in cells expressing the constructs at low level. (B) Similar fragmentation of the Golgi was observed in live cells expressing high levels of the FAPP1-PH-mRFP (arrows in bottom row), but this effect was not so prominent with the OSBP-PH-mRFP (top row).
Arf-1 Is Necessary for Localization of the OSBP and FAPP1-PH Domains to the Golgi
The localization of the OSBP- or FAPP1-PH domains to the Golgi area raised the possibility that Arf1 has an important role (direct or indirect) in determining the localization of the PH domain GFP fusion proteins. This has already been suggested by previous studies using the same PH domains (Levine and Munro, 2002; Godi et al., 2004), and it has been shown that PI4K-IIIβ also associates with and is regulated by Arf1 in mammalian cells (Godi et al., 1999). Therefore, we investigated whether inhibition of Arf1 GDP/GTP exchange by BFA has an effect on the localization of the OSBP- or FAPP1-PH domains. As shown in Figure 3, addition of BFA (5 μg/ml) led to the rapid release of both the FAPP1- and OSBP-PH domains from the Golgi in most of the cells. Importantly, the BFA-induced dissociation of the OSBP-PH was consistently faster than that of the FAPP1-PH (Figure 3). In contrast to the Golgi, OSBP-PH associated with small intracellular vesicles was resistant to BFA treatment and so were the large vesicles present in cells that expressed high concentrations of the FAPP1-PH domain (unpublished data).
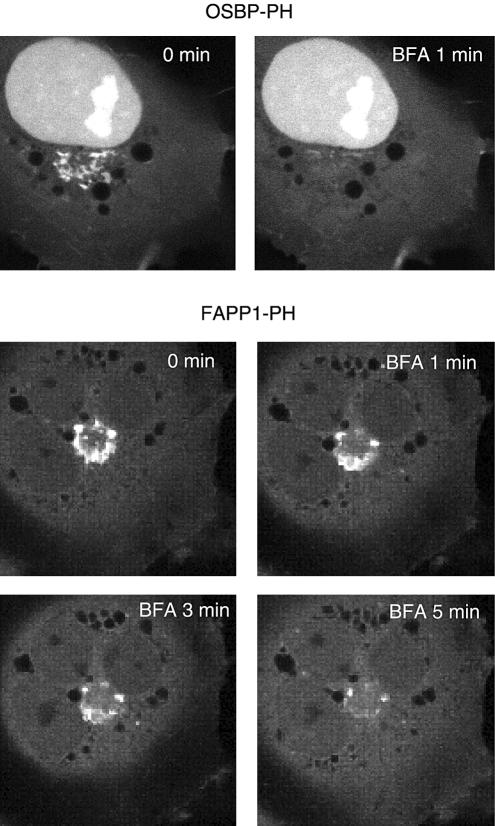
Figure 3.
Effects of brefeldin A (BFA) treatment on the localization of OSBP- and FAPP1-PH-GFP in COS-7 cells. Cells were transfected with the indicated constructs and live cells were examined by laser confocal microscopy 24 h later at 35°C. (A) After addition of BFA (5 μg/ml) OSBP-PH-GFP dissociated from the Golgi very rapidly (within 1 min), whereas the FAPP1-PH domain required a longer period (3–5 min).
Effects of PI4K Inhibition or Down-regulation on PH Domain Localization
To inhibit the activity of the type-III PI 4-kinases, we treated the cells with high concentrations (up to 10 μM) of Wm. Pretreatment of cells with 10 μM Wm at 37°C decreased the number of cells showing prominent localization at the Golgi and changed the localization to a more dispersed vesicular pool. Nonetheless, there were still a significant number of cells in which the OSBP- or FAPP1-PH domains were associated with the Golgi area after treatment of the cells for 1 h with up to 10 μM Wm and even the formation of tubular structures could be observed (Figure 4A). In some cases it was possible to find the same cell before and after Wm administration or to monitor the change in the PH domain distribution during Wm treatment (Figures 4B and 5). Overall, there was a great variability in the Wm-sensitive fraction of the Golgi localization with both PH domains.
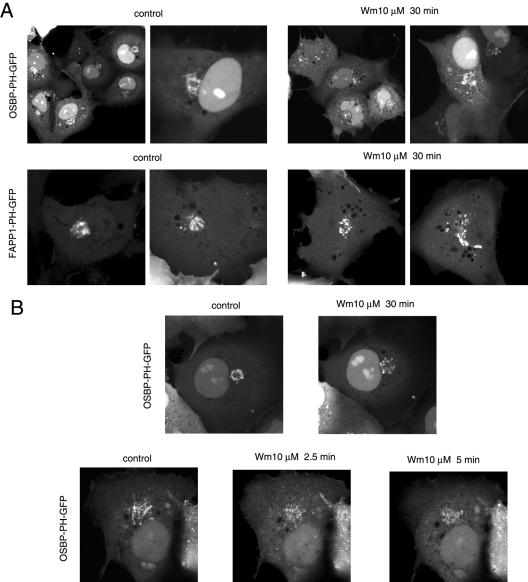
Figure 4.
Effects of wortmannin (Wm) treatment on the localization of the OSBP or FAPP1-PH-GFP fusion proteins in COS-7 cells. COS-7 cells were transfected with the indicated PH-GFP construct for 24 h and studied as live cells by confocal microscopy. (A) Cells were treated with 10 μM Wm for 30 min (at 37°C) to fully inhibit the type-III PI4Ks. There was a decrease in the Golgi localization of both PH domains after Wm treatment, but there was still a significant localization in vesicular compartments, often associated with the Golgi area. In most cases these vesicles were more dispersed than those in untreated cells. (B) The difference between pre- and post-Wm treatment is illustrated in a cell that was recorded both before and after the Wm treatment. The bottom panel shows a cell that rapidly loses some of its OSBP-PH domain localization after Wm treatment. Figure 7 shows additional examples of the effects of Wm treatment on the Golgi localization.
Figure 5.
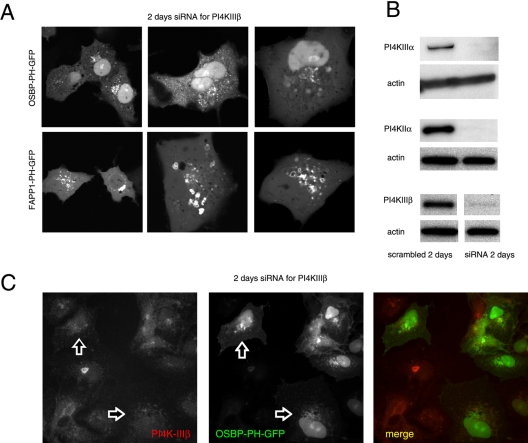
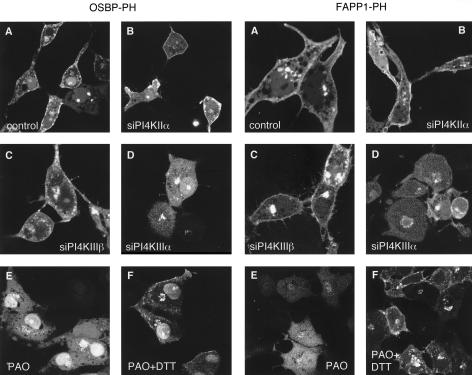
Effects of down-regulation of the type-IIIβ PI4K by siRNA treatment on the localization of OSBP- and FAPP1-PH-GFP in live (A) and OSBP-PH-GFP in fixed (C) COS-7 cells. COS-7 cells were treated with double-stranded RNA designed to interfere with expression of PI4KIIIβ for 2 consecutive days and transfected with the PH-GFP constructs on the third day for an additional day as described in Materials and Methods. Cells were either studied live at 35 C (A) or analyzed after fixation and immunostaining with a polyclonal anti-PI4KIIIβ antibody (C). The arrows point to two cells in which PI4KIIIβ is largely eliminated. To determine the effect of siRNA treatment, cells treated identically with the various siRNAs directed against the three PI4K enzymes were subjected to Western blot analysis using antibodies against the respective kinases and the blots were also analyzed for actin (B). In the picture showing the effect of PI4KIIIβ knockdown the control and siRNA-treated samples were not loaded on adjacent lanes, indicated by the gap between the images. Nevertheless the images are derived from the same gel scanned and processed with identical settings.
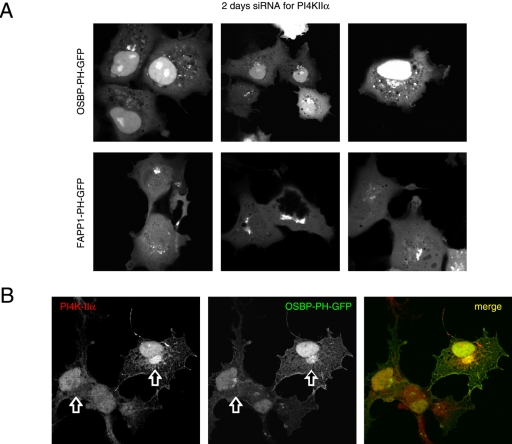
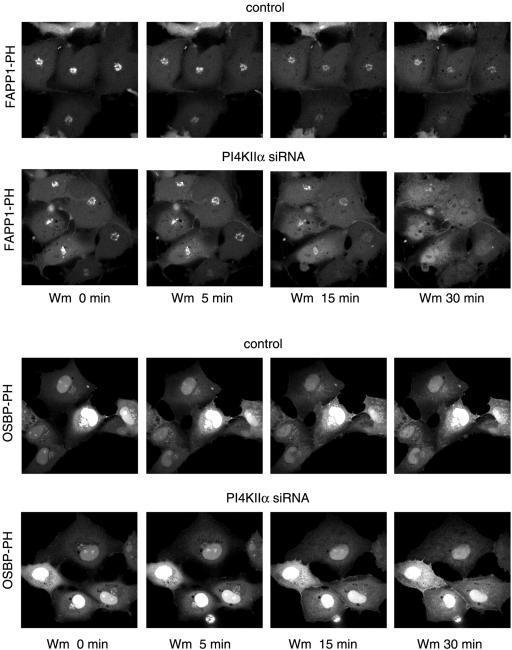
We also used small interfering (si)RNA to evaluate the importance of the specific PI4Ks in providing the PI(4)P for localization of the PH domains. Cells were treated for 2 d with siRNA designed against PI4KIIIβ, PI4KIIα, or PI4KIIIα and transfected for 1 d with the PH-GFP constructs. PI4KIIIα knock-down cells showed no major alteration in their PH domain localization patterns (unpublished data). In case of PI4KIIIβ knock-down, a significant number of cells showed no or little Golgi localization with either PH domains and the remaining localization was found in dispersed vesicular structures that did not show the usual Golgi morphology (Figure 5A). Knock-down of the PI4KIIα also impaired Golgi localization, but its effect was not as dramatic with the FAPP1-PH domain as with OSBP-PH in the live cells (Figure 6A). When cells were fixed and immunostained for the respective kinases, it was also confirmed that in many— but not all— cells in which the kinases were largely knocked down, the Golgi localization of the PH domains was greatly reduced or eliminated (Figures 5C and 6B). However, it was important to note that in many cells after 2 d of siRNA treatment, in spite of the reduced immunostaining of either kinases, there was still localization of the PH domains (unpublished data), suggesting that in a significant fraction (∼50%) of cells either of the kinases can be eliminated without a dramatic effect on the steady state distribution of the PH domains. However, when cells were chosen in which the Golgi localization of the PH domains was relatively well preserved after knock-down of PI4KIIα, this residual localization was completely eliminated by Wm (10 μM) treatment (Figure 7).
Figure 6.
Effects of down-regulation of the type-IIα PI4K by siRNA treatment on the localization of OSBP- and FAPP1-PH-GFP in live (A) and OSBP-PH-GFP in fixed (B) COS-7 cells. COS-7 cells were treated with double-stranded RNA designed to interfere with expression of PI4KIIα for 2 consecutive days and transfected with the PH-GFP constructs for an additional day as described in Materials and Methods. Cells were either studied live at 35°C (A) or analyzed after fixation and immunostaining with a polyclonal anti-PI4KIIα antibody (B; Guo et al., 2003). The arrows point to the Golgi area of two cells, one in which the PI4KIIα is largely eliminated and another in which it is still readily detectable.
Figure 7.
Effect of wortmannin treatment on the localization of the OSBP- and FAPP1-PH-GFP after 2-d treatment with siRNA against PI4KIIα. COS-7 cells were treated with siRNA and transfected with the PH domain constructs as described in the legend to Figure 5. Live cells were studied at 35°C and treated with 10 μM Wm for the indicated times. Note the complete loss of localization of the PH domains in the siRNA-treated cells but not in the controls.
Manipulations of Cellular Ca2+ Affects the Distribution of OSBP and FAPP1-PH-GFP Proteins
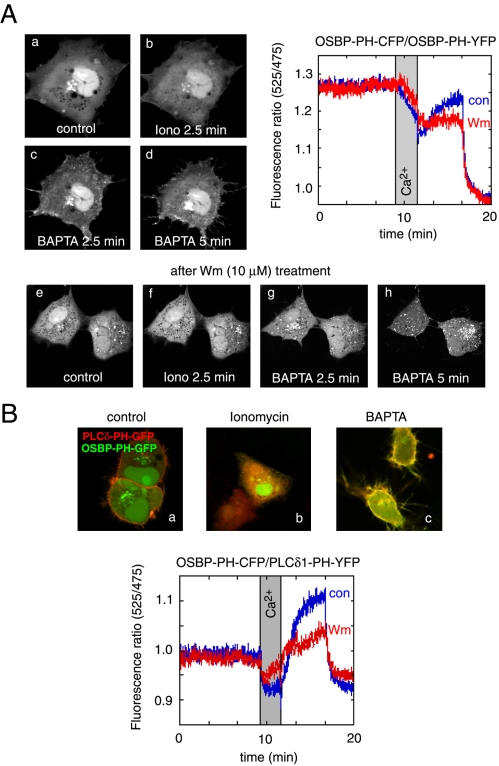
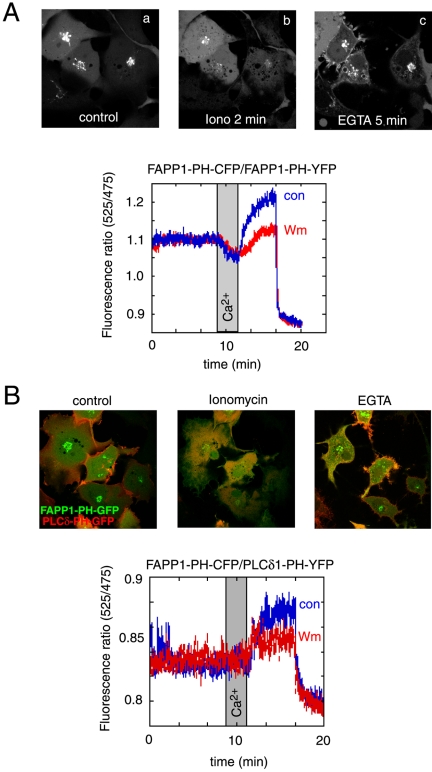
The experiments described so far gave information on the steady state distribution of the PH domains. To determine whether rapid elimination of the inositol lipids have an effect on the distribution of the PH domains, we used activation of the endogenous PLC enzymes so that all of the PI(4,5)P2 and PI(4)P is acutely eliminated from the cells. This was achieved by ionomycin treatment that has been shown to be an effective way of hydrolyzing all of the PI(4)P and PI(4,5)P2 in the membranes (Várnai and Balla, 1998). As shown in Figure 8A, ionomycin treatment induced the release of the OSBP-PH domain from the membranes, including the Golgi membranes with a parallel increase in the cytoplasmic fraction of the protein. Chelation of the Ca2+ that decreases PLC activity, hence allowing the replenishment of the lipids via resynthesis, caused a reappearance of the PH domains at the Golgi, as well as on the surface of small vesicles in the cytoplasm and importantly, at the plasma membrane. The plasma membrane association of the PH domain was significantly stronger after the Ca2+ increase and chelation than prior to the ionomycin treatment (Figure 8, A and B). This plasma membrane association was much more prominent with the OSBP- than with the FAPP1-PH domain (compare Figures 8 and 9). When these experiments were performed in cells after treatment with high concentrations of Wm (1–10 μM), the association of the OSBP-PH domain with the plasma membrane was almost completely abolished, whereas some reassociation of the domain with the Golgi and the intracellular vesicles were still observed (Figure 8A). It is important to note that no prominent translocation of either PI4KIIIα or PI4KIIIβ to the plasma membrane was observed under the same conditions (unpublished data).
Figure 8.
Effects of cytoplasmic [Ca2+] increases on the distribution of the OSBP-PH-GFP fusion protein. Cells were transfected with the indicated constructs and live cells were examined by confocal microscopy (Zeiss LSM410) at 35°C. (A) Addition of ionomycin (10 μM) in the presence of 2 mM external Ca2+ caused the translocation of the PH domain from the Golgi to the cytoplasm. Chelation of Ca2+ by BAPTA restores the localization in the Golgi and induces a prominent plasma membrane localization of the PH domain. Wortmannin (Wm) pretreatment prevents the plasma membrane localization but not the vesicular localization of the PH domain after ionomycin/BAPTA treatment (A, lower series). FRET measurements between CFP- and YFP-fused OSBP-PH domains coexpressed in COS-7 cells show a high initial FRET value (probably because of the high nuclear accumulation of the constructs), which decreases after ionomycin addition, but shows a rapid increase after Ca2+ chelation, which is mostly sensitive to Wm (10 μM) treatment (A, graph). (B) Simultaneous monitoring of the OSBP-PH and PLCδ1-PH domain translocations by coexpression of GFP and mRFP fused PH domains, respectively. Note the prominent colocalization of the red and green signals after (but not before) the iono/BAPTA treatment. FRET measurements between the same domains fused to CFP and YFP (OSBP-PH-CFP and PLCδ1-PH-YFP) shows a small initial FRET that is abolished after ionomycin treatment and returns well above baseline after Ca2+ chelation. This FRET analysis shows the plasma membrane component of the OSBP-PH movement that is significantly reduced after Wm treatment.
Figure 9.
Effects of cytoplasmic [Ca2+] increases on the distribution of the FAPP1-PH-GFP fusion protein. See legend to Figure 8 for experimental details. (A) Addition of ionomycin (10 μM) in the presence of 2 mM external Ca2+ caused the translocation of the PH domain from the Golgi to the cytoplasm. Chelation of Ca2+ by EGTA restores the localization in the Golgi but the plasma membrane localization of the FAPP1-PH domain is less prominent that that of the OSBP-PH. FRET measurements between CFP- and YFP-fused FAPP1-PH domains coexpressed in COS-7 cells show a rapid decrease after ionomycin addition and show a rapid increase above basal after Ca2+ chelation. About 50% of this increase is sensitive to Wm (10 μM) treatment (A, graph). (B) Simultaneous monitoring of the FAPP1-PH and PLCδ1-PH domain translocations by coexpression of GFP and mRFP fused PH domains, respectively. The colocalization of the red and green signals after the iono/EGTA treatment is not as prominent as with the OSBP-PH domain. FRET measurements between the same domains fused to CFP and YFP (FAPP1-PH-CFP and PLCδ1-PH-YFP) shows no initial FRET and no change after ionomycin treatment. FRET values; however, increase above baseline after Ca2+ chelation. This FRET analysis shows the plasma membrane component of the FAPP1-PH movement that is smaller than with the OSBP-PH and is also significantly reduced after Wm treatment.
Because these changes were relatively robust, we tried to quantitate them using the FRET approach that we have used before (van Der Wal et al., 2001; Varnai et al., 2002). For this purpose, the CFP- and YFP-tagged versions of the respective PH domains were created and coexpressed in COS-7 cells. Cells were then removed from the culture plates with mild trypsinization and incubated in a fluorescence spectrophotometer for ratiometric measurements of YFP/CFP ratios using CFP excitation at 425 nm. As shown before (van Der Wal et al., 2001), when the PH domains with the CFP and YFP fluorophores are bound to the membrane they are within FRET distance, whereas being in the cytoplasm they are not. Therefore, the FRET signal is a good reflection of the localization of the domains to the cellular membranes. As shown in the graph in Figure 8A, ionomycin treatment slowly decreased the FRET signal between the OSBP-PH-YFP/OSBP-PH-CFP pairs and similarly for the FAPP1-PH-YFP/FAPP1-PH-CFP pairs (Figure 8A, graph). This signal was restored once the Ca2+ was removed with the Ca2+ chelator BAPTA or EGTA. There was clearly a larger FRET signal after the Ca2+ chelation observed with the FAPP1-PH domains than before ionomycin addition (Figure 9A, graph inset). This was obscured in the case of the OSBP-PH domain by the high initial FRET values probably originating from the highly concentrated presence of the domains in the nucleus (Figure 8A, graph). There was no increase in the FRET signal after BAPTA (or EGTA) addition without ionomycin treatment (unpublished data). Importantly, in both cases, a significant fraction (∼50–60%) of the FRET increase after ionomycin and BAPTA treatment was abolished by treatment of the cells with 10 μM Wm (Figures 8A and 9A, graphs). At lower concentrations of Wm (10–100 nM) that inhibit only the PI 3-kinases, there were no inhibitory effects observed (unpublished data).
To assess the fraction of the PH domains associated with the plasma membrane, we performed FRET measurements by pairing the OSBP- or the FAPP1-PH domain CFP chimera with the YFP-fused PLCδ1PH domain. Because the PLCδ1PH-YFP protein localizes primarily to the plasma membrane and follows a translocation to and from the cytosol in response to ionomycin and BAPTA treatment, respectively (Várnai and Balla, 1998), the FRET between the PLCδ1PH-YFP and CFP-fused OSBP- or FAPP1-PH domain reflects the interaction between these domains at the plasma membrane. This is illustrated in Figures 8 and 9 (panels B), where the translocation of the PLCδ1PH-mRFP and GFP-OSBP-PH proteins were followed simultaneously during ionomycin and BAPTA treatment. As shown in Figure 8, the FRET signal between the CFP-OSBP-PH and PLCδ1PH-YFP is relatively modest under basal conditions and decreases in response to ionomycin treatment, reflecting the disappearance of the small initial association of the OSBP-PH with the plasma membrane. After BAPTA treatment, there was a large increase observed in the FRET signal, reflecting the increased association of the two PH domains at the plasma membrane. This response was, again, greatly inhibited by Wm treatment (Figure 8B, graph). Consistent with the morphological findings, the FRET signal was much smaller between the FAPP1-PH-CFP and PLCδ1PH-YFP constructs (hence the noisier trace in Figure 9B, graph), indicating that FAPP1-PH barely associates with the plasma membrane under basal steady state conditions. There was also very little change after ionomycin treatment, and only after Ca2+ chelation was an increase in the FRET signal observed. This increase was, again, greatly inhibited by Wm treatment. These data together suggested that after a large Ca2+ transient, there is a significant type-III PI 4-kinase–dependent association of the OSBP-PH and to a lesser degree of the FAPP1-PH domain with the plasma membrane.
The Role of PI4K Enzymes in the Redistribution of the PH Domains after Ionomycin Treatment and Ca2+ Chelation
To assess whether any of the two type-III PI 4-kinases was responsible for the Wm-sensitive PH domain redistribution to the plasma membrane, similar experiments were performed in COS-7 cells after down-regulation of the various PI4K isoforms by 2-d treatment with siRNA. These experiments were performed either in live cells (Figure 10) or in cells that were fixed after the ionomycin/EGTA treatment and processed for immunocytochemistry to determine the extent of knock-down (unpublished data). Down-regulation of either PI4KIIIβ or PI4KIIα failed to impair the relocalization of the PH domains to the plasma membrane after ionomycin/EGTA treatment and variably affected the Golgi localization (Figure 10, B and C). In some cells the Golgi localization was minimal; in others it was still detectable after the treatment. In contrast, down-regulation of PI4KIIIα almost completely abolished the localization of the PH domains to the small peripheral vesicles and to the plasma membrane (Figure 10D), but did not affect their relocalization to the Golgi in most cells. As an alternative to down-regulation, we also used phenylarsineoxide (PAO) at a concentration (10 μM) that preferentially inactivates the type-IIIα PI4K isoform (Balla et al., 2002). Pretreatment of the cells with 10 μM PAO for 10 min did not have any effect on steady state localization (unpublished data), but completely prevented the relocalization of the either PH domains to the plasma membrane (Figure 10E). In the case of the FAPP1- but not OSBP-PH, PAO also impaired the relocalization of the PH domain to the Golgi. The effect of PAO was reversed by simultaneous incubation with 1 mM dithiothreitol (DTT; Figure 10F) but not with β-mercaptoethanol (unpublished data). Taken together, these experiments indicated that PI4KIIIα is the enzyme that is mainly responsible for the production of PI(4)P that reaches the plasma membrane during active phosphoinositide resynthesis.
Figure 10.
Relocalization of OSBP- and FAPP1PH-GFP in COS-7 cells in which PI4K isoforms were down-regulated or inhibited. Cells were treated with siRNA and transfected with the plasmid DNA as described in the legend to Figure 5. Cells were studied live on the temperature-controlled stage of a Zeiss 410 laser confocal microscope at 35°C. Cells were stimulated with ionomycin (10 μM) until most of their localized PH domains were released from the Golgi (2–3 min), at which point Ca2+ was chelated by the addition of EGTA (7 mM) and images were recorded after 3–7 min. When indicated, PAO (10 μM) was added 10 min before ionomycin either in the presence or absence of 1 mM DTT. Note that the prominent plasma membrane localization of both PH domains was abolished in cells treated with PAO or in which the PI4KIIIα was down-regulated.
DISCUSSION
The present experiments were designed to explore whether PI 4-kinase function can be assessed at the single cell level using PH domains that recognize PI(4)P in vitro. Recent studies in yeast have concluded that the Golgi localization of the OSBP- and FAPP1-PH domains, the two domains used in the present study, are dependent both on the function of the yeast PI 4-kinase, Pik1, and on an Arf1-dependent additional component (Levine and Munro, 2002). Our present data on COS-7 cells are in complete agreement with those conclusions, in that both the lipid component and Arf1 in its GTP-bound form are necessary for efficient Golgi localization of these PH domains in steady state. However, our observations also showed that the OSBP-PH domain is also found in small vesicular compartments outside the Golgi, and the FAPP1-PH domain was also observed in a vesicular pool that became quite enlarged in cells expressing high levels of the fusion protein. Interestingly, at these extra-Golgi sites neither constructs showed the requirement for the Arf1 protein function.
An important finding of the present study was the prominent redistribution of both PH domains after Ca2+-induced PLC activation followed by Ca2+ removal. Both domains translocated to the cytosol in the presence of high intracellular [Ca2+], and reassociated with the Golgi after Ca2+ chelation. However, a significant fraction of the PH domains were found in the plasma membrane after this manipulation, and this response was largely abolished by concentrations of Wm that are consistent with the involvement of type-III PI4Ks in the process. It was also noteworthy that parallel to the appearance of the PH domains at the plasma membrane, large number of small vesicles were also visible just beneath the plasma membrane. These data indicated that PI(4)P is produced near and perhaps within the plasma membrane under these conditions and that both the OSBP- and FAPP1-PH-GFP are able to report on these changes. It is not clear at present why the localization of OSBP- or FAPP1-PH at the plasma membrane is low in resting cells. Whether this reflects low steady state PI(4)P levels at the plasma membrane or masking of this lipid by more stable interaction(s) with other endogenous proteins are questions to be further investigated.
Given the presence of multiple PI4K isoforms in eukaryotic cells, an important question to address was whether they all contributed to the membrane localization of the OSBP- and FAPP1-PH domains. This question was approached by pharmacological means as well as by using siRNA to down-regulate the respective PI4K enzymes. These experiments showed that both the type-IIα and the Wm-sensitive type-IIIβ enzyme was involved in the Golgi localization of the PH domains. Although the endogenous type-IIIβ enzyme shows the closest colocalization with either PH domains and also with the cis-Golgi marker gm130, the distribution of the type-IIα enzyme is only partially overlapping with the PH domains, consistent with the reported trans-Golgi and endosomal localization of this protein (Balla et al., 2002; Wei et al., 2002; Wang et al., 2003). In addition, the type-II enzyme (both endogenous and expressed) shows a significant amount of vesicular localization outside the Golgi, but at these sites it does not attract either of the PH domains (Balla, A., and Balla, T., unpublished observation). Based on the partial Wm sensitivity and the knock-down studies, the function of the type-IIIβ enzyme is clearly important for the Golgi localization of the PH domains. However, our data also show that the type-IIα enzyme also contributes to PI(4)P formation in the Golgi, as also have been indicated by its involvement in AP-1 recruitment (Wang et al., 2003). The relative contribution of the two PI 4-kinases appeared to show a great deal of variability in the COS-7 cells, and we could not find any morphological clues to predict the predominance of one over the other.
Remarkably, however, none of these enzymes was found to be necessary for the plasma membrane localization of the PH domains during active acute PI(4)P resynthesis after a strong, Ca2+-mediated PLC activation. The plasma membrane recruitment of the PH domains was Wm sensitive, was inhibited by 10 μM PAO and was largely eliminated in cells in which the PI4KIIIα was down-regulated. These data together suggest that the type-IIIα PI 4-kinase is responsible for the generation of this plasma membrane pool of PI(4)P, at least under these extreme conditions. This enzyme has been reported to localize primarily to the ER (Wong et al., 1997), a finding also confirmed by our studies, and plasma membrane localization of the enzyme was not demonstrable either in unstimulated cells or after inomycin/EGTA treatment (Balla, A., and Balla, T., unpublished observation). Nevertheless, we cannot rule out the function of PI4KIIIα at the plasma membrane, and in Saccharomyces cerevisiae the homologue of this enzyme, Stt4 has also been shown to generate PI(4)P at the plasma membrane for subsequent conversion to PI(4,5)P2 by the yeast PIP kinase, Mss4 (Audhya and Emr, 2002). However, it is also possible that PI(4)P is produced in a sub–plasma membrane vesicular pool of ER origin and the lipid is either transported to the PM or reaches the PM through dynamic exchange of these vesicles with the PM. Answering these questions will require further analysis and could reveal important new details about the generation of phosphoinositide pools at the plasma membrane.
Recently, Godi et al. (2004) have reported on the localization and functions of the FAPP1 and FAPP2 proteins and the importance of the PH domains in their localization. Several findings reported in that study is relevant to the discussion of our current results. Godi et al. (2004) have reported that the PH domains showed only partial colocalization with the cis-Golgi marker and a better colocalization with the TGN. In our studies, using COS-7 cells, both PH domains showed almost perfect colocalization with the cis-Golgi marker gm130 at low expression levels, but this colocalization was not maintained in cells expressing higher levels of the proteins. Godi et al. (1999) also showed that Arf1 is important for the localization of both PH domains and demonstrated a direct interaction between recombinant Arf1 and the PH domains. This could explain the data obtained with BFA, but it remains to be seen whether direct interaction is the sole mechanism by which Arf1 can regulate PH domain association, because Arf1 had also been shown earlier by the same authors to be important in the recruitment of the type-IIIβ PI4K. Although the nature of the PI4Ks that contribute to the recruitment of the FAPP1 and FAPP2 proteins was not the main focus of the above study, some of the data indicated the importance of the type-IIIβ PI4K. Our data are also consistent with the involvement of the type-IIIβ PI4K enzyme in the membrane recruitment of the PH domains.
In summary, our data demonstrate that PI(4)P generation at distinct membrane compartments is regulated by distinct PI 4-kinase enzymes and this process can be visualized by the FAPP1- and OSBP-PH domains. These studies also show the importance of Ca2+ in regulating the dynamics of the interaction of the PH domains with the various membranes. Clearly, the restrictive, Arf1-dependent steady state Golgi localization of the PH domains was dramatically changed after a cytoplasmic Ca2+ challenge. Whether the localization of the PH domains in the extra-Golgi compartments requires additional proteins is yet to be determined. PI4Ks are clearly emerging as important enzymes with multifaceted functions that reach far beyond the production of PI(4)P as a simple lipid precursor of plasma membrane PI(4,5)P2 and it may be more than coincidence that all of the known PH domains that recognize PI(4)P are found in lipid-transfer proteins, such as OSBP, FAPP2, or in the recently described CERT (Hanada et al., 2003). Exploration of the identity of the kinase(s) that regulate the synthesis of PI(4)P in distinct cellular compartments should aid further studies to understand the complex functions of phosphoinositides.
Acknowledgments
We thank Drs. Jun Guo and Pietro DeCamilli for the anti-PI4KIIα antibody, Drs. Paul Randazzo and Julie Donaldson for the Arf171QL construct, and Dr. Roger Y. Tsien for the monomeric red fluorescent protein. Part of the microscopy work (on the Zeiss 510 system) was performed at the Microscopy & Imaging Core (National Institute of Child Health and Development, National Institutes of Health) with the assistance of Drs. Vincent Schram and James T. Russell. The expert advice of Dr. Vincent Schram on the FRAP analysis is greatly appreciated. V.P. was partially supported by a grant from the Hungarian Science Foundation (OTKA T-034606).
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E04-07-0578) on January 5 2005.
Abbreviations used: BFA, brefeldin A; GFP, enhanced green fluorescent protein; mRFP, monomeric red fluorescent protein; OSBP, oxysterol-binding protein; PI, phosphatidylinositol; PI4K, phosphatidylinositol 4-kinase; PI(4)P, phosphatidylinositol 4-phosphate; PI(4,5)P2, phosphatidylinositol 4,5-bisphosphate; PH, pleckstrin homology; Wm, wortmannin.
References
- Audhya, A., Foti, M., and Emr, S. D. (2000). Distinct roles for the yeast phosphatidylinositol 4-kinases, stt4p and pik1p, in secretion, cell growth, and organelle membrane dynamics. Mol. Biol. Cell 11, 2673-2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audhya, A., and Emr, S. D. (2002). Stt4 PI 4-kinase localizes to the plasma membrane and functions in the Pkc1-mediated MAP kinase cascade. Dev. Cell 2, 593-605. [DOI] [PubMed] [Google Scholar]
- Balla, A., Tuymetova, G., Barshishat, M., Geiszt, M., and Balla, T. (2002). Characterization of type II phosphatidylinositol 4-kinase isoforms reveals association of the enzymes with endosomal vesicular compartments. J. Biol. Chem. 277, 20041-20050. [DOI] [PubMed] [Google Scholar]
- Balla, T., Bondeva, T., and Varnai, P. (2000). How accurately can we image inositol lipids in live cells? Trends Pharmacol. Sci. 21, 238-241. [DOI] [PubMed] [Google Scholar]
- Balla, T. and Varnai, P. (2002). Visualizing cellular phosphoinositide pools with GFP-fused protein-modules. Sci. STKE 125, PL3, 1-16. [DOI] [PubMed] [Google Scholar]
- Barylko, B., Gerber, S. H., Binns, D. D., Grichine, N., Khvotchev, M., Sudhof, T. C., and Albanesi, J. P. (2001). A novel family of phosphatidylinositol 4-kinases conserved from yeast to humans. J. Biol. Chem. 276, 7705-7708. [DOI] [PubMed] [Google Scholar]
- Berditchevski, F., Tolias, K. F., Wong, K., Carpenter, C. L., and Hemler, M. E. (1997). A novel link between integrins, transmembrane-4 superfamily proteins (CD63 and CD81), and phosphatidylinositol 4-kinase. J. Biol. Chem. 272, 2595-2598. [DOI] [PubMed] [Google Scholar]
- Berridge, M. J. (1984). Inositol trisphosphate and diacylglycerol as intracellular messengers. Biochem. J. 220, 345-360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell, R. E., Tour, O., Palmer, A. E., Steinbach, P. A., Baird, G. S., Zacharias, D. A., and Tsien, R. Y. (2002). A monomeric red fluorescent protein. Proc. Natl. Acad. Sci. USA 99, 7877-7882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowler, S., Currie, R. A., Campbell, D. G., Deak, M., Kular, G., Downes, C. P., and Alessi, D. R. (2000). Identification of pleckstrin-homology-domain-containing proteins with novel phosphoinositide-binding specificities. Biochem. J. 351, 19-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downing, G. J., Kim, S., Nakanishi, S., Catt, K. J., and Balla, T. (1996). Characterization of a soluble adrenal phosphatidylinositol 4-kinase reveals wortmannin-sensitivity of Type III phosphatidylinositol 4-kinases. Biochemistry 35, 3587-3594 [DOI] [PubMed] [Google Scholar]
- Flanagan, C. A., Schnieders, E. A., Emerick, A. W., Kunisawa, R., Admon, A., and Thorner, J. (1993). Phosphatidylinositol 4-kinase: gene structure and requirement for yeast cell viability. Science 262, 1444-1448. [DOI] [PubMed] [Google Scholar]
- Fruman, D. A., Meyers, R. E., and Cantley, L. C. (1998). Phosphoinositide kinases. Annu. Rev. Biochem. 67, 481-507. [DOI] [PubMed] [Google Scholar]
- Garcia-Bustos, J. F., Marini, F., Stevenson, I., Frei, C., and Hall, M. N. (1994). PIK1, an essential phosphatidylinositol 4-kinase associated with the yeast nucleus. EMBO J. 13, 2352-2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godi, A., Pertile, P., Meyers, R., Marra, P., Di Tullio, G., Iurisci, C., Luini, A., Corda, D., and De Matteis, M. A. (1999). ARF mediates recruitment of PtdIns-4-OH kinase-β and stimulates synthesis of PtdIns(4,5)P2 on the Golgi complex. Nat. Cell Biol. 1, 280-287. [DOI] [PubMed] [Google Scholar]
- Godi, A., Di Campi, A., Konstantakopoulos, A., Di Tullio, G., Alessi, D. R., Kular, G. S., Daniele, T., Marra, P., Lucocq, J. M., and De Matteis, M. A. (2004). FAPPs control Golgi-to-cell-surface membrane traffic by binding to ARF and PtdIns(4)P. Nat. Cell Biol. 6, 393-404. [DOI] [PubMed] [Google Scholar]
- Guo, J., Wenk, M. R., Pellegrini, L., Onofri, F., Benfenati, F., and De Camilli, P. (2003). Phosphatidylinositol 4-kinase type IIalpha is responsible for the phosphatidylinositol 4-kinase activity associated with synaptic vesicles. Proc. Natl. Acad. Sci. USA 100, 3995-4000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hama, H., Schnieders, E. A., Thorner, J., Takemoto, J. Y., and DeWald, D. B. (1999). Direct involvement of phosphatidylinositol 4-phosphate in secretion in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 274, 34294-34300. [DOI] [PubMed] [Google Scholar]
- Hanada, K., Kumagai, K., Yasuda, S., Miura, Y., Kawano, M., Fukasawa, M., and Nishijima, M. (2003). Molecular machinery for non-vesicular trafficking of ceramide. Nature 426, 803-809. [DOI] [PubMed] [Google Scholar]
- Hurley, J. H., and Meyer, T. (2001). Subcellular targeting by membrane lipids. Curr. Opin. Cell Biol. 13, 146-152. [DOI] [PubMed] [Google Scholar]
- Kauffmann-Zeh, A., Klinger, R., Endemann, G., Waterfield, M. D., Wetzker, R., and Hsuan, J. J. (1994). Regulation of human type II phosphatidylinositol kinase activity by EGF-dependent phosphorylation and receptor association. J. Biol. Chem. 269, 31243-31251. [PubMed] [Google Scholar]
- Lemmon, M. A., and Ferguson, K. M. (2000). Signal-dependent membrane targeting by pleckstrin homology (PH) domains. Biochem. J. 350, 1-18. [PMC free article] [PubMed] [Google Scholar]
- Lemmon, M. A. (2003). Phosphoinositide recognition domains. Traffic 4, 201-213. [DOI] [PubMed] [Google Scholar]
- Levine, T. P., and Munro, S. (1998). The pleckstrin-homology domain of oxysterol-binding protein recognizes a determinant specific to Golgi membranes. Curr. Biol. 8, 729-739. [DOI] [PubMed] [Google Scholar]
- Levine, T. P., and Munro, S. (2001). Dual targeting of Osh1p, a yeast homologue of oxysterol-binding protein, to both the Golgi and the nucleus-vacuole junction. Mol. Biol. Cell 6, 1633-1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine, T. P., and Munro, S. (2002). Targeting of Golgi-specific pleckstrin homology domains involves both PtdIns 4-kinase-dependent and -independent components. Curr. Biol. 12, 695-704. [DOI] [PubMed] [Google Scholar]
- Martin, T. F. (1997). Phosphoinositides as spatial regulators of membrane traffic. Curr. Opin. Neurobiol. 7, 331-338. [DOI] [PubMed] [Google Scholar]
- Michell, R. H. (1975). Inositol phospholipids and cell surface receptor function. Biochim. Biophys. Acta 415, 81-147. [DOI] [PubMed] [Google Scholar]
- Minogue, S., Anderson, J. S., Waugh, M. G., dosSantos, M., Corless, S., Cramer, R., and Hsuan, J. J. (2001). Cloning of a human type II phosphatidylinositol 4-kinase reveals a novel lipid kinase family. J. Biol. Chem. 276, 16635-16640. [DOI] [PubMed] [Google Scholar]
- Nakagawa, T., Goto, K., and Kondo, H. (1996). Cloning, expression and localization of 230 kDa phosphatidylinositol 4-kinase. J. Biol. Chem. 271, 12088-12094. [DOI] [PubMed] [Google Scholar]
- Nakanishi, S., Catt, K. J., and Balla, T. (1995). A wortmannin-sensitive phosphatidylinositol 4-kinase that regulates hormone-sensitive pools of inositolphospholipids. Proc. Natl. Acad. Sci. USA 92, 5317-5321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odorizzi, G., Babst, M., and Emr, S. D. (2000). Phosphoinositide signaling and the regulation of membrane trafficking in yeast. Trends Biochem. Sci. 25, 229-235. [DOI] [PubMed] [Google Scholar]
- van Der Wal, J., Habets, R., Varnai, P., Balla, T., and Jalink, K. (2001). Monitoring Phospholipase C activation kinetics in live cells by FRET. J. Biol. Chem. 276, 15337-15344. [DOI] [PubMed] [Google Scholar]
- Varnai, P., Lin, X., Lee, S. B., Tuymetova, G., Bondeva, T., Spat, A., Rhee, S. G., Hajnoczky, G., and Balla, T. (2002). Inositol lipid binding and membrane localization of isolated pleckstrin homology (PH) domains. Studies on the PH domains of phospholipase C delta 1 and p130. J. Biol. Chem. 277, 27412-27422. [DOI] [PubMed] [Google Scholar]
- Várnai, P., and Balla, T. (1998). Visualization of phosphoinositides that bind pleckstrin homology domains: calcium-and agonist-induced dynamic changes and relationship to myo-[3H]inositol-labeled phosphoinositide pools. J. Cell Biol. 143, 501-510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walch-Solimena, C., and Novick, P. (1999). The yeast phosphatidylinositol-4-OH kinase Pik1 regulates secretion at the Golgi. Nat. Cell Biol. 1, 523-525. [DOI] [PubMed] [Google Scholar]
- Wang, Y. J., Wang, J., Sun, H. Q., Martinez, M., Sun,. Y. X., Macia, E., Kirschhausen, T., Albanesi, J.P., Roth, M.G., and Yin, H.L. (2003). Phosphatidylinositol 4 phosphate regulates targeting of clathrin adaptor AP-1 complexes to the Golgi. Cell 114, 299-310. [DOI] [PubMed] [Google Scholar]
- Waugh, M. G., Minogue, S., Anderson, J. S., Balinger, A., Blumenkrantz, D., Calnan, D. P., Cramer, R., and Hsuan, J. J. (2003). Localization of a highly active pool of type II phosphatidylinositol 4-kinase in a p97/valosin-containing-protein-rich fraction of the endoplasmic reticulum. Biochem. J. 373, 57-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, Y. J., Sun, H. Q., Yamamoto, M., Wlodarski, P., Kunii, K., Martinez, M., Barylko, B., Albanesi, J. P., and Yin, H. L. (2002). Type II phosphatidylinositol 4-kinase beta is a cytosolic and peripheral membrane protein that is recruited to the plasma membrane and activated by Rac-GTP. J. Biol. Chem. 277, 46586-46593. [DOI] [PubMed] [Google Scholar]
- Wong, K., Meyers, R., and Cantley, L. C. (1997). Subcellular localization of phosphatidylinositol 4-kinase isoforms. J. Biol. Chem. 272, 13236-13241. [DOI] [PubMed] [Google Scholar]
- Yamaguchi, N., and Fukuda, M. N. (1995). Golgi retention mechanism of β-1,4-galactosyltransferase. Membrane-spanning domain-dependent homodimerization and association with α- and β-tubulins. J. Biol. Chem. 270, 12170-12176. [DOI] [PubMed] [Google Scholar]
- Yoshida, S., Ohya, Y., Goebl, M., Nakano, A., and Anrakur, Y. (1994). A novel gene, STT4, encodes a phosphatidylionositol 4-kinase in the PKC1 protein kinase pathway of Saccharomyces cerevisiae. J. Biol. Chem. 269, 1166-1171. [PubMed] [Google Scholar]
- Yu, J. W., Mendrola, J. M., Audhya, A., Singh, S., Keleti, D., DeWald, D. B., Murray, D., Emr, S. D., and Lemmon, M. A. (2004). Genome-wide analysis of membrane targeting by S. cerevisiae pleckstrin homology domains. Mol. Cell 13, 677-688. [DOI] [PubMed] [Google Scholar]