Abstract
Deubiquitinating enzymes (Dubs) are potential regulators of ubiquitination-dependent processes. Here, we focus on a member of the yeast ubiquitin-specific processing protease (Ubp) family, the Ubp1 protein. We could show that Ubp1 exists in two forms: a longer membrane-anchored form (mUbp1) and a shorter soluble form (sUbp1) that seem to be independently expressed from the same gene. The membrane-associated mUbp1 variant could be localized to the endoplasmic reticulum (ER) membrane by sucrose density gradient centrifugation and by immunofluorescence microscopy. Overexpression of the soluble Ubp1 variant stabilizes the ATP-binding cassette-transporter Ste6, which is transported to the lysosome-like vacuole for degradation, and whose transport is regulated by ubiquitination. Ste6 stabilization was not the result of a general increase in deubiquitination activity, because overexpression of Ubp1 had no effect on the degradation of the ER-associated degradation substrate carboxypeptidase Y* and most importantly on Ste6 ubiquitination itself. Also, overexpression of another yeast Dub, Ubp3, had no effect on Ste6 turnover. This suggests that the Ubp1 target is a component of the protein transport machinery. On Ubp1 overexpression, Ste6 accumulates at the cell surface, which is consistent with a role of Ubp1 at the internalization step of endocytosis or with enhanced recycling to the cell surface from an internal compartment.
INTRODUCTION
Many cellular proteins are modified by the attachment of the 76-amino acid polypeptide ubiquitin (Hochstrasser, 1996; Hershko and Ciechanover, 1998). The main role of ubiquitination is to target proteins for degradation either directly by acting as a degradation signal that is recognized by the 26S proteasome or indirectly by sorting membrane proteins into the lysosomal/vacuolar degradation pathway (Hicke, 1999). Ubiquitination of substrate proteins, which occurs by a cascade of enzymatic reactions, is reversible. Ubiquitin can again be removed from proteins by deubiquitinating enzymes (Dubs). A large number of Dubs have been identified in various organisms that are either cysteine proteases or metalloproteases (Verma et al., 2002; Yao and Cohen, 2002). The more classical cysteine proteases can again be divided into two groups: the ubiquitin C-terminal hydrolases (Uchs) that preferentially cleave ubiquitin from peptides and small adducts (e.g., Yuh1 in yeast) and the extremely divergent family of ubiquitin-specific processing proteases (Ubps) that cleave ubiquitin from protein substrates (Hochstrasser, 1996). Of the 17 cysteine-protease–type Dubs in yeast 16 belong to the Ubp class.
The degree of ubiquitination seems to be mainly regulated at the level of ubiquitin attachment. However, evidence is accumulating that deubiquitination also can be important for the regulation of ubiquitination levels. Evidence has been presented that the deubiquitinating enzyme Fat facets (Faf), which is involved in eye development in Drosophila, acts as a substrate-specific regulator of ubiquitination of the epsin homologue Liquid facets (Lqf). Faf seems to prevent proteolysis of Lqf by counteracting its ubiquitination (Chen et al., 2002). Another example is the Dub mUBPy from mouse that may play a role in controlling degradation of the Ras nucleotide exchange factor CDC25Mm. Coexpression of mUBPy with CDC25Mm reduces ubiquitination of CDC25Mm and increases its half-life (Gnesutta et al., 2001). Another potential regulator of ubiquitination is UCH-L1, one of the most abundant proteins in brain. Mutations in UCH-L1 may either increase or decrease the susceptibility for Parkinson's disease by affecting the turnover of α-synuclein (Leroy et al., 1998; Liu et al., 2002).
In yeast, evidence for a regulatory role of Dubs has been presented for Ubp3. It has been shown that Ubp3 interacts with Sir4, a component of a complex required for silencing at the silent mating-type loci and at telomeres (Moazed and Johnson, 1996). Silencing is enhanced in Δubp3 mutants, suggesting that Ubp3 acts as an inhibitor of silencing. Furthermore, Ubp3 seems to have a role in the pheromone response pathway in yeast (Wang and Dohlman, 2002). In addition, Ubp3 together with an additional factor Bre5 seems to regulate the stability of components of the COPI and COPII complexes in endoplasmic reticulum (ER)-to-Golgi trafficking (Cohen et al., 2003a,b).
We are mainly interested in the role of ubiquitination in membrane trafficking. Many ubiquitin-dependent processes, such as internalization of cell surface proteins (Hicke, 1999), targeting of membrane proteins to multivesicular bodies (MVBs) (Katzmann et al., 2001; Losko et al., 2001; Reggiori and Pelham, 2001), retrovirus budding (Patnaik et al., 2000; Schubert et al., 2000; Strack et al., 2000), or ER-associated degradation (ERAD) (Hiller et al., 1996; Wiertz et al., 1996) occur at cellular membranes. To identify potential regulators of ubiquitin-dependent processes at membranes, we looked for membrane-associated Dubs among the 16 yeast members of the Ubp family (Kinner and Kölling, 2003). Although the Ubps are in general hydrophilic proteins devoid of potential membrane spanning segments, a varying degree of membrane association was detected for most of them. Only one Ubp protein, Ubp16, was completely membrane associated. We could show that this protein is localized to the outer mitochondrial membrane. The function of Ubp16 at mitochondria, however, is still unclear. Here, we focus on the Ubp1 protein, which showed a remarkable fractionation pattern in subcellular fractionation experiments. We demonstrate that Ubp1 exists in two forms, a membrane-anchored form and a soluble form, which seem to be expressed independently from the same gene. The membrane-anchored form could be localized to the ER membrane. Our studies further suggest that Ubp1 plays a role in protein trafficking in the early endocytic pathway.
MATERIALS AND METHODS
Strains and Plasmids
Yeast strains are listed in Table 1. Gene deletion or gene tagging was accomplished by insertion of a polymerase chain reaction (PCR)-generated cassette into the yeast genome (Longtine et al., 1998). The changes were verified by PCR. The UBP1 gene was amplified by PCR with PfuUltra (Stratagene, La Jolla, CA) from chromosomal yeast DNA and inserted upstream of the 13myc sequence into the 2 μ-vector pRK722 (based on YEplac195; Gietz and Sugino, 1988) and into the CEN/ARS plasmid pRK717 (based on YCplac33; Gietz and Sugino, 1988) to give plasmids pRK805 and pRK806. The 13myc fragment was obtained from pFA6a-13myc-His3MX6 (Longtine et al., 1998). The correctness of the UBP1 sequence was confirmed by sequencing. Based on these plasmids, UBP1 mutants were generated by QuikChange PCR mutagenesis (Stratagene). In pRK851 (based on pRK806), the serine residues 530 and 531 were changed to alanine; in pRK911 (based on pRK805) and pRK912 (based on pRK806), methionine 67 was changed to serine; and in pRK982 (based on pRK806) and pRK983 (based on pRK805), cysteine 110 was changed to serine. The correctness of the mutagenized sequences was confirmed by sequencing. In pRK857, a PCR-generated UBP3 fragment was cloned into the 2 μ-vector YEplac195. Plasmid pRK879 was generated from pRK805 by insertion of a stop codon between UBP1 ORF and the 13myc sequence. To construct pRK989, the UBP1 fragment of pRK879 was inserted into the 2 μ-vector YEplac112 (Gietz and Sugino, 1988). To construct pRK590, a 5.5-kb HindIII/SacI STE6 fragment from pRK182 (Losko et al., 2001) was cloned into YEplac112. YEp112 contains a hemagglutinin (HA)-tagged ubiquitin gene under the control of the CUP1 promoter (Hochstrasser et al., 1991). The STE2 gene was amplified by PCR with PfuUltra (Stratagene) from chromosomal yeast DNA and inserted upstream of the 13myc sequence into the CEN/ARS plasmid pRK717 to give plasmid pRK929. Plasmids pRK806-PGAL and pRK929-PGAL were generated by in vivo recombination with a PCR-generated cassette (template, pFA6a-His3-PGAL1; Longtine et al., 1998). pRK806-PGAL contains the GAL1 promoter directly upstream of the second ATG codon in the UBP1-13myc sequence.
Table 1.
Yeast strains
| Strain | Genotype | Reference |
|---|---|---|
| JD52 | MATa ura3-52 his3-Δ200 leu2-3,112 trp1-Δ63 lys2-801 | J. Dohmen, Universität Köln, Köln, Germany |
| RKY959 | MATa ura3-52 his3-Δ200 leu2-3,112 trp1-Δ63 lys2-801 Δste6::LEU2 | Losko et al., 2001 |
| RKY1894 | MATa ura3-52 his3-Δ200 leu2-3,112 trp1-Δ63 lys2-801 UBP1-13myc::HIS3 | This study |
| RKY1932 | MATa ura3-52 his3-Δ200 leu2-3,112 trp1-Δ63 lys2-801 Δubp1::kan | This study |
| RKY2008 | MATa ura3-52 his3-Δ200 leu2-3,112 trp1-Δ63 lys2-801 kan::PGAL1-UBP1-13myc::HIS3 | This study |
| RKY2019 | MATa ura3-52 his3-Δ200 leu2-3,112 trp1-Δ63 lys2-801 prc1-1 | This study |
| RKY2020 | MATa ura3-52 his3-Δ200 leu2-3,112 trp1-Δ63 lys2-801 Δubp1::kan prc1-1 | This study |
| RKY2026 | MATa ura3-52 his3-Δ200 leu2-3,112 trp1-Δ63 lys2-801 prc1-1 Δubc7::kan | This study |
Differential Centrifugation
Ten A600 units of cells growing exponentially (A600 = 0.4–0.7, 2–4 × 107 cells/ml) in YPD medium (1% yeast extract, 2% bacto peptone, 2% glucose) were harvested, washed in water, and resuspended in lysis buffer (0.3 M sorbitol, 50 mM HEPES, pH 7.5, and 10 mM NaN3 + protease inhibitor cocktail). Cells were lysed by vortexing with glass beads for 5 min at 4°C. Intact cells and cell debris were removed by centrifugation at 500 × g for 5 min. The cell extract was first spun at 13,000 × g for 10 min to give the P13 pellet fraction and the S13 supernatant. The S13 fraction was then centrifuged at 100,000 × g for 1 h to give the P100 pellet and the S100 supernatant. To test for solubility, equal aliquots of the extract were treated with 1% Triton X-100, 1 M NaCl, or 0.1 M Na2CO3, pH 11, for 30 min on ice before centrifugation. Equal portions of the fractions were assayed for the presence of proteins by Western blotting.
Flotation Gradients
Flotation gradients were essentially performed as described in Bagnat et al. (2000). Briefly, cells were grown to logarithmic phase in YPD. Ten A600 units were harvested by centrifugation, washed with water, and lysed by agitation with glass beads in TNE buffer (50 mM Tris, pH 7.4, 150 mM NaCl, and 5 mM EDTA + protease inhibitors). After removal of cell debris and intact cells by centrifugation at 500 × g for 5 min, 125 μl of the extract were mixed with 250 μl of 60% Optiprep solution (Axis-Shield, Oslo, Norway). The resulting 40% Optiprep fraction was transferred to the bottom of a centrifugation tube and overlaid with 600 μl of 30% Optiprep in TNE buffer and 100 μl of TNE. Then, the gradients were centrifuged at 77,000 rpm and 4°C for 2 h in a TLA 100.2 rotor in a Beckman Tabletop ultracentrifuge. Six equal fractions were collected and assayed for the presence of proteins by Western blotting.
Sucrose Density Gradient Centrifugation
One hundred milliliters of an exponentially growing YPD culture was harvested by vacuum filtration onto nitrocellulose filters and resuspended in 250 μl of STED10 [10% (wt/wt) sucrose, 10 mM Tris, pH 7.6, 1 mM EDTA, pH 8.0, and 1 mM dithiothreitol + protease inhibitors]. Cells were lysed by vortexing with glass beads for 5 min at 4°C and diluted with 1 ml of STED10. Intact cells and cell debris were removed by centrifugation at 500 × g for 5 min. Cell extracts were loaded onto the top of a sucrose density gradient ranging from 20 to 40% sucrose (in STED buffer) and centrifuged for 14 h at 30,000 rpm in a Beckman SW40 rotor at 4°C. Eighteen gradient fractions were collected and assayed for the presence of proteins by Western blotting.
Immunofluorescence
The immunofluorescence experiments were performed as described previously (Kölling and Hollenberg, 1994). Ste6-c-myc and Ubp1-13myc were detected with 9E10 antic-myc primary antibodies (1:200; Covance, Berkeley, CA) and with fluorescein isothiocyanate (FITC)-conjugated anti-mouse secondary antibodies (1:300; Dianova, Hamburg, Germany). Fluorescence was observed with a Zeiss Axioskop (Carl Zeiss, Göttingen, Germany) equipped with a Zeiss AxioCam digital camera by using an FITC filter set.
In Vivo Labeling of Cells and Immunoprecipitation
For the phosphorylation experiments, cells were grown overnight in SD-low phosphate medium (30 μM KH2PO4) to an A600 ≤ 0.8 (4 × 107 cells/ml). Cells (5 × 107) were labeled with 200 μCi of [32P]orthophosphate (PBS11; Amersham Biosciences, Piscataway, NJ) for 30 min at 30°C in 0.5 ml of SD-low phosphate medium. The cells were washed in 10 mM NaN3, resuspended in 110 μl of lysis buffer (50 mM HEPES, 0.3 M sorbitol, 10 mM NaN3, pH 7.5, protease inhibitor cocktail, and 50 μg/ml RNase A) and vortexed for 3 min with 400 mg of glass beads. Pulse-chase experiments and immunoprecipitation were essentially performed as described previously (Losko et al., 2001).
RESULTS
Ubp1 Exists in Two Forms That Can Be Separated by Cell Fractionation
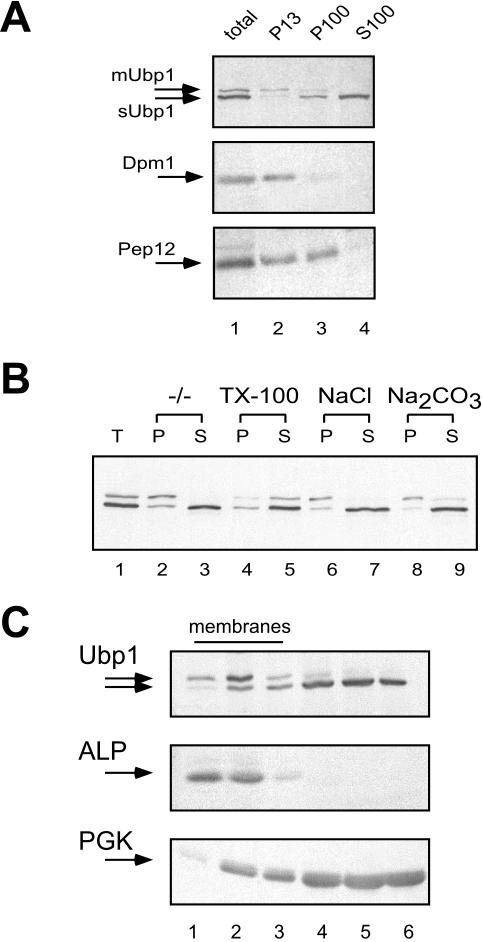
For detection, Ubp1 was C-terminally tagged with a 13myc-epitope by integration of a PCR-generated cassette at the 3′ end of the chromosomal copy of the UBP1 gene. Ubp1 was examined for membrane association by differential centrifugation experiments. Cell extracts of the UBP1-13myc strain RKY1894 were fractionated by two consecutive centrifugation steps at 13,000 and 100,000 × g into P13 and P100 pellet fractions and a S100 supernatant fraction. Interestingly, two closely spaced Ubp1 bands were observed by Western blotting that could be separated by differential centrifugation (Figure 1A). The upper, slower migrating band was almost exclusively found in the P13 pellet, whereas the lower, faster migrating band was mainly found in the S100 fraction, which contains the soluble proteins. In addition, a small amount of the lower form also was detected in the P100 pellet fraction (Figure 1A). This result suggests that Ubp1 exists in two forms, a pelletable presumably membrane-associated form (mUbp1) and a soluble form (sUbp1).
Figure 1.
Fractionation of Ubp1. (A) Cell extracts of the UBP1-13myc strain RKY1894 (lane 1, total cell extract) were centrifuged at 13,000 × g for 10 min to pellet the P13 fraction (lane 2). The supernatant was separated into P100 pellet fraction (lane 3) and S100 supernatant fraction (lane 4) by an additional centrifugation at 100,000 × g for 1 h. (B) Equal aliquots of the cell extract were treated with 1% Triton X-100 (lanes 4 and 5), 1 M NaCl (lanes 6 and 7), and 0.1 M Na2CO3, pH 11 (lanes 8 and 9) for 30 min on ice before a 1-h centrifugation at 100,000 × g. Lanes 1–3, untreated extract. Equal portions of the P100 (P) and S100 (S) fractions were analyzed for Ubp1-13myc. Lane 1, total cell extract. (C) Cell extracts of the UBP1-13myc strain RKY1894 were fractionated on Optiprep flotation gradients. Six fractions (lanes 1–6) were collected from the gradients (lanes 1–3, float fraction; lanes 4–6, nonfloat fraction). Fractions were analyzed by Western blotting with specific antibodies as indicated.
Depending on their intracellular localization, membrane proteins are typically distributed in a characteristic way among the P13 and P100 fractions as exemplified by the two marker proteins shown in Figure 1A, the ER marker protein Dpm1 and the endosomal marker protein Pep12. Proteins associated with larger organelles such as mitochondria, the vacuole, the ER, or the plasma membrane are mostly found in the P13 pellet, whereas proteins associated with the Golgi or with smaller vesicles are mostly found in the P100 pellet. Typically, the endosomal marker Pep12 is evenly distributed among the two pellet fractions (Becherer et al., 1996). This experiment, therefore, suggests that mUbp1 is localized to a larger intracellular compartment and argues against an endosomal or Golgi localization of mUbp1.
Membrane association of mUbp1 was further investigated by treating cell extracts with different reagents before centrifugation at 100,000 × g. As can be seen in Figure 1B, detergent treatment (1% Triton X-100) almost completely solubilized mUbp1 (shift from P100 to S100), whereas the distribution of sUbp1 was unaffected. Reagents that are typically used to strip off peripheral membrane proteins from membranes (1 M NaCl and Na2CO3, pH 11) had no effect on the distribution of mUbp1. Thus, mUbp1 behaves like an integral membrane protein. A small amount of sUbp1 also was detected in the P100 pellet fraction. This could be an indication that a certain fraction of sUbp1 also is membrane associated. However, the P100 fraction of sUbp1 proved to be resistant to detergent extraction. Thus, part of sUbp1 is either associated with detergent-resistant membranes (“rafts”) or with a larger protein complex.
Membrane association of mUbp1 is further supported by flotation analysis on Optiprep gradients. Cell extracts of RKY1894 were mixed with a high-density Optiprep solution, placed at the bottom of a centrifuge tube, and overlaid with Optiprep solutions of lower density. During centrifugation, membranes float to the top of the gradient due to their lower density, whereas soluble proteins remain behind in the lower part of the gradient. This is illustrated by the behavior of marker proteins (Figure 1C). The membrane protein alkaline phosphatase (ALP) was mainly found in the top two fractions of the gradient, whereas the soluble protein phosphoglycerate kinase (PGK) was mostly found in the lower three fractions. On these gradients, the two forms of Ubp1 showed a different fractionation pattern. Whereas sUbp1 cofractionated with the soluble marker PGK, mUbp1 floated to the top of the gradient like a typical membrane protein (Figure 1C). From these experiments, we conclude that Ubp1 exists in two forms, a soluble form and a membrane-associated form.
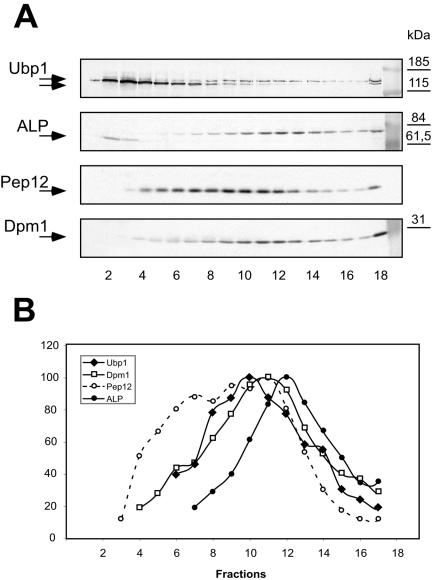
To obtain information about the intracellular localization of mUbp1, cell extracts of strain RKY1894 were fractionated on sucrose density gradients (Figure 2). Again, the two forms of Ubp1 were clearly separated on these gradients. sUbp1 cofractionated with the soluble proteins in the first few fractions of the gradient, whereas mUbp1 migrated further down the gradient. The distribution of mUbp1 was clearly distinct from the distribution of the vacuolar marker protein ALP and the plasma membrane marker Pma1, which was found at the bottom of the tube due to the high density of the plasma membrane (our unpublished data). The endosomal marker Pep12 displayed a complex fractionation pattern, which could reflect a localization of Pep12 to multiple compartments of different density. This is in line with the differential centrifugation experiment where Pep12 is distributed between low- and high-speed pellet fractions (Figure 1A). Although mUbp1 partially overlapped with the Pep12 distribution, the shape of the mUbp1 peak was clearly different from the shape of the Pep12 distribution. Therefore, mUbp1 does not seem to localize to the Pep12 compartment(s). The closest match was observed between mUbp1 and the ER marker protein Dpm1. The match, however, was not perfect because the peak fraction of mUbp1 was shifted by one fraction compared with Dpm1. But, this shift could be a quantification artifact because due to the close spacing of the bands, it was not possible to completely separate the mUbp1 from the sUbp1 signal. So, sUbp1 contributes to a certain extent to the measured mUbp1 signal, especially in the upper part of the gradient. Alternatively, mUbp1 could be localized to a subdomain of the ER with a slightly different density. Despite these limitations, the sucrose density fractionation is most compatible with an ER localization of mUbp1.
Figure 2.
Fractionation of Ubp1 by density gradient centrifugation. A whole cell extract of the UBP1-13myc strain RKY1894 was fractionated on a sucrose density gradient [20–40% (wt/wt) sucrose; fraction 1, low sucrose density]. (A) Gradient fractions were analyzed for the presence of marker proteins by Western blotting with specific antibodies as indicated. (B) Densitometric quantification of the Western blot signals. The Western blot signal intensities were quantified with the program ImageJ. The strongest signals were set to 100%. Ubp1, diamonds; Dpm1, squares; Pep12, open circles; and ALP, filled circles.
Two Ubp1 Variants Are Encoded by the UBP1 Gene
Only a certain fraction of Ubp1 proved to be membrane associated. Notably, membrane association of Ubp1 was connected with altered mobility on SDS-PAGs. What is the basis of this membrane association? Ubp1 could be anchored to membranes by posttranslational modifications such as prenylation or palmitoylation. Prenylation could be excluded because no consensus sequences for prenylation (CAAX, CXC, CC, and CCXX) are found at the C terminus of Ubp1. Sensitivity to weak nucleophiles, including thiols, is a hallmark of the thioester linkage of palmitoylation (Roth et al., 2002). Membrane association of mUbp1, however, proved to be thiol resistant (our unpublished data). Thus, palmitoylation as the basis of membrane association of mUbp1 could be discounted as well. We also considered the possibility that monoubiquitination of Ubp1 could account for the slower migrating Ubp1 band; however, we were not able to detect an ubiquitin signal for Ubp1 (our unpublished data).
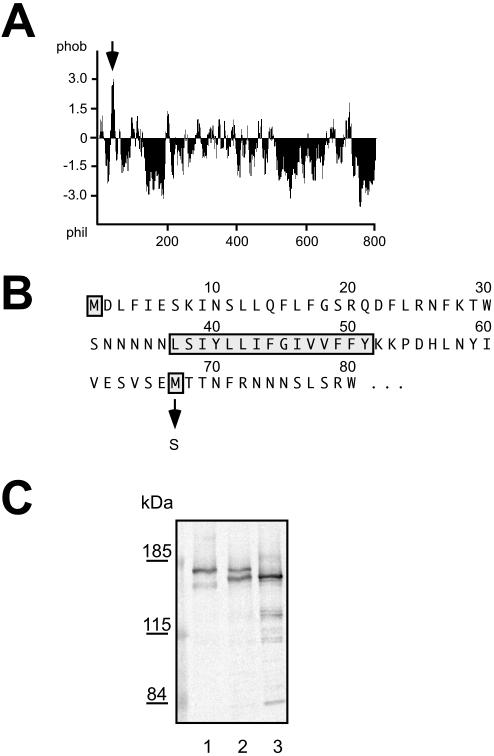
Because the most prevalent ways of posttranslational membrane attachment do not seem to be pertinent to mUbp1 membrane association, we looked for membrane anchors in the protein sequence itself. As can be seen from the hydropathy profile of Ubp1 (Figure 3A), a short hydrophobic stretch of ∼15 amino acids, which could potentially function as a membrane anchor, is found close to the N terminus of the Ubp1 sequence (Figure 3B). But, if Ubp1 contains a membrane anchor why is only a certain fraction of Ubp1 membrane associated? Two possibilities can be envisioned: either the membrane anchor is proteolytically cleaved from a certain fraction of Ubp1 or two different variants, with and without membrane anchor, are synthesized from the same gene. As a corollary to the latter prediction, there should be two different promoters in front of the UBP1 gene directing the expression of the two Ubp1 variants. If the second ATG codon in the UBP1 sequence is used as a translational start point, a protein 7883 Da smaller than full-length Ubp1 will be generated. This corresponds exactly to the size difference (8 kDa) observed on Western blots. The smaller Ubp1 variant should be soluble because it contains no longer the potential membrane anchor (Figure 3B). To test these predictions, we mutagenized the second ATG codon to a serine codon (TCG, M67S). If we remove the second ATG codon by mutagenesis, a shorter variant can no longer be synthesized because the next ATG triplet in the sequence is not in frame with the original UBP1 open reading frame (ORF). In line with this interpretation, only a single band was observed on Western blots for the Ubp1 M67S variant, which runs exactly at the position of the upper Ubp1 band (Figure 3C). We also performed the complementary experiment. The GAL1 promoter was inserted in front of the second ATG codon thereby preventing expression of the full-length UBP1 ORF. Under these conditions, only one band was observed that ran exactly at the position of the lower Ubp1 band (Figure 3C). Together, these experiments strongly suggest that two different variants of Ubp1 are independently expressed from the UBP1 gene, a longer membrane anchored form (mUbp1) and a shorter soluble form (sUbp1).
Figure 3.
Two Ubp1 variants are encoded by the UBP1 gene. (A) Hydropathy profile of Ubp1. The hydrophobic peak corresponding to the potential TMD is marked by an arrow. (B) N-terminal part of the Ubp1 sequence. The putative TMD and the first and second methionine in the sequence are highlighted. The M67S mutation is indicated. (C) Cell extracts of strain JD52 transformed with different plasmids expressing different versions of the UBP1 gene were examined for Ubp1 by Western blotting. Lane 1, pRK912 (UBP1-13myc M67S); lane 2, pRK806 (UBP1-13myc); and lane 3, pRK806-PGAL (PGAL-UBP1-13myc). Lanes 1 and 2, glucose medium; lane 3, galactose medium. In lane 3, only one-fifth of the normal amount of cell extract was loaded.
The Membrane-anchored mUbp1 Variant Is Localized to the ER
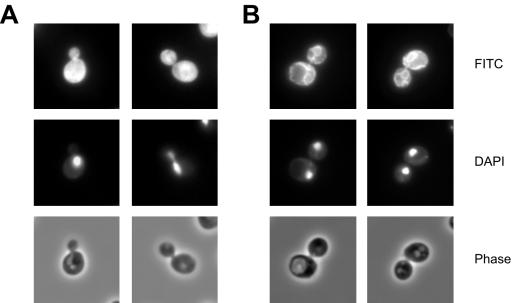
The intracellular localization of 13myc tagged Ubp1 was studied by immunofluorescence microscopy. With wild-type UBP1, a bright cytoplasmic staining was observed (Figure 4A), in agreement with results obtained from a genome-wide protein localization study (Huh et al., 2003). We were, however, especially interested in the localization of the membrane-associated mUbp1 variant. But obviously, the strong cytoplasmic signal of the sUbp1 variant obscured the mUbp1 signal. With the constructs described in the preceding section, we are now in a position to express the two Ubp1 variants separately. This allowed us to study the intracellular localization of mUbp1 by immunofluorescence microscopy independently from the sUbp1 background. With the 13myc-tagged Ubp1 M67S variant, expressed from a 2μ plasmid, a typical ER staining was observed (Figure 4B). The staining surrounded the nuclei, visualized by 4,6-diamidino-2-phenylindole (DAPI) staining, and extended further from the nuclei to the cell periphery. A similar, faint perinuclear staining also was observed with the single-copy Ubp1 M67S variant (our unpublished data). These results are in agreement with the cell fractionation experiments that also point to an ER localization of mUbp1.
Figure 4.
Localization of Ubp1 by immunofluorescence. Ubp1 variants, encoded by pRK805 (UBP1-13myc) (A) and pRK911 (UBP1-13myc M67S) (B) were detected with anti-myc primary antibodies (9E10) and FITC-conjugated anti-mouse secondary antibodies. Top, FITC fluorescence; middle, staining of nuclei with DAPI; and bottom, phase contrast image.
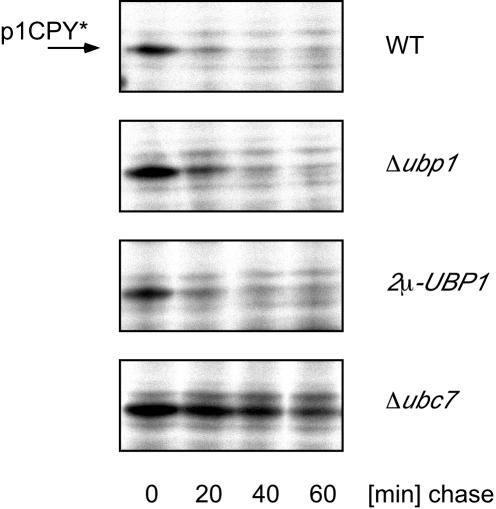
What could be the function of mUbp1 at the ER membrane? The most prominent ubiquitination-dependent process at the ER membrane is the ERAD pathway (Hampton, 2002). To test whether Ubp1 is involved in ERAD, we examined the effect of UBP1 deletion or overexpression on the turnover of the ERAD substrate carboxypeptidase Y* (CPY*). CPY* is a mutant form of CPY that is retained in the ER in its core-glycosylated p1 form and degraded via the ERAD pathway (Hiller et al., 1996). CPY* turnover was examined by pulse-chase experiments. In the wild-type strain JD52, CPY* was degraded quickly with a half-life of ∼15 min (Figure 5), as reported previously (Hiller et al., 1996). A similar turnover rate was observed in the UBP1 deletion strain RKY1932 and upon overproduction of UBP1 from a 2μ plasmid (RKY1932/pRK879). The Ubp1 protein level was approximately fivefold higher in UBP1-overexpressing cells compared with single-copy UBP1 (our unpublished data). As a positive control, we used a strain (RKY2015) that is deleted for a central component of the ERAD pathway, the ubiquitin-conjugating enzyme Ubc7 (Biederer et al., 1996). As expected, CPY* was strongly stabilized in this strain. From these experiments, we therefore conclude that Ubp1 probably does not play a general role in ERAD.
Figure 5.
CPY* turnover is unaffected by Ubp1. The turnover of CPY* in different strains was examined by pulse-chase analysis. Cells were labeled with [35S] Translabel for 15 min and chased with an excess of cold methionine and cysteine for the time intervals indicated. CPY* was precipitated from cell extracts with polyclonal antibodies directed against CPY. Precipitated CPY* was detected by autoradiography. The p1-CPY* band is marked with an arrow. Strains used (from top to bottom) are RKY2019 (WT), RKY2020 (Δubp1), RKY2019/pRK879 (2μ-UBP1), and RKY2026 (Δubc7). All strains contain the prc1-1 allele (CPY*).
UBP1 Overexpression Affects Ste6 Trafficking
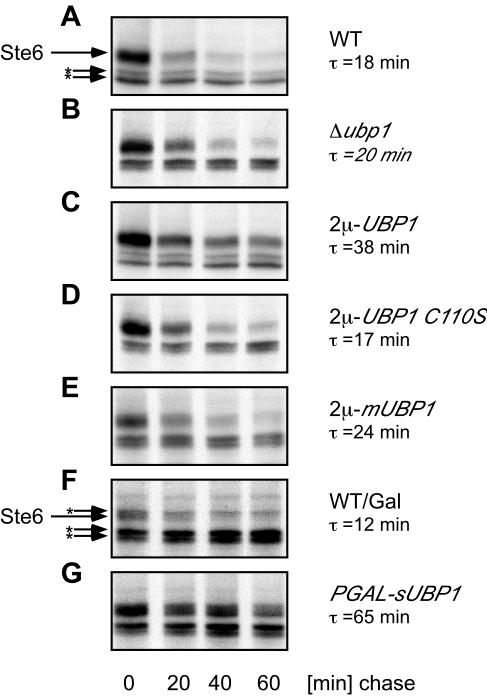
The ABC-transporter Ste6 is transported to the yeast vacuole for degradation via the MVB pathway (Losko et al., 2001). Trafficking of Ste6 and other cell surface proteins to the vacuole is regulated by ubiquitination (Kölling and Hollenberg, 1994; Hicke, 1999). As a deubiquitinating enzyme, Ubp1 is a potential regulator of membrane protein trafficking in the endocytic pathway. To test whether Ubp1 is involved in the regulation of Ste6 trafficking, we examined the effect of UBP1 deletion or overexpression on Ste6 turnover. Again, Ste6 turnover was investigated by pulse-chase experiments. In the wild-type strain, JD52 Ste6 was quickly degraded with a half-life of 18 min (Figure 6A), as reported previously (Kölling and Hollenberg, 1994). Although Ste6 turnover was unaffected by UBP1 deletion (Figure 6B, τ = 20 min), Ste6 was stabilized approximately twofold by UBP1 overexpression (Figure 6C, τ = 38 min). Ste6 turnover was not affected by overexpression of a catalytically inactive Ubp1 variant (C110S), where the active site cysteine had been replaced by serine (Figure 6D, τ = 17 min). This indicates that the deubiquitinating activity of Ubp1 is required for Ste6 stabilization. To find out which of the two Ubp1 forms is responsible for the observed stabilization, the 13myc-tagged mUbp1 and the sUbp1 variants were overexpressed separately. To overexpress mUbp1, a 2μ plasmid carrying the UBP1 M67S variant under the control of its own promoter was introduced into the UBP1 deletion strain RKY1932. mUbp1 was overexpressed approximately five-fold under these conditions (our unpublished data). The sUbp1 variant was expressed from the strong GAL1 promoter integrated into the chromosomal copy of the UBP1 gene (RKY2008), which leads to ∼10-fold overexpression (our unpublished data). As can be seen from Figure 6E, overexpression of mUbp1 had no significant effect on Ste6 turnover (τ = 24 min). In contrast, overexpression of sUbp1 strongly stabilized Ste6 (Figure 6G, τ = 65 min). On galactose containing media, which are required to induce the GAL1 promoter, Ste6 turnover was approximately twice as fast as on glucose-containing media (Figure 6F, half-life 12 min). Ste6 turnover was not affected by overexpression of another deubiquitinating enzyme, UBP3, from a 2μ plasmid (our unpublished data), indicating that the observed UBP1 effects may be specific.
Figure 6.
Effect of Ubp1 on Ste6 turnover. The turnover of Ste6 in different strains was examined by pulse-chase analysis. Cells were labeled with [35S] Translabel for 15 min and chased with an excess of cold methionine and cysteine for the time intervals indicated. Ste6 was precipitated from cell extracts with polyclonal antibodies directed against Ste6. Precipitated Ste6 was detected by autoradiography. The Ste6 band is marked with an arrow. Background bands are labeled with asterisks. Strains used were JD52 (WT) (A), RKY1932 (Δubp1) (B), JD52/pRK805 (2μ-UBP1–13myc) (C), JD52/pRK983 (2μ-UBP1-13myc C110S) (D), JD52/pRK911 (2μ-UBP1-13myc M67S) (E), JD52 (WT/Gal) (F), and RKY2008 (PGAL-sUBP1-13myc) (G). Cells were either grown on glucose medium (A–E) or on galactose medium (F and G).
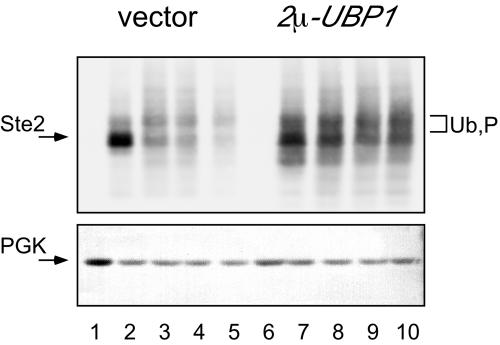
To see whether UBP1 overexpression affects other endocytic cargo proteins in addition to Ste6, we examined the effect of UBP1 overexpression on the degradation of the α-factor receptor Ste2. On binding of its ligand, α-factor, Ste2 is internalized from the cell surface and degraded in the vacuole with a half-life of <10 min (Hicke and Riezman, 1996). Full-length Ste2, marked with a 13myc-tag at its C terminus, was expressed under the control of the GAL1 promoter from a single-copy plasmid. Expression from the GAL1 promoter is induced by galactose and repressed by glucose in the growth media. A specific Ste2 signal was detected on galactose-grown cells, whereas no signal was detectable on glucose medium (Figure 7). On addition of α-factor, Ste2 becomes hyperphosphorylated and ubiquitinated (Hicke et al., 1998). Thus, in addition to the main Ste2 band, phosphorylated and ubiquitinated forms with slower mobility can be detected. Ste2 turnover was analyzed by a “Gal-depletion” experiment, i.e., cells were first grown on galactose medium and then transferred to glucose medium to turn off expression form the GAL1 promoter. After transfer to glucose, α-factor was added to the culture (t0), and then samples were taken at 10-min intervals. As a loading control, the glycolytic enzyme PGK was detected by Western blotting with specific antibodies. As can be seen in Figure 7, after α-factor addition, Ste2, was quickly degraded in the control cells (lanes 2–5), whereas Ste2 was stable in cells overexpressing UBP1 (lanes 7–10). Thus, like Ste6, Ste2 is stabilized by overexpression of UBP1. This result suggests that UBP1 overexpression has a general effect on the trafficking of cargo proteins in the endocytic pathway.
Figure 7.
Effect of Ubp1 on Ste2 turnover. Ste2 turnover was examined by a “Gal-depletion” experiment. JD52 transformed with pRK929-PGAL (PGAL1-STE2-13myc) and with YEplac112 (vector) (lanes 2–5) or pRK989 (2μ-UBP1) (lanes 7–10) were pregrown on galactose and then transferred to glucose medium. At t0, α-factor was added to 5 μM. Samples were taken at 10-min intervals and analyzed for Ste2 (top) by Western blotting with 9E10 antibodies and for PGK (bottom). The Ste2 and PGK bands are marked by arrows; phosphorylated (P) and ubiquitinated (Ub) forms are marked by bracket. Lanes 1 and 6, JD52/pRK929-PGAL/YEplac112 (lane 1) and JD52/pRK929-PGAL/pRK989 (lane 6) grown on medium containing 5% glucose.
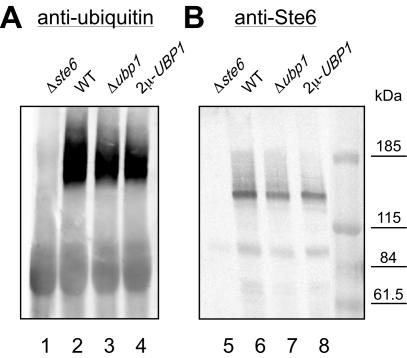
Ubiquitination of Ste6 is required for its rapid degradation in the vacuole (Kölling and Hollenberg, 1994). Obviously, UBP1 overexpression could stabilize Ste6 by reducing its ubiquitination. To test this assumption, we examined the effect of UBP1 deletion or overexpression on Ste6 ubiquitination. To detect ubiquitination, Ste6 was immunoprecipitated from cell extracts prepared from different strains expressing STE6 from a 2μ plasmid. In addition, the strains were either transformed with an empty vector or with a plasmid overexpressing UBP1. The immunoprecipitates were examined for the presence of Ste6 and ubiquitin by Western blotting (Figure 8). A ubiquitin signal can only be detected in the immunoprecipitates, if ubiquitin is covalently attached to Ste6. The ubiquitin signals were normalized to the amount of Ste6 present in the immunoprecipitates. A clear ubiquitin signal could be detected in the wild-type strain that was absent in a strain that did not express Ste6. But, neither UBP1 deletion (92% of WT) nor UBP1 overexpression (119% of WT) had a significant effect on the Ste6 ubiquitin signal. UBP1 overexpression, therefore, does not seem to exert its effect on Ste6 turnover through deubiquitination of Ste6.
Figure 8.
Effect of Ubp1 on Ste6 ubiquitination. Cell extracts were prepared from different strains transformed with a STE6 overexpressing plasmid (pRK590) or a vector control (YEplac112). In addition, the strains contained either an empty vector (YEplac195) or an UBP1-overexpressing plasmid (pRK805). Proteins immunoprecipitated with Ste6 antibodies were analyzed by Western blotting with anti-ubiquitin antibody (P4D1; Covance) (A) and with anti-Ste6 antibodies (B). Lanes 1 and 5, RKY959 (Δste6)/YEplac112/YEplac195; lanes 2 and 6, JD52 (WT)/pRK590/YEplac195; lanes 3 and 7, RKY1932 (Δubp1)/pRK590/YEplac195; and lanes 4 and 8, JD52/pRK590/pRK805.
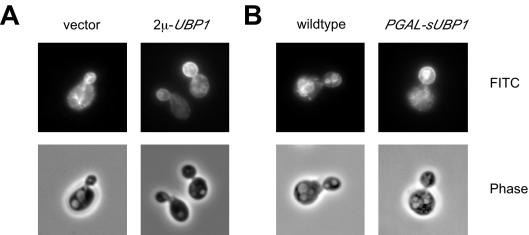
We were interested to know at which step UBP1 overexpression interferes with Ste6 trafficking to the vacuole. To gain information about the step at which Ste6 accumulates upon UBP1 overexpression, Ste6 localization was examined by immunofluorescence microscopy. Under wild-type conditions, internal dots or patches were observed that occasionally surrounded the vacuole (Figure 9A). These dots or patches presumably correspond to endosomal structures. On UBP1 overexpression, a striking, polar cell surface staining was observed. This suggests that UBP1 overexpression either interferes with internalization from the cell surface or leads to enhanced recycling to the cell surface from internal compartments. The same pattern was observed upon overexpression of the sUbp1 variant from the GAL1 promoter (Figure 9B).
Figure 9.
Effect of UBP1 overexpression on Ste6 localization. Ste6 localization was examined by immunofluorescence. (A) JD52 was transformed with the 2μ plasmid pYKS2 (Kuchler et al., 1993) expressing c-myc–tagged Ste6. In addition, the strain contained either an empty vector (YEplac195) or an UBP1-overexpressing plasmid (pRK879). (B) JD52 or RKY2008 (PGAL-sUBP1-13myc), transformed with the 2μ plasmid pRK736 expressing an HA-tagged Ste6 variant, were grown on galactose medium to induce expression of sUbp1. Ste6 was detected with anti-myc or anti-HA primary antibodies and FITC-conjugated anti-mouse secondary antibodies. Top, FITC-fluorescence; bottom, phase contrast images.
Ubp1 Is Phosphorylated
By phosphoproteome analysis, a phosphorylated peptide was detected through mass spectrometry that was assigned to Ubp1 (Ficarro et al., 2002). To confirm this finding, Ubp1 was directly examined for phosphorylation. To test for phosphorylation, 13myc-tagged Ubp1 was immunoprecipitated from cell extracts prepared from [32P]orthophosphate-labeled cells and examined by autoradiography. Two phosphorylated bands, which were absent in the negative control, with spacing and intensity characteristic for the two Ubp1 forms, could be detected on the autoradiogram (our unpublished data). Thus, both forms of Ubp1 seem to be phosphorylated. To explore the role of phosphorylation for Ubp1 activity, we wanted to eliminate Ubp1 phosphorylation by mutagenizing the two predicted phosphorylation acceptor sites (serine 530, serine 531). However, when we examined the S530, 531A mutant for phosphorylation, it turned out that phosphorylation was unaffected. Thus, either these two serine residues constitute only minor phosphorylation sites or the identified phosphopeptide was erroneously assigned to Ubp1.
DISCUSSION
Here, we present evidence for a role of the deubiquitinating enzyme Ubp1 in membrane protein trafficking in the endocytic pathway. We further show that the UBP1 gene codes for two variants, a longer membrane-anchored form (mUbp1) and a slightly smaller soluble form (sUbp1). Although not rigorously proven, our results strongly suggest that these two forms are independently expressed from the same gene. The two Ubp1 forms differ in size by ∼8 kDa. If we assume that translation of the two forms starts at the first and the second ATG codon in the UBP1 ORF, two proteins with exactly this size difference will be produced. This interpretation is supported by our mutagenesis experiment where we changed the second ATG codon into a serine codon. With this mutation, the short form of Ubp1 was no longer produced. However, we cannot exclude the formal possibility that a specific protease cleaves at the position of the second methionine and that the M67S mutation inactivates this cleavage site. But, we consider this interpretation unlikely. There is precedent for a similar arrangement in yeast. Two forms of invertase are produced from the SUC2 gene: a longer secreted form with a hydrophobic signal sequence and a shorter intracellular form without signal sequence (Carlson and Botstein, 1982). The two forms are expressed from two different promoters that are regulated differently. It remains to be shown whether the two Ubp1 forms also are subjected to different kinds of control. Alternatively, the two forms could be expressed from a single mRNA by a leaky scanning mechanism of the ribosome initiating translation at either the first or the second AUG codon. There also is precedent for such a mechanism (Kozak, 1999).
By several criteria, we have localized the mUbp1 variant to the ER membrane. In differential centrifugation experiments, mUbp1 was found exclusively in the P13 pellet fraction together with the ER marker Dpm1; on sucrose density gradients, its fractionation pattern was most compatible with the distribution of Dpm1; and in immunofluorescence experiments, mUbp1 showed a typical ER pattern with perinuclear staining and tubular structures. The unusually short putative transmembrane domain (TMD) of 15 amino acids also fits into this picture. According to the bilayer-mediated sorting model (Pelham and Munro, 1993), the length of the TMD, at least in part, mediates the localization of transmembrane proteins. As a general rule, the length of the TMDs increases from inside to outside, if one moves along the secretory pathway. The shortest TMDs are observed in ER proteins (∼16 aa) and the longest in plasma membrane proteins (∼25 aa) (Rayner and Pelham, 1997).
What could be the function of mUbp1 at the ER membrane? It has been suggested that the quite extensive family of deubiquitinating enzymes in yeast could simply provide a high but relatively nonspecific ubiquitin-hydrolyzing activity (Amerik et al., 2000). Their main function would be to recycle ubiquitin from substrates destined for degradation by the proteasome or the vacuole. Indeed, an important role in ubiquitin homeostasis has been demonstrated for two Ubps, Ubp4/Doa4 (Swaminathan et al., 1999) and Ubp6 (Chernova et al., 2003; Hanna et al., 2003). UBP4 and UBP6 mutants are sensitive against various stress conditions, show a lowered free ubiquitin level, and a reduced amount of large ubiquitin conjugates. Most of the phenotypes can be suppressed by overexpression of ubiquitin. All other UBP mutants, however, display only mild phenotypes or no obvious phenotype at all (Amerik et al., 2000). This makes it seem unlikely that Ubps other than Ubp4 and Ubp6 play a major role in ubiquitin homeostasis. With UBP1 mutants, we tested various growth conditions but could not detect any obvious phenotype (our unpublished data). Also, the pattern of ubiquitin conjugates from UBP1 mutants was indistinguishable from the wild-type pattern. We, therefore, consider it more likely that mUbp1 performs a very specific function at the ER membrane. Although, Ubp1 does not seem to play a general role in ERAD, it could regulate the turnover of individual ERAD substrates. An example for such a control is provided by von Hippel-Lindau protein-interacting deubiquitinating enzyme-1 (VDU1). VDU1 regulates the turnover of type 2 iodothyronine deiodinase (D2), an ER-localized integral membrane protein that is involved in the generation of biological active thyroid hormone (Curcio-Morelli et al., 2003). D2 has a short half-life due to ubiquitination and proteasomal degradation and is stabilized by VDU1 through deubiquitination.
The two Ubp1 forms seem to carry out distinct functions. Although no phenotype could be detected for overexpression of mUbp1, overexpression of sUbp1 strongly stabilized Ste6. Transport of the a-factor transporter Ste6 to the yeast vacuole for degradation is regulated by ubiquitination (Kölling and Hollenberg, 1994). In principle, a deubiquitinating enzyme such as Ubp1 could prevent degradation by removing the ubiquitin tag from Ste6. Is Ubp1 specifically involved in the regulation of Ste6 trafficking, or is Ste6 stabilization upon UBP1 overexpression simply the result of a general increase in deubiquitinating activity? Our findings argue for a specific role of Ubp1 in Ste6 trafficking. First, overexpression of UBP1 had no effect on the turnover of the ERAD substrate CPY*, which is dependent on ubiquitination and second, overexpression of another deubiquitinating enzyme, Ubp3, did not affect Ste6 turnover. Finally and most importantly, the Ste6 ubiquitination level was unaffected by UBP1 overexpression. This suggests that the Ubp1 target is a component of the machinery required for Ste6 trafficking to the vacuole. Which step in the trafficking pathway to the vacuole could be affected by Ubp1? Ubiquitination has been implicated in the internalization step of endocytosis at the plasma membrane (Hicke, 1999) and in sorting of membrane proteins into the MVB pathway (Katzmann et al., 2001; Losko et al., 2001; Reggiori and Pelham, 2001; Urbanowski and Piper, 2001). On UBP1 overexpression, Ste6 accumulates at the plasma membrane, which is consistent with a role of Ubp1 at the internalization step of endocytosis. A different localization would have been expected, if Ubp1 affected sorting into the MVB pathway (Losko et al., 2001). A block at this stage usually leads to an accumulation of the affected membrane proteins at the vacuolar membrane.
Another possibility, which is also compatible with the observed cell surface accumulation, is enhanced recycling of Ste6 from an internal compartment. On UBP1 overexpression, Ste6 accumulates at the cell surface in a polar manner, i.e., it is mainly localized to the surface of the newly emerging daughter cell, the bud. A similar distribution has been observed for the v-SNARE Snc1 (Lewis et al., 2000). For Snc1, it has been proposed that this asymmetric distribution is achieved through “kinetic polarization,” i.e., through endocytic recycling and localized exocytosis (Valdez-Taubas and Pelham, 2003). A similar mechanism may be responsible for the polar cell surface localization of Ste6 upon UBP1 overexpression. This notion is supported by our recent findings on the trafficking of ubiquitination-deficient Ste6 variants (our unpublished data).
What are the candidate proteins for Ubp1 deubiquitination? In mammalian cells, several proteins that are involved in clathrin-mediated endocytosis, such as epsin, Eps15, and Hrs, have been shown to be ubiquitinated (van Delft et al., 1997; Klapisz et al., 2002; Polo et al., 2002). These proteins carry ubiquitin-interacting motifs (Hofmann and Falquet, 2001) that also seem to be intimately linked with their ubiquitination. There is evidence that deubiquitination may be involved in the regulation of the early endocytic pathway. The deubiquitinating enzyme Faf of Drosophila specifically regulates the turnover of the epsin homologue Lqf by counteracting its ubiquitination (Chen et al., 2002), and another deubiquitinating enzyme, mouse UBPY, interacts with Hrs-binding protein, a protein that tightly associates with Hrs (Kato et al., 2000). In yeast, proteins corresponding to epsin (Ent1, Ent2), Eps15 (Ede1), and Hrs (Vps27) exist. However, so far no ubiquitination of these proteins has been reported. Still, they are prime candidates for Ubp1 targets. These targets remain to be identified.
Acknowledgments
We thank Thomas Sommer for the CPY* integration plasmid. We also are grateful to Karin Krapka for technical assistance and to Stefanie Huppert and Agnes Pawelec for help. This work was supported by Deutsche Forschungsgemeinschaft grant Ko 963/3-2 (to R. K.).
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E04-05-0425) on January 5, 2005.
References
- Amerik, A. Y., Li, S. J., and Hochstrasser, M. (2000). Analysis of the deubiquitinating enzymes of the yeast Saccharomyces cerevisiae. Biol. Chem. 381, 981-992. [DOI] [PubMed] [Google Scholar]
- Bagnat, M., Keranen, S., Shevchenko, A., and Simons, K. (2000). Lipid rafts function in biosynthetic delivery of proteins to the cell surface in yeast. Proc. Natl. Acad. Sci. USA 97, 3254-3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becherer, K. A., Rieder, S. E., Emr, S. D., and Jones, E. W. (1996). Novel syntaxin homologue, Pep12p, required for the sorting of lumenal hydrolases to the lysosome-like vacuole in yeast. Mol. Biol. Cell 7, 579-594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biederer, T., Volkwein, C., and Sommer, T. (1996). Degradation of subunits of the Sec61p complex, an integral component of the ER membrane, by the ubiquitin-proteasome pathway. EMBO J. 15, 2069-2076. [PMC free article] [PubMed] [Google Scholar]
- Carlson, M., and Botstein, D. (1982). Two differentially regulated mRNAs with different 5′ ends encode secreted with intracellular forms of yeast invertase. Cell 28, 145-154. [DOI] [PubMed] [Google Scholar]
- Chen, X., Zhang, B., and Fischer, J. A. (2002). A specific protein substrate for a deubiquitinating enzyme: liquid facets is the substrate of Fat facets. Genes Dev. 16, 289-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernova, T. A., Allen, K. D., Wesoloski, L. M., Shanks, J. R., Chernoff, Y. O., and Wilkinson, K. D. (2003). Pleiotropic effects of Ubp6 loss on drug sensitivities and yeast prion are due to depletion of the free ubiquitin pool. J. Biol. Chem. 278, 52102-52115. [DOI] [PubMed] [Google Scholar]
- Cohen, M., Stutz, F., Belgareh, N., Haguenauer-Tsapis, R., and Dargemont, C. (2003a). Ubp3 requires a cofactor, Bre5, to specifically de-ubiquitinate the COPII protein, Sec23. Nat. Cell Biol. 5, 661-667. [DOI] [PubMed] [Google Scholar]
- Cohen, M., Stutz, F., and Dargemont, C. (2003b). Deubiquitination, a new player in Golgi to endoplasmic reticulum retrograde transport. J. Biol. Chem. 278, 51989-51992. [DOI] [PubMed] [Google Scholar]
- Curcio-Morelli, C., Zavacki, A. M., Christofollete, M., Gereben, B., de Freitas, B. C., Harney, J. W., Li, Z., Wu, G., and Bianco, A. C. (2003). Deubiquitination of type 2 iodothyronine deiodinase by von Hippel-Lindau protein-interacting deubiquitinating enzymes regulates thyroid hormone activation. J. Clin. Investig. 112, 189-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ficarro, S. B., McCleland, M. L., Stukenberg, P. T., Burke, D. J., Ross, M. M., Shabanowitz, J., Hunt, D. F., and White, F. M. (2002). Phosphoproteome analysis by mass spectrometry and its application to Saccharomyces cerevisiae. Nat. Biotechnol. 20, 301-305. [DOI] [PubMed] [Google Scholar]
- Gietz, R. D., and Sugino, A. (1988). New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene 74, 527-534. [DOI] [PubMed] [Google Scholar]
- Gnesutta, N., Ceriani, M., Innocenti, M., Mauri, I., Zippel, R., Sturani, E., Borgonovo, B., Berruti, G., and Martegani, E. (2001). Cloning and characterization of mouse UBPy, a deubiquitinating enzyme that interacts with the Ras guanine nucleotide exchange factor CDC25(Mm)/Ras-GRF1. J. Biol. Chem. 276, 39448-39454. [DOI] [PubMed] [Google Scholar]
- Hampton, R. Y. (2002). ER-associated degradation in protein quality control and cellular regulation. Curr. Opin. Cell Biol. 14, 476-482. [DOI] [PubMed] [Google Scholar]
- Hanna, J., Leggett, D. S., and Finley, D. (2003). Ubiquitin depletion as a key mediator of toxicity by translational inhibitors. Mol. Cell. Biol. 23, 9251-9261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershko, A., and Ciechanover, A. (1998). The ubiquitin system. Annu. Rev. Biochem. 67, 425-479. [DOI] [PubMed] [Google Scholar]
- Hicke, L. (1999). Gettin'down with ubiquitin: turning off cell-surface receptors, transporters and channels. Trends Cell. Biol. 9, 107-112. [DOI] [PubMed] [Google Scholar]
- Hicke, L., and Riezman, H. (1996). Ubiquitination of a yeast plasma membrane receptor signals its ligand-stimulated endocytosis. Cell 84, 277-287. [DOI] [PubMed] [Google Scholar]
- Hicke, L., Zanolari, B., and Riezman, H. (1998). Cytoplasmic tail phosphorylation of the α-factor receptor is required for its ubiquitination and internalization. J. Cell Biol. 141, 349-358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiller, M. M., Finger, A., Schweiger, M., and Wolf, D. H. (1996). ER degradation of a misfolded luminal protein by the cytosolic ubiquitin-proteasome pathway. Science 273, 1725-1728. [DOI] [PubMed] [Google Scholar]
- Hochstrasser, M. (1996). Ubiquitin-dependent protein degradation. Annu. Rev. Genet. 30, 405-439. [DOI] [PubMed] [Google Scholar]
- Hochstrasser, M., Ellison, M. J., Chau, V., and Varshavsky, A. (1991). The short-lived MATα2 transcriptional regulator is ubiquitinated in vivo. Proc. Natl. Acad. Sci. USA 88, 4606-4610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann, K., and Falquet, L. (2001). A ubiquitin-interacting motif conserved in components of the proteasomal and lysosomal protein degradation systems. Trends Biochem. Sci. 26, 347-350. [DOI] [PubMed] [Google Scholar]
- Huh, W. K., Falvo, J. V., Gerke, L. C., Carroll, A. S., Howson, R. W., Weissman, J. S., and O'Shea, E. K. (2003). Global analysis of protein localization in budding yeast. Nature 425, 686-691. [DOI] [PubMed] [Google Scholar]
- Kato, M., Miyazawa, K., and Kitamura, N. (2000). A deubiquitinating enzyme UBPY interacts with the Src homology 3 domain of Hrs-binding protein via a novel binding motif PX(V/I)(D/N)RXXKP. J. Biol. Chem. 275, 37481-37487. [DOI] [PubMed] [Google Scholar]
- Katzmann, D. J., Babst, M., and Emr, S. D. (2001). Ubiquitin-dependent sorting into the multivesicular body pathway requires the function of a conserved endosomal protein sorting complex, ESCRT-I. Cell 106, 145-155. [DOI] [PubMed] [Google Scholar]
- Kinner, A., and Kölling, R. (2003). The yeast deubiquitinating enzyme Ubp16 is anchored to the outer mitochondrial membrane. FEBS Lett. 549, 135-140. [DOI] [PubMed] [Google Scholar]
- Klapisz, E., Sorokina, I., Lemeer, S., Pijnenburg, M., Verkleij, A. J., and van Bergen en Henegouwen, P. M. (2002). A ubiquitin-interacting motif (UIM) is essential for Eps15 and Eps15R ubiquitination. J. Biol. Chem. 277, 30746-30753. [DOI] [PubMed] [Google Scholar]
- Kölling, R., and Hollenberg, C. P. (1994). The ABC-transporter Ste6 accumulates in the plasma membrane in a ubiquitinated form in endocytosis mutants. EMBO J. 13, 3261-3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak, M. (1999). Initiation of translation in prokaryotes and eukaryotes. Gene 234, 187-208. [DOI] [PubMed] [Google Scholar]
- Kuchler, K., Dohlman, H. G., and Thorner, J. (1993). The a-factor transporter (STE6 gene product) and cell polarity in the yeast Saccharomyces cerevisiae. J. Cell Biol. 120, 1203-1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leroy, E., et al. (1998). The ubiquitin pathway in Parkinson's disease. Nature 395, 451-452. [DOI] [PubMed] [Google Scholar]
- Lewis, M. J., Nichols, B. J., Prescianotto-Baschong, C., Riezman, H., and Pelham, H. R. (2000). Specific retrieval of the exocytic SNARE Snc1p from early yeast endosomes. Mol. Biol. Cell 11, 23-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y., Fallon, L., Lashuel, H. A., Liu, Z., and Lansbury, P. T., Jr. (2002). The UCH-L1 gene encodes two opposing enzymatic activities that affect a-synuclein degradation and Parkinson's disease susceptibility. Cell 111, 209-218. [DOI] [PubMed] [Google Scholar]
- Longtine, M. S., McKenzie, A., 3rd, Demarini, D. J., Shah, N. G., Wach, A., Brachat, A., Philippsen, P., and Pringle, J. R. (1998). Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14, 953-961. [DOI] [PubMed] [Google Scholar]
- Losko, S., Kopp, F., Kranz, A., and Kölling, R. (2001). Uptake of the ATP-Binding Cassette (ABC) transporter Ste6 into the yeast vacuole is blocked in the doa4 mutant. Mol. Biol. Cell 12, 1047-1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moazed, D., and Johnson, D. (1996). A deubiquitinating enzyme interacts with SIR4 and regulates silencing in S. cerevisiae. Cell 86, 667-677. [DOI] [PubMed] [Google Scholar]
- Patnaik, A., Chau, V., and Wills, J. W. (2000). Ubiquitin is part of the retrovirus budding machinery. Proc. Natl. Acad. Sci. USA 97, 13069-13074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelham, H. R., and Munro, S. (1993). Sorting of membrane proteins in the secretory pathway. Cell 75, 603-605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polo, S., Sigismund, S., Faretta, M., Guidi, M., Capua, M. R., Bossi, G., Chen, H., De Camilli, P., and Di Fiore, P. P. (2002). A single motif responsible for ubiquitin recognition and monoubiquitination in endocytic proteins. Nature 416, 451-455. [DOI] [PubMed] [Google Scholar]
- Rayner, J. C., and Pelham, H. R. (1997). Transmembrane domain-dependent sorting of proteins to the ER and plasma membrane in yeast. EMBO J. 16, 1832-1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reggiori, F., and Pelham, H. R. (2001). Sorting of proteins into multivesicular bodies: ubiquitin-dependent and -independent targeting. EMBO J. 20, 5176-5186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth, A. F., Feng, Y., Chen, L., and Davis, N. G. (2002). The yeast DHHC cysteine-rich domain protein Akr1p is a palmitoyl transferase. J. Cell Biol. 159, 23-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert, U., Ott, D. E., Chertova, E. N., Welker, R., Tessmer, U., Princiotta, M. F., Bennink, J. R., Krausslich, H. G., and Yewdell, J. W. (2000). Proteasome inhibition interferes with gag polyprotein processing, release, and maturation of HIV-1 and HIV-2. Proc. Natl. Acad. Sci. USA 97, 13057-13062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strack, B., Calistri, A., Accola, M. A., Palu, G., and Gottlinger, H. G. (2000). A role for ubiquitin ligase recruitment in retrovirus release. Proc. Natl. Acad. Sci. USA 97, 13063-13068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaminathan, S., Amerik, A. Y., and Hochstrasser, M. (1999). The Doa4 deubiquitinating enzyme is required for ubiquitin homeostasis in yeast. Mol. Biol. Cell 10, 2583-2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbanowski, J. L., and Piper, R. C. (2001). Ubiquitin sorts proteins into the intralumenal degradative compartment of the late-endosome/vacuole. Traffic 2, 622-630. [DOI] [PubMed] [Google Scholar]
- Valdez-Taubas, J., and Pelham, H. R. (2003). Slow diffusion of proteins in the yeast plasma membrane allows polarity to be maintained by endocytic cycling. Curr. Biol. 13, 1636-1640. [DOI] [PubMed] [Google Scholar]
- van Delft, S., Govers, R., Strous, G. J., Verkleij, A. J., and van Bergen en Henegouwen, P. M. (1997). Epidermal growth factor induces ubiquitination of Eps15. J. Biol. Chem. 272, 14013-14016. [DOI] [PubMed] [Google Scholar]
- Verma, R., Aravind, L., Oania, R., McDonald, W. H., Yates, J. R., 3rd, Koonin, E. V., and Deshaies, R. J. (2002). Role of Rpn11 metalloprotease in deubiquitination and degradation by the 26S proteasome. Science 298, 611-615. [DOI] [PubMed] [Google Scholar]
- Wang, Y., and Dohlman, H. G. (2002). Pheromone-dependent ubiquitination of the mitogen-activated protein kinase kinase Ste7. J. Biol. Chem. 277, 15766-15772. [DOI] [PubMed] [Google Scholar]
- Wiertz, E. J., Tortorella, D., Bogyo, M., Yu, J., Mothes, W., Jones, T. R., Rapoport, T. A., and Ploegh, H. L. (1996). Sec61-mediated transfer of a membrane protein from the endoplasmic reticulum to the proteasome for destruction. Nature 384, 432-438. [DOI] [PubMed] [Google Scholar]
- Yao, T., and Cohen, R. E. (2002). A cryptic protease couples deubiquitination and degradation by the proteasome. Nature 419, 403-407. [DOI] [PubMed] [Google Scholar]