Abstract
Intraflagellar transport (IFT) is a bidirectional process required for assembly and maintenance of cilia and flagella. Kinesin-2 is the anterograde IFT motor, and Dhc1b/Dhc2 drives retrograde IFT. To understand how either motor interacts with the IFT particle or how their activities might be coordinated, we characterized a ts mutation in the Chlamydomonas gene encoding KAP, the nonmotor subunit of Kinesin-2. The fla3-1 mutation is an amino acid substitution in a conserved C-terminal domain. fla3-1 strains assemble flagella at 21°C, but cannot maintain them at 33°C. Although the Kinesin-2 complex is present at both 21 and 33°C, the fla3-1 Kinesin-2 complex is not efficiently targeted to or retained in the basal body region or flagella. Video-enhanced DIC microscopy of fla3-1 cells shows that the frequency of anterograde IFT particles is significantly reduced. Anterograde particles move at near wild-type velocities, but appear larger and pause more frequently in fla3-1. Transformation with an epitope-tagged KAP gene rescues all of the fla3-1 defects and results in preferential incorporation of tagged KAP complexes into flagella. KAP is therefore required for the localization of Kinesin-2 at the site of flagellar assembly and the efficient transport of anterograde IFT particles within flagella.
INTRODUCTION
Cilia and flagella perform essential motile and sensory functions for a variety of eukaryotic organisms. In vertebrates, defects in ciliary and flagellar motility (primary ciliary dyskinesia or PCD) result in randomization of left/right body asymmetry during embryonic development, chronic respiratory disease, and male sterility (Ibañez-Tallon et al., 2003). Defects in the assembly of primary cilia or cilia-associated proteins have also been linked to polycystic kidney disease (PKD), retinal degeneration, hearing loss, and human obesity disorders (Snell et al., 2004). Studies in Chlamydomonas, Caenorhabditis elegans, and mice have demonstrated that the machinery required for ciliary assembly is highly conserved (Rosenbaum and Witman, 2002). In most cases, the assembly and maintenance of these organelles depends on a bidirectional, intraflagellar transport (IFT) system of large protein particles driven by two distinct microtubule motors, a heterotrimeric Kinesin-2 and a novel cytoplasmic dynein (cDhc1b or Dhc2; Cole, 2003). Formerly known as kinesin-II (Lawrence et al., 2004), Kinesin-2 is responsible for transport from the basal body region to the tip of the axoneme (anterograde IFT; Kozminski et al., 1995; Piperno and Mead, 1997; Cole et al., 1998), whereas the cDhc1b/LIC complex is required for transport from the flagellar tip to the basal body region (retrograde IFT; Pazour et al., 1999; Porter et al., 1999; Signor et al., 1999a; Wicks et al., 2000; Perrone et al., 2003; Schafer et al., 2003).
The Kinesin-2 complex consists of two distinct kinesin-related motor subunits and a third kinesin-associated protein (KAP). This complex was first described in sea urchin eggs and subsequently identified in the mouse, Chlamydomonas, C. elegans, Drosophila, Xenopus, and Tetrahymena (Cole et al., 1993, 1998; Yamazaki et al., 1995, 1996; Tuma et al., 1998; Brown et al., 1999; Ray et al., 1999; Signor et al., 1999b). A role for Kinesin-2 in flagellar assembly was first indicated by the study of temperature sensitive, flagellar assembly (fla) mutations in Chlamydomonas and the identification of fla10 as defect in the motor domain of one of the kinesin-related subunits (Walther et al., 1994). Inactivation of the FLA10 kinesin disrupted IFT in Chlamydomonas (Kozminski et al., 1995), and microinjection of antibodies against a Kinesin-2 subunit blocked ciliary assembly in sea urchin embryos (Morris and Scholey, 1997). Mutations in Kinesin-2–related sequences also inhibit the assembly of motile cilia in Tetrahymena (Brown et al., 1999) and sensory cilia in C. elegans (Shakir et al., 1993), and Drosophila (Sarpal et al., 2004).
Defects in the Kinesin-2 complex and ciliary assembly have profound, pleiotropic effects in mammals. Knockouts of the motor subunits in mice (KIF3A or KIF3B) disrupt the assembly of cilia in the embryonic node, leading to randomization of the left-right body-axis, and other embryological abnormalities (Nonaka et al., 1998; Marszalek et al., 1999; Takeda et al., 1999). The neurological defects appear to be due in part to defects in sonic hedgehog signaling (Huangfu et al., 2003). Tissue-specific knockouts have further demonstrated a requirement for the Kinesin-2 complex in the assembly of primary cilia in the eye and kidney; such mutations ultimately led to photoreceptor degeneration and polycystic kidney disease by complex mechanisms (Marsalek et al., 2000; Lin et al., 2003).
Kinesin-2 is also involved in other forms of microtubule-based transport. In Drosophila, the complex is required for neuronal transport of choline acetyltransferase (Ray et al., 1999) and the transport or localization of components required for the signaling of cell fates during oogenesis (Pflanz et al., 2004), in addition to its role in the maintenance of ciliated sensory neurons (Sarpal et al., 2004). Dominant negative studies in Xenopus have indicated that Kinesin-2 is involved in the dispersion of pigment granules (Tuma et al., 1998), ER-to-Golgi transport (Le Bot et al., 1998), and most recently Vg1 mRNA localization (Betley et al., 2004). Finally, Kinesin-2 may also be involved in the establishment of neuronal polarity via association with the PAR-3 complex (Nishimura et al., 2004).
Despite all of the evidence indicating that Kinesin-2 plays a critical role in microtubule-based transport, the mechanisms that target this complex to a discrete subcellular location or regulate its association with specific cargo(es) are poorly understood. Considerable speculation has been focused on the possible function(s) of the highly conserved KAP subunit, which forms a large globular domain at the base of the Kinesin-2 complex (Wedaman et al., 1996; Yamazaki et al., 1996). The KAP subunit contains 10 armadillo domains, which are degenerate, 42 amino acid repeats that form superhelical structures thought to mediate protein-protein interactions (Gindhart and Goldstein, 1996). Yeast two-hybrid assays have identified the KAP subunit as a potential binding partner of a number of different molecules in vitro, but the functional significance of many of these interactions remains unclear (Shimizu et al., 1996, 1998; Nagata et al., 1998; Takeda et al., 2000; Jimbo et al., 2002).
As an alternative strategy to study the function of the KAP subunit in the Kinesin-2 complex, we have initiated a genetic analysis of KAP in Chlamydomonas. We have found that KAP is the gene product of the FLA3 locus and that the fla3-1 mutation results in an amino acid substitution within a conserved domain in the C-terminal region of KAP. This mutation disrupts both the localization of the Kinesin-2 complex at the site of flagellar assembly in the basal body region and the assembly and transport of anterograde IFT particles within the flagella. The fla3-1 mutant phenotype indicates that the C-terminal domain of the KAP subunit is critical for the interaction of the Kinesin-2 complex with other IFT components.
MATERIALS AND METHODS
Cell Culture and Mutant Strains
All strains were maintained as described in Perrone et al. (2000). The fla3 (CC1391), fla4 (CC1392), and fla10-1 (CC1919) strains were obtained from the Chlamydomonas Genetics Center (Duke University, Durham, NC). fla4 strain was crossed to an arg7 strain for use in cotransformation experiments. The fla3 strain from the stock center did not show a clear flagellar assembly defect at the restrictive temperature. CC1391 was therefore backcrossed to a wild-type strain (21gr) to remove potential modifiers or suppressors of the fla phenotype. The resulting tetrad progeny were scored after growth overnight at 33°C in liquid media, and several progeny with a clear fla phenotype were recovered. One of these progeny (1B) was designated as the representative allele of the fla3-1 mutation.
Cloning and Characterization of the KAP Gene
A cDNA clone (CL70g01, accession number AV396858) encoding the Chlamydomonas KAP sequence was obtained from the Kazusa DNA Research Institute (Chiba, Japan). A 3.4-kb insert was released by EcoRI/XhoI digestion and used to obtain six BAC clones (21k7, 10b10, 7j10, 35k13, 19m19, 2p23) containing the KAP gene. The complete transcription unit was identified within an 8.6-kb BamHI fragment from BAC clone 10b10 and sequenced by primer walking. The predicted amino acid sequence is similar but not identical to the gene model currently in the Chlamydomonas genome database (version 2, model 620048).
Epitope-tagged versions of KAP were generated by the insertion of either a triple HA tag or a GFP tag into a BbvcI site located near the 3′ end of the KAP gene. The triple HA tag was PCR amplified from the p3HA plasmid provided by M. Lavoie and C. Silflow (University of Minnesota, St. Paul, MN) using primers with BbvcI restriction sites. The GFP tag was PCR amplified from the pHisCrGFP plasmid (Fuhrmann et al., 1999). Sequence analysis confirmed that both epitope tags were inserted into the last exon between amino acid residues 839 and 840 in the proper orientation and reading frame. Both constructs were made using the genomic clone to ensure that the resulting tagged sequences would be expressed at approximately wild-type levels.
DNA and RNA isolation, agarose gels, Southern blots, and Northern blots were performed as previously described (Perrone et al., 2000). To place KAP on the genetic map, the cDNA clone was used to identify an EcoRI/XhoI restriction fragment length polymorphism (RFLP) between two Chlamydomonas strains, 137c and S1-D2. The KAP cDNA was then hybridized to a series of mapping filters containing EcoRI/XhoI-digested genomic DNA isolated from the tetrad progeny of crosses between multiply marked Chlamydomonas strains and S1-D2. The segregation of the KAP RFLP was analyzed relative to the segregation of >50 genetic and molecular markers as described in Porter et al. (1996). The resulting parental ditype:nonparental ditype:tetratype ratios indicated linkage to the genetic marker nic13 (9:0:6) and the molecular marker Tcr1-A (22:0:4). Centromere distance was estimated based on segregation with respect to the genetic markers ac17 and y1 (25:25:16).
The KAP genes present in fla3 and fla4 were recovered by PCR with gene-specific primers and sequenced directly. A single base pair mutation was observed in fla3. A 790-base pair region covering the site of the mutation was also PCR amplified from nine randomly chosen tetrad progeny. The same mutation was observed in four progeny exhibiting the fla phenotype, but no mutation was observed in five progeny with a wild-type phenotype.
Transformation of fla Mutants with Wild-type and Epitope-tagged KAP Genes
To test whether any of the clones could rescue the flagellar assembly defects in fla3 or fla4, each strain was cotransformed with a selectable marker and the clone in question. The KAP clones were linearized with SspI. The fla4 arg7 strain was cotransformed with the KAP clones and pARG7.8 (containing a wild-type copy of the ARG7 gene; Debuchy et al., 1989). Transformants were selected on solid media lacking arginine. The fla3 progeny were cotransformed with the KAP clones and EcoRI digested pSI103 (containing the aphVIII gene from Streptomyces rimosus; Sizova et al., 2001). Transformants were selected on solid media containing 10 μg/ml paromomycin (P-8692, Sigma Chemical Co., St. Louis, MO). Individual colonies were resuspended in liquid medium and then screened for rescue of the fla phenotype after overnight incubation at 33°C.
Protein Isolation, SDS-PAGE, and Western Blotting
Small- and large-scale culture of vegetative cells, the isolation and extraction of flagella, and sucrose density gradient centrifugation of flagellar extracts were performed as described in Perrone et al. (2000, 2003). Polypeptides were separated by SDS-PAGE on 5–15% polyacrylamide, 0.25 M glycerol gradient gels and blotted to polyvinylidene fluoride. Western blots were probed as described by Perrone et al. (1998, 2000) using either rat polyclonal antibodies to the HA epitope (clone 3F10, Roche Molecular Biochemicals, Indianapolis, IN), rabbit polyclonal antibodies to the FLA10 motor subunit of the Kinesin-2 complex (Cole et al., 1998), or mouse monoclonal antibodies to alpha tubulin (Sigma T-5168). The rat polyclonal antisera to the Chlamydomonas KAP subunit was generated against purified soluble fusion protein that was made by ligating 369 base pairs encoding the NH2-terminal 123 amino acids of KAP into the pTrcHis vector (Invitrogen, Carlsbad, CA). To compare the relative amounts of KAP and FLA10 in whole cells of different strains, equivalent numbers of total cells were loaded (between 1 and 5 × 106 cells per lane). Blots were also stained with a tubulin antibody to control for variations in protein loading. Multiple exposures were scanned and analyzed using NIH Image to compare the relative intensity of different samples (normalized to tubulin) to ensure that signals were within the linear range.
Immunofluorescence Light Microscopy and Live Cell Imaging
Wild-type and mutant strains were prepared for immunofluorescence light microscopy using ice-cold methanol fixation (Sanders and Salisbury, 1995) and stained with the primary antibodies described above and Alexafluor-488–conjugated secondary antibodies (Molecular Probes, Eugene, OR) as described in Perrone et al. (2003). Additional primary antibodies included an affinity-purified rabbit polyclonal antibody against the dynein LIC (Perrone et al., 2003) and a mouse monoclonal antibody against the IFT particle subunit p139 (Cole et al., 1998).
For analysis of IFT at 21°C, wild-type, fla3, and rescued strains were immobilized in 0.75% low-melting point agarose to inhibit flagellar motility and then imaged by video-enhanced DIC microscopy using a 100×, 1.3 NA Plan Neofluor lens, a 4× TV tube, and a C2400 Newvicon camera (Hamamatsu, Bridgewater, NJ). Video frames (30/s) were converted to digital images, and digital kymograms were created with MetaMorph software (Universal Imaging, Downingtown, PA) using methods similar to those described in Iomini et al. (2001). Light intensity profiles along a length of flagellum and across a width of 12 pixels were obtained from sequential video images and used to generate the kymograms. To generate kymograms that accurately measure flagellar lengths, all images were rotated so that flagella were aligned along the horizontal axis. This was required because the kymogram program estimates distance using pixel width but does not account for variations in sample orientation. The movements of individual IFT particles along the length of the flagellum versus time are represented as diagonal tracks on the resulting kymograms. The velocities of IFT particles were determined by measuring the slope of the tracks. Particle frequencies were determined by counting the total number of anterograde and retrograde particles in a randomly chosen 10-s interval for each cell. Because particle frequencies can be highly variable from cell to cell, we also determined the anterograde-to-retrograde frequency ratio for each cell; the frequency ratio is much less variable (Iomini et al., 2001). Particle pausing was estimated by counting the total number of anterograde particles present in a randomly chosen 3-μm section of the flagellum within a 10-s interval and then noting how many of these particles were observed to stop moving within the same time frame. The data are presented as mean ± SD (sample number). Data were compared using the Student's unpaired t test and Excel software. Samples were judged to be significantly different for values of p < 0.001.
The movement of the GFP-tagged KAP subunit was also visualized using confocal fluorescence microscopy. Time-lapse images were acquired with a Nikon TE200 inverted scope (Garden City, NY), a 100× 1.4 NA plan apo lens, a Yokogawa confocal scanner (CSU10; Shenandoah, GA) at a wavelength of 488 nm, and a Hamamatsu Orca-ER camera using an exposure time of 0.5 s.
Flagellar Length Measurements
To ensure maximal flagellar assembly, wild-type and mutant strains were grown in liquid TAP medium with aeration for 2–4 d before measurements of flagellar length. Cells were either maintained at 21°C or shifted to 33°C, and aliquots of cells were removed at 60-min time points and fixed with 1% glutaraldehyde. Cells were imaged using phase contrast microscopy with a 40× lens, 4× TV tube, and C2400 Newvicon (Hamamatsu) camera. The lengths of individual flagella were measured using the Metamorph software package (Universal Imaging). In some experiments, the vegetatively growing cells were collected by centrifugation and transferred to M-N/5 medium for 4–24 h to generate gametic cells. Data sets were compared using the Student's t test as described above.
RESULTS
Cloning and Characterization of the Chlamydomonas KAP Gene
To identify the gene encoding the Chlamydomonas homologue of KAP, we screened the EST databases with the sea urchin sequence (U38655) and identified a cDNA clone with high sequence homology to sea urchin KAP. An ∼3.4-kb insert was isolated from this clone to use as a hybridization probe. Southern blot analysis of wild-type and S1D2 genomic DNA demonstrated that the KAP sequence is a single copy gene (Figure 1A). Northern blot analysis of wild-type RNA isolated before and after deflagellation also revealed that the KAP sequence hybridized to an ∼3.4-kb transcript that is up-regulated in response to deflagellation, as expected for a gene involved in flagellar assembly or motility (Figure 1B).
Figure 1.
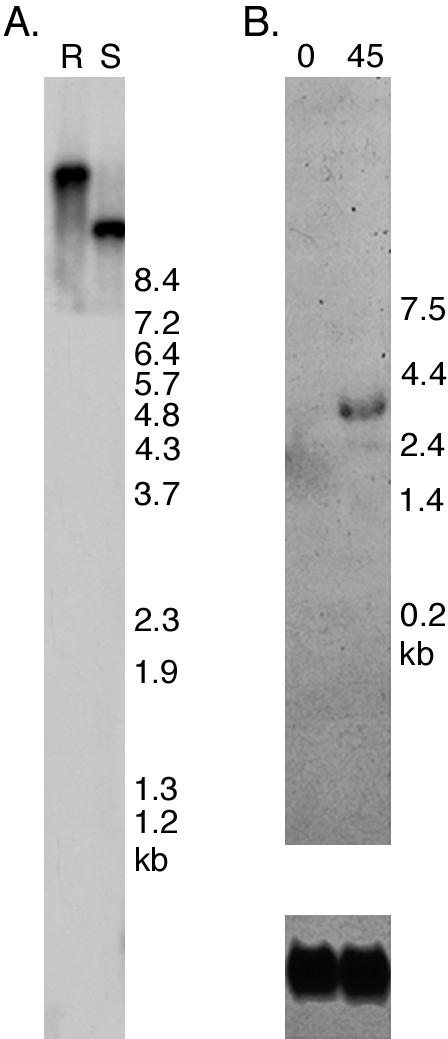
Expression of the Chlamydomonas KAP gene in response to deflagellation. (A) A Southern blot of wild-type (R) and S1D2 (S) genomic DNA was hybridized with the ∼3.4-kb insert from the KAP cDNA clone CL70g01. A single EcoRI/XhoI restriction fragment was seen in wild-type, and a polymorphic fragment was seen in S1D2. (B) A Northern blot of total RNA isolated from wild-type cells before (0) and 45 min after deflagellation (45) was hybridized with the KAP cDNA (top panel). Expression of the ∼3.4-kb KAP transcript was enhanced by deflagellation, as compared with a probe (Cry1) for a ribosomal protein subunit used as a loading control (bottom panel).
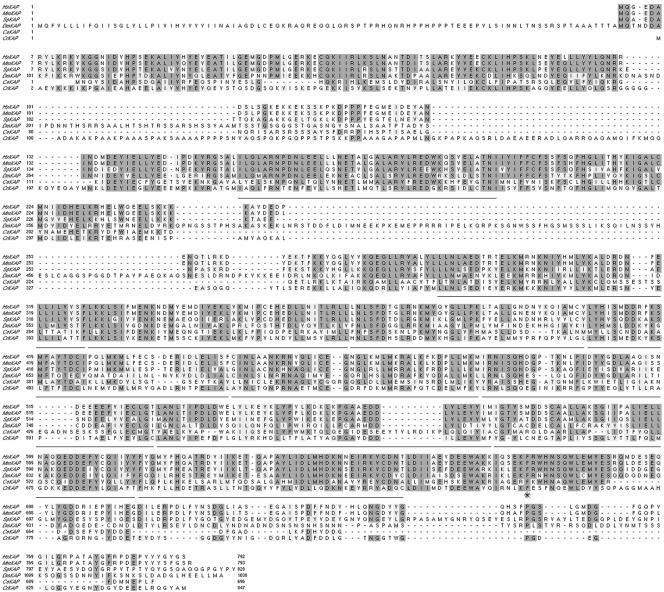
Sequence analysis of the ∼3.4-kb insert indicated the presence of a full-length cDNA encoding a polypeptide of 847 amino acid residues with a predicted molecular weight of ∼95 kDa and a pI of ∼5.21 (GenBank accession number AY739907). Sequence alignment programs indicate that CrKAP is closely related (e.g., ∼44% identity and ∼64% similarity) to KAP sequences identified in other organisms over most of its length (Figure 2). The regions of homology include the 10 armadillo repeats first described in the sea urchin sequence (Gindhart and Goldstein, 1996).
Figure 2.
Chlamydomonas KAP is similar to other members of the KAP family. A Clustal W alignment of the predicted amino acid sequence of the Chlamydomonas KAP polypeptide (CrKAP) with orthologues from humans (Hs), mouse (Mm), sea urchin (Sp), flies (Dm), and worms (Ce) is shown here. The regions containing the conserved armadillo repeats (residues 229–270, 274–314, 360–400, 401–441, 442–480, 481–523, 525–561, 562–604, 605–648, 649–690, 691–733) are underlined. The conserved phenylalanine residue that is mutated in fla3 is indicated by an asterisk.
Two nonconserved regions were noted in the Chlamydomonas KAP sequence. The first is basic domain located between amino acids 95–200 that is also alanine and proline rich. This region is present in other Chlamydomonas KAP ESTs, the genomic clone, and related RT-PCR products, but its function is unknown. The second nonconserved region corresponds to the C-terminal 83 amino acids, which display only limited similarity with other KAP sequences. The function of this region is unknown, but it has been useful site for the introduction of epitope tags to follow the expression of the KAP polypeptide (see below).
Identification of a Mutation in the KAP Gene
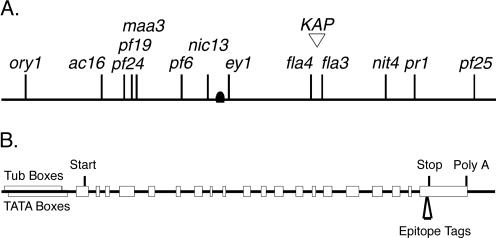
To identify candidate mutations in the KAP gene, we used RFLP mapping procedures to place the sequence on the genetic map of Chlamydomonas. The EcoRI/XhoI RFLP shown in Figure 1A was used as a molecular marker to follow the cosegregation of the KAP gene with respect to a series of molecular and genetic markers. The KAP gene was mapped to the right arm of linkage group X, based on linkage to the genetic marker nic13 and the molecular marker Tcr1-A (see Materials and Methods and Figure 3A). The KAP cDNA was used to screen a Chlamydomonas BAC library and recover six clones containing the KAP transcription unit. Two of these BACs (21k7 and 2p23) also hybridized with the molecular marker GP145, which has recently been mapped to linkage group X (Kathir et al., 2003). The mapping data place the KAP gene in the vicinity of two temperature-sensitive mutations, fla3 and fla4, representing two distinct loci involved in flagellar assembly (Adams et al., 1982).
Figure 3.
Linkage of the KAP gene to flagellar assembly mutations and diagram of constructs used for rescue by transformation. (A) RFLP mapping procedures were used to place the KAP gene on the right arm linkage group X, based on linkage to the genetic marker nic13 (∼20 cM) and the molecular marker Tcr1-A (∼7.7 cM), ∼12.5 cM from the centromere (see Materials and Methods for details). These data placed the KAP gene in the vicinity of two flagellar assembly mutations, fla3 and fla4 (Adams et al., 1982). (B) Schematic diagram of the intron-exon structure of the KAP transcription unit used in transformation experiments. The KAP gene is contained within an ∼8.6-kb BamHI fragment. The open boxes represent exons and the solid lines represent introns. The positions of predicted Tub boxes, TATA boxes, translation start and stop sites, and the site of polyadenylation (Poly A) are also indicated. The site of insertion of either the HA or GFP epitope tag into the last exon is also shown here.
To determine if either fla3 or fla4 might be a mutation in the KAP gene, we subcloned and sequenced the full-length genomic clone encoding the wild-type gene from BAC 10b10 (Figure 3B, GenBank accession number AY739906) and then used the genomic clone in cotransformation experiments with the fla3 and fla4 mutant strains. Both fla3 and fla4 assemble flagella at the permissive temperature of 21°C, but lose their flagella at the restrictive temperature of 33°C (Adams et al., 1982). Transformants were therefore screened for the presence of flagella after overnight incubation at the restrictive temperature. No rescue of the flagellar assembly defect was observed with any of the fla4 transformants (n = 264). Transformation of fla3 cells with the wild-type KAP gene resulted in the rescue of the flagellar assembly defects at the restrictive temperature in ∼10% (24/282) of the transformants, a value typical for cotransformation frequencies.
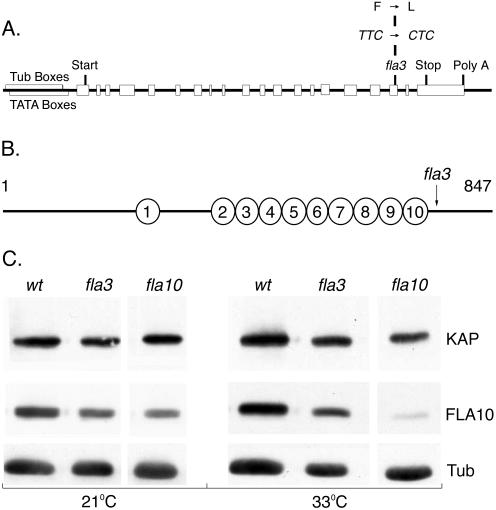
To confirm the transformation results and identify the site of the mutation in fla3, we used PCR to recover the KAP gene from both fla3 and fla4 genomic DNA. Sequence analysis indicated the presence of a single base pair change in the fla3 KAP gene but no changes in fla4. The mutation cosegregated with the temperature-sensitive, flagellar assembly phenotype in progeny of a backcross to a wild-type strain (see Materials and Methods). In each case, nucleotide 6853 was changed from T to C, resulting in the change of a phenylalanine to a leucine at amino acid residue 753 (Figure 4A). This phenylalanine residue is invariant in all KAP sequences described thus far, but its function is unknown. However, the fla3 mutation does alter a conserved domain within the C-terminal region, just after the last armadillo repeat (Figures 2 and 4B). This region has been predicted to be a potential cargo-binding site (Deacon et al., 2003).
Figure 4.
Identification of a KAP mutation in fla3-1. (A) Sequence analysis of the KAP gene in fla3-1 revealed a point mutation at nucleotide 6853 in exon 17 that changes amino acid residue 753 from a phenylalanine to a leucine. (B) Diagram of the KAP polypeptide showing the position of fla3-1 mutation relative to the armadillo repeats. (C) The fla3-1 mutation does not alter the level of the KAP subunit in whole cells. Western blots of wild-type, fla3, and fla10 cells grown at 21°C or overnight at 33°C were probed with antibodies against KAP, FLA10, and tubulin. Each lane was loaded with extracts prepared from ∼5 × 106 cells. The relative intensities of the KAP and FLA10 signals were normalized to the tubulin signals to control for small variations in protein loading. No significant changes in the levels of the KAP subunit were observed in the fla3-1 mutant at either the permissive or restrictive temperature.
The Effects of the fla3 Mutation on the Stability and Localization of the Kinesin-2 Complex
To assess whether the KAP mutation might affect the stability of the Kinesin-2 complex in fla3 cells, we analyzed whole-cell extracts on Western blots. Previous studies of the temperature-sensitive fla10-1 mutation have indicated that the FLA10 motor subunit of the Kinesin-2 complex is less stable in fla10-1 cells (Walther et al., 1994; Kozminski et al., 1995; Cole et al., 1998). Wild-type and mutant strains were maintained at 21°C or grown overnight at 33°C, and then whole-cell extracts were analyzed on Western blots probed with antibodies to FLA10, KAP, and tubulin. No significant changes in the levels of the KAP subunit or the FLA10 subunit were observed in fla3 cells at either the permissive or restrictive temperature (Figure 4C). In contrast, an obvious decrease in the FLA10 subunit was seen in fla10-1 cells at 33°C, consistent with earlier reports (Walther et al., 1994; Kozminski et al., 1995; Cole et al., 1998). Thus the fla3 mutation differs from the fla10-1 mutation in that it does not have a dramatic effect on the stability of its mutant gene product. In addition, fla3 does not appear to have a significant effect on the stability of FLA10.
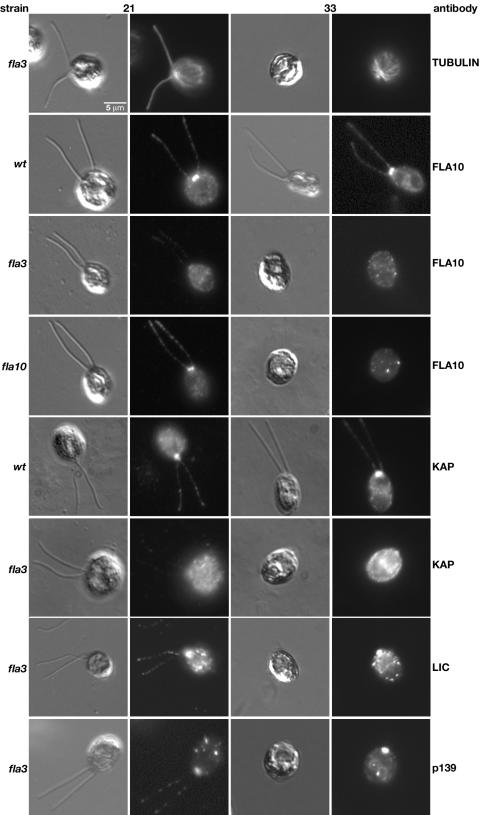
To determine if the fla3 mutation alters the subcellular localization of the Kinesin-2 motor, we analyzed the distribution of the Kinesin-2 complex and other IFT components in fixed wild-type and mutant cells by immunofluorescence microscopy. Staining of fla3 cells with an antibody against α-tubulin demonstrates the clear presence of the two flagella at 21°C and their complete absence at 33°C (Figure 5). Moreover, the cytoplasmic microtubule array appears to be relatively normal at both temperatures, similar to that reported previously for FLA10 null strains (Matsuura et al., 2002). Staining of wild-type cells with a FLA10 antibody confirms that the FLA10 subunit is concentrated in the basal body region, at the anterior end of the cell, and also present in punctate spots along the length of the two flagella (Figure 5), as reported previously (Vashishtha et al., 1996; Cole et al., 1998). However, analysis of fla3 cells grown at 21°C revealed that FLA10 staining of both the basal body region and the two flagella was significantly reduced (Figure 5). Interestingly, the defect in the localization of FLA10 subunit at the permissive temperature was more severe in fla3 cells than that observed in fla10-1 cells (Figure 5). Staining with an antibody to KAP confirmed that the KAP subunit is concentrated in the basal body region and flagella of wild-type cells, but dispersed in fla3 cells (Figure 5). The simplest interpretation is that the fla3-1 mutation is altering either the targeting or retention of the Kinesin-2 complex at the site of flagellar assembly.
Figure 5.
The fla3 mutation disrupts the localization of the Kinesin-2 complex in the basal body region. Wild-type and mutant strains were fixed at both 21 and 33°C, stained with a specific antibody (indicated on the right), and then imaged using both DIC and fluorescence microscopy. The top row shows fla3 cells stained with an antibody to tubulin. The cytoplasmic microtubule array is wild-type, but flagellar microtubules are absent at the restrictive temperature. The second row shows wild-type cells stained with an antibody to the FLA10 motor subunit, which concentrates in the basal body region and in a punctate pattern along the length of the two flagella. The third row shows fla3 cells stained with the FLA10 antibody; note how FLA10 staining is dispersed at both temperatures. The fourth row shows fla10-1 cells strained with the FLA10 antibody. Although FLA10 is reduced in the fla10-1 mutant, basal body localization can still be observed at 21°C. The fifth row shows wild-type cells stained with an antibody to KAP; note that KAP is concentrated in the basal body region and in a punctate pattern in the flagella, similar to FLA10 above. The sixth row shows fla3 cells stained with the KAP antibody; note how KAP staining is dispersed in the mutant strain. The seventh and eighth rows are fla3 cells stained with antibodies against other IFT components, the dynein LIC and the IFT particle subunit p139. The basal body and flagellar localization of these components appears to be relatively wild-type at 21°C. These components remain concentrated in the basal body region of the aflagellate cells at 33°C.
At 21°C, the fla3 defect appears to be limited to the aberrant localization of the Kinesin-2 complex. The staining seen with antibodies to other IFT components, such as the retrograde IFT motor or IFT particles, is similar to that observed in wild-type cells (Figure 5 and Perrone et al., 2002). For example, both the dynein LIC and the IFT particle subunit p139 are found in the basal body region and in punctate spots along the length of the flagella. Western blots of isolated flagella also showed that other IFT components are present at approximately wild-type levels (Mueller and Porter, unpublished results). Thus low levels of a partially functional Kinesin-2 complex can apparently maintain flagellar assembly at 21°C. Consistent with this hypothesis, measurements of steady state flagellar lengths under optimal growth conditions (see Materials and Methods) indicate that fla3 flagella are only slightly shorter than wild-type flagella (Table 1). However, under certain experimental conditions, such as flagellar regeneration or maintenance at higher temperature, the effectiveness of the Kinesin-2 complex to promote flagellar assembly is clearly compromised in fla3 cells. Previous studies have indicated that wild-type cells grown at 33°C require higher levels of Kinesin-2 activity for flagellar maintenance (Kozminski et al., 1995). When fla3 cells are grown at 33°C, this requirement appears to exceed the levels of functional motor complex available, and flagellar assembly is inhibited. Staining with the FLA10 and KAP antibodies confirms that localization of the Kinesin-2 complex to the basal body region is completely disrupted (Figure 5). In contrast, the dynein LIC and IFT subunit p139 are still present in the anterior region of the cell, even though there is no flagellar assembly at the restrictive temperature (Figure 5).
Table 1.
Effects of the fla3 mutation on flagellar length
| Strain | Average length (μm) | Average length excluding zero length flagella (μm) | % Aflagellate cells |
|---|---|---|---|
| Vegetative cells | |||
| Wild-type (137c) | 11.1 ± 3.5 (105) | 11.6 ± 2.6 (100) | 4.8 |
| fla3 | 8.0 ± 4.2 (107) | 8.9 ± 3.5 (97) | 9.3 |
| fla3::KAP-GFP | 10.7 ± 3.1 (102) | 11.1 ± 2.3 (98) | 3.9 |
| Gametic cells | |||
| Wild-type (137c) | 12.6 ± 3.8 (107) | 13.2 ± 2.5 (102) | 4.7 |
| fla3 | 9.4 ± 3.7 (85) | 10.6 ± 1.3 (75) | 11.8 |
Flagellar length measurements are expressed as mean ± SD (sample size). Cells were grown with aeration in TAP medium (vegetative growth) or transferred to M-N/5 for 4 h to induce gametogenesis. All measurements were made at 21°C. The mean flagellar lengths of fla3 cells are significantly shorter (p < 0.001) than those of wild-type cells for both vegetative and gametic cells. Rescue of fla3 cells with the epitope-tagged KAP restores flagellar lengths to wild-type levels.
Rescue of the fla3 Defects by Transformation with Epitope-tagged KAP Genes
To follow the wild-type KAP sequence in fla3 rescued cells, we tested two epitope-tagged versions of the wild-type KAP gene for their ability to incorporate into the Kinesin-2 complex, restore localization of Kinesin-2 to the basal body region, and thereby rescue the flagellar assembly defects. The epitope tags were inserted into the nonhomologous C-terminal region of the polypeptide, between amino acid residues 839 and 840, followed by the last seven amino acid residues. The fla3 mutant was transformed with a selectable marker and genomic clones encoding either the HA-tagged KAP or the GFP-tagged KAP subunit. Transformants were screened for rescue of the flagellar assembly defects at 33°C. Both constructs rescued the flagellar assembly defect at frequencies similar to that observed with the untagged KAP gene (i.e., 17 rescues out of 424 transformants with the HA-tagged KAP and 10 rescues out of 480 transformants with the GFP-tagged KAP). Measurements of flagellar lengths at 21°C also demonstrated the flagella of the rescued strains are similar in length to wild-type and longer than those of fla3 (Table 1).
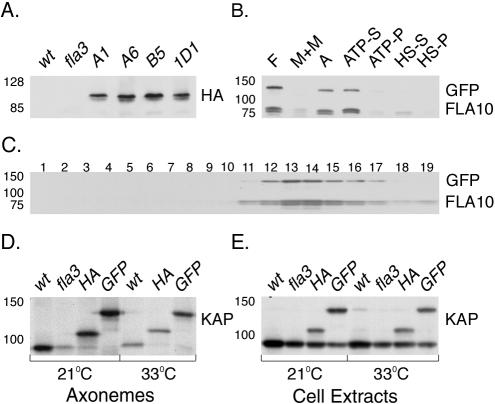
To determine if the epitope-tagged subunits are present in the flagella of the rescued strains, several rescued strains were grown at both 21 and 33°C, and then isolated flagella were analyzed on Western blots. As shown in Figure 6A, flagella isolated from fla3 strains rescued with the HA-tagged KAP gene contained an ∼100-kDa polypeptide that was not observed in either wild-type or fla3 cells. Similarly, flagella isolated from GFP rescued strains contained an ∼122-kDa polypeptide (Figure 6B). In both cases, the increase in the apparent size of the KAP polypeptide is consistent with the size of the inserted epitope tag. The tagged KAP polypeptides and FLA10 subunit were released from isolated axonemes by extraction with 10 mM MgATP (Figure 6B), similar to that reported previously for the wild-type Kinesin-2 complex (Cole et al., 1998). Sucrose density gradient centrifugation of the ATP extracts revealed that both the HA-tagged KAP (unpublished results) and the GFP-tagged KAP (Figure 6C) cosedimented with the FLA10 subunit at ∼10S. Western blots probed with a KAP antibody further demonstrated that the epitope-tagged KAP subunits assemble into flagella more efficiently than the fla3 KAP subunit (Figure 6D), even though they are expressed at similar levels in whole cells (Figure 6E). To verify that the tagged KAP subunit is fully functional in IFT, we also monitored the behavior of the GFP-KAP subunit in living cells by confocal fluorescence microscopy (see Supplementary Video). The bidirectional movement of GFP-labeled particles is clearly visible in the flagella of the rescued strain.
Figure 6.
Epitope-tagged KAPs are incorporated into the Kinesin-2 complex. (A) A Western blot of flagella isolated from wild-type, fla3, and four rescued fla3 strains (A1, A6, B5, 1D1, obtained by transformation with the KAP-HA construct) was probed with an antibody against the HA epitope. The KAP-HA subunit contains 892 amino acids and migrates at ∼100 kDa. (B) A Western blot showing the sequential extraction of isolated flagella (F) prepared from the KAP-GFP–rescued strain was probed with antibodies against GFP and FLA10. (M+M) is the membrane plus matrix fraction obtained by extraction of isolated flagella with 1.0% Nonidet-P-40, and A is the resulting pellet of axonemes. Most of the Kinesin-2 complex is released by extraction of axonemes with 10 mM MgATP (ATP-S); a small amount remains in the ATP extracted pellet (ATP-P). Treatment with 0.6 M NaCl releases the remainder into a high salt extract (HS-S) and high salt pellet (HS-P). Note the cofractionation of the KAP-GFP subunit with the FLA10 subunit. The KAP-GFP contains 1089 amino acids and migrates at ∼122 kDa. (C) The 10 mM MgATP extract shown in B was fractionated by sucrose density gradient centrifugation. A Western blot of the resulting fractions was probed with antibodies against GFP and FLA10. Note the cosedimentation of KAP-GFP with FLA10 at ∼10S in fractions 13 and 14; the top of the gradient is on the right. Reprobing of the blot with the KAP antibody failed to detect any of the endogenous fla3 KAP subunit in the 10S complex. (D) A Western blot of isolated axonemes prepared from wild-type (wt), fla3 (fla3), KAP-HA–rescued (HA), and KAP-GFP–rescued (GFP) cells at 21 and 33°C was probed with an antibody against KAP. Note the preferential assembly of the larger, epitope-tagged KAP subunits in axonemes of the rescued strains at both temperatures. (E) A Western blot of whole cell extracts prepared from wild-type, fla3, KAP-HA–rescued, and KAP-GFP–rescued strains was probed with an antibody against KAP. Note that the epitope-tagged KAP subunits are expressed at levels similar to the endogenous fla3 KAP subunit in the rescued cells.
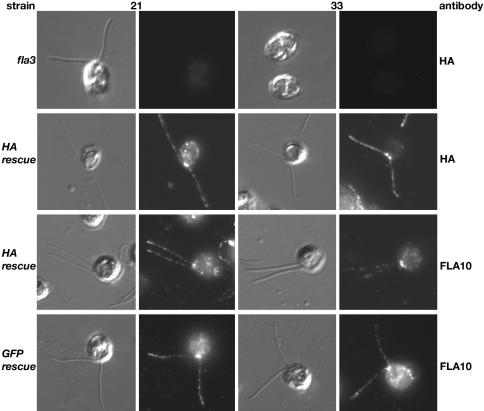
The ability of the tagged KAP subunits to restore the localization of the Kinesin-2 complex to the basal body region and flagella was assessed by immunofluorescence microscopy of fixed cells. Staining of fla3 cells with the HA antibody resulted in only faint background fluorescence of the cell bodies (Figure 7), whereas staining of KAP-HA–rescued cells demonstrated that the HA-tagged KAP was concentrated in the anterior region of the cell and in punctate spots along the length of the two flagella (Figure 7). Staining of either KAP-HA– or KAP-GFP–rescued cells with the FLA10 antibody indicated that the Kinesin-2 complex is now reconcentrated in the basal body region and present along the length of the two flagella. The pattern of FLA10 staining seen in the rescued strains was strikingly different from that observed previously in fla3 mutant cells (see Figure 5), at both 21 and 33°C. These results strongly suggest that the defect in localization of the Kinesin-2 complex to the basal body region is the basis of fla3 flagellar assembly defects and that this defect is rescued by the epitope-tagged KAP subunits. Staining of rescued cells with antibodies against other IFT components (such as the dynein LIC or the IFT subunit p139) confirmed that these components are now present in flagella at the restrictive temperature, as expected for Kinesin-2 cargoes (Perrone and Porter, unpublished results). The simplest interpretation of these observations is that the fla3-1 KAP subunit fails to be effectively coupled to its cargo(s) at the site of flagellar assembly, and as a result the Kinesin-2 complex becomes mis-localized.
Figure 7.
Epitope-tagged KAPs restore localization of the FLA10 Kinesin-2 complex to the basal body region and flagella. fla3 and fla3 rescued cells were fixed at both 21 and 33°C, stained with the antibodies as indicated on the right, and then imaged using both DIC and fluorescence microscopy. The top row shows fla3 cells stained with an antibody against the HA epitope; only background staining is observed. The second row shows KAP-HA–rescued cells stained with the HA-antibody, note the concentration of KAP-HA in the basal body region and along the length of the flagella at both temperatures, similar to wild-type pattern observed with the FLA10 and KAP antibodies in Figure 5. KAP-HA–rescued cells (third row) and KAP-GFP–rescued cells (bottom row) were also stained with the FLA10 antibody. Note that the FLA10 motor subunit is now concentrated in the basal body region and the flagella of the rescued strains.
The fla3 Mutation Affects IFT and Flagellar Assembly at the Permissive Temperature
Recent studies of other fla mutant strains have indicated that the loss of flagella at 33°C is a complex phenomenon involving both resorption due to defects in flagellar maintenance and active deflagellation by flagellar excision at the transition zone (Parker and Quarmby, 2003). In addition, many fla mutants have defects in different phases of IFT at 21°C (Iomini et al., 2001). Because there has been comparatively little study of fla3, we analyzed the behavior of fla3 and fla3 rescued cells at both 21 and 33°C. These studies revealed that fla3 cells also exhibit significant flagellar assembly defects at 21°C. For instance, when fla3 cells are resuspended from solid medium into liquid medium or subjected to deflagellation by pH shock, flagellar regeneration requires several hours, compared with wild-type or rescued cells, which regenerate full-length flagella within 60–90 min. These results and the defects in Kinesin-2 localization described above (Figure 5) suggested that intraflagellar transport (IFT) might be compromised in fla3 cells at 21°C.
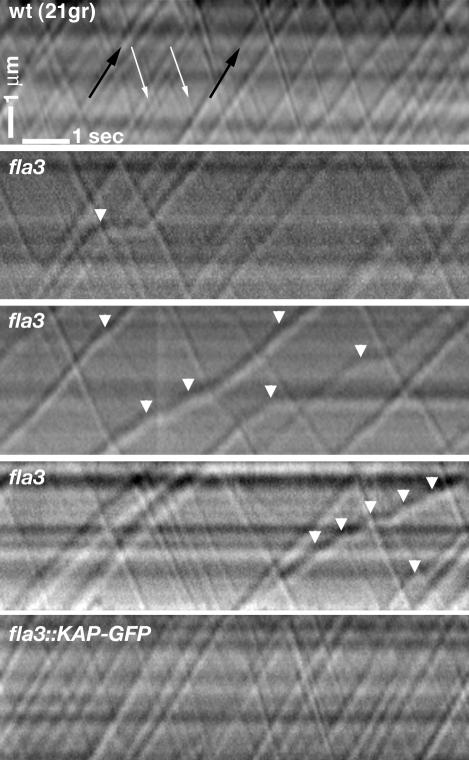
To determine the effect of the fla3 mutation on IFT, we analyzed the movement of individual IFT particles in wild-type, fla3, and the KAP-GFP–rescued strains using video-enhanced DIC light microscopy. Previous studies have shown that specific defects in IFT particle velocities and frequencies can be observed in several fla mutants, but the fla3 strain was not analyzed (Iomini et al., 2001). These earlier studies also used a paralyzed flagellar mutant background (pf15) to immobilize the fla cells and facilitate particle tracking. However, we were able to immobilize the flagella by mounting cells in 0.75% low-melting-point agarose, which allowed us to analyze each strain directly. As shown in Figure 8 and Table 2, the fla3 cells are specifically defective in the movement of anterograde IFT particles at 21°C. The most significant difference noted between the strains was the presence of fewer anterograde particles per second in fla3 flagella, compared with either the wild-type or the KAP-GFP–rescued strains (p < 0.0001). In contrast, no significant differences in the number of retrograde particles per second were observed between the strains. The effect on particle frequency is also illustrated in the anterograde-to-retrograde particle frequency ratio, which is much lower for fla3 than for either the wild-type or rescued strains (Table 2). Interestingly, we observed only a slight decrease in the velocity of moving anterograde particles in fla3 flagella. However, the anterograde IFT particles were observed to pause much more frequently and for significantly longer periods in fla3 cells than in either wild-type or rescued strains. More than 50% of the anterograde particles were observed to pause or stop in fla3 cells, compared with <19% in wild-type or rescued cells. In addition, the apparent sizes of the anterograde particles appeared to be larger in fla3 than in either wild-type or the rescued cells. (Figure 8). These results clearly indicate that both the number and the overall velocity of anterograde IFT particles are decreased in fla3 cells. In addition, all of these defects are completely rescued by the GFP-tagged KAP subunit.
Figure 8.
Anterograde IFT is altered in fla3 cells at 21°C. Representative kymograms illustrating the movements of individual IFT particles within a single flagellum are shown here. Individual IFT particles are seen as diagonal tracks. Time elapsed is plotted along the lower horizontal axis of the kymogram with time zero starting at the left. The length of the flagellum analyzed is depicted along the vertical axis of the kymograms. Anterograde particles are larger and move from the bottom left (proximal region) to the top right (distal region) of the kymograms (see black arrows in top panel). Retrograde particles are smaller and move from the top left to the bottom right (see white arrows in top panel). Note that many of the anterograde IFT particles in fla3 are larger than anterograde particles in either wild-type or the KAP-GFP–rescued strain. Anterograde particles also pause more frequently in fla3 flagella, as indicated by the white arrowheads.
Table 2.
Velocity and frequency of particles undergoing anterograde and retrograde IFT
| Strain | Anterograde frequency (p/s) | Retrograde frequency (p/s) | Frequency ratio | Total frequency (p/s) | Anterograde velocity (μ/s) | Retrograde velocity (μ/s) |
|---|---|---|---|---|---|---|
| Wild-type (21gr) | 1.3 ± 0.5 (21) | 2.2 ± 0.6 (21) | 0.6 ± 0.1 (21) | 3.7 ± 1.0 (21) | 1.7 ± 0.3 (416) | 2.8 ± 0.6 (360) |
| Wild-type (137c) | 1.3 ± 0.5 (11) | 2.2 ± 0.6 (11) | 0.6 ± 0.1 (11) | 3.5 ± 1.0 (11) | 1.8 ± 0.3 (176) | 2.6 ± 0.7 (180) |
| fla3 | 0.5 ± 0.2 (22) | 1.9 ± 0.5 (22) | 0.2 ± 0.1 (22) | 2.3 ± 0.6 (22) | 1.6 ± 0.3 (377) | 2.8 ± 0.5 (420) |
| fla3::KAP-GFP | 1.5 ± 0.4 (13) | 2.8 ± 0.6 (13) | 0.6 ± 0.1 (13) | 4.2 ± 0.8 (13) | 2.2 ± 0.3 (220) | 3.4 ± 0.7 (259) |
All measurements are expressed as mean ± SD (sample size). For frequency measurements, n = total number of cells, and for velocity measurements, n = total number of particles. Frequency is expressed as particles per second (p/s), and velocity is expressed as microns per second (μ/s). The frequency ratio is the frequency of anterograde particles divided by the frequency of retrograde particles for each cell. Because the fla3 mutation was originally isolated in the 137c background, but backcrossed into 21gr, we analyzed particle movement in both strains. No significant differences in particle frequency or velocity were observed between the two wild-type strains. For fla3 cells, the frequency of anterograde particles is significantly reduced, but anterograde particle velocity is only slightly decreased relative to wild-type cells. No statistically significant differences (p < 0.001) were observed for retrograde particle frequency or velocity. Transformation of fla3 with the GFP-tagged KAP construct (fla3::KAP-GFP) restores the frequency of anterograde particles (and the frequency ratio) to wild-type levels.
The fla3 Mutation Leads to Deflagellation at the Restrictive Temperature
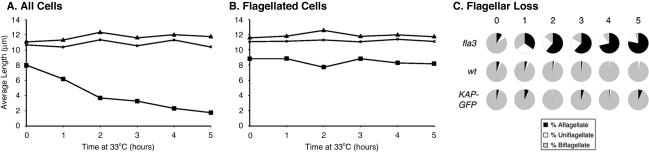
Previous studies of other fla mutants at 33°C have indicated that there is significant variability between different strains with respect to the time course of flagellar loss (Adams et al., 1982, Lux and Dutcher, 1991). Furthermore, depending on the external calcium concentration of the media, flagellar loss or disassembly appears to proceed by a combination of active flagellar excision at the transition zone and flagellar resorption via disassembly at the tip (Parker and Quarmby, 2003; Pan et al., 2004). We have found that fla3 cells display a similar response. After a shift to from 21 to 33°C, the average lengths of fla3 flagella gradually decrease, whereas wild-type and rescued cells maintain a relatively constant flagellar length over a 6-h time course (Figure 9A). However, the decrease in flagellar length in fla3 is primarily due to flagellar loss by active excision, as the relative numbers of both aflagellate and uniflagellate cells increases dramatically following the shift to 33°C (Figure 9C). When one excludes the aflagellate cells from determination of the average flagellar length, the lengths of the remaining flagella decrease only slightly over the same time course (Figure 9B, see Discussion). Significantly, the presence of a wild-type or epitope-tagged KAP transgene in the rescued fla3 cells prevents the rapid disassembly seen at 33°C. The total number of aflagellate and uniflagellate cells is similar to wild-type cells, and the average flagellar lengths are also relatively constant. These results suggest the presence of a signaling pathway that promotes active flagellar excision in fla3 cells when conditions are unfavorable for flagellar assembly. This pathway has recently been described in other fla mutants that are compromised with respect to flagellar assembly (Parker and Quarmby, 2003).
Figure 9.
Flagellar disassembly in fla3 occurs by active excision. Wild-type (triangles), fla3 (squares), and fla3::KAP-GFP rescued cells (circles) were shifted from 21 to 33°C at the indicated times. The lengths of flagella on at least 100 cells were measured for each time point. (A) The average flagellar lengths including the contribution of bald and aflagellate cells. (B) The same samples as in A, but the average flagellar lengths excluding the contribution of zero-length flagella. (C) Pie diagrams illustrating the percent of aflagellate (black), uniflagellate (white), and biflagellate cells at each time point.
DISCUSSION
Identification of a Temperature-sensitive, Flagellar Assembly Mutation in the KAP Gene
Several studies have demonstrated the essential role of the Kinesin-2 complex in the assembly of eukaryotic cilia and flagella, but the specific function of the KAP subunit in this process has been largely unexplored. In this report, we characterize the KAP gene, FLA3, in Chlamydomonas and demonstrate that it encodes a highly conserved polypeptide whose sequence is very similar to KAP subunits that have been characterized in other organisms (Figures 1, 2, 3). The observation that a mutation in FLA3 can disrupt flagellar assembly and maintenance demonstrates that KAP is an essential subunit of the heterotrimeric Kinesin-2 complex. Moreover, the complete inhibition of flagellar assembly at 33°C suggests that the Kinesin-2 complex is the primary anterograde IFT motor in Chlamydomonas. Studies in Tetrahymena and C. elegans have indicated that an OSM-3–related kinesin complex also contributes to anterograde IFT in these species (Shakir et al., 1993; Awan et al., 2004; Snow et al., 2004). Our findings are consistent with recent studies in Drosophila indicating that null mutations in the DrKAP gene disrupt the assembly of sensory cilia involved in hearing, proprioception, taste, and olfaction (Sarpal et al., 2003). However, an unexpected result of the Drosophila studies was that neither Kinesin-2 nor IFT is required for the assembly of Drosophila sperm flagella (Han et al., 2003; Sarpal et al., 2003). This apparent discrepancy is likely due to the fact that Drosophila sperm develop within a syncitium and become individualized only after flagellar assembly is completed (Fuller, 1993). Given the conservation of the heterotrimeric Kinesin-2 complexes seen in other organisms, it is likely that KAP is required for anterograde IFT in nearly all ciliated cells, with a few exceptions as noted above, but its specific function within the complex is not well understood. We have been able to use the conditional phenotype of the temperature-sensitive fla3-1 mutation to probe the function of the KAP subunit in Chlamydomonas.
Defects in the Localization of the Kinesin-2 Complex in fla3 Cells
Contrary to initial expectation, we found that the fla3 mutation does not appear to significantly alter the level of either KAP or FLA10 in whole cells (Figures 4C and 6E). This is distinct from previous studies of the FLA10 locus, which have shown that the different fla10 alleles have variable effects on the stability of the FLA10 protein and the extent of flagellar assembly (Huang et al., 1977; Adams et al., 1982; Lux and Dutcher, 1991; Walther et al., 1994; Kozminski et al., 1995; Vashishtha et al., 1996; Cole et al., 1998). The fla3 mutation does however have a very dramatic effect on the localization of the FLA10 Kinesin-2 complex in the basal body region. The fla3 mutation appears to reduce either the targeting or the retention of the FLA10 complex at the site of flagellar assembly (Figure 5). This defect is completely rescued by transformation of fla3 with wild-type or epitope-tagged versions of the KAP gene (Figure 7). The effect of fla3 on the localization of FLA10 is therefore distinct from that observed with mutations in other IFT components, such as the cDhc1b complex or IFT particle subunits. These mutants still concentrate FLA10 in the basal body region (Pazour et al., 1999; Porter et al., 1999; Deane et al., 2001; Perrone et al., 2002). Thus the concentration of the FLA10 Kinesin-2 complex in the basal body region is critically dependent on the KAP subunit, but independent of the retrograde motor and at least some IFT particle subunits. The dispersal of the Kinesin-2 complex throughout the cell has also been observed in the bld2-1 mutant (Cole et al., 1998). BLD2 encodes an epsilon tubulin required for basal body assembly and morphogenesis (Dutcher et al., 2002). As a result, bld2-1 cells have abnormal basal bodies and disorganized rootlet microtubules (Ehler et al., 1995). Taken together, these results suggest that the KAP subunit may facilitate the interaction of the Kinesin-2 complex with an unidentified component(s) of basal body apparatus or IFT machinery that is missing or disorganized in bld2-1 cells. These may include components of the transitional fibers that have recently been identified as potential docking sites for IFT particles (Deane et al., 2001) or another IFT component that is anchored in the basal body region and mediates interactions between KAP and the IFT particle. Future study of other mutants with defects in different stages of basal body assembly may prove useful in identifying other components required for Kinesin-2 localization. In addition, transformation with different KAP constructs should further define domains within the polypeptide sequence required for the efficient targeting or retention of the complex within the basal body region.
The fla3 Mutation Is Associated with Defects in Anterograde IFT
At 21°C, fla3 cells assemble flagella that are only slightly shorter than those of their wild-type counterparts (Table 1). However, we noted that the time course of flagellar regeneration was much slower with fla3 cells than with wild-type cells. We therefore analyzed the movement of IFT particles directly using video-enhanced DIC microscopy. This analysis revealed decreases in both the frequency and overall velocity of anterograde IFT particles in fla3, but no statistically significant changes in the frequency and/or velocity of retrograde IFT particles (Figure 8, Table 2). Pausing of anterograde IFT particles was also observed much more frequently in fla3 cells.
Previous studies have suggested that IFT can be subdivided into at least four distinct phases (Iomini et al., 2001). In phase I, anterograde particles are assembled in the basal body region by the remodeling and recycling of retrograde particles and by the addition of IFT motors and flagellar cargoes. In phase II, the anterograde particles are transported from the base to the tip of the flagellum by the Kinesin-2 complex. In phase III, cargoes are unloaded at the flagellar tips, and anterograde particles are remodeled into retrograde particles. In phase IV, retrograde particles are transported by cytoplasmic dynein from the tip to the proximal end of the flagellum and eventually to the basal body region. Mutant strains with lower anterograde to retrograde particle frequencies are thought to represent defects in phase I. Similarly, mutant strains with lower anterograde particle frequencies and decreased anterograde velocities are considered defective in phase I and phase II.
Using this classification system, fla3 is clearly defective in both phases I and II of IFT. Reductions in anterograde particle frequency are thought to arise from deficiencies in the loading of IFT particles into the flagella from the basal body region (Iomini et al., 2001). In the case of fla3, such defects are consistent with observations that FLA10 staining is reduced in the basal body region (Figure 5), even though other IFT components appear to be present at wild-type levels. Reduced Kinesin-2 activity in fla3 flagella may also account for the increase in the size of the anterograde particles. Increases in the apparent size of the anterograde particles have also been noted in other phase I-II mutants (Iomini et al., 2001). Interestingly, at least two of these mutants have been identified as mutations in the motor domains of the kinesin-related subunits (Vashishtha et al., 1996; Cole and Dutcher; personal communication). Whether these increases in particle size result from defects in particle assembly at the basal body region or from the merging of particles during IFT remains to be determined.
Decreases in anterograde particle velocities (phase II defects) can also arise from mutations in the Kinesin-2 complex. For instance, in fla10-1 cells, anterograde particles are transported at wild-type velocities at 21°C, but at decreased velocities at 32°C, until all IFT particle movement stops (Iomini et al., 2001). In the case of fla3, the effect on particle velocity appears to be more complex. In fla3, the velocities of the moving anterograde particles are similar to wild-type at 21°C, but anterograde particles stop or pause much more frequently in fla3 flagella than in either wild-type or KAP-GFP–rescued flagella (Figure 8). The frequent pausing of particles has not previously been reported in other fla mutants (Piperno et al., 1998; Iomini et al., 2001) and so it is unclear if this is a unique feature of the fla3 mutation. However, it seems highly likely that the pausing of the particles may be caused by their dissociation from the Kinesin-2 motor, particularly if the number of Kinesin-2 complexes per particle is relatively small. Indeed, a recent study of Kinesin-2 motor subunits in vitro has demonstrated that the complex is highly processive and therefore capable of transporting cargoes at a constant velocity that is largely independent of motor density (Zhang and Hancock, 2004). Furthermore, the rescue of the pausing defects seen in the fla3::KAP-GFP strain correlates very clearly with the observation that the KAP-GFP subunit assembles into flagella much more efficiently than the fla3 KAP subunit, even though both are expressed at similar levels in the cytoplasm (Figure 6, D and E). These results strongly suggest that the fla3 mutation alters the interaction between the Kinesin-2 complex and the IFT particle. Whether this effect is direct (i.e., due to a specific interaction between the KAP subunit and an IFT particle subunit) or indirect (i.e., due to an interaction with another component that regulates cargo binding) is still unknown. Further characterization of other fla mutations (e.g., fla1, fla8, fla18, fla27) that also alter anterograde IFT (Iomini et al., 2001) may help resolve this question.
Why Do fla3 Cells Deflagellate at 33°C?
Recent studies have demonstrated that several fla mutants disassemble their flagella at the restrictive temperature by a process of active deflagellation (Pan et al., 2004; Parker and Quarmby, 2003). fla3 cells display a similar response. After a shift from 21 to 32°C, the majority of cells disassemble their flagella by deflagellation, but the remaining cells resorb their flagella relatively slowly (Figure 9). One hypothesis is that inactivation of IFT at the restrictive temperature is a signal that initiates the deflagellation pathway (Parker and Quarmby, 1994). We therefore examined IFT in fla3 cells that had been held at 32°C but still retained their flagella. Contrary to expectation, both anterograde and retrograde IFT were observed in the flagella of fla3 cells that had been held at 32°C for as long as 6 h, even though IFT is completely inhibited in fla10-1 cells under the same conditions (Perrone and Porter, unpublished results). Iomini et al. (2001) have reported that IFT continues for at least 90 min in several other fla mutants that also exhibit defects in different phases of IFT (fla2, fla11, fla12, fla16, fla17, fla21, fla24, fla28). These observations suggest that although defects in IFT are likely to contribute to the deflagellation response seen in fla mutants, the signal that initiates flagellar excision precedes the complete inhibition of IFT.
The fla3 Mutation Identifies the C-terminal Region of KAP as Critical Functional Domain
Our studies also demonstrate that the conserved C-terminal region is a critical functional domain of the KAP subunit. Sequence analysis revealed that fla3-1 is a point mutation that results in the substitution of a leucine for a conserved phenylalanine at amino acid residue 753, 20 amino acids downstream from the last armadillo repeat (Figures 2 and 4). The specific function of this residue is unknown, but it is contained within a highly conserved region of the KAP sequence that has been identified in other species as the potential binding site for the mixed lineage protein kinase MLK2 (Nagata et al., 1998), the tumor suppressor APC (Jimbo et al., 2002), and the p150 subunit of the dynactin complex (Deacon et al., 2003). Our current working hypothesis is that the fla3 mutation disrupts an interaction between the KAP subunit and another component that regulates its association with the IFT particle, but the identity of this component is presently unknown.
Recent studies in mammalian cells have provided some new clues on the nature of the interaction between the Kinesin-2 complex and the IFT particles. Yeast two-hybrid assays have indicated that the mouse IFT20 subunit interacts directly with one of Kinesin-2 motor subunits, KIF3B, via coiled coil domains (Baker et al., 2003). Interestingly, this interaction appears to be regulated by ATP (Baker et al., 2003). As others have speculated that phosphorylation of the C-terminal region of the KAP subunit might regulate cargo interactions (Nagata et al., 1998), we analyzed the region around the site of the fla3 mutation for potential phosphorylation sites, but thus far none of the predicted sites appear to be universally conserved in other species. A second yeast two-hybrid study has identified an interaction between the human homologue of FLA10 (KIF3A) and a regulatory coiled-coil region of the protein kinase NEK1 (Surpili et al., 2003). Whether any of the Kinesin-2 subunits are substrates of the NEK1 kinase remains to be determined, but it is intriguing to note that mutations in the mouse Nek1 gene have a complex phenotype that includes late onset PKD, male sterility, and facial dysmorphism (Upadhya et al., 2000). A possible defect in flagellar assembly as the basis of the mouse Nek1 phenotype has not been explored. Intriguingly however, recent studies have identified a large gene family encoding NEK-related kinases in Chlamydomonas (Bradley et al., 2004), and at least two family members have been localized to flagella (Mahjoub et al., 2004; Bradley and Quarmby, personal communication). One of these kinases is essential for calcium-induced microtubule severing during deflagellation (Mahjoub et al., 2004), and defects in another have been correlated with changes in flagellar length (Bradley and Quarmby, personal communication). Further study of the Nek gene family in Chlamydomonas may therefore provide important insights into the pathways that regulate Kinesin-2 activity.
Rescue of fla3 Defects with Epitope-tagged KAP Sequences
Because the fla3 KAP subunit is relatively stable, we used epitope-tagged versions of the wild-type gene to rescue the mutant phenotype and localize the wild-type gene product in rescued cells. The epitope tags were placed in a nonconserved domain close to carboxy terminus, and by all criteria tested, both the HA- and GFP-tagged KAP sequences fully rescued the fla3 mutant phenotypes. Confocal microscopy of living cells confirmed that KAP-GFP–labeled particles can be observed to move in both directions along the length of the flagella, similar to that reported previously for KAP-GFP in the sensory cilia of the nematode, C. elegans (Signor et al., 1999). KAP-GFP signals have also been observed in blastula cilia of sea urchin embryos (Morris et al., 2004). Interestingly, the Chlamydomonas KAP-GFP subunit is more efficiently incorporated into the Kinesin-2 complex of the flagella than the fla3 mutant subunit, even though both are present at similar levels in the cell body. The tagged proteins are clearly part of a functional motor complex and serve as faithful reporters of the KAP subunit. These constructs will therefore be useful tools for future experiments designed to identify domains of the KAP sequence required for localization of the Kinesin-2 complex and to monitor the behavior of the Kinesin-2 complex in living cells under different experimental conditions.
Supplementary Material
Acknowledgments
We gratefully acknowledge our colleague Susan Dutcher (Washington University) for helpful suggestions and critical comments throughout this project. We also thank the Chlamydomonas Genetics Center and Elizabeth Harris for providing mutant strains. We also thank our colleagues at the University of Minnesota for assistance and advice, including Tom Hays, Dick Linck, Pete Lefebvre, Carolyn Silflow, and Meg Titus and Samantha Reed at the University of Idaho for technical assistance with fusion protein purification. This work was supported by National Institutes of Health grants R01 GM-55667 (to M.E.P.) and R01 GM-61920 (to D.G.C.)
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E04-10-0931) on December 22, 2004.
Abbreviations used: BAC, bacterial artificial chromosome; Dhc, dynein heavy chain; GFP, green fluorescent protein; HA, hemagglutin; IFT, intraflagellar transport; KAP, Kinesin-2 associated protein; LIC, light intermediate chain; M-N/5, minimal medium minus ammonium; PCD, primary ciliary dyskinesia; PKD, polycystic kidney disease; RFLP, restriction fragment length polymorphism; TAP, trisacetate phosphate.
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
References
- Adams, G.M.W., Huang, B., and Luck, D. J. (1982). Temperature-sensitive, assembly-defective flagella mutants of Chlamydomonas reinhardtii. Genetics 100, 579-586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awan, A., Bernstein, M., Hamasaki, T., and Satir, P. (2004). Cloning and characterization of Kin5, a novel Tetrahymenaciliary kinesin II. Cell Motil. Cytoskelet. 58, 1-9. [DOI] [PubMed] [Google Scholar]
- Baker, S. A., Freeman, K., Luby-Phelps, K., Pazour, G. J., and Besharse, J. C. (2003). IFT20 links kinesin II with a mammalian intraflagellar transport complex that is conserved in motile flagella and sensory cilia. J. Biol. Chem. 278, 34211-34218. [DOI] [PubMed] [Google Scholar]
- Berman, S. A., Wilson, N. F., Haas, N. A., and Lefebvre, P. A. (2003). A novel MAP kinase regulates flagellar length in Chlamydomonas. Curr. Biol. 13, 1145-1149. [DOI] [PubMed] [Google Scholar]
- Bradley, B. A., Wagner, J. J., and Quarmby, L. M. (2004). Identification and sequence analysis of six new members of the NIMA-related kinase family in Chlamydomonas. J. Eukaryot. Microbiol. 51, 66-72. [DOI] [PubMed] [Google Scholar]
- Brown, J. M., Marsala, C., Kosoy, R., and Gaertig, J. (1999). Kinesin-II is preferentially targeted to assembling cilia and is required for ciliogenesis and normal cytokinesis in Tetrahymena. Mol. Biol. Cell 10, 3081-3096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole, D. G. (2003). The intraflagellar transport machinery of Chlamydomonas reinhardtii. Traffic 4, 435-442. [DOI] [PubMed] [Google Scholar]
- Cole, D. G., Chinn, S. W., Wedaman, K. P., Hall, K., Vuong, T., and Scholey, J. M. (1993). Novel heterotrimeric kinesin-related protein purified from sea urchin eggs. Nature 366, 268-270. [DOI] [PubMed] [Google Scholar]
- Cole, D. G., Diener, D. R., Himelblau, A. L., Beech, P. L., Fuster, J. C., and Rosenbaum, J. L. (1998). Chlamydomonas kinesin-II-dependent intraflagellar transport (IFT): IFT particles contain proteins required for ciliary assembly in Caenorhabditis elegans sensory neurons. J. Cell Biol. 141, 993-1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deacon, S. W., Serpinskaya, A. S., Vaughan, P. S., Lopez Fanarraga, M., Vernos, I., Vaughan, K. T., and Gelfand, V. I. (2003). Dynactin is required for bidirectional organelle transport. J. Cell Biol. 160, 297-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deane, J. A., Cole, D. G., Seeley, E. S., Diener, D. R., and Rosenbaum, J. L. (2001). Localization of intraflagellar transport protein IFT52 identifies basal body transitional fibers as the docking site for IFT particles. Curr. Biol. 11, 1586-1590. [DOI] [PubMed] [Google Scholar]
- Dutcher, S. K., Morrissette, N. S., Preble, A. M., Rackley, C., and Stanga, J. (2002). Epsilon-tubulin is an essential component of the centriole. Mol. Biol. Cell 13, 3859-3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehler, L. L., Holmes, J. A., and Dutcher, S. K. (1995). Loss of spatial control of the mitotic spindle apparatus in a Chlamydomonas reinhardtii mutant strain lacking basal bodies. Genetics 141, 945-960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuhrmann, M., Oertel, W., and Hegemann, P. (1999). A synthetic gene coding for the green fluorescent protein (GFP) is a versatile reporter in Chlamydomonas reinhardtii. Plant J. 19, 353-361. [DOI] [PubMed] [Google Scholar]
- Fuller, M. (1993). Spermatogenesis. In: The Development of Drosophila melanogaster, Vol. 2, ed. M. Bate and A. Martinez Arias, Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press, 71-147. [Google Scholar]
- Gindhart, J. G., Jr., and Goldstein, L. S. (1996). Armadillo repeats in the SpKAP115 subunit of kinesin-II. Trends Cell Biol. 6, 415-416. [DOI] [PubMed] [Google Scholar]
- Huang, B., Rifkin, M. R., and Luck, D. J. (1977). Temperature-sensitive mutations affecting flagellar assembly and function in Chlamydomonas reinhardtii. J. Cell Biol. 72, 67-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huangfu, D., Liu, A., Rakeman, A. S., Murcia, N. S., Niswander, L., and Anderson, K. V. (2003). Hedgehog signalling in the mouse requires intraflagellar transport proteins. Nature 426, 83-87. [DOI] [PubMed] [Google Scholar]
- Ibañez-Tallon, I., Heintz, N., and Omran, H. (2003). To beat or not to beat: roles of cilia in development and disease. Hum. Mol. Genet. 12(Spec No 1), R27-R35. [DOI] [PubMed] [Google Scholar]
- Iomini, C., Babaev-Khaimov, V., Sassaroli, M., and Piperno, G. (2001). Protein particles in Chlamydomonas flagella undergo a transport cycle consisting of four phases. J. Cell Biol. 153, 13-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimbo, T., Kawasaki, Y., Koyama, R., Sato, R., Takada, S., Haraguchi, K., and Akiyama, T. (2002). Identification of a link between the tumor suppressor APC and the kinesin superfamily. Nat. Cell Biol. 4, 323-327. [DOI] [PubMed] [Google Scholar]
- Kozminski, K. G., Beech, P. L., and Rosenbaum, J. L. (1995). The Chlamydomonas kinesin-like protein FLA10 is involved in motility associated with the flagellar membrane. J. Cell Biol. 131, 1517-1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence, C. J. et al. (2004). A standardized kinesin nomenclature. J. Cell Biol. 167, 19-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, F., Hiesberger, T., Cordes, K., Sinclair, A. M., Goldstein, L. S., Somlo, S., and Igarashi, P. (2003). Kidney-specific inactivation of the KIF3A subunit of kinesin-II inhibits renal ciliogenesis and produces polycystic kidney disease. Proc. Natl. Acad. Sci. USA 100, 5286-5291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lux, F. G., 3rd, and Dutcher, S. K. (1991). Genetic interactions at the FLA10 locus: suppressors and synthetic phenotypes that affect the cell cycle and flagellar function in Chlamydomonas reinhardtii. Genetics 128, 549-561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahjoub, M. R., Rasi, M. Q., and Quarmby, L. M. (2004). A NIMA-related kinase, Fa2p, localizes to a novel site in the proximal cilia of Chlamydomonas and mouse kidney cells. Mol. Biol. Cell 15, 5172-5186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marszalek, J. R., and Goldstein, L. S. (2000). Understanding the functions of kinesin-II. Biochim. Biophys. Acta 1496, 142-150. [DOI] [PubMed] [Google Scholar]
- Marszalek, J. R., Ruiz-Lozano, P., Roberts, E., Chien, K. R., and Goldstein, L. S. (1999). Situs inversus and embryonic ciliary morphogenesis defects in mouse mutants lacking the KIF3A subunit of kinesin-II. Proc. Natl. Acad. Sci. USA 96, 5043-5048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuura, K., Lefebvre, P. A., Kamiya, R., and Hirono, M. (2002). Kinesin-II is not essential for mitosis and cell growth in Chlamydomonas. Cell Motil. Cytoskelet. 52, 195-201. [DOI] [PubMed] [Google Scholar]
- Morris, R. L., English, C. N., Lou, J. E., Dufort, F. J., Nordberg, J., Terasaki, M., and Hinkle, B. (2004). Redistribution of the kinesin-II subunit KAP from cilia to nuclei during the mitotic and ciliogenic cycles in sea urchin embryos. Dev. Biol. 274, 56-69. [DOI] [PubMed] [Google Scholar]
- Morris, R. L., and Scholey, J. M. (1997). Heterotrimeric kinesin-II is required for the assembly of motile 9+2 ciliary axonemes on sea urchin embryos. J. Cell Biol. 138, 1009-1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagata, K., Puls, A., Futter, C., Aspenstrom, P., Schaefer, E., Nakata, T., Hirokawa, N., and Hall, A. (1998). The MAP kinase kinase kinase MLK2 co-localizes with activated JNK along microtubules and associates with kinesin superfamily motor KIF3. EMBO J. 17, 149-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura, T., Kato, K., Yamaguchi, T., Fukata, Y., Ohno, S., and Kaibuchi, K. (2004). Role of the PAR-3-KIF3 complex in the establishment of neuronal polarity. Nat. Cell Biol. 6, 328-334. [DOI] [PubMed] [Google Scholar]
- Nonaka, S., Tanaka, Y., Okada, Y., Takeda, S., Harada, A., Kanai, Y., Kido, M., and Hirokawa, N. (1998). Randomization of left-right asymmetry due to loss of nodal cilia generating leftward flow of extraembryonic fluid in mice lacking KIF3B motor protein. Cell 95, 829-837. [DOI] [PubMed] [Google Scholar]
- Pan, J., Wang, Q., and Snell, W. J. (2004). An aurora kinase is essential for flagellar disassembly in Chlamydomonas. Dev. Cell 6, 445-451. [DOI] [PubMed] [Google Scholar]
- Parker, J. D., and Quarmby, L. M. (2003). Chlamydomonas fla mutants reveal a link between deflagellation and intraflagellar transport. BioMed Central Cell Biol. 4, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazour, G. J., Dickert, B. L., and Witman, G. B. (1999). The DHC1b (DHC2) isoform of cytoplasmic dynein is required for flagellar assembly. J. Cell Biol. 144, 473-481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrone, C. A., Myster, S. H., Bower, R., O'Toole, E. T., and Porter, M. E. (2000). Insights into the structural organization of the I1 inner arm dynein from a domain analysis of the 1beta dynein heavy chain. Mol. Biol. Cell 11, 2297-2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrone, C. A., Tritschler, D., Taulman, P., Bower, R., Yoder, B. K., and Porter, M. E. (2003). A novel dynein light intermediate chain colocalizes with the retrograde motor for intraflagellar transport at sites of axoneme assembly in Chlamydomonas and mammalian cells. Mol. Biol. Cell 14, 2041-2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrone, C. A., Yang, P., O'Toole, E., Sale, W. S., and Porter, M. E. (1998). The Chlamydomonas IDA7 locus encodes a 140-kDa dynein intermediate chain required to assemble the I1 inner arm complex. Mol. Biol. Cell 9, 3351-3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pflanz, R., Peter, A., Schafer, U., and Jackle, H. (2004). Follicle separation during Drosophila oogenesis requires the activity of the Kinesin II-associated polypeptide Kap in germline cells. EMBO Rep. 5, 510-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piperno, G., and Mead, K. (1997). Transport of a novel complex in the cytoplasmic matrix of Chlamydomonas flagella. Proc. Natl. Acad. Sci. USA 94, 4457-4462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piperno, G., Siuda, E., Henderson, S., Segil, M., Vaananen, H., and Sassaroli, M. (1998). Distinct mutants of retrograde intraflagellar transport (IFT) share similar morphological and molecular defects. J. Cell Biol. 143, 1591-1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter, M. E., Bower, R., Knott, J. A., Byrd, P., and Dentler, W. (1999). Cytoplasmic dynein heavy chain 1b is required for flagellar assembly in Chlamydomonas. Mol. Biol. Cell 10, 693-712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray, K., Perez, S. E., Yang, Z., Xu, J., Ritchings, B. W., Steller, H., and Goldstein, L. S. (1999). Kinesin-II is required for axonal transport of choline acetyltransferase in Drosophila. J. Cell Biol. 147, 507-518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbaum, J. L., and Witman, G. B. (2002). Intraflagellar transport. Nat. Rev. Mol. Cell. Biol. 3, 813-825. [DOI] [PubMed] [Google Scholar]
- Sanders, M. A., and Salisbury, J. L. (1995). Immunofluorescence microscopy of cilia and flagella. Methods Cell Biol. 47, 163-169. [DOI] [PubMed] [Google Scholar]
- Sarpal, R., Todi, S. V., Sivan-Loukianova, E., Shirolikar, S., Subramanian, N., Raff, E.C., Erickson, J. W., Ray, K., and Eberl, D. F. (2003). Drosophila KAP interacts with the kinesin II motor subunit KLP64D to assemble chordotonal sensory cilia, but not sperm tails. Curr. Biol. 13, 1687-1696. [DOI] [PubMed] [Google Scholar]
- Schafer, J. C., Haycraft, C. J., Thomas, J. H., Yoder, B. K., and Swoboda, P. (2003). XBX-1 encodes a dynein light intermediate chain required for retrograde intraflagellar transport and cilia assembly in Caenorhabditis elegans. Mol. Biol. Cell 14, 2057-2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shakir, M. A., Fukushige, T., Yasuda, H., Miwa, J., and Siddiqui, S. S. (1993). C. elegans osm-3 gene mediating osmotic avoidance behaviour encodes a kinesin-like protein. Neuroreport 4, 891-894. [DOI] [PubMed] [Google Scholar]
- Shimizu, K., Kawabe, H., Minami, S., Honda, T., Takaishi, K., Shirataki, H., and Takai, Y. (1996). SMAP, a Smg GDS-associating protein having arm repeats and phosphorylated by Src tyrosine kinase. J. Biol. Chem. 271, 27013-27017. [DOI] [PubMed] [Google Scholar]
- Shimizu, K., Shirataki, H., Honda, T., Minami, S., and Takai, Y. (1998). Complex formation of SMAP/KAP3, a KIF3A/B ATPase motor-associated protein, with a human chromosome-associated polypeptide. J. Biol. Chem. 273, 6591-6594. [DOI] [PubMed] [Google Scholar]
- Signor, D., Wedaman, K. P., Orozco, J. T., Dwyer, N. D., Bargmann, C. I., Rose, L. S., and Scholey, J. M. (1999a). Role of a class DHC1b dynein in retrograde transport of IFT motors and IFT raft particles along cilia, but not dendrites, in chemosensory neurons of living Caenorhabditis elegans. J. Cell Biol. 147, 519-530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Signor, D., Wedaman, K. P., Rose, L. S., and Scholey, J. M. (1999b). Two heteromeric kinesin complexes in chemosensory neurons and sensory cilia of Caenorhabditis elegans. Mol. Biol. Cell 10, 345-360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sizova, I., Fuhrmann, M., and Hegemann, P. (2001). A Streptomyces rimosus aphVIII gene coding for a new type phosphotransferase provides stable antibiotic resistance to Chlamydomonas reinhardtii. Gene 277, 221-229. [DOI] [PubMed] [Google Scholar]
- Snell, W. J., Pan, J., and Wang, Q. (2004). Cilia and flagella revealed: from flagellar assembly in Chlamydomonas to human obesity disorders. Cell 117, 693-697. [DOI] [PubMed] [Google Scholar]
- Snow, J. J., Ou, G., Gunnarson, A. L., Walker, M.R.S., Zhou, H. M., Brust-Macher, I., and Scholey, J. M. (2004). Two anterograde intraflagellar transport motors cooperate to build sensory cilia on C. elegans neurons. Nat. Cell Biol. 6, 1109-1113. [DOI] [PubMed] [Google Scholar]
- Surpili, M. J., Delben, T. M., and Kobarg, J. (2003). Identification of proteins that interact with the central coiled-coil region of the human protein kinase NEK1. Biochemistry 42, 15369-15376. [DOI] [PubMed] [Google Scholar]
- Takeda, S., Yonekawa, Y., Tanaka, Y., Okada, Y., Nonaka, S., and Hirokawa, N. (1999). Left-right asymmetry and kinesin superfamily protein KIF3A: new insights in determination of laterality and mesoderm induction by kif3A-/- mice analysis. J. Cell Biol. 145, 825-836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda, S., Yamazaki, H., Seog, D. H., Kanai, Y., Terada, S., and Hirokawa, N. (2000). Kinesin superfamily protein 3 (KIF3) motor transports fodrin-associating vesicles important for neurite building. J. Cell Biol. 148, 1255-1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuma, M. C., Zill, A., Le Bot, N., Vernos, I., and Gelfand, V. (1998). Heterotrimeric kinesin II is the microtubule motor protein responsible for pigment dispersion in Xenopus melanophores. J. Cell Biol. 143, 1547-1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upadhya, P., Birkenmeier, E. H., Birkenmeier, C. S., and Barker, J. E. (2000). Mutations in a NIMA-related kinase gene, Nek1, cause pleiotropic effects including a progressive polycystic kidney disease in mice. Proc. Natl. Acad. Sci. USA 97, 217-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vashishtha, M., Walther, Z., and Hall, J. L. (1996). The kinesin-homologous protein encoded by the Chlamydomonas FLA10 gene is associated with basal bodies and centrioles. J. Cell Sci. 109(Pt 3), 541-549. [DOI] [PubMed] [Google Scholar]
- Walther, Z., Vashishtha, M., and Hall, J. L. (1994). The Chlamydomonas FLA10 gene encodes a novel kinesin-homologous protein. J. Cell Biol. 126, 175-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wedaman, K. P., Meyer, D. W., Rashid, D. J., Cole, D. G., and Scholey, J. M. (1996). Sequence and submolecular localization of the 115-kD accessory subunit of the heterotrimeric kinesin-II (KRP85/95) complex. J. Cell Biol. 132, 371-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicks, S. R., de Vries, C. J., van Luenen, H. G., and Plasterk, R. H. (2000). CHE-3, a cytosolic dynein heavy chain, is required for sensory cilia structure and function in Caenorhabditis elegans. Dev. Biol. 221, 295-307. [DOI] [PubMed] [Google Scholar]
- Wilson, N. F., and Lefebvre, P. A. (2004). Regulation of flagellar assembly by glycogen synthase Kinase 3 in Chlamydomonas reinhardtii. Eukaryot. Cell 3, 1307-1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki, H., Nakata, T., Okada, Y., and Hirokawa, N. (1995). KIF3A/B: a heterodimeric kinesin superfamily protein that works as a microtubule plus end-directed motor for membrane organelle transport. J. Cell Biol. 130, 1387-1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki, H., Nakata, T., Okada, Y., and Hirokawa, N. (1996). Cloning and characterization of KAP3, a novel kinesin superfamily-associated protein of KIF3A/3B. Proc. Natl. Acad. Sci. USA 93, 8443-8448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y., and Hancock, W. O. (2004). The two motor domains of KIF3A/B coordinate for processive motility and move at different speeds. Biophys. J. 87, 1795-1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.