Abstract
Aim:
Bacterial vaginosis may lead to preterm birth through epigenetic programming of the inflammatory response, specifically via miRNA expression.
Methods:
We quantified bacterial 16S rRNA, cytokine mRNA and 800 miRNA from cervical swabs obtained from 80 women at 16–19 weeks’ gestation. We generated bacterial and cytokine indices using weighted quantile sum regression and examined associations with miRNA and gestational age at delivery.
Results & discussion:
Each decile of the bacterial and cytokine indices was associated with shorter gestations (p < 0.005). The bacterial index was associated with miR-494, 371a, 4286, 185, 320e, 888 and 23a (p < 0.05). miR-494 remained significant after false discovery rate correction (q < 0.1). The cytokine index was associated with 27 miRNAs (p < 0.05; q < 0.01).
Conclusion:
Future investigation into the role of bacterial vaginosis- and inflammation-associated miRNA and preterm birth is warranted.
Keywords: : bacterial vaginosis, cervix, cytokines, miRNA, preterm birth
Over 15 million infants are born preterm (<37 weeks of gestation) each year worldwide [1]. Over 1 million of those infants die, and survivors are at risk for a multitude of adverse sequelae including lung disease and neurodevelopmental disability [2]. With the exception of the modest efficacy of progesterone administration [3] and cerclage/pessary placement [4,5], efforts to prevent preterm birth by treating modifiable risk factors have not succeeded [2]. Progress in preventing preterm birth is stalled in part by a lack of understanding of the molecular mechanisms underlying spontaneous preterm birth.
Bacterial vaginosis (BV) is a strong risk factor for preterm birth [6]. BV is the most common infection of the vagina in women of reproductive age [7]. BV is caused by an imbalance of the normal bacterial flora with a preponderance of Gardnerella, Mobiluncus, Mycoplasma and anaerobic species [8]. Pregnant women with BV have 40–200% increased risk of preterm birth compared with women without BV [9,10]. However, several randomized controlled trials of antibiotic therapy for BV in pregnancy have not led to reductions in preterm birth risk [6,11]. These trials have questioned the causal relationship between BV and preterm birth; however, it is possible that molecular events are underway, and that treatment with antibiotics does not disrupt that cascade. The mechanism by which BV may lead to preterm birth is not well understood but is thought to be due to inflammatory pathways including elevation of cytokine levels induced by dysbiosis [12] that can lead to preterm labor [13–15]. BV may lead to local tissue programming through epigenetic processes. miRNAs represent one form of epigenetic gene regulation. miRNAs are short (20–24 nucleotides) noncoding RNA sequences that regulate gene expression and translation [16]. Several miRNAs are more abundant in inflammatory states [17]. Additionally, the miR-200 family of miRNAs regulates contraction-associated genes in labor and delivery [18]. It follows that miRNAs may be involved in early spontaneous parturition. With respect to preterm delivery, two studies have shown that differentially expressed miRNAs in the cervix are associated with preterm birth and shorter gestation [19,20]. In a previous study, we showed the levels of six miRNA (miR-21, miR-30e, miR-142, miR-148b, miR-29b and miR-223) measured in the cervix mid-pregnancy were associated with shorter subsequent gestations [20], three of which replicated previous investigators’ findings (miR-223, miR-30e and miR-148b) [19,21].
Extending this work, our hypothesis was that a higher abundance of organisms that can cause BV (analyzed as a mixture) as well as a mixture of cytokines would be associated with both altered miRNA expression and shorter gestation. We quantified the abundance of miRNAs, cytokine mRNA abundance, candidate organism rRNA in the second trimester, 4–5 months prior to delivery. Given the concurrent presence of multiple bacterial species as well as the correlated expression of cytokines, we selected a rigorous modeling approach, weighted quantile sum (WQS) regression, which accounts for correlation among the components of a mixture [22]. This approach reduces the dimensionality of complex correlated data into individual indices that can be used in regression models to predict gestational age at delivery and miRNA levels.
Materials & methods
Study design
We studied 80 Mexican women ages 18–40 years participating in the PROGRESS birth cohort in Mexico City [23]. These women provided written informed consent for an additional cervical swab during pregnancy thereby participating in the PROGRESS Cervix Study. Full details of enrollment for the parent cohort and subcohort are published elsewhere [20,24]. Briefly, women in their second trimester were recruited between 2007 and 2011 through the Mexican social security system (Instituto Mexicano del Seguro Social). The parent cohort consists of 1054 mothers enrolled in the second trimester. Because the PROGRESS Cervix Study was developed after the majority of recruitment had occurred, only the last 100 women enrolled in the parent cohort were approached. Eighty agreed to participate and provided written informed consent for an obstetrician to obtain a cervical swab mid-pregnancy (16–19 weeks gestation). The Institutional Review Boards of the participating institutions approved this study. All 80 samples were analyzed for cytokine mRNA expression and bacterial 16S rRNA profiling. For funding reasons, 60 (75%) of the 80 samples collected were randomly selected for miRNA profiling analysis.
Participant data collection
Demographics and birth outcomes were collected as part of the parent study, including maternal age, parity, second trimester BMI, household smoke exposure and years of education. No participants reported smoking during pregnancy. Gestational age was calculated in units of ‘weeks’ starting with the maternally reported last menstrual period to the date of delivery. The Capurro method (i.e., an infant physical exam) was used as a secondary confirmatory estimate of gestational age [25]. When the gestational age estimates differed by >3 weeks, the Capurro method-derived estimate replaced the last menstrual period estimate (n = 3) [20].
Cervical sample collection & extraction
Cervical cells were collected using a standard Pap smear protocol, wherein a cotton swab was used to collect cells from the endocervix just within external os. The sample was immediately placed in RNALater (Qiagen, CA, USA) and the specimen was frozen at -80°C until subsequent analysis. Total RNA was extracted using the Exiqon miRCURY kit (Exiqon, MA, USA) according to the manufacturer's protocol. A cleanup step was then performed using an Amicon Ultra 0.5 ml clean up kit (EMD Millipore, MA, USA). Total mRNA was quantified using a NanoPhotometer P-300 (Implent GmbH, CA, USA).
NanoString nCounter assay for mRNA & 16S rRNA expression
Bacterial 16S rRNA abundance and cytokine mRNA were quantified using the NanoString nCounter system (NanoString Technologies, WA, USA). This method enables multiplexed direct digital counting of 16S rRNA, mRNA and miRNA molecules [26]. Bacterial 16S rRNA was assessed simultaneously for the following organisms: Bacteroides spp., Gardnerella vaginalis, Mycoplasma spp., Ureaplasma urealyticum and Mobiluncus spp. Cytokines assessed on the gene expression panel included IL2, IL6, IL8 and TNF. The raw count data from the 80 samples were normalized using the NanoStringNorm R package [27]. The data were background-corrected using the mean plus two standard deviations of the negative controls included on the platform, and positive control normalized using the geometric mean of the positive controls to account for assay to assay variability. To account for variation in initial RNA concentrations, the data were normalized using two housekeeping genes with consistent expression in the cervical samples: HPRT1 and TUBB. After normalization, probes that were below the LOD were assigned an imputed value of the probe-specific minimum detected expression level divided by two.
NanoString nCounter assay for miRNA expression
miRNA expression was assessed using the NanoString nCounter system (NanoString Technologies). This platform measured a total of 800 probes that were available for analysis at the time of this study, and included both endogenous human-associated miRNAs as well as viral miRNAs that are expressed in human cells [28–30]. We performed a feasibility pilot with ten initial samples, including two technical replicates, which we previously reported showed strong correlation (r = 0.98) [20].
The raw count data from the 60 samples were normalized using the NanoStringNorm R package [27]. As previously reported, data were background-corrected by subtracting the mean of the six negative controls included on the platform, and normalized using the geometric mean of the ten probes with the lowest coefficients of variation – which were used to calculate a scaling factor as suggested by the package guidelines. A priori we required that probes be detectable in at least 60% of the samples. This resulted in 74 probes that were included in the analyses. Individual probes with expression levels below the LOD were assigned a value equal to the probe-specific LOD (minimum expression level measured for a specific miRNA). We report the proportion of samples below detect for each miRNA in the results. We also performed a sensitivity analyses that excluded probes below the LOD. The distributions of miRNA expression and model residuals showed that our selection of a linear model was appropriate for these data.
Bacterial/cytokine mixture WQS index
We created two indices to account for mixtures using the normalized expression of the five bacteria and four cytokines using WQS regression. To satisfy linear model assumptions, bacterial and cytokine expression levels were log2 transformed. Developed by our team, WQS regression [31,32] estimates a weighted sum (here, across microbe or cytokine expression levels) of quantiles most associated with a given health outcome and performs inference on the regression coefficient that characterizes the association between the outcome and this weighted sum. Details of the WQS model are presented in the Supplementary Material. We then applied the indices developed with respect to gestational age at delivery in a separate model with a different dependent variable (miRNA expression).
Statistical analysis
To examine the association between the bacterial and cytokine indices, and subsequent gestational age at delivery, we used linear regression models. To satisfy linear model assumptions, we applied the untransformed WQS index and gestational age (in completed weeks of gestation) since each was normally distributed. We chose covariates a priori and included maternal age and BMI during the second trimester, education, parity and smoke exposure inside the home. Both adjusted and unadjusted regression models were performed to examine the association between the WQS indices and subsequent gestational age at delivery. The reported β-coefficients represent the change in gestational age (weeks) per decile of the index. Estimates are converted to days per decile by multiplying the β by 7. We also examined the individual bacteria and cytokine levels and gestational age at delivery using models adjusted for the same covariates. The reported β-coefficients represent the change in gestational age (weeks) per fold change (doubling or halvings) in mRNA or 16S rRNA expression.
To examine the association between the WQS indices and miRNA expression, we used linear regression models. We used separate models to estimate the mean doubling of expression (log2 unit increase) of each miRNA associated with a decile increase in the WQS index. We included the same set of a priori covariates as above. Both adjusted and unadjusted regression models were performed to examine the association between the bacterial mixture index and the 74 probes’ log2-transformed miRNA expression levels. The reported β-coefficients represent the fold change (doubling or halvings) in miRNA expression per decile increment of the index. Because expression data are conventionally interpreted in log2 we present the β-coefficients that are easily interpreted in doublings or halvings. To transform from fold change into raw expression change, the β-coefficients can be back transformed using the antilog of 2. For example, the equation (2beta - 1) × 10 yields the percent change in raw miRNA expression per decile of the index. p-values and Storey's false discovery rate (FDR) q-values were calculated to estimate significance for the miRNAs to address multiple testing [33]; p < 0.05 was considered statistically significant. MiRNAs with an FDR q-value <0.1 that met these requirements in the adjusted model were retained for downstream target prediction and pathways enrichment analyses.
Prediction of miRNA targets, functional pathway & network enrichment analysis
To predict downstream mRNA targets, the set of miRNAs for each index, which passed p < 0.05 and FDR q-value <0.1 in the adjusted models, were uploaded into the Ingenuity Pathway Analysis (IPA) tool (Ingenuity® Systems, CA, USA). Putative miRNA–mRNA relationships were identified using the IPA microRNA Target Filter, based on a knowledgebase of predicted and experimentally observed relationships. We selected for either the experimentally observed or highly predicted miRNA–mRNA relationships, and the resulting target gene list was analyzed for functional network and pathway analysis. Details of functional pathway and network enrichment analyses are presented in the Supplementary Material.
Selected miRNA–mRNA relationships
To confirm miRNA–mRNA relationships, we applied the linear regression models, adjusting for an identical set of covariates as above. For each of the top miRNAs identified as associated with bacterial or cytokine indices, we used separate models to estimate the mean doubling of expression (log2 unit increase) of each miRNA associated with a fold change or doubling of individual bacterial/cytokine components of each index.
Results
Characteristics of the cervix study
The study demographics are presented in Table 1. Four women (5%) were lost to follow-up. The mean (standard deviation) gestational age at delivery of the remaining 76 women was 38 (1.3) weeks and ranged between 34 and 42 weeks. Four women (5%) delivered preterm at <37 weeks gestation. Descriptive statistics of the five candidate bacteria and four cytokines are shown in Table 2. Gardnerella vaginalis was detected in the majority of samples, followed by Bacteroides, U. urealyticum, Mobiluncus and Mycoplasma (detected in only six samples). The 16S rRNA counts of several species were significantly correlated (Supplementary Table 1).
Table 1. . Maternal demographics for 80 pregnant women participating in the PROGRESS cervix study.
| n (%) | |
|---|---|
|
Education | |
| <12 years |
22 (27.5) |
| ≥12 years |
58 (72.5) |
|
Smoke in home | |
| No |
57 (71) |
| Yes |
23 (29) |
|
Parity | |
| Multiparous |
50 (62.5) |
| Nulliparous |
30 (37.5) |
| |
Mean ± SD (range) |
|
Maternal age (years) |
28.0 ± 5.7 (18–41) |
|
Maternal BMI (kg/m2) |
26.8 ± 4.5 (18–43) |
| Gestational age at delivery (weeks)† | 38.4 ± 1.3 (34–42) |
†Four individuals were lost to follow-up at delivery and information on gestational age is not available.
SD: Standard deviation.
Table 2. . Descriptive statistics of the abundance of five candidate bacteria and four cytokines measured in the cervix during pregnancy.
| Bacterial species/genus | Samples with detected RNA n (%) | Average ± SD (range) Log2 expression count |
|---|---|---|
|
Bacteroides |
66 (83) |
7.0 ± 4.7 (0.01–18.1) |
|
Gardnerella vaginalis |
72 (90) |
14.3 ± 6.1 (6.2–25.2) |
|
Mobiluncus |
38 (48) |
8.2 ± 4.7 (0.5–22.7) |
|
Mycoplasma |
6 (8) |
3.7 ± 2.6 (0.5–7.7) |
|
Ureaplasma urealyticum |
66 (83) |
10.6 ± 5.2 (1.0–23.0) |
|
Cytokines | ||
|
IL-2 |
8 (10) |
2.2 ± 1.3 (0.01–3.5) |
|
IL-6 |
24 (30) |
4.8 ± 2.7 (0.4–10.7) |
|
IL-8 |
79 (99) |
15.8 ± 2.4 (10.8–22.0) |
| TNF | 74 (93) | 7.0 ± 2.5 (2.15–13.1) |
SD: Standard deviation.
Among the cytokines with measured mRNA expression, IL-8 was detected in 79 (99%) samples, followed by TNF in 74 (93%), and IL-6 in 24 (30%) samples. IL-2 was detected in just eight samples (10%). Examining the pairwise correlations, only TNF and IL-8 expressions were strongly correlated (r = 0.7, p < 0.0001) (Supplementary Table 1>). Notably, both TNF and IL-8 were also positively correlated with Bacteroides and G. vaginalis (p < 0.0001), whereas TNF, but not IL-8, was also positively correlated with Mobiluncus (r > 0.7; p < 0.0001) and U. urealyticum.
The relative weights derived from the WQS regression of the individual components in the bacterial and cytokine indices are shown in Table 3. The bacterial index gave higher weights to the contributions of U. urealyticum, G. vaginalis and Mycoplasma. The cytokine index gave higher weights to the contributions of TNF, IL-8 and IL-6. The bacterial and cytokine indices were correlated with one another with a Spearman's rank correlation of 0.60 (p < 0.0001).
Table 3. . Estimated weights for individual components of the weighted quantile sum bacterial and cytokine indices associated with gestational age at delivery (n = 76).
| Bacterial WQS index | |
|---|---|
|
Bacterial species/genus |
Weight |
|
Bacteroides |
0.05 |
|
Gardnerella vaginalis |
0.25 |
|
Mobiluncus |
<0.00001 |
|
Mycoplasma |
0.20 |
|
Ureaplasma urealyticum |
0.49 |
|
Cytokine WQS index | |
|
Cytokine |
Weight |
|
IL-2 |
0.12 |
|
IL-6 |
0.23 |
|
IL-8 |
0.33 |
| TNF | 0.33 |
WQS: Weighted quantile sum.
Bacterial & cytokine indices were both associated with shorter gestation
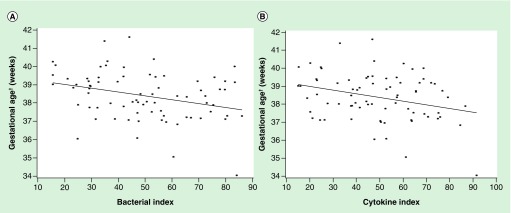
In adjusted models, the bacterial index was associated with shorter gestation (Figure 1A; Table 4). Specifically, per decile increment of the bacterial index gestations was 0.23 weeks (95% CI: 0.08, 0.4) (1.6 days) shorter. This estimate corresponds to an approximately 5.3-day gestational age difference when comparing the 25th to 75th percentiles (interquartile range) of the index. Compared with the individual bacteria, the bacterial index yielded the greatest statistical significance with respect to gestational age at delivery (Table 4). Unadjusted results were similar and are shown in Supplementary Material (Supplementary Table 2).
Figure 1. . Shorter gestational age is associated with each exposure index.
(A and B) Association of the bacterial and cytokine indices with gestational age at delivery (n = 76).
†Gestational age is adjusted for maternal age, BMI, education, smoke exposure and parity.
Table 4. . Adjusted linear regression estimates† (β-values and 95% CIs) per decile of the respective indices, or individual bacterium or cytokine expression with gestational age at delivery.
| WQS indices | β-value (95% CI) | p-value |
|---|---|---|
| WQS bacterial index |
-0.23 (-0.40 to -0.083) |
0.002 |
| WQS cytokine index |
-0.24 (-0.40 to -0.084) |
0.003 |
|
Bacterial species/genus | ||
|
Bacteroides |
-0.05 (-0.11–0.008) |
0.09 |
|
Gardnerella vaginalis |
-0.05 (-0.10 to -0.002) |
0.04 |
|
Mobiluncus |
-0.03 (-0.08–0.03) |
0.4 |
|
Mycoplasma |
-0.12 (-0.40–0.15) |
0.4 |
|
Ureaplasma urealyticum |
-0.07 (-0.12 to -0.02) |
0.004 |
|
Cytokine | ||
|
IL-2 |
-0.26 (-0.63–0.12) |
0.18 |
|
IL-6 |
-0.14 (-0.26 to -0.03) |
0.01 |
|
IL-8 |
-0.20 (-0.32 to -0.08) |
0.001 |
| TNF | -0.15 (-0.27–0.03) | 0.01 |
†βs derived from the individual regressions represent shorter gestation per log2 expression of 16S bacterial rRNA or cytokine mRNA whereas βs in the regressions using the WQS indices are per decile of the index.
WQS: Weighted quantile sum.
In adjusted models, the cytokine index was associated with shorter gestation (Figure 1B; Table 4). Specifically, per decile increase in the cytokine index gestations was 0.24 weeks (95% CI: 0.08–0.4) (1.7 days) shorter (p = 0.003). This estimate corresponds to an approximately 4.6-day gestational age difference when comparing the 25th to 75th percentiles (interquartile range) of the index. Compared with the individual cytokines, the cytokine index yielded the greatest statistical significance with respect to gestational age at delivery than all but one cytokine, IL-8 (Table 4). Unadjusted results were similar and are shown in Supplementary Material (Supplementary Table 3).
Bacterial index was associated with seven miRNAs
We identified seven miRNAs including miRs 494, 371a, 4286, 185, 888, 320e and 23a (p < 0.05) that were significantly associated with the bacterial index (Table 5). Notably, miRs 494, 185 and 320e were negatively associated with the bacterial index. Meaning that, for example, per decile increment of the bacterial index there was a 0.35 decrease (0.35 halvings or 22% decrease in raw expression) in miR-494 expression (Supplementary Figure 1A). Whereas miRs 371a, 4286, 888 and 23a were positively associated with the index.
Table 5. . Seven miRNAs associated with the bacterial mixture index (p < 0.05).
| miRNA | Samples with miRNA detected n (%) | β-value (95% CI) | p-value | FDR q-value |
|---|---|---|---|---|
| miR-494 |
56 (100) |
-0.35 (-0.57 to -0.14) |
0.001 |
0.1 |
| miR-720† |
56 (100) |
-0.25 (-0.44 to -0.06) |
0.01 |
0.3 |
| miR-371a-3p |
56 (100) |
0.10 (0.02–0.18) |
0.01 |
0.3 |
| miR-4286 |
50 (89) |
0.34 (0.04–0.63) |
0.03 |
0.4 |
| miR-185-5p |
55 (98) |
-0.22 (-0.42 to -0.01) |
0.04 |
0.4 |
| miR-320e |
55 (98) |
-0.19 (-0.38 to -0.00) |
0.04 |
0.4 |
| miR-888-5p |
56 (100) |
0.11 (0.00–0.22) |
0.04 |
0.4 |
| miR-23a-3p | 53 (95) | 0.22 (0.00–0.43) | 0.048 | 0.5 |
†miR-720 is no longer annotated as a miRNA
FDR: False discovery rate.
Bacteria-associated miRNA (miR-494) has known & predicted mRNA targets
Only miR-494 passed a stringent FDR correction (q-value < 0.1), and was retained for subsequent mRNA target and pathway analyses. MiRNA target analysis showed that miR-494 had six experimentally observed mRNA targets including FGF16, HMOX1, KIT, PTEN, SCN3A and VSNL1, as well as 576 highly predicted targets (data not shown; PTEN is annotated as both experimentally observed and highly predicted). Pathway analysis showed that the 581 experimentally observed and highly predicted downstream mRNA targets of miR-494 were enriched for canonical pathways including molecular mechanisms of cancer (p = 9.3 × 10-6); and AMP-activated protein kinase signaling (p = 3.9 × 10-6). Interestingly, miR-494 is downregulated in tissue from cervical squamous cell carcinoma [34].
Cytokine index was associated with 27 miRNAs
We identified 27 miRNAs including miRs 494, 142, 223, 15a, 25, 23a, 4454, 93, 193b, 15b, 16, 21, 30e, 29b, 1234, 514b, 19b, 125b, 222, 148a, 374a, 191, 29a, 148b, 548aa, let-7g and 1277 (p < 0.05; q < 0.1) that were significantly associated with the WQS cytokine index (Table 6). Once again, miR-494 was negatively associated with the cytokine index (Supplementary Figure 1B), whereas miR-142 and miR-223 among others were positively associated with the index. Pathway analysis of the 27 miRNAs identified two significantly associated networks of miRNAs: cancer, organismal injury and abnormalities, and reproductive system disease (p = 1 × 10-30); and inflammatory response, endocrine system disorders and gastrointestinal disease (p = 1 × 10-13). Interestingly, the most significantly associated disease was early-stage invasive cervical cancer (overlap p = 1.6 × 10-17), wherein nine cytokine index-associated miRNAs share sequence homology with miRNAs previously altered in cervical carcinoma tissue (miRs-142, 19b, 92a, 21, 16, 17, 29b, 30c and let-7a) [35].
Table 6. . Twenty-seven miRNAs associated with the cytokine mixture (p < 0.05).
| miRNA | Samples with miRNA detected n (%) | β-value (95% CI) | p-value | FDR q-value |
|---|---|---|---|---|
| miR-494 |
56 (100) |
-0.48 (-0.68 to -0.29) |
1.3 × 10-6 |
5.7 × 10-5 |
| miR-142-3p |
55 (98) |
0.60 (0.34–0.86) |
6.6 × 10-6 |
0.0001 |
| miR-223-3p |
55 (98) |
0.65 (0.36–0.93) |
7.6 × 10-6 |
0.0001 |
| miR-720† |
56 (100) |
-0.39 (-0.56 to -0.21) |
1.2 × 10-5 |
0.0001 |
| miR-15a-5p |
52 (93) |
0.55 (0.29–0.80) |
2.3 × 10-5 |
0.0001 |
| miR-25-3p |
56 (100) |
0.22 (0.10–0.34) |
0.0005 |
0.003 |
| miR-23a-3p |
53 (95) |
0.35 (0.15–0.55) |
0.0005 |
0.003 |
| miR-4454 |
56 (100) |
-0.44 (-0.71 to -0.16) |
0.002 |
0.008 |
| miR-93-5p |
51 (91) |
0.52 (0.19–0.85) |
0.002 |
0.008 |
| miR-193b-3p |
48 (86) |
-0.63 (-1.03 to -0.23) |
0.002 |
0.008 |
| miR-15b-5p |
52 (93) |
0.41 (0.14–0.67) |
0.003 |
0.009 |
| miR-16-5p |
56 (100) |
0.46 (0.16–0.76) |
0.003 |
0.009 |
| miR-21-5p |
46 (82) |
0.47 (0.16–0.79) |
0.003 |
0.009 |
| miR-30e-5p |
49 (88) |
0.36 (0.12–0.61) |
0.003 |
0.009 |
| miR-29b-3p |
51 (91) |
0.42 (0.14–0.7) |
0.003 |
0.009 |
| miR-1234 |
47 (84) |
-0.35 (-0.60 to -0.10) |
0.005 |
0.01 |
| miR-514b-5p |
56 (100) |
-0.12 (-0.20 to -0.03) |
0.006 |
0.02 |
| miR-19b-3p |
42 (75) |
0.44 (0.11–0.77) |
0.009 |
0.02 |
| miR-125b-5p |
51 (91) |
-0.21 (-0.37 to -0.05) |
0.009 |
0.02 |
| miR-222-3p |
56 (100) |
0.10 (0.03–0.18) |
0.009 |
0.02 |
| miR-148a-3p |
48 (86) |
0.46 (0.08–0.84) |
0.02 |
0.04 |
| miR-374a-5p |
45 (80) |
0.45 (0.08–0.82) |
0.02 |
0.04 |
| miR-191-5p |
55 (98) |
0.35 (0.05–0.66) |
0.02 |
0.04 |
| miR-29a-3p |
53 (95) |
0.34 (0.04–0.65) |
0.03 |
0.05 |
| miR-148b-3p |
34 (61) |
0.24 (0.02–0.45) |
0.03 |
0.05 |
| miR-548aa |
44 (79) |
-0.38 (-0.74 to -0.03) |
0.03 |
0.05 |
| let-7g-5p |
53 (95) |
0.30 (0.02–0.58) |
0.03 |
0.05 |
| miR-1277-3p | 36 (64) | -0.22 (-0.43 to -0.01) | 0.04 | 0.06 |
†miR-720 is no longer annotated as a miRNA.
FDR: False discovery rate.
Cytokine-associated miRNAs have known mRNA targets
Together, the cytokine-associated miRNAs targeted 7190 highly predicted or experimentally observed mRNAs. When restricted to only experimentally observed miRNA–mRNA relationships, we found that 17 miRNAs target 653 mRNAs. Notably, among the experimentally observed relationships annotated in IPA, miR-93 and miR-21 are known to target TNF, and miR-191 is known to target IL-6. Pathway analysis of the 653 predicted mRNA targets identified several significant networks of related molecules (Supplementary Table 4). The top three networks included neurological disease, cardiovascular diseases and organismal injury and abnormalities (p = 1 × 10-44); cell death and survival, cell cycle and cancer (p = 1 × 10-44); and cell death and survival, cellular growth and proliferation and hereditary disorder (p = 1 × 10-37).
Selected miRNA–mRNA relationships
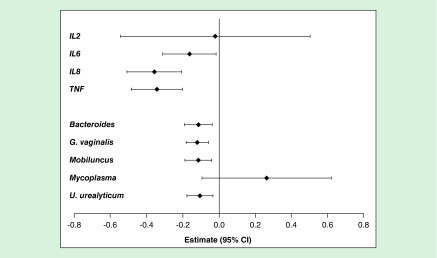
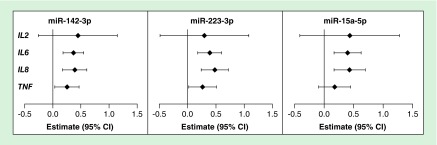
Last, given the direct link between miRNA regulation of mRNA expression, we examined the adjusted regressions of the nine candidate microbes or cytokines with the most differentially expressed miRNA: miR-494 (Figure 2; Supplementary Table 5). MiR-494 was negatively associated with seven of the candidate cytokines/organisms: IL-6, IL-8, TNF, Bacteroides, U. urealyticum, G. vaginalis and Mobiluncus. The miRNA–mRNA relationship with the strongest magnitude of association was miR-494 with IL-8. Wherein, per doubling of miR-494 expression there was a 0.36 decrease (0.36 halvings or 22% decrease in raw expression) in IL-8 expression. We examined the levels of individual cytokines with the top three other identified miRNAs, miR-142, 223 and 15a (Figure 3; Supplementary Table 6). IL-8 had the greatest magnitude and was positively associated with all three miRNAs.
Figure 2. . Estimates of association of individual bacteria and cytokines with miR-494, adjusted for maternal age, BMI, education, smoke exposure and parity.
Estimates (95% CIs) derived from the individual regressions represent the fold change in miR-494 expression per doubling of bacterial 16S rRNA or cytokine mRNA expression.
Figure 3. . Estimates of association of individual cytokines with miR-142, 223 and 15a, adjusted for maternal age, BMI, education, smoke exposure and parity.
Estimates (95% CIs) derived from the individual regressions represent the fold change in miRNA expression per doubling of cytokine mRNA expression.
Discussion
We found that a mixture of bacteria and a mixture of cytokines obtained from the cervix between 16 and 19 weeks of gestation were both associated with lower gestational age at delivery and with the expression of several miRNAs. Four of these miRNAs (miR-223, miR-30e, miR-148b and miR-93) we and others have shown to be predictive of gestational age at delivery or preterm birth [19,20]. Additionally, miR-223 is associated with spontaneous term labor [21]. Identifying new predictors of preterm birth, timing of delivery and potential molecular mediators between well-established risk factors for preterm birth is critical to moving the field of preterm birth prevention forward.
Our study builds on previous investigations demonstrating that both BV [10] and proinflammatory cytokines [13] predict shorter gestations. Our study demonstrates that differential abundance of microorganisms detected using molecular techniques, may be associated with the length of gestation. Culture-based studies have demonstrated that women heavily colonized with anaerobic bacteria are at higher risk of delivering preterm [36]. However, few studies have used sequence-based techniques to evaluate the role of colonization of the genitourinary tract and preterm birth. Romero et al. recently reported in a nested case–control study of 18 spontaneous preterm cases (<34 weeks of gestation) and 72 term controls that maternal vaginal microbiota during pregnancy did not differ with respect to taxa or relative abundance of bacterial communities [37]. Our study differs from this one in several respects. First, we used a targeted, hypothesis-driven approach to ascertain the burden of known pathogenic bacteria and inflammatory cytokines selected a priori. We selected five of the most common organisms implicated in BV [7]. We analyzed proinflammatory cytokines previously studied with respect to BV [38–40], although we recognize that other proinflammatory cytokines may play a role. The cytokines we measured are not tissue specific and although we sampled the cervix may have come from vaginal or vascular sources. We did not ascertain the abundance of potentially beneficial commensal organisms such as lactobacillus, which would be an important future direction for understanding the regulation of both cytokine and miRNA expression. Last, given the low prevalence of participants who delivered preterm (5%), we did not examine spontaneous preterm birth in a case–control study design. Instead we analyzed gestational age at delivery as a continuous outcome, a study design which benefits from improved power. Further, it should be noted that all of our participants were ≥34 weeks of gestation.
One of the most common culture-based BV classification methods is the Nugent score, a Gram stain-based arithmetic equation for which BV classified as ‘BV-positive’, ‘intermediate’ or ‘BV-negative’ [41]. In our study we did not quantify BV clinically but measured 16S rRNA for organisms known to be associated with BV. Moreover, our study was prospective, meaning that the bacteria/cytokine measurement was conducted in the early second trimester, 4–5 months before labor. This suggests that the bacterial flora present early in pregnancy may create low-grade inflammation that increases the probability of early labor induction. Because we used a mixtures statistical approach, we did not make a priori hypotheses that individual bacterial species predict earlier labor, but instead treated the overall flora as the predictor. We believe this is a major strength over approaches that treat each species independently. We developed our indices using WQS regression to create potentially informative composite measures of organisms and cytokines. WQS regression applies weights to the individual components of the mixtures according to the strength of their association with shorter gestations. Thus, our findings that such indices were associated with shorter gestations were not surprising given that many of the individual components were associated with shorter gestations. The findings indicate that among the individual components of the two mixtures, U. urealyticum and IL-8 were most strongly associated with shorter gestations. Note that the effect estimates for bacterial 16S rRNA quantification and cytokine mRNA expression cannot be compared directly. Despite being the current gold standard for organism identification, sequence-based detection of 16S rRNA cannot differentiate between viable and unviable organisms nor can it account for organisms with multiple copies of the 16S rRNA gene [42]. Nevertheless, compared with the clinical standard for diagnosis of BV, the Nugent Gram strain-based method, which is in fact an arithmetically derived index of mixed bacterial morphotypes, our use of 16S rRNA quantification in this study is appropriate given our novel statistical approach. Despite the reliability and specificity of the Nugent score but moderate sensitivity [43], new methods propose to improve clinical diagnosis of BV by including real-time quantitative PCR-based techniques [44,45]. Our statistical approach herein is novel in that we then used those bacterial and cytokine indices as independent variables to model associations with miRNAs in the cervix. In doing so, we identified miRNAs that might be important in elucidating mechanisms by which vaginal dysbiosis and inflammation could lead to preterm birth.
Most notably, miR-494 was the top-identified miRNA for both the bacterial and cytokine indices. Previous evidence shows that miR-494 is directly upregulated by TNF, and both molecules contribute to apoptosis signaling [46,47]. In vitro evidence that antagomir-494 can prevent TNF-induced apoptosis [46] suggests that miR-494 may be a promising therapeutic target in the inflammatory cascade that leads to preterm birth. Additionally, we found several cytokine index-associated miRNAs that have been shown to have pathophysiologic roles in prior studies. First, miR-15a/16 regulates macrophage phagocytosis after bacterial infection [48]. Second, inflammation triggers specific miRNA profiles in human macrophages and supernatants [49]. miR-223 regulates macrophages/inflammatory response by suppressing proinflammatory activation of macrophages [50].
Our study has several strengths. We used a novel statistical approach to address correlated mixtures of exposures of bacteria, cytokines and miRNA. We also analyzed miRNA from a target tissue critical to labor (the cervix), whereas many epigenetic studies use surrogate tissues such as blood or buccal swabs and thus may have limited relevance to the health outcome of interest. Our study also has several limitations. Our study was nested in a larger cohort designed to answer questions about metal exposures and neurodevelopment. We had no data to ascertain whether a woman had clinical BV during her pregnancy, nor did we have data on antibiotic use during pregnancy. We did not have ultrasound measures of gestational age, nor detailed data as to why a woman delivered early. Ideally WQS techniques are developed in one cohort and applied to another. Our sample size was too small to divide our data and there are no current analogous cohorts to replicate our findings. However, we did apply our weighted indices to individual-dependent variables (miRNA) after their derivation based on gestational age at delivery, implying their potential utility as composite biomarkers. We did not have enough RNA to analyze miRNAs using a second platform and thus our findings may only be generalizable to other studies that would use the NanoString platform. Additionally, although we performed technical replicates for the miRNA [20], we did not analyze technical replicates for the bacteria or cytokine RNA due to limited RNA yields. Last, due to the simultaneous collection of all three types of RNA, it is not possible to discern whether bacterial RNA induced changes to cytokine RNA and miRNA or whether miRNA alters a woman's susceptibility to colonization. Longitudinal sampling would be required to determine the order of events.
A paradox in the field of preterm birth prevention has been the tight correlation between BV and preterm birth but the failure of BV treatment to prevent preterm birth. While antibiotic therapy successfully treats BV, in several large randomized trials no reduction in preterm birth risk was observed among women treated in the second or third trimesters versus untreated [6,11]. Preterm delivery was reduced only among a subgroup of women with a previous preterm birth who had a clinical diagnosis of BV in the current pregnancy. Several potential explanations exist, one is that removing the organisms does not interrupt the cascade of pathophysiologic events either associated with or caused by BV. Another is that BV itself is not a causative agent, but is merely the marker of a different upstream cause that coincidentally induces BV. For example, the microbiome would be affected by alterations in stress level, diet or metabolism, which may induce altered flora, and could predispose to BV [51]. If the altered factor or dietary constituent is the true cause of prematurity, treating BV would not reduce risk. We know that antibiotics do not prevent preterm labor or premature rupture of membranes that lead to preterm birth, despite a significant decline in Nugent score [52]. Other explanations as to why treating BV does not prevent preterm birth are possible. One is that some other factor that is highly associated with BV causes preterm birth (confounding). The second is that BV causes changes to the local genitourinary tract that persist after treatment of BV. In other words, there is a set of events that leads to preterm birth that are immutable once antibiotic therapy is started, which would explain why antibiotics do not interrupt this cascade. Our data support considering miRNA expression as one potential mediator between a local environmental factor (BV) and earlier delivery. Our analysis of tissue-specific epigenetic signals (cervical miRNA) supports a biologically plausible possibility that traditional risk factors, in this case BV, may lead to earlier delivery through local tissue programming. Future work exploring epigenetic factors that result from other well-known preterm risk factors including smoking or prior spontaneous preterm birth is warranted. The discovery of new biomarkers of preterm birth risk is critical to preterm birth prevention because of the potential for better targeting of prevention strategies such as progesterone or cerclage and management of high-risk patients to connect them to tertiary delivery centers. New biomarkers can also shed light on potential new therapeutic targets that could potentially interrupt the cellular cascade that leads to preterm birth.
Conclusion
Our findings suggest that a mixture of cervical bacteria and cytokines in pregnancy are associated with shorter gestation and miRNA expression. Our work suggests that investigation into the role of miRNA in mediating bacterial/inflammation-associated preterm birth is warranted. Such discoveries could lead to interventions targeting the molecular cascade that results in preterm birth.
Executive summary.
Whether bacterial vaginosis (BV) may lead to shorter gestations through epigenetic programming of the inflammatory response, specifically via miRNA expression, is unknown.
We quantified bacterial 16S rRNA, cytokine mRNA and 800 miRNA from cervical swabs obtained from 80 women at 16–19 weeks’ gestation using the Nanostring nCounter Analysis System.
We then developed an index to analyze the mixture of five microbes that contribute to BV using a weighted quantile sum approach. We also created an inflammatory cytokine index to analyze the mixture of mRNA expression of IL-2, IL-6, IL-8 and TNF.
Using linear regression models, we examined associations of these indices with each miRNA and with gestational age at delivery. Models were adjusted for maternal age and BMI, education, parity and smoke exposure inside the home.
Each decile of the bacterial and cytokine indices was associated with shorter gestations (p < 0.005). The bacterial index was associated with miR-494, 371a, 4286, 185, 320e, 888 and 23a (p < 0.05). miR-494 remained significant after false discovery rate correction (q < 0.1). The cytokine index was associated with 27 miRNAs (p < 0.05; q < 0.01).
A mixture of bacteria and a mixture of cytokines obtained from the cervix between 16 and 19 weeks of gestation were both associated with lower gestational age at delivery and with the expression of several miRNAs.
Future investigation into the role of BV- and inflammation-associated miRNA and preterm birth is warranted.
Supplementary Material
Acknowledgements
The authors thank E Reuckert at NanoString Technologies for his assistance with the miRNA/RNA expression profiling and analyses. They thank the ABC Medical Center in Mexico City for providing facilities during data collection.
Footnotes
Financial and competing interests disclosures
This work was supported in part by Pilot Project funding from the HSPH-NIEHS Center for Environmental Health (ES000002) and NIH/NIEHS: K23ES022242, P30ES23515, R01ES013744, R01ES014930, R01ES020268, R01ES021357, the Klarman Scholars Program at Beth Israel Deaconess Medical Center, the Harvard Catalyst D-MaPS Program and the National Institute of Public Health/Ministry of Health of Mexico. The funding organizations had no role in study design, data collection, analysis or interpretation, or preparation of the manuscript for publication. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Ethical conduct of research
The International Review Boards of the participating institutions approved this study: Brigham and Women's Hospital #2006-P-001416 and P001792, Icahn School of Medicine at Mount Sinai human subjects management #12-00751 and Instituto Nacional de Salud Publica project #560. Written informed consent was obtained from women participating in the PROGRESS study.
References
Papers of special note have been highlighted as: • of interest; •• of considerable interest
- 1.Blencowe H, Cousens S, Oestergaard MZ, et al. National, regional, and worldwide estimates of preterm birth rates in the year 2010 with time trends since 1990 for selected countries: a systematic analysis and implications. Lancet. 2012;379(9832):2162–2172. doi: 10.1016/S0140-6736(12)60820-4. [DOI] [PubMed] [Google Scholar]
- 2.Behrman RE, Butler AS Institute of Medicine (US) Preterm Birth: Causes, Consequences, and Prevention. National Academies Press; Washington, DC, USA: 2007. Committee on understanding premature birth and assuring healthy outcomes. [PubMed] [Google Scholar]
- 3.Dodd JM, Jones L, Flenady V, Cincotta R, Crowther CA. Prenatal administration of progesterone for preventing preterm birth in women considered to be at risk of preterm birth. Cochrane Database Syst. Rev. 2013;7 doi: 10.1002/14651858.CD004947.pub3. CD004947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Abdel-Aleem H, Shaaban OM, Abdel-Aleem MA. Cervical pessary for preventing preterm birth. Cochrane Database Syst. Rev. 2013;5 doi: 10.1002/14651858.CD007873.pub3. CD007873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alfirevic Z, Stampalija T, Roberts D, Jorgensen AL. Cervical stitch (cerclage) for preventing preterm birth in singleton pregnancy. Cochrane Database Syst. Rev. 2012;4 doi: 10.1002/14651858.CD008991.pub2. CD008991. [DOI] [PubMed] [Google Scholar]
- 6.Brocklehurst P, Hannah M, Mcdonald H. Interventions for treating bacterial vaginosis in pregnancy. Cochrane Database Syst Rev. 2000;2 doi: 10.1002/14651858.CD000262. CD000262. [DOI] [PubMed] [Google Scholar]
- 7.CDC. Bacterial vaginosis – CDC fact sheet. 2015. https://www.cdc.gov/std/bv/bv-fact-sheet-march-2014.pdf
- 8.Catlin BW. Gardnerella vaginalis: characteristics, clinical considerations, and controversies. Clin. Microbiol. Rev. 1992;5(3):213–237. doi: 10.1128/cmr.5.3.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kimberlin DF, Andrews WW. Bacterial vaginosis: association with adverse pregnancy outcome. Semin. Perinatol. 1998;22(4):242–250. doi: 10.1016/s0146-0005(98)80012-8. [DOI] [PubMed] [Google Scholar]
- 10.Hillier SL, Nugent RP, Eschenbach DA, et al. Association between bacterial vaginosis and preterm delivery of a low-birth-weight infant. The vaginal infections and prematurity study group. N. Engl. J. Med. 1995;333(26):1737–1742. doi: 10.1056/NEJM199512283332604. [DOI] [PubMed] [Google Scholar]
- 11.Thinkhamrop J, Hofmeyr GJ, Adetoro O, Lumbiganon P, Ota E. Antibiotic prophylaxis during the second and third trimester to reduce adverse pregnancy outcomes and morbidity. Cochrane Database Syst. Rev. 2015;6 doi: 10.1002/14651858.CD002250.pub3. CD002250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Platz-Christensen JJ, Mattsby-Baltzer I, Thomsen P, Wiqvist N. Endotoxin and interleukin-1 alpha in the cervical mucus and vaginal fluid of pregnant women with bacterial vaginosis. Am. J. Obstet. Gynecol. 1993;169(5):1161–1166. doi: 10.1016/0002-9378(93)90274-m. [DOI] [PubMed] [Google Scholar]
- 13.Taylor BD, Holzman CB, Fichorova RN, et al. Inflammation biomarkers in vaginal fluid and preterm delivery. Hum. Reprod. 2013;28(4):942–952. doi: 10.1093/humrep/det019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ferguson KK, Mcelrath TF, Chen YH, Mukherjee B, Meeker JD. Longitudinal profiling of inflammatory cytokines and C-reactive protein during uncomplicated and preterm pregnancy. Am. J. Reprod. Immunol. 2014;72(3):326–336. doi: 10.1111/aji.12265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mcelrath TF, Hecht JL, Dammann O, et al. Pregnancy disorders that lead to delivery before the 28th week of gestation: an epidemiologic approach to classification. Am. J. Epidemiol. 2008;168(9):980–989. doi: 10.1093/aje/kwn202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]; • Identifies the role of miR-200 in regulating genes involved in uterine quiescence and contractility.
- 17.Pauley KM, Chan EK. MicroRNAs and their emerging roles in immunology. Ann. N. Y. Acad. Sci. 2008:226–239. doi: 10.1196/annals.1443.009. 1143. [DOI] [PubMed] [Google Scholar]
- 18.Renthal NE, Chen CC, Williams KC, Gerard RD, Prange-Kiel J, Mendelson CR. miR-200 family and targets, ZEB1 and ZEB2, modulate uterine quiescence and contractility during pregnancy and labor. Proc. Natl Acad. Sci. USA. 2010;107(48):20828–20833. doi: 10.1073/pnas.1008301107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Elovitz MA, Brown AG, Anton L, Gilstrop M, Heiser L, Bastek J. Distinct cervical microRNA profiles are present in women destined to have a preterm birth. Am. J. Obstet. Gynecol. 2014;210(3):e221–e211. doi: 10.1016/j.ajog.2013.12.043. 221. [DOI] [PubMed] [Google Scholar]
- 20.Sanders AP, Burris HH, Just AC, et al. microRNA expression in the cervix during pregnancy is associated with length of gestation. Epigenetics. 2015;10(3):221–228. doi: 10.1080/15592294.2015.1006498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hassan SS, Romero R, Pineles B, et al. MicroRNA expression profiling of the human uterine cervix after term labor and delivery. Am. J. Obstet. Gynecol. 2010 doi: 10.1016/j.ajog.2009.08.016. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 22.Carrico C, Gennings C, Wheeler DC, Factor-Litvak P. Characterization of weighted quantile sum regression for highly correlated data in a risk analysis setting. J. Agric. Biol. Environ. Stat. 2014;20(1):100–120. doi: 10.1007/s13253-014-0180-3. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Profiled miRNAs, and notably miR-223, in the cervix following term delivery and pregnancy.
- 23.Burris HH, Baccarelli AA, Motta V, et al. Association between length of gestation and cervical DNA methylation of PTGER2 and LINE 1-HS. Epigenetics. 2014;9(8):1083–1091. doi: 10.4161/epi.29170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Burris HH, Braun JM, Byun HM, et al. Association between birth weight and DNA methylation of IGF2, glucocorticoid receptor and repetitive elements LINE-1 and Alu. Epigenomics. 2013;5(3):271–281. doi: 10.2217/epi.13.24. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• The previous case–control study of preterm and term pregnancies quantified 16S rRNA abundance from vaginal microbial species.
- 25.Capurro H, Konichezky S, Fonseca D, Caldeyro-Barcia R. A simplified method for diagnosis of gestational age in the newborn infant. J. Pediatr. 1978;93(1):120–122. doi: 10.1016/s0022-3476(78)80621-0. [DOI] [PubMed] [Google Scholar]
- 26.Geiss GK, Bumgarner RE, Birditt B, et al. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat. Biotechnol. 2008;26(3):317–325. doi: 10.1038/nbt1385. [DOI] [PubMed] [Google Scholar]
- 27.Waggott D, Chu K, Yin S, Wouters BG, Liu FF, Boutros PC. NanoStringNorm: an extensible R package for the pre-processing of NanoString mRNA and miRNA data. Bioinformatics. 2012;28(11):1546–1548. doi: 10.1093/bioinformatics/bts188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stern-Ginossar N, Elefant N, Zimmermann A, et al. Host immune system gene targeting by a viral miRNA. Science. 2007;317(5836):376–381. doi: 10.1126/science.1140956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cullen BR. Five questions about viruses and microRNAs. PLoS Pathog. 2010;6(2):e1000787. doi: 10.1371/journal.ppat.1000787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grundhoff A, Sullivan CS. Virus-encoded microRNAs. Virology. 2011;411(2):325–343. doi: 10.1016/j.virol.2011.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carrico C. Characterization of a weighted quantile score approach for highly correlated data in risk analysis scenarios [PhD thesis] Department of Biostatistics, Virginia Commonwealth University; VA, USA: 2013. [Google Scholar]
- 32.Gennings C, Carrico C, Factor-Litvak P, Krigbaum N, Cirillo PM, Cohn BA. A cohort study evaluation of maternal PCB exposure related to time to pregnancy in daughters. Environ. Health. 2013;12(1):66. doi: 10.1186/1476-069X-12-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc. Natl Acad. Sci. USA. 2003;100(16):9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang Y, Dai Y, Huang Y, et al. Microarray profile of micro-ribonucleic acid in tumor tissue from cervical squamous cell carcinoma without human papillomavirus. J. Obstet. Gynaecol. Res. 2009;35(5):842–849. doi: 10.1111/j.1447-0756.2009.01055.x. [DOI] [PubMed] [Google Scholar]
- 35.Lee JW, Choi CH, Choi JJ, et al. Altered microRNA expression in cervical carcinomas. Clin. Cancer Res. 2008;14(9):2535–2542. doi: 10.1158/1078-0432.CCR-07-1231. [DOI] [PubMed] [Google Scholar]
- 36.Donders GG, Van Calsteren K, Bellen G, et al. Predictive value for preterm birth of abnormal vaginal flora, bacterial vaginosis and aerobic vaginitis during the first trimester of pregnancy. BJOG. 2009;116(10):1315–1324. doi: 10.1111/j.1471-0528.2009.02237.x. [DOI] [PubMed] [Google Scholar]; • Identified a number of Hg- and Pb-associated miRNAs measured in the cervix during pregnancy.
- 37.Romero R, Hassan SS, Gajer P, et al. The vaginal microbiota of pregnant women who subsequently have spontaneous preterm labor and delivery and those with a normal delivery at term. Microbiome. 2014;2:18. doi: 10.1186/2049-2618-2-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Beigi RH, Yudin MH, Cosentino L, Meyn LA, Hillier SL. Cytokines, pregnancy, and bacterial vaginosis: comparison of levels of cervical cytokines in pregnant and nonpregnant women with bacterial vaginosis. J. Infect. Dis. 2007;196(9):1355–1360. doi: 10.1086/521628. [DOI] [PubMed] [Google Scholar]
- 39.Thurman AR, Kimble T, Herold B, et al. Bacterial vaginosis and subclinical markers of genital tract inflammation and mucosal immunity. AIDS Res. Hum. Retroviruses. 2015;31(11):1139–1152. doi: 10.1089/aid.2015.0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fan SR, Liu XP, Liao QP. Human defensins and cytokines in vaginal lavage fluid of women with bacterial vaginosis. Int. J. Gynaecol. Obstet. 2008;103(1):50–54. doi: 10.1016/j.ijgo.2008.05.020. [DOI] [PubMed] [Google Scholar]
- 41.Nugent RP, Krohn MA, Hillier SL. Reliability of diagnosing bacterial vaginosis is improved by a standardized method of gram stain interpretation. J. Clin. Microbiol. 1991;29(2):297–301. doi: 10.1128/jcm.29.2.297-301.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kang YJ, Cheng J, Mei LJ, Hu J, Piao Z, Yin SX. Multiple copies of 16s rRNA gene affect the restriction patterns and DGGE profile as revealed by analysis of genome database. Mikrobiologiia. 2010;79(5):664–671. [PubMed] [Google Scholar]
- 43.Chaijareenont K, Sirimai K, Boriboonhirunsarn D, Kiriwat O. Accuracy of Nugent's score and each Amsel's criteria in the diagnosis of bacterial vaginosis. J. Med. Assoc. Thai. 2004;87(11):1270–1274. [PubMed] [Google Scholar]
- 44.Rodrigues FS, Peixoto S, Adami F, et al. Proposal of a new cutoff for Nugent criteria in the diagnosis of bacterial vaginosis. J. Microbiol. Methods. 2015;115:144–146. doi: 10.1016/j.mimet.2015.05.006. [DOI] [PubMed] [Google Scholar]
- 45.Cox C, Mckenna JP, Watt AP, Coyle PV. New assay for Gardnerella vaginalis loads correlates with Nugent scores and has potential in the diagnosis of bacterial vaginosis. J. Med. Microbiol. 2015;64(9):978–984. doi: 10.1099/jmm.0.000118. [DOI] [PubMed] [Google Scholar]
- 46.Wang T, Li P, Ma X, et al. MicroRNA-494 inhibition protects nucleus pulposus cells from TNF-alpha-induced apoptosis by targeting JunD. Biochimie. 2015;115:1–7. doi: 10.1016/j.biochi.2015.04.011. [DOI] [PubMed] [Google Scholar]
- 47.Lee H, Jee Y, Hong K, Hwang GS, Chun KH. MicroRNA-494, upregulated by tumor necrosis factor-alpha, desensitizes insulin effect in C2C12 muscle cells. PLoS ONE. 2013;8(12):e83471. doi: 10.1371/journal.pone.0083471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Moon HG, Yang J, Zheng Y, Jin Y. miR-15a/16 regulates macrophage phagocytosis after bacterial infection. J. Immunol. 2014;193(9):4558–4567. doi: 10.4049/jimmunol.1401372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ortega FJ, Moreno M, Mercader JM, et al. Inflammation triggers specific microRNA profiles in human adipocytes and macrophages and in their supernatants. Clin. Epigenetics. 2015;7(1):49. doi: 10.1186/s13148-015-0083-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhuang G, Meng C, Guo X, et al. A novel regulator of macrophage activation: miR-223 in obesity-associated adipose tissue inflammation. Circulation. 2012;125(23):2892–2903. doi: 10.1161/CIRCULATIONAHA.111.087817. [DOI] [PubMed] [Google Scholar]
- 51.Culhane JF, Rauh V, Mccollum KF, Hogan VK, Agnew K, Wadhwa PD. Maternal stress is associated with bacterial vaginosis in human pregnancy. Matern. Child Health J. 2001;5(2):127–134. doi: 10.1023/a:1011305300690. [DOI] [PubMed] [Google Scholar]
- 52.Anderson B, Zhao Y, Andrews WW, et al. Effect of antibiotic exposure on Nugent score among pregnant women with and without bacterial vaginosis. Obstet. Gynecol. 2011;117(4):844–849. doi: 10.1097/AOG.0b013e318209dd57. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.