Abstract
The molecular mechanisms underlying the aging process have garnered much attention in recent decades because aging is the most significant risk factor for many chronic diseases such as type 2 diabetes and cancer. Until recently, the aging process was not considered to be an actively regulated process; therefore, discovering that the insulin/insulin-like growth factor-1 signaling pathway is a lifespan-regulating genetic pathway in Caenorhabditis elegans was a major breakthrough that changed our understanding of the aging process. Currently, it is thought that animal lifespans are influenced by genetic and environmental factors. The genes involved in lifespan regulation are often associated with major signaling pathways that link the rate of aging to environmental factors. Although many of the major mechanisms governing the aging process have been identified from studies in short-lived model organisms such as yeasts, worms and flies, the same mechanisms are frequently observed in mammals, indicating that the genes and signaling pathways that regulate lifespan are highly conserved among different species. This review summarizes the lifespan-regulating genes, with a specific focus on studies in C. elegans.
Introduction
Aging is an inevitable process in nearly all organisms, and the aging process was previously regarded as a passive entropic process of tissue deterioration caused by damage to macromolecules of the cell, such as genomic DNA, proteins and lipids. In the mid-nineteenth century, Darwin proposed that all wild species arise and develop through the natural selection of small inherited variations that increase the individual’s ability to survive and reproduce to ensure the welfare of the species. Because aging is a process that occurs after reproduction, biologists have presumed that the regulation of aging was not a critical factor for the evolution of life. However, hybrid mice generated from long- and short-lived animals inherit longevity,1 suggesting that lifespan is a genetically regulated trait. Since the isolation of the first long-lived mutant in Caenorhabditis elegans (C. elegans), a number of reports have described various genes and signaling pathways that regulate longevity in model organisms such as yeasts, worms, flies and mice.
Since Brenner first introduced the C. elegans model in the 1960s, this free-living soil nematode has been widely used as a model organism in many areas of research, including aging. C. elegans is a self-fertilizing hermaphrodite that lives for a few weeks when cultured at 20 °C, and it was first used as a model organism to study aging in the 1970s.2–6 In 1983, Klass reported a method for isolating longevity mutants in C. elegans, and it was used to isolate eight long-lived mutants for which an increased lifespan was most likely associated with reduced calorie intake.7 Johnson and his colleagues identified the long-lived mutant called age-1.8,9 Subsequent studies have led to the identification of the insulin/insulin-like growth factor-1 signaling (IIS) pathway as the first established lifespan-regulating signaling pathway.10 Since then, a number of genetic factors have been shown to have an important role in regulating the aging process, and these genetic factors may link environmental factors to the rate of aging. Here we summarize the biological factors associated with lifespan regulation, including signaling transduction pathways, epigenetic factors, sensory perceptions and other physiological processes.
Lifespan-regulating genes
Insulin/insulin-like growth factor-1 signaling
As previously mentioned, the first pathway implicated in the regulation of the aging process in animals was the IIS pathway.10 In C. elegans, mutations that decrease the activity of DAF-2, the C. elegans homolog of the insulin/insulin-like growth factor-1 receptor, more than double the lifespan of the animal,11 and mutations that affect the activity of the IIS downstream target AGE-1, the C. elegans homolog of phosphatidylinositol 3-kinase, are also associated with increased longevity.8,9 Longevity induced by a reduction in IIS signaling is entirely dependent on DAF-16, the C. elegans homolog of the forkhead box FoxO transcription factor.11 DAF-2, AGE-1 and DAF-16 constitute the three key components of the IIS pathway.12–15 Previous studies have demonstrated that modest inhibition of IIS promotes stress resistance and lifespan extensions in multiple species.16–18 Low IIS activity leads to the translocation of DAF-16 to the nucleus, where DAF-16 either activates or represses the genes involved in the cellular stress response (e.g., heat-shock proteins, superoxide dismutase and catalase), metabolism and autophagy. The combined effects of DAF-16-mediated transcriptional changes cause lifespan extension.19–21
Although DAF-16 has a pivotal role in regulating lifespan, several lines of evidence suggest that it does not necessarily act alone. First, the overexpression of DAF-16 in wild-type animals only slightly increases their lifespan, indicating that solely increasing levels of DAF-16 is not sufficient to significantly extend longevity.22 Second, the nuclear localization of DAF-16, which is necessary for its transcriptional activity, is not sufficient to extend lifespan.23 Third, although the canonical DAF-16-binding element24 is present in the 5-kb upstream region of 78% of C. elegans genes,25 only a small number of these genes are activated in young adult animals.21 Thus, there may be other factors that assist DAF-16 in activating particular genes in the appropriate context. Indeed, JNK-1 (the C. elegans homolog of c-Jun N-terminal kinase) and CST-1 (the C. elegans homolog of mammalian ste20-like kinase (MST)) have been shown to regulate DAF-16 activity via post-translational modification.26,27 Overexpression of JNK-1 or CST-1 promotes lifespan extension in a DAF-16-dependent manner,26,27 suggesting that JNK-1 and CST-1 stimulate DAF-16 activity. Lifespan extension induced by mutations in daf-7 (a gene encoding a member of the C. elegans transforming growth factor-β family) is dependent on DAF-16, suggesting that the transforming growth factor-β pathway is an upstream regulator of IIS-mediated longevity.28 The ubiquitin proteasome system (UPS) also regulates DAF-16 activity. The loss of rle-1, a gene encoding an E3 ubiquitin ligase, increases lifespan,29 whereas the loss of math-33, a gene encoding deubiquitylase, suppresses the extended lifespan of daf-2 mutants.30 There are several additional transcription factors that also function as cofactors of DAF-16. Similar to mutations in daf-16, mutations in hsf-1 (a gene encoding the C. elegans homolog of heat-shock transcription factor), skn-1 (a gene encoding the C. elegans homolog of nuclear respiration factor 2 (Nrf2)) or pqm-1 (a gene encoding the C2H2-type zinc finger and leucine zipper-containing protein) suppress the lifespan extension phenotype of daf-2 mutants.31–33 These findings suggest that HSF-1, SKN-1 and PQM-1 cooperate with DAF-16 to regulate the overlapping pro-longevity genes, although have the distinct target genes. SKN-1 has also been shown to be activated by MPK-1, the C. elegans homolog of ERK MAP kinase, and to regulate DAF-16 activity.34 It was recently reported that the transcription factor AP-1 collaborates with DAF-16 downstream of KGB-1 (one of the C. elegans JNK homologs) under fasting conditions.35 Although a number of additional genes that influence DAF-16 activity have been identified, the mechanisms by which these genes regulate the lifespan are not entirely understood.
C. elegans contains several tissue types, and the tissue-specific requirements of the IIS pathway with respect to longevity have been studied; however, a number of these studies have demonstrated conflicting results. Tissue-specific restoration experiments in daf-2 and age-1 mutants have revealed that the restoration of DAF-2 and AGE-1 expression in neurons, respectively, but not in the intestine is sufficient to decrease the lifespan of the long-lived daf-2 and age-1 mutants, respectively.36 However, a recent report demonstrated that AGE-1 expression in the intestine can decrease the long lifespan of the age-1 mutant.37 Moreover, tissue-specific restoration experiments in a daf-2;daf-16 mutant demonstrated that restoration of DAF-16 expression in the intestine extends the lifespan, whereas the restoration of DAF-16 expression in neurons exhibited only modest effects on lifespan.38 It has also been reported that the IIS pathway acts in both a cell-autonomous and non-autonomous manner,36–40 which might partly account for the complexity associated with lifespan regulation by the IIS pathway.
It has been reported that DAF-16 mediates the longevity effect associated with certain dietary restriction regimens. Although DAF-16 is dispensable for the longevity induced by chronic calorie restriction (e.g., eat-2 mutant),41 it is required for the longevity induced by calorie restriction in middle-aged animals.42 DAF-16 is also required for intermittent fasting-induced longevity43 but not for continuous fasting-induced longevity.44,45 These results suggest that DAF-16 activation is dependent on a particular stimulus, as well as the timing or duration of the exposure to that stimulus.
TOR signaling
TOR is a mechanistic target of rapamycin or mammalian target of rapamycin, and a serine/threonine kinase that regulates cell growth, proliferation, motility and survival, as well as protein synthesis, autophagy and transcription.46 TOR is activated under nutrient- and energy-sufficient conditions, which in turn stimulate growth and block salvage pathways, such as autophagy.46 Thus, a reduction in TOR activity is thought to mimic nutrient- and energy-deficient conditions. Consistent with this hypothesis, inhibiting TOR signaling increases the lifespan in C. elegans in a DAF-16-dependent manner.47,48 The effects on longevity induced by the inhibition of TOR signaling in C. elegans are mediated by the transcription factor PHA-4/FoxA.49 PHA-4 has been shown to regulate autophagy, a process that has a significant role in lifespan regulation.50,51 A combination of two lifespan-extending genetic manipulations, the inhibition of daf-2 and inhibition of rsks-1 (a C. elegans S6 kinase and a TOR target) additively extend the lifespan in C. elegans. 52 Thus, the IIS pathway and the TOR pathway act together to mediate the distinct manner in which lifespan is regulated in C. elegans.
It was recently reported that food restriction-induced TOR inhibition promotes longevity by inducing PHA-4.43,53 Interestingly, the TOR pathway promotes longevity by inhibiting the IIS pathway under fasting conditions.43 These observations suggest that the TOR pathway exerts both anti- and pro-longevity effects in a context-dependent manner.
Sirtuin
Sirtuins are members of the nicotinamide adenine dinucleotide (NAD)-dependent protein deacetylase family of molecules, and directly linked to cellular nutrient signaling through NAD+.54 Sir2 is a positive regulator of lifespan in Saccharomyces cerevisiae,55 and in C. elegans, overexpression of SIR-2.1, the C. elegans homolog of Sir2, has been reported to extend lifespan56 in a DAF-16-dependent manner.57 SIR-2.1 has been reported to bind to DAF-16 in a 14-3-3-dependent manner in response to stress and promote DAF-16 activation.57 The pro-longevity activity of SIR-2.1 has been associated with a degree of controversy because a recent report demonstrated that the increased longevity, which was initially considered to result from sir-2.1 overexpression, was caused by a background mutation in the transgenic animals used in these experiments.58 However, subsequent studies have demonstrated that the overexpression of sir-2.1 in mutants outcrossed to a different genetic background significantly increased lifespan, albeit to a lesser extent than what was initially observed in the transgenic animals carrying the background mutation.59–61 Moreover, sir-2.1 mutations suppress a variety of lifespan extension phenotypes, suggesting that sirtuins have a positive role in lifespan regulation.60,62
Studies have demonstrated that Sir2 can mediate mild dietary restriction-induced longevity63 but not severe dietary restriction-induced longevity64 in S. cerevisiae, although Tsuchiya et al. 65 reported that dietary restriction-induced longevity in S. cerevisiae is not dependent on sirtuins. In C. elegans, the increased longevity induced by SIR-2.1 overexpression and eat-2-induced longevity do not exhibit an additive effect,66 suggesting that SIR-2.1 has a role in dietary restriction-induced longevity similar to that of eat-2. However, sir-2.1 has been shown to be dispensable for the increase in longevity induced by fasting and calorie restriction in middle-aged animals.43–45,67 Thus, sirtuins might exert pro-longevity effects under particular conditions of dietary restriction.
AMP-activated protein kinase
AMP-activated protein kinase (AMPK) is a conserved cellular energy sensor that helps cells adapt to low-energy conditions.68 AMPK restores energy homeostasis by stimulating catabolic processes and blocking energy-consuming processes.69 The loss of aak-2, the gene encoding the C. elegans AMPK protein, decreases lifespan, and the increased expression of aak-2 increases lifespan.41 Furthermore, overexpression of a constitutively active form of AMPK has been reported to increase lifespan to a greater extent than aak-2 upregulation.42,70 Metformin is an indirect AMPK agonist that also extends lifespan in C. elegans. 71 AAK-2 is required for the longevity effects mediated by reduced IIS,41 and DAF-16 is required for the longevity effects mediated by activated AMPK, suggesting that DAF-16 and AMPK act together in a feedback loop. Indeed, AAK-2 activates DAF-16,42 and DAF-16 regulates the expression of aakg-4, a gene encoding a regulatory subunit of AMPK.52 Both AMPK and sirtuins are cellular energy sensors, as well as pro-longevity effectors, suggesting that these factors could interact with each other. Indeed, the increased longevity observed in animals overexpressing sir-2.1 is dependent on aak-2,72 although these results were derived from experiments with transgenic worms harboring the confounding background mutation that was previously mentioned.58 CRTC (cAMP response element binding protein-regulating transcriptional coactivator) has been reported to mediate the lifespan extension induced by AMPK activation.70 Recently, neuronal AMPK activation was shown to regulate organismal lifespan via catecholamine signaling in C. elegans. 73
An AMPK–DAF-16 signaling pathway is activated by calorie restriction during middle age and mediates lifespan extension.42 However, this pathway is dispensable to lifespan extensions induced by chronic calorie restriction, such as that observed in the eat-2 mutant.47 Moreover, the transcription factors that are required for lifespan extension induced by chronic calorie restriction (e.g., PHA-4 and SKN-1) are dispensable for longevity induced by calorie restriction in middle-aged animals.67 Thus, the timing and quantity of food might be important factors in specifying which signaling pathway is activated.
Mitochondria
Mitochondria are essential cell organelles that provide functions that are central to cellular metabolism and apoptosis. Mitochondria are also a major source of reactive oxidative species (ROS). ROS are responsible for damage to macromolecules such as DNA, protein and lipids, and they also promote the deterioration of cells, tissues and, ultimately, the entire organism. Mitochondrial ROS are thought to be the primary cause of aging, and this hypothesis is referred to as the mitochondrial free radical theory of aging. Indeed, non-biased large-scale RNAi screens for genes associated with lifespan extension have revealed that most of the genes that promote lifespan extension are involved in metabolic pathways and components of the mitochondrial electron transport chain.74–77 These comprehensive analyses have also demonstrated that mitochondria have an important role in aging. However, the SOD-2 (C. elegans mitochondrial superoxide dismutase) loss-of-function mutation, which is predicted to increase oxidative damage, extends lifespan.78 The same research group demonstrated that treatment with the pro-oxidant paraquat also extended the lifespan of C. elegans, 79 suggesting that mild mitochondrial stress renders organisms less susceptible to subsequent perturbations. Furthermore, inhibiting respiration can extend lifespan by inducing a moderate increase in ROS, and this effect is mediated by HIF-1, AAK-2,80 CHE-23,81 CEP-182 and SKN-1.83 Moreover, the intrinsic apoptosis axis has also been shown to mediate longevity.84
Because mitochondria have a central role in cellular energy production, it is plausible that mitochondria also have an important role in regulating lifespan in response to dietary restrictions. Indeed, lifespan extension by glucose restriction is thought to be mediated by mitohormesis (the specific type of hormesis that is caused by increased formation of ROS within the mitochondria) in C. elegans. 85 Restricting glucose promotes the formation of ROS, which in turn may elicit the mitohormesis response.85 SKN-1 and AAK-2 mediate both calorie restriction-induced longevity42,86 and mitohormesis-mediated longevity,80,83 suggesting that SKN-1 and AAK-2 might function as a link between dietary restriction and mitochondria function.
Epigenetic mechanisms
Epigenetic mechanisms, such as DNA methylation, histone modification and noncoding RNAs, regulate the interpretation of genetic information and are linked to numerous biological processes. In humans, epigenetic changes correlate with older age in normal individuals and are a hallmark of patients with progeria syndrome; however, the significance of epigenetics in regulating the rate of aging is not well understood.
Histone-modifying enzymes
It has been reported that histone-modifying enzymes also have a role in lifespan regulation. A genome-wide RNAi screen identified two SET domain proteins (SET-9 and SET-15) that accelerate aging.76 Greer et al. 87 demonstrated that inhibition of the histone H3K4 methylation complex (composed of ASH-2, WDR-5 and SET-2) and overexpression of RBR-2, the enzyme that mediates H3K4 demethylation, resulted in lifespan extension. The lifespan extension associated with H3K4-modifying enzymes is dependent on the production of mature eggs.87 Surprisingly, although these genetic manipulations were present only in the parent animals, up to four generations of descendants presented extended lifespans.88 However, the molecular mechanisms for the transgenerational inheritance of longevity remain unknown. In addition, the inhibition of H3K27 demethylase UTX-1 has also been shown to extend lifespan.89,90
MicroRNA
Noncoding RNAs also have an important role in lifespan regulation. MicroRNA (miRNA) is a class of noncoding RNA molecules that regulate the expression of target mRNAs in a sequence-dependent manner. The miRNA lin-4 was the first discovered miRNA, and it regulates the lifespan of C. elegans. 91 Lifespan regulation associated with lin-4 and its target LIN-14 is mediated by the IIS pathway.91 Following the identification of lin-4, a number of additional miRNAs have been implicated in lifespan regulation.92 Recently, a long noncoding RNA named tts-1 was shown to regulate lifespan by modulating ribosomal activity.93
Although dietary restrictions and epigenetic mechanisms are both known to be involved in lifespan regulation, the potential links between epigenetics and dietary restriction-induced longevity remain largely unknown. miR-228 and miR-71 were recently reported to be necessary for calorie restriction-induced longevity.94 These miRNAs regulate lifespan by interacting with the dietary restriction responsive transcription factors PHA-4/FoxA and SKN-1/Nrf2.94 Pandit et al. 95 demonstrated that PHA-4 regulates the expression of miRNAs in calorie-restricted animals. These studies suggest that miRNAs are involved in at least some of the mechanisms that mediate dietary restriction-induced longevity.
Proteostasis
Protein homeostasis, or proteostasis, is essential to life. The loss of proteostasis is often involved in protein aggregation, a cellular process that is associated with many age-related disorders and often observed in aged organisms.96 The loss of proteostasis results in the deterioration in cellular function; therefore, it may be one of the main causes of organismal aging. Proteostasis is maintained by a complex interplay among protein synthesis, degradation and quality control. Polysome profiling analyses have revealed that the rate of protein synthesis in C. elegans is markedly decreased in 10-day-old animals compared with 4-day-old animals.97 These findings suggest that the loss of proteostasis during old age does not necessarily result from excess protein synthesis. Paradoxically, the C. elegans lifespan can be extended by decreasing protein synthesis. The long-lived daf-2 mutant exhibits a reduced rate of protein translation.93 The inhibition of eukaryotic initiation factors, ribosomal proteins and TOR signaling molecules, which is responsible for decreased translation in these animals, has been reported to account for the increased lifespan.98–100 The protein degradation pathway is also used as a method for reshaping proteome function. The UPS and autophagy are major protein degradation mechanisms. One study used in vivo imaging techniques (chimeric green fluorescent protein fused to a non-cleavable ubiquitin moiety) to demonstrate that UPS activity declines soon after animals complete development.101 Moreover, an increase rather than a decrease in UPS activity is associated with lifespan extension.29,30,35,102 These results suggest that decreased UPS activity contributes to the collapse of proteostasis in old age. Autophagy is another cellular process that has an important role in lifespan regulation. The protein quality control system, which consists of the heat-shock response, the unfolded protein response (UPR) of the endoplasmic reticulum (ER) (UPRER) and the mitochondrial UPR (UPRmt), is also important for proteostasis. The various genes involved in the protein quality control system are regulated by designated transcription factors (HSF-1 for heat-shock response, XBP-1 and ATF6 for UPRER, and ATFS-1 for UPRmt).96 Animals utilize these protein quality control pathways to manage a wide range of acute and chronic stress conditions during the aging process.96 The activation of heat-shock response or UPRER by overexpression of hsf-1 or xbp-1, respectively, extends lifespan,101,103 suggesting that heat-shock response and UPRER also have a role in lifespan regulation. Although the longevity induced by the inhibition of respiration requires UBL-5, a coactivator of UPRmt,104 the activation of UPRmt does not always extend lifespan.105 Thus, although the link between longevity and UPRmt is plausible, it is still not clear whether the activation of UPRmt alone is sufficient for lifespan extension.
Autophagy is an evolutionarily conserved intracellular degradation system that delivers cytoplasmic components to the lysosome.106 The autophagy system has been shown to be involved in several longevity pathways. The first study to evaluate the role of autophagy in lifespan regulation in C. elegans revealed that RNAi targeting bec-1, a gene encoding the C. elegans ortholog of yeast ATG6, significantly suppresses the long lifespan of daf-2 mutants.20 Subsequent studies have demonstrated that autophagy genes are also required for the longevity induced by reduced IIS and indicated that autophagy is induced in daf-2 Dauer larvae and adults.20,50,107,108 However, one study reported that RNAi targeting autophagy genes can extend lifespan under certain conditions.109 In addition, autophagy can regulate calorie restriction-induced lifespan extensions. Accordingly, RNAi targeting autophagy genes abolishes the lifespan extension induced by the eat-2 mutation or TOR signaling inhibition.50,108 Moreover, autophagy activity is enhanced in both TOR RNAi-treated animals and animals subjected to dietary restriction in a PHA-4-dependent manner.50 Autophagy also facilitates lipid storage in the intestine.110 Thus, autophagy might provide energy to the animal by stimulating lipogenesis in response to dietary restrictions. Consistent with this hypothesis, fasting induces both lipase genes and autophagy genes via activation of the HLH-30 transcription factor.110 Inhibition of mitochondrial respiration extends lifespan in wild-type animals but not in autophagy mutants,108 suggesting that autophagy is also required for the longevity induced by the inhibition of mitochondrial respiration. A recent report demonstrated that mitophagy, a selective type of autophagy targeting mitochondria for degradation, has an important role in maintaining mitochondrial homeostasis during aging.111 Together, these observations indicate that multiple lifespan-extending pathways require autophagy.
Hypoxia inducible factor-1
Hypoxia inducible factor 1 (HIF-1) is considered to be the master transcriptional regulator mediating the cellular response to hypoxia.112 HIF-1 is degraded by the E3 ubiquitin ligase von Hippel Lindau under normal oxygen conditions, and HIF-1 is stabilized under low-oxygen conditions.112 Stabilization of HIF-1 by the downregulation of von Hippel Lindau-1 or overexpression of HIF-1 significantly increases lifespan.113,114 Consistent with these observations, hypoxia induces lifespan extension in a HIF-1-dependent manner.115 HIF-1 also mediates the lifespan extension induced by the increase in ROS following paraquat treatment or respiration inhibition.80,116 Neuronal HIF-1 was recently demonstrated to mediate lifespan extensions by regulating the expression of intestinal flavin-containing monooxygenase (fmo-2) via serotonin secretion.117
Lifespan regulation by the interplay of different tissues
Gonads
The gonad consists of the germline and somatic gonad, and it has an important role in regulating lifespan in C. elegans. 118 The removal of germline cells by laser microsurgery or genetic manipulation (e.g., glp-1 mutants) increases lifespan, whereas the removal of the gonad (both the germline and somatic gonad) does not.119,120 Because the ablation of both the germline and somatic gonad results in sterility, the longevity induced by germline elimination is not only solely related to a resource trade-off but also to endocrine-mediated lifespan regulation (Figure 1a). In fact, certain endocrine signaling pathways have been shown to regulate longevity in animals without germlines118 (Figure 1a).
Figure 1.
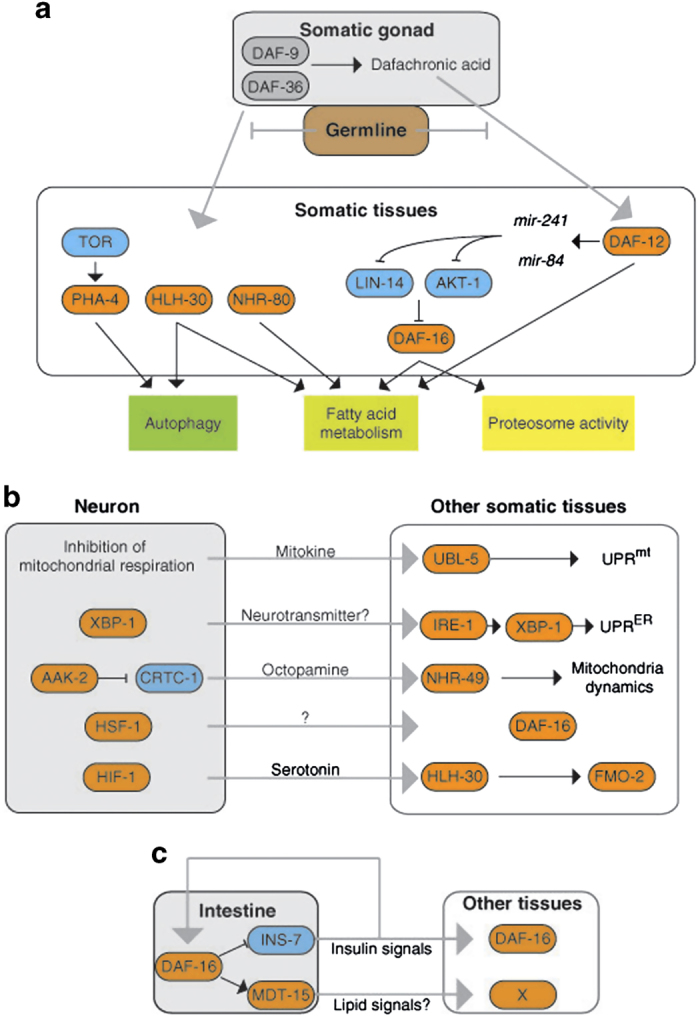
Tissue interplay that regulates lifespan. (a) Genes that mediate germline elimination-induced lifespans in C. elegans. Germline elimination enhances the steroid signal dafachronic acid, which is produced in somatic gonads. DAF-12 regulates the expression of mir-84 and mir-241, and increased mir-84 and mir-241 expression results in the downregulation of two DAF-16 inhibitors (AKT-1 and LIN-14) and promotes DAF-16 nuclear localization and activation. Germline elimination also activates the transcription factors PHA-4, HLH-30 and NHR-80. The activation of these transcription factors leads to changes in fatty acid metabolism, as well as enhanced autophagy and proteasome activity. (b) The genetic manipulations in neuronal cells are sufficient for increasing organismal lifespan. The RNAi of cco-1 or overexpression of xbp-1, aak-2, hsf-1 or hif-1 extends lifespans by regulating other tissues. (c) Intestinal DAF-16 regulates both DAF-16 and a non-DAF-16 transcription factor (referred to as X here) in other tissues via INS-7- and MDT-15-dependent lipid signals, respectively.
The nuclear hormone receptor DAF-12, which responds to the dafachronic acid (DA) ligand, is a key regulator of germline elimination-induced longevity.119 DAF-9 and DAF-36 are also components of the steroid hormone-signaling pathway that contributes to gonad-associated longevity.121–123 DA supplementation restores the longevity triggered by ablation of both germline and somatic gonads in a DAF-12-dependent manner, suggesting that the somatic gonad is involved in DA production.124 However, the location of DA production is not important because the overexpression of DAF-9 in other tissues is sufficient to restore the longevity of animals lacking gonads.124 The tissues in which DAF-12 functions to increase lifespan have yet to be determined. DAF-16 is an effector of the IIS pathway and a critical regulator of germline elimination-induced longevity.119 The DA–DAF-12 axis regulates DAF-16 nuclear localization and activity.123,125 DAF-12 regulates the expression of mir-84 and mir-241, and the increased expression of these two miRNAs leads to the downregulation of two inhibitors of DAF-16 (AKT-1 and LIN-14), thereby promoting DAF-16 nuclear localization and activation.126 DAF-12 also regulates the expression of the fatty acid reductase fard-1 and the lipase lips-17. 127 DAF-16 regulates the expression of the lipase lipl-4 128 and rnp-6, a subunit of the 19S proteasome.129 The upregulation of these genes contributes to germline elimination-induced longevity.
Germline elimination-induced longevity requires other transcription factors in addition to DAF-12 and DAF-16, including NHR-80 and PHA-4.51,130 NHR-80 links FAT-6 (a gene encoding an acyl-CoA desaturase)-induced fatty acid desaturation to longevity in germline-less animals in a DAF-16-independent manner.130 TOR expression is reduced in glp-1 mutants and results in enhanced autophagy in the intestine via the activity of PHA-4.51 It should be noted that germline elimination regulates DAF-16 in a manner distinct from that of the IIS pathway. Although reduced IIS induces DAF-16 nuclear translocation in all tissues,22 germline elimination induces DAF-16 nuclear translocation only in the intestine of young adult animals.125 In summary, germline elimination extends lifespan by regulating several signaling pathways in somatic tissues, such as the intestine, via endocrine pathways.
Somatic tissues
Animals sense and process environmental signals to help them prepare for environmental changes. Lifespan regulation by sensory perception was initially identified in C. elegans in studies using mutants defective in sensory perception,131 and the longevity of these mutants was largely but not entirely dependent on DAF-16.131 Gustatory, olfactory and thermosensory neurons were all subsequently shown to influence lifespan.132,133 Together, these studies suggest that neurons that sense environmental cues influence lifespan. Thus, certain sensory neurons shorten lifespan and others extend lifespan in an environmental context-dependent manner.133–137 Food availability is one of the most important environmental factors that influence lifespan. Indeed, certain mutants with defective sensory neurons exhibit a lifespan phenotype only when they eat certain bacteria as food.134 Interestingly, diffusible bacterial products suppress lifespan extension by promoting fasting,138 suggesting that not only food consumption but also food sensing is important for lifespan regulation. In addition, neuronal SKN-1 has been shown to mediate dietary restriction-induced longevity.86 Recent studies have demonstrated that genetic manipulations in neuronal cells only are sufficient to increase organismal lifespan (Figure 1b). The inhibition of respiration by neuron-specific cco-1 RNAi is sufficient to increase lifespan, and it induces the intestinal UTRmt response,104 suggesting that a systemic factor secreted from neurons might regulate global cellular responses. The overexpression of active AAK-2 or stable HIF-1 in neurons extends lifespan via the biogenic monoamine neurotransmitter serotonin or octopamine, respectively.73,117 The overexpression of a spliced form of XBP-1 in neurons extends lifespan in a neurosecretion-dependent manner,101 and the overexpression of HSF-1 increases thermotolerance and lifespan.103 The thermosensory circuit is required for the increase in thermotolerance induced by HSF-1 overexpression but not for the lifespan extension effect.103 However, intestinal DAF-16 is required for the lifespan extension induced by HSF-1 overexpression but not for the increase in thermotolerance.103 These observations suggest that neurons might be a key source of extracellular signals that regulate organismal lifespan.
The intestine has an important role in regulating organismal lifespan. There are several lines of evidence suggesting that the intestine receives signals from other cells that modulate organismal lifespan. First, intestinal restoration of DAF-16 significantly increases the lifespan of daf-2;daf-16 mutants.38 Second, intestinal DAF-16 is required for the lifespan extension induced by neuronal HSF-1 overexpression.103 Third, an increase in intestinal fmo-2 expression is required for neuronal HIF-1 activation.117 Fourth, intestinal UPRmt or UPRER is induced in response to neuronal cco-1 knockdown or the overexpression of spliced xbp-1, respectively.101,103 These observations suggest that the intestine is a key target organ responsible for regulating lifespan at the organismal level. Intestinal DAF-16 regulates both DAF-16 and a non-DAF-16 transcription factor in other tissues via INS-7-139 and MDT-15-dependent lipid signals,40 respectively, suggesting that the intestine can act as a sender tissue as well as a receiver tissue (Figure 1c).
Conclusions and future challenges
Although genetic factors have important roles in the regulation of organismal lifespans, environmental factors are also important in lifespan regulation. Following the groundbreaking studies that first identified lifespan-regulating genes in the 1980s and 1990s, a large number of genes have been shown to regulate organismal lifespan. Thus, one of the next major challenges is to determine how these genes link environmental factors to lifespan regulation. As previously mentioned, the relationship between genes and diet is the most important lifespan-regulating environmental cue, and it has been examined in many organisms. Environmental temperature and oxidative status are additional environmental cues that influence lifespan,4,79 and studies examining the relationship between genes and these specific environmental cues have recently been initiated.
One of the goals of aging research is to not only extend lifespan but also extend healthspan. Worms display certain age-associated characteristics that resemble those observed in humans;140 therefore, worms also serve as a useful model for healthspan studies. Healthspan studies in worms include those that have examined mobility declines141 and pharyngeal pumping,142 fluorescent compound dynamics (including lipofuscin and advanced glycosylation end products),143,144 and neuromuscular changes.141,145,146 In addition, the identification of small molecules that potentially slow down aging and extend lifespan in multiple species is another major challenge and goal associated with aging research. C. elegans has multiple advantages that make it a useful model for identifying and evaluating chemical compounds that extend lifespan.147
Aging induces declines in the integrity and function of tissues throughout the organismal body. If aging affects each tissue independently, the mechanism underlying organismal aging could be elucidated by dissecting the aging process associated with each cell type. If certain tissues have a role in coordinating the aging process among different tissues, researchers might focus on the systemic factors used by these tissues to communicate with other tissues. There are several lines of evidence for the presence of an aging control center. Thus, the location and mechanism associated with the regulation of organismal aging by lifespan-regulating genes are additional issues that merit further investigation. These studies will facilitate the development of therapeutic strategies that target the aging control center to help promote health spans in humans.
Acknowledgments
We thank members of our laboratory for helpful comments on the manuscript. This work was supported by JSPS KAKENHI Grant Number 26221101 (Grant-in-Aid for Scientific Research S).
Footnotes
The authors declare no conflict of interest.
References
- Goodrick, C. L. Life-span and the inheritance of longevity of inbred mice. J. Gerontol. 30, 257–263 (1975). [DOI] [PubMed] [Google Scholar]
- Gershon, D. Studies on aging in Nematodes. I. The nematode as a model organism for aging research. Exp. Gerontol. 5, 7–12 (1970). [DOI] [PubMed] [Google Scholar]
- Klass, M. & Hirsh, D. Non-ageing developmental variant of Caenorhabditis elegans. Nature 260, 523–525 (1976). [DOI] [PubMed] [Google Scholar]
- Klass, M. R. Aging in the nematode Caenorhabditis elegans: major biological and environmental factors influencing life span. Mech. Ageing Dev. 6, 413–429 (1977). [DOI] [PubMed] [Google Scholar]
- Johnson, T. E. & Wood, W. B. Genetic analysis of life-span in Caenorhabditis elegans. Proc. Natl Acad. Sci. USA 79, 6603–6607 (1982). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, T. E. Molecular and genetic analyses of a multivariate system specifying behavior and life span. Behav. Genet. 16, 221–235 (1986). [DOI] [PubMed] [Google Scholar]
- Klass, M. R. A method for the isolation of longevity mutants in the nematode Caenorhabditis elegans and initial results. Mech. Ageing Dev. 22, 279–286 (1983). [DOI] [PubMed] [Google Scholar]
- Friedman, D. B. & Johnson, T. E. A mutation in the age-1 gene in Caenorhabditis elegans lengthens life and reduces hermaphrodite fertility. Genetics 118, 75–86 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman, D. B. & Johnson, T. E. Three mutants that extend both mean and maximum life span of the nematode, Caenorhabditis elegans, define the age-1 gene. J. Gerontol. 43, B102–B109 (1988). [DOI] [PubMed] [Google Scholar]
- Kenyon, C. The first long-lived mutants: discovery of the insulin/IGF-1 pathway for ageing. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 366, 9–16 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenyon, C. , Chang, J. , Gensch, E. , Rudner, A. & Tabtiang, R. A C. elegans mutant that lives twice as long as wild type. Nature 366, 461–464 (1993). [DOI] [PubMed] [Google Scholar]
- Morris, J. Z. , Tissenbaum, H. A. & Ruvkun, G. A phosphatidylinositol-3-OH kinase family member regulating longevity and diapause in Caenorhabditis elegans. Nature 382, 536–539 (1996). [DOI] [PubMed] [Google Scholar]
- Kimura, K. D. , Tissenbaum, H. A. , Liu, Y. & Ruvkun, G. daf-2, an insulin receptor-like gene that regulates longevity and diapause in Caenorhabditis elegans. Science 277, 942–946 (1997). [DOI] [PubMed] [Google Scholar]
- Ogg, S. et al. The Fork head transcription factor DAF-16 transduces insulin-like metabolic and longevity signals in C. elegans. Nature 389, 994–999 (1997). [DOI] [PubMed] [Google Scholar]
- Lin, K. , Dorman, J. B. , Rodan, A. & Kenyon, C. daf-16: An HNF-3/forkhead family member that can function to double the life-span of Caenorhabditis elegans. Science 278, 1319–1322 (1997). [DOI] [PubMed] [Google Scholar]
- Clancy, D. J. et al. Extension of life-span by loss of CHICO, a Drosophila insulin receptor substrate protein. Science 292, 104–106 (2001). [DOI] [PubMed] [Google Scholar]
- Tatar, M. et al. A mutant Drosophila insulin receptor homolog that extends life-span and impairs neuroendocrine function. Science 2001; 292: 107–110. [DOI] [PubMed] [Google Scholar]
- Bluher, M. , Kahn, B. B. & Kahn, C. R. Extended longevity in mice lacking the insulin receptor in adipose tissue. Science 299, 572–574 (2003). [DOI] [PubMed] [Google Scholar]
- Lee, S. S. , Kennedy, S. , Tolonen, A. C. & Ruvkun, G. DAF-16 target genes that control C. elegans life-span and metabolism. Science 300, 644–647 (2003). [DOI] [PubMed] [Google Scholar]
- Melendez, A. et al. Autophagy genes are essential for dauer development and life-span extension in C. elegans. Science 2003; 301: 1387–1391. [DOI] [PubMed] [Google Scholar]
- Murphy, C. T. et al. Genes that act downstream of DAF-16 to influence the lifespan of Caenorhabditis elegans. Nature 424, 277–283 (2003). [DOI] [PubMed] [Google Scholar]
- Henderson, S. T. & Johnson, T. E. daf-16 integrates developmental and environmental inputs to mediate aging in the nematode Caenorhabditis elegans. Curr. Biol. 11, 1975–1980 (2001). [DOI] [PubMed] [Google Scholar]
- Lin, K. , Hsin, H. , Libina, N. & Kenyon, C. Regulation of the Caenorhabditis elegans longevity protein DAF-16 by insulin/IGF-1 and germline signaling. Nat. Genet. 28, 139–145 (2001). [DOI] [PubMed] [Google Scholar]
- Furuyama, T. , Nakazawa, T. , Nakano, I. & Mori, N. Identification of the differential distribution patterns of mRNAs and consensus binding sequences for mouse DAF-16 homologues. Biochem. J. 349, 629–634 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenyon, C. & Murphy, C. T. Enrichment of regulatory motifs upstream of predicted DAF-16 targets. Nat. Genet 38, 397–398 (2006); Author reply 398. [DOI] [PubMed] [Google Scholar]
- Oh, S. W. et al. JNK regulates lifespan in Caenorhabditis elegans by modulating nuclear translocation of forkhead transcription factor/DAF-16. Proc. Natl Acad. Sci. USA 2005; 102: 4494–4499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehtinen, M. K. et al. A conserved MST-FOXO signaling pathway mediates oxidative-stress responses and extends life span. Cell 125, 987–1001 (2006). [DOI] [PubMed] [Google Scholar]
- Shaw, W. M. , Luo, S. , Landis, J. , Ashraf, J. & Murphy, C. T. The C. elegans TGF-beta Dauer pathway regulates longevity via insulin signaling. Curr. Biol. 17, 1635–1645 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, W. , Gao, B. , Lee, S. M. , Bennett, K. & Fang, D. RLE-1, an E3 ubiquitin ligase, regulates C. elegans aging by catalyzing DAF-16 polyubiquitination. Dev. Cell 12, 235–246 (2007). [DOI] [PubMed] [Google Scholar]
- Heimbucher, T. et al. The Deubiquitylase MATH-33 controls DAF-16 stability and function in metabolism and longevity. Cell Metab. 22, 151–163 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu, A. L. , Murphy, C. T. & Kenyon, C. Regulation of aging and age-related disease by DAF-16 and heat-shock factor. Science 300, 1142–1145 (2003). [DOI] [PubMed] [Google Scholar]
- Tullet, J. M. et al. Direct inhibition of the longevity-promoting factor SKN-1 by insulin-like signaling in C. elegans. Cell 132, 1025–1038 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tepper, R. G. et al. PQM-1 complements DAF-16 as a key transcriptional regulator of DAF-2-mediated development and longevity. Cell 2013; 154: 676–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuyama, T. et al. The ERK-MAPK pathway regulates longevity through SKN-1 and insulin-like signaling in Caenorhabditis elegans. J. Biol. Chem. 285, 30274–30281 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uno, M. et al. A fasting-responsive signaling pathway that extends life span in C. elegans. Cell Rep. 3, 79–91 (2013). [DOI] [PubMed] [Google Scholar]
- Wolkow, C. A. , Kimura, K. D. , Lee, M. S. & Ruvkun, G. Regulation of C. elegans life-span by insulinlike signaling in the nervous system. Science 290, 147–150 (2000). [DOI] [PubMed] [Google Scholar]
- Iser, W. B. & Wolkow, C. A. DAF-2/insulin-like signaling in C. elegans modifies effects of dietary restriction and nutrient stress on aging, stress and growth. PLoS ONE 2, e1240 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libina, N. , Berman, J. R. & Kenyon, C. Tissue-specific activities of C. elegans DAF-16 in the regulation of lifespan. Cell 115, 489–502 (2003). [DOI] [PubMed] [Google Scholar]
- Apfeld, J. & Kenyon, C. Cell nonautonomy of C. elegans daf-2 function in the regulation of diapause and life span. Cell 95, 199–210 (1998). [DOI] [PubMed] [Google Scholar]
- Zhang, P. , Judy, M. , Lee, S. J. & Kenyon, C. Direct and indirect gene regulation by a life-extending FOXO protein in C. elegans: roles for GATA factors and lipid gene regulators. Cell Metab. 17, 85–100 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apfeld, J. , O’Connor, G. , McDonagh, T. , DiStefano, P. S. & Curtis, R. The AMP-activated protein kinase AAK-2 links energy levels and insulin-like signals to lifespan in C. elegans. Genes Dev. 18, 3004–3009 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer, E. L. et al. An AMPK-FOXO pathway mediates longevity induced by a novel method of dietary restriction in C. elegans. Curr. Biol. 17, 1646–1656 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honjoh, S. , Yamamoto, T. , Uno, M. & Nishida, E. Signalling through RHEB-1 mediates intermittent fasting-induced longevity in C. elegans. Nature 457, 726–730 (2009). [DOI] [PubMed] [Google Scholar]
- Kaeberlein, T. L. et al. Lifespan extension in Caenorhabditis elegans by complete removal of food. Aging Cell 5, 487–494 (2006). [DOI] [PubMed] [Google Scholar]
- Lee, G. D. et al. Dietary deprivation extends lifespan in Caenorhabditis elegans. Aging Cell 5, 515–524 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wullschleger, S. , Loewith, R. & Hall, M. N. TOR signaling in growth and metabolism. Cell 124, 471–484 (2006). [DOI] [PubMed] [Google Scholar]
- Vellai, T. et al. Genetics: influence of TOR kinase on lifespan in C. elegans. Nature 426, 620 (2003). [DOI] [PubMed] [Google Scholar]
- Jia, K. , Chen, D. & Riddle, D. L. The TOR pathway interacts with the insulin signaling pathway to regulate C. elegans larval development, metabolism and life span. Development 131, 3897–3906 (2004). [DOI] [PubMed] [Google Scholar]
- Sheaffer, K. L. , Updike, D. L. & Mango, S. E. The target of Rapamycin pathway antagonizes pha-4/FoxA to control development and aging. Curr. Biol. 18, 1355–1364 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen, M. et al. A role for autophagy in the extension of lifespan by dietary restriction in C. elegans. PLoS Genet. 4, e24 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapierre, L. R. , Gelino, S. , Melendez, A. & Hansen, M. Autophagy and lipid metabolism coordinately modulate life span in germline-less C. elegans. Curr. Biol. 21, 1507–1514 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, D. et al. Germline signaling mediates the synergistically prolonged longevity produced by double mutations in daf-2 and rsks-1 in C. elegans. Cell Rep. 5, 1600–1610 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panowski, S. H. , Wolff, S. , Aguilaniu, H. , Durieux, J. & Dillin, A. PHA-4/Foxa mediates diet-restriction-induced longevity of C. elegans. Nature 447, 550–555 (2007). [DOI] [PubMed] [Google Scholar]
- Imai, S. , Armstrong, C. M. , Kaeberlein, M. & Guarente, L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature 403, 795–800 (2000). [DOI] [PubMed] [Google Scholar]
- Kaeberlein, M. , McVey, M. & Guarente, L. The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes Dev. 13, 2570–2580 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tissenbaum, H. A. & Guarente, L. Increased dosage of a sir-2 gene extends lifespan in Caenorhabditis elegans. Nature 410, 227–230 (2001). [DOI] [PubMed] [Google Scholar]
- Berdichevsky, A. , Viswanathan, M. , Horvitz, H. R. & Guarente, L. C. elegans SIR-2.1 interacts with 14-3-3 proteins to activate DAF-16 and extend life span. Cell 125, 1165–1177 (2006). [DOI] [PubMed] [Google Scholar]
- Burnett, C. et al. Absence of effects of Sir2 overexpression on lifespan in C. elegans and Drosophila. Nature 477, 482–485 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viswanathan, M. & Guarente, L. Regulation of Caenorhabditis elegans lifespan by sir-2.1 transgenes. Nature 477, E1–E2 (2011). [DOI] [PubMed] [Google Scholar]
- Mouchiroud, L. et al. The NAD(+)/Sirtuin pathway modulates longevity through activation of mitochondrial UPR and FOXO signaling. Cell 154, 430–441 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmeisser, K. et al. Role of sirtuins in lifespan regulation is linked to methylation of nicotinamide. Nat. Chem. Biol. 9, 693–700 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludewig, A. H. et al. Pheromone sensing regulates Caenorhabditis elegans lifespan and stress resistance via the deacetylase SIR-2.1. Proc. Natl Acad. Sci. USA 110, 5522–5527 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, S. J. et al. Calorie restriction extends Saccharomyces cerevisiae lifespan by increasing respiration. Nature 418, 344–348 (2002). [DOI] [PubMed] [Google Scholar]
- Kaeberlein, M. , Kirkland, K. T. , Fields, S. & Kennedy, B. K. Sir2-independent life span extension by calorie restriction in yeast. PLoS Biol. 2, E296 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchiya, M. et al. Sirtuin-independent effects of nicotinamide on lifespan extension from calorie restriction in yeast. Aging Cell 5, 505–514 (2006). [DOI] [PubMed] [Google Scholar]
- Wang, Y. & Tissenbaum, H. A. Overlapping and distinct functions for a Caenorhabditis elegans SIR2 and DAF-16/FOXO. Mech. Ageing Dev. 127, 48–56 (2006). [DOI] [PubMed] [Google Scholar]
- Greer, E. L. & Brunet, A. Different dietary restriction regimens extend lifespan by both independent and overlapping genetic pathways in C. elegans. Aging Cell 8, 113–127 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardie, D. G. & Hawley, S. A. AMP-activated protein kinase: the energy charge hypothesis revisited. Bioessays 23, 1112–1119 (2001). [DOI] [PubMed] [Google Scholar]
- Hardie, D. G. , Ross, F. A. & Hawley, S. A. AMP-activated protein kinase: a target for drugs both ancient and modern. Chem. Biol. 19, 1222–1236 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mair, W. et al. Lifespan extension induced by AMPK and calcineurin is mediated by CRTC-1 and CREB. Nature 470, 404–408 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onken, B. & Driscoll, M. Metformin induces a dietary restriction-like state and the oxidative stress response to extend C. elegans Healthspan via AMPK, LKB1, and SKN-1. PLoS ONE 5, e8758 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis, R. , O’Connor, G. & DiStefano, P. S. Aging networks in Caenorhabditis elegans: AMP-activated protein kinase (aak-2) links multiple aging and metabolism pathways. Aging Cell 5, 119–126 (2006). [DOI] [PubMed] [Google Scholar]
- Burkewitz, K. et al. Neuronal CRTC-1 governs systemic mitochondrial metabolism and lifespan via a catecholamine signal. Cell 160, 842–855 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillin, A. et al. Rates of behavior and aging specified by mitochondrial function during development. Science 298, 2398–2401 (2002). [DOI] [PubMed] [Google Scholar]
- Lee, S. S. et al. A systematic RNAi screen identifies a critical role for mitochondria in C. elegans longevity. Nat. Genet. 33, 40–48 (2003). [DOI] [PubMed] [Google Scholar]
- Hamilton, B. et al. A systematic RNAi screen for longevity genes in C. elegans. Genes Dev. 19, 1544–1555 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen, M. , Hsu, A. L. , Dillin, A. & Kenyon, C. New genes tied to endocrine, metabolic, and dietary regulation of lifespan from a Caenorhabditis elegans genomic RNAi screen. PLoS Genet. 1, 119–128 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Raamsdonk, J. M. & Hekimi, S. Deletion of the mitochondrial superoxide dismutase sod-2 extends lifespan in Caenorhabditis elegans. PLoS Genet. 5, e1000361 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, W. & Hekimi, S. A mitochondrial superoxide signal triggers increased longevity in Caenorhabditis elegans. PLoS Biol. 8, e1000556 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang, A. B. et al. Feedback regulation via AMPK and HIF-1 mediates ROS-dependent longevity in Caenorhabditis elegans. Proc. Natl Acad. Sci. USA 111, E4458–E4467 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter, L. , Baruah, A. , Chang, H. W. , Pace, H. M. & Lee, S. S. The homeobox protein CEH-23 mediates prolonged longevity in response to impaired mitochondrial electron transport chain in C. elegans. PLoS Biol. 9, e1001084 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baruah, A. et al. CEP-1, the Caenorhabditis elegans p53 homolog, mediates opposing longevity outcomes in mitochondrial electron transport chain mutants. PLoS Genet. 10, e1004097 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmeisser, S. et al. Mitochondrial hormesis links low-dose arsenite exposure to lifespan extension. Aging Cell 12, 508–517 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yee, C. , Yang, W. & Hekimi, S. The intrinsic apoptosis pathway mediates the pro-longevity response to mitochondrial ROS in C. elegans. Cell 157, 897–909 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz, T. J. et al. Glucose restriction extends Caenorhabditis elegans life span by inducing mitochondrial respiration and increasing oxidative stress. Cell Metab. 6, 280–293 (2007). [DOI] [PubMed] [Google Scholar]
- Bishop, N. A. & Guarente, L. Two neurons mediate diet-restriction-induced longevity in C. elegans. Nature 447, 545–549 (2007). [DOI] [PubMed] [Google Scholar]
- Greer, E. L. et al. Members of the H3K4 trimethylation complex regulate lifespan in a germline-dependent manner in C. elegans. Nature 466, 383–387 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer, E. L. et al. Transgenerational epigenetic inheritance of longevity in Caenorhabditis elegans. Nature 479, 365–371 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, C. et al. Histone demethylase UTX-1 regulates C. elegans life span by targeting the insulin/IGF-1 signaling pathway. Cell Metab. 14, 161–172 (2011). [DOI] [PubMed] [Google Scholar]
- Maures, T. J. , Greer, E. L. , Hauswirth, A. G. & Brunet, A. The H3K27 demethylase UTX-1 regulates C. elegans lifespan in a germline-independent, insulin-dependent manner. Aging Cell 10, 980–990 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehm, M. & Slack, F. A developmental timing microRNA and its target regulate life span in C. elegans. Science 310, 1954–1957 (2005). [DOI] [PubMed] [Google Scholar]
- Smith-Vikos, T. & Slack, F. J. MicroRNAs and their roles in aging. J. Cell Sci. 125, 7–17 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essers, P. B. et al. A long noncoding RNA on the ribosome is required for lifespan extension. Cell Rep. 10, 339–345 (2015). [DOI] [PubMed] [Google Scholar]
- Smith-Vikos, T. et al. MicroRNAs mediate dietary-restriction-induced longevity through PHA-4/FOXA and SKN-1/Nrf transcription factors. Curr. Biol. 24, 2238–2246 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandit, A. , Jain, V. , Kumar, N. & Mukhopadhyay, A. PHA-4/FOXA-regulated microRNA feed forward loops during Caenorhabditis elegans dietary restriction. Aging (Albany NY) 6, 835–855 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labbadia, J. & Morimoto, R. I. Proteostasis and longevity: when does aging really begin? F1000Prime Rep. 6, 1–7 (2014), eCollection 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirstein-Miles, J. , Scior, A. , Deuerling, E. & Morimoto, R. I. The nascent polypeptide-associated complex is a key regulator of proteostasis. EMBO J. 32, 1451–1468 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen, M. et al. Lifespan extension by conditions that inhibit translation in Caenorhabditis elegans. Aging Cell 6, 95–110 (2007). [DOI] [PubMed] [Google Scholar]
- Pan, K. Z. et al. Inhibition of mRNA translation extends lifespan in Caenorhabditis elegans. Aging Cell 6, 111–119 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syntichaki, P. , Troulinaki, K. & Tavernarakis, N. Protein synthesis is a novel determinant of aging in Caenorhabditis elegans. Ann. N. Y. Acad. Sci. 1119, 289–295 (2007). [DOI] [PubMed] [Google Scholar]
- Taylor, R. C. & Dillin, A. XBP-1 is a cell-nonautonomous regulator of stress resistance and longevity. Cell 153, 1435–1447 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, G. , Rogers, J. , Murphy, C. T. & Rongo, C. EGF signalling activates the ubiquitin proteasome system to modulate C. elegans lifespan. EMBO J. 30, 2990–3003 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas, P. M. et al. Heterotypic signals from neural HSF-1 separate thermotolerance from longevity. Cell Rep. 12, 1196–1204 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durieux, J. , Wolff, S. & Dillin, A. The cell-non-autonomous nature of electron transport chain-mediated longevity. Cell 144, 79–91 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett, C. F. et al. Activation of the mitochondrial unfolded protein response does not predict longevity in Caenorhabditis elegans. Nat. Commun. 5, 3483 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine, B. & Klionsky, D. J. Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev. Cell 6, 463–477 (2004). [DOI] [PubMed] [Google Scholar]
- Hars, E. S. et al. Autophagy regulates ageing in C. elegans. Autophagy 3, 93–95 (2007). [DOI] [PubMed] [Google Scholar]
- Toth, M. L. et al. Longevity pathways converge on autophagy genes to regulate life span in Caenorhabditis elegans. Autophagy 4, 330–338 (2008). [DOI] [PubMed] [Google Scholar]
- Hashimoto, Y. , Ookuma, S. & Nishida, E. Lifespan extension by suppression of autophagy genes in Caenorhabditis elegans. Genes Cells 14, 717–726 (2009). [DOI] [PubMed] [Google Scholar]
- Lapierre, L. R. et al. The TFEB orthologue HLH-30 regulates autophagy and modulates longevity in Caenorhabditis elegans. Nat. Commun. 4, 2267 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palikaras, K. , Lionaki, E. & Tavernarakis, N. Coordination of mitophagy and mitochondrial biogenesis during ageing in C. elegans. Nature 521, 525–528 (2015). [DOI] [PubMed] [Google Scholar]
- Semenza, G. L. Targeting HIF-1 for cancer therapy. Nat. Rev. Cancer 3, 721–732 (2003). [DOI] [PubMed] [Google Scholar]
- Mehta, R. et al. Proteasomal regulation of the hypoxic response modulates aging in C. elegans. Science 324, 1196–1198 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y. , Shao, Z. , Zhai, Z. , Shen, C. & Powell-Coffman, J. A. The HIF-1 hypoxia-inducible factor modulates lifespan in C. elegans. PLoS ONE 4, e6348 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leiser, S. F. , Fletcher, M. , Begun, A. & Kaeberlein, M. Life-span extension from hypoxia in Caenorhabditis elegans requires both HIF-1 and DAF-16 and is antagonized by SKN-1. J. Gerontol. A Biol. Sci. Med. Sci. 68, 1135–1144 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, S. J. , Hwang, A. B. & Kenyon, C. Inhibition of respiration extends C. elegans life span via reactive oxygen species that increase HIF-1 activity. Curr. Biol. 20, 2131–2136 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leiser, S. F. et al. Cell nonautonomous activation of flavin-containing monooxygenase promotes longevity and health span. Science 350, 1375–1378 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antebi, A. Steroid regulation of C. elegans diapause, developmental timing and longevity. Curr. Top. Dev. Biol. 105, 181–212 (2013). [DOI] [PubMed] [Google Scholar]
- Hsin, H. & Kenyon, C. Signals from the reproductive system regulate the lifespan of C. elegans. Nature 399, 362–366 (1999). [DOI] [PubMed] [Google Scholar]
- Arantes-Oliveira, N. , Apfeld, J. , Dillin, A. & Kenyon, C. Regulation of life-span by germ-line stem cells in Caenorhabditis elegans. Science 295, 502–505 (2002). [DOI] [PubMed] [Google Scholar]
- Gerisch, B. , Weitzel, C. , Kober-Eisermann, C. , Rottiers, V. & Antebi, A. A hormonal signaling pathway influencing C. elegans metabolism, reproductive development and life span. Dev. Cell 1, 841–851 (2001). [DOI] [PubMed] [Google Scholar]
- Rottiers, V. et al. Hormonal control of C. elegans dauer formation and life span by a Rieske-like oxygenase. Dev. Cell 10, 473–482 (2006). [DOI] [PubMed] [Google Scholar]
- Gerisch, B. et al. A bile acid-like steroid modulates Caenorhabditis elegans lifespan through nuclear receptor signaling. Proc. Natl Acad. Sci. USA 104, 5014–5019 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamawaki, T. M. et al. The somatic reproductive tissues of C. elegans promote longevity through steroid hormone signaling. PLoS Biol. 8, e1000468 10.1371/journal.pbio.1000468 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman, J. R. & Kenyon, C. Germ-cell loss extends C. elegans life span through regulation of DAF-16 by kri-1 and lipophilic-hormone signaling. Cell 124, 1055–1068 (2006). [DOI] [PubMed] [Google Scholar]
- Shen, Y. , Wollam, J. , Magner, D. , Karalay, O. & Antebi, A. A steroid receptor-microRNA switch regulates life span in response to signals from the gonad. Science 338, 1472–1476 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormick, M. , Chen, K. , Ramaswamy, P. & Kenyon, C. New genes that extend Caenorhabditis elegans’ lifespan in response to reproductive signals. Aging Cell 11, 192–202 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, M. C. , O’Rourke, E. J. & Ruvkun, G. Fat metabolism links germline stem cells and longevity in C. elegans. Science 322, 957–960 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilchez, D. et al. RPN-6 determines C. elegans longevity under proteotoxic stress conditions. Nature 489, 263–268 (2012). [DOI] [PubMed] [Google Scholar]
- Goudeau, J. et al. Fatty acid desaturation links germ cell loss to longevity through NHR-80/HNF4 in C. elegans. PLoS Biol. 9, e1000599 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apfeld, J. & Kenyon, C. Regulation of lifespan by sensory perception in Caenorhabditis elegans. Nature 402, 804–809 (1999). [DOI] [PubMed] [Google Scholar]
- Alcedo, J. & Kenyon, C. Regulation of C. elegans longevity by specific gustatory and olfactory neurons. Neuron 41, 45–55 (2004). [DOI] [PubMed] [Google Scholar]
- Lee, S. J. & Kenyon, C. Regulation of the longevity response to temperature by thermosensory neurons in Caenorhabditis elegans. Curr. Biol. 19, 715–722 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier, W. , Adilov, B. , Regenass, M. & Alcedo, J. A neuromedin U receptor acts with the sensory system to modulate food type-dependent effects on C. elegans lifespan. PLoS Biol. 8, e1000376 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao, R. et al. A genetic program promotes C. elegans longevity at cold temperatures via a thermosensitive TRP channel. Cell 152, 806–817 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, T. & Cai, D. Counterbalance between BAG and URX neurons via guanylate cyclases controls lifespan homeostasis in C. elegans. EMBO J. 32, 1529–1542 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maures, T. J. et al. Males shorten the life span of C. elegans hermaphrodites via secreted compounds. Science 343, 541–544 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, E. D. et al. Age- and calorie-independent life span extension from dietary restriction by bacterial deprivation in Caenorhabditis elegans. BMC Dev. Biol. 8, 49 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy, C. T. , Lee, S. J. & Kenyon, C. Tissue entrainment by feedback regulation of insulin gene expression in the endoderm of Caenorhabditis elegans. Proc. Natl Acad. Sci. USA 104, 19046–19050 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tissenbaum, H. A. Genetics, life span, health span and the aging process in Caenorhabditis elegans. J. Gerontol. A Biol. Sci. Med. Sci. 67, 503–510 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herndon, L. A. et al. Stochastic and genetic factors influence tissue-specific decline in ageing C. elegans. Nature 419, 808–814 (2002). [DOI] [PubMed] [Google Scholar]
- Croll, N. A. , Smith, J. M. & Zuckerman, B. M. The aging process of the nematode Caenorhabditis elegans in bacterial and axenic culture. Exp. Aging Res. 3, 175–189 (1977). [DOI] [PubMed] [Google Scholar]
- Hosokawa, H. et al. Rapid accumulation of fluorescent material with aging in an oxygen-sensitive mutant mev-1 of Caenorhabditis elegans. Mech. Ageing Dev. 74, 161–170 (1994). [DOI] [PubMed] [Google Scholar]
- Ulrich, P. & Cerami, A. Protein glycation, diabetes and aging. Recent Prog. Horm. Res. 56, 1–21 (2001). [DOI] [PubMed] [Google Scholar]
- Pan, C. L. , Peng, C. Y. , Chen, C. H. & McIntire, S. Genetic analysis of age-dependent defects of the Caenorhabditis elegans touch receptor neurons. Proc. Natl Acad. Sci. USA 108, 9274–9279 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, J. et al. Functional aging in the nervous system contributes to age-dependent motor activity decline in C. elegans. Cell Metab. 18, 392–402 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucanic, M. , Lithgow, G. J. & Alavez, S. Pharmacological lifespan extension of invertebrates. Ageing Res. Rev. 12, 445–458 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]


