Abstract
Ras, a small GTPase found primarily on the inner leaflet of the plasma membrane, is an important signaling node and an attractive target for anticancer therapies. Lateral organization of Ras on cellular membranes has long been a subject of intense research; in particular, whether it forms dimers on membranes as part of its regulatory function has been a point of great interest. Here we report Ras dimer formation on membranes by Type II photosensitization reactions, in which molecular oxygen mediates the radicalization of proteins under typical fluorescence experimental conditions. The presence of Ras dimers on membranes was detected by diffusion-based fluorescence techniques including fluorescence correlation spectroscopy and single particle tracking, and molecular weights of the stable covalently coupled species were confirmed by gel electrophoresis. Fluorescence spectroscopy implicates interprotein dityrosine as one of the dimerization motifs. The specific surface tyrosine distribution on Ras renders the protein especially sensitive to this reaction, and point mutations affecting surface tyrosines are observed to alter dimerization potential. The photosensitization reactions are reflective of physiological oxidative stress induced by reactive oxygen species, suggesting such processes may occur naturally and influence signaling pathways in cells.
Ras is a critical protein that serves as a molecular switch in cellular signaling, cycling between GDP- and GTP-bound states on membranes as a part of many signaling pathways.1 Its importance is underscored by the fact that its oncogenic mutants are found in 30% of human cancers.2 As with many other membrane-bound signaling proteins, the lateral organization of Ras on membrane surfaces such as dimerization, clustering, and partitioning into different membrane regions is thought to be an integral part of Ras signal regulation.3 Manipulation of the lateral organization of Ras has also emerged as an alternative strategy for anticancer therapies, since traditional small-molecule approaches to directly inhibit Ras have been largely unsuccessful to date.4 Dimerization of Ras has been predicted based on its structure5 as well as interactions with regulators and effectors (e.g., Raf kinase and SOS) in the cellular context,6 but the intrinsic potential of Ras to form molecular dimers on membranes or its effect on signaling in cells remain unclear.
Ras dimers have been detected using optical techniques such as superresolution microscopy, dynamic light scattering (DLS), Förster resonant energy transfer (FRET), fluorescence correlation spectroscopy (FCS), photon counting histogram (PCH), and single particle tracking (SPT).5,7 While these methods identify the presence of Ras dimers and clusters, the identity of the dimeric or clustered species has not been revealed. Through efforts to determine the dimerization kinetics of Ras on membranes using fluorescence methods, we have found that Ras exhibits a distinctively high potential to form covalent dimers through oxidative reactions. A clue came from SPT experiments that followed long trajectories of single Ras molecules diffusing on the membrane. Dimer association or dissociation events were never observed in trajectories lasting up to several seconds (Figure S1). As fast kinetics would typically be expected for a weak dimerization interaction, this suggested that Ras dimers were not in dynamic equilibrium with the monomeric species; rather, the lack of any association or dissociation events was more consistent with cross-linked products. Fluorescence spectroscopy further reveals covalent dityrosine formation as one of the dimerization motifs, which may explain why Ras dimerization exhibits sensitivity to the mutation of protein surface tyrosine residues. Although such oxidative reactions on proteins are well-known,8 the high reactivity and sequence specificity exhibited by Ras in comparison to other proteins appear to be unappreciated aspects in the multifaceted behavior of this protein. Such reactions could be produced in a physiological setting by oxidative stress induced by reactive oxygen species (ROS).9 More broadly, photoredox reactions have emerged recently as a mechanism with applications in catalysis and small molecule activation.10
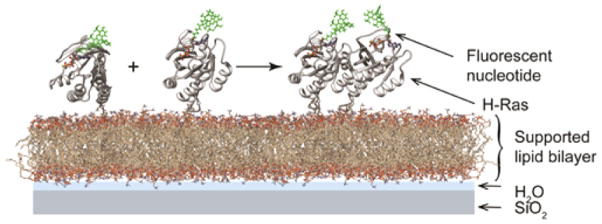
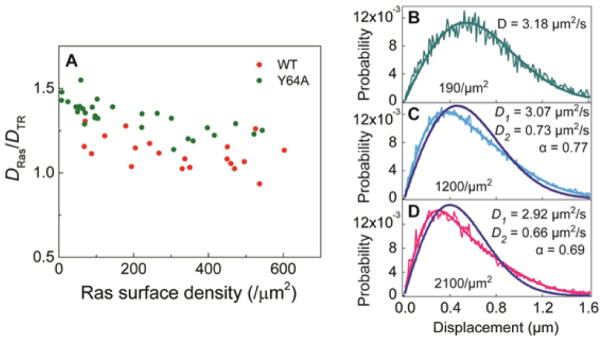
To quantitatively characterize the extent of Ras dimerization on membranes, Ras was reconstituted in vitro on a supported lipid bilayer (SLB) (Figure 1). The SLB platform mirrors the cellular membrane environment while providing a planar geometry amenable for quantitative fluorescence experiments.11 In this setup, Ras was coupled to the membrane via a maleimide–thiol reaction between the terminal cysteine (C181) and the headgroup of MCC-DOPE lipid. Fluorescently labeled nucleotide ATTO 488-GDP or ATTO 488-GppNHp was used to track the protein. When Ras dimerizes, it diffuses significantly more slowly, as the diffusion coefficient for doubly anchored species is appreciably lower than that for singly anchored species on an SLB.12 The mobility of Ras on the surface monitored by both FCS and SPT shows an increasing fraction of a slower-diffusing population as the overall Ras density on the membrane increases, indicating a higher fraction of Ras dimers. Figure 2A shows overall decreasing diffusion measured by FCS with increased surface density for the wild-type (WT) and Y64A mutant H-Ras, although the extent of the effect differs between the constructs. We also observe a similar density dependence in diffusion in SPT (Figure 2B–D, see also Figure S2). At low surface densities (Figure 2B), the step-size distribution acquired from WT Ras diffusion trajectories is adequately described by the two-dimensional (2D), single-species Brownian diffusion model. However, a second, more slowly diffusing species is present at higher surface densities (Figure 2C,D). The fraction of slow species increases with the Ras surface density, while the diffusion coefficients of both fast and slow species remain constant. These results indicate a surface concentration-dependent dimerization and resemble a 2D equilibrium dimerization reaction.7c However, in efforts to characterize the dynamic dimerization equilibrium by measuring the Ras dimer kinetic lifetime, we found evidence for long-lived species that were more suggestive of a nonequilibrium state and covalent dimers (Figure S1).
Figure 1.
Experimental schematic of reconstituted Ras on a supported lipid bilayer (structure from PDB ID 4EFL). The globular domain of Ras was covalently bound to the bilayer, and fluorescently labeled nucleotides were used to track Ras. See SI for detailed methods.
Figure 2.
Density-dependent diffusion of H-Ras on SLB. (A) Relative diffusion of fully labeled Ras compared to fluorescent lipids (Texas Red-DHPE, TR) measured by FCS shows apparent decrease with increasing Ras surface density. (B–D) Step-size distribution of fully labeled wild-type Ras at low (B), medium (C), and high surface densities (D). For B, the data were fit to the single-species Brownian diffusion model, and C and D were fit to two-species model to extract diffusion coefficients of the fast species (D1) and the slow species (D2), and the fraction of fast species (α). The blue lines represent ill-fitting single-species model.
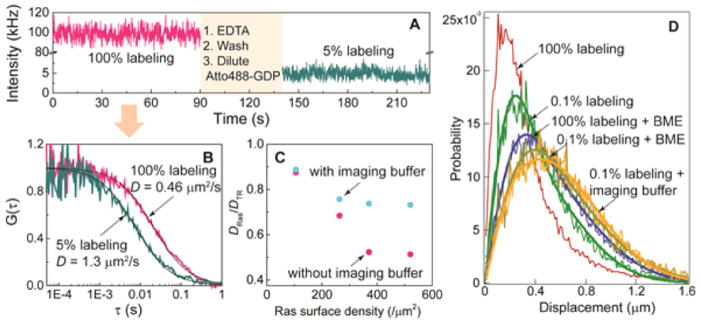
We hypothesized that covalent cross-linking driven by oxidative stress, specifically by fluorophore-mediated photosensitization in this case, was involved in the Ras dimer formation. To test this hypothesis, two conditions expected to reduce these oxidative effects were tested: (1) reducing the total number of fluorescent molecules in the system by substoichiometric labeling (where only a small fraction Ras is bound to fluorescent nucleotides and the rest to dark nucleotides), and (2) using an enzymatic oxygen scavenging imaging buffer (IB, see Methods in SI). Figure 3A,B shows the effect of under-labeling in FCS by comparing the mobility of fully labeled Ras (magenta traces) and the same sample at 5% labeling (green traces), after the removal of fluorescent nucleotide by EDTA and relabeling with dilute fluorescent nuecloetide. For the same sample, underlabeling not only proportionately decreases the average intensity of fluorescence (Figure 3A) but also substantially increases the diffusion coefficient of Ras (Figure 3B). Similarly, when IB is used instead of plain HEPES buffer during FCS data acquisition, Ras shows faster diffusion at the same densities, although IB does not completely eliminate the reduction in mobility (Figure 3C).
Figure 3.
Photosensitized cross-linking of H-Ras on membranes. (A) Fluorescence intensity fluctuation of a high-density His6-Ras sample (~1600/μm2) in an underlabeling experiment. Initially, the sample is fully labeled (magenta trace). Then, the sample is washed and underlabeled (green trace). (B) Autocorrelation of intensity fluctuations. (C) Diffusion coefficients of fully labeled His6-Ras samples with (blue) and without imaging buffer (magenta) measured by FCS. (D) Step-size distributions of high density Ras (~4500/μm2) under the various imaging conditions.
The results from analogous SPT experiments are consistent with the FCS observations. At a very high Ras surface density (~4500/μm2, Figure 3D), the step-size distribution of fully labeled Ras shows complex diffusion behavior that requires a diffusion model with more than two species (Figure 3D, red curve). The addition of BME significantly alleviates the slowing of diffusion, as thiols serve not only as a reducing agent but also as a singlet oxygen quencher, thus mitigating molecular oxygen-mediated cross-linking.13 However, substantial cross-linking occurs in SPT even at an extremely underlabeled condition (0.1% labeling, Figure 3D, green curve).
The SPT underlabeling experiment reveals important mechanistic aspects of the oxidative cross-linking. Considering the duration of an SPT experiment and the diffusion rate of Ras on SLB, it is unlikely that a labeled protein will encounter another labeled protein at 0.1% labeling. Cross-linking despite such low collision probability between labeled proteins suggests that it involves a catalytic intermediate rather than direct dye–dye interactions. This is consistent with Type II photosensitized reaction, in which the photosensitizer, e.g., fluorescent dye, is quenched from the triplet state (T1) to ground state (S0) by molecular oxygen. The oxygen then becomes excited to the singlet electronic state (1O2), a highly unstable and reactive species that may react with the protein of interest, resulting in structural changes, e.g., fragmentation, dimerization, aggregation, or denaturation.14 The aromatic amino acids tyrosine, histidine, and tryptophan as well as sulfur-containing methionine and cysteine are especially vulnerable to oxidative reactions.15 A single dye molecule could visit the excited triplet state and relax back to the ground state multiple times, catalytically producing one singlet oxygen molecule in each cycle, until the dye is eventually irreversibly oxidized.16 This mechanism is illustrated in Figure 4.
Figure 4.
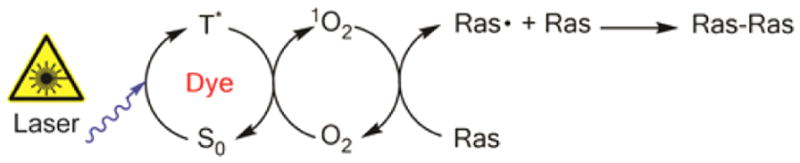
Type II photosensitization cross-linking of Ras. Proposed mechanism of nonspecific cross-linking reaction involves the dye photosensitizer cycling between the ground (S0) and triplet states (T*). The energy transfers to molecular oxygen (O2), which becomes excited to the singlet state (1O2). This ROS reacts with Ras, which forms a free radical species and cross-links with another Ras.
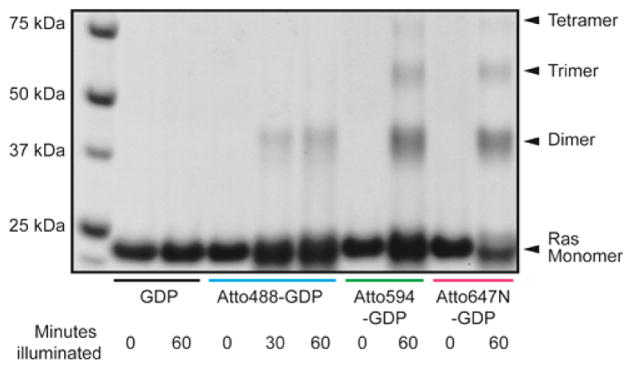
To confirm the formation of Ras covalent dimers through oxidative reactions, globular H-Ras (21 kDa) was irradiated in the presence of different sensitizers (GDP labeled with ATTO series dyes: 488, 594, and 647N) in solution with cw diode laser sources at appropriate wavelengths (485, 561, 640 nm). The irradiance for this experiment was kept near 0.03 kW/cm2, well below the typical irradiance used during FCS (0.4–4 kW/cm2) or SPT (0.1–0.25 kW/cm2) experiments. SDS-PAGE electrophoresis (Figure 5) shows that illumination in the presence of a sensitizer produces covalent Ras dimers. On the other hand, Ras is strictly monomeric in the absence of either irradiation or fluorescent sensitizer bound to the protein, indicating that photochemistry, not other side effects such as heating, is responsible for the dimer formation. The degree of cross-linking is dependent on the type of dye as well as the illumination dosage. Dimers formed with all dyes tested, and trimers and tetramers are clearly visible in some cases. This is consistent with the SPT observations (Figure 3D) indicating the presence of more than two species at high Ras density. The cross-linked Ras species generated by photosensitization were compared to the product of direct excitation of aromatic amino acid side chains by UV irradiation and to that of enzymatic cross-linking by horseradish peroxidase (HRP) and H2O2. Dimers are also formed by UV irradiation and HRP catalysis (Figure S3), indicating that these well-known mechanisms involving ROS can also produce cross-linked products.
Figure 5.
SDS-PAGE gel electrophoresis of laser-irradiated H-Ras. In the presence of GDP labeled with various photosensitizers, Ras forms covalent dimers and higher-order oligomers.
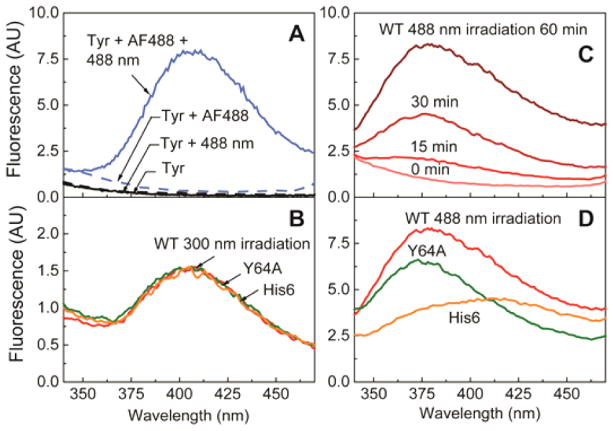
To gain insight into the location of the cross-linking site and the structural differences of dimers formed by different mechanisms, the fluorescence spectra of dityrosine in cross-linked Ras dimers were monitored. Dityrosine formation is a commonly studied reaction in the context of protein degradation due to oxidative stress.17 Oxidation of tyrosine produces tyrosyl free radicals, which can react with each other to generate three isomers of dityrosine.18 The presence of dityrosine can be detected by fluorescence emission at 400 nm, resulting from a shift in fluorescence wavelength of tyrosine at 300 nm upon the extended conjugation of the molecule.19 Figure 6A shows the fluorescence emission spectrum of the dityrosine generated from free L-tyrosine amino acids in HEPES buffer (pH 7.4) by irradiating a dye sensitizer (Alexa Fluor 488). The well-defined band at 400 nm signifies the formation of dityrosine. The fluorescence spectra of dityrosine in cross-linked H-Ras, which has nine surface Tyr, by direct excitation of tyrosine (Type I photosensitization reaction) are similar to that of free dityrosine and identical among the three H-Ras constructs (Figure 6B). This suggests that dimers formed by direct radicalization of tyrosine are uniform in cross-linking sites and its overall structure.
Figure 6.
Dityrosine fluorescence emission spectra (λex = 294 nm). (A) Free tyrosine in solution forms dityrosine in the presence of dye sensitizer (AF488) and laser illumination. (B) Cross-linked Ras products via UV illumination display aspectrum similar to that of free dityrosine. (C) Wild-type Ras in solution has an illumination dosage-dependent growth of the dityrosine band. (D) Three Ras constructs display different spectra, indicating that significantly different structures result from laser illumination.
Ras dimers formed by a Type II photosensitized reaction, where they are produced by fluorescent dye sensitization and laser irradiation, exhibit a fluorescence emission band slightly blue-shifted from the free dityrosine band, and three different H-Ras constructs showed distinct spectral lineshapes. There is a significant broadening for the His6-tagged H-Ras, and even a single point mutation of Y64A results in an appreciably different spectrum from that of the WT (Figure 6D). This indicates that oxidative reactions can be sensitive to small differences in the amino acid sequence and produce significantly different structures. This is not surprising, as Type II photosensitization reactions are nonspecific in the sense that there is no single defined cross-linking site; rather, an ensemble of cross-linked dimers generated by different combinations of multiple possible cross-linking sites is expected. Furthermore, the position of the dye sensitizer relative to the protein may introduce additional structural sensitivity absent in Type I reactions, in which tyrosine is directly excited. These results indicate that cross-linking via tyrosyl free radicals contributes to covalent dimer formation. Similar intermolecular dimerization reactions via dityrosine were observed in other proteins, e.g., calmodulin.20
Several factors may cause membrane-bound Ras to behave differently from solvated Ras, e.g., effectively higher concentration, reduced orientational degrees of freedom, and interactions with the membrane, which has a higher O2 concentration at the hydrocarbon core. In light of the structural sensitivity of the Type II photosensitized Ras dimerization reaction reported above, one may assume that the membrane can introduce more structural identity and enhance the differential susceptibility of Ras to these reactions. Indeed, such effects may underlie the observed differences between WT and the Y64A mutant on membrane surfaces.7c
To provide context to the vulnerability of Ras to oxidative reactions, the degree of covalent cross-linking in other proteins in similar environments was evaluated. Density-dependent diffusion was measured by FCS for H-Ras, the cytosolic domain of linker of activated T-cells (LAT), and nonfluorescent mCherry (mCherry S136C), all tethered to the membrane by His tag chelated to Ni-NTA modified lipid (Figure S4). LAT and nonfluorescent mCherry were labeled with Alexa Fluor 555 and 488, respectively. Only H-Ras showed significant density-dependent diffusion change, suggesting that Ras may be particularly sensitive to oxidative stress-driven dimerization. Its pronounced sensitivity may be attributed to the unusually high density of reactive residues on the surface: H-Ras has nine Tyr, four His, and one Cys, which is significantly higher in density compared to LAT or mCherry. Incidentally, other small GTPases of similar sizes, e.g., Rac1, RhoA, Rab1A, and Rap1A, also have substantially lower numbers of exposed Tyr, His, Trp, and Cys (Table S1). In addition to the number of surface-exposed reactive residues, their relative orientation with respect to each other may influence a protein’s sensitivity to oxidative cross-linking.
We have shown intermolecular cross-linking of Ras on membranes through a mechanism most consistent with Type II photosensitization. Reactive singlet oxygen species, catalytically generated by repeated excitation and quenching of a dye, create free radicals and generate cross-linked proteins via susceptible residues such as tyrosine. These photosensitized oxidative reactions have been known since the beginning of biological fluorescence experiments.21 High-illumination experimental methods such as FCS, TIRF, confocal microscopy, and superresolution techniques all provide sufficient exposure to drive such processes. These reactions, though inherently nonspecific, are remarkably sensitive, and even a point mutation can significantly alter the product distribution. This last point should be taken with a cautionary note, since comparative studies between point mutations are often regarded as the most definitive demonstrations of specificity in experimental results. In regard to the question of Ras dimerization on membranes, these findings may explain discrepancies in recent literature. Within the past few years, there have been reports of N- and H-Ras dimers in in vitro supported membrane with fluorescence methods such as FRET, FCS, PCH, and single molecule TIRF,5,7c and K-Ras dimers in solution with DLS7b and in fixed cells with superresolution imaging.7a Analysis of crystal structures with conserved dimer contacts made a case for transient dimerization of Ras as well.5 On the other hand, another study concluding that Ras lacks intrinsic dimerization using bulk fluorescence anisotropy and NMR,22 and a live cell two-photon PCH study which only found monomeric H-Ras.23 Covalent Ras dimerization was not taken into account in any of these studies. Considering the findings reported here, we would expect that conditions for covalent Ras dimerization on membranes by photosensitized oxidative processes are achieved, to varying degrees, in some of the experiments cited above. More importantly, the distinct predisposition of Ras to such reactions raises the possibility that Ras experiences similar reactions in the cellular context, where ROS are abundant and may even participate in the signaling network at various points.24
Supplementary Material
Acknowledgments
Supported by NIH P01AI091580, NCI U01CA202241, and the Croucher Foundation fellowship (to H.Y.M.L.). We thank Profs. J. Kuriyan and D. Fletcher and Dr. P. Bieling for plasmids, and Drs. S. Hansen and L. Oltrogge for helpful discussions.
Footnotes
Notes
The authors declare no competing financial interest.
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/jacs.5b12648.
Detailed methods; supporting SPT and FCS data (PDF)
References
- 1.Bollag G, McCormick F. Annu Rev Cell Biol. 1991;7:601. doi: 10.1146/annurev.cb.07.110191.003125. [DOI] [PubMed] [Google Scholar]
- 2.(a) Karnoub A, Weinberg RA. Nat Rev Mol Cell Biol. 2008;9:517. doi: 10.1038/nrm2438. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Pylayeva-Gupta Y, Grabocka E, Bar-Sagi D. Nat Rev Cancer. 2011;11:761. doi: 10.1038/nrc3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.(a) Omerovic J, Prior IA. FEBS J. 2009;276:1817. doi: 10.1111/j.1742-4658.2009.06928.x. [DOI] [PubMed] [Google Scholar]; (b) Abankwa D, Gorfe AA, Hancock JF. Semin Cell Dev Biol. 2007;18:599. doi: 10.1016/j.semcdb.2007.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.(a) Gysin S, Salt M, Young A, McCormick F. Genes Cancer. 2011;2:359. doi: 10.1177/1947601911412376. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Santos E. Sci Signaling. 2014;7:pe12. doi: 10.1126/scisignal.2005318. [DOI] [PubMed] [Google Scholar]; (c) Ledford H. Nature. 2015;520:278. doi: 10.1038/520278a. [DOI] [PubMed] [Google Scholar]
- 5.Guldenhaupt J, Rudack T, Bachler P, Mann D, Triola G, Waldmann H, Kotting C, Gerwert K. Biophys J. 2012;103:1585. doi: 10.1016/j.bpj.2012.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.(a) Iversen L, et al. Science. 2014;345:50. [Google Scholar]; (b) Gureasko J, Galush WJ, Boykevisch S, Sondermann H, Bar-Sagi D, Groves JT, Kuriyan J. Nat Struct Mol Biol. 2008;15:452. doi: 10.1038/nsmb.1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.(a) Nan X, et al. Proc Natl Acad Sci USA. 2015;112:7996. doi: 10.1073/pnas.1509123112. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Muratcioglu S, et al. Structure. 2015;23:1325. doi: 10.1016/j.str.2015.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Lin WC, et al. Proc Natl Acad Sci USA. 2014;111:2996. [Google Scholar]
- 8.Davies MJ. Biochem Biophys Res Commun. 2003;305:761. doi: 10.1016/s0006-291x(03)00817-9. [DOI] [PubMed] [Google Scholar]
- 9.Ray PD, Huang BW, Tsuji Y. Cell Signalling. 2012;24:981. doi: 10.1016/j.cellsig.2012.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Prier CK, Rankic DA, MacMillan DWC. Chem Rev. 2013;113:5322. doi: 10.1021/cr300503r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.(a) Yu CH, Groves JT. Med Biol Eng Comput. 2010;48:955. doi: 10.1007/s11517-010-0634-x. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Groves JT, Parthasarathy R, Forstner MB. Annu Rev Biomed Eng. 2008;10:311. doi: 10.1146/annurev.bioeng.10.061807.160431. [DOI] [PubMed] [Google Scholar]
- 12.(a) Knight JD, Lerner MG, Marcano-Velazquez JG, Pastor RW, Falke JJ. Biophys J. 2010;99:2879. doi: 10.1016/j.bpj.2010.08.046. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Groves JT, Wulfing C, Boxer SG. Biophys J. 1996;71:2716. doi: 10.1016/S0006-3495(96)79462-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.(a) Rasnik I, McKinney SA, Ha T. Nat Methods. 2006;3:891. doi: 10.1038/nmeth934. [DOI] [PubMed] [Google Scholar]; (b) Devasagayam TPA, Sundquist AR, Di Mascio P, Kaiser S, Sies H. J Photochem Photobiol, B. 1991;9:105. doi: 10.1016/1011-1344(91)80008-6. [DOI] [PubMed] [Google Scholar]
- 14.Wright A, Bubb WA, Hawkins CL, Davies MJ. Photochem Photobiol. 2002;76:35. doi: 10.1562/0031-8655(2002)076<0035:sompoe>2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 15.Michaeli A, Feitelson J. Photochem Photobiol. 1994;59:284. doi: 10.1111/j.1751-1097.1994.tb05035.x. [DOI] [PubMed] [Google Scholar]
- 16.Kasche V, Lindqvist L. J Phys Chem. 1964;68:817. [Google Scholar]
- 17.Heinecke JW, Li W, Daehnke HL, Goldstein JA. J Biol Chem. 1993;268:4069. [PubMed] [Google Scholar]
- 18.Shen HR, Spikes JD, Smith CJ, Kopecek J. J Photochem Photobiol, A. 2000;133:115. [Google Scholar]
- 19.Malencik DA, Anderson SR. Amino Acids. 2003;25:233. doi: 10.1007/s00726-003-0014-z. [DOI] [PubMed] [Google Scholar]
- 20.Malencik DA, Anderson SR. Biochemistry. 1987;26:695. doi: 10.1021/bi00377a006. [DOI] [PubMed] [Google Scholar]
- 21.(a) Sheetz MP, Koppel DE. Proc Natl Acad Sci USA. 1979;76:3314. doi: 10.1073/pnas.76.7.3314. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Goosey JD, Zigler J, Kinoshita JH. Science. 1980;208:1278. doi: 10.1126/science.7375939. [DOI] [PubMed] [Google Scholar]; (c) Niedermayer T, et al. Proc Natl Acad Sci USA. 2012;109:10769. doi: 10.1073/pnas.1121381109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kovrigina EA, Galiakhmetov AR, Kovrigin EL. Biophys J. 2015;109:1000. doi: 10.1016/j.bpj.2015.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smith EM, MacDonald PJ, Chen Y, Mueller JD. Biophys J. 2014;107:66. doi: 10.1016/j.bpj.2014.04.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thannickal VJ, Fanburg BL. Am J Physiol: Lung Cell Mol Physiol. 2000;279:L1005. doi: 10.1152/ajplung.2000.279.6.L1005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.