Abstract
Activation of small GTPases of the Ras superfamily by guanine nucleotide exchange factors (GEFs) is a key step in numerous cell signaling processes. Unveiling the detailed molecular mechanisms of GEF-GTPase signaling interactions is of great importance due to their central roles in cell biology, including critical disease states, and their potential as therapeutic targets. Here we present an assay to monitor individual Ras activation events catalyzed by single molecules of the GEF Son of Sevenless (SOS) in the natural membrane environment. The assay employs zero-mode waveguide (ZMW) nanostructures containing a single Ras-functionalized liposome. The ZMWs facilitate highly localized excitation of fluorophores in the vicinity of the liposome membrane, allowing direct observation of individual Ras activation events as single SOS enzymes catalyze exchange of unlabeled nucleotides bound to Ras with fluorescently labeled nucleotides from solution. The system is compatible with continuous recording of long sequences of individual enzymatic turnover events over hour-long time scales. The single turnover waiting time sequence is a molecular footprint that details the temporal characteristics of the system. Data reported here reveal long-lived activity states that correspond to well-defined conformers of SOS at the membrane. Liposome functionalized ZMWs allow for studies of nucleotide exchange reactions at single GTPase resolution, providing a platform to gauge the mechanisms of these processes.
Keywords: GTPase, GEF, Ras, SOS, zero-mode waveguides, single molecule
Graphical abstract
Membrane-bound small GTPases of the Ras superfamily function as regulatory toggle switches in many signal transduction pathways and are implicated in a number of severe pathologies, including cancer.1–3 GTPases shuttle between an inactive GDP-bound state and a GTP-bound state that promotes downstream signaling. Guanine nucleotide exchange factors (GEFs) activate their target GTPases by facilitating release of GDP from the nucleotide binding cleft, which is followed by uptake of a GTP nucleotide from the cell’s cytosol. Activation of Ras by the GEF Son of Sevenless (SOS) relays signals from transmembrane receptors downstream to the mitogen-activated protein kinase (MAPK) cascade.4 In the cell, Ras is anchored to the inner leaflet of the plasma membrane through palmitoyl and farnesyl moieties5 whereas SOS resides in the cytosol and gets recruited to the membrane upon activation of transmembrane receptors.6,7 At the membrane, SOS catalyzes the exchange of Ras-bound GDP with GTP, thus activating Ras and triggering downstream signaling.8,9 Pathological mutations in Ras map to more than 30% of human cancers,10,11 whereas several mutations in Son of Sevenless (SOS) are implicated in the developmental disorders Noonan’s12,13 and CFC syndrome.14 For this reason, extensive resources have been invested in developing inhibitors of Ras activation15,16 and toward blocking the Ras–SOS interaction.17–19 However, clinical success to this end has been limited. Shedding light on the molecular scale functional properties of Ras activation by SOS can provide insights into this problem, possibly exposing new pharmacalogical strategies.
Here we present a fluorescence microscopy assay that enables detection of single GEF-catalyzed Ras GTPase activation events reconstituted at a lipid membrane surface. Single turnover recording is the ultimate resolution limit in enzyme activity assays and has radically widened our view of enzyme kinetics, most prominently by uncovering the mechanistically important concept of memory effects,20–22 that is, dynamic spontaneous fluctuations between distinct active conformers. In addition, studies of single enzyme turnover statistics have provided a means for inferring complex enzyme reaction landscapes that cannot be mapped from ensemble experiments.20,23 Single turnover assays have been established for a number of enzyme classes (e.g., cholesterol oxidase,24 lysozyme,25 and metabolic lipases21) but at this stage no such method is available for the broad class of GEF-mediated activation of small GTPases, including Ras.
We recently reported that Ras binding to the allosteric pocket within the catalytic module of SOS (SOSCat) is sufficient for recruiting SOS to lipid bilayers in a sustained manner.26,27 This finding enables isolation of single molecules of SOS using micropatterned supported lipid bilayers and subsequent assay of their time-averaged nucleotide exchange activity.26 These studies revealed that Ras activation by SOS does not occur at a constant pace; SOS activity fluctuates between discrete long-lived (up to minutes) activity states with lifetimes comparable to that of cellular signaling responses. A key insight from these experiments was that alteration of the activity fluctuation pattern of SOS has the capacity to alter the integrated output of an entire signaling pathway. Most importantly, this modulation exists in the time sequence of activity and is not detectable in the averaged behavior of SOS. However, due to the limited temporal resolution in the supported bilayer assay it is unclear whether fast (second scale) activity fluctuations are also present in the system or if the observed states correspond to discrete structures with well-defined catalytic rates. Here, by monitoring the time sequence of single GTPase activation events we show that there are no systematic rapid fluctuations in the kinetics of Ras activation by SOS, thus demonstrating the observed long-lived activity states of SOS correspond to well-defined conformations of the enzyme at the membrane surface.
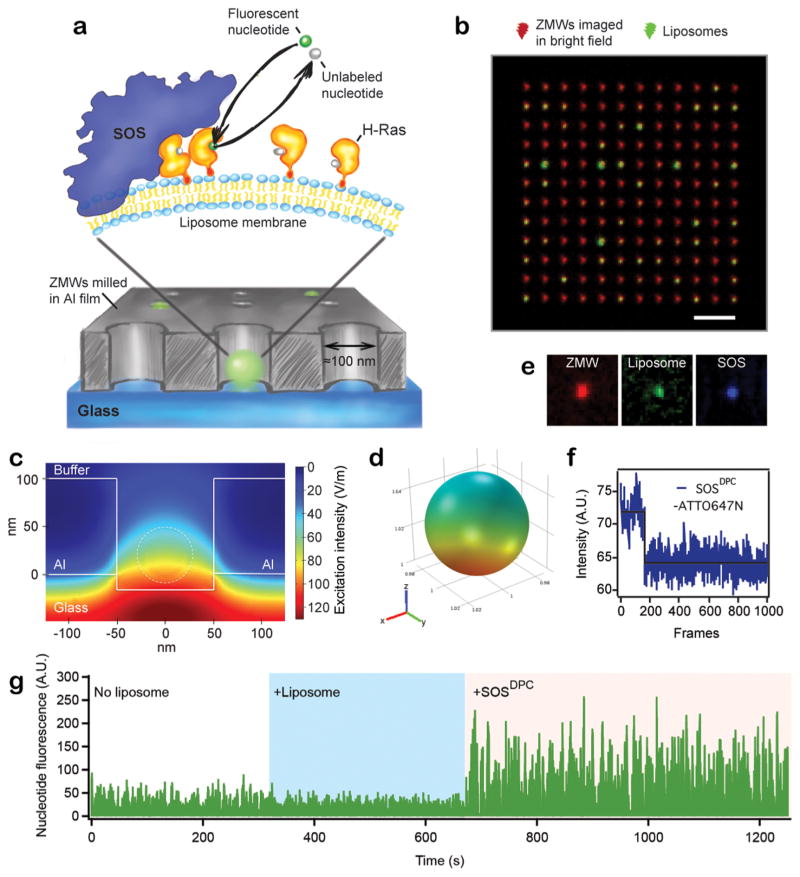
We reconstituted SOS-mediated activation of membrane-coupled H-Ras (henceforth simply Ras) at the surface of liposomes that have been incorporated within arrays of zero-mode waveguides (ZMWs) (Figure 1a). ZMWs are nanoscopic apertures (ø ~ 100 nm) in thin metal films (Supplementary Figure 1) that upon illumination with light produce a highly confined evanescent field with an effective focal volume in the atto- to zeptoliter range (10−18–10−21 L).28 We exploit this property to minimize background signal from fluorescently labeled nucleotide analogs, distributed in solution and functioning as reporters of single Ras activation events (Figure 1a). The good match between the physical size of individual ZMWs and small unilamellar liposomes29 facilitates stoichiometric 1:1 loading of these two components simply by incubation and spontaneous adsorption (Figure 1b).
Figure 1.
ZMWs functionalized with Ras-SOS decorated liposomes. (a) ZMWs with diameters of ~100 nm loaded with liposomes of compatible dimensions (average ø ~ 30 nm, see Methods in the Supporting Information). H-Ras is chemically anchored via a lipid to the liposome surface. The liposomes also bear a single copy of the Ras-GEF SOS, which is stably associated with the lipid membrane via binding of Ras at an allosteric binding pocket. SOS-catalyzed nucleotide exchange on Ras is observed by following the acquisition of fluorescently labeled GTP from solution. (b) Micrograph of a ZMW array (red, imaged in bright field) and colocalized liposomes (green, fluorescence image of membrane marker). (c) Simulation of the electromagnetic excitation field inside a ZMW with a liposome bound at the glass interface. Dashed line indicates the position of the liposome. (d) World map representation of the excitation field on the surface of the liposome. (e) Micrographs showing a ZMW and the associated liposome and SOS signal. (f) Bleaching of the SOS label (ATTO647N) in a single step signifies that the liposome harbors a single SOS molecule (trace corresponding to the micrographs in (e)). (g) Intensity trace of ATTO488-GTP from a single ZMW. The first part of the trace reflects the signal from a single ZMW without a Ras decorated liposome present. In the second part of the trace, a Ras loaded liposome was immobilized in the same ZMW. Finally, SOS was added from solution, resulting in apparent recruitment of fluorescent nucleotides (observed as a marked signal increase). The fluorescent nucleotide, ATTO488-GTP, was incubated at the ZMW grid at 10 μM. See also Supplementary Figure 3.
The highly confined excitation volume inside a ZMW allows for single molecule imaging at μM concentration of fluorescently labeled reactants in solution, a concentration range relevant for numerous protein–protein and protein–ligand interactions natively occurring in cells.30 ZMW technology has enabled a number of single enzyme applications, including DNA sequencing by tracing of polymerase activity,31 real-time monitoring of protein translation by the ribosome32 and probing the lifetime of protein–protein interactions in the GroEl–GroES chaperonin system.33 A fundamental requirement in these experiments is that the enzyme of interest is tethered at the floor of the ZMWs, thus confining a single enzyme to a single waveguide. However, this strategy is incompatible with studying enzymes that either are embedded in or functionally coupled to lipid membranes. Our use of surface supported liposomes34 as the reconstitution scaffold circumvents this issue. A crucial strength of this approach is that proteins and ligands bound to the liposome membrane retain lateral fluidity, and thus their ability to collide and react, while being confined to individual ZMWs. We note that continuous supported lipid bilayers can be formed on glass surfaces35 and have previously been interfaced with arrays of ZMWs to study diffusion of membrane associated fluorescent molecules.36,37 In those studies, however, proteins and ligands were not trapped within the ZMW and as such that configuration is incompatible with extended single molecule observation. While supported membranes could conceivably be formed in confined ZMWs in combination with membrane-patterning methods26,38,39 we chose to work with whole liposomes as a simpler approach. Liposomes have the additional advantage of accommodating transmembrane proteins, which are notoriously problematic in the supported membrane configuration.40
The evanescent field inside a ZMW exhibits a nontrivial decay profile, which potentially could complicate observation of fluorescence from dyes diffusing at the membrane of immobilized liposomes. We performed finite element simulations of electromagnetic wave propagation in ZMWs30 to illuminate this aspect (see Methods in the Supporting Information). Figure 1c shows the result of a simulation where a liposome (dashed line) was situated at the center of a ZMW. The excitation field intensity at the membrane varies with approximately a factor of 2 between the top and the bottom of the liposome (see also world map representation in Figure 1d). However, molecules at the membrane are not fixed but undergo Brownian motion in the course of a camera exposure at the microscope. A direct consequence is that fluorophores sample an ensemble of excitation intensities during each exposure. Indeed, for the imaging settings used in the reported work (≈20 ms per frame), we find that Brownian motion is sufficient to average out any experimentally relevant heterogeneity in the excitation field (see Methods in the Supporting Information and Supplementary Figure 2). This is a critical feature, because it allows the detection of single fluorophore binding and bleaching events as, respectively, abrupt step increases and step decreases in fluorescence intensity.
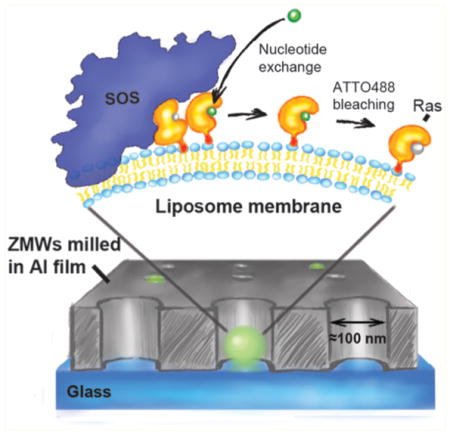
We monitored successive single Ras activation events by observing SOS mediated exchange of unlabeled nucleotides bound to Ras with a fluorescent nucleotide analog (GTP-ATTO488) from solution (Figure 1a). Ras was chemically linked to the liposome bilayer via coupling of a C-terminal cysteine to maleimide functionalized lipids27 (see Methods in the Supporting Information). Prior to immobilization, the Ras decorated liposomes were incubated with SOS, resulting in stable anchoring of SOS to the liposome upon binding of Ras to the allosteric pocket.26 The liposomes serve as a vector for docking the reactant complex in the ZMWs and provide an authentic membrane environment reminiscent of the native signaling platform of these enzymes.
The experimental design allows direct imaging of the location of ZMWs (bright field), liposomes (via a membrane marker), and SOS (labeled with a single dye molecule) (Figure 1e). Experiments were run under conditions where each active liposome is likely to bind only a single molecule of SOS, which was confirmed by step-photobleaching of the SOS label (Figure 1f). Control experiments in which fluorescent nucleotide is incubated on the same ZMW array before and after immobilization of Ras liposomes and, ultimately, in the presence of SOS verify that nucleotide exchange activity (observed as increased fluorescence intensity) is evident only in the presence of SOS (Figure 1g and Supplementary Figure 3).
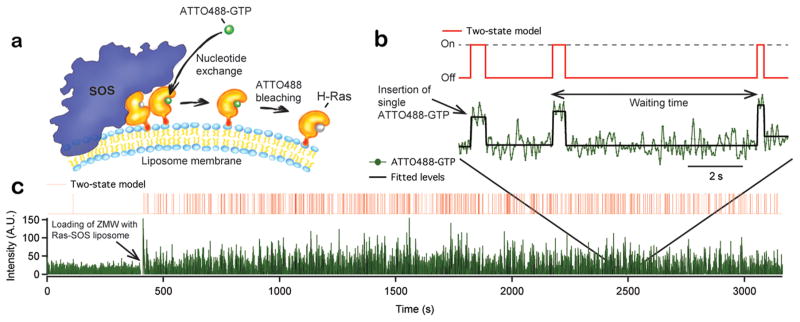
Upon activation of single Ras proteins, the fluorescence intensity recorded from individual ZMWs exhibits an abrupt increase, followed by a plateau, and finally step photobleaching of the nucleotide label (Figure 2a,b). Following bleaching, the fluorescence intensity remains at baseline level until the next Ras activation event, resulting in telegram-type sequence of activation events, as shown in Figure 2b. We fit entire hour-long trajectories with hundreds of activation events, employing a change point algorithm and a set of level assignment criteria to sort the trace into “on” (corresponding to the scenario where a fluorescing nucleotide is bound to Ras at the liposome) and “off” (no fluorescing nucleotide bound) states (Figure 2b,c see also Methods in the Supporting Information). In this way, the waiting times between consecutive Ras activation events are extracted (Figure 2b). The resulting single turnover waiting time sequence is a molecular footprint that details the temporal characteristics of the system comprised by a single SOS enzyme catalyzing Ras activation at a particular liposome.
Figure 2.
Single turnover recording of reconstituted Ras activation by SOS. (a) Assay principle: SOS exchanges unlabeled nucleotide on Ras for ATTO488-GTP. Shortly after binding of the nucleotide to Ras, ATTO488 bleaches and the fluorescence signal reverses to baseline before the next insertion event. This gives rise to telegram-like kinetic traces, where individual insertion events stand out as transient step increases of the fluorescence nucleotide signal. (b) Nucleotide exchange activity trace (green) showing single Ras activation events characterized by abrupt step increases in ATTO488-GTP fluorescence intensity followed by single step bleaching of the nucleotide label. To quantify waiting times, steps were located using a change point detection algorithm (black trace) and then fitted to a two-state on/off model (red trace). (c) Full nucleotide exchange trace from a single ZMW before and after immobilization of a Ras-SOS liposome. Same trace coloring as in (b).
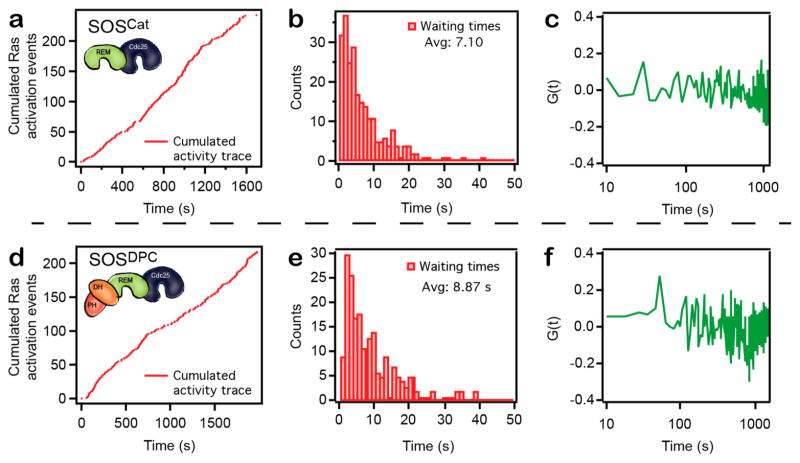
Figure 3a,d illustrates cumulated nucleotide exchange events as a function of time for a SOSCat (construct containing only the catalytic core of the protein, comprised by the CDC25 and REM domains) and a SOSDPC enzyme (construct that, in addition to the catalytic core, has a DH-PH module). For each trace, the associated histogram of waiting times (respectively, Figure 3 panel b and e) and the autocorrelation function of the waiting times (respectively, Figure 3 pane c and f) are calculated. The SOSDPC trace exhibits slightly longer average waiting times (corresponding to slower catalysis) that is in qualitative agreement with previous results showing that the domains N-terminal to the catalytic module of SOS dampens the nucleotide exchange activity26,27,41 (Supplementary Figure 4). Under the conditions of these measurements, the observed nucleotide exchange kinetics in the ZMW were slower compared to our previous study with planar supported membranes26 (see also Supplementary Figure 4). While this effect could conceivably be due to membrane curvature, other differences in the assay format, such as nucleotide concentration (10 μM in the ZMW versus 120 μM in the planar supported bilayer measurements), cannot be ruled out. However, we do observe distinct features among the recorded traces indicating kinetics in the lipsome–ZMW system are not dominated by limited diffusion.
Figure 3.
Single turnover analysis reveals that SOS catalytic states correspond to well-defined conformations of the enzyme. (a) Cumulated Ras activation events as a function of time for an activity trace acquired with the catalytic core of SOS (SOSCat). (b) Waiting time histogram corresponding to data in a. (c) Autocorrelation (G(t) = 〈Δτ(0)Δτ(m)〉 / 〈Δτ2〉, Δτ(m) = τ(m) − 〈τ〉20) of the waiting times corresponding to data shown in a. (d–f) Same data format as in a–c but for an activity trace acquired with SOSDPC, a construct containing the N-terminal DH-PH domains of SOS in addition to the catalytic core. It should be noted that this type of data are intrinsically stochastic and therefore no two traces are the same. Importantly, the overall method is reproducible in its ability to capture these long sequences of Ras activation events.
An enzyme working at a fixed catalytic rate (characteristic of a well-defined protein conformation) throughout a finite time interval exhibits no correlation between consecutive waiting times20 (see also Supplementary Figure 5). On the contrary, dynamic fluctuations in enzyme catalytic rate manifests as a nonzero autocorrelation function with a decay time characteristic to the time scale of the underlying fluctuations.20 Extremely long time scale (e.g., hundreds to thousands of seconds) dynamic heterogeneity in the catalytic rate of Ras activation by SOS have been observed directly in single molecule membrane assays.26 Those earlier studies, however, lack the temporal resolution to determine if faster dynamic heterogeneity exists, as would be expected if the structure of SOS on the membrane is very flexible, or if the long-lived activity states correspond to well-defined structures. Data reported in Figure 3 exhibit no indication of systematic fluctuations in SOS activity on shorter time scales. Thus, we conclude that individual SOS catalytic states exist in well-defined conformations with highly regular catalytic cycles on the membrane. We did also observe the extremely long time scale state transitions,26 as illustrated by a trace from SOSDPC with correlated waiting times at time scales of ≈1300 s. (Figure 4a,b).
Figure 4.
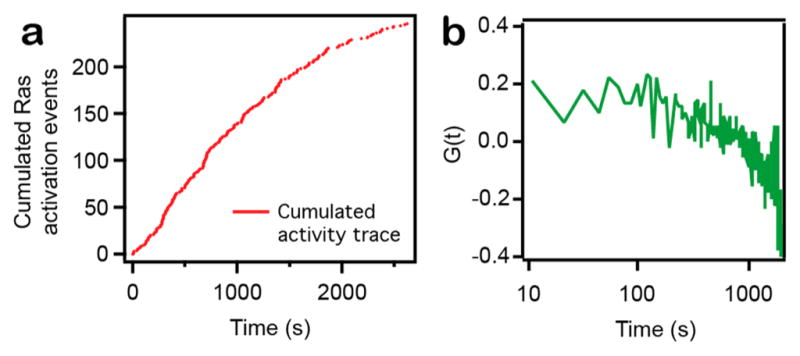
Evidence for long time scale state transition in SOS activity. (a) Cumulated Ras activation events as a function of time (trace acquired with SOSDPC). Note the decreased turnover activity toward the end of the trace. (b) Normalized autocorrelation of waiting times for a SOSDPC trace exhibiting a nonzero G(t), indicative of dynamically disordered enzymatic activity.
We have introduced a liposome-interfaced ZMW assay employed here to measure time sequences of single Ras activation events catalyzed by SOS on a lipid membrane surface. To the best of our knowledge, this is the first example of continuously resolving single GTPase activation kinetics mediated by an individual GEF enzyme. The assay principle with Ras and SOS can be readily generalized to study a multitude of biologically important GEF–GTPase signaling reactions at lipid membranes. We anticipate that studies of single GTPase activation kinetics will make important contributions toward deciphering the detailed molecular mechanisms of GEF–GTPase interactions. The introduced single molecule assay based on liposome-interfaced ZMW nanostructures enables experimental assessment of the fundamental kinetic properties of these critically important regulatory enzymes.
Supplementary Material
Acknowledgments
We thank Professor John Kuriyan for generous access to his laboratory. We thank Dr. Wan-Li for helpful discussions and assistance and Dr. Scott Hansen for purifying Ras. We thank Fang Fang for preparing the displayed cartoon representations. Major support included NIH P01 AI091580. The work was supported in part by an individual postdoc grant to S.M.C. (Danish Council for Independent Research – Natural Sciences). D.S. was supported by a grant from the Danish Council for Independent Research – Natural Sciences.
Footnotes
Author Contributions
S.M.C. and J.T.G. conceived the experiments. S.M.C. and M.G.T. performed the experiments and analyzed the data. C.R., M.G.T., and S.M.C. prepared the ZMW substrates. M.G.T. and C.R. performed finite element simulations. S.M.C. wrote the software for data analysis. H.L.T. assisted with the experiment design and with the SOS labeling. J.S.I. purified the proteins. S.M.C. and J.T.G. wrote the paper with assistance from M.G.T. All authors discussed and commented on the manuscript.
Notes
The authors declare no competing financial interest.
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.nano-lett.6b00969.
Description of methods and five supplementary figures. (PDF)
References
- 1.Cherfils J, Zeghouf M. Nat Chem Biol. 2011;7:493–495. doi: 10.1038/nchembio.608. [DOI] [PubMed] [Google Scholar]
- 2.Barr F, Lambright DG. Curr Opin Cell Biol. 2010;22(4):461–470. doi: 10.1016/j.ceb.2010.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pylayeva-Gupta Y, Grabocka E, Bar-Sagi D. Nat Rev Cancer. 2011;11:761–774. doi: 10.1038/nrc3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mor A, Philips MR. Annu Rev Immunol. 2006;24:771–800. doi: 10.1146/annurev.immunol.24.021605.090723. [DOI] [PubMed] [Google Scholar]
- 5.Ahearn IM, Haigis K, Bar-Sagi D, Philips MR. Nat Rev Mol Cell Biol. 2011;13:39–51. doi: 10.1038/nrm3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McCormick F. Nature. 1993;363(6424):15–6. doi: 10.1038/363015a0. [DOI] [PubMed] [Google Scholar]
- 7.Jun JE, Li M, Chen H, Chakraborty AK, Roose JP. Mol Cell Biol. 2013;33:2470. doi: 10.1128/MCB.01593-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bos JL, Rehmann H, Wittinghofer A. Cell. 2007;129(5):865–77. doi: 10.1016/j.cell.2007.05.018. [DOI] [PubMed] [Google Scholar]
- 9.Findlay GM, Pawson T. Nat Struct Mol Biol. 2008;15(6):538–40. doi: 10.1038/nsmb0608-538. [DOI] [PubMed] [Google Scholar]
- 10.Karnoub AE, Weinberg RA. Nat Rev Mol Cell Biol. 2008;9(7):517–31. doi: 10.1038/nrm2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pylayeva-Gupta Y, Grabocka E, Bar-Sagi D. Nat Rev Cancer. 2011;11(11):761–74. doi: 10.1038/nrc3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tartaglia M, Pennacchio LA, Zhao C, Yadav KK, Fodale V, Sarkozy A, Pandit B, Oishi K, Martinelli S, Schackwitz W, Ustaszewska A, Martin J, Bristow J, Carta C, Lepri F, Neri C, Vasta I, Gibson K, Curry CJ, Siguero JP, Digilio MC, Zampino G, Dallapiccola B, Bar-Sagi D, Gelb BD. Nat Genet. 2007;39(1):75–9. doi: 10.1038/ng1939. [DOI] [PubMed] [Google Scholar]
- 13.Roberts AE, Araki T, Swanson KD, Montgomery KT, Schiripo TA, Joshi VA, Li L, Yassin Y, Tamburino AM, Neel BG, Kucherlapati RS. Nat Genet. 2007;39(1):70–4. doi: 10.1038/ng1926. [DOI] [PubMed] [Google Scholar]
- 14.Tumurkhuu M, Saitoh M, Takita J, Mizuno Y, Mizuguchi M. J Recept Signal Transduction Res. 2013;33(2):124–8. doi: 10.3109/10799893.2013.779279. [DOI] [PubMed] [Google Scholar]
- 15.Thompson H. Nat Med. 2013;19(8):949–950. doi: 10.1038/nm0813-949. [DOI] [PubMed] [Google Scholar]
- 16.Stephen AG, Esposito D, Bagni RK, McCormick F. Cancer Cell. 2014;25:272–281. doi: 10.1016/j.ccr.2014.02.017. [DOI] [PubMed] [Google Scholar]
- 17.Wang W, Fang G, Rudolph J. Mol Med. 2012;18:1. [Google Scholar]
- 18.Patgiri A, Yadav KK, Arora PS, Bar-Sagi D. Nat Chem Biol. 2011;7(9):585–7. doi: 10.1038/nchembio.612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maurer T, Garrenton LS, Oha A, Pitts K, Anderson DJ, Skelton NJ, Fauber BP, Pan B, Malek S, Stokoe D, Ludlam MJC, Bowmana KK, Wue J, Giannettic AM, Starovasnik MA, Mellman I, Jackson PK, Rudolph J, Wang W, Fang G. Proc Natl Acad Sci U S A. 2012;109(14):5299–304. doi: 10.1073/pnas.1116510109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.English BP, Min W, van Oijen AM, Lee KT, Luo G, Sun H, Cherayil BJ, Kou SC, Xie XS. Nat Chem Biol. 2006;2:87–94. doi: 10.1038/nchembio759. [DOI] [PubMed] [Google Scholar]
- 21.Engelkamp H, Hatzakis NS, Hofkens J, De Schryver FC, Nolte RJM, Rowan AE. Chem Commun. 2006:935–940. doi: 10.1039/b516013h. [DOI] [PubMed] [Google Scholar]
- 22.Lu HP. Chem Soc Rev. 2014;43:1118–1143. doi: 10.1039/c3cs60191a. [DOI] [PubMed] [Google Scholar]
- 23.Moffitt JR, Chemla YR, Bustamante C. Methods Enzymol. 2010;475:221–257. doi: 10.1016/S0076-6879(10)75010-2. [DOI] [PubMed] [Google Scholar]
- 24.Lu HP, Xun L, Xie XS. Science. 1998;282:1877–1882. doi: 10.1126/science.282.5395.1877. [DOI] [PubMed] [Google Scholar]
- 25.Choi Y, Moody IS, Sims PC, Hunt SR, Corso BL, Perez I, Weiss GA, Collins PG. Science. 2012;335:319–324. doi: 10.1126/science.1214824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Iversen L, Tu HL, Lin WC, Christensen SM, Abel SM, Iwig J, Wu HJ, Gureasko J, Rhodes C, Petit RS, Hansen SD, Thill P, Yu CH, Stamou D, Chakraborty AK, Kuriyan J, Groves JT. Science. 2014;345(6192):50–4. doi: 10.1126/science.1250373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gureasko J, Galush WJ, Boykevisch S, Sondermann H, Bar-Sagi D, Groves JT, Kuriyan J. Nat Struct Mol Biol. 2008;15(5):452–61. doi: 10.1038/nsmb.1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhu P, Craighead HG. Annu Rev Biophys. 2012;41:269–293. doi: 10.1146/annurev-biophys-050511-102338. [DOI] [PubMed] [Google Scholar]
- 29.Kunding AH, Mortensen MW, Christensen SM, Stamou D. Biophys J. 2008;95:1176–1188. doi: 10.1529/biophysj.108.128819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Levene MJ, Korlach J, Turner SW, Foquet M, Craighead HG, Webb WW. Science. 2003;299:682–686. doi: 10.1126/science.1079700. [DOI] [PubMed] [Google Scholar]
- 31.Eid J, Fehr A, Gray J, Luong K, Lyle J, Otto G, Peluso P, Rank D, Baybayan P, Bettman B, Bibillo A, Bjornson K, Chaudhuri B, Christians F, Cicero R, Clark S, Dalal R, de Winter A, Dixon J, Foquet M, Gaertner A, Hardenbol P, Heiner C, Hester K, Holden D, Kearns G, Kong X, Kuse R, Lacroix Y, Lin S, Lundquist P, Ma C, Marks P, Maxham M, Murphy D, Park I, Pham T, Phillips M, Roy J, Sebra R, Shen G, Sorenson J, Tomaney A, Travers K, Trulson M, Vieceli J, Wegener J, Wu D, Yang A, Zaccarin D, Zhao P, Zhong F, Korlach J, Turner S. Science. 2009;323:133–138. doi: 10.1126/science.1162986. [DOI] [PubMed] [Google Scholar]
- 32.Uemura S, Aitken CE, Korlach J, Flusberg BA, Turner SW, Puglisi JD. Nature. 2010;464:1012–1017. doi: 10.1038/nature08925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sameshima T, Iizuka R, Ueno T, Wada J, Aoki M, Shimamoto N, Ohdomari I, Tanii T, Funatsu T. J Biol Chem. 2010;285:23159–23164. doi: 10.1074/jbc.M110.122101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Christensen SM, Stamou D. Soft Matter. 2007;3:828–836. doi: 10.1039/b702849k. [DOI] [PubMed] [Google Scholar]
- 35.Tamm LK, Groves JT. J Struct Biol. 2009;168:1–2. doi: 10.1016/j.jsb.2009.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wenger J, Rigneault H, Dintinger J, Marguet D, Lenne PF. J Biol Phys. 2006;32:SN1–SN4. doi: 10.1007/s10867-006-2909-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Samiee KT, Moran-Mirabal JM, Cheung YK, Craighead HG. Biophys J. 2006;90:3288–3299. doi: 10.1529/biophysj.105.072819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Groves JT, Ulman N, Boxer SG. Science. 1997;275(5300):651–653. doi: 10.1126/science.275.5300.651. [DOI] [PubMed] [Google Scholar]
- 39.Groves JT, Boxer SG. Acc Chem Res. 2002;35(3):149–57. doi: 10.1021/ar950039m. [DOI] [PubMed] [Google Scholar]
- 40.Salafsky J, Groves JT, Boxer SG. Biochemistry. 1996;35(40):14773–14781. doi: 10.1021/bi961432i. [DOI] [PubMed] [Google Scholar]
- 41.Gureasko J, Kuchment O, Makino DL, Sondermann H, Bar-Sagi D, Kuriyan J. Proc Natl Acad Sci U S A. 2010;107(8):3430–5. doi: 10.1073/pnas.0913915107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.