Abstract
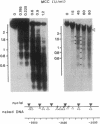
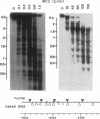
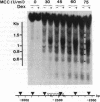
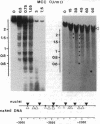
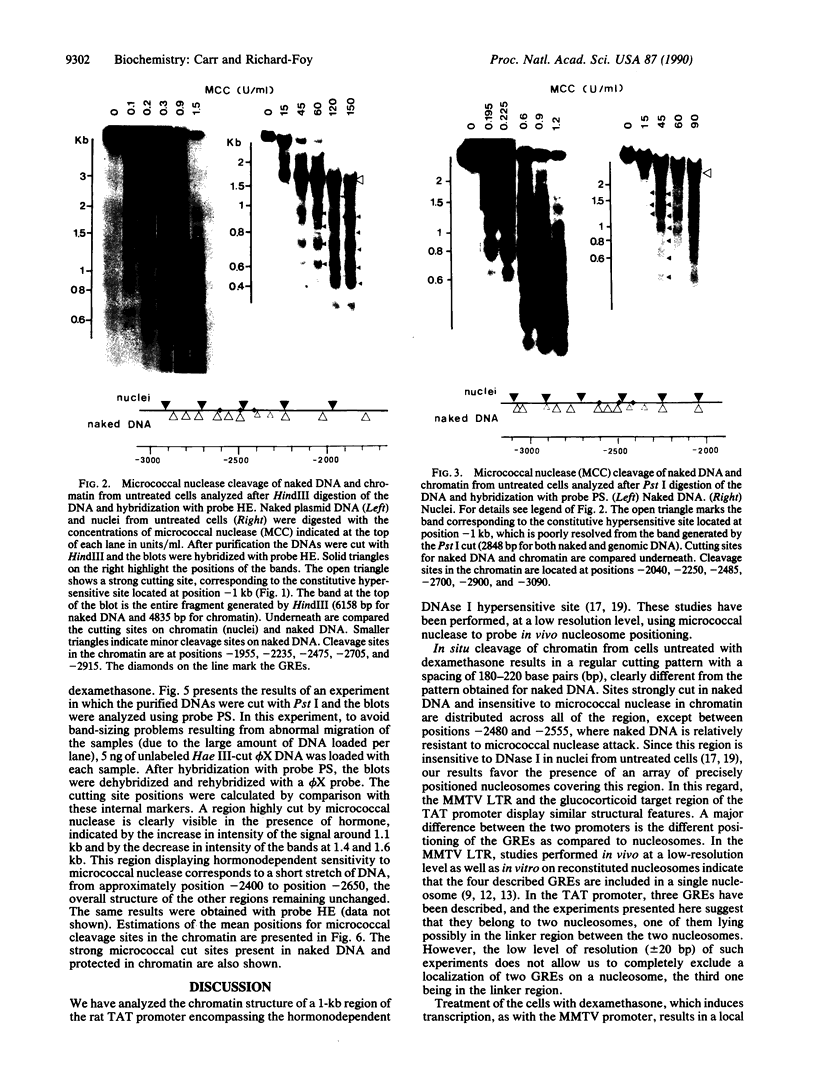
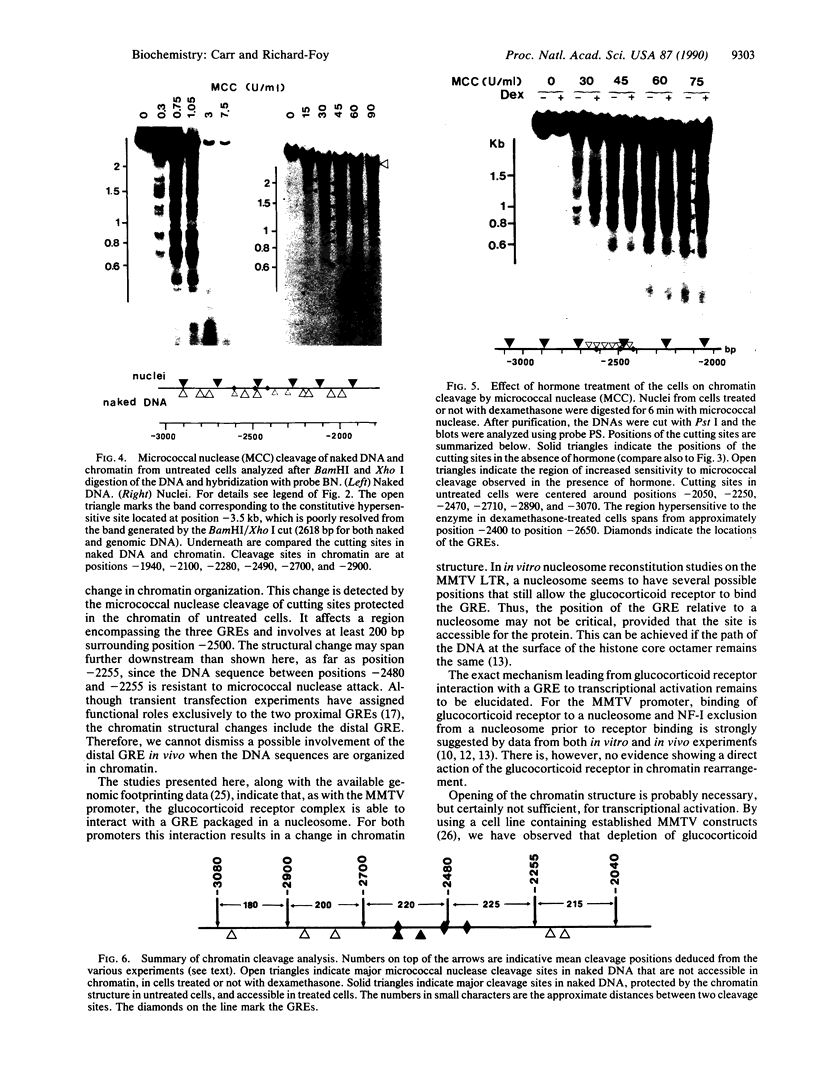
Transcriptional activation by steroid hormones is often associated with the appearance of a DNase I hypersensitive site resulting from a local alteration of the nucleoprotein structure of the promoter. For the mouse mammary tumor virus long terminal repeat, a viral promoter under glucocorticoid control, a model has been proposed: the appearance of the hormonodependent DNase I hypersensitive site reflects the displacement of a single precisely positioned nucleosome associated with the glucocorticoid responsive elements. To determine if such a mechanism is of general relevance in transcriptional activation by steroid hormones, we have investigated the nucleosomal organization of the rat tyrosine aminotransferase promoter over a 1-kilobase region that contains the glucocorticoid regulatory target. This region displays a hormonodependent DNase I hypersensitive site. In the absence of hormone, micrococcal nuclease digestion of nuclei demonstrates the presence of positioned nucleosomes, with cutting sites centered around positions -3080, -2900, -2700, -2800, -2255, and -2040. Treatment of the cells with dexamethasone induces a disruption of the chromatin structure over a relatively short stretch of DNA (approximately positions -2400 to -2650) that overlaps two nucleosomes. These observations suggest a strong similarity in the role of chromatin structure in glucocorticoid-dependent transcriptional activation of mouse mammary tumor virus and tyrosine aminotransferase promoters.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Almer A., Rudolph H., Hinnen A., Hörz W. Removal of positioned nucleosomes from the yeast PHO5 promoter upon PHO5 induction releases additional upstream activating DNA elements. EMBO J. 1986 Oct;5(10):2689–2696. doi: 10.1002/j.1460-2075.1986.tb04552.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker P. B., Gloss B., Schmid W., Strähle U., Schütz G. In vivo protein-DNA interactions in a glucocorticoid response element require the presence of the hormone. Nature. 1986 Dec 18;324(6098):686–688. doi: 10.1038/324686a0. [DOI] [PubMed] [Google Scholar]
- Benezra R., Cantor C. R., Axel R. Nucleosomes are phased along the mouse beta-major globin gene in erythroid and nonerythroid cells. Cell. 1986 Mar 14;44(5):697–704. doi: 10.1016/0092-8674(86)90835-4. [DOI] [PubMed] [Google Scholar]
- Cordingley M. G., Riegel A. T., Hager G. L. Steroid-dependent interaction of transcription factors with the inducible promoter of mouse mammary tumor virus in vivo. Cell. 1987 Jan 30;48(2):261–270. doi: 10.1016/0092-8674(87)90429-6. [DOI] [PubMed] [Google Scholar]
- Dingwall C., Lomonossoff G. P., Laskey R. A. High sequence specificity of micrococcal nuclease. Nucleic Acids Res. 1981 Jun 25;9(12):2659–2673. doi: 10.1093/nar/9.12.2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elgin S. C. The formation and function of DNase I hypersensitive sites in the process of gene activation. J Biol Chem. 1988 Dec 25;263(36):19259–19262. [PubMed] [Google Scholar]
- Grange T., Guénet C., Dietrich J. B., Chasserot S., Fromont M., Befort N., Jami J., Beck G., Pictet R. Complete complementary DNA of rat tyrosine aminotransferase messenger RNA. Deduction of the primary structure of the enzyme. J Mol Biol. 1985 Jul 20;184(2):347–350. doi: 10.1016/0022-2836(85)90386-9. [DOI] [PubMed] [Google Scholar]
- Grange T., Roux J., Fromont-Racine M., Pictet R. Positive and negative regulation of a transfected chimeric tyrosine aminotransferase gene: effect of copy number. Exp Cell Res. 1989 Jan;180(1):220–233. doi: 10.1016/0014-4827(89)90226-7. [DOI] [PubMed] [Google Scholar]
- Grange T., Roux J., Rigaud G., Pictet R. Two remote glucocorticoid responsive units interact cooperatively to promote glucocorticoid induction of rat tyrosine aminotransferase gene expression. Nucleic Acids Res. 1989 Nov 11;17(21):8695–8709. doi: 10.1093/nar/17.21.8695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han M., Kim U. J., Kayne P., Grunstein M. Depletion of histone H4 and nucleosomes activates the PHO5 gene in Saccharomyces cerevisiae. EMBO J. 1988 Jul;7(7):2221–2228. doi: 10.1002/j.1460-2075.1988.tb03061.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hörz W., Altenburger W. Sequence specific cleavage of DNA by micrococcal nuclease. Nucleic Acids Res. 1981 Jun 25;9(12):2643–2658. doi: 10.1093/nar/9.12.2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jantzen H. M., Strähle U., Gloss B., Stewart F., Schmid W., Boshart M., Miksicek R., Schütz G. Cooperativity of glucocorticoid response elements located far upstream of the tyrosine aminotransferase gene. Cell. 1987 Apr 10;49(1):29–38. doi: 10.1016/0092-8674(87)90752-5. [DOI] [PubMed] [Google Scholar]
- Morse R. H., Simpson R. T. DNA in the nucleosome. Cell. 1988 Jul 29;54(3):285–287. doi: 10.1016/0092-8674(88)90190-0. [DOI] [PubMed] [Google Scholar]
- Nedospasov S. A., Georgiev G. P. Non-random cleavage of SV40 DNA in the compact minichromosome and free in solution by micrococcal nuclease. Biochem Biophys Res Commun. 1980 Jan 29;92(2):532–539. doi: 10.1016/0006-291x(80)90366-6. [DOI] [PubMed] [Google Scholar]
- Nitsch D., Stewart A. F., Boshart M., Mestril R., Weih F., Schütz G. Chromatin structures of the rat tyrosine aminotransferase gene relate to the function of its cis-acting elements. Mol Cell Biol. 1990 Jul;10(7):3334–3342. doi: 10.1128/mcb.10.7.3334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oddos J., Grange T., Carr K. D., Matthews B., Roux J., Richard-Foy H., Pictet R. Nucleotide sequence of 10 kilobases of rat tyrosine aminotransferase gene 5' flanking region. Nucleic Acids Res. 1989 Nov 11;17(21):8877–8878. doi: 10.1093/nar/17.21.8877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrowski M. C., Richard-Foy H., Wolford R. G., Berard D. S., Hager G. L. Glucocorticoid regulation of transcription at an amplified, episomal promoter. Mol Cell Biol. 1983 Nov;3(11):2045–2057. doi: 10.1128/mcb.3.11.2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payvar F., DeFranco D., Firestone G. L., Edgar B., Wrange O., Okret S., Gustafsson J. A., Yamamoto K. R. Sequence-specific binding of glucocorticoid receptor to MTV DNA at sites within and upstream of the transcribed region. Cell. 1983 Dec;35(2 Pt 1):381–392. doi: 10.1016/0092-8674(83)90171-x. [DOI] [PubMed] [Google Scholar]
- Perlmann T., Wrange O. Specific glucocorticoid receptor binding to DNA reconstituted in a nucleosome. EMBO J. 1988 Oct;7(10):3073–3079. doi: 10.1002/j.1460-2075.1988.tb03172.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piña B., Brüggemeier U., Beato M. Nucleosome positioning modulates accessibility of regulatory proteins to the mouse mammary tumor virus promoter. Cell. 1990 Mar 9;60(5):719–731. doi: 10.1016/0092-8674(90)90087-u. [DOI] [PubMed] [Google Scholar]
- Richard-Foy H., Hager G. L. Sequence-specific positioning of nucleosomes over the steroid-inducible MMTV promoter. EMBO J. 1987 Aug;6(8):2321–2328. doi: 10.1002/j.1460-2075.1987.tb02507.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheidereit C., Geisse S., Westphal H. M., Beato M. The glucocorticoid receptor binds to defined nucleotide sequences near the promoter of mouse mammary tumour virus. Nature. 1983 Aug 25;304(5928):749–752. doi: 10.1038/304749a0. [DOI] [PubMed] [Google Scholar]
- Shinomiya T., Scherer G., Schmid W., Zentgraf H., Schütz G. Isolation and characterization of the rat tyrosine aminotransferase gene. Proc Natl Acad Sci U S A. 1984 Mar;81(5):1346–1350. doi: 10.1073/pnas.81.5.1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas G. H., Elgin S. C. Protein/DNA architecture of the DNase I hypersensitive region of the Drosophila hsp26 promoter. EMBO J. 1988 Jul;7(7):2191–2201. doi: 10.1002/j.1460-2075.1988.tb03058.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C. The 5' ends of Drosophila heat shock genes in chromatin are hypersensitive to DNase I. Nature. 1980 Aug 28;286(5776):854–860. doi: 10.1038/286854a0. [DOI] [PubMed] [Google Scholar]