Abstract
As the fundamental packing units of DNA in eukaryotes, nucleosomes play a central role in governing DNA accessibility in a variety of cellular processes. Our understanding of the mechanisms underlying this complex regulation has been aided by unique structural and dynamic perspectives offered by single molecule techniques. Recent years have witnessed remarkable advances achieved using these techniques, including the generation of a detailed histone-DNA energy landscape, elucidation of nucleosome disassembly processes, and real-time monitoring of molecular motors interacting with nucleosomes. These and other highlights of single molecule nucleosome studies will be discussed in this review.
Introduction
In eukaryotic cells, chromatin is responsible for compacting and organizing DNA within the nucleus and for regulating complex and dynamic patterns of gene expression. The ability of chromatin to accomplish these vital and diverse tasks lies in part in its fundamental unit, the nucleosome core particle (NCP). The NCP consists of approximately 147 base pairs (bp) of DNA wrapped 1.65 times around an octamer of histone proteins containing two copies each of H2A, H2B, H3 and H4 [1]. With the aid of linker histones, arrays of NCPs are further compacted into higher-order structures of varying degrees of complexity. Thus, nucleosomes present a significant obstacle for many processes involving DNA, including transcription, replication, and repair.
DNA accessibility is modulated by nucleosome structural dynamics and positioning [2, 3]. Specifically, transient thermal fluctuations (breathing), which temporarily loosen localized DNA-histone interactions, and spontaneous thermal repositioning (sliding) of the histone octamer along the DNA both serve to regulate access to protein binding sites [3]. In addition, a variety of chaperones [4–6] and ATP- dependent enzymes known as chromatin remodelers [7, 8] have been found to reposition, restructure, or remove nucleosomes. A further layer of complexity and control is provided by a wide array of histone posttranslational modifications (PTMs) and histone variants that alter the structure and stability of nucleosomes, as well as the chromatin higher-order structure [9–11]. Understanding how these factors act individually and collectively to regulate DNA-based processes is a significant goal of current biological, biochemical, and biophysical research.
As an important part of this scientific endeavor, single-molecule (SM) techniques are making a pronounced impact on the field through their ability to provide a unique perspective into biological processes. While some SM methods enable the precise and controlled application and measurement of force and torque, others are able to directly measure the kinetics of biomolecular systems. They all share the advantage of removing the issue of ensemble averaging common in bulk studies. Real-time measurements of dynamic events on the millisecond to minute timescales and the capacity to resolve population heterogeneities allow for a more complete understanding of protein function and regulation. However, these techniques are usually implemented in vitro and often require modifying the system of interest, e.g. by attaching a microsphere or a fluorophore. SM techniques are best suited for well-defined molecular systems, and other approaches may be more appropriate to investigate more complex systems. Ultimately, to gain a full understanding of intracellular processes, SM and bulk methods should be used as complementary approaches.
In this review, we focus on notable studies of nucleosome structure, dynamics, remodeling, and interactions with RNA polymerase that have been made in the last several years utilizing optical or magnetic tweezers as well as fluorescence-based SM techniques. Although other SM techniques have made significant strides in the field, we have narrowed the scope of this paper to cover the most commonly used methods and aim to highlight how they are driving important advances in nucleosome studies.
Histone-DNA Interactions within a Nucleosome
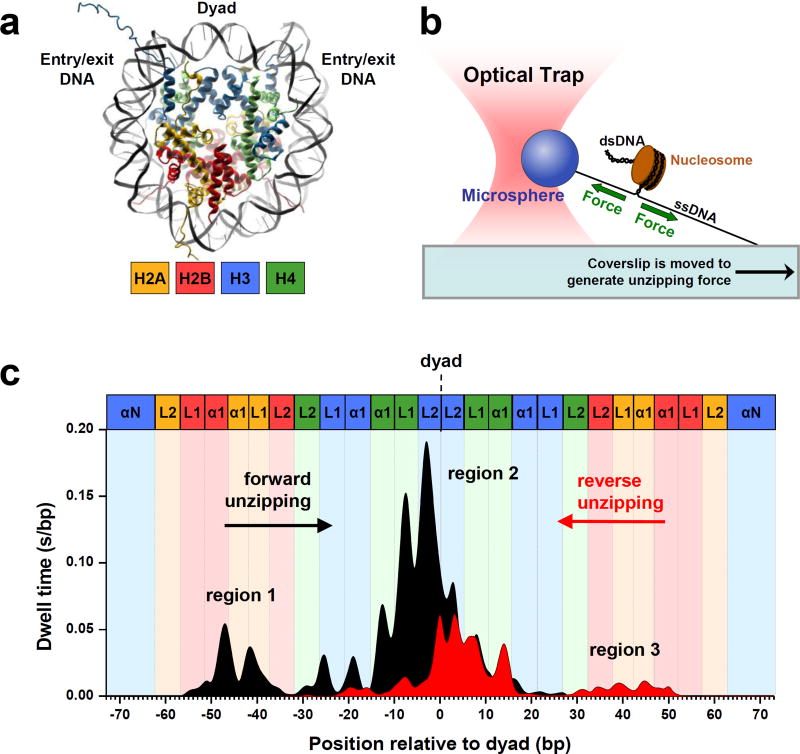
Histone-DNA interactions in a nucleosome represent a fundamental level of gene regulation through which nucleosome stability, DNA accessibility, transcription, and chromatin higher order structures are all controlled [3]. The high-resolution crystal structure of the nucleosome [1, 12] (Figure 1a) shows the locations of these interactions, while SM techniques are capable of directly measuring not only those locations, but their strengths and dynamic behaviors as well. Hall et al. [13] produced a detailed map of the DNA-histone interaction landscape using a powerful optical tweezers-based DNA unzipping technique (Figures 1b & 1c). These experiments revealed highly non-uniform strengths of histone-DNA interactions within a nucleosome and three broad regions of interactions: a region of strong interactions at the dyad and two regions of relatively weaker interactions ~40 bp from either side of the dyad. These results are in agreement with previous SM results from stretching nucleosomal DNA using optical tweezers [14, 15]. In addition, a distinctive 5-bp periodicity was observed throughout all three regions. According to the crystal structure of the nucleosome, histone core domains were expected to make strong contacts with the DNA minor groove every 10 bp [1, 12]. The observed 5 bp periodicity demonstrated that two distinct interactions at each minor groove contact, one from each strand, could be disrupted sequentially rather than simultaneously (Figure 1b). Furthermore, Hall et al. demonstrated that as histone-DNA interactions were disrupted beyond the dyad region, the entire histone octamer dissociated from the DNA. Beyond quantifying key features of the histone-DNA interaction landscape, this study has significant implications for the mechanism of transcription through a nucleosome which we will address later in this review.
Figure 1. Dynamic mapping of histone-DNA interactions in a nucleosome.
(a) The crystal structure [12] of the nucleosome core particle (NCP) revealed 147 (bp) of duplex DNA wrapped in 1.65 turns around an octameric histone core in a left-handed superhelix. The octameric core consists of two H2A/H2B dimers and one (H3/H4)2 tetramer. The tetramer and the dimers are arranged around a two-fold symmetry axis (the dyad).
(b) Experimental configuration of the optical tweezers-based DNA unzipping technique [13]. An optical trap utilizes a highly focused laser beam to generate a gradient force to trap a micron-sized particle. The same laser beam can also be used to precisely measure the position of the particle in the trap. In this experiment, the nucleosomal template was suspended between the glass coverslip surface and a microsphere. An optical trap was used to apply a force necessary to unzip through the nucleosomal DNA as the coverslip was moved away from the trapped microsphere.
(c) The histone-DNA interaction map within a nucleosome, generated from dwell time histograms of the unzipping fork held under a constant force of ~ 28 pN. The nucleosomal DNA was unzipped in both the forward (black) and reverse (red) unzipping directions. Each peak corresponds to an individual histone-DNA interaction and their heights are indicative of their relative strengths. Three regions of strong interactions are indicated: one located at the dyad (region 2) and two located ~40 bp away from the dyad (regions 1 and 3). Colored boxes indicate predictions from the crystal structure where individual histone binding motifs as shown in (a) are expected to interact with DNA. (Adapted from Hall et al. [13] with publisher’s permission)
DNA unzipping can provide important structural information when a crystal structure is not available. In a recent study, Dechassa et al. [16] employed bulk methods combined with DNA unzipping to probe the structure of the centromeric nucleosome to differentiate between several proposed models [17–19]. They reconstituted budding yeast centromeric nucleosomes (with histone H3 replaced by its yeast variant, Cse4). Results from both bulk and SM methods pointed towards a canonical octameric structure with weakened outer turn DNA interactions for Cse4-containing nucleosomes. The crystal structure of the centromeric nucleosome containing the human analog of Cse4 (CENP-A) [20] published several months later was consistent with these findings.
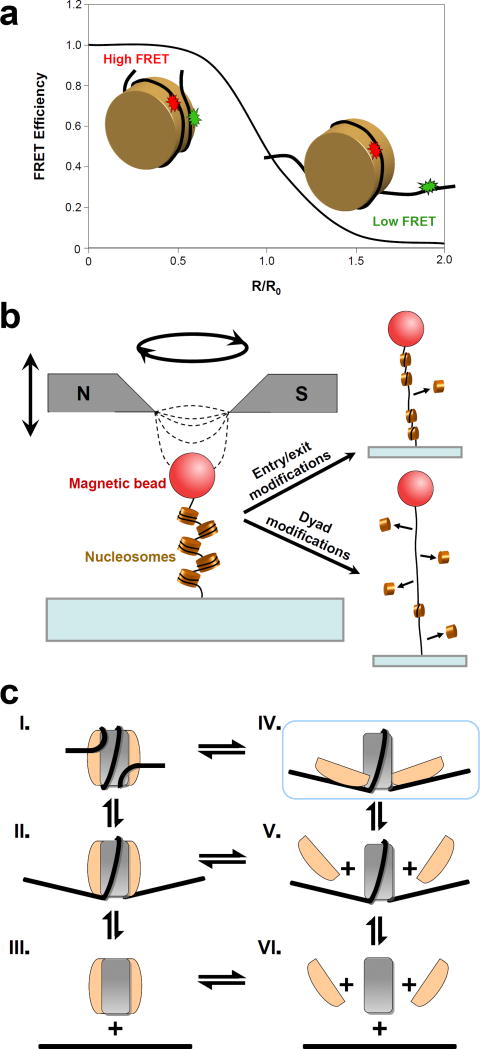
These studies have shown that histone-DNA interactions create a significant energy barrier [13, 14, 21] that limits the accessibility of nucleosomal DNA. Certain PTMs, most notably acetylation and phosphorylation, have been implicated to play a role in the modulation of these interactions and in altering chromatin higher-order structure [11, 22, 23]. Two recent SM studies coupled with novel protein engineering techniques involving histone core domain modifications took a closer look at how this regulation is accomplished. Neumann et al. [24] genetically encoded a site-specific acetyl-lysine modification (H3K56Ac) and used a combination of biochemical methods and SM fluorescence resonance energy transfer (smFRET) (Figure 2a) to investigate the effects of this modification on nucleosome breathing, chromatin higher-order structure, and chromatin remodeling. A 7-fold increase in nucleosomal DNA breathing in the acetylated samples was detected, while chromatin compaction and remodeling were largely unaffected, implying the presence of other mechanisms contributing to the role of H3K56Ac in gene silencing [25]. Another study by Simon et al. [26] used magnetic tweezers (Figure 2b) and FRET to examine the effects of several site-specific PTMs created by chemical ligation. They found that certain PTMs near the dyad affected nucleosome stability and promoted disassembly, whereas other PTMs near the DNA entry/exit sites tended to increase unwrapping rates and enhance transcription factor binding without affecting nucleosome stability. The apparent ability of distinct PTMs to independently regulate unwrapping rates and nucleosome stability is an intriguing result that has implications for processes involving nucleosome disassembly. In addition to histone PTMs, modification of the underlying chemical structure of DNA has been recently implicated as a factor in nucleosome stability. In particular, by monitoring nucleosome breathing on CpG-methylated DNA using smFRET, Choy et al. [27] demonstrated that DNA methylation increases nucleosome compaction and rigidity. This discovery may point towards the mechanistic basis for the contribution of CpG methylation to gene repression [28].
Figure 2. Single molecule studies of nucleosome stability and disassembly.
(a) FRET efficiency is shown as a function of the relative distance between a donor (green) and an acceptor (red) dyes attached to a nucleosomal DNA. FRET is typically used as a molecular ruler on the scale of 1–10 nm, making it an ideal technique to detect and quantify nucleosome dynamics (nucleosome radius ~5 nm). As an example, increased DNA breathing in a nucleosome containing H3K56Ac from that of an unmodified nucleosome was manifested as a reduced FRET efficiency in experiments by Neumann et al. [24
(b) Schematic of the magnetic tweezers experiments on a nucleosomal array. A pair of permanent magnets produces a gradient of the magnetic field, thus exerting a force on the super-paramagnetic bead. Magnetic tweezers are also potentially capable of inducing rotational motion. In the experiments by Simon et al. [26] the authors exerted forces up to 29 pN and observed partial dissociation of the nucleosomes that had been acetylated near the dyad location (lower right), while acetylation near the entry/exit site did not significantly impact nucleosome stability (upper right).
(c) Pathways for nucleosome disassembly. The (H3/H4)2 tetramer is shown in grey and the H2A/H2B dimer is shown in beige. Disassembly has been previously proposed [30, 31] to occur via the dissociation of the octamer as a whole (III-VI) or H2A/H2B dissociation followed by the loss of the tetramer (V-VI). Böhm et al. [32] have additionally found evidence for a novel intermediate (IV, highlighted in blue) in which dimer-tetramer interactions are partially disrupted, while dimer-DNA contacts are maintained. The authors suggest that nucleosome disassembly proceeds sequentially through this intermediate (I-IV-V-VI). Future studies are necessary to differentiate between these possible pathways of disassembly. (Adapted from Böhm et al. [32] with publisher’s permission)
Nucleosome Disassembly
While the results of the aforementioned studies are beginning to elucidate how histone-DNA interactions regulate nucleosomal DNA accessibility, other studies are revealing the mechanism by which nucleosomes disassemble. Two recent studies used FRET to examine salt-induced nucleosome disassembly pathways as a model for in vivo chaperone-mediated disassembly processes. Gansen et al. 29] identified the loss of H2A/H2B dimers as an intermediate disassembly step, in agreement with bulk data [30, 31]. The authors also found evidence that disassembly occurred in 5-bp steps, corroborating the histone-DNA interaction map [13]. Subsequently, researchers created histone-DNA FRET-labeled pairs to better distinguish various intermediates. This study [32] demonstrated conclusively that the H2A-H2B dimer dissociates prior to the tetramer during salt-induced disassembly, and also discovered a previously unobserved intermediate state in which the dimer-tetramer interface separates while the dimer remains attached to the DNA (Figure 2c). The authors estimate that this open conformation is populated at 0.2 to 3% under physiological conditions. The biological significance of this alternate disassembly pathway is unknown; however, it is interesting to note that a similar structure had already been proposed in a magnetic tweezers study performed several years prior [33]. That study found evidence for a dramatic nucleosome structural rearrangement induced by positive torsion, with important implications for transcription [33, 34].
Nucleosome Positioning
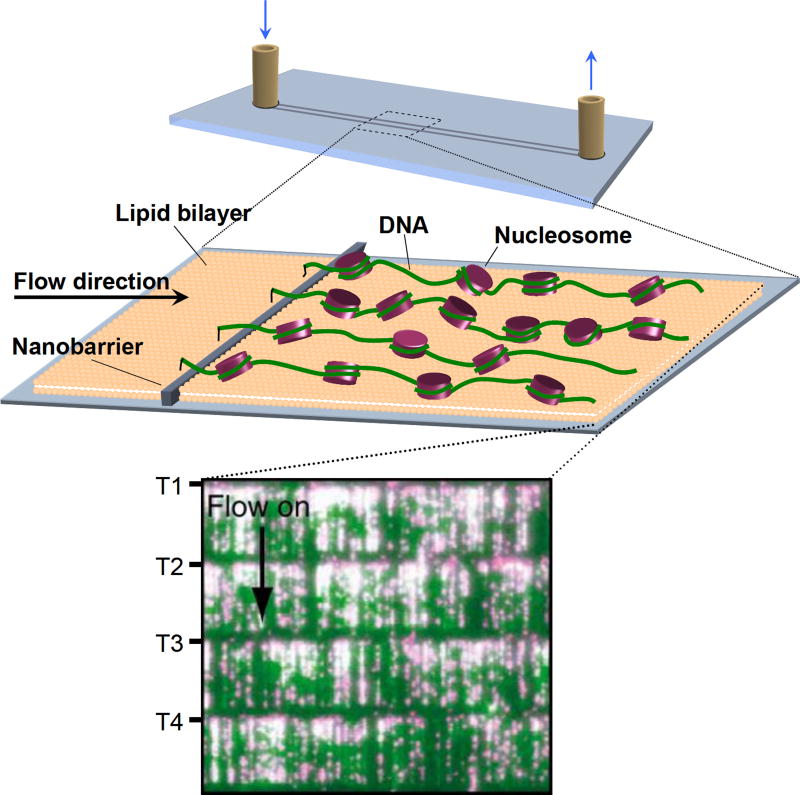
It is widely known that the nucleosome distribution throughout the genome has a significant correlation with gene expression, and consequently, this important topic has been approached by numerous biochemical and theoretical studies [35, 36]. A novel high-throughput SM technique called DNA curtains [37] was recently employed by Visnapuu and Greene [38], enabling the direct SM visualization of nucleosome positioning on model DNA substrates (Figure 3). Their results were in agreement with recent theoretical and bulk studies of nucleosome positioning in vitro [35, 36]. They then examined nucleosomal particles containing only Cse4, H4, and centromeric protein Scm3 [17]. The authors observed that these particles preferentially localized towards poly(dA-dT) tracts such as those found in centromeric DNA [39], thus providing an example of an unusual nucleosome with a strongly altered DNA sequence preference. One should note that whether Scm3 is a part of the centromeric nucleosome or acts as a chaperone is still under debate [19, 40–42].
Figure 3. High-throughput single molecule visualization through DNA curtains [38].
Nucleosome arrays are anchored to a lipid bilayer on the surface of a fused silica microfluidic sample chamber. A hydrodynamic force is applied to direct the molecules towards the leading edges of nanofabricated barriers. The alignment of nucleosomal arrays produced by these barriers makes it possible to visualize thousands of individual molecules simultaneously in real time using total internal reflection fluorescence microscopy (TIRFM).
(Inset) A TIRFM image of DNA curtains. DNA molecules were stained with the YOYO1 intercalating dye (green) and nucleosomes were labeled with fluorescent quantum dots (magenta). Four curtains are shown, with T1-T4 indicating the tethered ends of the curtains and the arrow indicating the direction of the flow used to apply the hydrodynamic force.
(Adapted from Visnapuu et al. [38] with publisher’s permission)
Higher-Order Structures
While most SM studies so far have focused on single nucleosomes, several groups have investigated the behavior of nucleosomal arrays. Of particular interest is the 30-nm fiber: the compacted form adopted by regular nucleosomal arrays in the presence of physiological amounts of divalent cations and linker histones [43]. Despite decades of research, its structure is still controversial and various models have been supported by recent experimental and theoretical data [44–46]. Kruithof et al. [47] used magnetic tweezers to probe the elastic response of the 30-nm fiber and found that its properties changed depending on the linker DNA length (in agreement with [45]). At shorter linker DNA lengths, the fiber adopted a zigzag structure, while longer lengths produced a solenoidal configuration.
Another study by Recouvreux et al. [48] investigated the behavior of arrays of nucleosomes with linker histones (chromatosomes) under torsion, extending previous work by the same lab [33]. Interestingly, the nucleosome and chromatosome arrays behaved similarly under the application of force and torsion, and linker histones were not ejected during this process. The authors also presented a quantitative model of the chromatosome array that predicts a chiral transition to occur around 20 pN∙nm of torque; it will be interesting to test this prediction using optical or magnetic tweezers with torque- measuring capability [49–51].
Transcription through the nucleosome
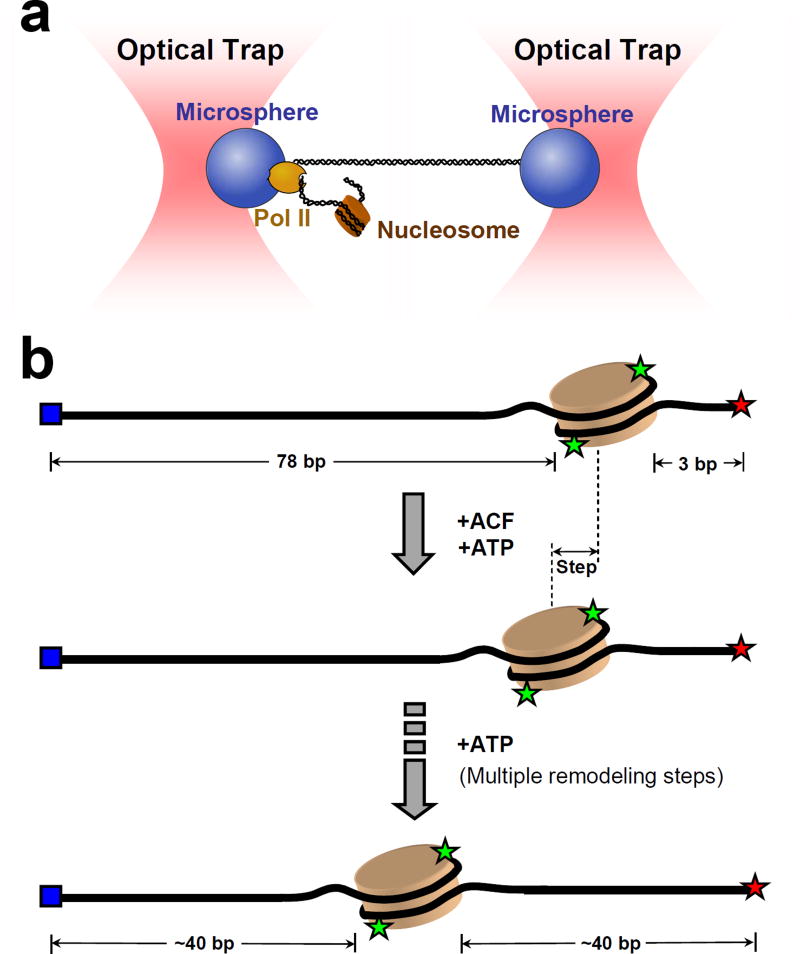
The aforementioned studies provide a foundation for understanding important processes, such as transcription, which are at least in part regulated by nucleosomes. Two recent SM studies on transcription have aimed to address how polymerases handle the significant energy barrier imposed by a nucleosome and the fate of said nucleosome after transcription. Hodges et al. [52] used a high-resolution dual optical trap (Figure 4a) to examine in real time how a transcribing Pol II interacts with a nucleosome. The study demonstrated that Pol II likely acts as a passive thermal ratchet to rectify nucleosome fluctuations, moving forward only when histone-DNA interactions momentarily subside during breathing. In addition, the authors found that nucleosome-like structures were present on the DNA after transcription under zero force, while they were largely absent after transcription under tension, which supports the looping model of histone transfer [53]. It will be interesting to conduct further studies on the identity of these nucleosome-like structures.
Figure 4. Single molecule studies of nucleosome interactions with molecular motors.
(a) Ultra-stable dual-beam optical tweezers setup to monitor Pol II transcription through the nucleosome [52]. DNA was attached to one bead via digoxigenin-antidigoxygenin linkage and to the other bead via a stalled Pol II. Addition of NTPs restarted Pol II movement in the direction of the downstream nucleosome.
(b) Schematic of the ACF-catalyzed nucleosome remodeling monitored by single molecule FRET [60]. The nucleosome is assembled close to the end of a 228 bp DNA template. H2A histones are labeled with Cy3 (green). DNA is labeled with Cy5 (red) at one end and with a biotin (blue) at the other. Upon addition of ACF and ATP, the nucleosome is translocated from the end of the DNA towards the center in discrete steps. This process is monitored in real time by the change of the FRET efficiency between Cy3 and Cy5 using TIRFM.
Another study by Jin et al. [54] employed E. coli RNA polymerase as a model system [55] to precisely determine the polymerase locations during various stages of transcription through a nucleosome. Using the DNA unzipping technique described above, the authors found that polymerase pauses with a distinct 10-bp periodicity when invading the nucleosome but progresses unimpeded when the dyad interactions have been disrupted, in good agreement with the predictions made from the histone-DNA interaction map [13]. Nucleosome-induced pausing was also found to be associated with polymerase backtracking by 10-15 bps. Preventing backtracking by using either RNase or a trailing polymerase enhanced transcription by a factor of 5, a finding that was corroborated by a parallel biochemical study using Pol II [56]. These studies suggest that the presence of multiple transcribing RNA polymerases could enhance transcription efficiency by reducing backtracking in vivo.
Nucleosome remodeling
Single molecule studies have just begun to provide insights into the mechanisms of nucleosome remodelers. Earlier studies employed optical and magnetic tweezers to examine the locations and structures of nucleosomes remodeled by SWI/SNF [57], and the kinetics of loop formation by SWI/SNF and RSC [58, 59]. Recently, Blosser et al. [60] investigated the human ACF complex (Figure 4b), a chromatin remodeler of the ISWI family, known to produce regularly spaced nucleosomes [61]. This study observed three distinct ATP-dependent steps during remodeling: ACF binding, pausing, and translocation. Kinetic pauses occurred after binding, after an initial 7 bp step, and between subsequent 3 to 4 bp steps. The authors hypothesized that these pauses reflect an ATP-dependent conformational change of the enzyme that prepares it for the next round of remodeling. The study also found ACF to exhibit remarkable processivity and bidirectionality, most probably due to it functioning as a dimer. This conclusion was supported by bulk measurements [62]. Studies on related members of the ISWI family have presented evidence for alternative remodeling mechanisms [63–65], and future work will undoubtedly shed light on the basis for these differences as well as on remaining questions in this exciting area of investigation.
Conclusion and Outlook
This review highlights how single molecule optical and magnetic tweezers and fluorescence- based methods have provided detailed, real-time, and molecular-level descriptions of biological processes. Recent years have witnessed several technical advances whose promise for the study of nucleosomes has yet to be fully realized. These techniques include DNA curtain methods for high throughput visualization of single molecules [37], direct biological torque measurements using an angular optical trap [49, 66] or magnetic tweezers [50, 51] , and the combination of optical trapping with fluorescence [67, 68]. Future SM studies using both established and newly developed techniques will undoubtedly expand and extend into many areas of nucleosome research.
Highlights.
-
➢
We review single molecule studies of nucleosome structure and stability.
-
➢
We review single molecule studies of distinct nucleosome disassembly pathways
-
➢
We review single molecule studies of chromatin higher order structures.
-
➢
We review the nucleosome positioning study using the DNA curtain technique.
-
➢
We review the study of the interactions of nucleosomes and related motor proteins.
Acknowledgments
We thank Dr. S. Fellman and Dr. R.M. Fulbright for critical comments of the manuscript. We wish to acknowledge support from National Institutes of Health grants (GM059849 to M.D.W.), National Science Foundation grant (MCB-0820293 to M.D.W.) and Cornell’s Molecular Biophysics Training Grant (T32GM008267) Traineeship (to J.L.K.).
Citations
Of outstanding interest **
Of special interest *
- 1.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 2.Bell O, Tiwari VK, Thoma NH, Schubeler D. Determinants and dynamics of genome accessibility. Nat Rev Genet. 2011;12:554–564. doi: 10.1038/nrg3017. [DOI] [PubMed] [Google Scholar]
- 3.Andrews AJ, Luger K. Nucleosome structure(s) and stability: variations on a theme. Annu Rev Biophys. 2011;40:99–117. doi: 10.1146/annurev-biophys-042910-155329. [DOI] [PubMed] [Google Scholar]
- 4.Park YJ, Luger K. Histone chaperones in nucleosome eviction and histone exchange. Curr Opin Struct Biol. 2008;18:282–289. doi: 10.1016/j.sbi.2008.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ransom M, Dennehey BK, Tyler JK. Chaperoning histones during DNA replication and repair. Cell. 2010;140:183–195. doi: 10.1016/j.cell.2010.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Das C, Tyler JK, Churchill ME. The histone shuffle: histone chaperones in an energetic dance. Trends Biochem Sci. 2010;35:476–489. doi: 10.1016/j.tibs.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clapier CR, Cairns BR. The biology of chromatin remodeling complexes. Annu Rev Biochem. 2009;78:273–304. doi: 10.1146/annurev.biochem.77.062706.153223. [DOI] [PubMed] [Google Scholar]
- 8.Bowman GD. Mechanisms of ATP-dependent nucleosome sliding. Curr Opin Struct Biol. 2010;20:73–81. doi: 10.1016/j.sbi.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Campos EI, Reinberg D. Histones: annotating chromatin. Annu Rev Genet. 2009;43:559–599. doi: 10.1146/annurev.genet.032608.103928. [DOI] [PubMed] [Google Scholar]
- 10.Ray-Gallet D, Almouzni G. Nucleosome dynamics and histone variants. Essays Biochem. 2010;48:75–87. doi: 10.1042/bse0480075. [DOI] [PubMed] [Google Scholar]
- 11.Bannister AJ, Kouzarides T. Regulation of chromatin by histone modifications. Cell Res. 2011;21:381–395. doi: 10.1038/cr.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Davey CA, Sargent DF, Luger K, Maeder AW, Richmond TJ. Solvent mediated interactions in the structure of the nucleosome core particle at 1.9 a resolution. J Mol Biol. 2002;319:1097–1113. doi: 10.1016/S0022-2836(02)00386-8. [DOI] [PubMed] [Google Scholar]
- 13**.Hall MA, Shundrovsky A, Bai L, Fulbright RM, Lis JT, Wang MD. High-resolution dynamic mapping of histone-DNA interactions in a nucleosome. Nat Struct Mol Biol. 2009;16:124–129. doi: 10.1038/nsmb.1526. This work employed DNA unzipping to obtain with near basepair precision a detailed map of histone-DNA interactions in a nucleosome. It revealed coarse (three regions of interactions) as well as fine (5 bp periodicity) details of this energy landscape. The paper also corroborated the view that histone-DNA interactions near the dyad are crucial for nucleosome stability. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brower-Toland B, Wacker DA, Fulbright RM, Lis JT, Kraus WL, Wang MD. Specific contributions of histone tails and their acetylation to the mechanical stability of nucleosomes. J Mol Biol. 2005;346:135–146. doi: 10.1016/j.jmb.2004.11.056. [DOI] [PubMed] [Google Scholar]
- 15.Brower-Toland BD, Smith CL, Yeh RC, Lis JT, Peterson CL, Wang MD. Mechanical disruption of individual nucleosomes reveals a reversible multistage release of DNA. Proc Natl Acad Sci U S A. 2002;99:1960–1965. doi: 10.1073/pnas.022638399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dechassa ML, Wyns K, Li M, Hall MA, Wang MD, Luger K. Structure and Scm3-mediated assembly of budding yeast centromeric nucleosomes. Nat Commun. 2011;2:313. doi: 10.1038/ncomms1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mizuguchi G, Xiao H, Wisniewski J, Smith MM, Wu C. Nonhistone Scm3 and histones CenH3-H4 assemble the core of centromere-specific nucleosomes. Cell. 2007;129:1153–1164. doi: 10.1016/j.cell.2007.04.026. [DOI] [PubMed] [Google Scholar]
- 18.Furuyama T, Henikoff S. Centromeric nucleosomes induce positive DNA supercoils. Cell. 2009;138:104–113. doi: 10.1016/j.cell.2009.04.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Camahort R, Shivaraju M, Mattingly M, Li B, Nakanishi S, Zhu D, Shilatifard A, Workman JL, Gerton JL. Cse4 is part of an octameric nucleosome in budding yeast. Mol Cell. 2009;35:794–805. doi: 10.1016/j.molcel.2009.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tachiwana H, Kagawa W, Shiga T, Osakabe A, Miya Y, Saito K, Hayashi-Takanaka Y, Oda T, Sato M, Park SY, et al. Crystal structure of the human centromeric nucleosome containing CENP-A. Nature. 2011;476:232–235. doi: 10.1038/nature10258. [DOI] [PubMed] [Google Scholar]
- 21.Ranjith P, Yan J, Marko JF. Nucleosome hopping and sliding kinetics determined from dynamics of single chromatin fibers in Xenopus egg extracts. Proc Natl Acad Sci U S A. 2007;104:13649–13654. doi: 10.1073/pnas.0701459104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shogren-Knaak M, Ishii H, Sun JM, Pazin MJ, Davie JR, Peterson CL. Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science. 2006;311:844–847. doi: 10.1126/science.1124000. [DOI] [PubMed] [Google Scholar]
- 23.North JA, Javaid S, Ferdinand MB, Chatterjee N, Picking JW, Shoffner M, Nakkula RJ, Bartholomew B, Ottesen JJ, Fishel R, et al. Phosphorylation of histone H3(T118) alters nucleosome dynamics and remodeling. Nucleic Acids Res. 2011;39:6465–6474. doi: 10.1093/nar/gkr304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Neumann H, Hancock SM, Buning R, Routh A, Chapman L, Somers J, Owen-Hughes T, van Noort J, Rhodes D, Chin JW. A method for genetically installing site-specific acetylation in recombinant histones defines the effects of H3 K56 acetylation. Mol Cell. 2009;36:153–163. doi: 10.1016/j.molcel.2009.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu F, Zhang K, Grunstein M. Acetylation in histone H3 globular domain regulates gene expression in yeast. Cell. 2005;121:375–385. doi: 10.1016/j.cell.2005.03.011. [DOI] [PubMed] [Google Scholar]
- 26*.Simon M, North JA, Shimko JC, Forties RA, Ferdinand MB, Manohar M, Zhang M, Fishel R, Ottesen JJ, Poirier MG. Histone fold modifications control nucleosome unwrapping and disassembly. Proc Natl Acad Sci U S A. 2011;108:12711–12716. doi: 10.1073/pnas.1106264108. By combining smFRET and MT, this study showed how separate PTMs can independently regulate nucleosome unwrapping rates and stability. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Choy JS, Wei S, Lee JY, Tan S, Chu S, Lee TH. DNA methylation increases nucleosome compaction and rigidity. J Am Chem Soc. 2010;132:1782–1783. doi: 10.1021/ja910264z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 29.Gansen A, Valeri A, Hauger F, Felekyan S, Kalinin S, Toth K, Langowski J, Seidel CA. Nucleosome disassembly intermediates characterized by single-molecule FRET. Proc Natl Acad Sci U S A. 2009;106:15308–15313. doi: 10.1073/pnas.0903005106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yager TD, McMurray CT, van Holde KE. Salt-induced release of DNA from nucleosome core particles. Biochemistry. 1989;28:2271–2281. doi: 10.1021/bi00431a045. [DOI] [PubMed] [Google Scholar]
- 31.Andrews AJ, Chen X, Zevin A, Stargell LA, Luger K. The histone chaperone Nap1 promotes nucleosome assembly by eliminating nonnucleosomal histone DNA interactions. Mol Cell. 2010;37:834–842. doi: 10.1016/j.molcel.2010.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32*.Bohm V, Hieb AR, Andrews AJ, Gansen A, Rocker A, Toth K, Luger K, Langowski J. Nucleosome accessibility governed by the dimer/tetramer interface. Nucleic Acids Res. 2011;39:3093–3102. doi: 10.1093/nar/gkq1279. Provided evidence for the existence of a novel nucleosome disassembly intermediate which involves breaking of the tetramer/dimer interactions while maintaining intact histone-DNA contacts. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bancaud A, Wagner G, Conde ESN, Lavelle C, Wong H, Mozziconacci J, Barbi M, Sivolob A, Le Cam E, Mouawad L, et al. Nucleosome chiral transition under positive torsional stress in single chromatin fibers. Mol Cell. 2007;27:135–147. doi: 10.1016/j.molcel.2007.05.037. [DOI] [PubMed] [Google Scholar]
- 34.Becavin C, Barbi M, Victor JM, Lesne A. Transcription within condensed chromatin: Steric hindrance facilitates elongation. Biophys J. 2010;98:824–833. doi: 10.1016/j.bpj.2009.10.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Field Y, Kaplan N, Fondufe-Mittendorf Y, Moore IK, Sharon E, Lubling Y, Widom J, Segal E. Distinct modes of regulation by chromatin encoded through nucleosome positioning signals. PLoS Comput Biol. 2008;4:e1000216. doi: 10.1371/journal.pcbi.1000216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kaplan N, Moore IK, Fondufe-Mittendorf Y, Gossett AJ, Tillo D, Field Y, LeProust EM, Hughes TR, Lieb JD, Widom J, et al. The DNA-encoded nucleosome organization of a eukaryotic genome. Nature. 2009;458:362–366. doi: 10.1038/nature07667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Greene EC, Wind S, Fazio T, Gorman J, Visnapuu ML. DNA curtains for high-throughput singlemolecule optical imaging. Methods Enzymol. 2010;472:293–315. doi: 10.1016/S0076-6879(10)72006-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38*.Visnapuu ML, Greene EC. Single-molecule imaging of DNA curtains reveals intrinsic energy landscapes for nucleosome deposition. Nat Struct Mol Biol. 2009;16:1056–1062. doi: 10.1038/nsmb.1655. A high-throughput SM study of nucleosome positioning, made possible by the DNA curtains techniques. Besides validating previous bulk studies with canonical nucleosomes, this work also showed that non-histone protein Scm3 alters the intrinsic landscape of centromeric nucleosomes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Black BE, Bassett EA. The histone variant CENP-A and centromere specification. Curr Opin Cell Biol. 2008;20:91–100. doi: 10.1016/j.ceb.2007.11.007. [DOI] [PubMed] [Google Scholar]
- 40.Xiao H, Mizuguchi G, Wisniewski J, Huang Y, Wei D, Wu C. Nonhistone Scm3 binds to AT-rich DNA to organize atypical centromeric nucleosome of budding yeast. Mol Cell. 2011;43:369–380. doi: 10.1016/j.molcel.2011.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cho US, Harrison SC. Recognition of the centromere-specific histone Cse4 by the chaperone Scm3. Proc Natl Acad Sci U S A. 2011;108:9367–9371. doi: 10.1073/pnas.1106389108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhou Z, Feng H, Zhou BR, Ghirlando R, Hu K, Zwolak A, Miller Jenkins LM, Xiao H, Tjandra N, Wu C, et al. Structural basis for recognition of centromere histone variant CenH3 by the chaperone Scm3. Nature. 2011;472:234–237. doi: 10.1038/nature09854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li G, Reinberg D. Chromatin higher-order structures and gene regulation. Curr Opin Genet Dev. 2011;21:175–186. doi: 10.1016/j.gde.2011.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schalch T, Duda S, Sargent DF, Richmond TJ. X-ray structure of a tetranucleosome and its implications for the chromatin fibre. Nature. 2005;436:138–141. doi: 10.1038/nature03686. [DOI] [PubMed] [Google Scholar]
- 45.Routh A, Sandin S, Rhodes D. Nucleosome repeat length and linker histone stoichiometry determine chromatin fiber structure. Proc Natl Acad Sci U S A. 2008;105:8872–8877. doi: 10.1073/pnas.0802336105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Depken M, Schiessel H. Nucleosome shape dictates chromatin fiber structure. Biophys J. 2009;96:777–784. doi: 10.1016/j.bpj.2008.09.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47*.Kruithof M, Chien FT, Routh A, Logie C, Rhodes D, van Noort J. Single-molecule force spectroscopy reveals a highly compliant helical folding for the 30-nm chromatin fiber. Nat Struct Mol Biol. 2009;16:534–540. doi: 10.1038/nsmb.1590. A SM measurement of the elastic properties of the 30-nm chromatin fiber. This study was able to differentiate between two proposed structural models of the fiber. [DOI] [PubMed] [Google Scholar]
- 48.Recouvreux P, Lavelle C, Barbi M, Conde ESN, Le Cam E, Victor JM, Viovy JL. Linker histones incorporation maintains chromatin fiber plasticity. Biophys J. 2011;100:2726–2735. doi: 10.1016/j.bpj.2011.03.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Deufel C, Forth S, Simmons CR, Dejgosha S, Wang MD. Nanofabricated quartz cylinders for angular trapping: DNA supercoiling torque detection. Nat Methods. 2007;4:223–225. doi: 10.1038/nmeth1013. [DOI] [PubMed] [Google Scholar]
- 50.Celedon A, Nodelman IM, Wildt B, Dewan R, Searson P, Wirtz D, Bowman GD, Sun SX. Magnetic tweezers measurement of single molecule torque. Nano Lett. 2009;9:1720–1725. doi: 10.1021/nl900631w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lipfert J, Kerssemakers JW, Jager T, Dekker NH. Magnetic torque tweezers: measuring torsional stiffness in DNA and RecA-DNA filaments. Nat Methods. 2010;7:977–980. doi: 10.1038/nmeth.1520. [DOI] [PubMed] [Google Scholar]
- 52**.Hodges C, Bintu L, Lubkowska L, Kashlev M, Bustamante C. Nucleosomal fluctuations govern the transcription dynamics of RNA polymerase II. Science. 2009;325:626–628. doi: 10.1126/science.1172926. A SM study of Pol II transcription through the nucleosome. Pol II was found to act as a passive ratchet that rectifies nucleosomal DNA fluctuations. This work also provided support for the looping model of histone transfer upon the passage of Pol II. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kulaeva OI, Gaykalova DA, Pestov NA, Golovastov VV, Vassylyev DG, Artsimovitch I, Studitsky VM. Mechanism of chromatin remodeling and recovery during passage of RNA polymerase II. Nat Struct Mol Biol. 2009;16:1272–1278. doi: 10.1038/nsmb.1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54*.Jin J, Bai L, Johnson DS, Fulbright RM, Kireeva ML, Kashlev M, Wang MD. Synergistic action of RNA polymerases in overcoming the nucleosomal barrier. Nat Struct Mol Biol. 2010;17:745–752. doi: 10.1038/nsmb.1798. By using E. coli RNAP as a model system, this work illustrated how histone-DNA interactions dictate the motion of the polymerase as it transcribes through a nucleosome, and highlighted how cooperativity between RNAPs can enhance transcription. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Walter W, Kireeva ML, Studitsky VM, Kashlev M. Bacterial polymerase and yeast polymerase II use similar mechanisms for transcription through nucleosomes. J Biol Chem. 2003;278:36148–36156. doi: 10.1074/jbc.M305647200. [DOI] [PubMed] [Google Scholar]
- 56.Kulaeva OI, Hsieh FK, Studitsky VM. RNA polymerase complexes cooperate to relieve the nucleosomal barrier and evict histones. Proc Natl Acad Sci U S A. 2010;107:11325–11330. doi: 10.1073/pnas.1001148107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shundrovsky A, Smith CL, Lis JT, Peterson CL, Wang MD. Probing SWI/SNF remodeling of the nucleosome by unzipping single DNA molecules. Nat Struct Mol Biol. 2006;13:549–554. doi: 10.1038/nsmb1102. [DOI] [PubMed] [Google Scholar]
- 58.Lia G, Praly E, Ferreira H, Stockdale C, Tse-Dinh YC, Dunlap D, Croquette V, Bensimon D, Owen-Hughes T. Direct observation of DNA distortion by the RSC complex. Mol Cell. 2006;21:417–425. doi: 10.1016/j.molcel.2005.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang Y, Smith CL, Saha A, Grill SW, Mihardja S, Smith SB, Cairns BR, Peterson CL, Bustamante C. DNA translocation and loop formation mechanism of chromatin remodeling by SWI/SNF and RSC. Mol Cell. 2006;24:559–568. doi: 10.1016/j.molcel.2006.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60**.Blosser TR, Yang JG, Stone MD, Narlikar GJ, Zhuang X. Dynamics of nucleosome remodelling by individual ACF complexes. Nature. 2009;462:1022–1027. doi: 10.1038/nature08627. A real-time smFRET study of nucleosome remodeling. This work revealed the distinct steps taken by human ACF during remodeling, and suggested that it may work as a dimer to achieve remarkable processivity and bidirectionality. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.LeRoy G, Loyola A, Lane WS, Reinberg D. Purification and characterization of a human factor that assembles and remodels chromatin. J Biol Chem. 2000;275:14787–14790. doi: 10.1074/jbc.C000093200. [DOI] [PubMed] [Google Scholar]
- 62.Racki LR, Yang JG, Naber N, Partensky PD, Acevedo A, Purcell TJ, Cooke R, Cheng Y, Narlikar GJ. The chromatin remodeller ACF acts as a dimeric motor to space nucleosomes. Nature. 2009;462:1016–1021. doi: 10.1038/nature08621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zofall M, Persinger J, Kassabov SR, Bartholomew B. Chromatin remodeling by ISW2 and SWI/SNF requires DNA translocation inside the nucleosome. Nat Struct Mol Biol. 2006;13:339–346. doi: 10.1038/nsmb1071. [DOI] [PubMed] [Google Scholar]
- 64.Gangaraju VK, Bartholomew B. Dependency of ISW1a chromatin remodeling on extranucleosomal DNA. Mol Cell Biol. 2007;27:3217–3225. doi: 10.1128/MCB.01731-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yamada K, Frouws TD, Angst B, Fitzgerald DJ, DeLuca C, Schimmele K, Sargent DF, Richmond TJ. Structure and mechanism of the chromatin remodelling factor ISW1a. Nature. 2011;472:448–453. doi: 10.1038/nature09947. [DOI] [PubMed] [Google Scholar]
- 66.Forth S, Deufel C, Sheinin MY, Daniels B, Sethna JP, Wang MD. Abrupt buckling transition observed during the plectoneme formation of individual DNA molecules. Phys Rev Lett. 2008;100:148301. doi: 10.1103/PhysRevLett.100.148301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lang MJ, Fordyce PM, Engh AM, Neuman KC, Block SM. Simultaneous, coincident optical trapping and single-molecule fluorescence. Nat Methods. 2004;1:133–139. doi: 10.1038/nmeth714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Comstock MJ, Ha T, Chemla YR. Ultrahigh-resolution optical trap with single-fluorophore sensitivity. Nat Methods. 2011;8:335–340. doi: 10.1038/nmeth.1574. [DOI] [PMC free article] [PubMed] [Google Scholar]