Single molecule techniques reveal short- and long-range dynamics of supercoiled DNA
Perhaps the reader can remember the good old days of wired phones, their cords so prone to absent-minded twisting that eventually produced a multitude of small coiled coils. Wired phones are quickly retreating to the ash heap of history, but their cords still serve as inspiration to those interested in the coiling process of the most famous molecule of the cell - DNA. The striking beauty of DNA coiling (aptly named supercoiling) was first illustrated by Vinograd and coworkers (1) in their electron microscope (EM) images of a circular polyoma virus DNA, which revealed the existence of multiple intertwined loops. These loops, also called plectonemes, can play an important role in gene regulation by bringing together distant DNA elements, such as enhancers and promoters (2). On p. XXX of this issue, van Loenhout et al. use powerful single molecule techniques to uncover the rich dynamics of plectoneme formation and movement.
While EM images of plectonemes have long captured scientists’ imagination, they provided only static snapshots of these DNA structures. Dynamic torsional manipulation of DNA was first demonstrated by Strick et al. in the mid-1990s (3). Using magnetic tweezers, a DNA molecule was torsionally constrained via multiple tags between a glass slide and a super-paramagnetic bead. A pair of permanent magnets placed above the sample chamber pulled the bead vertically towards the magnets until the magnetic force was balanced by the restoring elastic force from the DNA. DNA was then supercoiled by the rotation of the bead. Plectoneme formation manifested itself as a steady decrease in the DNA length as each turn added was converted to the growth of the plectoneme. Subsequent experiments using an angular optical trap demonstrated that the initial plectoneme formation is abrupt, as revealed by a sudden extension drop followed by a torque plateau (4). However, many questions still remain regarding the nature of plectonemes. How many plectonemes coexist on a single DNA molecule? Do plectonemes remain at the same locations or move about dynamically? How and where do they nucleate, grow, shrink, and disappear?
In order to visually localize structures along the DNA, van Loenhout et al. first supercoiled the DNA with a pair of permanent magnets as described above, and then replaced them with a side magnet, so that the supercoiled DNA tether could be pulled sideways. A 21 kbp DNA molecule, sparsely labeled with fluorophores, was subsequently visualized using fluorescence. While relaxed DNA appeared as a band of uniform fluorescence intensity in the images, bright fluorescent spots were detected in supercoiled DNA. These spots reflected high local DNA density, indicative of the existence of plectonemes. Interestingly, under certain conditions the researchers observed the presence of multiple plectonemes along the DNA. Their number varied from a single plectoneme at high force and salt (3.2 pN, 300 mM NaCl) to about three at low force and salt (0.4 pN, 20 mM NaCl). The authors explain this trend as being due to an interplay between entropic gain provided by multiple plectoneme domains and enthalpic cost due to the formation of initial plectonemic loops. Notably, the experimental results are in good qualitative agreement with two recent theoretical models (5, 6), which underscore the significant progress made in the quantitative understanding of DNA mechanics.
Plectoneme visualization was only the first step – several seconds of imaging time permitted the authors to observe the long-coveted plectoneme dynamics. And here some intriguing observations were made. First, the researchers found that each plectoneme diffused along the DNA. The diffusion constant of an average plectoneme varied between 0.1 μm2/s at 0.8 pN to < 0.01 μm2/s at 3.2 pN, significantly smaller than expected based on a simple hydrodynamic model. This discrepancy is explained by the observation of van Loenhout et al. that plectonemes were not evenly distributed along the DNA, but instead preferentially localized near certain positions. This suggested that diffusion takes place on a rugged energy landscape, modulated by the intrinsic curvature and bendability of the underlying DNA sequence. The authors found that roughness on the order of 1–2 kBT was sufficient to explain their diffusion data. Interestingly, this energy landscape roughness is of the same order of magnitude as has been estimated for the analogous energy barrier of plectoneme formation in DNA with intrinsic bends (7).
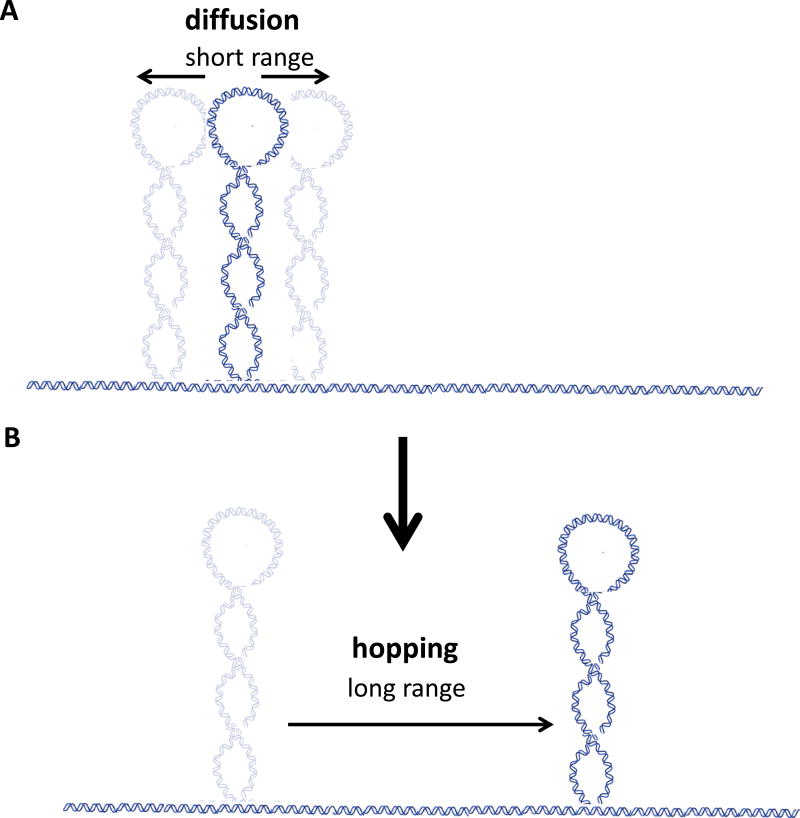
However, plectoneme dynamics turns out not to be limited to diffusion. Unexpectedly, van Loenhout et al. observed that plectonemes could hop, suddenly disappearing and rapidly reappearing thousands of basepairs away from their original locations. In fact, this novel behavior was found to be one of the defining features of plectoneme dynamics, as the mean lifetime of a plectoneme under most experimental conditions was less than 1 second. Thus, while diffusional motion during that time was limited to a few hundreds of basepairs, hopping could transplant the plectoneme several kilobasepairs within tens of milliseconds.
The demonstrated visualization of plectonemes is a significant experimental achievement. However, the story does not end here, and questions remain for further research. For example, the current experimental approach allowed for the detection of plectonemes with a minimum size of ~0.5 kb, which is larger than the size of an initial plectonemic loop. Recent theoretical work (5) suggested that single loops can coexist with plectonemes; a more sensitive detection method would be needed to test this hypothesis. It would also be of interest to further investigate the hopping process. It appears from the data that it is possible for the twist generated from the shrinking of one plectoneme to redistribute to other plectonemes in addition to nucleating a new one. Hopping can thus be viewed as a special case of exchanges of length between plectonemes.
The novel findings from van Loenhout et al. have significant implications for processes that take place over DNA. The observation of preferential plectoneme localization suggests new ways in which DNA sequence can regulate genetic transactions. Indeed, one could imagine that certain DNA sequences are designed to “pin down” plectonemes and thus bring neighboring regulatory DNA elements into close proximity. More research is required to identify the sequences that can localize plectonemes and analyze their distribution genome-wide. Plectoneme hopping, a dramatic long-range rearrangement of the DNA conformation on a millisecond timescale, could permit fast searching during DNA recombination or enhancer-activated gene expression. The next challenge will be to identify instances of plectoneme hopping in vivo and relate them to specific biological functions.
Figure. Dynamics of DNA plectonemes.
A) Plectonemes can undergo short-range diffusive motion along DNA. The hydrodynamic drag on a plectoneme is small – it is sequence preferences that slow down the rate of such excursions.
B) Plectonemes can also display hopping behavior, disappearing from one location and reappearing thousands of basepairs away within tens of milliseconds.
References
- 1.Vinograd J, Lebowitz J, Radloff R, Watson R, Laipis P. Proc Natl Acad Sci U S A. 1965 May;53:1104. doi: 10.1073/pnas.53.5.1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu Y, Bondarenko V, Ninfa A, Studitsky VM. Proc Natl Acad Sci U S A. 2001 Dec 18;98:14883. doi: 10.1073/pnas.261477898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Strick TR, Allemand JF, Bensimon D, Bensimon A, Croquette V. Science. 1996 Mar 29;271:1835. doi: 10.1126/science.271.5257.1835. [DOI] [PubMed] [Google Scholar]
- 4.Forth S, et al. Phys Rev Lett. 2008 Apr 11;100:148301. doi: 10.1103/PhysRevLett.100.148301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marko JF, Neukirch S. Phys Rev E Stat Nonlin Soft Matter Phys. 2012 Jan;85:011908. doi: 10.1103/PhysRevE.85.011908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Emanuel GLM, Schiessel H. arXiv. 2012;1204:1324v2. [Google Scholar]
- 7.Daniels BC, Sethna JP. Physical Review E. 2011 Apr 26;83 doi: 10.1103/PhysRevE.83.041924. [DOI] [PubMed] [Google Scholar]