Abstract
Bacillus subtilis biofilms have a fundamental role in shaping the soil ecosystem. During this process, they unavoidably interact with neighbour bacterial species. We studied the interspecies interactions between biofilms of the soil-residing bacteria B. subtilis and related Bacillus species. We found that proximity between the biofilms triggered recruitment of motile B. subtilis cells, which engulfed the competing Bacillus simplex colony. Upon interaction, B. subtilis secreted surfactin and cannibalism toxins, at concentrations that were inert to B. subtilis itself, which eliminated the B. simplex colony, as well as colonies of Bacillus toyonensis. Surfactin toxicity was correlated with the presence of short carbon-tail length isomers, and synergistic with the cannibalism toxins. Importantly, during biofilm development and interspecies interactions a subpopulation in B. subtilis biofilm lost its native plasmid, leading to increased virulence against the competing Bacillus species. Overall, these findings indicate that genetic programs and traits that have little effect on biofilm development when each species is grown in isolation have a dramatic impact when different bacterial species interact.
Introduction
Soil bacteria have a central role in shaping soil ecology. The soil microorganismic community is composed of diverse populations of bacterial species that dramatically affect the availability of soil nutrients and plant diversity.1–4 The immense bacterial diversity within the soil leads to unavoidable interspecies interactions, which ultimately form a structured microbial community.5,6 Generation of antagonistic and mutualistic behaviours, mediated by exchange of small diffusible secondary metabolites, enables bacterial adaptation to the complex communal life.6,7 Such communication can induce resistance to various antibiotics, or can eliminate rival bacterial species competing for limited nutrients.
Most bacteria in nature live in structurally and dynamically complex biological systems called biofilms. Biofilms are multicellular communities of surface-associated bacteria enveloped in a self-produced extracellular matrix.8 The extracellular matrix isolates the bacteria in the biofilm from the external environment and protects them from antibiotics, sterilising agents and the immune system.9 Here we studied the interactions between two robust biofilm formers—Bacillus subtilis and the closely related Bacillus simplex. Both bacterial species reside in and compete for the same ecological niche—the soil.10,11
B. subtilis is a master of differentiation, displaying a multitude of distinct cell types within its biofilms. B. subtilis can differentiate into cells capable of taking up DNA from the environment.12–14 During the development of genetic competence, the production of a small cyclic lipopeptide named surfactin is induced.15 The machinery for surfactin synthesis is encoded within the srfAA-AB-AC-AD operon. Surfactin is also a powerful surfactant that shows a wide range of biological activities such as antibacterial,16 antiviral and antifungal actions.17 It is composed of an amphipathic, cyclic heptapeptide head group that is interlinked with a hydrophobic β-hydroxy fatty acid tail, comprising 12–16 carbon atoms.18–20 These features enable the surfactin molecule to act on cellular membranes and disrupt the membrane integrity.21 The production of surfactin is sensed together with additional environmental signals, by a portion of the biofilm population that then produces the extracellular matrix.8,22,23 In addition, this subpopulation produces an extracellular killing factor SkfA and SdpC that function to kill (or cannibalise) cells that have not yet commenced to sporulation.24 Importantly, the production of cannibalism toxins was only contributing to biofilm development in mutants where genes predicted to have a role either in the production of, or in the sensitivity to, cannibalism toxins were deleted.25 Thus, it is still unclear, what is the exact function of the cannibal cells in wild-type (WT) biofilms. In a biofilm, an additional distinct subpopulation of cells express sigD, the sigma factor necessary for flagella production, resulting in motile heterogeneity.26 The role of motile cells in biofilms that are formed over solid surfaces remains unknown.
The main components of the B. subtilis extracellular matrix are exopolysaccharides, synthesised by the epsA-O operon-encoded genes, TasA, a functional amyloid, encoded in the three-gene operon yqxM\TapA-sipW-tasA27,28 and BslA, that forms a hydrophobic coat over the biofilm.29 The master regulator controlling the switch to a biofilm lifestyle is the repressor SinR.30 The commitment to the biofilm state requires the phosphorylation of Spo0A. Spo0A-P activates SinI, which directly binds and represses SinR.31 Recently, an additional regulatory gene for biofilm formation, rapP, was found on the native 80-kb pBS32 plasmid, which was lost during B. subtilis domestication.32 RapP encodes a phosphatase that dephosphorylates the intermediate response regulator Spo0F, and thus indirectly represses Spo0A activity.33 The decreased phosphorylation of Spo0A results in an altered biofilm formation.32,33 In addition, RapP regulates genetic competence.34
Here, we demonstrate that upon intercolony contact, motile B. subtilis cells emerged from the mature biofilm to engulf the competing colony. Surfactin, Skf and Sdp then synergistically mediated the elimination of the competing B. simplex colony. The secretion of all relevant virulence factors was reinforced by a subpopulation of B. subtilis biofilm cells that lost their native pBS32 plasmid and exhibited increased aggressiveness against B. simplex cells. Similarly to B. subtilis, the fitness of naturally evolved genetic variants within B. simplex biofilms also changed during the interaction.
Importantly, the molecular mechanisms underlie the antagonistic interaction of B. subtilis and B. simplex turned out to be quite general. The synergistic and selective effect of the cannibalism toxins and surfactin towards competing Bacillus species, as well as a robust negative control of a native plasmid on the virulence of these effectors occurred under several conditions. It was evident on several biofilm growth media and during interspecies interaction between B. subtilis and the soilborne bacterium Bacillus toyonensis.
Results
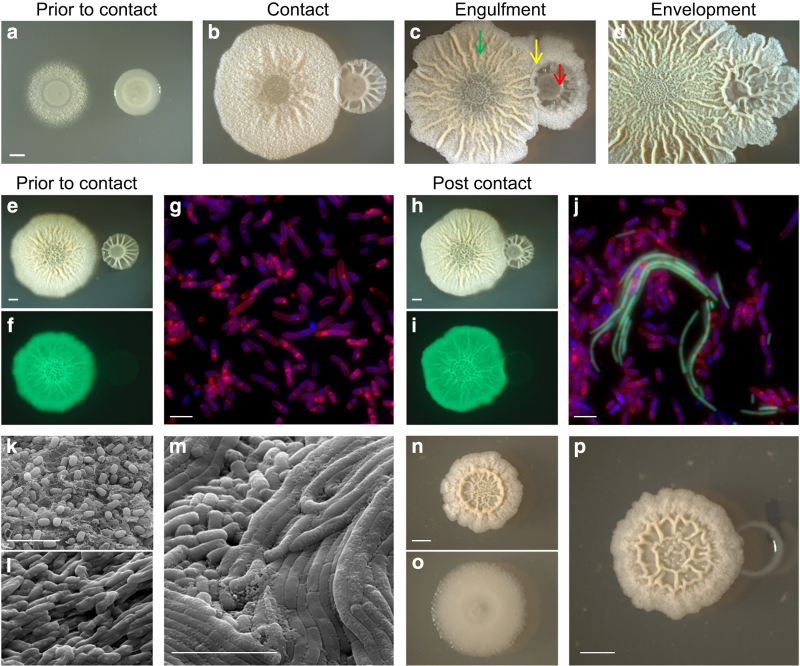
When grown on biofilm-inducing medium, both B. subtilis and B. simplex formed architecturally complex biofilms. B. subtilis created highly wrinkled complex biofilms, with a defined centre and dense ridges. B. simplex, isolated from soils, formed a wrinkled colony, with significant gaps between the thick wrinkles, and with a well-defined, smooth centre (Figure 1a–m and Supplementary Figure S1). When the two complex colonies came into proximity, and within a few hours of initial contact, a fragile ring of B. subtilis formed around B. simplex biofilm. The B. subtilis ring thickened with time, and engulfed the B. simplex biofilm (Figure 1a–d). Eventually, the entire surface of the engulfed colony was covered with B. subtilis matrix. To monitor this interaction at a single-cell resolution, the WT B. subtilis strain labelled with GFP was inoculated in proximity to B. simplex biofilms. Upon first contact, B. subtilis chains, significantly varying in length and number, invaded the B. simplex biofilm (Figure 1e–j). This variation might imply that B. subtilis cells are capable of replicating inside the B. simplex biofilm.
Figure 1.
Antagonistic interaction between B. subtilis biofilms and competing Bacillus colonies. (a–m) Biofilms of B. subtilis and B. simplex grown at 30 °C on MSgg biofilm-inducing medium. (a–d) Biofilms of B. simplex were inoculated next to a B. subtilis biofilm at a distance of 0.8 cm. (a) Before contact (day 1), (b) contact (day 2), (c) engulfment (day 3) and (d) envelopment (day 4). Arrows indicate the different regions of the interaction: B. subtilis area (green), interphase (yellow) and B. simplex area (red), Scale bar represent 2 mm. (e–j) Biofilms of B. simplex were inoculated next to a biofilm of expressing GFP B. subtilis harbouring Phyperspank-gfp at a distance of 1 cm (before contact) and 0.8 cm (post contact), and grown for 2 days. (e,h) Bright field colonies images, scale bar represents 2 mm, (f,i) GFP fluorescence images of the colonies in e and h. (g,j) florescent microscope image: green—B. subtilis strain expressing GFP, red—membrane stain FM4–64, blue—4,6-diamidino-2-phenylindole DNA stain. Scale bar represents 5 μm. (k–m) environmental scanning electron microscopy images of B. subtilis and B. simplex biofilms grown for 3 days. Scale bars represent 5 μm. (k) B. subtilis grown separately. (l) B. simplex grown separately. (m) Interaction area of B. subtilis and B. simplex interacting biofilms in the engulfment stage. (n–p) B. subtilis and B. toyonensis colonies grown for 3 days at 30 °C on B4 biofilm medium. (n) B. subtilis grown separately. (o) B. toyonensis grown separately. (p) Interaction between B. subtilis and B. toyonensis, inoculated 0.3 cm apart. Scale bar represents 2 mm.
Using environmental scanning electron microscopy, the two biofilms could be clearly distinguished by their different extracellular matrix and the 3D organisation of their constituent cells (Figure 1k–m). The B. subtilis extracellular matrix was characterized by a fibrous web-like structure, while the B. simplex biofilm contained a thick extracellular matrix coating each cell in the biofilm. In addition, B. simplex biofilm cells were approximately twofold larger than B. subtilis biofilm cells. Upon direct contact, the B. subtilis and B. simplex interaction zone took on a different morphology than each isolated biofilm. This interaction zone was characterized by both an extremely dense extracellular matrix that heavily coated each of the interacting cells in the biofilm and by unique cell organisation patterns, and especially long cell chains (Figure 1m). Notably, B. subitlis cells form short-lived chains during biofilm growth, also producing the extracellular matrix.35,36 These cell chains disassemble within 48 h (Supplementary Figure 1 and ref. 35). As the scanning electron microscope revealed the presence of cell chains in 72 h biofilms, it is feasible that the invasion process into B. simplex induces chaining within aging B. subitlis biofilms.
We then asked whether B. subtilis biofilms can antagonise other Bacillus species. Thus, we studied the interaction between B. subtilis and the soil-bacterium B. toyonensis, highly related to Bacillus cereus. As B. toyonensis is not capable of growing on defined biofilm media, we chose to study the interaction on top of a rich solid biofilm media (B4).37 We found that within 3 days, B. toyonensis colony is mostly eradicated by B. subtilis (Figure 1n–p). These results led us to the conclusion that B. subtilis biofilms may be allopathic to competing Bacillus species. Thus, we decided to explore the interspecies interaction between B. subtilis colonies and competing colonies by evaluating the viability of the interacting partners.
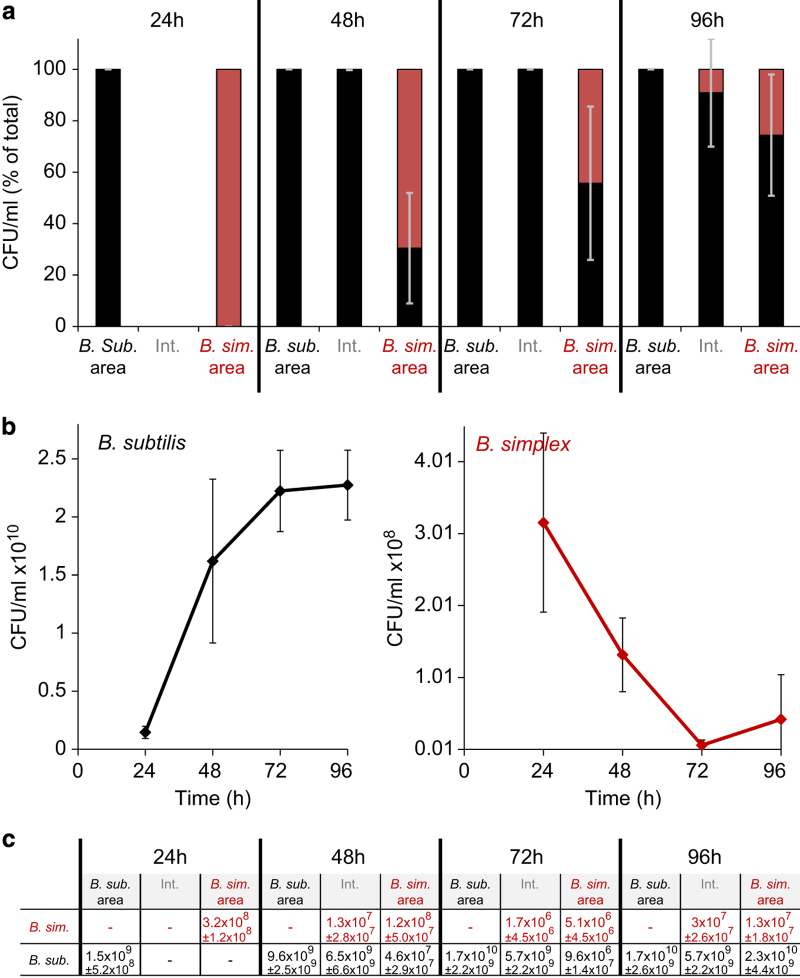
Once the biofilms of B. subtilis and B. simplex contacted each other, a massive invasion of the B. subtilis cells into the B. simplex biofilm occurred, gradually leading to a nearly complete extinction of the B. simplex population (Figure 2). In contrast, when grown in isolation, the B. simplex population increased steadily (Supplementary Figure S2). Finally, after the envelopment stage, the absolute number of B. subtilis cells comprised over 90% of the total population within the former B. simplex colony area. Importantly, in the interaction zone, the number of multiplying viable B. simplex cells mildly increased, demonstrating that a subpopulation of B. simplex cells is resistant to B. subtilis killing (Figures 2b and c). This interaction occurred both on defined (Figures 1 and 2) and rich (Supplementary Figure S3) biofilm media.
Figure 2.
B. subtilis engulfment of and invasion into B. simplex biofilm mediates eradication of B. simplex. (a) Percentage of B. subtilis and B. simplex colony forming units at each stage of the interaction: the interacting colonies were divided into three sections: B. sub. area—B. subtilis section, Int.—interaction zone and B. sim area—B. simplex section. Each section was collected, sonicated and plated to determine the number of replicative cells of each species. In each section, percentage of each species from total cell number is presented (n=6). (b) Sum of the cell numbers from all sections as described in a, during 4 days of interaction (n=4). (c) Cell number from all sections as described in a, during 4 days of interaction (n=4). Error bars represent s.d.
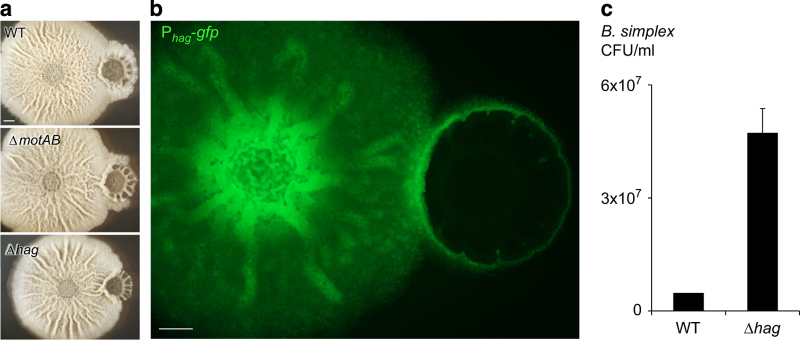
We then asked whether flagellated motility or flagella production have a role in the formation of the B. subtilis engulfment ring around B. simplex. Mature B. subtilis biofilms contain a small subpopulation of motile cells.38 When grown alone, deletion of the flagellin gene (Δhag),39 yields somewhat smaller B. subtilis biofilms that are highly similar to the parental WT strain. In contrast, when we examined the ability of this non-motile mutant to engulf the B. simplex biofilms, the mutant froze in the engulfment ring stage (Figure 3). Characterisation of the motility expression, using a transcriptional fusion to the hag promoter, revealed that the B. subtilis cells that engulf B. simplex have an increased expression of motility, as judged by the expression of hag (Figure 3b and Supplementary Figure S4C). Furthermore, the ability of the non-motile Δhag mutant to overcome and eliminate B. simplex was significantly reduced (Figure 3c). We then asked whether the motor unit proteins, MotA and MotB of the flagella are also important for engulfment of B. simplex. We found that a mutant in motAB, similarly to a flagellin mutant, had an apparent delay in engulfing the B. simplex colony. The delay was more modest than of a flagellin mutant, but highly reproducible (Figure 3a). These findings suggest that the spreading of B. subtilis structured colonies relies primarily on flagella production, while the rotation of the flagella has a secondary role when engulfing competing colonies.
Figure 3.
Expression of the motility genes is required for B. subtilis engulfment and killing of B. simplex biofilms. (a) upper—B. subtilis WT strain inoculated on MSgg biofilm-inducing medium at a distance of 0.8 cm from B. simplex. middle—B. subtilis ΔmotAB mutant inoculated at distance of 0.8 cm from B. simplex. lower—B. subtilis Δhag mutant inoculated at distance of 0.6 cm from B. simplex. Biofilms were grown for 3 days at 30 °C. Scale bars represent 2 mm. (b) Phag-gfp expression. B. subtilis Phag -gfp strain inoculated on biofilm-inducing medium at a distance of 0.8 cm from a B.simplex biofilm grown for 2 days at 30 °C. Scale bars represent 2 mm. (c) Colony forming units counts of B. simplex taken 3 days post inoculation next to B. subtilis WT (distance of 0.8 cm) or B. subtilis Δhag mutant (distance of 0.6 cm), as in a. The interacting colonies were collected, sonicated and plated to determine the number of replicative cells. n=3, error bars represent s.d.
Engulfment did not require a fully functional chemotaxis apparatus, as demonstrated using the ΔcheA, and ΔcheY B. subtilis mutants.40 Those mutants could successfully form an engulfment ring around B. simplex. The engulfment was also independent of swarming, as a mutant in degU, which is required for effective swarming,41,42 had little or no defect in the engulfment stage (Supplementary Figure S4A).
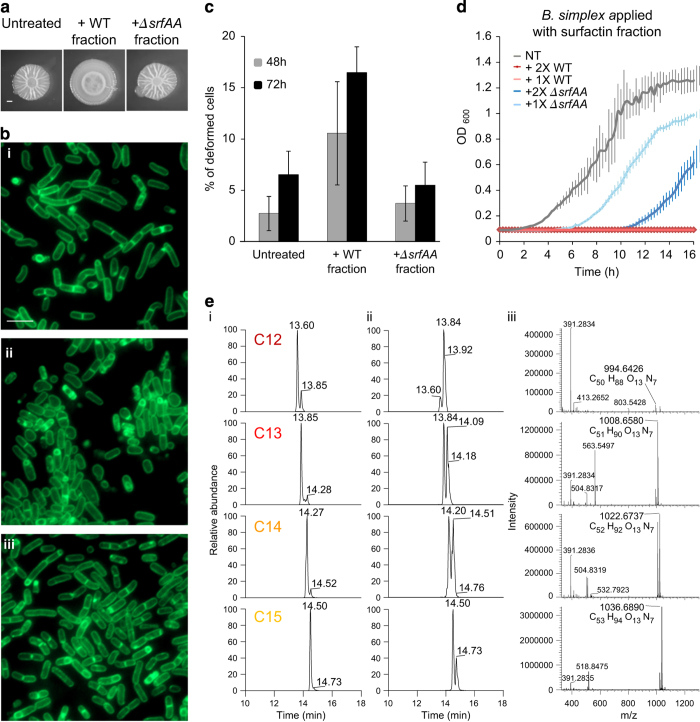
The invasion of B. subtilis into B. simplex biofilms led to elimination of the cells within the invaded colony. As we found that the supernatants of post-logarithmic and stationary planktonic B. subtilis cultures led to complete inhibition of B. simplex growth (Supplementary Figure S5, Panel A), we attempted to purify bioactive B. subtilis-derived molecules that mediate the killing (for details, see Materials and Methods section). Importantly, the killing activity of the supernatant was density dependent, consistent with the idea that the killing factor(s) production can be regulated by quorum sensing, a phenomenon where bacterial behaviours are modulated in accordance with population density, through the synthesis and perception of small signalling molecules.43 When separating and purifying supernatant components on a gradient of organic solvent on a C-18 Sep-Pak cartridge as done previously for isolation of small bioactive compounds,44,45 we found that the most hydrophobic fraction, eluted in 100% methanol, had a strong inhibitory effect on B. simplex biofilm development, growth and biofilm-forming capacity (Figure 4a and Supplementary Figures S6A–C). The 80% methanol fraction showed a mild effect on B. simplex growth and development, while the rest of the fractions had no impact. When the cells treated with the bioactive 100% methanol fraction were examined under a florescence microscope, we found that the treated cells had severe morphological changes and abnormalities (Figure 4b). The cells were oval and deformed, in contrast to the elongated rod-shaped untreated cells. In addition, the cell membranes were severely damaged. We used liquid chromatography-mass spectrometry of the different fractions, and looked for known antibacterial compounds produced by B. subtilis. The liquid chromatography-mass spectrometry analysis demonstrated that the active 100% methanol fraction contained a high concentration of surfactin (Supplementary Figure S7). A lower concentration of surfactin was found in the slightly active 80% methanol fraction and only traces of surfactin were found in the non-active fraction. Unlike surfactin, the concentrations of iturin, fengycin,46 bacitracin,47 bacillibactin,48 plipastatin,49 subtilosin50 or bacillomycin F51 were not correlated with the killing activity. Consistent with our finding that the killing activity of the media is density dependent, surfactin production depends in the accumulation above a threshold of a peptide pheromone named ComX, accumulated late in the logarithmic stage.15 Indeed surfactin presence in the biofilm growth media was only evident after 8 h (Supplementary Figures S8 and 9).
Figure 4.
Surfactin mediates growth inhibition and cell deformation of B. simplex. (a) Biofilm formation of B. simplex grown for 3 days on biofilm-inducing plates, in the presence or absence of ×2 of B. subtilis supernatant fraction from either the WT or srfAA mutant, eluted in 100% methanol. Scale bars represent 2 mm. (b) Fluorescence microscope images of B. simplex biofilm cells grown for 3 days in the presence or absence of ×2 B. subtilis supernatant fraction from either the WT or srfAA mutant, eluted in 100% methanol, and stained with FM1–43. (i) Control, (ii) +WT fraction, (iii) +srfAA mutant fraction. Scale bar represents 2 μm. (c) Quantification of deformed B. simplex cells in B. simplex biofilms treated with or without ×2 B. subtilis supernatant fractions from WT or ΔsrfAA mutant strains, eluted with 100% methanol. (n=5000). Error bars represent the s.d. (d) Growth curves of B. simplex in 96-well plates with shaking in MSgg. Cells were supplemented with supernatant fractions derived from either WT or srfAA mutant strains. (e) Liquid chromatography-mass spectrometry analysis of the varying tail length surfactin molecules found in the active surfactin fraction. (i) Chromatographs of surfactin standard (SIGMA S3523), (ii) chromatographs of the active surfactin fraction, (iii) mass spectrometry analysis of the varient surfactin molecules found in the active fraction.
The surfactin molecule with a C12 tail was only found in the active fractions, implying that the length of the tail affects the killing properties of the molecule. In order to verify the liquid chromatography-mass spectrometry results, supernatant from ΔsrfAA, a B. subtilis mutant incapable of producing surfactin, were collected and fractionated. The 100% methanol fraction of the mutant supernatants had no effect on B. simplex growth and biofilm formation (Figure 4a,b). In addition, single cells treated with this fraction had no abnormalities in cell shape, in striking contrast to cells treated with the WT fraction (Figure 4a–c). In addition, the WT fraction fully inhibited B. simplex growth, while the equivalent fraction eluted from the ΔsrfAA mutant only induced a moderate lag in the initiation of B. simplex growth (Figure 4d).
We then quantified the amount of surfactin in the supernatant. Using a commercially available standard, we showed that the purified fractions from B. subtilis are significantly more potent (Figure 4e and Supplementary Figure S10) than the commercially available surfactin. Consistent with our previous finding that short-tail isomers of surfactin are correlated with increased toxicity towards B. simplex, the ratio of short length (C12) to full length (C14–15) surfactin isomers was increased significantly in the purified surfactin fractions. Owing to the significantly increased potency of the surfactin purified from the conditioned media and the subtle effect on B. simplex growth observed upon exposure to high concentrations of the ΔsrfAA mutant supernatant fractions, we speculated that additional bioactive killing molecules are secreted by B. subtilis.
The characterisation of these molecules was important to fully comprehend the attack mechanisms participating in this interspecies interaction. To this end, active proteins and/or peptides were eluted from the toxic supernatant using a Centricon Dialysis kit (Millipore). Concentrated supernatant products larger than 3 kDa had a strong inhibitory effect on B. simplex growth (Supplementary Figure S11). Mass spectrometry analysis of the protein fraction identified SdpC, a secreted toxin involved in bacterial cannibalism,24 as the active protein (Supplementary Figure S11B). An equivalent purified supernatant from a strain defective in production of both SdpC and SkfA (ΔsdpCΔskfA), the two B. subtilis cannibalism toxins, had no effect on B. simplex growth (Figure 5a). Consistent with our finding that the killing activity of the supernatant of B. subtilis in a biofilm media was only significant after 8 h of growth, we found that the secretion of the cannibalism toxins was enhanced between 6 and 8 h of growth (Supplementary Figure S11C). Notably, a small residual activity of the protein fraction was evident after 6 h, consistent with our observation that the cannibalism toxins contribution to elimination of the competing B. simplex cells is more subtle that the contribution of surfactin.
Figure 5.
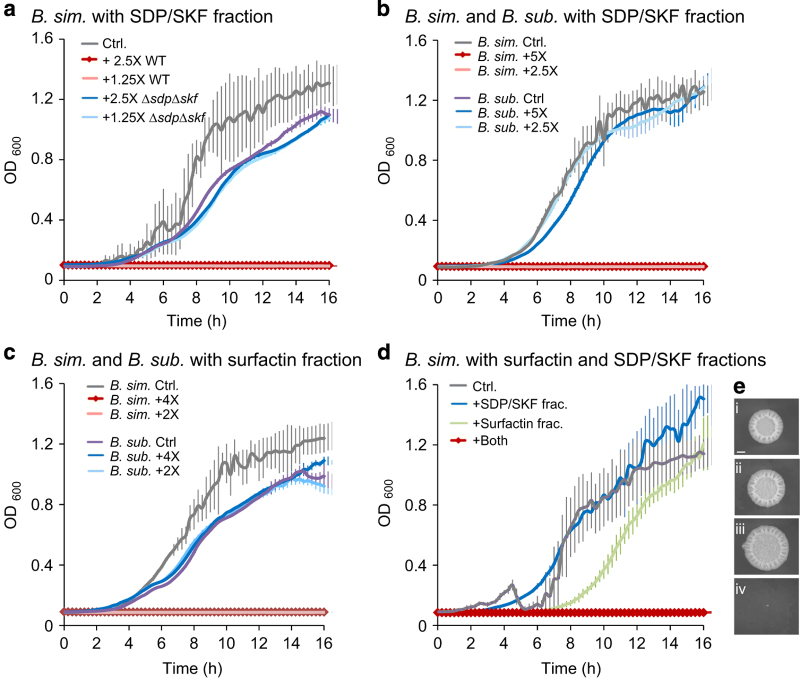
Surfactin and the cannibalism toxins inhibit B. simplex growth and biofilm formation at concentrations inert to B. subtilis itself and show synergistic cooperation. Growth curves of B. simplex or B. subtilis in liquid MSgg biofilm-inducing medium grown in 96-well plates, with shaking, at 30 °C. (a) Growth curves of B. simplex. Media supplemented with ×1.25 or ×2.5 B. subtilis supernatant protein fractions derived from WT or ΔsdpCΔskfA mutant strain cultures, enriched using a 3 kDa Centricon. n=6 wells, error bars represent s.d. (b) Growth curves of B. simplex and B. subtilis. Media supplemented with ×2.5 or ×5 B. subtilis supernatant protein fractions, enriched using a 3 kDa centricon. n=6 wells, Error bars represent the s.d. (c) Growth curves of B. simplex and B. subtilis. Media supplemented with ×2 or ×4 B. subtilis supernatant surfactin fraction. n=6 wells, Error bars represent the s.d. (d) Growth curves of B. simplex. Media supplemented with: (i) sub-toxic concentrations of ×0.31 B. subtilis supernatant protein fractions, enriched using a 3 KDa Centricon, (ii) ×0.1 B. subtilis supernatant fraction eluted in 100% methanol, or (iii) a combination of both of these fractions. n=6 wells, Error bars represent the s.d. (e) Biofilm formation of B. simplex grown for 2 days on biofilm-inducing medium plates: (i) untreated, (ii) treated with ×0.625 B. subtilis active protein fraction, (iii) treated with ×0.5 B. subtilis supernatant fraction eluted with 100% methanol and (iv) treated with a combination of both fractions. Scale bar represents 2 mm.
We used a strain carrying the fusion of the sdp promoter with a luciferase gene to monitor the activity of this promoter in the presence and absence of B. simplex. Quite interestingly the expression of sdpA-C, encoding for SDP was found to be increased during the interaction with B. simplex. When the effect of a cell-free supernatant was compared with the effect of co-culturing, it became evident that co-culturing induces the expression of the sdpA-C promoter more strongly than the secretome of B. simplex (Supplementary Figure S12). These results can be consistent with our finding (Figure 2), that B. simplex colonies eradication is mediated by direct contact between the competing colonies.
To date, B. subtilis has mostly been thought to produce self-targeted cannibalism toxins, which act to enable survival and delay sporulation upon nutrient starvation. The cannibalism toxins were described previously as toxic for different bacterial species.52,53 However, a direct comparison of the potency of purified cannibalism toxins on B. subtilis and versus competing Bacillus species was the best of our knowledge never directly compared. To assess whether the B. subtilis-generated SdpC and SkfA cannibalism toxins and surfactin, induce a stronger effect against foreign Bacillus species than on B. subtilis, we grew B. subtilis and B. simplex in several concentrations of the SkfA and SdpC eluate (Figure 5b) and the surfactin fraction (Figure 5c). Both the surfactin fraction and the cannibalism toxins elute fully inhibited B. simplex growth at concentrations that failed to affect B. subtilis. It is still feasible that the cannibalism toxins can inhibit the growth of B. subtilis in much higher concentrations, when applied to a biofilm medium. In addition, surfactin, Skf and Sdp, had a dramatic synergistic effect on B. simplex; when added separately, low concentrations of the surfactin fraction and of the SKF-SDP eluate had only a mild effect on B. simplex growth (Figure 5d). However, when applied as a mixture they caused complete inhibition of B. simplex growth (Figure 5d). The surfactin fraction and the cannibalism toxins elute also synergistically inhibited biofilm development of B. simplex (Figure 5e).
Furthermore, mutants deleted for either sdp,skf operons or srfAA displayed a severely impaired attack process. These mutants partially took over the B. simplex biofilm, and showed a massive decrease in their ability to invade the B. simplex biofilm and a subtle defect in overcoming B. simplex. However a triple mutant for srfAA, skf and sdp had the most substantial defect in killing B. simplex consistent with the idea that SKF, SDP and surfactin have synergistic killing activity during the interaction (Supplementary Figure S13).
We then asked whether surfactin, SDP and SKF were also at the core of the antagonistic interaction with additional Bacillus species. When we examined the effect of the B. subtilis supernatant on the bacterium B. toyonensis we found it is extremely toxic, when collected from the WT, and lacks killing activity when purified from a triple mutant for the production of surfactin, SDP and SKF (Supplementary Figure S14C). These results imply that similar mechanisms enable the allopathic interaction of B. subtilis biofilms with B. toyonensis (Figure 1n–p). These killing mechanisms were also used by B. subtilis when we tested the interaction with B. simplex on top of a rich solid biofilm media (B4) (Supplementary Figures S14A and B).
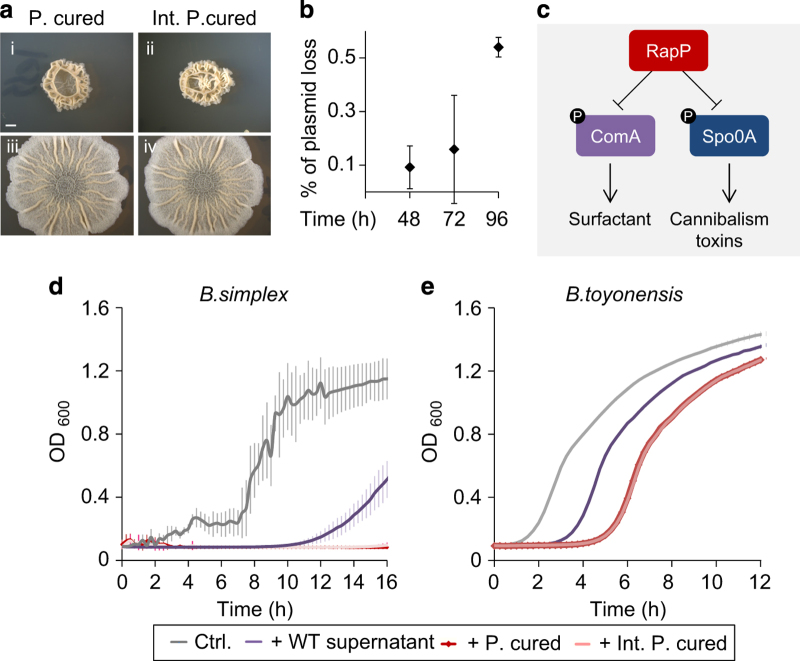
Strikingly, in the interaction with B. simplex we found an increasing occurrence of spontaneous mutations in the B. subtilis biofilm. Significantly more mutants were formed in interacting colonies in the final stages of the interaction compared with the initial stages (Figure 6b and Supplementary Figure S15A). In addition, all B. subtilis mutants were characterized by hyperrugose biofilms, reminiscent of biofilms formed by B. subtilis strains cured from their natural plasmid (Figure 6a). A whole-genome sequencing analysis showed that all the hyperrugose biofilm mutants did not carry any mutation in their genome, indicating that the hyperrugose phenotype may be an outcome of B. subtilis plasmid loss. A PCR analysis demonstrated that the evolved strains lacked rapP, a phosphatase whose encoding gene is carried on the plasmid pBS32 (Supplementary Figure S15B), and dephosphorylates Spo0F, resulting in activation of extracellular matrix production (Figure 6c). As we suspected that the hyperrugose phenotype is due to loss of RapP, we complemented the mutants for the rapP gene (Figure 6a), which led to full restoration of the biofilm morphology.
Figure 6.
Loss of the natural B. subtilis plasmid increases the production of B. subtilis killing factors that inhibit rival Bacillus species growth (a) (i) B. subtilis plasmid-cured strain. (ii) B. subtilis plasmid-cured strains isolated from interacting B. subtilis-B. simplex biofilms. (iii, iv) Genetic Complimentation with the rapP for the strains in (i) and (ii) respectively. (b) Percentage of B. subtilis biofilms cells that lost their natural plasmid during the interaction with B. simplex. n=6, error bars represent s.d. (c) The role of RapP in regulation of the killing factors that inhibit rival Bacillus species growth. (d, e) Growth curves of Bacillus species exposed to supernatant of B. subtilis WT, plasmid-cured, and interaction evolved strain. Bacteria were grown in 96-well plates, with shaking, at 30 °C. (b) B. simplex growth curves. MSgg medium supplemented with 8-h ×0.25 B. subtilis supernatants. n=6, error bars represent s.d. (d) Growth curves of B. toyonensis. Liquid LB broth medium, supplemented with 8-h ×0.25 B. subtilis supernatants. n=6, error bars represent the s.d.
The supernatants of plasmid-cured mutants demonstrated increased killing capacity toward B. simplex, when compared with those of the WT parental strain. The same phenotype was observed in a WT strain was artificially cured from the plasmid (Figure 6d). Strikingly, the plasmid loss also increased the toxicity of B. subtilis supernatant towards B. toyonensis (Figure 6e).
Importantly, an increase in Spo0F and ComA phosphorylation were previously observed in the plasmid-cured strain,33,54 thus indicating that a significant increase in the levels of Surfactin, SKF and SDP also elevates the fitness of plasmid-cured strains during interspecies competition.
Importantly, interspecies competition also affected the rate and the nature of the acquired mutated phenotypes in B. simplex biofilms. When B. simplex is grown alone, mutants can be isolated from mature biofilms, which show different levels of biofilm defects (Supplementary Figure S16). Quite strikingly, during the interspecies interaction, only mutants in which biofilm formation was fully inhibited were detected, suggesting that their competitive advantage was greater than those forming partially defective biofilms (Identified mutations are demonstrated in Supplementary Figure S16). Many of the biofilm inhibited B. simplex mutants enriched during interspecies interactions were associated with the loss of Spo0A (Supplementary Figure S16).
Discussion
Soil bacteria have developed diverse mechanisms to ensure their survival in harsh competitive soil environments. Some activities, such as generation and secretion of small active secondary metabolites, and the development of structurally complex colonies, present new therapeutic targets. Understanding interspecies interactions is also an important tool in comprehending bacterial developmental programs.8,55 The bacterial physiology is greatly affected by its surrounding environment, where the interaction between species can modify bacterial gene expression patterns and induce the secretion of various antimicrobial molecules.55,56
We chose to study the interaction between B. subtilis and B. simplex, two soil bacteria that form structured communities. These closely related bacteria can generate three dimensional complex biofilm structures while competing for the same ecological niche. Competition between the two species leads to B. subtilis engulfment and elimination of the B. simplex colony and enables B. subtilis to take over the ecological niche. The contact between the two biofilms leads to changes in the morphology and composition of the cells in the interaction zone, suggesting interspecies signalling.
Importantly, engulfment requires functional flagella, a finding that is consistent with the abundance of motile cells within mature B. subtilis biofilms (up to 10%, (ref. 26) data not shown). Although a non-motile mutant in the flagellin protein had little or no biofilm defect when grown in isolation, it was incapable of engulfing and overcoming neighbouring B. simplex colony. These finding imply that a reservoir of flagellated cells is actively maintained in B. subtilis biofilms, overcoming the negative-feedback loops downregulating motility in the single-cell level, and improving the fitness of the biofilm population during interspecies competition. Interestingly, the accumulation of motile cells in the interaction area was correlated with the formation of a thick wrinkle in the interphase (Figure 1d,m and Figure 3b). It was suggested previously that channels exist within B. subtilis wrinkles,57 and facilitate the transport of liquids57 and of motile cells.58 Thus, it is feasible that the transport of B. subtilis bioactive killing factors into competing Bacillus colonies and cross-species interactions may be enhanced by localised formation of channels.
Three secreted factors were shown to have a central role in elimination of competing Bacillus colonies: the small biosurfactant surfactin, the SdpC protein and the SkfA peptide. All three are considered autoinducers, affecting the behaviour of B. subtilis itself. It has been previously shown that B. subtilis changes its membrane phospholipid composition during secretion of surfactin, in order to defend itself against the membrane disruption activity of surfactin.59 Our results imply that the C-12-long surfactin carbon tail is correlated with the strongest effect on B. simplex cells. The length of the surfactin tail may act differently on different bacterial species, and surfactin synthesis patterns may be dictated by the specific bacterial species interacting with B. subtilis.
Both the SdpC protein and SkfA peptide are autoinducers that allow a subpopulation of B. subtilis cells expressing the master-regulator Spo0A, to eliminate those B. subtilis cells that did not commit to Spo0A-dependent programs. Thus, the primary role of these factors has been associated with postponement of sporulation during nutrient starvation.60 Alternatively, cannibal cells were suggested to eliminate ‘cheater’ cells not expressing extracellular matrix components during biofilm formation in mutants defective in cell-wall modulation, but had no phenotype in a WT background.25 We found that these molecules are orders of magnitude more potent towards competing Bacillus species, such as B. simplex, than to B. subtilis itself. Potency towards planktonically growing B. subtilis cells, biofilm state B. subtilis, and stationary B. subtilis cells remained relatively low. Similar results were observed with surfactin. Thus, it is highly feasible that the primary role of SdpC, SkfA and surfactin is to act against rival colonies in the soil. We suggest that the various adaptations of B. subtilis, such as the ability to both change its membrane characteristics to gain surfactin resistance, and to generate immunity factors against the cannibalism toxins, provide B. subtilis a unique ability to use autoinducers dually—as an weapon as well as self-regulatory mechanism. Importantly, we found that these three active factors are much more potent when working together, which may explain why they are all temporally co-expressed during biofilm maturation. The synergistic killing properties of these effectors may be mediated by the pore-forming surfactin, which facilitates its penetration into the rival biofilm extracellular matrix. This penetration can enable the diffusion of SdpC and SkfA into various layers of the rival biofilm that then mediate bacterial killing in these areas. In addition, it is feasible that surfactin promotes the entry of the polar SKF and SDP peptides into rival biofilm cells.
In numerous studies, plasmid acquisition was shown to be an effective means of gaining virulence properties, improving fitness during an interspecies competition.61,62 Quite strikingly, a reverse strategy is used by B. subtilis when encountering competing Bacillus species. During interspecies interaction, B. subtilis showed increased loss of its natural plasmid, which led to increased secretion of virulence factors SdpC, SkfA and surfactin. The increased secretion is due to the loss of the phosphatase, RapP, encoded on the plasmid, and subsequent increase in the phosphorylation of the master regulators ComA, that activates genetic competence and surfactin secretion, and Spo0A, that regulates the production of SKF and SDP.33,54,63 As the loss only occurs in a subpopulation of the biofilm cell community, it is highly feasible that plasmid loss is a strategy to temporary increase the overall fitness of the multicellular community during interspecies interactions, until the plasmid is re-gained following alleviation of the selective pressure. The mechanisms underlying plasmid loss and its regulation remain to be determined.
Overall, our results demonstrate how interspecies interactions contribute to the shaping of the expression patterns and genetic organisation of rival bacterial species. More specifically, we show that B. subtilis is much more dependent on its motile cells subpopulation, autoinducers, and on its capacity to lose a plasmid-encoded master regulator, in order to overcome a rival biofilm during an interspecies interaction, compared with the dependence on the same factors when grown in isolation.
Materials and Methods
Strains and media
All experiments were performed with B. subtilis NCIB 3610 (ref. 64) strain, B. simplex WT strain and their derivatives. For cloning purposes, we used B. subtilis strain PY79 and Escherichia coli strain DH5α. A complete list is shown in Supplementary Table S1.
Strains and plasmids were constructed using standard methods.64,65 Oligonucleotides used for PCR in this study are listed in Supplementary Table S2. All deletion mutations were generated by long-flanking homology PCR mutagenesis.66 DNA was first introduced by transformation into strain PY79 and the deletion further integrated into NCIB 3610 by transformation as described previously.67
The strains were routinely manipulated in LB broth (Difco, Le Pont de Claix, France), B4 was prepared as described previously,37 mLB solid medium (modified LB)68 or MSgg medium (5 m mol/l potassium phosphate, 100 m mol/l MOPS pH 7, 2 m mol/l MgCl2, 50 μ mol/l MnCl2, 125 μ mol/l FeCl3, 700 μ mol/l CaCl2, 1 μ mol/l ZnCl2, 2 μ mol/l thiamine, 0.5% glycerol, 0.5% glutamate, 50 μg/ml threonine, tryptophan and phenylalanine). Solid medium contained 1.5% bacto agar (Difco). Note that the iron concentration in the solid MSgg medium was 2.5-fold higher compared with the original recipe.64 Selective media were prepared as described in Supplementary Materials and Methods.
Interaction assay
For details, please refer to the Supplementary Text and Supplementary Materials and Methods.
Quantification of plasmid-cured cells
To analyse the percentage of cells that lost their natural plasmid during the interaction between B. subtilis and B. simplex, interaction plates were incubated for the required time period. The biofilms were collected, inserted into 200 μl phosphate-buffered saline and mildly sonicated. The cells were then diluted, plated and incubated at 37 °C overnight to allow the formation of colonies. Colonies were then further incubated at 23 °C overnight. The plasmid-cured colonies were recognised by their colony morphology.32 The suspected colonies were confirmed on MSgg-agar plates. PCR analysis confirmed that the evolved colonies lacked the rapP gene.
Supernatant production
For details, please refer to the Supplementary Text and Supplementary Materials and Methods.
Separation and identification of surfactin
The growth inhibiting, 100% methanol elute of the 8-h-old B. subtilis conditioned medium was eluted from a C-18 SPE column (Waters, Milford, MA, USA). A single B. subtilis WT colony, isolated on a solid LB plate, was inoculated into 3 ml of LB broth, grown overnight at room temperature. The overnight culture (100 μl) were inoculated into 100 ml MSgg and grown in a 300 l flask at 37 °C with shaking for 8 h. The conditioned medium was centrifuged at 8,000 r.p.m. for 10 min; supernatant was removed and filtered through a 0.22-μm filter. For further purification the supernatant was fractionated on a C-18 Sep-Pak cartridge using stepwise elution of 0–100% methanol with steps of 10%–20%–30%–40%–60%–80%–100%. The methanol fractions were evaporated by speed-vac. To test the activity of the extracts, the evaporated samples were dehydrated with dW to a final concentration of 20×. The active fraction was further analysed using liquid chromatography-mass spectrometry as described in Supplementary Materials and Methods.
Separation of SdpC and SkfA peptides
Protein fraction of the conditioned medium from 8-h-old B. subtilis was fractionised by Cellulose Amicon Ultra Centrifugal Filter Unit with Ultracel-3 Membrane, 15 ml Capacity, 3 kDa nominal molecular weight limit (NMWL; Millipore, Cork, Ireland). For details please refer to Supplementary Materials and Methods.
Growth measurements
A single colony of B. subtilis or B. simplex, isolated on a solid LB plate, was inoculated into 3 ml of LB broth and grown to a mid-logarithmic phase of growth (B. subtilis 4 h, B. simplex 5 h at 37 °C with shaking). Cells were diluted 1:100 in 150 μl liquid MSgg medium of each well of a 96-well microplate (Thermo Scientific, Roskilde, Denmark). Cells were grown with agitation at 30 °C for 16 h in a microplate reader (Synergy 2, BioTek, Winooski, VT, USA), and the optical density at 600 nm (OD600) was measured every 15 min.
Assessing the effects of B. subtilis bioactive effectors on biofilm development and cell growth
For details, please refer to the Supplementary Text and Supplementary Materials and Methods.
Fluorescence microscopy
To analyse the effect of contact between the two biofilms, B. subtilis WT or its indicated derivatives and B. simplex, interaction plates were prepared as described, and incubated for the required time period. The interacting biofilms were divided to three areas as mentioned in the analysis of bacterial population from the interaction. The B. simplex area was separated from the plate and suspended in 200 μl phosphate-buffered saline. The B. simplex biofilm was roughly disassembled by pipetting. Samples were centrifuged briefly, and re-suspended in 10 μl of 1× phosphate-buffered saline supplemented with the membrane stain FM4–64 (Molecular Probes, Eugene, OR, USA) at 1 μg/ml and the DNA stain 4,6-diamidino-2-phenylindole (Sigma, Saint-Louis, MO, USA) at 2 μg/ml. This concentrated cell suspension (3 μl) was placed on a microscope slide. A freshly prepared poly l-lysine-treated (Sigma) coverslip. Treated coverslip was used to monitor the B. subtilis GFP marked strain invasion to the B. simplex biofilm area. The cells were observed by Axio microscope (Zeiss, Goettingen, Germany) images were analysed by Zen-10 software (Zeiss).
To analyse the effect of the bioactive fraction eluted with methanol on B. simplex biofilms in the single-cell level, the biofilm was roughly disassembled by pipetting. Samples were centrifuged briefly, and re-suspended in 10 μl of 1× phosphate-buffered saline supplemented with the membrane stain FM1–43 (Molecular Probes) at 1 μg/ml and the DNA stain 4,6-diamidino-2-phenylindole (Sigma) at 2 μg/ml. Importantly. This concentrated cell suspension (3 μl) was placed on a microscope slide. A freshly prepared poly l-lysine-treated (Sigma) coverslip was used to immobilise the cells for membrane visualisation. The cells were observed by Axio microscope (Zeiss) images were analysed by Zen-10 software.
Environmental scanning electron microscopy
Biofilms grown for 72 h at 30 °C were fixed overnight at 4 °C with 2% glutaraldehyde, 3% papaformaldehyde, 0.1 mol/l Sodium Cacodylate (pH 7.4), 5 m mol/l CaCl2. After two 15 min washes with double distilled water, samples were dehydrated through series of ethanol washes. Subsequently, samples were dried on filter paper (Whatman) overnight at room temperature, mounted and stored under vacuum. Mounted samples were sputter coated with gold–palladium shortly before examination with a scanning electron microscope ×L30 with field emission gun.
Estimating the effect of interspecies interaction on bacterial evolution
Interacted biofilms, treated as described in analysis of bacterial populations in the interaction, were divided, diluted and plated on LB-agar plates. The bacterial colonies from the LB-agar plates were inoculated into 3 ml of LB broth and grown to a mid-logarithmic phase of growth. A measure of 2 μl of each culture was inoculated onto MSgg-agar plates and incubated at 30 °C for 48 h. The bacterial biofilm were screened for biofilm defects. Biofilms with hereditable morphological defects upon re-inoculation were characterized as biofilm mutants. The mutants generated as a result of biofilms interactions were compared with mutants from of B. subtilis and B. simplex biofilms that were grown in isolation.
RapP complementation
For details, please refer to the Supplementary Text and Supplementary Materials and Methods.
Alignment of B. subtilis reads and identification of mutations
For details, please refer to the Supplementary Text and Supplementary Materials and Methods.
B. simplex genome assembly and mutation identification
For details, please refer to the Supplementary Text and Supplementary Materials and Methods.
Acknowledgments
We thank Professor Ed Bayer for his helpful advices, and for suggesting the title for our manuscript. We thank Dr Assaf Vardi and his lab members for many helpful discussions. This research is supported by ISF-icore grant 152/1, by a research grant from Ayala Benjamin-Mashat, KAMIN program for R & D and by Angel-Fiavovich fund for ecological research, a research grant from Mr. and Mrs. Dan Kane; Ms. Lois Rosen; the Larson Charitable Foundation; and Ruth and Herman Albert Scholars Program for New Scientists and Clore Krenner Katz grant. IK-G is a recipient of Rowland and Sylvia Career Development Chair.
Footnotes
Supplementary Information accompanies the paper on the npj Biofilms and Microbiomes website (http://www.nature.com/npjbiofilms)
The authors declare no conflict of interest.
References
- Tringe, S. G. et al. Comparative metagenomics of microbial communities. Science 308, 554–557 (2005). [DOI] [PubMed] [Google Scholar]
- Torsvik, V. , Goksøyr, J. & Daae, F. L. High diversity in DNA of soil bacteria. Appl. Environ. Microbiol. 56, 782–787 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wardle, D. A. et al. Ecological linkages between aboveground and belowground biota. Science 304, 1629–1633 (2004). [DOI] [PubMed] [Google Scholar]
- Lynch, J. M. et al. Microbial diversity in soil: ecological theories, the contribution of molecular techniques and the impact of transgenic plants and transgenic microorganisms. Biol. Fertil. Soils 40, 363–385 (2004). [Google Scholar]
- Kolter R. & Greenberg E. P. Microbial sciences: the superficial life of microbes. Nature 441, 300–302 (2006). [DOI] [PubMed] [Google Scholar]
- Elias, S. & Banin, E. Multi-species biofilms: living with friendly neighbors. FEMS Microbiol. Rev. 36, 990–1004 (2012). [DOI] [PubMed] [Google Scholar]
- Little, A. E. F. , Robinson, C. J. , Peterson, S. B. , Raffa, K. F. & Handelsman, J. Rules of engagement: interspecies interactions that regulate microbial communities. Ann. Rev. Microbiol. 62, 375–401 (2008). [DOI] [PubMed] [Google Scholar]
- Vlamakis, H. , Chai, Y. , Beauregard, P. , Losick, R. & Kolter, R. Sticking together: building a biofilm the Bacillus subtilis way. Nat. Rev. Microbiol. 11, 157–168 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beloin, C. , Renard, S. , Ghigo, J.-M. & Lebeaux, D. Novel approaches to combat bacterial biofilms. Curr. Opin. Pharmacol. 18, 61–68 (2014). [DOI] [PubMed] [Google Scholar]
- Earl, A. M. , Losick, R. & Kolter, R. Ecology and genomics of Bacillus subtilis. Trends Microbiol. 16, 269–275 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski, J. & Nevo, E. Patterns of thermal adaptation of Bacillus simplex to the microclimatically contrasting slopes of 'Evolution Canyons' I and II, Israel. Environ. Microbiol. 9, 716–726 (2007). [DOI] [PubMed] [Google Scholar]
- Dubnau, D. Genetic competence in Bacillus subtilis. Microbiol. Rev. 55, 395–424 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubnau, D. & Provvedi, R. Internalizing DNA. Res. Microbiol. 151, 475–480 (2000). [DOI] [PubMed] [Google Scholar]
- Roggiani, M. & Dubnau, D. ComA, a phosphorylated response regulator protein of Bacillus subtilis, binds to the promoter region of srfA. J. Bacteriol. 175, 3182–3187 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnuson, R. , Solomon, J. & Grossman, A. D. Biochemical and genetic characterization of a competence pheromone from B. subtilis. Cell 77, 207–216 (1994). [DOI] [PubMed] [Google Scholar]
- Gonzalez, D. J. et al. Microbial competition between Bacillus subtilis and Staphylococcus aureus monitored by imaging mass spectrometry. Microbiology 157, 2485–2492 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falardeau, J. , Wise, C. , Novitsky, L. & Avis, T. J. Ecological and mechanistic insights into the direct and indirect antimicrobial properties of Bacillus subtilis lipopeptides on plant pathogens. J. Chem. Ecol. 39, 869–878 (2013). [DOI] [PubMed] [Google Scholar]
- Avigad, L. S. & Bernheimer, A. W. Nature and properties of a cytolytic agent produced by Bacillus subtilis. J. Gen Microbiol. 61, 361–369 (1970). [DOI] [PubMed] [Google Scholar]
- Hoefler B. C. , Gorzelnik K. V. , Yang J. Y. , Hendricks N. & Dorrestein P. C . Enzymatic resistance to the lipopeptide surfactin as identified through imaging mass spectrometry of bacterial competition. Proc. Natl Acad. Sci. USA 109, 13082–13087 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watrous, J. et al. Mass spectral molecular networking of living microbial colonies. Proc. Natl Acad. Sci. USA 109, E1743–E1752 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grau, A. , Go, J. C. & Ortiz, A. A study on the interactions of surfactin with phospholipid vesicles. Biochim. Biophys. Acta 1418, 307–319 (1999). [DOI] [PubMed] [Google Scholar]
- Chai, Y. , Chu, F. , Kolter, R. & Losick, R. Bistability and biofilm formation in Bacillus subtilis. Mol. Microbiol. 67, 254–263 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez, D. , Fischbach, M. A. , Chu, F. , Losick, R. & Kolter, R. Structurally diverse natural products that cause potassium leakage trigger multicellularity in Bacillus subtilis. Proc. Natl Acad. Sci. USA 106, 280–285 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellermeier, C. D. , Hobbs, E. C. , Gonzalez-Pastor, J. E. & Losick, R. A three-protein signaling pathway governing immunity to a bacterial cannibalism toxin. Cell 124, 549–559 (2006). [DOI] [PubMed] [Google Scholar]
- Lopez, D. , Vlamakis, H. , Losick, R. & Kolter, R. Cannibalism enhances biofilm development in Bacillus subtilis. Mol. Microbiol. 74, 609–618 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlamakis, H. , Aguilar, C. , Losick, R. & Kolter, R. Control of cell fate by the formation of an architecturally complex bacterial community. Genes Dev. 22, 945–953 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branda, S. S. , Chu, F. , Kearns, D. B. , Losick, R. & Kolter, R. A major protein component of the Bacillus subtilis biofilm matrix. Mol. Microbiol. 59, 1229–1238 (2006). [DOI] [PubMed] [Google Scholar]
- Chu, F. , Kearns, D. B. , Branda, S. S. , Kolter, R. & Losick, R. Targets of the master regulator of biofilm formation in Bacillus subtilis. Mol. Microbiol. 59, 1216–1228 (2006). [DOI] [PubMed] [Google Scholar]
- Hobley, L. et al. BslA is a self-assembling bacterial hydrophobin that coats the Bacillus subtilis biofilm. Proc. Natl Acad. Sci. USA 110, 13600–13605 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearns, D. B. , Chu, F. , Branda, S. S. , Kolter, R. & Losick, R. A master regulator for biofilm formation by Bacillus subtilis. Mol. Microbiol. 55, 739–749 (2005). [DOI] [PubMed] [Google Scholar]
- Bai, U. , Mandic-Mulec, I. & Smith, I. SinI modulates the activity of SinR, a developmental switch protein of Bacillus subtilis, by protein-protein interaction. Genes Dev. 7, 139–148 (1993). [DOI] [PubMed] [Google Scholar]
- McLoon, A. L. , Guttenplan, S. B. , Kearns, D. B. , Kolter, R. & Losick, R. Tracing the domestication of a biofilm-forming bacterium. J. Bacteriol. 193, 2027–2034 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parashar, V. , Konkol, M. A. , Kearns, D. B. & Neiditch, M. B. A plasmid-encoded phosphatase regulates Bacillus subtilis biofilm architecture, sporulation, and genetic competence. J. Bacteriol. 195, 2437–2448 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omer Bendori, S. , Pollak, S. , Hizi, D. & Eldar, A. The RapP-PhrP quorum-sensing system of Bacillus subtilis strain NCIB3610 affects biofilm formation through multiple targets, due to an atypical signal-insensitive allele of RapP. J. Bacteriol. 197, 592–602 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai, Y. , Kolter, R. & Losick, R. Reversal of an epigenetic switch governing cell chaining in Bacillus subtilis by protein instability. Mol. Microbiol. 78, 218–229 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norman, T. M. , Lord, N. D. , Paulsson, J. & Losick, R. Memory and modularity in cell-fate decision making. Nature 503, 481–486 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barabesi, C. et al. Bacillus subtilis gene cluster involved in calcium carbonate biomineralization. J. Bacteriol. 189, 228–235 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houry, A. et al. Bacterial swimmers that infiltrate and take over the biofilm matrix. Proc. Natl Acad. Sci. USA 109, 13088–13093 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavallie, E. R. & Stahl, M. L. Cloning of the flagellin gene from bacillus subtilis and complementation studies of an in vitro-derived deletion mutation. J. Bacteriol. 171, 3085–3094 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao, C. V. , Glekas, G. D. & Ordal, G. W. The three adaptation systems of Bacillus subtilis chemotaxis. Trends. Microbiol. 16, 480–487 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearns, D. B. , Chu, F. , Rudner, R. & Losick, R. Genes governing swarming in Bacillus subtilis and evidence for a phase variation mechanism controlling surface motility. Mol. Microbiol. 52, 357–369 (2004). [DOI] [PubMed] [Google Scholar]
- Verhamme, D. T. , Kiley, T. B. & Stanley-Wall, N. R. DegU co-ordinates multicellular behaviour exhibited by Bacillus subtilis. Mol. Microbiol. 65, 554–568 (2007). [DOI] [PubMed] [Google Scholar]
- Ng, W. L. , Bassler, B. L. Bacterial quorum-sensing network architectures. Annual Rev. Genet. 43, 197–222 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolodkin-Gal, I. , Hazan, R. , Gaathon, A. , Carmeli, S. & Engelberg-Kulka, H. A linear pentapeptide is a quorum-sensing factor required for mazEF-mediated cell death in Escherichia coli. Science 318, 652–655 (2007). [DOI] [PubMed] [Google Scholar]
- Kumar, S. , Kolodkin-Gal, I. & Engelberg-Kulka, H. Novel quorum-sensing peptides mediating interspecies bacterial cell death. mBio 4, e00314–00313 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim P. I. , Ryu J. , Kim Y. H. & Chi Y. T. Production of biosurfactant lipopeptides iturin A, fengycin, and surfactin A from Bacillus subtilis CMB32 for control of colletotrichum gloeosporioides. J. Microbiol. Biotechnol. 20, 138–145 (2010). [PubMed] [Google Scholar]
- Ming, L. J. & Epperson, J. D. Metal binding and structure-activity relationship of the metalloantibiotic peptide bacitracin. J. Inorg. Biochem. 91, 46–58 (2002). [DOI] [PubMed] [Google Scholar]
- May, J. J. , Wendrich, T. M. & Marahiel, M. A. The dhb operon of Bacillus subtilis encodes the biosynthetic template for the catecholic siderophore 2,3-dihydroxybenzoate-glycine-threonine trimeric ester bacillibactin. J. Biol. Chem. 276, 7209–7217 (2001). [DOI] [PubMed] [Google Scholar]
- Volpon, L. , Besson, F. & Lancelin, J. M. NMR structure of antibiotics plipastatins A and B from Bacillus subtilis inhibitors of phospholipase A(2). FEBS Lett. 485, 76–80 (2000). [DOI] [PubMed] [Google Scholar]
- Babasaki, K. , Takao, T. , Shimonishi, Y. & Kurahashi, K. Subtilosin A, a new antibiotic peptide produced by Bacillus subtilis 168: isolation, structural analysis, and biogenesis. J. Biochem. 98, 585–603 (1985). [DOI] [PubMed] [Google Scholar]
- Mhammedi, A. , Peypoux, F. , Besson, F. & Michel, G. Bacillomycin F, a new antibiotic of iturin group: isolation and characterization. J. Antibiot. 35, 306–311 (1982). [DOI] [PubMed] [Google Scholar]
- Liu, W. T. et al. Imaging mass spectrometry of intraspecies metabolic exchange revealed the cannibalistic factors of Bacillus subtilis. Proc. Natl Acad. Sci. USA 107, 16286–16290 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamsa, A. , Liu, W. T. , Dorrestein, P. C. & Pogliano, K. The Bacillus subtilis cannibalism toxin SDP collapses the proton motive force and induces autolysis. Mol. Microbiol. 84, 486–500 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Y. et al. A plasmid-born Rap-Phr system regulates surfactin production, sporulation and genetic competence in the heterologous host, Bacillus subtilis OKB105. Appl. Microbiol. Biotechnol. 99, 7241–7252 (2015). [DOI] [PubMed] [Google Scholar]
- Seyedsayamdost, M. R.. , Traxler, M. F. , Clardy, J. & Kolter, R. Old meets new: using interspecies interactions to detect secondary metabolite production in actinomycetes. Methods Enzymol. 517, 89–109 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shank, E. A. et al. Interspecies interactions that result in Bacillus subtilis forming biofilms are mediated mainly by members of its own genus. Proc. Natl Acad. Sci. USA 108, E1236–E1243 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilking, J. N. et al. Liquid transport facilitated by channels in Bacillus subtilis biofilms. Proc. Natl Acad. Sci. USA 110, 848–852 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asally, M. et al. Localized cell death focuses mechanical forces during 3D patterning in a biofilm. Proc. Natl Acad. Sci. USA 109, 18891–18896 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seydlová, G. et al. Surfactin production enhances the level of cardiolipin in the cytoplasmic membrane of Bacillus subtilis. Biochim. Biophys. Acta 1828, 2370–2378 (2013). [DOI] [PubMed] [Google Scholar]
- Gonzalez-Pastor, J. E. , Hobbs, E. C. & Losick, R. Cannibalism by sporulating bacteria. Science 301, 510–513 (2003). [DOI] [PubMed] [Google Scholar]
- Goessweiner-Mohr, N. , Arends, K. , Keller, W. & Grohmann, E. Conjugative type IV secretion systems in Gram-positive bacteria. Plasmid 70, 289–302 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kienesberger, S. et al. Interbacterial macromolecular transfer by the Campylobacter fetus subsp. venerealis type IV secretion system. J. Bacteriol. 193, 744–758 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boguslawski, K. M. , Hill, P. A. & Griffith, K. L. Novel mechanisms of controlling the activities of the transcription factors Spo0A and ComA by the plasmid-encoded quorum sensing regulators Rap60-Phr60 in Bacillus subtilis. Mol. Microbiol. 96, 325–348 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branda, S. S. , Gonzalez-Pastor, J. E. , Ben-Yehuda, S. , Losick, R. & Kolter, R. Fruiting body formation by Bacillus subtilis. Proc. Natl Acad. Sci. USA 98, 11621–11626 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J. & Russell, D. W. (eds) Molecular cloning: a laboratory manual (Cold Spring Harbor Laboratory Press, 2001). [Google Scholar]
- Wach, A. PCR-synthesis of marker cassettes with long flanking homology regions for gene disruptions in S. cerevisiae. Yeast 12, 259–265 (1996). [DOI] [PubMed] [Google Scholar]
- Wilson, G. A. & Bott, K. F. Nutritional factors influencing the development of competence in the Bacillus subtilis transformation system. J. Bacteriol. 95, 1439–1449 (1968). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shemesh, M. & Chai, Y. A combination of glycerol and manganese promotes biofilm formation in Bacillus subtilis via histidine kinase KinD signaling. J. Bacteriol. 195, 2747–2754 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.