Short abstract
Recent sequencing of the genome and proteomic analysis of a model aerobic methanotrophic bacterium, Methylococcus capsulatus (Bath) has revealed a highly versatile metabolic potential. In parallel, environmental genomics has provided glimpses into anaerobic methane oxidation by certain archaea, further supporting the hypothesis of reverse methanogenesis.
Abstract
Recent sequencing of the genome and proteomic analysis of a model aerobic methanotrophic bacterium, Methylococcus capsulatus (Bath) has revealed a highly versatile metabolic potential. In parallel, environmental genomics has provided glimpses into anaerobic methane oxidation by certain archaea, further supporting the hypothesis of reverse methanogenesis.
Methane is a powerful greenhouse gas, and its atmospheric concentration has been steadily increasing over the past 300 years. There are two major ways in which methane is removed from the environment: aerobic oxidation by a specialized group of bacteria and anaerobic oxidation by a specialized group of archaea. The former is important for keeping methane concentrations balanced in freshwater sediments and soils, whereas the latter is the major process in anoxic marine environments. The biochemistry of aerobic methane oxidation is relatively well understood, following intensive research efforts with a number of model organisms, but the biochemistry of anaerobic methane oxidation is not yet fundamentally understood and no anaerobic methane-oxidizer has been isolated in pure culture so far. Three recent studies using global approaches [1-3] have shed new light on both aerobic and anaerobic systems. Here, we first review background information on the two metabolic systems involving methane and then discuss the insights revealed through the three recent studies [1-3], as well as a fourth [4] that is useful for interpreting the new results on anaerobic methane oxidation [3].
Aerobic and anaerobic methanotrophs
Three types of aerobic methanotrophs are recognized. Type I methanotrophs are γ-proteobacteria that have stacked membranes harboring methane monooxygenase (pMMO), the enzyme for primary methane oxidation, and that use the ribulose monophosphate (RuMP) cycle, which converts formaldehyde into multicarbon compounds, for building cell biomass [5]. Type II methanotrophs belong to the α-proteobacteria, have rings of pMMO-harboring membranes at the periphery of the cells, and use the serine cycle, an alternative pathway for converting formaldehyde into biomass; these bacteria also often contain a soluble (s) MMO in addition to pMMO [5]. The third type, type X methanotrophs, belong to the genus Methylococcus (γ-proteobacteria) and combine features characteristic of the other two types: they have stacked membranes and the RuMP cycle, but they also have elements of the serine cycle and sMMO [5]. The type X methanotroph Methylococcus capsulatus has been a favorite model for research because of its robust growth on methane and its relative ease of use as a genetic system [6-9]. Two almost identical gene clusters have been identified encoding the subunits of pMMO, which are expressed simultaneously and are functionally redundant [7,8], and another gene cluster encodes the subunits of sMMO [9]. Copper has been shown to play an essential role in expression of the pMMO operons, whereas the sMMO operon appears to be expressed only in low-copper conditions [10]. The catalytic mechanisms for both pMMO and sMMO [11,12] are understood on a sophisticated level, but until recently no whole-genome sequence has been available for M. capsulatus or for any other methanotroph. Two recent studies [1,2] have used a whole-genome-shotgun sequencing approach to complement the mounting dataset on the biochemistry and regulation of aerobic methane oxidation.
In contrast, understanding of the process of anaerobic methane oxidation is in its infancy. Geochemical evidence points strongly towards a coupling of anaerobic methane oxidation with sulfate reduction [13]. Microbes involved in this process have been identified recently as archaea related to Methanosarcinales that fall phylogenetically into two distinct groups, ANME-I and ANME-II; these are normally found in association with sulfate-reducing bacteria [13]. There is no clear concept of how methane oxidation is linked to sulfate reduction; Figure 1 shows a possible model. This co-metabolism has to be viewed in the light of the thermodynamic constraints, however; the free energy (ΔG) for anaerobic methane oxidation in situ is estimated at -20 to -40 kJ/mol), the lowest value described that enables microbial growth [13,14].
Figure 1.
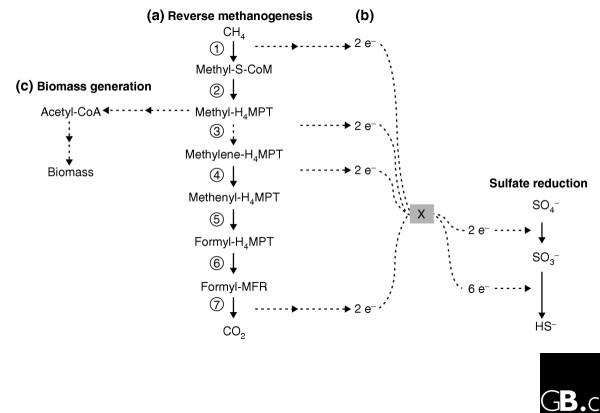
A proposed pathway for anaerobic oxidation of methane involving the homolog of methyl-CoM reductase and a novel methylene-tetrahydromethanopterin (H4MPT) reductase (Mer), and its connection with the sulfate reduction pathway. (a) The reverse methanogenesis pathway. Solid arrows represent enzymes predicted from the sequences found by Hallam et al. [3]; the dotted arrow represents the one enzyme that was not predicted, methylene H4MPT-reductase (Mer). Enzymes performing steps 1-7: 1, Methyl-CoM reductase-like protein (MCR); 2, Methyl-H4MPT:coenzyme M (CoM) methyl-transferase (Mtr); 3, Methylene-H4MPT reductase (Mer); 4, F420-dependent methylene-H4MPT dehydrogenase (Mtd); 5, Methenyl-H4MPT cyclohydrolase (Mch); 6, Formyl-MFR:H4MPT formyltransferase (Ftr); 7, Formyl-MFR dehydrogenase (Fmd). (b) Reverse methanogenesis is thought to be connected to sulfate reduction through an unknown intermediate (X); e- represents an electron. Hallam et al. [3] suggest that steps 1 and 2 in (a) function in the down direction and methyl-H4MPT is used for biomass generation (c), while steps 4 to 7 function in the up direction and the methylene-H4MPT produced is either converted to biomass through the serine cycle or is oxidized to CO2. We suggest that Mer or an analogous enzyme probably performs step 3 instead.
There is agreement on the hypothesis that reverse methanogenesis plays a key role in the methane oxidation process [13,14]: most enzymes of methanogenesis are easily reversible, and part of the methanogenesis pathway operates in reverse for energy generation in Methanosarcina species growing on such substrates as methanol or methylamine [15,16]. But the last step of methanogenesis and presumably the first in anaerobic methane oxidation (step 1 in Figure 1), catalyzed by methyl-coenzyme M reductase (MCR), presents a mechanistic challenge given the fact that methane is chemically unreactive. Nevertheless, data have been obtained showing that methanotrophic archaea have homologs of the genes for all three subunits of MCR, suggesting that MCR or a similar enzyme may indeed be responsible for anaerobic methane oxidation [17]. Two recent studies [3,4] describe efforts to establish the roles of mcr homologs and of other genes potentially involved in reverse methanogenesis by directly assessing environmental DNA and protein pools.
Genomic insights into the aerobic methanotrophy of M. capsulatus
In a paper recently published in PLoS Biology, Ward et al. [1] describe the complete genomic sequence of Methylococcus capsulatus (Bath). They annotate the genome in terms of the specific adaptations this organism has evolved in order to succeed at a lifestyle solely dependent on utilization of methane. The genome of M. capsulatus (3.3 megabases, Mb) is much smaller than the genome of a model facultative methylotroph, Methylobacterium extorquens AM1 (7 Mb), a bacterium with a much more versatile lifestyle [18], but is comparable in size to the genome of another obligate methylotroph, Methylobacillus flagellatus (2.9 Mb) [19], suggesting that the degree of specialization in methylotrophy may correlate with genome size. The cause of the obligate methylotrophy of M. capsulatus remains unresolved, however. The tricarboxylic acid (TCA) cycle is the pathway that converts acetyl-CoA to CO2 and is the major source of reducing equivalents during growth on multicarbon compounds; the long-held hypothesis that M. capsulatus lacks a complete TCA cycle [20] has not been proven true by genome sequencing, as putative genes for all the enzymes of the cycle were identified in the recent study [1]. In addition, the organism seems to encode an array of enzymes that could metabolize sugars, so the inability of M. capsulatus to grow on sugars remains enigmatic.
Analysis of the genes encoding enzymes involved in the metabolism of single-carbon compounds in M. capsulatus (Figure 2) has been greatly simplified by the addition of data available from pre-genomic analyses [7-9,21] and from the initial analysis of the genome of M. extorquens [18]. As expected, all the genes encoding enzymes of the RuMP pathway have been identified. In accordance with previous observations, most of the genes for the serine cycle were also found, as were the genes for the Calvin-Benson-Bassham (CBB) cycle, the pathway that reduces CO2 and converts it into biomass (Figure 2f) [5,20]. The potential to operate all three known pathways for the assimilation of single-carbon compounds that are found in various methylotrophs makes this organism unique, but further analysis involving knockout mutations is needed to understand the functions of each of the three pathways.
Figure 2.
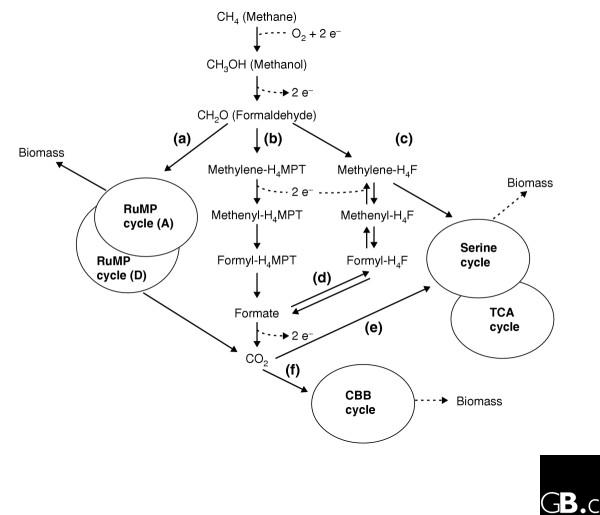
Pathways in the aerobic methanotrophic bacterium Methylococcus capsulatus involved in the metabolism of single-carbon compounds, as determined by genome sequencing and proteome analysis. Formaldehyde produced from methane can be metabolized in the following alternative ways: (a) through the ribulose monophosphate (RuMP) cycle, which can either generate biomass (via the assimilatory (A) RuMP cycle) or CO2 (via the dissimilatory (D) RuMP cycle); (b) by conversion to formate via intermediates containing tetrahydromethanopterin (H4MPT); (c) via methylene-tetrahydrofolate (methylene-H4F) to the serine cycle and from there into biomass. Under certain conditions, there can be an excess of formaldehyde and formate; the former can be used up through pathway (c) and the latter by reduction to methylene-H4F (d) and thus directed into the serine cycle. CO2 produced in any of these reactions can be converted to biomass by either (e) the serine cycle or (f) the Calvin-Benson-Bassham (CBB) cycle.
Proteomics of M. capsulatus
The first glimpses into the expression patterns of pathways enabling methanotrophy are coming from a proteomic analysis of M. capsulatus by a group that has independently sequenced the M. capsulatus genome to 8X coverage [2]. In this work [2], quantitative proteomic analysis was performed in order to compare the response of M. capsulatus to low-copper and high-copper conditions. Kao et al. [2] identified a total of 682 differentially expressed proteins using a cleavable isotope-coded affinity tag (cICAT) technique. The authors [2] demonstrated that, as expected, pMMO is overexpressed in conditions of high copper whereas sMMO is expressed at low copper levels. Equally interesting data from this work concern the expression of proteins other than MMOs, indicating that, indeed, all three assimilatory pathways are simultaneously expressed. The oxidative pathway linked to tetrahydromethanopterin (H4MPT) is one of the pathways by which formaldehyde can be oxidized to CO2 (Figure 2b); all the enzymes in this pathway were identified [2], pointing to the importance of this pathway, as suggested previously by enzyme-activity measurements [22]. Peptides for the oxidative branch of the RuMP cycle were also identified [2], suggesting that it is operational in M. capsulatus (Figure 2a).
Some of the major serine-cycle enzymes were found to be overexpressed under high-copper conditions [2]. It is unlikely, however, that their expression would be directly regulated by copper; it is more likely that they are responding to the higher flux of formaldehyde that occurs during growth under high-copper conditions. It is important to note that the serine cycle cannot operate as a major assimilatory pathway in M. capsulatus unless the two-carbon compound glyoxylate that is depleted during the cycle can be regenerated [20], but no genes have been identified in the genome that potentially encode either of the enzyme systems that can convert acetyl-CoA into glyoxylate: the isocitrate lyase and the glyoxylate-regeneration cycle [23].
Given these considerations, what might the function of the serine cycle (and the interconnected TCA cycle) be in M. capsulatus? We suggest that a possible role for this pathway could be to handle the extra flux of formaldehyde that the organism may encounter under certain growth conditions (Figure 2c). The excess of formate generated in the H4MPT-linked pathway (Figure 2b) could also be redirected into the serine cycle after reduction to methylene-tetrahydrofolate (methylene-H4F; Figure 2d). Acetyl-CoA and other intermediates generated in this way could serve as building blocks for cell biomass.
The role of the CBB cycle in M. capsulatus (Figure 2f) is not clear at present. Given that the fixation of CO2 is a far less efficient mechanism of carbon sequestration than the RuMP or serine cycles, a significant amount of carbon shunted through the CBB cycle would be predicted to decrease growth yield. It is possible, however, that it serves to reduce the local concentration of CO2 and/or to generate intermediates for biomass production. Once again, further experiments are needed to establish the validity of these hypotheses.
A novel MCR-like enzyme and anaerobic methane oxidation
To provide support for the hypothesis that reverse methanogenesis is important in anaerobic methanotrophy, a consortium of researchers focused on identifying the enzyme potentially involved in the initial step of anaerobic methane oxidation; this enzyme is hypothesized to be similar to the bacterial MCR (Figure 1, step 1). A microbial mat in the Black Sea largely consisting of ANME-1-type archaea was chosen as a source of this hypothetical enzyme. As described in Nature in 2003 by Krüger et al. [4], a conspicuous protein consisting of three subunits similar to the α, β, and γ subunits of MCR is abundantly present in this microbial mat (7% of the total extracted protein), suggesting that it has an important role in anaerobic methane oxidation. The protein contains a variant of F430, a cofactor used by the classical MCR, but the two cofactors differ in molecular weight as determined by mass spectrometry. The genes encoding this protein were sequenced as a part of an insert detected in an environmental DNA library [4]. Alignment of amino-acid sequences translated from these genes with the respective sequences of methanogen MCR subunits showed that residues involved in active-site formation in the β and γ subunits were conserved, but one of the important residues in the active site of the α subunit was substituted. It is interesting to speculate that this modification of the active site and the use of a modified F430 cofactor could provide a mechanism for the biochemical activation of methane and could make the first step of reverse methanogenesis thermodynamically and kinetically possible. Further in-depth mechanistic studies of this enzyme will be of great interest.
The environmental genomics of reverse methanogenesis
In a recent paper published in Science, Hallam et al. [3] describe a large environmental sequencing effort which aimed to provide further evidence for the hypothesis of reverse methanogenesis. The group [3] isolated DNA from a 52O-meter-deep sediment of Eel River Basin in California, known for a high abundance of ANME-1 and ANME-II archaea, and used it for both whole-genome shotgun analysis and fosmid 'walking' (fosmids are large-insert plasmids). A total of 111.3 Mb of non-redundant sequence was generated by shotgun sequencing and another 4.6 Mb more were generated by fosmid-end sequencing. Fosmids containing either 16S rRNA genes belonging to ANME-I or ANME-II archaea or homologs of the mcrA gene were analyzed in detail, producing an additional 7.4 Mb of sequence.
The main conclusion from this work [3] is that ANME archaea contain most of the genes involved in methanogenesis, with one exception: mer, the gene encoding methylene-H4MPT reductase (step 3 of reverse methanogenesis; see Figure 1) [15]. On the basis of the apparent lack of mer, the authors propose a model in which parts of the methanogenesis pathway function in two opposite directions: a novel MCR-like enzyme oxidizes methane to methyl-CoM (step 1), and methyl-H4MPT:CoM methyl-transferase catalyzes a reverse reaction to produce methyl-H4MPT (step 2), while the rest of the enzymes reduce CO2 to methylene-H4MPT (steps 4 to 7 in reverse); that is, contrary to previous models [13,14], methane is not oxidized to CO2 by ANME archaea. This proposed scenario creates some metabolic difficulties, however. Firstly, the model aggravates the thermodynamic constraints mentioned earlier, given that reduction of CO2 to formyl-methanofuran (step 7) is an energy-consuming reaction (ΔG° = +16 kJ/mol) [15]. Secondly, the fate of the methylene-H4MPT produced in steps 4 to 7 is proposed to involve either the assimilatory serine cycle or formaldehyde oxidation, but the high energy cost of such schemes would suggest they could operate only as minor pathways, not as major assimilatory or detoxification pathways. Thirdly, there is no discussion by Hallam et al. [3] of how net CO2 would be produced from methane.
Thus, although the schemes presented by Hallam et al. [3] are an attempt to explain how methane metabolism might function in the absence of mer, they highlight the many aspects of this metabolic mode that are still unknown. Two different explanations might be that either mer has simply not been detected because of incomplete sequence data, or that the function of Mer is fulfilled by a novel enzyme (a non-homologous substitution), possibly involving a cofactor different from F420, so the reverse-methanogenesis pathway might in fact be complete (as in Figure 1). An example of such a non-homologous substitution is seen in methylotrophic bacteria, in which a version of the 'reverse methanogenesis' pathway has been found to operate where an NAD(P)-linked methylene-H4MPT dehydrogenase acts in place of unrelated F420-linked or H2-forming enzymes [24].
In conclusion, recent studies involving both organismal and environmental genomics shed new light on the biochemical details of the two processes important for methane balance on Earth - aerobic and anaerobic methane oxidation - and suggest that these processes have more in common than just the substrate, methane, and the final oxidation product, CO2. Both processes involve common cofactors, such as H4MPT, common single-carbon intermediates bound to H4MPT, and common or similar enzymes for core reactions. Although some enzymes involved in reactions that shift single-carbon compounds between different levels of oxidation are evolutionarily related in both processes, the primary methane oxidation enzymes, MMO and the newly identified MCR homolog, must have evolved independently and are fundamentally different.
Acknowledgments
Acknowledgements
L.C. and M.E.L. acknowledge support from the NSF Microbial Observatories program. J.A.V. acknowledges support from the CNRS and the MPG.
References
- Ward N, Larsen O, Sakwa J, Bruseth L, Khouri H, Durkin AS, Dimitrov G, Jiang L, Scanlan D, Kang KH, et al. Genomic insights into methanotrophy: the complete genome sequence of Methylococcus capsulatus (Bath). PLoS Biol. 2004;2:e303. doi: 10.1371/journal.pbio.0020303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao WC, Chen YR, Yi EC, Lee H, Tian Q, Wu KM, Tsai SF, Yu SS, Chen YJ, Aebersold R, Chan SI. Quantitative proteomic analysis of metabolic regulation by copper ions in Methylococcus capsulatus (Bath). J Biol Chem. 2004;279:51554–51560. doi: 10.1074/jbc.M408013200. [DOI] [PubMed] [Google Scholar]
- Hallam SJ, Putnam N, Preston CM, Detter JC, Rokhsar D, Richardson PM, DeLong EF. Reverse methanogenesis: testing the hypothesis with environmental genomics. Science. 2004;305:1457–1462. doi: 10.1126/science.1100025. [DOI] [PubMed] [Google Scholar]
- Krüger M, Meyerdierks A, Glöckner FO, Amann R, Widdel F, Kube M, Reinhardt R, Kahnt J, Böcher R, Thauer RK, Shima S. A conspicuous nickel protein in microbial mats that oxidize methane anaerobically. Nature. 2003;426:878–881. doi: 10.1038/nature02207. [DOI] [PubMed] [Google Scholar]
- Lidstrom ME. Aerobic methylotrophic prokaryotes. In: Stackebrandt E, editor. The Prokaryotes. 3. New York: Springer-Verlag; 2001. [Google Scholar]
- Yu SS, Chen KH, Tseng MY, Wang YS, Tseng CF, Chen YJ, Huang DS, Chan SI. Production of high-quality particulate methane monooxygenase in high yields from Methylococcus capsulatus (Bath) with a hollow-fiber membrane bioreactor. J Bacteriol. 2003;185:5915–5924. doi: 10.1128/JB.185.20.5915-5924.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolyar S, Costello AM, Peeples TL, Lidstrom ME. Role of multiple gene copies in particulate methane monooxygenase activity in the methane-oxidizing bacterium Methylococcus capsulatus Bath. Microbiology. 1999;145:1235–1244. doi: 10.1099/13500872-145-5-1235. [DOI] [PubMed] [Google Scholar]
- Stolyar S, Franke M, Lidstrom ME. Expression of individual copies of Methylococcus capsulatus (Bath) particulate methane monooxygenase genes. J Bacteriol. 2001;183:1810–1812. doi: 10.1128/JB.183.5.1810-1812.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csaki R, Bodrossy L, Klem J, Murrell JC, Kovacs KL. Genes involved in the copper-dependent regulation of soluble methane monooxygenase of Methylococcus capsulatus (Bath): cloning, sequencing and mutational analysis. Microbiology. 2003;149:1785–1795. doi: 10.1099/mic.0.26061-0. [DOI] [PubMed] [Google Scholar]
- Murrell JC, Gilbert B, McDonald IR. Molecular biology and regulation of methane monooxygenase. Arch Microbiol. 2000;173:325–332. doi: 10.1007/s002030000158. [DOI] [PubMed] [Google Scholar]
- Lipscomb JD. Biochemistry of the soluble methane monooxygenase. Annu Rev Microbiol. 1994;48:371–399. doi: 10.1146/annurev.mi.48.100194.002103. [DOI] [PubMed] [Google Scholar]
- Chan SI, Chen KH, Yu SS, Chen CL, Kuo SS. Toward delineating the structure and function of the particulate methane monooxygenase from methanotrophic bacteria. Biochemistry. 2004;43:4421–4430. doi: 10.1021/bi0497603. [DOI] [PubMed] [Google Scholar]
- Valentine DL. Biogeochemistry and microbial ecology of methane oxidation in anoxic environments: a review. Antonie Van Leeuwenhoek. 2002;81:271–282. doi: 10.1023/A:1020587206351. [DOI] [PubMed] [Google Scholar]
- Strous M, Jetten MSM. Anaerobic oxidation of methane and ammonium. Annu Rev Microbiol. 2004;58:99–117. doi: 10.1146/annurev.micro.58.030603.123605. [DOI] [PubMed] [Google Scholar]
- Thauer RK. Biochemistry of methanogenesis: a tribute to Majory Stephenson. Microbiology. 1998;144:2377–2406. doi: 10.1099/00221287-144-9-2377. [DOI] [PubMed] [Google Scholar]
- Deppenmeier U. The membrane-bound electron transport system of Methanosarcina species. J Bioenerg Biomembr. 2004;36:55–64. doi: 10.1023/B:JOBB.0000019598.64642.97. [DOI] [PubMed] [Google Scholar]
- Hallam SJ, Girguis PR, Preston CM, Richardson PM, DeLong EF. Identification of methyl coenzyme M reductase A (mcrA) genes associated with methane-oxidizing archaea. Appl Environ Microbiol. 2003;69:5483–5491. doi: 10.1128/AEM.69.9.5483-5491.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chistoserdova L, Chen SW, Lapidus A, Lidstrom ME. Methylotrophy in Methylobacterium extorquens AM1 from a genomic point of view. J Bacteriol. 2003;185:2980–2987. doi: 10.1128/JB.185.10.2980-2987.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Methylobacillus flagellatus draft genome http://genome.jgi-psf.org/draft_microbes/metfl/metfl.home.html
- Anthony C. Biochemistry of methylotrophs. Academic Press, London; 1982. [Google Scholar]
- Baxter NJ, Hirt RP, Bodrossy L, Kovacs KL, Embley TM, Prosser JI, Murrell JC. The ribulose-1,5-bisphosphate carboxylase/oxygenase gene cluster of Methylococcus capsulatus (Bath). Arch Microbiol. 2002;177:279–289. doi: 10.1007/s00203-001-0387-x. [DOI] [PubMed] [Google Scholar]
- Vorholt JA, Chistoserdova L, Stolyar SM, Thauer RK, Lidstrom ME. Distribution of tetrahydromethanopterin-dependent enzymes in methylotrophic bacteria and phylogeny of methenyl tetrahydromethanopterin cyclohydrolases. J Bacteriol. 1999;181:5750–5757. doi: 10.1128/jb.181.18.5750-5757.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korotkova N, Chistoserdova L, Kuksa V, Lidstrom ME. Glyoxylate regeneration pathway in the methylotroph Methylobacterium extorquens AM1. J Bacteriol. 2002;184:1750–1758. doi: 10.1128/JB.184.6.1750-1758.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagemeier CH, Chistoserdova L, Lidstrom ME, Thauer RK, Vorholt JA. Characterization of a second methylene tetrahydromethanopterin dehydrogenase from Methylobacterium extorquens AM1. Eur J Biochem. 2000;267:3762–3769. doi: 10.1046/j.1432-1327.2000.01413.x. [DOI] [PubMed] [Google Scholar]


