Abstract
Cell division (mitosis) and gamete production (meiosis) are fundamental requirements for normal organismal development. The mammalian cell cycle is tightly regulated by different checkpoints ensuring complete and precise chromosomal segregation and duplication. In recent years, researchers have become increasingly interested in understanding how O-GlcNAc regulates the cell cycle. The O-GlcNAc post-translation modification is an O-glycosidic bond of a single β-N-acetylglucosamine sugar to serine/threonine residues of intracellular proteins. This modification is sensitive toward changes in nutrient levels in the cellular environment making O-GlcNAc a nutrient sensor capable of influencing cell growth and proliferation. Numerous studies have established that O-GlcNAcylation is essential in regulating mitosis and meiosis, while loss of O-GlcNAcylation is lethal in growing cells. Moreover, aberrant O-GlcNAcylation is linked with cancer and chromosomal segregation errors. In this review, we will discuss how O-GlcNAc controls different aspects of the cell cycle with a particular emphasis on mitosis and meiosis.
Introduction
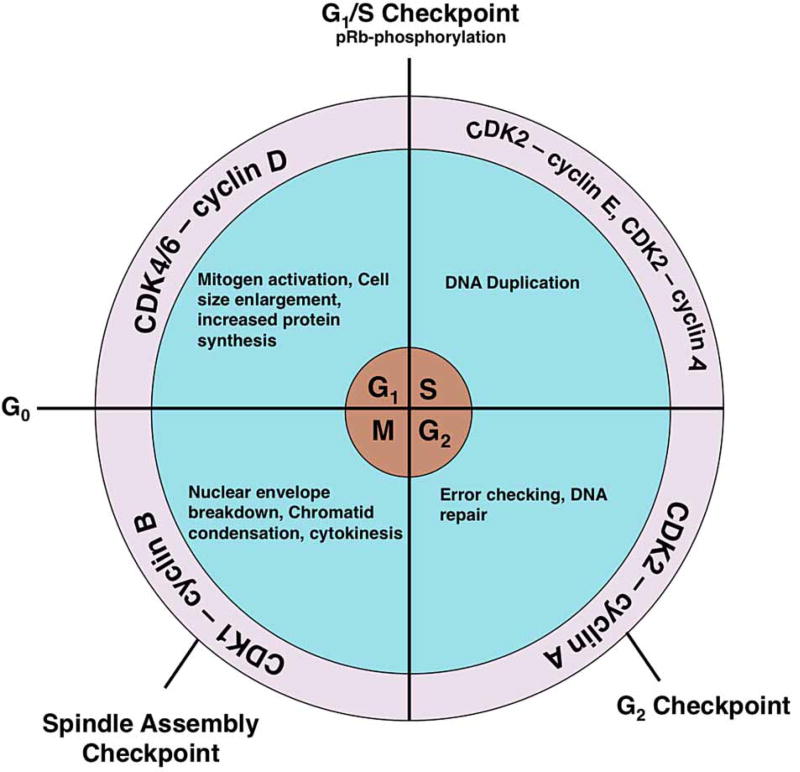
The cell cycle consists of four phases, namely G1 (gap phase 1), S (DNA synthesis), G2 (gap phase 2), and M (mitosis/meiosis), and successful completion of the cell cycle results in the generation of two daughter cells [1]. At the G1 phase, the cell increases in size, begins transcription of cell cycle control genes (cyclins for example), and synthesizes proteins while conducting a series of checks before DNA synthesis. Then, in the S-phase, the entire genome of the cell is replicated. At the G2 phase, the cell prepares for division and checks for size and DNA duplication errors. During M-phase, the nuclear envelope breaks down, chromatin is condensed, and the 4n DNA is segregated into two diploid, daughter cells. Upon completion of cell division, the cells may either renter G1 phase to resume the next round of the cell cycle or remain quiescent in G0 phase (Figure 1) [1]. To ensure that cell division occurs without errors, the cell cycle is regulated in both a temporal and spatial manner. The cell cycle regulatory system consists of checkpoints that pause during phase transitions to assess whether cellular conditions are proper for growth and division [2]. Currently, cell cycle regulation includes protein phosphorylation by cyclin-dependent kinase complexes (CDKCs) and timed expression of cyclins (Figure 1) [3]. However, emerging evidence demonstrates that O-GlcNAc cycling, which is the addition and removal of O-GlcNAc, is an important regulator of the cell cycle.
Figure 1. After mitogen activation, quiescent cells enter the cell cycle from the G0 phase.
During the G1 phase, activation of CDK4/6 by cyclin D causes the cell size to increase, induces transcription of cell cycle genes, and leads to organelle duplication. The phosphorylation of the pRb protein promotes escape from the G1/S checkpoint and into the S-phase. Activation of CDK2 by cyclin E induces DNA synthesis, whereas CDK2 activation by cyclin A leads to the completion of DNA synthesis and promotes entry into the G2 phase. After the cell checks for error-free DNA replication, activation of CDK1 by cyclin B promotes M-phase entry. At the M-phase, the nuclear envelope breaks down, chromatids condense, the spindle forms, and the cell separates at cytokinesis into two daughter cells.
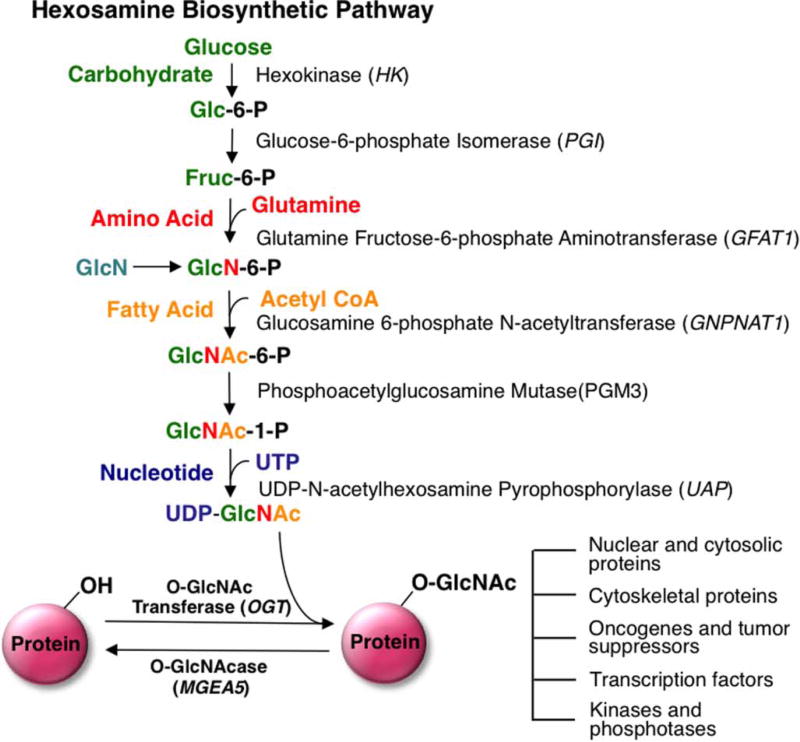
O-GlcNAcylation is a post-translational modification (PTM) involving the attachment of a single β-N-acetylglucosamine to serine/threonine amino acid residues of nuclear, cytoplasmic, and mitochondrial proteins. The modification is dynamically regulated by the opposing functions of two specific enzymes; O-GlcNAc transferase (OGT) adds the modification, whereas O-GlcNAcase (OGA) removes it. Furthermore, the OGT metabolic substrate, UDP-GlcNAc, is synthesized via the hexosamine biosynthetic pathway (HBP) linking several metabolic inputs with O-GlcNAcylation (Figure 2) [4]. Ultimately, these metabolic inputs make O-GlcNAc a nutrient sensor capable of influencing many cellular processes, including transcription, cell growth, and proliferation [5]. Extensive cross-talk exists between O-GlcNAcylation and phosphorylation. The influence of O-GlcNAcylation on phosphorylation is complicated because alterations in O-GlcNAcylation can both increase and decrease phosphorylation [5–8]. For example, elevation of O-GlcNAcylation through OGA inhibition in NIH 3T3 cells increases 148 phosphorylation sites and decreases 280 phosphorylation sites, demonstrating the complex relationship between O-GlcNAcylation and phosphorylation [9]. Several mechanisms regulate the complex interplay between O-GlcNAc and phosphorylation. First, phosphorylation and O-GlcNAcylation are mutually exclusive from each other when they target the same amino acid. Such mutual exclusivity occurs at proteins like RNA polymerase II [10,11] and the c-Myc proto-oncogene product [12]. Alternatively, on other proteins such as calcium/calmodulin-dependent kinase IV (CaMKIV), O-GlcNAcylation is known to deactivate CaMKIV by blocking a proximal activating phosphorylation site [13]. Secondly, O-GlcNAc is known to modify and regulate many protein kinases and phosphatases [5]. Nonetheless, there are many O-GlcNAcylated sites that are not phosphorylation sites [14], suggesting that O-GlcNAc can modulate the cell cycle independently of phosphorylation.
Figure 2. Schematic representation of the HBP and O-GlcNAc modification.
Glutamine:fructose-6-phosphate (GFAT) is the rate-limiting step of the HBP pathway. GlcN treatment can bypass GFAT, causing an elevation in O-GlcNAcylation. The HBP is composed of various metabolic inputs including glucose, amino acid, fatty acid, and nucleotide metabolisms that ultimately serve to synthesize the donor substrate for OGT, UDP-GlcNAc. These metabolic inputs make O-GlcNAc a nutrient sensor capable of influencing many cellular processes including transcription, cell growth, and proliferation. Glc-6-P, glucose-6-phosphate; Fruc-6-P, fructose-6-phosphate; GlcN-6-P, glucosamine-6-phosphate; GlcNAc-6-P, N-acetylglucosamine-6-P; GlcNAc-1-P, N-acetylglucosamine-1-P. Arrow indicates that multiple steps are involved in the conversion.
Indication that O-GlcNAc regulates cell growth comes from initial studies demonstrating that an imbalance in the UDP-GlcNAc pool disrupts cell growth [15–17]. Subsequently, deletion of OGT in mouse embryonic fibroblasts [18,19] and cancer cell lines [20] resulted in increased expression of the cyclin inhibitor p27 Kip1 and cell growth arrest [18,19]. In contrast, OGA knockout mice exhibit developmental delays, perinatal lethality [21,22], and increased genomic instability leading to aneuploidy [22]. Recently, both OGT and OGA were found to be essential genes required for proliferation and survival in human cancer cell lines [23]. Importantly, aberrant O-GlcNAcylation is linked to proliferative diseases such as cancer; however, what is still unclear is the role of O-GlcNAcylation in cancer [4,16,23]. Evidence supports O-GlcNAc cycling as a global regulator of cell growth and proliferation, and delineating O-GlcNAc’s regulatory roles in the cell cycle is essential.
O-GlcNAc cycling in interphase
Interphase comprises G1, S, and early G2 phases of the cell cycle. These phases consist of checkpoints that prepare and ensure that the cell is ready for cell division. Interestingly, like phosphorylation, O-GlcNAcylation is regulated as metazoan cells progress through interphase. First, levels of O-GlcNAc on nuclear and cytoplasmic proteins rise when cells progress into the G1 phase [24,25], but decrease rapidly when cells enter the S-phase [24]. Both OGT and OGA protein levels increase as the cells progress through the S-phase [24]. Generally, changes in OGA protein levels are more pronounced relative to OGT. Moreover, OGA activity is also increased in the S-phase, which is consistent with the reduction in O-GlcNAcylation [24]. Glucosamine (GlcN) supplementation elevates O-GlcNAcylation. GlcN selectively enters the HBP, bypasses the rate-limiting enzyme glutamine:fructose-6-phosphate (GFAT1), and leads to elevations in UDP-GlcNAc concentrations and O-GlcNAcylation (Figure 1). Incubation of mesengial cells with GlcN induced hypertrophy and caused an accumulation at G0/G1 [26]. Levels of two cyclin-dependent kinase inhibitor (CDKI) proteins were altered; p21Waf1/Cip1 was increased, while p27Kip1 was decreased. Such alterations in the CDKI proteins may cause the cells to arrest at G1 [26]. Finally, the G1/S checkpoint protein, retinoblastoma (pRb), is heavily O-GlcNAcylated at the G1 phase [27]; however, phosphorylation of pRb is important in allowing progression through the G1/S-checkpoint into the S-phase. These data suggest the potential for the interplay between phosphorylation and O-GlcNAcylation to regulate pRB checkpoint control [28]. Taken together, these data argue that O-GlcNAc controls G1 progression and entry into the S-phase.
O-GlcNAc regulates DNA synthesis and probably influences cell cycle-dependent gene transcription. First, HeLa cells treated with 6-daizo-norleucine (a semi-selective GFAT1 inhibitor) have an accelerated S-phase and increased rate of DNA synthesis, whereas OGA inhibition causes a slight delay in S-phase progression and the DNA synthesis rate [29]. Interestingly, 3T3-L1 preadipocyte cells, which are a chemically independent but hormonally inducible model for cell cycle progression, also showed delays in S-phase progression after OGA inhibition [29]. In spite of this, the mechanisms in which O-GlcNAc regulates the S-phase are unclear and warrant further investigation.
During DNA synthesis in S-phase, the newly formed DNA strand is loaded onto histone proteins, and histones, including H1, H2A, H2B, H3, and H4, are O-GlcNAcylated [25,30–32]. A histone octamer is composed of two copies of the four core histone proteins, namely H2A, H2B, H3, and H4 [33]. The DNA wraps around these histone octameric complexes and forms nucleosomes [34]. This process is regulated by histone phosphorylation, methylation, and acetylation at different phases of the cell cycle. Interestingly, histone H3 O-GlcNAcylation steadily rises into G2; however, when the cell transitions into M-phase, the H3 O-GlcNAcylation level declines gradually [32]. Histone H1 O-GlcNAcylation increases at the beginning of M-phase only to decline in the late M-phase in Nicotiana tabacum [32]. Similarly in mammalian cells, histone H3 O-GlcNAcylation significantly declines at the G2/M transition [30]. Potentially, O-GlcNAcylation could influence the loading of newly replicated DNA on the histone and histone assembly or disassembly. For example, reconstituted synthetic histone nucleosomes show destabilized H2A–H2B dimers, which mimic an open chromatin state [35]. Furthermore, the histone chaperone complex FACT (Facilitates chromatin transcripts) interacts with synthetic nucleosomes containing a synthetic O-GlcNAc site at H2A-S112 [36], while OGT is known to interact with the histone chaperone HIRA complex promoting nucleosome assembly of H3.3 [37]. These data suggest a role for O-GlcNAcylation in modulating the composition and structure of the nucleosome and hint at an important role for S-phase histone O-GlcNAcylation. However, the O-GlcNAc stoichiometry on histones appears to be low, since some groups have reported difficulty in measuring histone O-GlcNAcylation [38]. Hence, the understanding of O-GlcNAc histone regulation would benefit from better tools to detect and probe the function of the modification.
O-GlcNAc is an essential regulator of mitosis (M-phase)
Mitosis (M-phase) consists of prophase, metaphase, anaphase, and telophase (cytokinesis). The end result of M-phase is the production of two daughter cells that are genetically identical with the parent cell. Alteration in M-phase progression can have serious consequences, leading to cell death or unregulated cell proliferation [39]. Numerous studies demonstrated that O-GlcNAc cycling is an important regulator of M-phase. Disrupting O-GlcNAc cycling via OGT or OGA overexpression severely alters M-phase progression causing a prolonged M-phase [29]. Additionally, OGA knockdown causes defects in M-phase progression and a higher incidence of delayed M-phase exit [40]. Taken together, these data point to critical roles for O-GlcNAcylation in the control of mitotic progression [29].
O-GlcNAc cycling is important in cell cycle progression because it influences cyclin expression and modulates mitotic phosphorylation. Overexpression of OGT or OGA alters both cyclin expression and mitotic phosphorylation by cyclin-dependent kinases (CDKs) [29]. For example, cyclin D expression increases in G1 phase and slowly declines in S- and M-phases. Expression of cyclins A and B peak during prophase but rapidly decline in metaphase. In synchronized HeLA cells, OGA overexpression delays cyclin D expression as cells transition from M- to G1 phase [29]. On the other hand, OGT overexpression lowers cyclin D expression in all stages of the cell cycle, probably caused by cells delayed in M-phase. Furthermore, cyclins A and B fail to decline after metaphase due to mitotic exit defects caused by the overexpression of OGT/OGA. Taken together, these data demonstrate that alterations in O-GlcNAc cycling promote defects in M-phase progression.
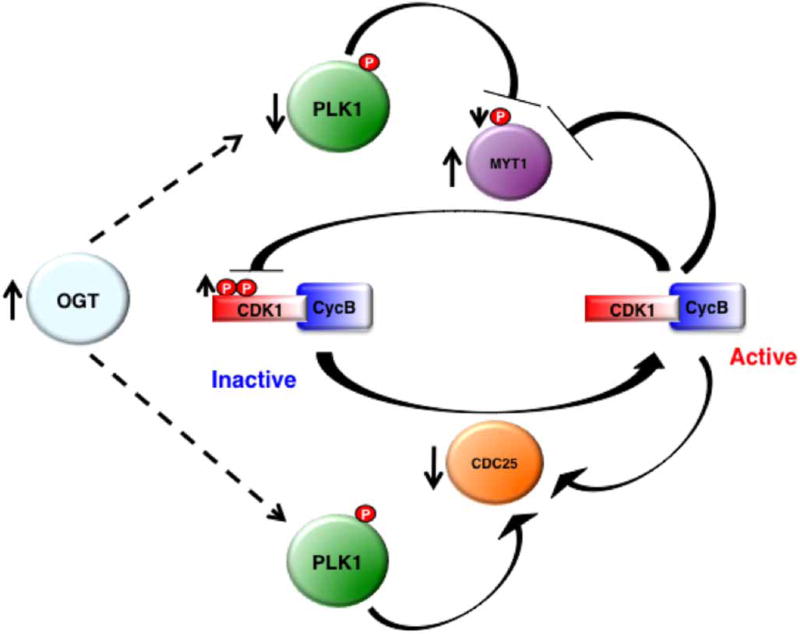
Alterations in O-GlcNAc cycling will disrupt the temporal expression of cyclins, in turn affecting mitotic phosphorylation. HeLa cells overexpressing OGA display a lag of maximal M-phase phosphorylation as detected by MPM-2, a monoclonal antibody that recognizes mitosis-specific phosphorylation. In contrast, OGT overexpressing cells never reach maximal mitotic phosphorylation [29]. The changes in mitotic phosphorylation caused by OGT/OGA overexpression are the result of altered CDK1 activity, the master regulator of the M-phase. OGT overexpression decreases Polo-like kinase 1 (PLK1) expression, a mitotic kinase. Loss of PLK1 activity reduces protein kinase Membrane Associated Tyrosine and Threonine cdc2 inhibitory Kinase (MYT1) phosphorylation and elevates MYT1 expression. Subsequently, MYT1 phosphorylates CDK1 and inhibits CDK1 function. PLK1 also activates Cdc25 (CDK1 phosphatase), promoting CDK1 dephosphorylation. However, OGT overexpression reduces Cdc25 mRNA levels (Figure 3) [14]. The additive effect of disrupted O-GlcNAc cycling is increased CDK1 inhibitory phosphorylations and delayed mitotic progression.
Figure 3. CDK1 signaling is affected by elevations of O-GlcNAcylation.
Overexpression of OGT causes PLK1 expression to decrease. Reduced PLK1 leads to decreased MYT1 phosphorylation and increased protein expression. Subsequently, elevation of MYT1 expression increases CDK1 inhibitory phosphorylation. Furthermore, CDC25 (CDK1 phosphatase) that is activated by PLK1 has a lower mRNA level after OGT overexpression.
Mitotic PTMs of histone tails play a crucial role in regulating chromosome condensation during M-phase [41]. OGT overexpression alters mitotic acetylation and methylation of Histone H3. One potential mechanism for the histone methylation changes is alteration in co-activator-associated arginine methyltransferase 1 (CARM1) activity [25]. OGT overexpression disrupts CARM1 cellular localization during mitosis and increases chromosomal bridge formation [25]. Furthermore, O-GlcNAc regulates histone mitotic phosphorylation. OGT/OGA overexpression and OGA knockdown reduce Ser10 Histone H3 phosphorylation by Aurora kinase B (AURKB), leading to distortions of the spindle architecture and impaired formation [40,42]. The loss of spindle fidelity increases the number of multipolar spindle cells [40,42]. Intriguingly, spindle fidelity can be rescued after inhibiting O-GlcNAc turnover in OGT/OGA overexpressing cells [42]. All these results strongly support O-GlcNAc cycling as a key regulator of mitotic histone PTMs, chromosomal condensation, and spindle function.
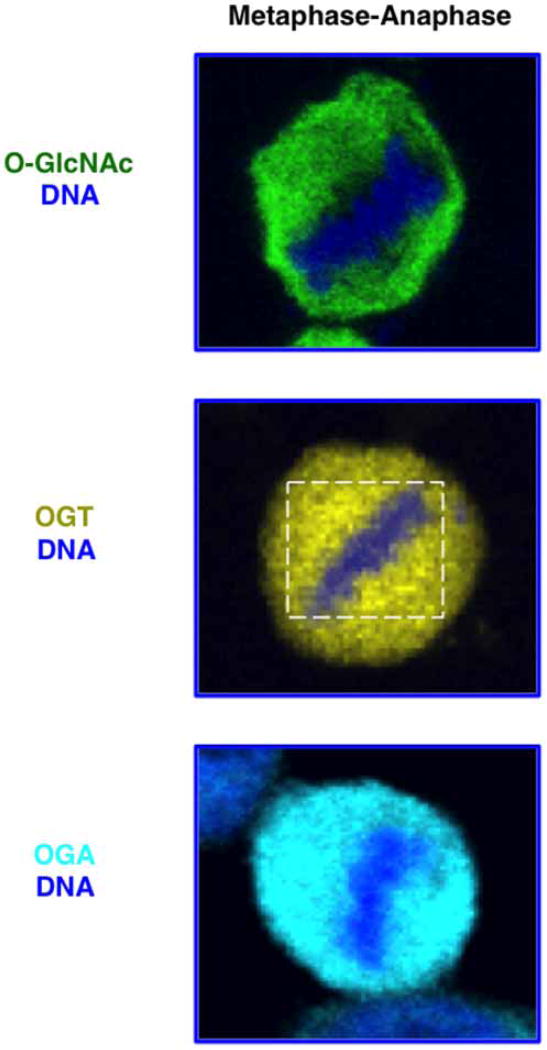
Clearly, OGT function is critical for the proper formation of the mitotic spindle and segregation of chromosomes [29]. OGT localizes with the mitotic spindle as the cell progresses from prophase through meta-anaphase (Figure 4). Then, OGT becomes concentrated at the mid-body as the cleavage furrow forms during cytokinesis [29,43]. On the other hand, OGA is localized throughout the cell in M-phase. Both OGT and OGA interact with spindle regulatory proteins AURKB and protein phosphatase 1 (PP1) [43]. The chromosomal passenger complex (CPC) is composed of AURKB, INCENP, Survivin, and Borealin. CPC kinase activity is crucial in controlling the assembly and disassembly of the spindle apparatus as the cell progresses through mitosis, whereas PP1 antagonizes complex activity [44]. The interaction of OGT/OGA with the CPC alters the post-translational state of CPC substrates. Vimentin, an intermediate filament protein, is a substrate for both AURKB [45,46] and OGT [43] during mitosis. Overexpression of both OGT and OGA alters CPC phosphorylation of vimentin during mitosis. Of note, AURKB inhibition disrupts OGT spindle localization [43]; however, it is unclear if OGT or OGA are substrates for AURKB. Altogether, these results support that OGT is an essential component at the mitotic spindle and the cross-talk between OGT/OGA and the CPC appears to modulate spindle dynamics.
Figure 4. In HeLA cells during metaphase–anaphase, a subset of OGT localizes to the spindle (white box) as determined by confocal microscopy.
OGA and total cellular O-GlcNAc show no specific metaphase–anaphase localization. DNA (blue), O-GlcNAc (green), OGT (yellow), and OGA (cyan).
Finally, O-GlcNAcylation of mitotic proteins could affect spindle architecture and affect M-phase progression. Numerous mitotic proteins are O-GlcNAcylated and several of their O-GlcNAc sites have been identified [14]. For example, the nuclear mitotic apparatus protein 1 (NuMA1), which is required for the maintenance and establishment of the mitotic spindle poles during cell division, is modified by O-GlcNAc [14]. OGT overexpression leads to mislocalization of NuMA1 away from the spindle pole, suggesting that NuMA1 O-GlcNAcylation controls localization [14]. In addition, nuclear pore protein 153 (Nup153), a component of the nuclear pore complex that is important in facilitating trafficking across the membrane and reformation of the nuclear pore envelope after anaphase, and EMSY, a protein involved in maintaining genomic stability during the M-phase, are mitotically O-GlcNAcylated [14]. All these data support that O-GlcNAc regulates mitotic protein functions, but how O-GlcNAc affects the function of these proteins is an active area of investigation.
O-GlcNAc in meiosis
Unlike mitosis that occurs in somatic cells, meiosis only occurs in reproductive cells. The goal of meiosis is to generate haploid gametes, allowing for the formation of diploid offspring upon fertilization [47]. Meiosis involves two rounds of cell division (meiosis I and meiosis II) without an intervening round of DNA replication. In mammals, fully grown oocytes in the ovary are arrested in prophase I, and it is only upon hormonal stimulation at the time of ovulation that they will resume meiosis, progress through meiosis I, and then arrest at metaphase of meiosis II [47]. Then, it is only following fertilization that the second meiotic arrest will be released and zygotes will enter early embryonic mitotic divisions [47]. Meiosis I includes homologous chromosome pairing, synapsis, and recombination and separation of homologous chromosomes. Meiosis II, on the other hand, involves separation of sister chromatids. As such, meiosis I is a unique cell division, whereas meiosis II is much more similar to mitosis. Emerging data in both Xenopus and mouse oocytes indicate that O-GlcNAcylation plays an important role during meiosis.
A key initial event in meiotic resumption is the breakdown of the oocyte nucleus (germinal vesicle) — also known as germinal vesicle breakdown (GVBD). In Xenopus oocytes, upon progesterone induction, the activation of maturation-promoting factor triggers GVBD, chromosome condensation, spindle formation, and progression to metaphase II [48]. O-GlcNAc regulates the onset of GVBD and meiotic progression. When O-GlcNAc cycling is inhibited in Xenopus oocytes via microinjection of β-galactosyltransferase, which caps terminal O-GlcNAc residues, GVBD is blocked and results in apoptosis [49]. Furthermore, O-GlcNAcylation of meiotic proteins is dynamic as the cell progresses through meiosis [50,51]. Pharmacological manipulation of O-GlcNAc levels alters progesterone-mediated maturation in Xenopus oocytes [50]. Additionally, microinjection of OGT into immature oocytes accelerates meiotic progression upon progesterone stimulation, whereas microinjection of anti-OGT antibodies impedes GVBD [52].
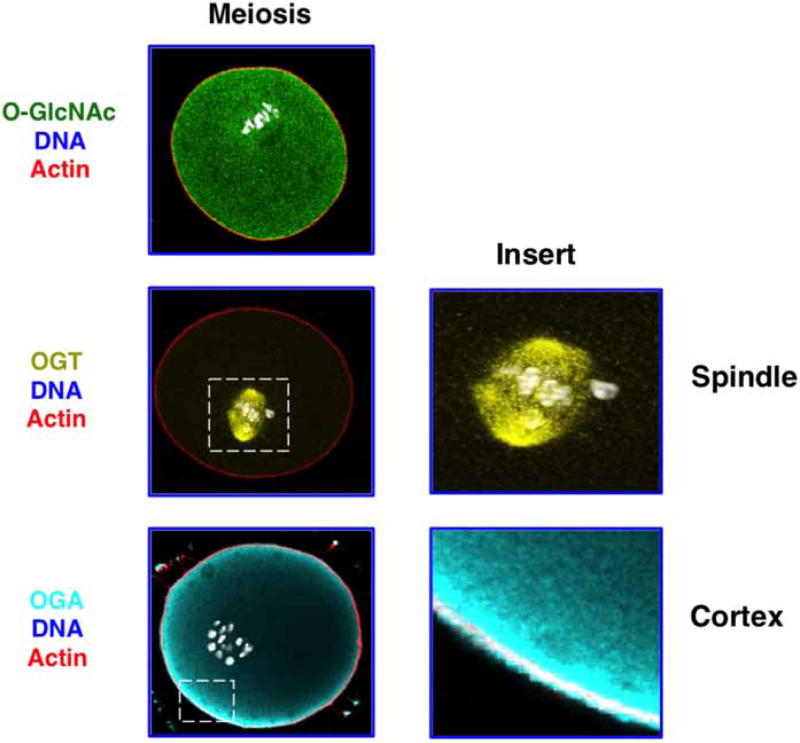
Correspondingly less is known about the role of O-GlcNAcylation during mammalian meiosis. However, OGT, OGA, and O-GlcNAcylated substrates are expressed in the mouse oocyte and have distinct localizations throughout meiosis (see Figure 5) [53]. OGT concentrates at the meiotic spindle at meiosis I and meiosis II. Although this is a similar distribution to somatic cells, the meiotic spindle enrichment is much more prominent. In contrast with mitosis, OGA is enriched at the oocyte cortex throughout meiosis. The distinct spatial distributions of OGT at the spindle and OGA away from the spindle suggest that the meiotic spindle may be an important target of O-GlcNAcylation. In support of this, O-GlcNAcylated proteins are found in the region of the spindle (Figure 5). Defining meiotic O-GlcNAc targets and determining how perturbation of O-GlcNAc cycling affect mammalian meiotic progression and aneuploidy are active areas of research. Owing to the fundamental mechanistic differences between meiosis and mitosis, O-GlcNAc regulation of mitosis and meiosis could be different, although these potential mechanistic differences have yet to be explored.
Figure 5. O-GlcNAc machinery localizes to discrete locations in mouse oocytes at the metaphase of meiosis I.
OGT localizes to the meiotic spindle (see enlarged insert), whereas a subset of OGA localizes to the cell cortex (see enlarged insert). Images were taken using confocal microscopy. O-GlcNAc (green), OGT (yellow), OGA (cyan), and actin (red).
Conclusion
O-GlcNAc cycling is an important regulator of the cell cycle. Disrupting O-GlcNAc cycling affects cell cycle progression and cell division. O-GlcNAc, OGT, and OGA localize and are expressed differentially in the cell as the cell progresses through the different stages of the cell cycle. In addition to regulating mitosis, O-GlcNAc cycling is also essential in controlling meiosis. Still, clear roles for O-GlcNAc control of the cell cycle await determination. For instance, how OGT is targeted to proteins during mitosis and meiosis is uncertain, and how O-GlcNAc affects the function of modified proteins is an active area of investigation. Since all the interventions currently used to manipulate O-GlcNAc levels also change the expression of OGT and OGA [54], making a clear conclusion from a given experiment is challenging. However, continuous effort to address these issues may result in the development of new approaches and fresh insights into how O-GlcNAc affects proliferation.
Acknowledgments
The authors are grateful to the Biochemical Society Transactions for the invitation to submit this review. Furthermore, we acknowledge our funding sources: National Institute of Diabetes and Digestive and Kidney Diseases R01DK100595 to C.S., Biomedical Research Training Program (BRTP) Fellowship, and Mabel A. Woodyard Fellowship to E.P.T. F.E.D. acknowledge the Centers of Biomedical Research Excellence [P20 GM104936] and the University of Kansas School of Medicine Start Up funds.
Abbreviations
- AURKB
Aurora kinase B
- CaMKIV
calcium/calmodulin-dependent kinase IV
- CARM1
co-activator-associated arginine methyltransferase 1
- CDK
cyclin-dependent kinase
- CDKCs
cyclin-dependent kinase complexes
- CDKI
cyclin-dependent kinase inhibitor
- CPC
chromosomal passenger complex
- GlcN
glucosamine
- GVBD
germinal vesicle breakdown
- HBP
hexosamine biosynthetic pathway
- NuMA1
nuclear mitotic apparatus protein 1
- OGA
O-GlcNAcase
- OGT
O-GlcNAc transferase
- PLK1
polo-like kinase 1
- PP1
protein phosphatase 1
- pRb
retinoblastoma
- PTM
post-translational modification
Footnotes
Competing Interests
The Authors declare that there are no competing interests associated with the manuscript.
References
- 1.Nurse P, Masui Y, Hartwell L. Understanding the cell cycle. Nat. Med. 1998;4:1103–1106. doi: 10.1038/2594. [DOI] [PubMed] [Google Scholar]
- 2.Satyanarayana A, Kaldis P. Mammalian cell-cycle regulation: several Cdks, numerous cyclins and diverse compensatory mechanisms. Oncogene. 2009;28:2925–2939. doi: 10.1038/onc.2009.170. [DOI] [PubMed] [Google Scholar]
- 3.Murray AW. Recycling the cell cycle: cyclins revisited. Cell. 2004;116:221–234. doi: 10.1016/s0092-8674(03)01080-8. [DOI] [PubMed] [Google Scholar]
- 4.Slawson C, Copeland RJ, Hart GW. O-GlcNAc signaling: a metabolic link between diabetes and cancer? Trends Biochem. Sci. 2010;35:547–555. doi: 10.1016/j.tibs.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hart GW, Slawson C, Ramirez-Correa G, Lagerlof O. Cross talk between O-GlcNAcylation and phosphorylation: roles in signaling, transcription, and chronic disease. Annu. Rev. Biochem. 2011;80:825–858. doi: 10.1146/annurev-biochem-060608-102511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Griffith LS, Schmitz B. O-linked N-acetylglucosamine levels in cerebellar neurons respond reciprocally to pertubations of phosphorylation. Eur. J. Biochem. 1999;262:824–831. doi: 10.1046/j.1432-1327.1999.00439.x. [DOI] [PubMed] [Google Scholar]
- 7.Haltiwanger RS, Grove K, Philipsberg GA. Modulation of O-linked N-acetylglucosamine levels on nuclear and cytoplasmic proteins in vivo using the peptide O-GlcNAc-β-N-acetylglucosaminidase inhibitor O-(2-acetamido-2-deoxy-d-glucopyranosylidene)amino-N-phenylcarbamate. J. Biol. Chem. 1998;273:3611–3617. doi: 10.1074/jbc.273.6.3611. [DOI] [PubMed] [Google Scholar]
- 8.Lefebvre T, Alonso C, Mahboub S, Dupire M-J, Zanetta J-P, Caillet-Boudin M-L, et al. Effect of okadaic acid on O-linked N-acetylglucosamine levels in a neuroblastoma cell line. Biochim. Biophys. Acta. 1999;1472:71–81. doi: 10.1016/s0304-4165(99)00105-1. [DOI] [PubMed] [Google Scholar]
- 9.Wang Z, Gucek M, Hart GW. Cross-talk between GlcNAcylation and phosphorylation: site-specific phosphorylation dynamics in response to globally elevated O-GlcNAc. Proc. Natl Acad. Sci. U.S.A. 2008;105:13793–13798. doi: 10.1073/pnas.0806216105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Comer FI, Hart GW. Reciprocity between O-GlcNAc and O-phosphate on the carboxyl terminal domain of RNA polymerase II. Biochemistry. 2001;40:7845–7852. doi: 10.1021/bi0027480. [DOI] [PubMed] [Google Scholar]
- 11.Kelly WG, Dahmus ME, Hart GW. RNA polymerase II is a glycoprotein. Modification of the COOH-terminal domain by O-GlcNAc. J. Biol. Chem. 1993;268:10416–10424. [PubMed] [Google Scholar]
- 12.Chou T-Y, Hart GW, Dang CV. c-Myc is glycosylated at threonine 58, a known phosphorylation site and a mutational hot spot in lymphomas. J. Biol. Chem. 1995;270:18961–18965. doi: 10.1074/jbc.270.32.18961. [DOI] [PubMed] [Google Scholar]
- 13.Dias WB, Cheung WD, Wang Z, Hart GW. Regulation of calcium/calmodulin-dependent kinase IV by O-GlcNAc modification. J. Biol. Chem. 2009;284:21327–21337. doi: 10.1074/jbc.M109.007310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang Z, Udeshi ND, Slawson C, Compton PD, Sakabe K, Cheung WD, et al. Extensive crosstalk between O-GlcNAcylation and phosphorylation regulates cytokinesis. Sci. Signal. 2010;3:ra2. doi: 10.1126/scisignal.2000526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boehmelt G, Wakeham A, Elia A, Sasaki T, Plyte S, Potter J, et al. Decreased UDP-GlcNAc levels abrogate proliferation control in EMeg32-deficient cells. EMBO. J. 2000;19:5092–5104. doi: 10.1093/emboj/19.19.5092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krug E, Zweibaum A, Schulz-Holstege C, Keppler D. d-glucosamine-induced changes in nucleotide metabolism and growth of colon-carcinoma cells in culture. Biochem. J. 1984;217:701–708. doi: 10.1042/bj2170701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wice BM, Trugnan G, Pinto M, Rousset M, Chevalier G, Dussaulx E, et al. The intracellular accumulation of UDP-N-acetylhexosamines is concomitant with the inability of human colon cancer cells to differentiate. J. Biol. Chem. 1985;260:139–146. [PubMed] [Google Scholar]
- 18.O’Donnell N, Zachara NE, Hart GW, Marth JD. Ogt-dependent X-chromosome-linked protein glycosylation is a requisite modification in somatic cell function and embryo viability. Mol. Cell Biol. 2004;24:1680–1690. doi: 10.1128/MCB.24.4.1680-1690.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shafi R, Iyer SPN, Ellies LG, O’Donnell N, Marek KW, Chui D, et al. The O-GlcNAc transferase gene resides on the X chromosome and is essential for embryonic stem cell viability and mouse ontogeny. Proc. Natl Acad. Sci. U.S.A. 2000;97:5735–5739. doi: 10.1073/pnas.100471497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kazemi Z, Chang H, Haserodt S, McKen C, Zachara NE. O-linked beta-N-acetylglucosamine (O-GlcNAc) regulates stress-induced heat shock protein expression in a GSK-3β-dependent manner. J. Biol. Chem. 2010;285:39096–39107. doi: 10.1074/jbc.M110.131102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Keembiyehetty C, Love DC, Harwood KR, Gavrilova O, Comly ME, Hanover JA. Conditional knock-out reveals a requirement for O-linked N-Acetylglucosaminase (O-GlcNAcase) in metabolic homeostasis. J. Biol. Chem. 2015;290:7097–7113. doi: 10.1074/jbc.M114.617779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang YR, Song M, Lee H, Jeon Y, Choi E-J, Jang H-J, et al. O-GlcNAcase is essential for embryonic development and maintenance of genomic stability. Aging Cell. 2012;11:439–448. doi: 10.1111/j.1474-9726.2012.00801.x. [DOI] [PubMed] [Google Scholar]
- 23.Wang T, Birsoy K, Hughes NW, Krupczak KM, Post Y, Wei JJ, et al. Identification and characterization of essential genes in the human genome. Science. 2015;350:1096–1101. doi: 10.1126/science.aac7041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Drougat L, Olivier-Van Stichelen S, Mortuaire M, Foulquier F, Lacoste A-S, Michalski J-C, et al. Characterization of O-GlcNAc cycling and proteomic identification of differentially O-GlcNAcylated proteins during G1/S transition. Biochim. Biophys. Acta. 2012;1820:1839–1848. doi: 10.1016/j.bbagen.2012.08.024. [DOI] [PubMed] [Google Scholar]
- 25.Sakabe K, Hart GW. O-GlcNAc transferase regulates mitotic chromatin dynamics. J. Biol. Chem. 2010;285:34460–34468. doi: 10.1074/jbc.M110.158170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Masson E, Wiernsperger N, Lagarde M, El Bawab S. Glucosamine induces cell-cycle arrest and hypertrophy of mesangial cells: implication of gangliosides. Biochem. J. 2005;388:537–544. doi: 10.1042/BJ20041506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wells L, Slawson C, Hart GW. The E2F-1 associated retinoblastoma-susceptibility gene product is modified by O-GlcNAc. Amino Acids. 2011;40:877–883. doi: 10.1007/s00726-010-0709-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bartek J, Bartkova J, Lukas J. The retinoblastoma protein pathway in cell cycle control and cancer. Exp. Cell Res. 1997;237:1–6. doi: 10.1006/excr.1997.3776. [DOI] [PubMed] [Google Scholar]
- 29.Slawson C, Zachara NE, Vosseller K, Cheung WD, Lane MD, Hart GW. Perturbations in O-linked β-N-acetylglucosamine protein modification cause severe defects in mitotic progression and cytokinesis. J. Biol. Chem. 2005;280:32944–32956. doi: 10.1074/jbc.M503396200. [DOI] [PubMed] [Google Scholar]
- 30.Fong JJ, Nguyen BL, Bridger R, Medrano EE, Wells L, Pan S, et al. β-N-acetylglucosamine (O-GlcNAc) is a novel regulator of mitosis-specific phosphorylations on histone H3. J. Biol. Chem. 2012;287:12195–12203. doi: 10.1074/jbc.M111.315804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang S, Roche K, Nasheuer H-P, Lowndes NF. Modification of histones by sugar β-N-acetylglucosamine (GlcNAc) occurs on multiple residues, including histone H3 serine 10, and is cell cycle-regulated. J. Biol. Chem. 2011;286:37483–37495. doi: 10.1074/jbc.M111.284885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Delporte A, De Zaeytijd J, De Storme N, Azmi A, Geelen D, Smagghe G, et al. Cell cycle-dependent O-GlcNAc modification of tobacco histones and their interaction with the tobacco lectin. Plant Physiol. Biochem. 2014;83:151–158. doi: 10.1016/j.plaphy.2014.07.021. [DOI] [PubMed] [Google Scholar]
- 33.Luger K, Dechassa ML, Tremethick DJ. New insights into nucleosome and chromatin structure: an ordered state or a disordered affair? Nat. Rev. Mol. Cell Biol. 2012;13:436–447. doi: 10.1038/nrm3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Eickbush TH, Moudrianakis EN. The compaction of DNA helices into either continuous supercoils or folded-fiber rods and toroids. Cell. 1978;13:295–306. doi: 10.1016/0092-8674(78)90198-8. [DOI] [PubMed] [Google Scholar]
- 35.Lercher L, Raj R, Patel NA, Price J, Mohammed S, Robinson CV, et al. Generation of a synthetic GlcNAcylated nucleosome reveals regulation of stability by H2A-Thr101 GlcNAcylation. Nat. Commun. 2015;6:7978. doi: 10.1038/ncomms8978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Raj R, Lercher L, Mohammed S, Davis BG. Synthetic nucleosomes reveal that GlcNAcylation modulates direct interaction with the FACT complex. Angew. Chem. Int. Ed. Engl. 2016;55:8918–8922. doi: 10.1002/anie.201603106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee J-S Lee, Zhang Z. O-linked N-acetylglucosamine transferase (OGT) interacts with the histone chaperone HIRA complex and regulates nucleosome assembly and cellular senescence. Proc. Natl Acad. Sci. U.S.A. 2016;113:E3213–E3220. doi: 10.1073/pnas.1600509113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gagnon J, Daou S, Zamorano N, Iannantuono NVG, Hammond-Martel I, Mashtalir N, et al. Undetectable histone O-GlcNAcylation in mammalian cells. Epigenetics. 2015;10:677–691. doi: 10.1080/15592294.2015.1060387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hayashi MT, Karlseder J. DNA damage associated with mitosis and cytokinesis failure. Oncogene. 2013;32:4593–4601. doi: 10.1038/onc.2012.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lanza C, Tan EP, Zhang Z, Machacek M, Brinker AE, Azuma M, et al. Reduced O-GlcNAcase expression promotes mitotic errors and spindle defects. Cell Cycle. 2016;15:1363–1375. doi: 10.1080/15384101.2016.1167297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Georgatos SD, Markaki Y, Christogianni A, Politou AS. Chromatin remodeling during mitosis: a structure-based code? Front. Biosci. 2009;14:2017–2027. doi: 10.2741/3360. [DOI] [PubMed] [Google Scholar]
- 42.Tan EP, Caro S, Potnis A, Lanza C, Slawson C. O-linked N-acetylglucosamine cycling regulates mitotic spindle organization. J. Biol. Chem. 2013;288:27085–27099. doi: 10.1074/jbc.M113.470187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Slawson C, Lakshmanan T, Knapp S, Hart GW. A mitotic GlcNAcylation/phosphorylation signaling complex alters the posttranslational state of the cytoskeletal protein vimentin. Mol. Biol. Cell. 2008;19:4130–4140. doi: 10.1091/mbc.E07-11-1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Carmena M, Wheelock M, Funabiki H, Earnshaw WC. The chromosomal passenger complex (CPC): from easy rider to the godfather of mitosis. Nat. Rev. Mol. Cell Biol. 2012;13:789–803. doi: 10.1038/nrm3474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Goto H, Kosako H, Tanabe K, Yanagida M, Sakurai M, Amano M, et al. Phosphorylation of vimentin by Rho-associated kinase at a unique amino-terminal site that is specifically phosphorylated during cytokinesis. J. Biol. Chem. 1998;273:11728–11736. doi: 10.1074/jbc.273.19.11728. [DOI] [PubMed] [Google Scholar]
- 46.Goto H, Yasui Y, Kawajiri A, Nigg EA, Terada Y, Tatsuka M, et al. Aurora-B regulates the cleavage furrow-specific vimentin phosphorylation in the cytokinetic process. J. Biol. Chem. 2003;278:8526–8530. doi: 10.1074/jbc.M210892200. [DOI] [PubMed] [Google Scholar]
- 47.Hörmanseder E, Tischer T, Mayer TU. Modulation of cell cycle control during oocyte-to-embryo transitions. EMBO J. 2013;32:2191–2203. doi: 10.1038/emboj.2013.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Masui Y, Clarke HJ. Oocyte maturation. Int. Rev. Cytol. 1979;57:185–282. doi: 10.1016/s0074-7696(08)61464-3. [DOI] [PubMed] [Google Scholar]
- 49.Fang B, Miller MW. Use of galactosyltransferase to assess the biological function of O-linked N-acetyl-d-glucosamine: a potential role for O-GlcNAc during cell division. Exp. Cell Res. 2001;263:243–253. doi: 10.1006/excr.2000.5110. [DOI] [PubMed] [Google Scholar]
- 50.Slawson C, Shafii S, Amburgey J, Potter R. Characterization of the O-GlcNAc protein modification in Xenopus laevis oocyte during oogenesis and progesterone-stimulated maturation. Biochim. Biophys. Acta. 2002;1573:121–129. doi: 10.1016/s0304-4165(02)00369-0. [DOI] [PubMed] [Google Scholar]
- 51.Lefebvre T, Baert F, Bodart J-F, Flament S, Michalski J-C, Vilain J-P. Modulation of O-GlcNAc glycosylation during Xenopus oocyte maturation. J. Cell. Biochem. 2004;93:999–1010. doi: 10.1002/jcb.20242. [DOI] [PubMed] [Google Scholar]
- 52.Dehennaut V, Hanoulle X, Bodart J-F, Vilain J-P, Michalski J-C, Landrieu I, et al. Microinjection of recombinant O-GlcNAc transferase potentiates Xenopus oocytes M-phase entry. Biochem. Biophys. Res. Commun. 2008;369:539–546. doi: 10.1016/j.bbrc.2008.02.063. [DOI] [PubMed] [Google Scholar]
- 53.Slawson C, Duncan FE. Sweet action: the dynamics of O-GlcNAcylation during meiosis in mouse oocytes. Mol. Reprod. Dev. 2015;82:915. doi: 10.1002/mrd.22577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang Z, Tan EP, VandenHull NJ, Peterson KR, Slawson C. O-GlcNAcase expression is sensitive to changes in O-GlcNAc homeostasis. Front. Endocrinol. 2014;5:206. doi: 10.3389/fendo.2014.00206. [DOI] [PMC free article] [PubMed] [Google Scholar]