Abstract
Rapeseed (Brassica napus L.) is one of the most important oil crops almost all over the world. Seed-related traits, including oil content (OC), silique length (SL), seeds per silique (SS), and seed weight (SW), are primary targets for oil yield improvement. To dissect the genetic basis of these traits, 192 recombinant inbred lines (RILs) were derived from two parents with distinct oil content and silique length. High-density linkage map with a total length of 1610.4 cM were constructed using 1,329 double-digestion restriction site associated DNA (ddRAD) markers, 107 insertion/deletions (INDELs), and 90 well-distributed simple sequence repeats (SSRs) markers. A total of 37 consensus quantitative trait loci (QTLs) were detected for the four traits, with individual QTL explained 3.1–12.8% of the phenotypic variations. Interestingly, one OC consensus QTL (cqOCA10b) on chromosome A10 was consistently detected in all three environments, and explained 9.8% to 12.8% of the OC variation. The locus was further delimited into an approximately 614 kb genomic region, in which the flanking markers could be further evaluated for marker-assisted selection in rapeseed OC improvement and the candidate genes targeted for map-based cloning and genetic manipulation.
Keywords: Brassica napus, restriction site associated DNA, quantitative trait locus, yield-related trait, oil content
Introduction
Rapeseed (Brassica napus L.) is one of the most important oil crops almost all over the world. It is not only a leading source for vegetable oil (Snowdon 2007), but also a major source for industrial materials such as biofuel and lubricants. In China, rapeseed provides about 40% of the vegetable oil supply (Sun et al. 2012). However, due to the great demands for human food and industrial materials in recent years, the production of rapeseed hardly meets the global consumption. Development of cultivars with high yield is one of the most important ways to solve the shortage of rapeseed oil supply. Yield-related traits such as thousand-seed weight (SW), seeds per silique (SS) and silique length (SL) are important determinants of yield and thus target traits in rapeseed breeding and quantitative trait locus (QTL) mapping. Many yield-related QTLs have been identified (Chen et al. 2007, 2011, Shi et al. 2009, Yang et al. 2012, Zhang et al. 2012). Of these, only one major QTL for SS (Li et al. 2015b, Zhang et al. 2011) and one major QTL for SL and SW were mapped and their underlying genes were identified recently (Li et al. 2014, Liu et al. 2015). Further understanding of the genetic basis of these traits in rapeseed remains essentially needed.
Restriction fragment length polymorphisms (RFLPs) and simple sequence repeats (SSRs) are the major types of markers for linkage map construction in previous QTL analysis. Due to limited RFLP and SSR markers, the density of molecular markers and the mapping resolutions for those oil content (OC) and yield-related QTLs in B. napus were relatively low (Chen et al. 2007, 2011, Shi et al. 2009, Yang et al. 2012, Zhang et al. 2012), which restrained further utilization in map-based cloning and marker-assisted selection. Those markers with short sequences are difficult to precisely align to the reference genome sequence of B. napus (Bus et al. 2012). Single nucleotide polymorphisms (SNPs) are currently taking the place of RFLP and SSR markers and becoming predominant in linkage mapping in many crop species (Rowe et al. 2011), which are more conducive to substantial improvement of marker density, statistical power, and QTL mapping precision (Jiang and Zeng 1995). In B. napus, next generation sequencing (NGS) technologies have been used to discover mass SNPs, in the form of genotyping by sequencing (GBS) and/or double-digest restriction-site associated DNA (ddRAD) (Bus et al. 2012, Liu et al. 2013, 2014, Trick et al. 2009). ddRAD-based high-density genetic linkage map SNPs can be used for dissecting the genetic basis of agronomically important traits in oilseed rape.
It is estimated that an increase of 1.0% seed OC is equivalent to an increase of 2.3–2.5% of seed yield in rapeseed (Wang 2004), suggesting seed OC improvement may be more effective to increase oil yield. In the past two decades, 3 to 63 QTLs controlling OC in B. napus have been identified in different studies (Chen et al. 2010, Delourme et al. 2006, Ecke et al. 1995, Javed et al. 2016, Wang et al. 2013). Javed et al. (2016) detected a common QTL on chromosome A10 in all four environments. Largely due to relatively long genetic distance, low marker density, or other genomic factors, none of these QTLs for OC has been delimited in a genomic region small enough to facilitate positional cloning.
The objectives of this study were (1) to develop a high-density genetic linkage map using ddRAD and other sequence-based markers; (2) to identify QTLs controlling OC, SL, SS and SW; (3) to delimit the genomic regions for stably expressed QTLs; and (4) to compare QTLs identified in this study with previous ones.
Materials and Methods
Plant materials
Two rapeseed inbred lines, ‘M201’ with high OC and long siliques, and ‘352’ with low OC and short siliques, were selected to develop a population of recombinant inbred lines (RILs) for detection of QTL controlling seed OC and yield-related traits including SL, SS, and SW. An F1 hybrid from the cross of the two inbred lines was self-pollinated to obtain F2 plants that were self-pollinated and advanced to the F6 generation using the single seed descent (SSD) method. A total of 192 recombinant inbred lines (RILs) were obtained as of 2010. Genomic DNA was extracted from 250 mg of young leaves collected from a single plant of each RIL using the cetyltrimethylammonium bromide (CTAB) method.
Experiment design and trait measurement
Using a randomized complete block design, the ‘352’ × ‘M201’ RIL population and their parents were planted in 2011 (Y11) at Hezhen, Gansu Province, China and in 2012 and 2013 (Y12 and Y13) at the experimental farm in Wuhan, Hubei Province, China, respectively. Each RIL or parent was grown in two row plots, with 10–12 plants in each row. Y11 had one replication and seeds were sown in mid-May and harvested at the end of September of the year. Y12 and Y13 each had three replications and seeds were sown in early October and harvested in early May of 2012 and 2013, respectively. Field management followed standard agricultural practices. The distance between the two rows was 30 cm, and the distance between any two neighboring plants was approximately 20 cm. Five mature plants from the center of each plot were selected for trait survey. SL of each plant was based on the average length of 10 well-developed siliques (not including the beak) from the middle of the main inflorescence. SS of each plant was defined as the average number of seeds from 10 well-developed siliques from the middle of the main inflorescence. SW of each plant was defined as the average weight of 1000 well-filled open-pollinated seeds. Threshed seeds were desiccated to minimize the moisture content. OC was measured by nuclear magnetic resonance (NMR) using a previously described method (Burns et al. 2003). The OC in each plot was based on an average of five plants.
Preparation and sequencing of ddRAD libraries
ddRAD libraries of the 192 RILs were prepared for paired-end sequencing as previously described (Chen et al. 2013), with the exceptions that only 50 ng of genomic DNA was used and inserts with size range of 270–320 bp were recovered. Briefly, 50 ng of genomic DNA was double-digested by MseI and SacI and indexed adaptors were ligated. After digestion and adapter ligation, ligates of 50 RILs were pooled together and separated on a 2% agarose gel. Fragments in the size range of 350–400 bp were recovered from the gel, which corresponded to 270–320 bp of unligated fragments. Approximately 50 ng of recovered DNA fragments was used as the template for PCR amplification in a 50 μl reaction with the two sequencing primers (Chen et al. 2013). The PCR products were separated on a 2% agarose gel and DNA fragments in the size range of 400–450 bp were recovered and sequenced on the HiSeq2000 platform (Illumina, San Diego, CA, USA) with paired-end (PE) reads of 90 bp.
SNP genotyping of the RIL population
The 90 bp PE reads were subjected for quality filtering. Sequence reads from the Illumina runs did not match one of the expected barcodes and sequence reads of poor overall quality (5% base quality score under Q30) were removed from the analysis. The clean data was then parsed into different individuals with the outermost barcodes and remnant restriction sites at both ends exactly matching the adaptors used. The 5 bp barcodes at the 5′ end and the 5 bp error-rich nucleotides at the 3′ end were removed from the PE reads. After trimming, the remaining 80 nucleotides of each PE read were kept for further analysis. A total of 192 RILs were initially used for sequencing and marker discovery. The parents, ‘M201’ and ‘352’, were sequenced together with a much higher coverage (approximately 6-fold) than that of the RILs in this study. The average depth of sequence of the parents were 10× and RILs were 4.5×.
SNP discovery and genotyping were performed exactly following the RFAPtools pipeline (Chen et al. 2013). First, a pseudo-reference sequence (PRS) was assembled using the reads from both ‘M201’ and ‘352’ and all RILs. Then, the 80 bp PE reads of each parent and RIL were aligned to the PRS with the SOAP software (Li et al. 2009). Three mismatches to the pseudo-reference sequence were allowed on each end of the PE reads. SNPs were identified for the two parents and all RILs. The threshold of maximum genotype missing rate was set to 25%. SNPs with missing genotypes in either parent were also excluded from further analysis. The sequence data of the RIL population were deposited in NCBI with the Sequence Read Archive accession number PRJNA345453.
SSR and INDEL marker screening and genotyping
Polymorphism between the parents was also screened using previously published 212 SSRs (Cheng et al. 2009, Xu et al. 2010, Yang et al. 2012), and 595 in-house INDELs developed from resequencing data of 22 rapeseed cultivars (Mahmood et al. 2016). The 595 INDELs were evenly distributed throughout the A and C subgenomes and uniquely mapped on the reference genomes of B. napus. PCR for SSR and INDEL detection was performed as previously described (Cheng et al. 2009), and the products were separated on 6% denatured polyacrylamide gels and stained with AgNO3 solution. Polymorphic SSRs evenly distributed on all the 19 linkage groups were selected as anchored markers to compare LGs and QTLs.
Construction of the genetic linkage map
JoinMap 4.0 (Van Ooijen 2006) was used for genetic linkage map construction. SSRs and INDELs were used as anchor markers to assign LGs to specific chromosomes. The maximum recombinant frequency was set to 0.4, and minimum logarithm of odds (LOD) scores of 3.0, and the threshold for goodness of fit was set to ≤5.0, which is a normalized difference of goodness-of-fit chi-squared used to decide whether or not a locus should remain in the linkage group during the process of building the linkage group. CentiMorgan distances were calculated by the Kosambi function for map distance (Kosambi 1943). Pearson’s chi-squared test was performed to examine the goodness of fit to the expected 1:1 segregation ratio (P < 0.05) for each marker.
Statistical analysis, QTL mapping, meta-analysis and interaction detection
Statistical analyses for all traits were carried out using SAS 9.2 (SAS Institute, Cary, NC, USA). Pearson’s phenotypic correlation coefficients were calculated among all traits across the three environments using the CORR procedure. The genetic correlation coefficient was calculated using the general linear model (GLM, Yijkl = μ + ri + bij + gk + eijkl, where Yijkl is an observation of genotype k in replication l of block j in environment i, μ is the general mean, ri is the effect of the environment i, bij is the effect of block j in environment i, gk is the effect of genotype k, and eijkl is the residual effect of observation, the residual variance is a combination of genotype × location interaction variance and the within location error variance.). The genetic correlation was calculated as: , where covGxy, , and were the genetic covariance and variances of a pair-wise traits, respectively. The significance of each genetic correlation was determined using a t-test of correlation coefficients (Kong 2005). The broad-sense heritability was calculated as: , where is genotypic variance, is interaction variance of the genotype with the environment, is error variance, n is the number of environments, and r is the number of blocks in each environment. The lines with missing phenotype data were ignored in each trait.
QTLs were detected by composite interval mapping using WinQTL cartographer 2.5 (Zeng 1994). The number of control markers, window size and walk speed were set to 5, 10 and 1 cM, respectively. The LOD threshold for each trait was determined by permutation test with 1000 repetitions t. A QTL was declared when the LOD score was greater than the threshold value, LOD scores corresponding to P < 0.05 were used to identify significant QTLs. To avoid missing QTLs with small genetic effects, loci with LOD scores larger than 2.0 but smaller than the threshold in multiple environments were treated as micro-real QTLs (Long et al. 2007). These micro-real QTLs might become major QTLs in different environments or in different segregating populations (Long et al. 2007). The nomenclature of QTLs followed previous descriptions (Udall et al. 2006) with minor modifications. A QTL was named starting with the lowercase letter q; followed by an uppercase two-letter designation for the trait name (OC, SL, SS, or SW); an uppercase chromosome set letter (A or C); a chromosome number; a dot; a 1, 2, or 3 (representing 2011representing 2012, or 2013, respectively, when the QTL was detected; and a lowercase letter (a, b, c, ...) for one of the multiple QTLs detected in the same linkage group and environment (Yang et al. 2012).
To merge and compare QTLs detected in different environments and located in the same chromosome region, either for the same or different trait, meta-analysis was conducted using BioMercator 3.0 (Sosnowski et al. 2012). If QTLs for the same trait were detected in multiple environments with overlapping confidence intervals, these QTLs were firstly merged as a consensus QTL and designated with initial letters “cq” followed by trait name and linkage group (Yang et al. 2012). QTLs for different traits having overlapping confidence intervals were further integrated into unique QTL and designated with initial letters “uq” followed by the linkage group. The algorithm of BioMercator software can help to determine the position of the overlapping QTLs based on the variance of these QTLs position and the confidence interval values (Arcade et al. 2004).
Epistatic interactions were analyzed based on mixed linear model approaches using QTLNetwork 2.0 (Yang et al. 2008). The testing windows, filtration windows and walk speed were set to 10, 10 and 1 cM, respectively. Both 1D and 2D searches were analyzed with 1000 permutations. P < 0.05 was set as the significance threshold.
Comparison of chromosomes between the RIL linkage maps and the B. napus genome sequences
The linkage map constructed in this study was aligned to the B. napus genome (version 4.1) (www.genoscope.cns.fr/brassicanapus) through BLAST of the mapped ddRAD sequences and INDEL and SSR amplicon sequences. The E-value was set to ≤1E-5 for all the markers. A Perl script was used to extract the position of best hit in the B. napus genome sequence. The orthologous gene(s) in B. rapa (http://brassicadb.org/brad/, version 1.5) and A. thaliana within the genomic regions delimited by the markers flanking a QTL interval were considered putatively to be associated with the QTL.
Results
Phenotypic variations and genetic correlations among traits
The two inbred lines, ‘M201’ and ‘352’, and their RILs were grown in three environments to compare OC, SL, SS, and SW. The OC of ‘M201’ was 42.7 ± 2.92%, 49.6 ± 2.77% and 44.9 ± 2.51% (Mean ± SD) in Y11, Y12, and Y13, respectively. The OC of ‘M201’ was significantly higher than that of ‘352’ in the three environments (P < 0.01), which were 29.8 ± 3.31%, 35.3 ± 2.60% and 30.7 ± 2.09% (Mean ± SD), respectively (Table 1). The SL of ‘M201’ was 5.8 ± 0.47 cm, 6.1 ± 0.41 cm, and 5.9 ± 0.39 cm in Y11, Y12, and Y13 (Mean ± SD), respectively. The SL of ‘352’ was 4.3 ± 0.49 cm, 4.7 ± 0.33 cm, and 4.5 ± 0.31 cm (Mean ± SD) in Y11, Y12, and Y13, respectively (Table 1), which were significantly shorter than those of ‘M201’ (P < 0.01). SS and SW did not show significant differences between the two parents in the three environments (Table 1).
Table 1.
Statistical analysis of oil content (OC), sileque length (SL), seeds per silique (SS), and seed weight (SS) for the parental lines and the recombinant inbreed lines (RILs)
| Years | Locations | Traits | Parents | RIL population | ||||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| M201(Mean ± SD) | 352 (Mean ± SD) | (P value) | Range | Mean ± SD | CV(%) | |||
| 2011 (Y11) | Gansu | OC (%) | 42.7 ± 2.92 | 29.8 ± 3.31 | <0.01 | 24.6–47.6 | 35.3 ± 4.22 | 12.3 |
| SL (cm) | 5.8 ± 0.47 | 4.3 ± 0.49 | <0.01 | 3.0–9.0 | 4.8 ± 0.68 | 15.9 | ||
| SS | 19 ± 3.13 | 15 ± 3.22 | >0.05 | 8–17 | 17 ± 3.27 | 19.3 | ||
| SW (g) | 2.98 ± 0.33 | 2.61 ± 0.29 | >0.05 | 1.36–4.22 | 2.70 ± 0.47 | 17.5 | ||
| 2012 (Y12) | Wuhan | OC (%) | 49.6 ± 2.77 | 35.3 ± 2.60 | <0.01 | 29.5–49.3 | 42.3 ± 3.02 | 7.0 |
| SL (cm) | 6.1 ± 0.41 | 4.7 ± 0.33 | <0.01 | 3.6–6.7 | 5.1 ± 0.57 | 11.4 | ||
| SS | 20 ± 2.18 | 17 ± 2.06 | >0.05 | 10–25 | 18 ± 2.90 | 16.2 | ||
| SW (g) | 3.47 ± 0.49 | 3.29 ± 0.28 | >0.05 | 2.38–4.58 | 3.36 ± 0.39 | 11.6 | ||
| 2013 (Y13) | Wuhan | OC (%) | 44.9 ± 2.51 | 30.7 ± 2.09 | <0.001 | 23.7–46.7 | 37.6 ± 3.95 | 10.4 |
| SL (cm) | 5.9 ± 0.39 | 4.5 ± 0.31 | <0.001 | 3.7–6.3 | 4.9 ± 0.54 | 11.2 | ||
| SS | 16 ± 2.95 | 14 ± 2.63 | >0.05 | 7–25 | 15 ± 3.15 | 21.2 | ||
| SW (g) | 3.33 ± 0.24 | 3.02 ± 0.48 | >0.05 | 2.26–4.61 | 3.30 ± 0.41 | 12.5 | ||
Y11–rapeseed growth environment in 2011 at Hezhen, Gansu Province, China; Y12 and Y13–rapeseed growth environments in 2012 and 2013 at the experimental farm in Wuhan, Hubei Province, China; SD–standard deviation; CV–coefficient of variation.
The t test significant level was 0.05.
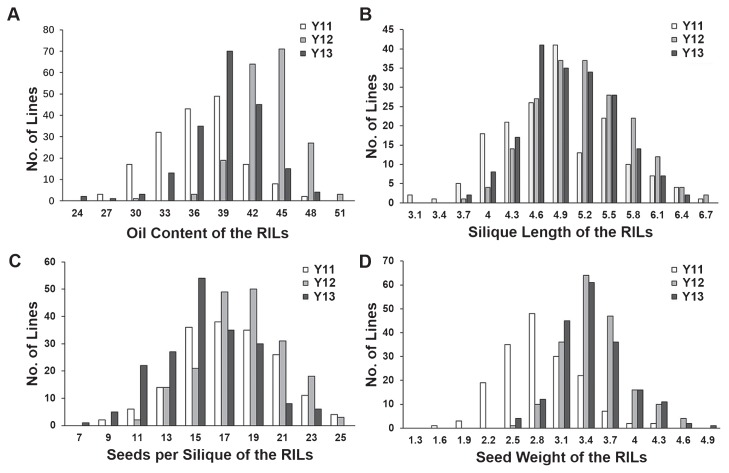
The distributions of the four traits in the RILs displayed a continuous distribution with transgressive segregation (Fig. 1). Normality test indicated that the segregation of the four traits fit a normal distribution model. Two-way ANOVA of individual traits across environments indicated that the genotypes of RILs (G), growing environment (E) and genotype-environment interactions (G × E) had significant effects on the four traits (Supplemental Table 1), suggesting that the variations of individual traits were caused by genotype differences among the RILs. In addition, the environments also had a significant effect on the performance of the four traits. The broad-sense heritability of OC, SL, SS and SW was 78.8%, 89.4%, 78.5% and 81.9%, respectively, suggesting that these traits are stable in different environments (Supplemental Table 1) and SL was the least affected by environmental variations.
Fig. 1.
Phenotypic variation of the four traits in the recombinant inbreed line (RIL) populations in a rapeseed growth environment in 2011 (Y11) at Hezhen, Gansu Province, China and two rapeseed growth environments in 2012 and 2013 (Y12 and Y13) at the experimental farm in Wuhan, Hubei Province, China. (A), (B), (C) and (D) show the distribution of oil content (%), silique length (cm), seeds per silique, and seed weight (gram), respectively.
Phenotypic correlation analyses were performed among the four traits. SS showed a negative correlation with SW in Y12, and Y13, but no correlation in Y11. SL showed a significant positive correlation with OC, SS and SW in all three environments (Supplemental Table 2). SW showed a significant positive correlation with OC in Y11 and Y12. The genetic correlations among the four traits were also evaluated (Supplemental Table 3). OC was positively correlated with SL, SS and SW, respectively. SS showed a weak negative correlation with SW, while SL showed a significant positive correlation with SS, suggesting that longer siliques could produce larger seeds.
ddRAD tag sequencing and SNP discovery in the RIL populations
A total of 68,450,311 high-quality PE90 reads were obtained for the 192 RILs after quality filtering. The number of reads varied from 0.04 and 1.07 million, with an average of 0.36 million reads per RIL (Supplemental Table 4). Four RILs with less than 0.10 million reads were excluded from further analysis, and the remaining 188 RILs were used for SNP discovery and genotyping. The reads from all RILs were collapsed and used to generate a total of 15 million unique ddRAD tags. A pseudo-reference sequence was assembled using RFAPtools with short reads obtained from sequencing reads of the parents and the whole RIL population as described (Chen et al. 2013). All sequence reads from the two parents (352,533 tags from ‘M201’ and 318,938 tags from ‘352’) and individual RILs were aligned to the pseudo-reference using SOAPsnp with a maximum of three mismatches (Li et al. 2008). A total of 1,812 polymorphic ddRAD tags were identified. A subset of 1,700 polymorphic ddRAD markers containing 3,765 SNPs was identified having a maximum missing genotype rate lower than 25%. Of these SNPs, 29.2% and 29.6% were C/T and A/G transitions respectively, and 11.6%, 11.7%, 9.0%, and 8.9% were A/C, G/T, A/T and C/G transversions respectively.
Construction of genetic linkage map
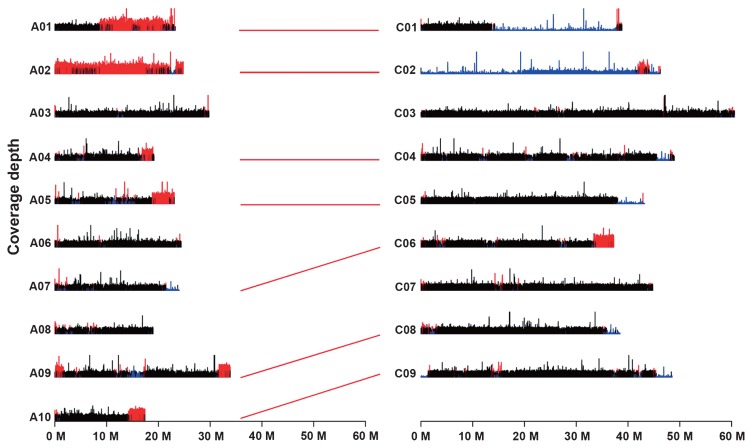
Of the 595 INDELs and 211 SSRs, 126 and 104 showed polymorphisms between ‘M201’ and ‘352’, respectively, and were genotyped in the RILs. A genetic linkage map containing 1,329 ddRAD markers, 107 INDELs and 90 SSRs was constructed. The linkage map covered a total length of 1610.4 cM and consisted of 19 linkage groups (LGs), with an average distance of 1.06 cM between adjacent loci. The length of LGs ranged from 30.8 (C01) to 181.4 cM (C03), and the average interval between markers ranged from 0.5 (A05) to 6.7 cM (A02). C03 had the maximum number of markers (201). A02 and C02 did not have any SNPs; A02 had 8 INDELs and SSRs and C02 had 11 INDELs and SSRs. We found that five out of the eight markers on A02 had a heterozygous rate ranging from 26% to 66%, which is much higher than the expected heterozygous rate (3.1%) at the F6 generation. Alignment of ‘352’ short reads to the B. napus reference genome indicated that the coverage depth on chromosome A02 was significantly higher (more than 3 fold, P < 0.05) than that on chromosome C02 and other chromosomes (Fig. 2), suggesting that homeologous nonreciprocal transposition (HNRT) (Zhao et al. 2006) or homeologous exchange (HE) (Chaloub et al. 2015) from A02 to C02 occurred in the genome of ‘352’. HNRTs or HEs were also identified between A01 and C01, A04 and C04, A05 and C05, A07 and C06, A09 and C08, and A10 and C09 (Fig. 2). In all of these transpositions, only one chromosomal segment was transposed from the C subgenome (C06) to the A subgenome (A07), the others were transposed from the A subgenome to the C subgenome (Fig. 2). HNRTs resulted in the presence of a duplication of a chromosomal region and loss of the corresponding homeologous region, which is a common phenomenon in B. napus (Chaloub et al. 2015, Zhao et al. 2006). These HNRTs increased the level of heterozygosity in the RILs (Zhao et al. 2006), and thus affected our discovery and genotyping of SNPs.
Fig. 2.
Homeologous nonreciprocal transpositions (HNRTs) or homeologous exchanges (HEs) between B. napus chromosomes A2 and C2. Coverage depth was obtained after mapping Illumina sequence reads to the reference genome of B. napus. Segmental HEs are revealed based on sequence read coverage analysis, where a duplication (red) is revealed by geater coverage for a given segment than the rest of the genome (black) and a deletion (blue) by little or no coverage for the corresponding homeologous segment. The X axis indicated the chromosome length, sizes of chromosomes are indicated in Mb. The Y axis showed the coverage depth of sequence reads mapped to the reference. The red lines indicated HNRT or HE occurred between the two chromosomes.
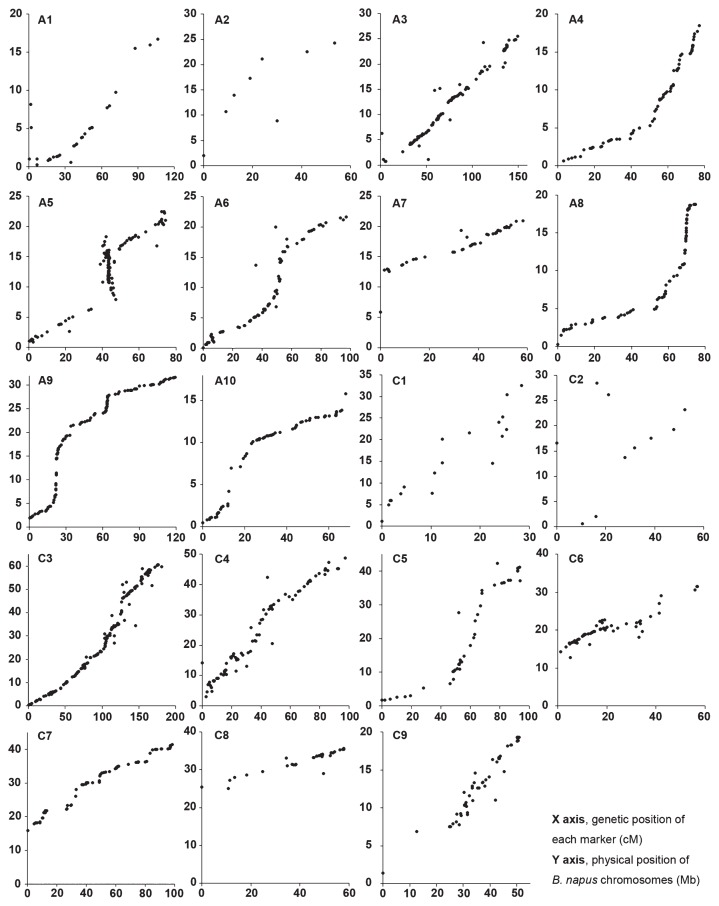
The sequences of all loci on the genetic linkage map were aligned to the B. napus reference genome sequence (Fig. 3). If a locus was mapped to multiple positions in the B. napus genome, only the location with the best hit was selected for colinearity analysis. Alignments indicated that all LGs had a good colinearity with the B. napus reference genome ‘Darmour-bzh’ with several minor inconsistencies (Fig. 3). The inconsistencies such as regions on chromosome A05, C01, C04 and C09 (Fig. 3) might be caused by chromosomal rearrangement in ‘352’, or caused by mis-assembly in the reference genome sequence.
Fig. 3.
Alignments between the recombinant inbreed line (RIL) linkage maps and the B. napus reference genome sequences. The X axis indicated the genetic position of each marker (cM), and the Y axis indicated the physical position of reference sequence of each corresponding B. napus chromosomes (Mb).
Analysis of the goodness of fit indicated that 411 (26.9%) markers showed a distorted segregation (P < 0.05). The distorted segregation markers were unevenly distributed across the 19 chromosomes. The majority of the distorted markers (63.75%) biased towards the parent ‘352’. Markers on A03, A04, A10, C01, C04 and C09 skewed to ‘M201’, while markers on A02, A05, A06, A08, C02, C03, C05, C06 and C07 skewed to ‘352’.
QTLs detected for the four traits
QTL analysis was performed using the phenotypic data collected in two winter environments and one spring environment. A total of 18, 11, 13 and 12 QTLs were detected for oil content, silique length, seeds per silique and seed weight, respectively (Supplemental Fig. 1, Supplemental Table 5). The 33 QTLs with overlapping confidence intervals were integrated into 15 consensus QTLs (Table 2), all of these identified 37 QTLs with 24 located in the A subgenome and 13 in the C subgenome. Most of the QTLs for these four traits individually explained a small fraction of the corresponding phenotypic variation. In addition, most of the 37 QTLs were only in the spring environment (Gansu, Y11) or in one or two winter environments (Wuhan, Y12 and Y13) and explained less 10% of the phenotypic variation (Table 2), suggesting that these four traits are controlled by multiple QTLs with minor effects and most of the QTLs are specific to environment. A total of nine SL QTLs were detected: two of them detected both in the spring and semi-winter environments, three only in the spring environment, and four only in the semi-winter environments. The number of the SL QTLs detected in the spring environment was close to that detected in the semi-winter environment. In contrast, much fewer OC, SS, and SW QTLs were detected in the spring environment than in the semi-winter environments, respectively. The differences in the numbers of QTLs detected between the two environments among the four traits might partly reflect the different trait heritability and allele contribution from the parents. For example, 11 of the 12 OC QTLs were identified only in one or two semi-winter environments, and the QTL alleles came from the semi-winter parent “M201”, suggesting that some of the 11 OC QTLs might play a role in regulating the adaptability of “M201” to the semi-winter growing conditions. It is worth noting that one consensus QTL for oil content (cqOCA10b) was repeatedly detected in the spring and the two winter environments and explaining 9.8–12.8% of the phenotypic variation.
Table 2.
Consensus quantitative trait loci (QTLs) for oil content (OC), sileque length (SL), seeds per silique (SS), and seed weight (SS) obtained by meta-analysis in all three environments in the recombinant inbreed lines (RILs)
| Trait | Consensus QTL | LG | Position | CI | LOD | R2 (%) | Add | Env | QTL in Ref |
|---|---|---|---|---|---|---|---|---|---|
| OC | cqOCA1b | A01 | 21.4 | 19.4–23.3 | 2.5–3.8 | 4.2–6.6 | 0.78–0.82 | Y12/Y13 | oilA1-1 Jiang et al. (2014) |
| cqOCA10a | A10 | 47.3 | 46.4–48.2 | 4.1–6.8 | 7.6–11.9 | 0.84–1.37 | Y12/Y13 | ||
| cqOCA10b | A10 | 57.7 | 56.9–58.4 | 5.3–7.6 | 9.8–12.8 | 1.08–1.41 | Y11/Y12/Y13 | ||
| cqOCC4 | C04 | 71.5 | 69.8–73.2 | 2.4–2.6 | 3.7–4.1 | 0.58–0.81 | Y12/Y13 | ||
| cqOCC8b | C08 | 37.8 | 35.9–39.7 | 2.2–4.5 | 3.5–7.4 | 0.74–0.83 | Y12/Y13 | ||
| SL | cqSLA1 | A01 | 25.7 | 22.5–29.0 | 2.2–4.1 | 4.2–7.4 | 0.14–0.16 | Y11/Y12 | sl1 Chen et al. (2007) |
| cqSLA6b | A06 | 41.5 | 40.9–42.1 | 4.2–4.8 | 7.0–9.2 | 0.15–0.21 | Y11/Y12 | ||
| SS | cqSSA1b | A01 | 44.1 | 43.6–44.7 | 2.9–3.6 | 5.5–6.3 | 0.68–0.80 | Y12/Y13 | |
| cqSSA6a | A06 | 5.8 | 5.4–6.5 | 3.6–3.9 | 6.7–7.6 | −0.86––0.81 | Y11/Y12 | ||
| cqSSC1 | C01 | 25.8 | 25.4–26.2 | 2.8–3.1 | 5.4–6.4 | 0.71–0.87 | Y11/Y12/Y13 | ||
| cqSSC2 | C02 | 38.4 | 36.8–40.0 | 3.6–3.8 | 6.6–6.7 | 0.81–0.87 | Y12/Y13 | cqSLC2 Yang et al. (2012) | |
| SW | cqSWA3c | A03 | 113.5 | 112.3–114.9 | 2.2–4.4 | 4.6–7.8 | −0.11––0.10 | Y11/Y12 | |
| cqSWA6a | A06 | 24.6 | 22.6–26.6 | 2.4–2.8 | 3.1–4.1 | 0.09 | Y12/Y13 | ||
| cqSWA6b | A06 | 31.6 | 29.6–33.6 | 2.3–2.4 | 3.8–4.2 | 0.08–0.09 | Y12/Y13 | ||
| cqSWA7b | A07 | 56.2 | 55.8–56.7 | 4.0–4.1 | 6.7–7.0 | 0.11 | Y12/Y13 | qSWA7.1 Yang et al. (2012) |
All QTLs were prefixed with “cq”, followed by the abbreviated trait name, mapped linkage group, and a small alphabet (a, b, ...) representing multiple QTLs if they are detected for the trait in the same linkage group and same environment. For example, cqOCA1a is one of the two QTLs for oil content (OC) detected on linkage A1, and cqOCA1b is another one.
CI: the flanking marker closest to the 95% confidence interval; R2: percentage of the phenotypic variation explained by the QTL.
Additive effects indicate the effects of “M201” allele.
Y11–rapeseed growth environment in 2011 at Hezhen, Gansu Province, China; Y12 and Y13–rapeseed growth environments in 2012 and 2013 at the experimental farm in Wuhan, Hubei Province, China.
QTL in Ref were the QTL detected in previous studies. The references for the QTL were also shown in the same column.
QTL of phenotypically correlated yield-related traits may co-localize (Tuberosa et al. 2002). We examined the confidence intervals of consensus QTL for different traits by meta–analysis. overlapped consensus QTLs were integrated into seven unique QTL (Table 3). These results indicated that these QTLs have pleiotropic effects or tightly linked.
Table 3.
Unique quantitative trait loci (QTLs) for oil content (OC), sileque length (SL), seeds per silique (SS), and seed weight (SS) obtained by meta-analysis in all three environments in the recombinant inbreed lines (RILs)
| Unique QTL | Consensus QTL | LG | Position | CI |
|---|---|---|---|---|
| uqA1 |
cqOCA1b cqSLA1 |
A1 | 22.5 | 20.8–24.1 |
| uqA3 |
cqSSA3 cqSWA3a |
A3 | 24.1 | 19.9–28.3 |
| uqA6a |
cqSLA6a cqSSA6b |
A6 | 35.4 | 34.6–36.2 |
| uqA6b |
cqSLA6b cqSSA6c |
A6 | 41.6 | 41.0–42.1 |
| uqA7 |
cqOCA7 cqSWA7a |
A7 | 50.2 | 49.4–51.0 |
| cqC1 |
cqOCC1b cqSSC1 |
C1 | 25.8 | 25.4–26.2 |
| uqC2 |
cqOCC2 cqSLC2 cqSSC2 |
C2 | 38.5 | 37.1–39.8 |
LG–linkage groups; CI–confidence interval for the unique QTLs.
Epistatic interactions between loci
Epistatic interactions were detected for OC, SL, SS and SW. Ten pairs of interactions involving 19 loci were detected for all traits (Table 4). These loci distributed on 12 linkage groups were identified to have effects on the four traits. Two loci on A06 (M505-M58 and M1572-M1423) were found to co-localize for cqSWA6a and cqSLA6b, respectively. These interactions f in total explained less than 1.5% of the phenotypic variation of each trait, suggesting that the effects of digenic interactions are very small and these four traits are primarily controlled by additive effect.
Table 4.
Epistatic interactions for the four traits
| Traita | Chromosome | Marker interval-i | Chromosome | Marker interval-j | Additive by additive effect | R2 (%) |
|---|---|---|---|---|---|---|
| OC | A03 | M301–M551 | A04 | M836–M394 | −4.54 | 0.77 |
| A08 | M1325–BRGMS2025 | C04 | M1153–M169 | −1.42 | 0.26 | |
| SL | A02 | ID9–ID7 | A06 | M1572–M1423 | 0.16 | 0.03 |
| A03 | M301–M551 | C07 | M463–M130 | −0.48 | 0.09 | |
| A05 | M528–M150 | C04 | M1179–M939 | −0.14 | 0.03 | |
| SS | A02 | ID108–BNGMS635 | C03 | M6–M618 | −1.73 | 0.25 |
| C03 | M660–M1619 | C03 | M1190–M118 | −1.84 | 0.31 | |
| SW | A07 | M316–M106 | C01 | M285–M1006 | −0.04 | 0.02 |
| A06 | M505–M58 | C04 | M1484–M682 | −0.11 | 0.02 | |
| A09 | M545–M1590 | C03 | M318–ID71 | 0.12 | 0.02 |
OC–oil content; SL–silique length; SS–seeds per silique; and SW–seed weight.
Discussion
OC, SL, SS, and SW are among the mostly studied traits in rapeseed (Chen et al. 2007, Javed et al. 2016, Qi et al. 2014, Wang et al. 2013, Yang et al. 2012). We assigned QTL regions onto the B. napus genome (Version 4.1) (Chalhoub et al. 2014) through BLAST analysis of markers linked to the QTLs, and compared our QTLs with those previously reported ones. Of the 12 OC QTLs in this study, cqOCA1b, cqOCA7 and cqOCC2 were co-localized with oilA1-1, oilA7-1 and oilC2-3 reported by Jiang et al. (2014), respectively; and cqOCC8a was co-localized with qOC-C8-2 in the KN population (Wang et al. 2013). cqOCA1a, cqOCA10a and cqOCA10b in A subgenome and cqOCC1a, cqOCC1b, cqOCC3, cqOCC4 and cqOCC8b in C subgenome apparently were new. Of our 9 SL QTLs, cqSLA1 was co-localized with sl1 (Chen et al. 2007). cqSLC2 and cqSLC3b were co-localized with cqSLC2 and cqSLC3b reported by Yang et al. (2012), respectively. The remaining six, cqSLA4a, cqSLA4b, cqSLA4c, cqSLA6a cqSLA6b, and cqSLC3a, appeared to be new. Only one pleiotropic SS QTL, cqSSC2, was co-localized with cqSLC2 in the study of Yang et al. (2012), and the other 7 QTLs were likely new. For SW, cqSWA3b, cqSWA7a and cqSWA7b in this study were co-localized with qSL.N3-3 (Zhang et al. 2011), TSWA7a-06 (Fan et al. 2010) and qSWA7.1 (Yang et al. 2012), respectively. The other 5 SW QTLs, including cqSWA3a, cqSWA3c, cqSWA6a and cqSWA6b, and cqSWC6 apparently were new. In this study three SS QTLs (cqSSA6a, cqSSA6b and cqSSA6c) were detected on A06 and one QTL (cqSSC1) on C01 whereas there was no SS QTL detected on A06 and C01 in the previous studies (Chen et al. 2011, Qi et al. 2014, Shi et al. 2009, Zhang et al. 2011). In this study, we identified 26 potential new QTLs and 11 QTLs co-localized with previous studies. And the two stably expressed QTLs are great value for breeding cultivars with wide flexibility in different environments. These co-localized and potentially new QTLs acquired in this study facilitate further understanding of these traits and utilization in genetic breeding in the furture.
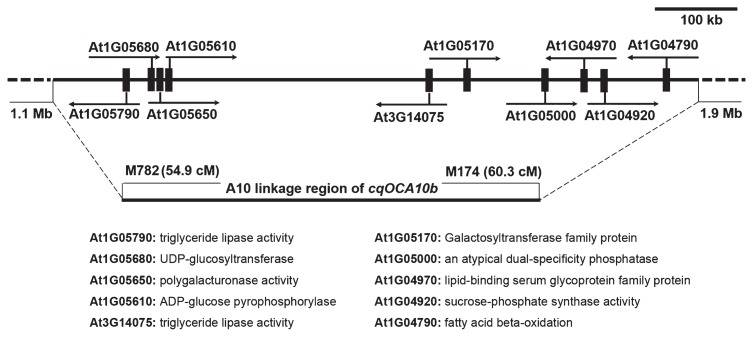
Large and stable effects of QTL are considered as the key factors for fine mapping and map-based cloning success. OC is most important agronomic trait for rapeseed, it is controlled by complex mechanisms and highly influenced by the environment. So far, only a gene increased oil and oleic-acid contents was cloned in maize (Zheng et al. 2008). In oilseed rape, many QTLs for OC have been detected. However, there has no genes controlling seed OC isolated, this is because, most of the detected OC QTLs explain less than 10% of the phenotypic variation (Jiang et al. 2014, Sun et al. 2012, Wang et al. 2013). In this study, a major QTL (cqOCA10b) was repeatedly detected in one spring and two winter environments, which may enable us to conduct the fine mapping of candidate genes for OC. The six markers (M174, ID52, M1601, M53, M730 and M782) linked to cqOCA10b will be useful for MAS (marker assisted selection) breeding. The interval of cqOCA10b between markers M782 and M174 (representing an approximately 6 cM region of chromosome A10) represented approximately 614 kb of the B. napus genome, a total of 124 genes (Supplemental Table 6) were identified in the region. Our primary candidate gene screening of B. rapa and A. thaliana orthologues found four genes were directly involved in lipid metabolism activities, i.e., fatty acid beta-oxidation (Wang et al. 2008), lipid-binding (Hanada 2011), mono-/di-acylglycerol lipase (Li-Beisson et al. 2010), triglyceride lipase activity (Saleh 2008). Six other genes were homologous to sucrose-phosphate synthase (Lutfiyya et al. 2007), dual-specificity phosphatase (Roma-Mateo et al. 2011), galactosyltransferase (Qu et al. 2008), ADP-glucose pyrophosphorylase (Schwarte et al. 2015), UDP-glucosyltransferase (Li et al. 2015a), and polygalacturonase (Lou et al. 2007). The two orthologues genes (AT3G14075 and AT1G05790) likely have directly relationship with the lipid synthesis, these two orthologues genes may be the best choice for the candidate gene (Fig. 4, Supplemental Table 6). Although more candidate genes may be found, these function-known candidate genes in the A10 OC QTL region are very encouraging. Fine mapping and map-based cloning have been demonstrated to be one of the most efficient ways to dissect these trait-related quantitative trait loci (Miura et al. 2011, Takeda and Matsuoka 2008). The QTL with strongly expressed and stable effects was highly suitable for map-based cloning. Thus, validating potential candidate genes is a reliable and feasible strategy for QTL cloning. The information obtained from this study demonstrates potentially novel roles for candidate genes in rapeseed oil accumulation. Future works will provide an opportunity to identify the genes that control seed OC in B. napus.
Fig. 4.
Linkage and genomic region of cqOCA10b on the B. rapa genome. The vertical black bars represent some candidate genes.
Supplementary Information
Acknowledgements
This work was supported by the National Hi-Tech R&D Program of China (2013AA102602).
Literature Cited
- Arcade, A., Labourdette, A., Falque, M., Mangin, B., Chardon, F., Charcosset, A. and Joets, J. (2004) BioMercator: integrating genetic maps and QTL towards discovery of candidate genes. Bioinformatics 20: 2324–2326. [DOI] [PubMed] [Google Scholar]
- Burns, M.J., Barnes, S.R., Bowman, J.G., Clarke, M.H., Werner, C.P. and Kearsey, M.J. (2003) QTL analysis of an intervarietal set of substitution lines in Brassica napus: (i) Seed oil content and fatty acid composition. Heredity 90: 39–48. [DOI] [PubMed] [Google Scholar]
- Bus, A., Hecht, J., Huettel, B., Reinhardt, R. and Stich, B. (2012) High-throughput polymorphism detection and genotyping in Brassica napus using next-generation RAD sequencing. BMC Genomics 13: 281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalhoub, B., Denoeud, F., Liu, S., Parkin, I.A., Tang, H., Wang, X., Chiquet, J., Belcram, H., Tong, C., Samans, B.et al. (2014) Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science 345: 950–953. [DOI] [PubMed] [Google Scholar]
- Chen, G., Geng, J., Rahman, M., Liu, X., Tu, J., Fu, T., Li, G., McVetty, P.B.E. and Tahir, M. (2010) Identification of QTL for oil content, seed yield, and flowering time in oilseed rape (Brassica napus). Euphytica 175: 161–174. [Google Scholar]
- Chen, W., Zhang, Y., Liu, X., Chen, B., Tu, J. and Tingdong, F. (2007) Detection of QTL for six yield-related traits in oilseed rape (Brassica napus) using DH and immortalized F2 populations. Theor. Appl. Genet. 115: 849–858. [DOI] [PubMed] [Google Scholar]
- Chen, W., Zhang, Y., Yao, J., Ma, C., Tu, J. and Tingdong, F. (2011) Quantitative trait loci mapping for two seed yield component traits in an oilseed rape (Brassica napus) cross. Plant Breed. 130: 640–646. [Google Scholar]
- Chen, X., Li, X., Zhang, B., Xu, J., Wu, Z., Wang, B., Li, H., Younas, M., Huang, L., Luo, Y.et al. (2013) Detection and genotyping of restriction fragment associated polymorphisms in polyploid crops with a pseudo-reference sequence: a case study in allotetraploid Brassica napus. BMC Genomics 14: 346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, X., Xu, J., Xia, S., Gu, J., Yang, Y., Fu, J., Qian, X., Zhang, S., Wu, J. and Liu, K. (2009) Development and genetic mapping of microsatellite markers from genome survey sequences in Brassica napus. Theor. Appl. Genet. 118: 1121–1131. [DOI] [PubMed] [Google Scholar]
- Delourme, R., Falentin, C., Huteau, V., Clouet, V., Horvais, R., Gandon, B., Specel, S., Hanneton, L., Dheu, J.E., Deschamps, M.et al. (2006) Genetic control of oil content in oilseed rape (Brassica napus L.). Theor. Appl. Genet. 113: 1331–1345. [DOI] [PubMed] [Google Scholar]
- Ecke, W., Uzunova, M. and Weissleder, K. (1995) Mapping the genome of rapeseed (Brassica napus L.). II. Localization of genes controlling erucic acid synthesis and seed oil content. Theor. Appl. Genet. 91: 972–977. [DOI] [PubMed] [Google Scholar]
- Fan, C., Cai, G., Qin, J., Li, Q., Yang, M., Wu, J., Fu, T., Liu, K. and Zhou, Y. (2010) Mapping of quantitative trait loci and development of allele-specific markers for seed weight in Brassica napus. Theor. Appl. Genet. 121: 1289–1301. [DOI] [PubMed] [Google Scholar]
- Hanada, K., Sawada, Y., Kuromori, T., Klausnitzer, R., Saito, K., Toyoda, T., Shinozaki, K., Li, W.-H. and Yokota Hirai, M. (2011) Functional compensation of primary and secondary metabolites by duplicate genes in Arabidopsis thaliana. Mol. Biol. Evol. 28: 377–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javed, N., Geng, J., Tahir, M., Mcvetty, P.B.E., Li, G. and Duncan, R.W. (2016) Identification of QTL influencing seed oil content, fatty acid profile and days to flowering in Brassica napus L. Euphytica 207: 191–211. [Google Scholar]
- Jiang, C. and Zeng, Z.B. (1995) Multiple trait analysis of genetic mapping for quantitative trait loci. Genetics 140: 1111–1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, C., Shi, J., Li, R., Long, Y., Wang, H., Li, D., Zhao, J. and Meng, J. (2014) Quantitative trait loci that control the oil content variation of rapeseed (Brassica napus L.). Theor. Appl. Genet. 127: 957–968. [DOI] [PubMed] [Google Scholar]
- Kong, F. (2005) Quantitaive Genetics in Plants. China Agricultural University Press, Beijing, China. [Google Scholar]
- Kosambi, D.D. (1943) The estimation of map distances from recombination values. Ann. Eugen. 12: 172–175. [Google Scholar]
- Li, N., Shi, J., Wang, X., Liu, G. and Wang, H. (2014) A combined linkage and regional association mapping validation and fine mapping of two major pleiotropic QTLs for seed weight and silique length in rapeseed (Brassica napus L.). BMC Plant Biol. 14: 114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, R., Li, Y., Kristiansen, K. and Wang, J. (2008) SOAP: short oligonucleotide alignment program. Bioinformatics 24: 713–714. [DOI] [PubMed] [Google Scholar]
- Li, R., Yu, C., Li, Y., Lam, T.W., Yiu, S.M., Kristiansen, K. and Wang, J. (2009) SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics 25: 1966–1967. [DOI] [PubMed] [Google Scholar]
- Li, S., Chen, L., Zhang, L., Li, X., Liu, Y., Wu, Z., Dong, F., Wan, L., Liu, K., Hong, D.et al. (2015a) BnaC9.SMG7b functions as a positive regulator of the number of seeds per silique in Brassica napus by regulating the formation of functional female gametophytes. Plant Physiol. 169: 2744–2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, W., Zhang, F., Chang, Y., Zhao, T., Schranz, M.E. and Wang, G. (2015b) Nicotinate O-glucosylation is an evolutionarily metabolic trait important for seed germination under stress conditions in Arabidopsis thaliana. Plant Cell 27: 1907–1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li-Beisson, Y., Shorrosh, B., Beisson, F., Andersson, M.X., Arondel, V., Bates, P.D., Baud, S., Bird, D., Debono, A., Durrett, T.P.et al. (2010) Acyl-lipid metabolism. Arabidopsis Book 8: e0133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, D., Ma, C., Hong, W., Huang, L., Liu, M., Liu, H., Zeng, H., Deng, D., Xin, H., Song, J.et al. (2014) Construction and analysis of high-density linkage map using high-throughput sequencing data. PLoS ONE 9: e98855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, J., Hua, W., Hu, Z., Yang, H., Zhang, L., Li, R., Deng, L., Sun, X., Wang, X. and Wang, H. (2015) Natural variation in ARF18 gene simultaneously affects seed weight and silique length in polyploid rapeseed. Proc. Natl. Acad. Sci. USA 112: E5123–5132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, L., Qu, C., Wittkop, B., Yi, B., Xiao, Y., He, Y., Snowdon, R.J. and Li, J. (2013) A high-density SNP map for accurate mapping of seed fibre QTL in Brassica napus L. PLoS ONE 8: e83052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, S., Liu, Y., Yang, X., Tong, C., Edwards, D., Parkin, I.A., Zhao, M., Ma, J., Yu, J., Huang, S.et al. (2014) The Brassica oleracea genome reveals the asymmetrical evolution of polyploid genomes. Nat. Commun. 5: 3930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long, Y., Shi, J., Qiu, D., Li, R., Zhang, C., Wang, J., Hou, J., Zhao, J., Shi, L., Park, B.S.et al. (2007) Flowering time quantitative trait loci analysis of oilseed brassica in multiple environments and genome-wide alignment with Arabidopsis. Genetics 177: 2433–2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou, Y., Gou, J.Y. and Xue, H.W. (2007) PIP5K9, an Arabidopsis phosphatidylinositol monophosphate kinase, interacts with a cytosolic invertase to negatively regulate sugar-mediated root growth. Plant Cell 19: 163–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutfiyya, L.L., N. Xu, R.L. D’Ordine, J.A. Morrell, P.W. Miller and S.M. Duff (2007) Phylogenetic and expression analysis of sucrose phosphate synthase isozymes in plants. J. Plant Physiol. 164: 923–933. [DOI] [PubMed] [Google Scholar]
- Mahmood, S., Li, Z., Yue, X., Wang, B., Chen, J. and Liu, K. (2016) Development of INDELs markers in oilseed rape (Brassica napus L.) using re-sequencing data. Mol. Breedi. 36: 79. [Google Scholar]
- Miura, K., Ashikari, M. and Matsuoka, M. (2011) The role of QTLs in the breeding of high-yielding rice. Trends Plant Sci. 16: 319–326. [DOI] [PubMed] [Google Scholar]
- Qi, L., Mao, L., Sun, C., Pu, Y., Fu, T., Ma, C., Shen, J., Tu, J., Yi, B. and Wen, J. (2014) Interpreting the genetic basis of silique traits in Brassica napus using a joint QTL network. Plant Breed. 133: 52–60. [Google Scholar]
- Qu, Y., Egelund, J., Gilson, P.R., Houghton, F., Gleeson, P.A., Schultz, C.J. and Bacic, A. (2008) Identification of a novel group of putative Arabidopsis thaliana β-(1,3)-galactosyltransferases. Plant Mol. Biol. 68: 43–59. [DOI] [PubMed] [Google Scholar]
- Roma-Mateo, C., Sacristan-Reviriego, A., Beresford, N.J., Caparros-Martin, J.A., Culianez-Macia, F.A., Martin, H., Molina, M., Tabernero, L. and Pulido, R. (2011) Phylogenetic and genetic linkage between novel atypical dual-specificity phosphatases from non-metazoan organisms. Mol. Genet. Genomics 285: 341–354. [DOI] [PubMed] [Google Scholar]
- Rowe, H.C., Renaut, S. and Guggisberg, A. (2011) RAD in the realm of next-generation sequencing technologies. Mol. Ecol. 20: 3499–3502. [DOI] [PubMed] [Google Scholar]
- Saleh, A., Alvarez-Venegas, R., Yilmaz, M., Le, O., Hou, G., Sadder, M., Al-Abdallat, A., Xia, Y., Lu, G., Ladunga, I.et al. (2008) The highly similar Arabidopsis homologs of trithorax ATX1 and ATX2 encode proteins with divergent biochemical functions. Plant Cell 20: 568–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarte, S., Wegner, F., Havenstein, K., Groth, D., Steup, M. and Tiedemann, R. (2015) Sequence variation, differential expression, and divergent evolution in starch-related genes among accessions of Arabidopsis thaliana. Plant Mol. Biol. 87: 489–519. [DOI] [PubMed] [Google Scholar]
- Shi, J., Li, R., Qiu, D., Jiang, C., Long, Y., Morgan, C., Bancroft, I., Zhao, J. and Meng, J. (2009) Unraveling the complex trait of crop yield with quantitative trait loci mapping in Brassica napus. Genetics 182: 851–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snowdon, R.J. (2007) Cytogenetics and genome analysis in Brassica crops. Chromosome Res. 15: 85–95. [DOI] [PubMed] [Google Scholar]
- Sosnowski, O., Charcosset, A. and Joets, J. (2012) BioMercator V3: an upgrade of genetic map compilation and quantitative trait loci meta-analysis algorithms. Bioinformatics 28: 2082–2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, M., Hua, W., Liu, J., Huang, S., Wang, X., Liu, G. and Wang, H. (2012) Design of new genome- and gene-sourced primers and identification of QTL for seed oil content in a specially high-oil Brassica napus cultivar. PLoS ONE 7: e47037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda, S. and Matsuoka, M. (2008) Genetic approaches to crop improvement: responding to environmental and population changes. Nat. Rev. Genet. 9: 444–457. [DOI] [PubMed] [Google Scholar]
- Trick, M., Long, Y., Meng, J. and Bancroft, I. (2009) Single nucleotide polymorphism (SNP) discovery in the polyploid Brassica napus using Solexa transcriptome sequencing. Plant Biotechnol. J. 7: 334–346. [DOI] [PubMed] [Google Scholar]
- Tuberosa, R., Salvi, S., Sanguineti, M.C., Landi, P., Maccaferri, M. and Conti, S. (2002) Mapping QTLs regulating morpho-physiological traits and yield: case studies, shortcomings and perspectives in drought-stressed maize. Ann. Bot. 89 Spec. No: 941–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udall, J.A., Quijada, P.A., Lambert, B. and Osborn, T.C. (2006) Quantitative trait analysis of seed yield and other complex traits in hybrid spring rapeseed (Brassica napus L.): 2. Identification of alleles from unadapted germplasm. Theor. Appl. Genet. 113: 597–609. [DOI] [PubMed] [Google Scholar]
- Van Ooijen, J.W. (2006) JoinMap4. Software for the calculation of genetic linkage maps in experimental populations. Kyazma BV, Wageningen, Netherlands. [Google Scholar]
- Wang, H. (2004) Strategy for rapeseed genetic improvement in China in the coming fifteen years. Chin. J. Oil Crop Sci. 26: 98–101. [Google Scholar]
- Wang, X., Wang, H., Long, Y., Li, D., Yin, Y., Tian, J., Chen, L., Liu, L., Zhao, W., Zhao, Y.et al. (2013) Identification of QTLs associated with oil content in a high-oil Brassica napus cultivar and construction of a high-density consensus map for QTLs comparison in B. napus. PLoS ONE 8: e80569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y., Zhang, W.Z., Song, L.F., Zou, J.J., Su, Z. and Wu, W. (2008) Transcriptome analyses show changes in gene expression to accompany pollen germination and tube growth in Arabidopsis. Plant Physiol. 148: 1201–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, J., Qian, X., Wang, X., Li, R., Cheng, X., Yang, Y., Fu, J., Zhang, S., King, G.J., Wu, J.et al. (2010) Construction of an integrated genetic linkage map for the A genome of Brassica napus using SSR markers derived from sequenced BACs in B. rapa. BMC Genomics 11: 594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, J., Hu, C., Hu, H., Yu, R., Xia, Z., Ye, X. and Zhu, J. (2008) QTLNetwork: mapping and visualizing genetic architecture of complex traits in experimental populations. Bioinformatics 24: 721–723. [DOI] [PubMed] [Google Scholar]
- Yang, P., Shu, C., Chen, L., Xu, J., Wu, J. and Liu, K. (2012) Identification of a major QTL for silique length and seed weight in oilseed rape (Brassica napus L.). Theor. Appl. Genet. 125: 285–296. [DOI] [PubMed] [Google Scholar]
- Zeng, Z. (1994) Precision mapping of quantitative trait loci. Genetics 136: 1457–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, L., Yang, G., Liu, P., Hong, D., Li, S. and He, Q. (2011) Genetic and correlation analysis of silique-traits in Brassica napus L. by quantitative trait locus mapping. Theor. Appl. Genet. 122: 21–31. [DOI] [PubMed] [Google Scholar]
- Zhang, L., Li, S., Chen, L. and Yang, G. (2012) Identification and mapping of a major dominant quantitative trait locus controlling seeds per silique as a single Mendelian factor in Brassica napus L. Theor. Appl. Genet. 125: 695–705. [DOI] [PubMed] [Google Scholar]
- Zhao, J., Udall, J.A., Quijada, P.A., Grau, C.R., Meng, J. and Osborn, T.C. (2006) Quantitative trait loci for resistance to Sclerotinia sclerotiorum and its association with a homeologous non-reciprocal transposition in Brassica napus L. Theor. Appl. Genet. 112: 509–516. [DOI] [PubMed] [Google Scholar]
- Zheng, P., Allen, W.B., Roesler, K., Williams, M.E., Zhang, S., Li, J., Glassman, K., Ranch, J., Nubel, D., Solawetz, W.et al. (2008) A phenylalanine in DGAT is a key determinant of oil content and composition in maize. Nat. Genet. 40: 367–372. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.