Short abstract
An analysis of nuclear-encoded oxidative phosphorylation genes in Drosophila and Anopheles reveals that pairs of duplicated genes have strikingly different expression patterns.
Abstract
Background
In eukaryotic cells, oxidative phosphorylation (OXPHOS) uses the products of both nuclear and mitochondrial genes to generate cellular ATP. Interspecies comparative analysis of these genes, which appear to be under strong functional constraints, may shed light on the evolutionary mechanisms that act on a set of genes correlated by function and subcellular localization of their products.
Results
We have identified and annotated the Drosophila melanogaster, D. pseudoobscura and Anopheles gambiae orthologs of 78 nuclear genes encoding mitochondrial proteins involved in oxidative phosphorylation by a comparative analysis of their genomic sequences and organization. We have also identified 47 genes in these three dipteran species each of which shares significant sequence homology with one of the above-mentioned OXPHOS orthologs, and which are likely to have originated by duplication during evolution. Gene structure and intron length are essentially conserved in the three species, although gain or loss of introns is common in A. gambiae. In most tissues of D. melanogaster and A. gambiae the expression level of the duplicate gene is much lower than that of the original gene, and in D. melanogaster at least, its expression is almost always strongly testis-biased, in contrast to the soma-biased expression of the parent gene.
Conclusions
Quickly achieving an expression pattern different from the parent genes may be required for new OXPHOS gene duplicates to be maintained in the genome. This may be a general evolutionary mechanism for originating phenotypic changes that could lead to species differentiation.
Background
The accessibility of whole-genome sequence data for several organisms, together with the development of efficient computer-based search tools, has revolutionized modern biology, allowing in-depth comparative analysis of genomes [1-4]. In many cases, comparisons among species at various levels of divergence have helped to define protein-coding genes, recognize nonfunctional genes, and find regulatory sequences and other functional elements in the genome. When applied to a set of genes correlated by function and/or subcellular localization of their products, intra- and interspecies comparative analyses can be especially efficient tools to obtain information on the functional constraints acting on the evolution of the gene set and on the mechanisms regulating its coordinate expression.
A set of genes present in all eukaryotic genomes and expected to be subject to peculiar evolutionary constraints is represented by the genes involved in oxidative phosphorylation (OXPHOS), the primary energy-producing process in all aerobic organisms [5]. To generate cellular ATP, OXPHOS uses the products of both nuclear and mitochondrial genes, organized in five large complexes embedded in the lipid bilayer of the inner mitochondrial membrane. Except for complex II, which is formed by four proteins encoded by nuclear genes, the other respiratory complexes depend on both mitochondrial and nuclear genomes; so, assembling the OXPHOS complexes and fine tuning their activity to satisfy cell- and tissue-specific energy demands requires specialized regulatory mechanisms and evolutionary strategies to optimize the cross-talk between the two genomes and ensure the coordinated expression of their relevant products.
Analysis of co-regulated mitochondrial and nuclear genes, and of the transcription factors regulating the functional network they constitute, might also be a useful approach to investigate the origin of mitochondrial dysfunction in humans. Disorders of mitochondrial oxidative phosphorylation are now recognized as the most common inborn errors of metabolism, affecting at least one in 5,000 newborn children [6]. In this context, the expanding spectrum of identified mitochondrial proteins provides an opportunity to test a whole new range of candidate genes whose mutations may be responsible for common human diseases. For example, a recent study by Mootha et al. [7] suggests a promising strategy for clarifying the molecular etiology of mitochondrial pathologies by profiling the tissue-specific expression pattern of candidate mitochondrial proteins.
Despite the long evolutionary divergence time, many key pathways that control development and physiology are conserved between Drosophila and humans, and about 70% of the genes associated with human disease have direct counterparts in the Drosophila genome [8,9]. For example, the potential role of Drosophila as a model system for understanding the molecular mechanisms involved in human genetic disease is validated by the recent identification of a Drosophila mutation causing a necrotic phenotype that mimics in detail the diseases that arise from serpin mutations in humans [10].
It has been suggested that comparisons between D. melanogaster and other species of the genus Drosophila could provide a model system for developing and testing new algorithms and strategies for the functional annotation of complex genomes [3]. To obtain new information on the evolution of a set of genes that control a basic biological function by encoding products targeted to a specific cellular compartment, we have performed a comparative analysis of the OXPHOS genes of D. melanogaster and D. pseudoobscura; the complete genome of the latter was recently made available by the Baylor Human Genome Sequencing Center. These two species are the only species of the Drosophila genus for which whole-genome sequence data exist at present [11-13]. We also took advantage of the complete sequence of the A. gambiae genome [14] to compare the Drosophila OXPHOS genes with those of this more distantly related dipteran (the divergence time between D. melanogaster and A. gambiae is thought to be approximately 250 million years, as compared to 46 million years between D. melanogaster and D. pseudoobscura [15,16]). Although extensive reshuffling within and between chromosomal regions is known to have occurred since the divergence of Anopheles from Drosophila [4,17,18], we show that in these organisms the conservation of the OXPHOS genes is still sufficient to permit their meaningful comparison.
Here we report the identification of 78 D. pseudoobscura and 78 A. gambiae genes representing the counterparts of D. melanogaster OXPHOS genes which, in turn, were previously identified as putative orthologs of human OXPHOS genes [19]. We have annotated these genes, taking into account conservation in amino-acid sequence, intron-exon structure, intron length, and the presence of duplications in the genome. The conservation of genomic organization and evidence from evolutionary trees based on sequence similarity suggest that these genes are one-to-one orthologs in the three species, and that in many cases they originated (produced?) duplicates by transpositional and/or recombinational events during evolution. We have identified in the three dipteran genomes a total of 47 genes that probably originated by duplication of the above-mentioned genes, and we show that the duplicate gene has usually acquired a pattern of expression strikingly different from that of the gene from which it derived. Moreover, when the comparison is possible, the gene duplicate almost always shows a strongly testis-biased expression, in contrast to the soma-biased expression of its parent gene.
Results and discussion
Identification and comparative annotation of D. pseudoobscura and A. gambiae OXPHOS genes
We have previously reported [19] the identification of 285 D. melanogaster nuclear genes encoding mitochondrial proteins that represent the counterparts of human peptides annotated in the Swiss-Prot database as mitochondrial [20]. On the basis of comparative evidence obtained by BLASTP analysis, 78 of these genes are involved in the OXPHOS system, encoding 66 proteins known to be components of the five large respiratory complexes and 12 proteins involved in oxidative phosphorylation as accessory proteins. To identify the putative counterparts of the D. melanogaster OXPHOS genes in D. pseudoobscura and A. gambiae we performed a TBLASTN search [13,21] on the whole genome sequences of these species using the amino-acid sequences of the 78 D. melanogaster peptides as queries. Sequences giving the best reciprocal BLAST hits were tentatively assumed to identify functional counterparts in two species if they could be aligned over at least 60% of the gene length and the BLAST E-score was less than 10-30. By these criteria, all the 78 D. melanogaster OXPHOS genes investigated have a counterpart both in D. pseudoobscura and in A. gambiae. To better compare the structure of the OXPHOS genes in the three dipteran species, we used the predicted coding sequences as queries for a search of expressed sequence tags (EST) [21], and used the retrieved sequences to annotate the transcribed noncoding sequences of the A. gambiae genes investigated. Although little EST information is available for D. pseudoobscura, it was still possible to predict unambiguously the exon-intron gene structure of the OXPHOS genes in this species, as well as the amino-acid sequence of their full-length products, by exploiting the high level of similarity with D. melanogaster. The results of BLAST analysis, together with the construction of phylogenetic trees that also include other genes that show lesser but still significant sequence similarity to the 78 genes assumed to be one-to-one orthologs in the three species investigated (see below), strongly suggest that the newly identified D. pseudoobscura and A. gambiae genes are the functional counterparts of the 78 D. melanogaster genes used as probes.
Table 1 lists the 78 putative orthologous OXPHOS genes in the three dipteran genomes and their cytological location. For each gene, a record showing the gene map and reporting the annotated genomic sequences as well as the mRNA and protein sequences is available and can be queried at the MitoComp website [22] (see also Additional data files). MitoComp also compares the structure of the D. melanogaster, D. pseudoobscura and A. gambiae putative orthologous genes and their duplications when present (see below), and aligns the orthologous coding sequences (CDS), and also aligns their deduced amino-acid products with the corresponding human protein.
Table 1.
Number of exons and chromosomal localization of the 78 orthologous D. melanogaster, D pseudoobscura and A. gambiae OXPHOS genes
| Cluster ID* | Protein name | D. melanogaster gene name | Number of exons† | Map position | FlyBase ID | D. pseudoobscura gene name | Number of exons† | Map position | A. gambiae gene name | Number of exons† | Map position |
| Complex I: NADH:ubiquinone oxidoreductase | |||||||||||
| NUMM | 13 kDa A subunit | CG8680 | 3 | 2L;25C6 | FBgn0031684 | Dpse\CG8680 | 3 | 4 | agEG14117 | 3 | 3R;33B |
| NUFM | 13 kDa B subunit | CG6463 | 3 | 3L;67E7 | FBgn0036100 | Dpse\CG6463 | 3 | XR | agEG15380 | 3 | 2L22E |
| NIPM | 15 kDa subunit | CG11455 | 2 | 2L;21B1-2 | FBgn0031228 | Dpse\CG11455 | 2 | 4 | agEG13302 | 2 | 3R;35C-D |
| NUYM | 18 kDa subunit | CG12203 | 3 | X;18C7 | FBgn0031021 | Dpse\CG12203 | 3 | XL | agEG18985 | 4 | 2L;27A |
| NUPM | 19 kDa subunit | CG3683 | 4 | 2R;60D13 | FBgn0035046 | Dpse\CG3683 | 4 | 3 | agEG19249 | 3 | 2L;26B |
| NUKM | 20 kDa subunit | CG9172 | 1 | X; 14A5 | FBgn0030718 | Dpse\CG9172 | 1 | ND | agEG16939 | 1 | X;4A |
| NUIM | 23 kDa subunit | ND23 | 3 | 3R;89A5 | FBgn0017567 | Dpse\CG3944 | 3 | 2 | agEG9698 | 2 | 2R;9A |
| NUHM | 24 kDa subunit | CG5703 | 3 | X; 16B10 | FBgn0030853 | Dpse\CG5703 | 3 | XL | agEG16953 | 5 | 2R;11A |
| NUGM | 30 kDa subunit | CG12079 | 3 | 3L;63B7 | FBgn0035404 | Dpse\CG12079 | 3 | XR | agEG11610 | 3 | 2L;24D |
| NUEM | 39 kDa subunit | CG6020 | 4 | 3L;77C6 | FBgn0037001 | Dpse\CG6020 | 4 | XR | agEG18760 | 3 | 3L;40A |
| NUDM | 42 kDa subunit | ND42 | 2 | 3R;94A1 | FBgn0019957 | Dpse\CG6343 | 2 | 2 | agEG10090 | 2 | 3L;41C |
| NUCM | 49 kDa subunit | CG1970 | 6 | 4;102C2 | FBgn0039909 | Dpse\CG1970 | 6 | ND | agEG18856 | 1 | X;1B |
| NUBM | 51 kDa subunit | CG9140 | 4 | 2L;26B6-7 | FBgn0031771 | Dpse\CG9140 | 4 | 4 | agEG9927 | 4 | 3R;36D |
| NUAM | 75 kDa subunit | ND75 | 5 | X;7E1 | FBgn0017566 | Dpse\CG2286 | 5 | XL | agEG19681 | 4 | 2R;8D |
| NI8M | B8 subunit | CG15434 | 3 | 2L;24F3 | FBgn0040705 | Dpse\CG15434 | 3 | 4 | agEG16251 | 3 | 2R;15B |
| NB2M | B12 subunit | CG10320 | 2 | 2R;57F6 | FBgn0034645 | Dpse\CG10320 | 2 | 3 | agEG9277 | 1 | 3L;46D |
| NB4M | B14 subunit | CG7712 | 3 | 2R;47C6 | FBgn0033570 | Dpse\CG7712 | 3 | 3 | agEG12033 | 2 | 2R;15A |
| N4AM | B14.5A subunit | CG3621 | 2 | X; 2D6-E1 | FBgn0025839 | Dpse\CG3621 | 2 | XL | agEG14707 | 4 | 2R;17A |
| N4BM | B14.5B subunit | CG12400 | 3 | 2L:23D3 | FBgn0031505 | Dpse\CG12400 | 3 | 4 | agEG16232 | 3 | 2R;13C |
| NB5M | B15 subunit | CG12859 | 2 | 2R;51C2 | FBgn0033961 | Dpse\CG12859 | 2 | 3 | agEG17759 | 2 | 3L;44C |
| NB6M | B16.6 subunit | CG3446 | 2 | X;5F2 | FBgn0029868 | Dpse\CG3446 | 2 | XL | agEG7829 | 3 | 3R;35A |
| NB7M | B17 subunit | l(2)35Di | 3 | 2L;35D | FBgn0001989 | Dpse\CG13240 | 3 | 4 | agEG18567 | 3 | 3R;34D |
| N7BM | B17.2 subunit | CG3214 | 4 | 2L;23A1 | FBgn0031436 | Dpse\CG3214 | 4 | 4 | agEG10758 | 4 | 3R;31A |
| NB8M | B18 subunit | CG5548 | 1 | X;13A8 | FBgn0030605 | Dpse\CG5548 | 1 | XL | agEG8436 | 3 | 2L;28C |
| NI2M | B22 subunit | CG9306 | 3 | 2L;34B8 | FBgn0032511 | Dpse\CG9306 | 3 | 4 | agEG12344 | 3 | 3R;35C-D |
| ACPM | Acyl carrier | mtacp1 | 4 | 3L;61F6 | FBgn0011361 | Dpse\CG9190 | 4 | XR | agEG11237 | 5 | 3L;38B |
| NIAM | ASHI subunit | CG3192 | 3 | X;6C5 | FBgn0029888 | Dpse\CG3192 | 3 | XL | agEG8821 | 3 | 2R;10A |
| NUML | MLRQ subunit | CG32230 | 3 | 3L;80E2 | FBgn0052230 | Dpse\CG32230 | 3 | XR | agEG12063 | 3 | 2R;15A |
| NINM | MNLL subunit | CG18624 | 1 | X;7C | FBgn0029971 | Dpse\CG18624 | 1 | XL | agEG22692 | 1 | X;5A |
| NIDM | PDSW subunit | Pdsw | 3 | 2L;23F3 | FBgn0021967 | Dpse\CG8844 | 3 | 4 | agEG7887 | 4 | 3R;29A |
| NISM | SGDH subunit | l(3)neo18 | 4 | 3L;68F5 | FBgn0011455 | Dpse\CG9762 | 4 | XR | agEG13573 | 2 | 2L;27D |
| NIGM | AGGG subunit | CG40002 | 3 | ND | FBgn0058002 | Dpse\CG40002 | 3 | XR | agEG18653 | 2R;12D | |
| Complex II: Succinate dehydrogenase | |||||||||||
| DHSA | Flavoprotein subunit | Scs-fp | 4 | 2R;56D3 | FBgn0017539 | Dpse\CG17246 | 4 | 3 | agEG7754 | 3 | 3L;38B |
| DHSB | Iron-sulfur protein | SdhB | 3 | 2R;42D3-4 | FBgn0014028 | Dpse\CG3283 | 3 | 3 | agEG13539 | 4 | 2L;27D |
| C560 | Cytochrome B560 subunit | CG6666 | 2 | 3R;86D7-8 | FBgn0037873 | Dpse\CG6666 | 2 | 2 | agEG14929 | 2 | 3L;39B |
| DHSD | Cytochrome b small subunit | CG10219 | 4 | 3R;95B1 | Fbgn0039112 | Dpse\CG10219 | 4 | XR | agEG16772 | 3 | X;1C |
| Complex III: Ubiquinol-cytochrome c reductase | |||||||||||
| UCRY | 6.4 kDa protein | CG14482 | 2 | 2R;54C9 | FBgn0034245 | Dpse\CG14482 | 2 | 3 | agEG12505 | 2 | 3L;43B |
| UCRX | 7.2 kDa protein | ox | 2 | 2R;49C2 | FBgn0011227 | Dpse\CG8764 | 2 | 3 | agEG15210 | 2 | 2L;20C |
| UCRH | 11 kDa protein | Ucrh | 2 | 3R | FBgn0066066 | Dpse\Ucrh | 2 | 2 | agEG19398 | 2 | 2R;11B |
| UCR6 | 14 kDa protein | CG3560 | 3 | X;14B10 | FBgn0030733 | Dpse\CG3560 | 3 | XL | agEG11611 | 3 | 3L;46A |
| UCRI | Iron-sulfur subunit | RFeSP | 3 | 2L;22A3 | FBgn0021906 | Dpse\CG7361 | 3 | 4 | agEG16975 | 4 | 3R;32C |
| CY1 | Cytochrome c1, heme protein | CG4769 | 6 | 3L;64C13 | FBgn0035600 | Dpse\CG4769 | 6 | XR | agEG19223 | 4 | 2L;26C |
| UCR1 | Core protein 1 | CG3731 | 6 | 3R;88D6 | FBgn0038271 | Dpse\CG3731 | 6 | 2 | agEG21302 | 3 | X;5C |
| UCR2 | Core protein 2 | CG4169 | 4 | 3L;73A10 | FBgn0036642 | Dpse\CG4169_1 | 4 | XR | agEG17930 | 4 | 2L;24A |
| UCRQ | Ubiquinone-binding protein QP- | CG7580 | 2 | 3L;74C3 | FBgn0036728 | Dpse\CG7580 | 2 | XR | agEG20223 | 2 | 3L;38C |
| Complex IV: Cytochrome c oxidase | |||||||||||
| CX41 | Polypeptide IV | CG10664 | 2 | 2L;38A8 | FBgn0032833 | Dpse\CG10664 | 2 | 4 | agEG13327 | 2 | 3R;31C |
| COXA | Polypeptide Va | CoVa | 1 | 3R;86F9 | FBgn0019624 | Dpse\CG14724 | 1 | 2 | agEG19581 | 1 | 3L;41D |
| COXB | Polypeptide Vb | CG11015 | 3 | 2L;26E3 | FBgn0031830 | Dpse\CG11015 | 3 | 4 | agEG8633 | 4 | 3R;31C |
| COXD | Polypeptide VIa | CG17280 | 2 | 2R;59E3 | FBgn0034877 | Dpse\CG17280 | 2 | 3 | agEG7821 | 2 | X;5A |
| COXG | Polypeptide VIb | CG18809 | 1 | X;18E5 | FBgn0042132 | Dpse\CG18809 | 1 | XL | agEG11043 | 1 | 2L;25A |
| COXH | Polypeptide VIc | cype | 2 | 2L;25D6 | FBgn0015031 | Dpse\CG14028 | 2 | 4 | EST357342 | 2 | 3R;29A |
| COXK | Polypeptide VIIa | CG9603 | 2 | 3R;84F13 | FBgn0040529 | Dpse\CG9603 | 2 | XR | agEG17423 | 3 | X;4B |
| COXO | Polypeptide VIIc | CG2249 | 2 | 2R;46D8-9 | FBgn0040773 | Dpse\CG2249 | 2 | 3 | agEG22887 | 2 | 2L;28C |
| Complex V: ATP synthase | |||||||||||
| ATPA | Alpha chain | blw | 4 | 2R;59B1-2 | FBgn0011211 | Dpse\CG3612 | 4 | 3 | agEG7500 | 4 | 2L;21E |
| ATPB | Beta chain | ATPsyn-beta | 3 | 4;102D1 | FBgn0010217 | Dpse\CG11154 | 3 | ND | agEG14379 | 1 | 3L;45C |
| ATPG | Gamma chain | ATPsyn-gamma | 1 | 3R;99B10 | FBgn0020235 | Dpse\CG7610 | 1 | 2 | agEG7678 | 2 | 3R;29C |
| ATPD | Delta chain | CG2968 | 3 | X;9B4 | FBgn0030184 | Dpse\CG2968 | 3 | ND | agEG16076 | 1 | 3R;29B |
| ATPE | Epsilon chain | sun | 4 | X;13F12 | FBgn0014391 | Dpse\CG9032 | 4 | ND | agEG10095 | 4 | X;3D |
| ATPF | B chain | ATPsyn-b | 3 | 3L;67C5 | FBgn0019644 | Dpse\CG8189 | 3 | XR | agEG9580 | 3 | 2R;7A |
| ATPQ | D chain | ATPsyn-d | 1 | 3R;91F | FBgn0016120 | Dpse\CG6030 | 1 | ND | agEG10180 | 3 | 3L;41C |
| ATPJ | E chain | CG3321 | 1 | 3R;88B4 | FBgn0038224 | Dpse\CG3321 | 1 | 2 | agEG10809 | 3 | 2L;26B |
| ATPK | F chain | CG4692 | 2 | 2R;60D8-9 | FBgn0035032 | Dpse\CG4692 | 2 | 3 | agEG1544 | 1 | ND |
| ATPN | G chain | l(2)06225 | 2 | 2L;32C1 | FBgn0010612 | Dpse\CG6105 | 2 | ND | agEG8590 | 2 | 3R;34B |
| ATPR | Coupling factor 6 | ATPsyn-Cf6 | 2 | 3R;94E13 | FBgn0016119 | Dpse\CG4412 | 2 | 2 | agEG19097 | 2 | 2R;19D |
| AT91 | Lipid-binding protein P1 | CG1746 | 3 | 3R;100B7 | FBgn0039830 | Dpse\CG1746 | 3 | 2 | agEG14837 | 3 | X;2B |
| ATPO | OSCP | Oscp | 3 | 3R;88E8-9 | FBgn0016691 | Dpse\CG4307 | 3 | 2 | agEG9393 | 3 | 2R;15D |
| Others | |||||||||||
| ATPW | ATP synthase coupling factor B | CG10731 | 1 | 2R;52F | FBgn0034081 | Dpse\CG10731 | 1 | 3 | agEG15185 | 1 | 2R;19B |
| CI30 | Complex I intermediate- associate protein 30 | CG7598 | 2 | 3R;99B9 | FBgn0039689 | Dpse\CG7598 | 2 | 2 | agEG7818 | 2 | X;5A |
| CYC | Cytochrome C | Cyt-c-p | 1 | 2L;36A11 | FBgn0000409 | Dpse\CG17903 | 1 | 4 | agEG17602 | 1 | 3R;34C |
| COXZ | Complex IV assembly protein COX11 | CG6922 | 1 | 2L;25E5 | FBgn0031712 | Dpse\CG6922 | 1 | 4 | agEG19985 | 2 | 3L;38B |
| COXS | Complex IV copper chaperone | CG9065 | 2 | X;13A9 | FBgn0030610 | Dpse\CG9065_1 | 2 | XL | agEG23169 | 1 | 3L;44C |
| OXA1 | Biogenesis protein OXA1 | CG6404 | 3 | 3L;67F1 | FBgn0027615 | Dpse\CG6404 | 3 | XR | agEG11581 | 3 | 2L;22C |
| ETFA | Electron transfer flavoprotein alpha subunit | wal | 3 | 2R;48C1-2 | FBgn0010516 | Dpse\CG8996 | 3 | 3 | agEG11798 | 2 | 2R;17B |
| ETFB | Electron transfer flavoprotein beta subunit | CG7834 | 2 | 3R;99C1 | FBgn0039697 | Dpse\CG7834 | 2 | 2 | agEG13614 | 2 | 2R;19D |
| ETFD | Electron transfer flavoprotein- ubiquinone oxidoreductase | CG12140 | 5 | 2R;46C4 | FBgn0033465 | Dpse\CG12140 | 5 | 3 | agEG10998 | 4 | 2L;23B |
| COXX | Protoheme IX farnesyltransferase | CG5037 | 4 | 2L;31D9 | FBgn0032222 | Dpse\CG5037 | 3 | ND | agEG11452 | 4 | 3R;32B |
| SCO1 | Sco1 protein homolog | CG8885 | 2 | 2L;25B5 | FBgn0031656 | Dpse\CG8885 | 2 | 4 | agEG10475 | 1 | 3R;31C |
| SUR1 | Surfeit locus protein 1 | Surf1 | 4 | 3L65D4 | FBgn0029117 | Dpse\CG9943 | 4 | XR | agEG8998 | 4 | 2L;25C |
*IDs in this column are taken from Swiss-Prot [20]. †Only coding exons were considered. ND, map position not determined. D. melanogaster, D. pseudoobscura and A. gambiae sequences used to determine intron-exon gene structures are available as supplementary material at the MitoComp website [22]
Amino-acid sequence comparison
For the products of the OXPHOS genes investigated, the D. melanogaster/D. pseudoobscura average amino-acid sequence identity is 88%, compared to 64% between D. melanogaster and A. gambiae. Figure 1 shows the frequency distribution of sequence identities, and Additional data file 1 lists all pairwise identity values between the products of the 78 OXPHOS genes when orthologous D. melanogaster/D. pseudoobscura, D. melanogaster/A. gambiae and D. melanogaster/human gene products are compared. A multiple alignment of each cluster of homologous proteins is shown at the MitoComp website [22].
Figure 1.
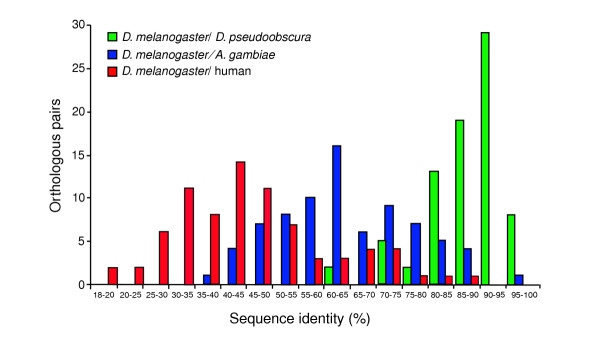
Histogram of pairwise sequence identities between the unprocessed products of 78 orthologous D. melanogaster, D. pseudoobscura, A. gambiae and human OXPHOS genes.
It should be kept in mind that identity values reported in Figure 1 and in the table in Additional data file 1 were calculated on the whole sequence of the predicted unprocessed proteins; they are much higher if the putative amino-terminal pre-sequences are excluded, since such sequences, possessed by most mitochondrion-targeted products, show little amino-acid sequence conservation [23,24], although they do share specific physicochemical properties [25,26]. When only the predicted mature protein is considered, the average percentage identity increases to 90% between D. melanogaster and D. pseudoobscura, and to 70% between D. melanogaster and A. gambiae.
A striking example of evolutionary conservation is provided by the genes encoding cytochrome c (an essential and ubiquitous protein found in all organisms) in the three dipteran species: the amino-acid sequences of the gene products are identical in D. melanogaster and D. pseudoobscura, whereas 96% identity is preserved between Drosophila and Anopheles. Coding sequences are also extremely conserved, suggesting that the nucleotide sequence itself is subject to strong evolutionary constraints, maybe due to codon usage bias. Only synonymous substitutions (21 out of 108 codons) were found on comparing D. melanogaster and D. pseudoobscura cytochrome c coding sequences, whereas 28 synonymous substitutions and only four nonsynonymous substitutions were observed between D. melanogaster and A. gambiae (see MitoComp website [22]).
Gene structure comparisons
It is well known that a given function may be supplied in different species by genes that are not directly derived from a common ancestor, that is, by paralogous, not orthologous, genes. Therefore, we thought it would be interesting to compare the structural organization of the OXPHOS genes in the three species investigated, on the principle that it should be possible to infer derivation from a common ancestor, that is, 'structural orthology', if an identical or very similar overall structure was preserved. As the introns of the putative orthologous OXPHOS genes in the three species are, as expected, too divergent in DNA sequence to be aligned, we used conservation of number of introns, conservation of their location in the coding sequence, and preservation of the reading frame with respect to the flanking exons as our primary criteria.
With the only exception of Dpse\CG5037, putatively encoding protoheme IX farnesyltransferase, whose 5' genomic sequence was impossible to find in the relevant contig assembly, all other investigated D. pseudoobscura genes show a structural organization almost identical to that of their D. melanogaster counterparts. Of the 78 Anopheles genes studied, 39 maintain the structural organization observed in Drosophila, whereas gain or loss of introns occurred in 33, and in six the location of introns is not preserved at all. In agreement with a previous report [4], the intron-exon structure of the gene appears to be conserved in all three dipteran species when splicing of alternative coding exons occurs: the alternative splice forms of both the Drosophila NADH-ubiquinone oxidoreductase acyl carrier protein (mtacp1, CG9160) [27] and the Drosophila ATP synthase epsilon chain (sun, CG9032) [19] have very similar counterparts in Anopheles, as shown by genomic structure comparison, alignment of splice variants and EST mapping (Figure 2).
Figure 2.
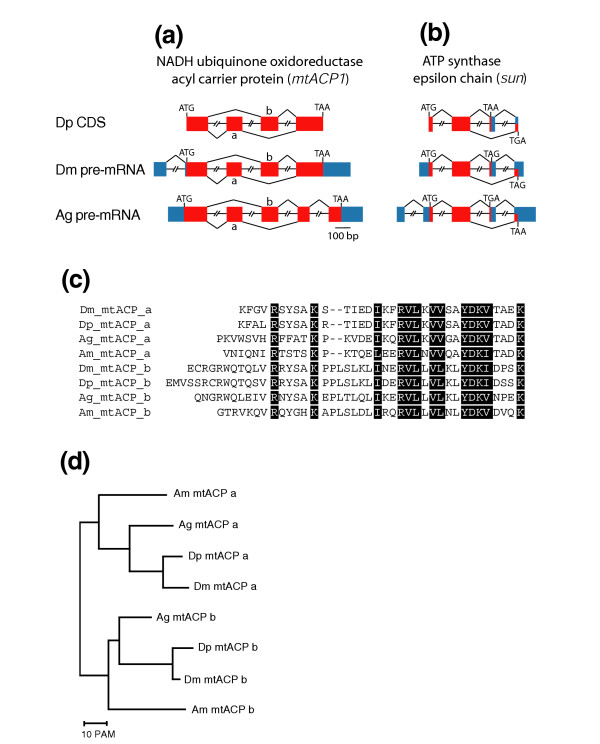
Conservation of alternative splice variants of two OXPHOS genes in D. melanogaster, D. pseudoobscura and A. gambiae. (a,b) Schematic representation and comparison of intron-exon structure of the genes encoding the NADH ubiquinone-oxidoreductase acyl carrier protein and the ATP synthase epsilon chain in D. pseudoobscura (Dp), D. melanogaster (Dm) and A. gambiae (Ag). Coding exons are represented by red boxes and untranslated UTRs by blue boxes. Introns are not drawn to scale. Because no sufficient information is available about the transcribed non coding sequences of D. pseudoobscura, only the coding exons of the D. pseudoobscura genes are shown. mtacp1 exons duplicated in tandem are labelled 'a' and 'b'. (c) alignment of the amino-acid sequences encoded by the duplicate a and b exons of the mtacp1 gene in D. melanogaster (Dm), D. pseudoobscura (Dp), A. gambiae (Ag) and A. mellifera (Am). Residues conserved in both exons are shown in white on a black background. (d) Dendrogram showing the phylogenetic relationships between the duplicated exon DNA sequences used for the alignment shown in (c). The neighbor-joining tree derived from distance matrix analysis was constructed using MultAlin [62]. Other tree-construction methods produced similar results. PAM, percent point accepted mutations.
Genes encoding the acyl carrier protein (mtacp1) in the three species are characterized by the mutually exclusive use of homologous exons that are repeated in tandem (Figure 2a). The duplicate exons occur at the same location in the aligned amino-acid sequences, and are flanked on both sides by a phase 1 intron. When the sequences of the duplicated exons are compared, they show the expected divergence pattern (that is, the similarity between duplicate exons within a gene is less than the similarity of each exon to its equivalent in the orthologous gene). Evidence from genomic and transcribed sequences (GenBank accession numbers BI510891 and BI508135) shows that the duplicated mtacp1exons are also preserved in the more distantly related insect Apis mellifera (honeybee) (Figure 2c,d), indicating a specific adaptive benefit for this gene structure, as also suggested by the evolutionary convergence leading to the occurrence of alternative splicing in members of three different ion-channel gene families from Drosophila to humans [28]. However, there is no evidence from ESTs that duplicated mtacp1 exons undergo alternative splicing in vertebrates and nematodes.
Analysis of intron length
Interspecies comparison of the introns of putative orthologous genes indicates that there is little constraint on their nucleotide sequence, which undergoes nucleotide substitutions at a rate comparable to that of pseudogenes [29]. However, several observations suggest that intron size is subject to natural selection. For example, in D. melanogaster and several other organisms the distribution of intron length has been shown to be asymmetrical, with a large group of introns falling into a narrow distribution around a 'minimal' length and the remaining showing a much broader length distribution, ranging from hundreds to thousands of base-pairs [30-32].
Of the introns that interrupt the coding sequence in the 78 OXPHOS genes investigated in the present study, 88 (64.7%) of 136 in D. melanogaster, 96 (70.5%) of 136 in D. pseudoobscura and 87 (67.9%) of 128 in A. gambiae fall into the short-size class (Figure 3a). However, in A. gambiae the length distribution of these introns appears slightly broader (62-150 bp, compared with 51-100 bp in both Drosophila species). The remaining introns show a broad length distribution, ranging from 151 to 4,702 bp with no clear boundary between classes.
Figure 3.
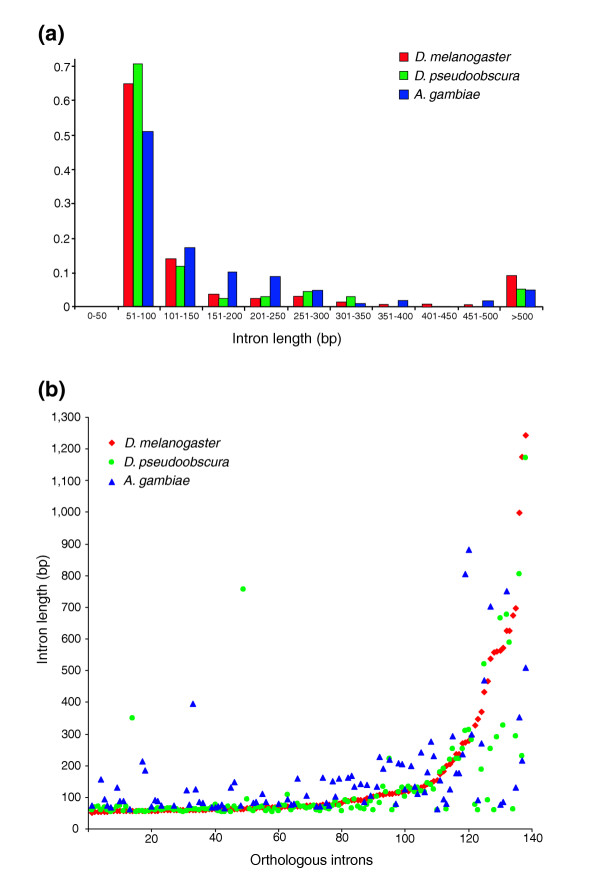
Length distribution of OXPHOS gene introns. (a) Length distribution of the 400 introns interrupting the coding sequence in the 78 D. melanogaster, D. pseudoobscura and A. gambiae OXPHOS genes investigated. (b) Comparison of the orthologous introns in the three species. Length of 138 D. melanogaster introns plotted in ascending length order was compared with the length of the 138 D. pseudoobscura orthologous introns and with the length of 98 orthologous A. gambiae introns. Note that length class shifts are rare.
A comparison of the length of introns in corresponding positions in the putative D. melanogaster, D. pseudoobscura and A. gambiae orthologs suggests that changes from the short-size to the long-size (more than 300 bp) intron class, or the converse, have been rare in the evolutionary history of these species: only seven class changes were observed comparing D. melanogaster and D. pseudoobscura introns, and six between D. melanogaster and A. gambiae (Figure 3b). On the whole, our data confirm the highly asymmetrical intron length distribution in D. melanogaster and extend this finding to the introns of the D. pseudoobscura and A. gambiae OXPHOS genes.
OXPHOS gene duplications
It is generally accepted that gene duplication is the basic process that underlies the diversification of genes and the origination of novel gene functions [33]; however, many features of this process are still elusive. To obtain more information on the molecular evolution of the genes involved in the OXPHOS system, we searched the genomes of D. melanogaster, D. pseudoobscura and A. gambiae for duplications of the 78 OXPHOS genes whose orthologs we have identified in the three species.
Duplicate gene pairs were tentatively identified within each genome as best reciprocal hits with an E-value of less than 10-20 in both directions in a TBLASTN search using the default parameters. Deciding whether two proteins may be considered homologous becomes difficult when their sequence identity is within the 20-30% range (the so-called 'twilight zone' [34]), and so the following additional criteria were used: first, the two sequences could be aligned over more than 60% of their length; second, the putative processed proteins encoded had to have more than 40% identity; and third, amino-acid percentage similarity had to be larger than percentage identity [35]. Even if meeting these criteria and reported as different genes in the ENSEMBL database [36], identical Anopheles nucleotide sequences were excluded from further analysis, as they are likely to reflect annotation artifacts.
Duplications, or in some instances triplications, of 24 OXPHOS genes were found. Overall, we identified 47 genes (20 in D. melanogaster, 19 in D. pseudoobscura and eight in A. gambiae) each of which shows significant similarity with one of the 78 OXPHOS genes reported above. When the structure of a member of a paralogous gene set indicates that it has been produced by retroposition, it seems reasonable to assume that it is derived from a pre-existing 'parent' gene. For duplicates not clearly originating by retroposition, we also assume, on the basis of the much higher level of conservation and expression, that the genes we find to be the structural orthologs in all three species are the parent ones, and in this case also we will henceforth refer to their paralogs as OXPHOS gene duplicates. The amino-acid percentage identity between the products of duplicate gene pairs ranges from 40% to 85%. For each of the OXPHOS gene duplicates, cytological localization, number of exons interrupting the coding sequence, and number of ESTs found in the D. melanogaster and A. gambiae EST databases are reported in Table 2. Neighbor-joining trees derived from distance matrix analysis and showing the inferred evolutionary relationship between members of each gene cluster are available at the MitoComp website [22].
Table 2.
OXPHOS gene duplications in the genomes of D. melanogaster, D. pseudoobscura and A. gambiae
| Protein name | D. melanogaster gene name | Number of exons | Number of ESTs* | Map position | D. pseudoobscura gene name | Number of exons | Map position | A. gambiae gene name | Number of exons | Number of ESTs | Map position |
| Complex I: NADH:ubiquinone oxidoreductase | |||||||||||
| 18 kDa subunit | CG12203 | 3 | X;18C7 | Dpse\CG12203.1 | 3 | XL | agEG18985 | 4 | 2L;27A | ||
| Dpse\CG12203.2 | 3 | 2 | |||||||||
| 20 kDa subunit | CG9172 | 1 | 36 (1) | X;14A5 | Dpse\CG9172 | 1 | ND | agEG16939 | 1 | 47 | X;4A |
| CG2014 | 1 | 0 | 3R;99B2 | Dpse\CG2014 | 1 | 2 | agEG12298 | 1 | 2 | 2R;14D | |
| 24 kDa subunit | CG5703 | 3 | 33 (2) | X;16B10 | Dpse\CG5703 | 3 | XL | agEG16953 | 5 | 2R;11A | |
| CG6485 | 1 | 4 (4) | 3L;74A4 | Dpse\CG6485 | 1 | XR | |||||
| 49 kDa subunit | CG1970 | 6 | 47 (0) | 4;102C2 | Dpse\CG1970 | 6 | ND | agEG18856 | 1 | 38 | X;1B |
| CG11913 | 2 | 0 | 3R;96D2 | Dpse\CG11913 | 2 | 2 | agEG19332 | 1 | 0 | 2L;26B | |
| 51 kDa subunit | CG9140 | 4 | 135 (2) | 2L;26B6-7 | Dpse\CG9140 | 4 | 4 | agEG9927 | 4 | 3R;36D | |
| CG11423 | 1 | 4 (4) | 2R;54C12 | Dpse\CG11423 | 1 | 3 | |||||
| CG8102 | 2 | 3 (3) | 2R;51F3-4 | Dpse\CG8102 | 2 | 3 | |||||
| B14.5A subunit | CG3621 | 2 | 16 (1) | X;2D6-E1 | Dpse\CG3621 | 2 | XL | agEG14707 | 4 | 2R;17A | |
| CG6914 | 1 | 3 (3) | 3L;79F2 | Dpse\CG6914 | 1 | XR | |||||
| Complex II: Succinate dehydrogenase | |||||||||||
| Flavoprotein subunit | Scs-fp | 4 | 54 (0) | 2R; 56D3 | Dpse\CG17246 | 4 | 3 | agEG7754 | 3 | 3L:38B | |
| CG5718 | 1 | 5 (14) | 3L;68E3 | Dpse\CG5718 | 1 | XR | |||||
| Iron-sulfur protein | SdhB | 3 | 83 (0) | 2R;42D3-4 | Dpse\CG3283 | 3 | 3 | agEG13539 | 4 | 2L;27D | |
| CG7349 | 3 | 14 (12) | X;17F3 | Dpse\CG7349 | 1 | XL | |||||
| Complex III: Ubiquinol-cytochrome c reductase | |||||||||||
| Cytochrome C1, heme protein | CG4769 | 6 | 246 (3) | 3L;64C13 | Dpse\CG4769 | 6 | XR | agEG19223 | 4 | 2L;26C | |
| CG14508 | 1 | 7 (7) | 3R;99A1 | Dpse\CG14508 | 1 | 2 | |||||
| 11 kDa protein | Ucrh | 2 | 16 (0) | 3R | Dpse\Ucrh | 2 | 2 | agEG19398 | 2 | 2R;11B | |
| CG30354 | 1 | 1 (1) | 2R;44E2 | ||||||||
| 14 kDa protein | CG3560 | 3 | 8 (0) | X;14B10 | Dpse\CG3560 | 3 | XL | agEG11611 | 2 | 3L;46A | |
| CG17856 | 1 | 0 | 3R;98C3 | ||||||||
| Core protein 1 | CG3731 | 6 | 3R;88D6 | Dpse\CG3731 | 6 | 2 | agEG21302 | 3 | 56 | X;5C | |
| agEG10358 | 1 | 2 | 2R;9A | ||||||||
| gEG15332 | 1 | 46 | 2L;22D | ||||||||
| Core protein 2 | CG4169 | 4 | 3L;73A10 | Dpse\CG4169.1 | 4 | XR | agEG17930 | 4 | 2L;24A | ||
| Dpse\CG4169.2 | 1 | XR | |||||||||
| Complex IV: Cytochrome c oxidase | |||||||||||
| Subunit IV | CG10664 | 2 | 138 (3) | 2L;38A8 | Dpse\CG10664 | 2 | 4 | agEG13327 | 2 | 3R;31C | |
| CG10396 | 1 | 9 (7) | 2R;41F3 | Dpse\CG10396.1 | 1 | 2 | |||||
| Dpse\CG10396.2 | 1 | XL | |||||||||
| Polypeptide VB | CG11015 | 3 | 41 (0) | 2L;26E3 | Dpse\CG11015 | 3 | 4 | agEG8633 | 4 | 3R;31C | |
| CG11043 | 2 | 4 (4) | 2L;26E3 | Dpse\CG11043 | 2 | 4 | |||||
| Polypeptide VIA | CG17280 | 2 | 90 (2) | 2R;59E3 | Dpse\CG17280 | 2 | 3 | agEG7821 | 2 | 63 | X;5A |
| CG30093 | 1 | 1 (1) | 2R;52D3 | agEG4851 | 1 | 0 | 3R;32A | ||||
| Polypeptide VIIA | CG9603 | 2 | 30 (0) | 3R;84F13 | Dpse\CG9603 | 2 | XR | agEG17423 | 3 | X;4B | |
| CG18193 | 2 | 4 (4) | 3R;84F13 | ||||||||
| Complex V: ATP synthase | |||||||||||
| Beta chain | ATPsyn-beta | 3 | 484 (6) | 4;102D1 | Dpse\CG11154 | 3 | ND | agEG14379 | 1 | 3L;45C | |
| CG5389 | 3 | 3 (3) | 3L;72D5-6 | Dpse\CG5389 | 3 | XR | |||||
| Epsilon chain | sun | 4 | 11 (0) | X;13F12 | Dpse\CG9032 | 4 | ND | agEG10095 | 4 | 15 | X;3D |
| CG12810 | 1 | 0 | 3R;85F11 | agEG20782 | 4 | 6 | 3R,34C | ||||
| agEG8173 | 1 | 0 | 2L;21D | ||||||||
| G chain | l(2)06225 | 2 | 90 (6) | 2L;32C1 | Dpse\CG6105 | 2 | ND | agEG8590 | 2 | 3R;34B | |
| CG7211 | 2 | 1 (1) | 2L;28C2 | Dpse\CG7211 | 2 | 4 | |||||
| Coupling factor 6 | ATPsyn-Cf6 | 2 | 55 (0) | 3R;94E13 | Dpse\CG4412 | 2 | 2 | agEG19097 | 2 | 2R;19D | |
| CG12027 | 2 | 2 (2) | 3L;64C4 | Dpse\CG12027 | 1 | XR | |||||
| Lipid-binding protein P1 | CG1746 | 3 | 3R;100B7 | Dpse\CG1746 | 3 | 2 | agEG14837 | 3 | 408 | X;2B | |
| agEG12441 | 3 | 4 | 3L;42A | ||||||||
| Others | |||||||||||
| Complex IV, copper chaperone | CG9065 | 2 | X;13A9 | Dpse\CG9065.1 | 2 | XL | agEG23169 | 1 | 3L;44C | ||
| Dpse\CG9065.2 | 1 | 4 | |||||||||
| Cytochrome c | Cyt-c-p | 1 | 134 (0) | 2L;36A11 | Dpse\CG17903 | 1 | 4 | agEG17602 | 1 | 3R;34C | |
| Cyt-c-d | 1 | 25 (25) | 2L;36A11 | Dpse\CG13263 | 1 | 4 | |||||
*The number of ESTs in testis-derived libraries is in parentheses. Because insufficient information on D. pseudoobscura ESTs is available in the public EST databases, only D. melanogaster and A. gambiae ESTs were considered. Bold type is used to identify the putative orthologous genes in the three species (see text). Only coding exons were considered. ND, location not determined. D. melanogaster, D. pseudoobscura and A. gambiae OXPHOS sequences used are available at the MitoComp website [22]
Duplications (or triplications) of 16 of the 78 OXPHOS genes investigated were found in both D. melanogaster and D. pseudoobscura. In such cases, to assign pairwise orthology, besides taking into account conservation of structural organization, given the general conservation of microsyntenic gene order in the two species, we used the products of D. melanogaster genes flanking the duplicate loci to search for homologous sequences also flanking the same genes in the D. pseudoobscura genome.
The genomic organization of many OXPHOS duplicates shows that they were originated by retropositional events, because they are intronless, or have only very few introns that are likely to have been inserted into the coding sequence after the duplication event. In other cases, duplication apparently resulted from transposition of genomic DNA sequences or from recombinational events, as duplicate genes maintain an identical or very similar structural organization.
On the basis of the presence of the duplication in both species, supported by evidence from evolutionary trees and conservation of microsyntenic gene order, it can be inferred that 15 of the duplications identified occurred before the D. melanogaster/D. pseudoobscura divergence (about 46 million years ago). On the other hand, five duplications were found only in D. melanogaster and four only in D. pseudoobscura; in these instances, if the duplication occurred before the divergence of the two species, it has been followed by loss of one of the copies in the lineage leading to the species in which the gene is no longer duplicated. On the assumption that the rate of gene duplication is constant over time, this translates to approximately 0.0014 duplications per gene per million years (4 or 5 duplications per 78 genes per 46 million years) that achieved fixation and long-term preservation in the genome. This value is about twofold lower than the 0.0023 value calculated by Lynch and Conery [37] for the 13,601 genes of the whole genome of D. melanogaster. However, it can be argued that the rate of long-term preservation in the genome of OXPHOS gene duplicates cannot be meaningfully compared with the general rate of preservation of duplicates in the whole genome since, while recent data suggest that in eukaryotic genomes there is preferential duplication of conserved proteins [38], duplicates of genes that encode subunits of multiprotein complexes, as most of the genes we have investigated do, negatively influence the fitness of an organism [39], and are therefore unlikely to become fixed in the population. In summary, it appears reasonable to assume that the preservation in the genome of OXPHOS gene duplicates should occur very infrequently, unless special mechanisms allowing their fixation in the population are present (see the next section).
In A. gambiae we found only four duplications and two triplications of the OXPHOS genes analyzed; of these, four involve genes also duplicated in one or both Drosophila species (Table 2). Pairwise orthology could not be assigned between Drosophila and Anopheles gene duplicates as neither microsynteny nor evolutionary trees provide sufficient evidence for the origin of the gene pairs from a single-copy gene before the Drosophila/Anopheles divergence.
Expression pattern of OXPHOS gene duplicates
The relative abundance of ESTs in a EST library may be assumed roughly to reflect the level of expression of each mRNA in the tissues from which the library was prepared. We therefore used the mRNA sequences predicted in silico to be transcribed from the OXPHOS duplicate genes investigated in this work as queries in a search of the public D. melanogaster and A. gambiae EST databases to infer the relative abundance of the mRNA copies from the hits scored. For each gene, the number of ESTs found in the databases is detailed in Table 2. With the exception of one of the paralogs of the A. gambiae gene encoding ubiquinol-cytochrome c reductase core protein 1, in all cases the search found the number of ESTs originating from the duplicate gene was strikingly lower than that originating from the putative parent gene, in both D. melanogaster and A. gambiae (in total, 100 versus 1,747 in D. melanogaster and 60 versus 687 in A. gambiae). A smaller number of ESTs originating from the OXPHOS gene duplicates was observed even in A. gambiae EST libraries that are normalized. Remarkably, and regardless of the mechanism of the duplication, in D. melanogaster, in which several organ-specific or developmental stage specific libraries are available, the search showed that the expression of the OXPHOS gene duplicates is strongly testis-biased, as 97 out of the 100 ESTs originating from them were found in testis-derived libraries, while only 27 out of the 1,769 ESTs originating from the parent genes were found in such libraries, the bulk of them being instead found in libraries derived from embryos or somatic tissues.
Our finding that the expression of the OXPHOS gene originated by duplication is strongly testis-biased is validated by the data obtained by Parisi et al. [40] using the FlyGEM microarray to identify D. melanogaster genes showing ovary-, testis- or soma-biased expression. With the exception of CG7349, CG30354, CG30093 and CG12810, for which no data were presented by Parisi et al. [40], all other genes reported in this work as OXPHOS gene duplicates were found in the genomic fraction showing testis-biased expression, whereas all the parent genes present in the dataset showed soma-biased expression. Additional data file 2 summarizes the relevant data extracted from Parisi et al. [40].
The pattern of strongly testis-biased expression of OXPHOS gene duplicates holds for a further sample of 40 duplications of genes annotated in the MitoDrome database [19] as encoding products that are mitochondrion-targeted but not involved in the OXPHOS system. For 15 of these no data are provided by Parisi et al. [40], but all the remaining 25 genes show a testes-biased expression (data not shown).
Duplications of genes encoding OXPHOS subunits, for which stoichiometry is important, are likely to be strongly deleterious owing to the negative consequences of an imbalance in the concentration of the respiratory complex constituents, unless, as proposed by Lynch and Force [41], 'subfunctionalization' and/or a differential expression pattern of duplicate copies occurs. In this case, the duplicate OXPHOS genes would have a reduced or absent capacity to functionally complement mutations in their parent genes, in contrast to what is generally assumed to be the main short-term advantage of gene duplication. In D. melanogaster at least there is evidence for this, as FlyBase [42] and BDGP P-Element Gene Disruption Project [43] searches for P-insertion mutants in the D. melanogaster OXPHOS genes found that lethal alleles for 11 out of 19 D. melanogaster parent genes are known (see the MitoComp website [22]), indicating that loss-of-function of the parent gene cannot be compensated for by the presence of the gene duplicate. P-insertion mutants with an abnormal phenotype, indicating a functional divergence, are known for only one of the D. melanogaster OXPHOS gene duplicates - Cyt-c-d, encoding cytochrome c). Interestingly, although Cyt-c-d is adjacent to its putative parent gene, Cyt-c-p, it shows a different pattern of expression, suggesting that the two genes must be regulated at individual gene level and not at chromatin domain level (see Table 2).
A systematic investigation of the expression pattern of other D. melanogaster duplicate genes will be necessary to answer the question of whether the testis-biased expression pattern reported here is specific to the duplicates of genes encoding mitochondrial proteins, or is a more general phenomenon. According to the balance hypothesis, validated by experimental results obtained on yeast [39], single gene duplications involving genes encoding components of multiprotein complexes are expected to severely affect fitness. Therefore, the expression pattern we have observed could be a necessary condition to maintain some gene duplicates in the D. melanogaster genome, at least until they evolve a new useful function. Finally, as nothing is known about the tissue-specific pattern of expression of the genes investigated in D. pseudoobscura and Anopheles, it also remains unclear whether the testis-biased expression of gene copies originated by duplication is specific to D. melanogaster, or is also to be found in other dipterans, and possibly in other organisms.
Codon usage in the OXPHOS genes
Because of the preferential use of codons ending in C or G, the D. melanogaster coding sequences have an average GC content higher than the genomic average [44,45]. This is also true for the 78 D. melanogaster OXPHOS coding sequences reported in this work and for their D. pseudoobscura and A. gambiae counterparts (68% of the codons in the OXPHOS genes end in C or G in D. pseudoobscura and 77% in A. gambiae, compared to 74% in D. melanogaster). In all three species, the coding sequences of OXPHOS gene duplicates show a lower percentage of codons ending in C or G, when compared to both the entire set of 78 orthologous OXPHOS genes and the gene subset including only their parent genes. In samples including all the OXPHOS gene duplicates annotated in this paper the aggregate percentage of C- or G-ending codons is 63%, 46% and 73% in D. melanogaster, D. pseudobscura and A. gambiae respectively, as compared with 70%, 64% and 88% in their parent genes. In D. pseudoobscura, the shift toward a higher percentage of A- or T-ending codons is also detected in the pattern of synonymous codon usage; for 12 of the 18 amino acids that are encoded by more than one codon, the most frequently used codon in the D. pseudoobscura gene duplicates is different from the one used in their parent genes (see Additional data file 3).
Chromosomal arm location, interarm homology and microsynteny
It has been reported that in many eukaryotes including yeast [46], C. elegans [47], D. melanogaster [48,49] and humans [50], genes with related functions and similar expression patterns tend to be clustered, suggesting that they share aspects of transcriptional regulation depending on their inclusion in the same chromatin domain. In particular, Boutanaev et al. [48] reported that in D. melanogaster clusters of three or more testis-specific genes are much more frequent than expected by chance. Therefore, we investigated the chromosomal distribution of the OXPHOS genes to determine whether clustering could be detected. In all three dipteran species considered, the 78 OXPHOS orthologous genes are randomly distributed on all chromosomal arms (Table 1). Two D. melanogaster genes (Ucrh, encoding the 11 kDa subunit of ubiquinol-cytochrome c reductase, and CG40002, encoding the AGGG subunit of NADH-ubiquinone oxidoreductase) have a heterochromatic location.
No evidence of OXPHOS gene duplicate clustering was found either, despite the common testis-biased expression of such genes. Moreover, no evidence of clustering with other testis-specific genes was found when an EST database search for such genes was performed in the regions flanking the investigated gene duplicates.
However, in accord with two studies reporting a significant deficit of genes with a male-biased expression on the D. melanogaster X chromosome [51,52], only one out of the 20 D. melanogaster OXPHOS gene duplicates, two out of 19 in D. pseudoobscura and none (out of eight) in A. gambiae were found to be X-linked (Table 2). It may be that duplications of X-linked genes encoding OXPHOS subunits would be especially deleterious because of the male X chromosome transcriptional hyperactivity, which allows dosage compensation.
In all three dipteran species, a disproportionately high fraction of OXPHOS gene duplicates appears to be constituted of autosomal genes derived from parent genes located on the X chromosome (Table 2). As suggested by recent work on the generation and preservation of functional genes produced by retroposition both in Drosophila [53] and in the human and mouse genomes [54], this may be explained by a selective advantage for duplicates of X-linked genes that move to an autosomal location and so escape the X inactivation in early spermatogenesis that occurs both in Drosophila [55] and in mammals [56].
We would like to speculate that such selective advantage may be especially significant for duplicates of OXPHOS genes, given the heavy reliance of sperm on mitochondrial function. In fact, the excess of autosomal duplicates of X-linked genes is not observed for MitoDrome annotated genes not involved in the OXPHOS system (see above). However, as the general pattern of much lower, testis-biased expression holds even for OXPHOS and other mitochondrial gene duplicates that apparently derive from autosomal parental genes, and even for X-linked duplicates, this pattern (and the explanation of the evolutionary preservation of such genes) cannot only be due to the selective advantage of escaping X inactivation during spermatogenesis.
With the exception of CG9603, all euchromatic D. melanogaster orthologs maintain their localization on the homologuos D. pseudoobscura chromosomal arm (Table 3). CG9603, encoding the VIIa polypeptide of cytochrome c oxidase, is located on the 3R chromosomal arm in D. melanogaster, whereas Dpse\CG9603, its counterpart in D. pseudoobscura, is located on XR; microsyntenic gene order with the flanking genes is conserved in both species, suggesting that a chromosomal rearrangement occurred after their divergence.
Table 3.
Chromosomal location and interarm homology of the orthologous D. melanogaster, D. pseudoobscura and A. gambiae OXPHOS genes
| D. pseudoobscura chromosomal arm | A. gambiae chromosomal arm | |||||||||||
| 2 | 3 | XL | XR | 4 | ND | 2L | 2R | 3L | 3R | X | ND | |
| D. mel. 2L 18→ | 16 | 2 | 1 | 1 | 16 | |||||||
| D. mel. 2R 15→ | 15 | 6 | 3 | 4 | 1 | 1 | ||||||
| D. mel. 3L 12→ | 12 | 7 | 2 | 3 | ||||||||
| D. mel. 3R 16→ | 15 | 1 | 1 | 5 | 4 | 1 | 5 | |||||
| D. mel. X 14→ | 11 | 3 | 3 | 4 | 2 | 2 | 3 | |||||
| D. mel. 4 2→ | 2 | 1 | 1 | |||||||||
| D. mel. ND 1→ | 1 | 1 | ||||||||||
The first column shows the distributions of the OXPHOS genes on D. melanogaster chromosomal arms (D. mel). Arrows show the direction of counting; D. melanogaster → D. pseudoobscura or D. melanogaster → A. gambiae. Bold type is used when inter arm homology is conserved betweeen two species. Note that Dm 2L, Ag 3R is the only correspondence between D. melanogaster and A. gambiae chromosomal arms. ND, location not determined.
OXPHOS gene duplicates also almost always maintain the same chromosomal location and microsyntenic gene order in D. melanogaster and in D. pseudoobscura. However, a more complex situation was observed with regard to the gene encoding subunit IV of cytochrome c oxidase, which is duplicated in D. melanogaster and triplicated in D. pseudoobscura (Table 2). On the basis of identical genomic organization, conserved chromosomal location and mycrosyntenic gene order Dpse\CG10664 is inferred to be the ortholog of D. melanogaster CG10664. Dm CG10396, Dpse\CG10396.1 and Dpse\CG10396.2 are intronless, and neither interarm homology nor microsyntenic order offer any clue to their phylogenetic relationship. The dendrogram based on sequence divergence (see the MitoComp website [22], complex IV, subunit IV) suggests, however, that a duplication event occurred before the D. melanogaster/D. pseudobscura speciation, originating the CG10664-CG10396 gene pair (Dpse\CG10664-Dpse\CG10396 in D. pseudoobscura). A further duplication event, occurring in the D. pseudoobscura lineage after the D. melanogaster/D. pseudoobscura divergence, probably created the Dpse\CG10396.1-Dpse\CG10396.2 gene pair.
In contrast to the maintained location of almost all investigated genes on homologous chromosomal arms in the two Drosophila species, when D. melanogaster and A. gambiae are compared the only meaningful correspondence found concerns the genes on the D. melanogaster 2L and the A. gambiae 3R chromosomal arms (Table 3). This result is consistent with previous reports that compared the location of homologous genes in D. melanogaster and A. gambiae, concluding that extensive reshuffling both within and between chromosomal regions has occurred since the divergence of the two species [4,17].
Conclusions
We have catalogued 78 nuclear genes that control oxidative phosphorylation in three dipteran species and compiled a web-based dataset, MitoComp [22], that contains all the data on which this article is based and which is available with the online version of this article. We have conducted only some basic comparative analyses of the many which are possible using such a dataset, and it is our hope that it will provide a valuable resource for those looking for information about nuclear genes encoding mitochondrion-targeted products in the context of functional genomics and proteomics. Future studies based on this information, especially if the comparative analysis is extended to other species, will surely allow a better understanding of the evolutionary history of a set of genes that control a basic biological function, and also offer interesting insights into the mechanisms of their coordinated expression. In fact, a first in silico analysis of the D. melanogaster and D. pseudoobscura nuclear energy gene sequences suggests that a genetic regulatory circuit, based on a single regulatory element, coordinates the expression of the whole set of energy-producing genes in Drosophila [57].
The comparative analysis of the 78 OXPHOS genes in the three dipteran species shows a high level of amino-acid sequence identity, as well as a substantial conservation of intron-exon structure, indicating that these genes are under strong selective constraints. An unexpected and intriguing result of this study is that in D. melanogaster, duplication-originated OXPHOS genes are expressed at a much lower level (or possibly not expressed at all) in most or all the tissues where their parent genes are expressed, as judged by the abundance of ESTs derived from their transcripts in all libraries other than those derived from testis. On the other hand, OXPHOS gene duplicates have a strongly testis-biased pattern of expression, a finding validated by other authors with a different approach based on the use of microarrays [40]. In A. gambiae, although no testis-specific ESTs databases are available, a pattern of expression of almost all duplicate OXPHOS genes different from that of the gene from which they originated, and possibly limited to specific tissues, is suggested by the fact that in all EST libraries available the abundance of the sequences originated from the duplicate genes is very low when compared with that of the sequences derived from their respective parent genes.
We suggest that, at least in D. melanogaster, the acquisition of a new, testis-biased pattern of expression may be required to maintain duplicates of certain genes in the genome. This may also allow rapid acquisition of new functions by the gene product(s), as it has recently been shown that proteins encoded by duplicated genes with a changed expression pattern often show accelerated evolution [58,59]. Subfunctionalization could then further favor the preservation of multiple paralogous genes.
No data are at present available to support the possibility that our findings could be extrapolated to other gene sets or even to the whole genome. However, we propose that duplication of the genes encoding products that are part of multiprotein complexes may be especially deleterious, unless sequence divergence allowing only testis-specific expression of one of the duplicate copies occurs. In turn, this could facilitate the development of novel functions, which is usually assumed to be the main evolutionary advantage of gene duplication, providing a general mechanism for originating phenotypic changes that might also lead to species differentiation.
Materials and methods
To identify orthologous OXPHOS genes and their duplications in D. pseudoobscura and A. gambiae, contigs from BCM [13] and scaffolds from AnoBase [21] were searched using TBLASTN with the D. melanogaster OXPHOS peptides listed in the MitoDrome database [19] as queries.
Amino-acid sequence identity and similarity values were obtained from pairwise alignments using the Needleman-Wunsch global alignment algorithm at the EMBL-EBI server [60]. Multiple sequence alignments of the OXPHOS amino-acid and coding sequences and visualization of the dendrograms were obtained using the MultAlin 5.4.1 software [61] from MultAlin server [62].
The genomic sequence of each gene was manually searched for intron-exon boundaries and the predicted mRNA sequence reconstructed in silico. A. gambiae mRNAs were assembled by overlapping ESTs extracted from AnoBase [21].
We have named each newly identified A. gambiae gene with the four-letter code 'agEG' followed by the last four or five digits of its Ensembl [36] gene number, excluding the multiple zeros of the prefix; the D. pseudoobscura genes were named with the code 'Dpse\CG' followed by the Celera number of their D. melanogaster counterparts.
The D. pseudoobscura OXPHOS genes investigated here were assigned a chromosomal location where possible, using the putative chromosomal assignments available at BCM [13] for the majority of the large D. pseudoobscura contigs. We also utilized the Ensembl mosquito genome server [36] to identify and visualize the chromosomal location of the A. gambiae annotated OXPHOS DNA sequences.
The D. melanogaster EST database, available from the National Center for Biotechnology Information (NCBI) contains ESTs from cDNA libraries obtained from different developmental stages and body parts. The relative abundance of the transcripts of duplicate or triplicate D. melanogaster OXPHOS genes was defined by counting their cognate ESTs in non-normalized cDNA libraries generated by the Berkeley Drosophila Genome Project (BDGP) [43] from embryos (LD), larvae/pupae (LP), and adult ovary (GM), head (GH) and testes (AT), and also the ESTs from adult testes generated at the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) [63]. ESTs from BDGP normalized EST libraries generated from head (RH) and embryos (RE) were also considered. The relative abundance of the transcripts of duplicate or triplicate A. gambiae OXPHOS genes was defined by counting their cognate ESTs in all libraries recovered from the Anobase server [21]. Since the number of sequences in the EST databases changes as new EST sequences are added, our values are calculated on the EST sequences present in the databases as of July 2004.
The list of D. melanogaster P-insertion OXPHOS mutants is reported in the MitoComp website [22] and was mostly compiled using information from FlyBase [42] and from the BDGP P-Element Gene Disruption Project [43].
Additional data files
A web-based dataset, MitoComp, contains all data on which this work is based and is available at [22]. It includes information on the cytological location of each gene, its genomic organization and the structure of its transcript(s). The genomic structures of the D. melanogaster, D. pseudoobscura and A. gambiae putative OXPHOS orthologs are shown and compared, and their deduced amino-acid products are aligned with the corresponding human protein. When paralogs of the gene exist, neighbor-joining trees derived from distance matrix analysis are also shown to visualize the evolutionary relationships between them. Additional data files available with the online version of this article are as follows. Additional data file 1 contains a table that reports pairwise amino-acid sequence conservation values between the D. melanogaster OXPHOS genes investigated and their D. pseudoobscura, A. gambiae and human counterparts. Additional data file 2 contains data extracted from the Parisi et al. dataset [40]. Additional data file 3 reports the codon usage in the orthologous and duplicate OXPHOS genes of D. melanogaster, D. pseudoobscura and A. gambiae.
Supplementary Material
A table that reports pairwise amino-acid sequence conservation values between the D. melanogaster OXPHOS genes investigated and their D. pseudoobscura, A. gambiae and human counterparts
Data extracted from the Parisi et al. dataset
The codon usage in the orthologous and duplicate OXPHOS genes of D. melanogaster, D. pseudoobscura and A. gambiae
Acknowledgments
Acknowledgements
This work was supported by grants from Centro Eccellenza (CE) and Ministero dell'Istruzione, dell'Università e della Ricerca (MIUR). We thank Cecilia Saccone and Graziano Pesole for critical reading of the manuscript.
Contributor Information
Gaetano Tripoli, Email: g.tripoli@biologia.uniba.it.
Domenica D'Elia, Email: domenica.delia@ba.itb.cnr.it.
Paolo Barsanti, Email: p.barsanti@biologia.uniba.it.
Corrado Caggese, Email: caggese@biologia.uniba.it.
References
- Boffelli D, McAuliffe J, Ovcharenko D, Lewis KD, Ovcharenko I, Pachter L, Rubin EM. Phylogenetic shadowing of primate sequences to find functional regions of the human genome. Science. 2003;299:1391–1394. doi: 10.1126/science.1081331. [DOI] [PubMed] [Google Scholar]
- Kellis M, Patterson N, Endrizzi M, Birren B, Lander ES. Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature. 2003;423:241–254. doi: 10.1038/nature01644. [DOI] [PubMed] [Google Scholar]
- Bergman CM, Pfeiffer BD, Rincon-Limas DE, Hoskins RA, Gnirke A, Mungall CJ, Wang AM, Kronmiller B, Pacleb J, Park S, et al. Assessing the impact of comparative genomic sequence data on the functional annotation of the Drosophila genome. Genome Biol. 2002;3:research0086.1–0086.20. doi: 10.1186/gb-2002-3-12-research0086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zdobnov EM, Von Mering C, Letunic I, Torrents D, Suyama M, Copley RR, Christophides GK, Thomasova D, Holt RA, Subramanian GM, et al. Comparative genome and proteome analysis of Anopheles gambiae and Drosophila melanogaster. Science. 2002;298:149–159. doi: 10.1126/science.1077061. [DOI] [PubMed] [Google Scholar]
- Saraste M. Oxidative phosphorylation at the fin de siècle. Science. 1999;283:1488–1493. doi: 10.1126/science.283.5407.1488. [DOI] [PubMed] [Google Scholar]
- Skladal D, Halliday J, Thorburn DR. Minimum birth prevalence of mitochondrial respiratory chain disorders in children. Brain. 2003;126:1905–1912. doi: 10.1093/brain/awg170. [DOI] [PubMed] [Google Scholar]
- Mootha VK, Bunkenborg J, Olsen JV, Hjerrild M, Wisniewski JR, Stahl E, Bolouri MS, Ray HN, Sihag S, Kamal M, et al. Integrated analysis of protein composition, tissue diversity, and gene regulation in mouse mitochondria. Cell. 2003;115:629–640. doi: 10.1016/S0092-8674(03)00926-7. [DOI] [PubMed] [Google Scholar]
- Rubin GM, Yandell MD, Wortman JR, Gabor Miklos GL, Nelson CR, Hariharan IK, Fortini ME, Li PW, Apweiler R, Fleischmann W, et al. Comparative genomics of the eukaryotes. Science. 2000;287:2204–2215. doi: 10.1126/science.287.5461.2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiter LT, Potocki L, Chien S, Gribskov M, Bier E. A systematic analysis of human disease-associated gene sequences in Drosophila melanogaster. Genome Res. 2001;11:1114–1125. doi: 10.1101/gr.169101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green C, Brown G, Dafforn TR, Reichhart JM, Morley T, Lomas DA, Gubb D. Drosophila necrotic mutations mirror disease-associated variants of human serpins. Development. 2003;130:1473–1478. doi: 10.1242/dev.00350. [DOI] [PubMed] [Google Scholar]
- Adams MD, Celniker SE, Holt RA, Evans CA, Gocayne JD, Amanatides PG, Scherer SE, Li PW, Hoskins RA, Galle RF, et al. The genome sequence of Drosophila melanogaster. Science. 2000;287:2185–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- Celniker SE, Wheeler DA, Kronmiller B, Carlson JW, Halpern A, Patel S, Adams M, Champe M, Dugan SP, Frise E, et al. Finishing a whole-genome shotgun: release 3 of the Drosophila euchromatic genome sequence. Genome Biol. 2002;3:research0079.1–0079.14. doi: 10.1186/gb-2002-3-12-research0079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Human Genome Sequencing Center at Baylor College of Medicine: Drosophila Genome Project http://www.hgsc.bcm.tmc.edu/projects/drosophila
- Holt RA, Subramanian GM, Halpern A, Sutton GG, Charlab R, Nusskern DR, Wincker P, Clark AG, Ribeiro JM, Wides R, et al. The genome sequence of the malaria mosquito Anopheles gambiae. Science. 2002;298:129–149. doi: 10.1126/science.1076181. [DOI] [PubMed] [Google Scholar]
- Powell JR. Progress and Prospects in Evolutionary Biology: The Drosophila Model. Oxford: Oxford University Press; 1997. [Google Scholar]
- Gaunt MW, Miles MA. An insect molecular clock dates the origin of the insects and accords with palaeontological and biogeographic landmarks. Mol Biol Evol. 2002;19:748–761. doi: 10.1093/oxfordjournals.molbev.a004133. [DOI] [PubMed] [Google Scholar]
- Bolshakov VN, Topalis P, Blass C, Kokoza E, della Torre A, Kafatos FC, Louis C. A comparative genomic analysis of two distant Diptera, the fruit fly, Drosophila melanogaster, and the malaria mosquito, Anopheles gambiae. Genome Res. 2002;12:57–66. doi: 10.1101/gr.196101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomasova D, Ton LQ, Copley RR, Zdobnov EM, Wang X, Hong YS, Sim C, Bork P, Kafatos FC, Collins FH. Comparative genomic analysis in the region of a major Plasmodium-refractoriness locus of Anopheles gambiae. Proc Natl Acad Sci USA. 2002;99:8179–8184. doi: 10.1073/pnas.082235599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sardiello M, Licciulli F, Catalano D, Attimonelli M, Caggese C. MitoDrome: a database of Drosophila melanogaster nuclear genes encoding proteins targeted to the mitochondrion. Nucleic Acids Res. 2003;31:322–324. doi: 10.1093/nar/gkg123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ExPASy - Swiss-Prot and TrEMBL http://us.expasy.org/sprot
- Blast server for Anopheles sequences http://www.anobase.org/cgi-bin/blast.pl
- mitoloc_index: MITOCOMP http://www.mitocomp.uniba.it
- Schatz G, Dobberstein B. Common principles of protein translocation across membranes. Science. 1996;271:1519–1526. doi: 10.1126/science.271.5255.1519. [DOI] [PubMed] [Google Scholar]
- Voos W, Martin H, Krimmer T, Pfanner N. Mechanisms of protein translocation into mitochondria. Biochim Biophys Acta. 1999;1422:235–254. doi: 10.1016/s0304-4157(99)00007-6. [DOI] [PubMed] [Google Scholar]
- Roise D, Schatz G. Mitochondrial presequences. J Biol Chem. 1988;263:4509–4511. [PubMed] [Google Scholar]
- von Heijne G, Steppuhn J, Herrmann RG. Domain structure of mitochondrial and chloroplast targeting peptides. Eur J Biochem. 1989;180:535–545. doi: 10.1111/j.1432-1033.1989.tb14679.x. [DOI] [PubMed] [Google Scholar]
- Ragone G, Caizzi R, Moschetti R, Barsanti P, De Pinto V, Caggese C. The Drosophila melanogaster gene for the NADH:ubiquinone oxidoreductase acyl carrier protein: developmental expression analysis and evidence for alternatively spliced forms. Mol Gen Genet. 1999;261:690–697. doi: 10.1007/s004380050012. [DOI] [PubMed] [Google Scholar]
- Copley RR. Evolutionary convergence of alternative splicing in ion channels. Trends Genet. 2004;20:171–176. doi: 10.1016/j.tig.2004.02.001. [DOI] [PubMed] [Google Scholar]
- Graur D, Li W-H. Fundamentals of Molecular Evolution. 2. Sunderland, MA: Sinauer Associates Inc; 2000. [Google Scholar]
- Mount SM, Burks C, Hertz G, Stormo GD, White O, Fields C. Splicing signals in Drosophila: intron size, information content, and consensus sequences. Nucleic Acids Res. 1992;20:4255–4262. doi: 10.1093/nar/20.16.4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutsch M, Long M. Intron-exon structures of eukaryotic model organisms. Nucleic Acids Res. 1999;27:3219–3228. doi: 10.1093/nar/27.15.3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Yang Z, Kibukawa M, Paddock M, Passey DA, Wong GK. Minimal introns are not "junk". Genome Res. 2002;12:1185–1189. doi: 10.1101/gr.224602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno S. Evolution by Gene Duplication. Heidelberg, Germany: Springer-Verlag; 1970. [Google Scholar]
- Doolittle RF. Of URFs and ORFs: A Primer on How to Analyze Derived Amino Acid Sequences. Mill Valley, CA: University Science Books; 1986. [Google Scholar]
- Rost R. Twilight zone of protein sequence alignments. Protein Eng. 1999;12:85–94. doi: 10.1093/protein/12.2.85. [DOI] [PubMed] [Google Scholar]
- Ensembl mosquito genome server http://www.ensembl.org/Anopheles_gambiae
- Lynch M, Conery JS. The evolutionary fate and consequences of duplicate genes. Science. 2000;290:1151–1155. doi: 10.1126/science.290.5494.1151. [DOI] [PubMed] [Google Scholar]
- Davis JC, Petrov DA. Preferential duplication of conserved proteins in eukaryotic genomes. PLoS Biol. 2004;2:e55. doi: 10.1371/journal.pbio.0020055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papp B, Pàl C, Hurst LD. Dosage sensitivity and the evolution of gene families in yeast. Nature. 2003;424:194–197. doi: 10.1038/nature01771. [DOI] [PubMed] [Google Scholar]
- Parisi M, Nuttall R, Edwards P, Minor J, Naiman D, Lu J, Doctolero M, Vainer M, Chan C, Malley J, et al. A survey of ovary-, testis-, and soma-biased gene expression in Drosophila melanogaster adults. Genome Biol. 2004;5:R40. doi: 10.1186/gb-2004-5-6-r40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M, Force A. The probability of duplicate gene preservation by subfunctionalization. Genetics. 2000;154:459–473. doi: 10.1093/genetics/154.1.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FlyBase http://flybase.bio.indiana.edu
- BDGP: Berkeley Drosophila Genome Project http://www.fruitfly.org/index.html
- Shields DC, Sharp PM, Higgins DG, Wright F. 'Silent' sites in Drosophila genes are not neutral: evidence of selection among synonymous codons. Mol Biol Evol. 1988;5:704–716. doi: 10.1093/oxfordjournals.molbev.a040525. [DOI] [PubMed] [Google Scholar]
- Laird CD. DNA of Drosophila chromosomes. Annu Rev Genet. 1973;7:177–204. doi: 10.1146/annurev.ge.07.120173.001141. [DOI] [PubMed] [Google Scholar]
- Cohen BA, Mitra RD, Hughes JD, Church GM. A computational analysis of whole-genome expression data reveals chromosomal domains of gene expression. Nat Genet. 2000;26:183–186. doi: 10.1038/79896. [DOI] [PubMed] [Google Scholar]
- Blumenthal T, Evans D, Link CD, Guffanti A, Lawson D, Thierry-Mieg J, Thierry-Mieg D, Chiu WL, Duke K, Kiraly M, Kim SK. A global analysis of Caenorhabditis elegans operons. Nature. 2002;417:851–854. doi: 10.1038/nature00831. [DOI] [PubMed] [Google Scholar]
- Boutanaev AM, Kalmykova AI, Shevelyov YY, Nurminsky DI. Large clusters of co-expressed genes in the Drosophila genome. Nature. 2002;420:666–669. doi: 10.1038/nature01216. [DOI] [PubMed] [Google Scholar]
- Spellman PT, Rubin GM. Evidence for large domains of similarly expressed genes in the Drosophila genome. J Biol. 2002;1:5. doi: 10.1186/1475-4924-1-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caron H, van Schaik B, van der Mee M, Baas F, Riggins G, van Sluis P, Hermus MC, van Asperen R, Boon K, Voute PA, et al. The human transcriptome map: clustering of highly expressed genes in chromosomal domains. Science. 2001;291:1289–1292. doi: 10.1126/science.1056794. [DOI] [PubMed] [Google Scholar]
- Parisi M, Nuttall R, Naiman D, Bouffard G, Malley J, Andrews J, Eastman S, Oliver B. Paucity of genes on the Drosophila X chromosome showing male-biased expression. Science. 2003;299:697–700. doi: 10.1126/science.1079190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranz JM, Castillo-Davis CI, Meiklejohn CD, Hartl DL. Sex-dependent gene expression and evolution of the Drosophila transcriptome. Science. 2003;300:1742–1745. doi: 10.1126/science.1085881. [DOI] [PubMed] [Google Scholar]
- Betran E, Thornton K, Long M. Retroposed new genes out of the X in Drosophila. Genome Res. 2002;12:1854–1859. doi: 10.1101/gr.6049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerson JJ, Kaessmann , Betran E, Long M. Extensive gene traffic on the mammalian X chromosome. Science. 2004;303:537–540. doi: 10.1126/science.1090042. [DOI] [PubMed] [Google Scholar]
- Lifschytz E, Lindsley DL. The role of X-chromosome inactivation during spermatogenesis (Drosophila-allocycly-chromosome evolution-male sterility-dosage compensation). Proc Natl Acad Sci USA. 1972;69:182–186. doi: 10.1073/pnas.69.1.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richler C, Soreq H, Wahrman J. X inactivation in mammalian testis is correlated with inactive X-specific transcription. Nat Genet. 1992;2:192–195. doi: 10.1038/ng1192-192. [DOI] [PubMed] [Google Scholar]
- Sardiello M, Tripoli G, Romito A, Minervini C, Viggiano L, Caggese C, Pesole G. Energy biogenesis: one key for coordinating two genomes. Trends Genet. 2005;21:12–16. doi: 10.1016/j.tig.2004.11.009. [DOI] [PubMed] [Google Scholar]
- Thornton K, Long M. Rapid divergence of gene duplicates on the Drosophila melanogaster X chromosome. Mol Biol Evol. 2002;19:918–925. doi: 10.1093/oxfordjournals.molbev.a004149. [DOI] [PubMed] [Google Scholar]
- Zhang J. Evolution by gene duplication: an update. Trends Ecol Evol. 2003;18:292–298. doi: 10.1016/S0169-5347(03)00033-8. [DOI] [Google Scholar]
- Pairwise alignments algorithm: Emboss-Align http://www.ebi.ac.uk/emboss/align
- Corpet F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 1988;16:10881–10890. doi: 10.1093/nar/16.22.10881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MultAlin http://prodes.toulouse.inra.fr/multalin/multalin.html
- Andrews J, Bouffard GG, Cheadle C, Lu J, Becker KG, Oliver B. Gene discovery using computational and microarray analysis of transcription in the Drosophila melanogaster testis. Genome Res. 2000;10:2030–2043. doi: 10.1101/gr.10.12.2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misra S, Crosby MA, Mungall CJ, Matthews BB, Campbell KS, Hradecky P, Huang Y, Kaminker JS, Millburn GH, Prochnik SE, et al. Annotation of the Drosophila euchromatic genome: a systematic review. Genome Biol. 2002;3:research0083.1–0083.22. doi: 10.1186/gb-2002-3-12-research0083. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A table that reports pairwise amino-acid sequence conservation values between the D. melanogaster OXPHOS genes investigated and their D. pseudoobscura, A. gambiae and human counterparts
Data extracted from the Parisi et al. dataset
The codon usage in the orthologous and duplicate OXPHOS genes of D. melanogaster, D. pseudoobscura and A. gambiae


