Abstract
Leaf color is an important indicator when evaluating plant growth and responses to biotic/abiotic stress. Acquisition of images by digital cameras allows analysis and long-term storage of the acquired images. However, under field conditions, where light intensity can fluctuate and other factors (shade, reflection, and background, etc.) vary, stable and reproducible measurement and quantification of leaf color are hard to achieve. Digital scanners provide fixed conditions for obtaining image data, allowing stable and reliable comparison among samples, but require detached plant materials to capture images, and the destructive processes involved often induce deformation of plant materials (curled leaves and faded colors, etc.). In this study, by using a lightweight digital scanner connected to a mobile computer, we obtained digital image data from intact plant leaves grown in natural-light greenhouses without detaching the targets. We took images of soybean leaves infected by Xanthomonas campestris pv. glycines, and distinctively quantified two disease symptoms (brown lesions and yellow halos) using freely available image processing software. The image data were amenable to quantitative and statistical analyses, allowing precise and objective evaluation of disease resistance.
Keywords: disease symptom, field and greenhouse, image analysis, plant leaf color
Introduction
Leaf color is of prime importance in diagnosing the physiological status of plants under various environmental, nutritional, and stress conditions (Ibaraki and Gupta 2015, Rahaman et al. 2015). Image analysis is very often applied to evaluate the qualitative and quantitative characteristics of leaf color (Goodwin and Hsiang 2010, Murakami et al. 2005). Digital cameras are used widely to obtain images, and have the advantage of producing data suitable for long-term storage and allowing various types of image analyses. However, analysis of image data acquired under natural light is always subject to fluctuating weather conditions, which can change the hue, brightness, and luster of a target. In addition, contingent and local environmental factors influencing the imaging targets, such as the relative angle of target surface, shadow, and background, are often nonuniform in an image and can disturb the reproducibility of analyses (Ibaraki and Gupta 2015, Wang et al. 2014). Flatbed scanners are alternative devices that can be used to obtain images under constant conditions (Goodwin and Hsiang 2010, Wijekoon et al. 2008). A disadvantage of using scanners, which are normally used indoors, is the requirement for detaching the target. Detached leaves, e.g., those of rice, often wither and deform quickly, making it hard to analyze large numbers of samples at a time. To our knowledge, no precedent study used flatbed scanners for imaging of plant leaves growing under natural light without detaching.
Fungal and bacterial diseases of plants manifest symptoms on infected leaves that are characteristic to each pathogen, and quantitative measurement of such symptoms is one of the most important diagnostic indicators of plant pathogen resistance. Evaluation of plant disease symptoms has traditionally relied on the human eye, but is highly dependent on the experience and subjectivity of individuals, and comparison between independent evaluators can give inconsistent results (Barbedo 2013). Digital image analysis has the potential to overcome such difficulties and allow quantitative measurement of specifically altered areas that can be subjected to statistical analyses.
In this study, to obtain color images of pathogen-infected leaves growing under natural light, we used a lightweight scanner connected to a mobile personal computer through a USB cable that provided electricity to the scanner from the computer. Leaf images of soybean infected with a pathogenic bacterium were obtained in a greenhouse without detaching the target leaves. Acquisition of images in greenhouses with the handheld scanner allowed nondestructive and quantitative analyses of leaf characteristics that can be used to evaluate susceptibility of soybean to a pathogenic bacterium.
Materials and Methods
Plant materials
Wild-type (cultivar Jack) and a susceptible line (JRX1) of soybean were grown in a greenhouse for 3 weeks under natural light. A strain of Xanthomonas campestris pv. glycines used in this study was isolated from a typical bacterial pustule lesion on a soybean plant grown in a field (Tsukubamirai, Ibaraki, Japan). JRX1 has a quantitative trait locus for susceptibility to X. campestris pv. glycines and infected leaves of JRX1 produce small pale green spots, and these spots gradually become brown pustules surrounded by yellow halos. For evaluation of leaf pustule lesions, leaves were sprayed with bacterial suspension (OD600 = 0.2 in 0.01% Tween 20) of X. campestris pv. glycines. The sprayed leaves were covered with a plastic bag overnight, then allowed to continue growing for 9 days before obtaining image data.
Image acquisition
A lightweight contact image sensor (CIS) scanner with RGB LEDs (CanoScan LiDE 500FV, Canon, Japan) connected to a mobile computer through a USB cable was used for image acquisition in the greenhouse. Images were obtained at 300 dpi resolution, and stored in TIFF format. A circular color panel (diameter 4.15 cm) was attached to the scanning surface of the scanner for setting scale and color intensity during image processing. The color panel was downloaded from https://lpixel.net/services/education/nourin186/.
Image processing
Images were processed with ImageJ (version 1.50i, https://imagej.nih.gov/ij/). If necessary, debris on images was erased using the Brush tool set to white color. Variabilities in color intensities among images were checked by analyzing the distribution of RGB color intensities using the Histogram command in ImageJ. If necessary, distribution in color intensities was adjusted using the Color Balance command in ImageJ. Petioles in images were erased with the Brush tool. The diameter of the circle in the color panel was measured using the Line tool, and pixels in images were converted to centimeters with the Set Scale command. After setting scale, the size indicator in an image was erased as above. To measure whole leaf area, we used Color Threshold with HSB color space, and thresholds values were set to select whole leaf area. Leaf area was then measured using the Measure command. Appropriate thresholds to specifically select brown lesions were set with HSB color space, and total area, number, and average size of brown lesions were measured. To analyze yellow halos, appropriate thresholds to specifically select yellow halos were set with HSB color space, and total halo area was measured using the Measure command. A default thresholding setup in ImageJ was used for all processes. Representative image in each step of the image processing method is shown in Supplemental Fig. 1.
Results
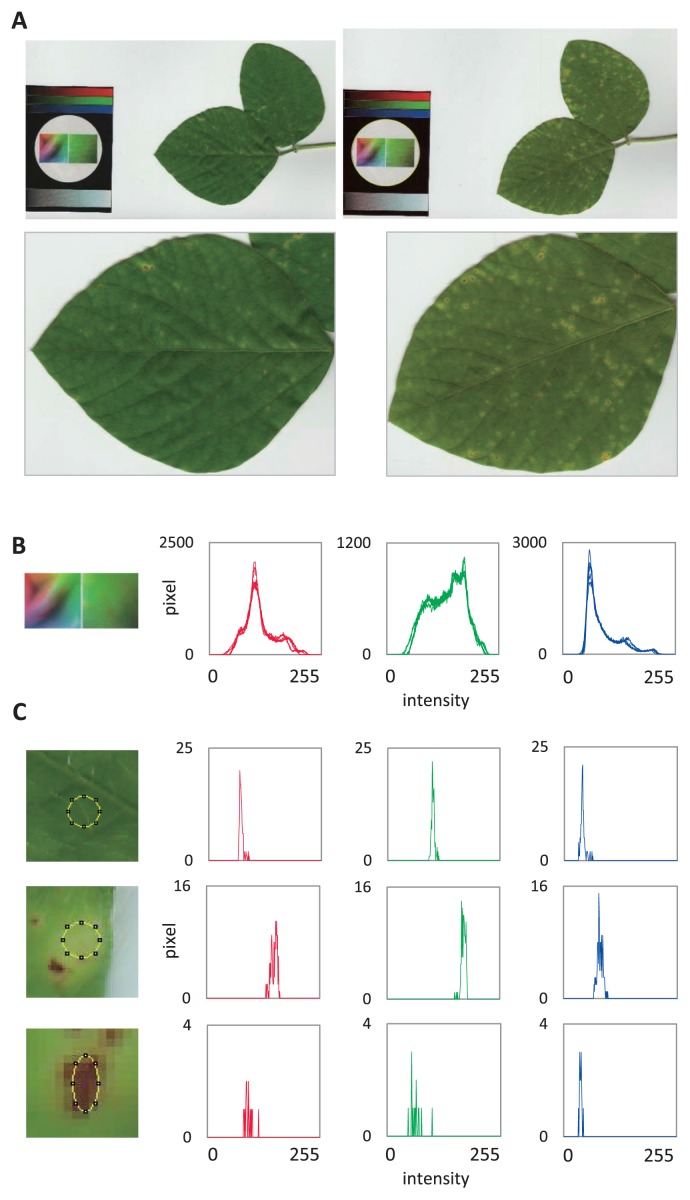
To quantify susceptibility of soybean to X. campestris, images of infected leaves of wild-type, and a line (JRX1) that shows disease symptoms upon infection, were acquired using a lightweight flatbed scanner (scanning size: 210 × 297 mm). The scanner was operated in a greenhouse via a small mobile computer, providing great flexibility in obtaining image data from intact plants growing under natural light. Two or three compound leaves were imaged together with a color panel (Fig. 1A), and variability among independent images was analyzed by dissecting the color intensities of the color plate into RGB channels (Fig. 1B). Although the scanner should shield imaging targets from external light, the thickness of the samples occasionally induced slight leakage of external light, affecting the resulting color intensities. RGB color intensities of the color panel in each sample were displayed using the Histogram command in ImageJ, and adjusted with the Color Balance command, resulting in uniform distributions of RGB color intensities in all samples (Fig. 1B). Distributions of RGB color intensities in selected leaf areas were similarly dissected into RGB color channels (Fig. 1C, top row, a healthy leaf region; middle row, a yellow halo region; bottom row, a brown lesion). Characteristic distributions in yellow halo and brown lesion were observed in red and green channels. The data indicated that using a lightweight scanner connected to a mobile computer allowed nondestructive image acquisition of leaves growing under natural light, and provided invariable image data that can be subjected to digital image analysis.
Fig. 1.
Color dissection of images acquired by a flatbed scanner. (A) Representative leaf images (upper left, wild-type (cultivar Jack); upper right, JRX1). Three-week-old seedlings of soybean were sprayed with Xanthomonas campestris pv. glycine, and images of infected leaves were taken in situ in the greenhouse using a flatbed scanner 9 days after spraying. A color panel containing a circle (diameter 4.15 cm) was placed in the scanner. Enlarged photos of one leaf of each line are shown below (lower). (B) RGB color histograms of color panels from six independent images are shown (left, red; center, green; right, blue channels). Vertical axes, pixels; horizontal axes, intensity in 256-grade scale. (C) Color distribution in a control (top row) and in a diseased leaf (middle row, yellow halo; bottom row, brown lesion). Color intensities in the areas indicated within the pictures (left panels) are shown in histogram form.
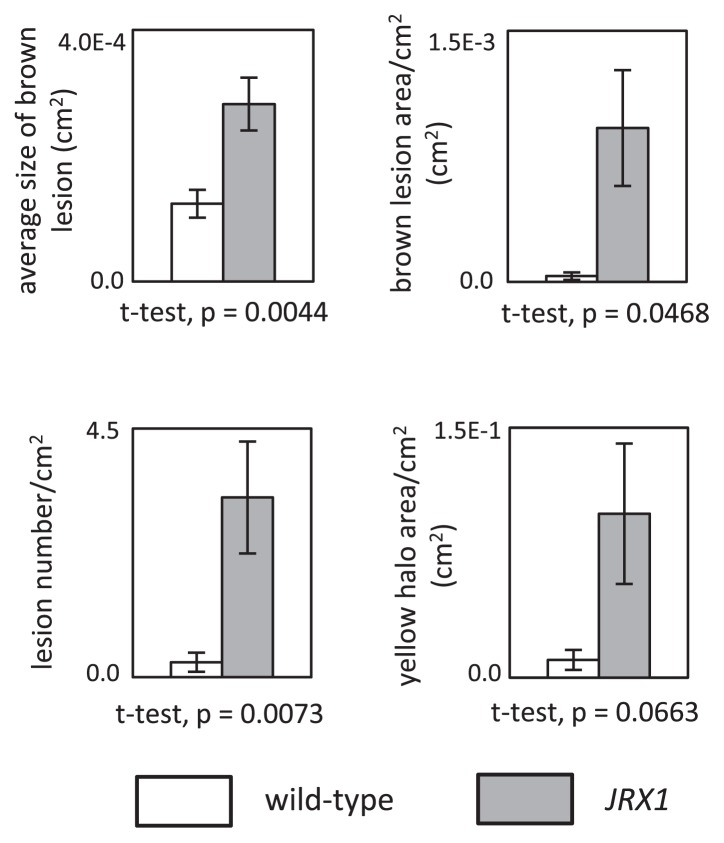
For color image analysis of disease symptoms, we tested various color spaces that are embedded in ImageJ and selected the HSB color space, which gave the best separation of yellow halos on green leaves. Petioles in images were erased manually using the Brush tool before quantification. Areas of whole leaf, yellow halos, and brown lesions were selected by applying appropriate threshold values in the Color Threshold command. Because infection of X. campestris induces various degree of leaf senescence and fading in leaf color, thresholds to discriminate yellow halos from the background leaf area were adjusted for each sample to obtain reliable measurements. In addition to areas of yellow halos and brown lesions, numbers of brown lesions per cm2 of leaf area, and average size of lesions were measured. The numerical data obtained were graphed; statistical tests applied to the data indicated significant differences in disease resistance between wild-type and JRX1 (Fig. 2).
Fig. 2.
Evaluation of susceptibility against bacterial leaf pustule of soybean by image analysis. Averages and standard deviations are shown (n = 3).
Discussion
Disease resistance is commonly evaluated by visual inspection of samples, which often leads to discrepancies among results from individual evaluators and difficulties in applying data to QTL analysis. Scanners are suitable for imaging of flat leaf samples, and are used frequently in quantifying disease symptoms appearing on leaves. However, samples to be imaged had to be limited to those keeping their original characteristics after being excised from growing plants, and the number of samples that could be analyzed at any one time was restricted. Image acquisition by digital cameras is a simple and easy mean to obtain data for image analysis, but environmental factors such as light intensity and shading commonly affect uniformity of images taken outdoors, resulting in severe disorder of color distribution (Supplemental Fig. 2).
Many software tools have been developed for small-scale plant image analysis that address specific or global purposes of plant phenotyping (Lobet et al. 2013, Rahaman et al. 2015). In this study, image analysis was performed with ImageJ, one of the most popular image processing softwares, which is free and used in many fields of science (Abràmoff et al. 2004). ImageJ provides four color spaces (HSB, RGB, Lab, and YUV), and each color space consists of a specific configuration of color distribution (Abràmoff et al. 2004, Pound and French 2015). A suitable color space must be selected to discriminate the objects to be measured from the background, and, in our current study, the HSB color space gave the best results in detecting brown lesions and yellow halos on green leaves. In addition to pustules, chlorosis and necrosis are other disease symptoms upon infection of pathogens. These symptoms are often similar to yellow halos and brown lesions, respectively, and therefore can be quantified by the HSB color space. Disease symptoms without visible lesions often exhibited fading in leaf color and such symptoms resemble those observed in senescent leaves. The HSB color space effectively discriminates color differences between young and senescent leaves growing under natural light (Supplemental Fig. 2). We also quantified damage to rice and maize plants under drought conditions by the same methods, and found that Lab color space gave the best discrimination between healthy and withered leaves (data not shown). In addition to color space, the thresholding method within ImageJ must be tested to give the best discrimination between object and background. For quantifying resistance of soybean to leaf pustule disease, the default thresholding method in ImageJ gave satisfactory discrimination from the background of both brown lesions and yellow halos.
Although scanners with a reduced optical system and charged-coupled devices have advantages in focus depth and scan speed, the simple structure of the CIS scanner, and the lack of a requirement for an external power supply, are preferred for image acquisition in the field and in greenhouses. Since objects analyzed by CIS scanners are required to be flat, analysis of three-dimensional objects growing under natural light represents a future challenge. Regardless of such limitations, acquisition of images in a stable, reproducible, and nondestructive manner in fields and greenhouses will be applicable to a number of purposes, not only to quantity resistance and tolerance to biotic and abiotic stresses, but also to monitor nutritional state and the ornamental value of various plants.
Supplementary Information
Acknowledgements
We thank Natsumaro Kutsuna and Helen Rothnie for comments on this study, LPixel Inc. for the color panel, and Rie Takahashi for technical assistance. This work was supported by a grant from JST/CREST (JPMJCR13B4) to S.M. and Y.H., and MXT/JSPS KAKENHI (15H05962) to S.M..
Literature Cited
- Abràmoff, M.D., Magalhães, P.J. and Ram, S.J. (2004) Image processing with ImageJ. Biophotonics Int. 11: 36–42. [Google Scholar]
- Barbedo, A.J.G. (2013) Digital image processing techniques for detecting, quantifying and classifying plant diseases. SpringerPlus. 2: 660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin, P.H. and Hsiang, T. (2010) Quantification of fungal infection of leaves with digital images and scion image software. In: Sharon, A. (ed.) Molecular and cell biology methods for fungi. Springer, New York, pp. 125–135. [DOI] [PubMed] [Google Scholar]
- Ibaraki, Y. and Gupta, S.D. (2015) Image analysis for plants: basic procedures and techniques. In: Gupta, S.D. and Ibaraki Y. (eds.) Plant Image Analysis, Fundamentals and Applications. CRC Press, Boca Raton, pp. 25–40. [Google Scholar]
- Lobet, G., Draye, X. and Périlleux, C. (2013) An online database for plant image analysis software tools. Plant Methods 9: 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami, P.F., Turner, M.R., van den Berg, A.K. and Schaberg, P.G. (2005) An instructional guide for leaf color analysis using digital imaging software. USDA Gen. Tech. Rep. NE-327. [Google Scholar]
- Pound, M.P. and French, A.P. (2015) An introduction to images and image analysis. In: Gupta, S.D. and Ibaraki Y. (eds.) Plant Image Analysis, Fundamentals and Applications. CRC Press, Boca Raton, pp. 1–24. [Google Scholar]
- Rahaman, M.M., Chen, D., Gillani, Z., Klukas, C. and Chen, M. (2015) Advanced phenotyping and phenotype data analysis for the study of plant growth and development. Front Plant Sci. 6: 619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y., Wang, D., Shi, P. and Omasa, K. (2014) Estimating rice chlorophyll content and leaf nitrogen concentration with a digital still color camera under natural light. Plant Methods 10: 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijekoon, C.P., Goodwin, P.H. and Hsiang, T. (2008) Quantifying fungal infection of plant leaves by digital image analysis using Scion Image software. J. Microbiol. Methods 74: 94–101. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.