Abstract
Seedling vigor is an important agricultural trait as direct-seeded rice technology becomes widely applied. In order to investigate the genetic mechanisms underlying seedling vigor in rice, seeds of 132 recombinant inbred lines (RILs) derived from 93-11 and PA64s, harvested from Lingshui and Hangzhou were cultivated in the nutrient solution, and four indices for seedling vigor were measured including seedling shoot length (SSL), seedling root length (SRL), seedling wet weight (SWW) and seedling dry weight (SDW). Significant correlations were observed among the indices, and also between 1000-seed weight (TSW) and SWW or SDW. Combined with a high-resolution genetic map generated from sequencing of the RILs, 65 quantitative trait loci (QTLs) were detected on all chromosomes with interval of 1.93 Mb on average. Among 57 QTLs for seedling vigor, 28 were detected from seeds harvested in both sites and 33 were first identified. With BC3F2 derived from 93-11 and a CSSL harboring segments from PA64s in 93-11 background, a major QTL for SSL, qSSL1b was fine mapped within 80.5 kb between two InDel markers. Our study provides a platform for further cloning of the QTL and dissecting the molecular basis for seedling vigor at early seedling stage in rice.
Keywords: rice, QTL, seedling vigor, qSSL1b, early seedling stage
Introduction
Rice direct seeding technology is widely applied in Southeast Asia these years as it requires lower labor costs compared to the conventional transplantation (Abe et al. 2012). However, there are many factors limiting the growth and development of direct-seeding rice, such as low emergence rate, difficult weeding and lodging in later growth period (Yang et al. 2015). Seedling vigor is the ability of a seed to emerge rapidly from soil or water, mainly reference to seed germination rate and early seedling growth (Huang et al. 2004). Seeds with high vigor is important for rice production because it can not only significantly enhance seedling establishment (Luo et al. 2007), but also improve the capability to compete against weeds at seedling stage (Rao et al. 2007). Therefore, seedling vigor has been paid more attention respect to cultivation techniques and genetic analysis in recent years.
Seedling vigor is a complex agronomic trait with several indicators, such as germination rate, final germination percentage and germination index during seed germination stage (Wang et al. 2010), and root length, shoot length, wet weight and dry weight in the early seedling growth process (Redoña and Mackill 1996, Regan et al. 1992). Quantitative trait loci (QTL) analysis has been demonstrated a effective way to study complex traits, for example, yield associated traits (Huang et al. 2009, Song et al. 2007, Wang et al. 2015). Recently, the method has also been employed to study seedling vigor in rice. Han et al. (2006) identified several QTLs for low-temperature vigor of germination at 14°C using the F2:3 populations and deduced that the gene action of low-temperature vigor was most likely to be partially dominant. Germination rate, final germination percentage and germination index were evaluated by Wang et al. (2010) with recombinant inbred line (RIL) population, and most of the detected QTLs for rice seedling vigor were found coincide with QTLs for seed weight, seed size or seed dormancy. In Dang’s study, 27 QTLs were identified for seedling vigor, and 15 elite parental combinations were designed to improve seedling vigor in rice (Dang et al. 2014). Xie et al. (2014) mapped eight quantitative trait loci (QTLs) for seedling vigor by using a RIL population, and narrowed two major QTLs, qSV-1 and qSV-5c to 1.13-Mbp and 400-kbp genomic regions, respectively. A major QTL for seedling height on the long arm of chromosome 3 was also fine mapped, including the candidate gene OsGA20ox1 (Abe et al. 2012). In all, these studies have provided useful information for seedling vigor, but there still more researches to be performed in this area.
In this study, we grown 132 RILs derived from 93-11 (Oryza sativa ssp. indica) × PA64s (Oryza sativa ssp. indica) in a nutrient solution for two weeks, and conducted seedling vigor evaluations by measurement of seedlings’ shoot length, root length, wet weight and dry weight. With the help of a high quality genetic map based on the SNPs generated from deep sequencing of the RIL genomes (Gao et al. 2013), a total of 57 QTLs were found. Several co-located QTLs share the same location with previous studies. The major QTL, qSSL1b was fine mapped within 80.5 kb between two insertion-deletion (InDel) markers. These results give more information of the genetic and molecular basis underlie seedling vigor in rice.
Materials and Methods
Plant materials
The recombinant inbred lines (RILs) presented in this study were developed by a cross between an elite paternal inbred Oryza sativa ssp. indica cv. 93-11 and the maternal inbred Oryza sativa ssp. indica cv. PA64s (a photo-thermo-sensitive male sterile line). The provided population was developed in the experimental fields at China National Rice Research Institute in Lingshui, Hainan Province, and in Hangzhou, Zhejiang Province, China. To develop a Chromosome segment substitution line (CSSL) containing the QTL for SSL, qSSL1b detected both in Lingshui and Hangzhou on chromosome 1, a line of RILs with PA64s genotype in the qSSL1b region was selected to backcross with recurrent parent 93-11. Two markers SNP1-202 and SNP1-257 (Table 4) were used for marker assisted selection (MAS) of each generation. As a result, a BC3F1 line, with 93-11 genetic background exhibiting heterozygous across the entire qSSL1b region, was constructed. After self-crossing, a BC3F2 population was obtained for fine mapping of qSSL1b.
Table 4.
Primers for InDel and SNP markers developed
| Primer | Forward (5′-3′) | Reverse (5′-3′) | Type |
|---|---|---|---|
| SNP1-202 | TGGATGGGAACTGTCAATTTG | TGTTCAAGGCAGCAAAGAGG | SNP |
| IND1-1 | ACATGGGCTAGCTGACAGGCGATC | TGGTGAGAACCCGGCACAACG | InDel |
| IND1-2 | GTGGGACAGACAGCCTCAGC | GCAATAATTCAGGAAATAATTGGG | InDel |
| IND1-3 | TTCCAGCTGACTGGTGACTGCTC | AGCTTGAGCTGCAATCCCACAG | InDel |
| IND1-4 | TGGTATCAATCGTTGATGGATGC | CAGATCCATGGATGTCTCCAAAC | InDel |
| SNP1-257 | GTTTGGACCAGGAGTACGAGG | TCAAGACCAGCATGAGCATATAGAG | SNP |
Phenotypic evaluation of rice seedling vigor
Mature seeds of parents and 132 core RILs were harvested in Lingshui (2012) and Hangzhou (2013) and soaked in deionized water overnight at 30°C in the dark. After germination, seeds were put into the holes of 96-well plates and transferred to 35 L plastic pots containing an nutrient solution (pH 5.6) containing 1.5 mM NH4NO3, 1.0 mM CaCl2, 1.6 mM MgSO4, 0.5 mM K2SO4, 0.3 mM NaH2PO4, 0.1 mM Fe-EDTA, 9.5 uM MnCl2, 20 uM H3BO3, 0.2 uM ZnSO4, 0.2 uM CuSO4, 0.05 uM Na2MoO4 and 0.01 uM Na2SiO3. The plants were grown in a greenhouse and the solution was changed once per 3 days. After 15 days, plants were collected and four indices were employed to evaluate seedling vigor: seedling shoot length (SSL), seedling root length (SRL), seedling wet weight (SWW) and seedling dry weight (SDW). For SSL and SRL, three plants of each line were measured. Six plants were gathered for measurement of SWW and SDW. Before measuring SDW, the samples were packed in individual paper bags and kept in an oven at 75°C for two days.
Data analyses and QTL detection
Statistical analysis of the parents and RILs was conducted for each index by SAS (version 8.01). Pearson correlation coefficients were calculated between each pair of all four indices and 1000-seed weight. A recombinant bin map with 2,262 high-quality polymorphic SNP markers was constructed by the re-sequencing parents and 132 core RIL lines (Gao et al. 2013). QTL analysis was performed with the R/qtl_1.26-14 (http://www.rqtl.org/) using Composite Interval Mapping (CIM). LOD threshold for each dataset was set based on a permutation test (1,000 permutation, P = 0.05). It was considered as a major effect QTL when its LOD score was larger than 2.5. PEV was estimated by ANOVA, and the QTL nomenclature followed the suggestion of McCouch et al. (2008).
Results
Phenotypic variations for seedling vigor and 1000-seed weight of the parents and RILs
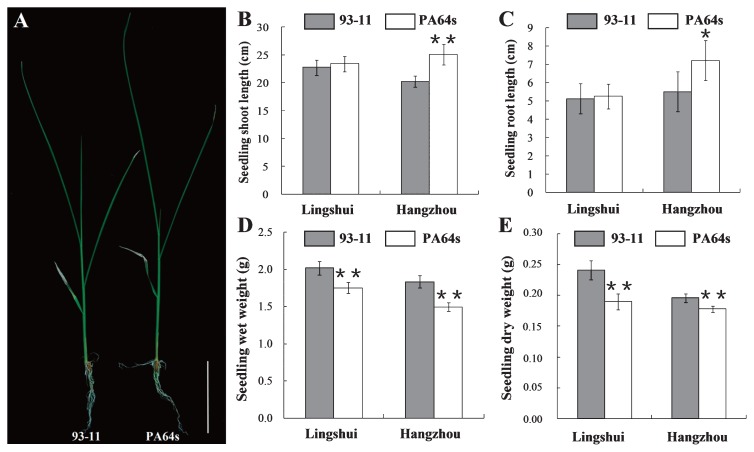
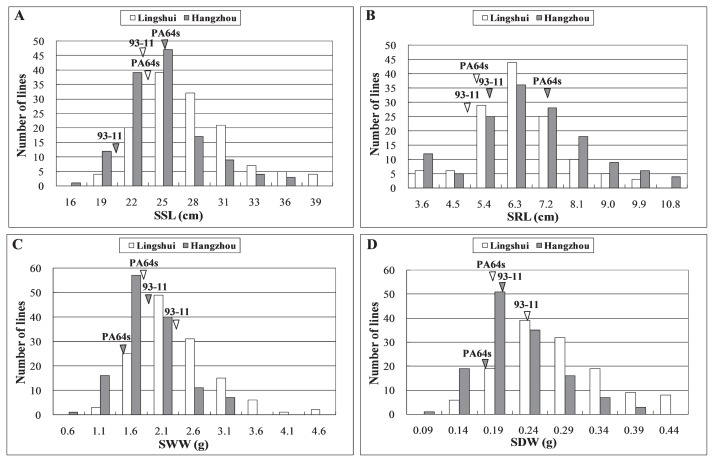
For SSL and SRL of seedlings, no significant difference existed between two parents from the seeds harvested in Lingshui while statistically significant difference was found in Hangzhou (Table 1, Fig. 1A–1C). The SWW and SDW of PA64s seedlings from seeds in both sites reached 80%–90% of 93-11, illustrating seedlings of 93-11 were stronger than PA64s (Table 1, Fig. 1D, 1E). The RIL population also showed significant divergence for all the evaluated traits. The mean value of SSL was about 24.50 cm, with the maximum value 38.52 cm and the minimum value 15.80 cm. The minimum value of SRL, SWW and SDW were around 20% of their mean value and the maximum value were about 170% of the mean value in the RIL population (Table 1). And the continuous distributions of four seedling vigor traits among the RILs showed significantly transgressive segregation with values either larger or smaller than those of the parents, which revealed that seedling vigor at early seedling stage was controlled by polygene (Table 1, Fig. 2).
Table 1.
Means (standard deviation), min, max, skewness and kurtosis for SSL, SRL, SWW and SDW during seedling stage in parents and RIL lines
| Harvested location | Trait | Parents | RIL population | |||||
|---|---|---|---|---|---|---|---|---|
|
|
|
|||||||
| 93-11 | PA64s | Mean ± SD | Min | Max | Skewness | Kurtosis | ||
| Lingshui | SSL | 22.74 ± 0.38 | 23.41 ± 0.39 | 25.83 ± 4.14 | 17.05 | 38.52 | 0.65 | 0.54 |
| SRL | 5.13 ± 0.03 | 5.26 ± 0.08 | 5.85 ± 1.20 | 3.82 | 9.56 | 0.67 | 0.56 | |
| SWW | 2.02 ± 0.09 | 1.75 ± 0.07 | 2.06 ± 0.58 | 0.95 | 4.43 | 1.12 | 2.22 | |
| SDW | 0.24 ± 0.02 | 0.19 ± 0.01 | 0.25 ± 0.08 | 0.11 | 0.61 | 1.33 | 3.80 | |
|
| ||||||||
| Hangzhou | SSL | 20.25 ± 0.37 | 25.10 ± 1.84 | 23.24 ± 3.56 | 15.80 | 33.04 | 0.69 | 0.43 |
| SRL | 5.51 ± 1.08 | 7.21 ± 1.08 | 6.55 ± 1.53 | 3.34 | 12.10 | 0.89 | 1.04 | |
| SWW | 1.84 ± 0.08 | 1.49 ± 0.06 | 1.60 ± 0.43 | 0.53 | 2.95 | 0.67 | 0.97 | |
| SDW | 0.20 ± 0.01 | 0.18 ± 0.01 | 0.19 ± 0.05 | 0.08 | 0.36 | 0.71 | 0.72 | |
SSL: seedling shoot length, SRL: seedling root length, SWW: seedling wet weight, SDW: seedling dry weight. The results of SSL and SRL in parents are presented as the means ± SD of triplicate samples. Results of SDW and SWW are shown as sum of 6 plants.
Fig. 1.
A. Seedling phenotype of 93-11 and PA64s at early seedling stage from Hangzhou. Bar = 5 cm. B–E. Four seedling vigor indices of 93-11 and PA64s from Lingshui and Hangzhou. Results of SDW and SWW are shown as a total of 6 plants. ** and * on the bars indicate significant difference at the 1% and 5% level, respectively according to t test.
Fig. 2.
Frequency distributions of seedling shoot length (SSL) (A), seedling root length (SRL) (B), seedling wet weight (SWW) (C) and seedling dry weight (SDW) (D) among RILs with seeds from Lingshui and Hangzhou in early seedling growth stage. Results of SDW and SWW are shown as a total of 6 plants. The white patterns represent phenotype collected in Lingshui and the grey ones represent those in Hangzhou. White triangles and grey triangles indicate SSL, SRL, SWW and SDW of 93-11 and PA64s from Lingshui and Hangzhou, respectively.
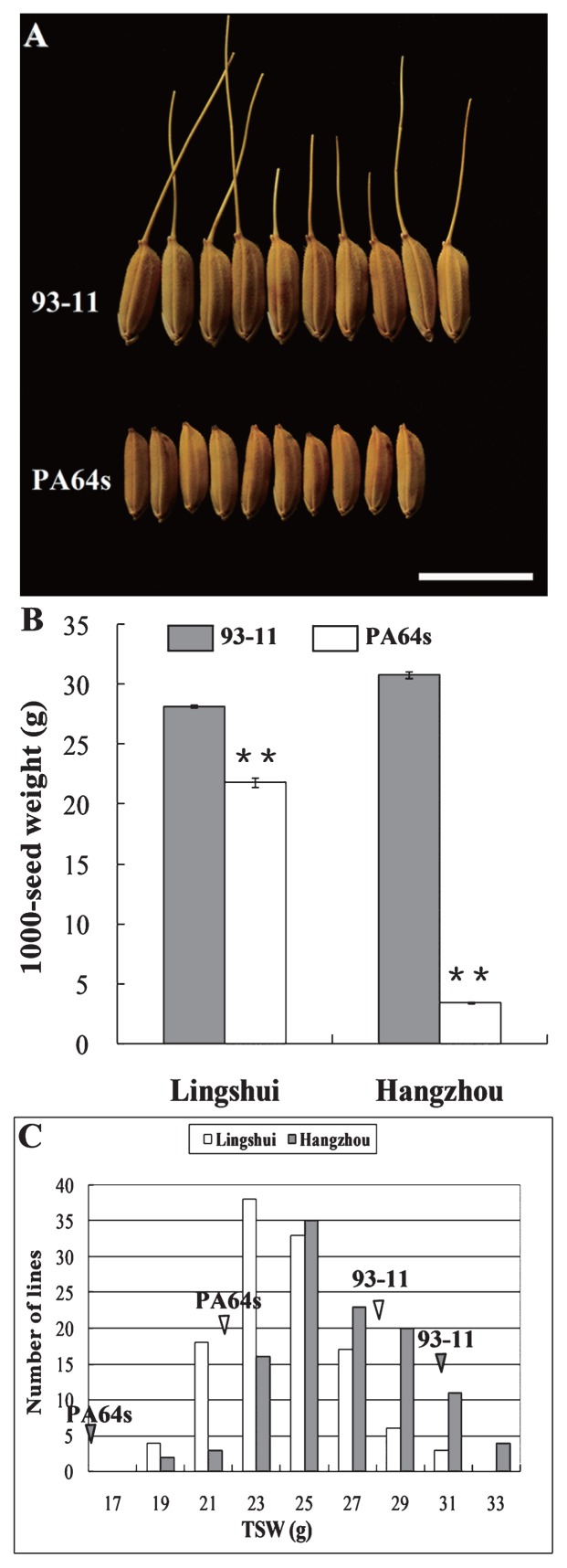
The differences in 1000-seed weight between 9311 and PA64s are displayed in Fig. 3A and 3B. Significant differences of seeds from Lingshui and Hangzhou were found between 9311 and PA64s with respect to 1000-seed weight. Nearly normal distributions were observed in the RIL population for 1000-seed weight from Lingshui and Hangzhou, respectively, indicating the trait was controlled by multi-genes (Fig. 3C).
Fig. 3.
A. Seeds of 93-11 and PA64s harvested from Lingshui. Bar = 10 mm. B. Comparison of 1000-seed weight of 93-11 and PA64s from Lingshui and Hangzhou. C. Frequency distributions of 1000-seed weight among RILs with seeds harvested from Lingshui and Hangzhou. White triangles and grey triangles indicate 1000-seed weight of 93-11 and PA64s in Lingshui and Hangzhou, respectively.
Correlation among the four indices and 1000-seed weight
Pair-wise correlation coefficients among the four indices and those indices with 1000-seed weight were presented in Table 2. Significant correlations were observed among the four parameters. Considering previous study suggested that seed weight might be correlated with seedling vigor (Cui et al. 2002), we also calculated correlation coefficients between 1000-seed weight with the four indices in the RILs and found significant correlations between 1000-seed weight and SSL, SWW, SDW from Hangzhou, and also between 1000-seed weight and SWW, SDW from Lingshui, indicating seed weight contribute to seedling vigor at early seedling stage in this population (Table 2).
Table 2.
Correlation coefficients among indices of seed vigor measured during seedling stage and 1000-seed weight in 132 RIL lines derived from 93-11 × PA64s
| Trait | SSL | SRL | SWW | SDW | TSW |
|---|---|---|---|---|---|
| SSL | 0.46** | 0.87** | 0.87** | 0.19 | |
| SRL | 0.60** | 0.51** | 0.50** | 0.07 | |
| SWW | 0.83** | 0.58** | 0.95** | 0.24* | |
| SDW | 0.85** | 0.57** | 0.94** | 0.25* | |
| TSW | 0.33* | 0.21 | 0.38** | 0.43** |
SSL: seedling shoot length, SRL: seedling root length, SDW: seedling dry weight, SWW: seedling wet weight. TSW: 1000-seed weight.
indicate the 5% and 1% significant level, respectively. Seeds from Lingshui are shown in above diagonal and Hangzhou in below diagonal.
QTL identification for seedling vigor
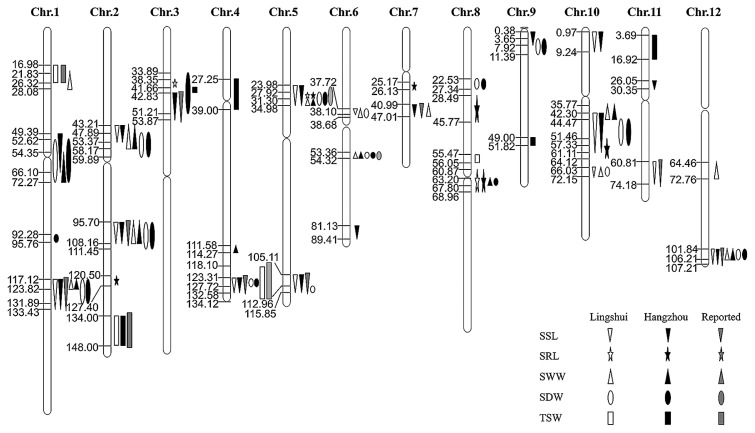
As shown in Table 3 and Fig. 4, combined with a high-resolution genetic map based on the SNPs generated from sequencing of the RILs (Gao et al. 2013), 57 QTLs well-distributed on all 12 chromosomes were identified for the four indices, including SSL, SRL, SWW and SDW. Among them, 28 QTLs were detected from seeds in both Lingshui and Hangzhou, and 33 QTLs were unreported so far.
Table 3.
QTLs for SSL, SRL, SDW and SWW during seedling stage and QTLs for TSW of seeds from Lingshui and Hangzhou
| Trait | QTL | Chr. | Genetic distance (cM) | Seeds from 2012 Lingshui | Seeds from 2013 Hangzhou | Reference | ||||
|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|||||||||
| LOD | %Var | Add. | LOD | %Var | Add. | |||||
| SSL | qSSL1a | 1 | 49.39–66.10 | 4.42 | 13.6 | 1.233 | ||||
| unnamed | 1 | http://archive.gramene.org/db/qtl/ | ||||||||
| qSSL1b | 1 | 117.12–133.43 | 5.48 | 10.5 | −1.039 | 3.77 | 11.2 | −1.193 | ||
| qCSH1 | 1 | Han et al. 2007 | ||||||||
| qSSL2a | 2 | 43.21–53.37 | 2.64 | 1.8 | 0.542 | 2.77 | 1.1 | 0.324 | ||
| qSSL2b | 2 | 95.70–108.16 | 3.00 | 3.6 | −0.789 | 3.23 | 2.1 | −0.374 | ||
| qCSH2 | 2 | Han et al. 2007 | ||||||||
| qSSL3 | 3 | 42.83–53.87 | 2.70 | 4.9 | 0.787 | |||||
| qSL3-2 | 3 | Cao et al. 2002 | ||||||||
| qSSL4 | 4 | 123.31–132.58 | 2.55 | 3.1 | 0.228 | 4.36 | 3.5 | 0.606 | ||
| qPHS4 | 4 | Abe et al. 2012 | ||||||||
| qSSL5a | 5 | 23.98–34.98 | 4.20 | 4.3 | −0.677 | 3.10 | 3.3 | −0.614 | ||
| qSV-5 | 5 | http://archive.gramene.org/db/qtl/ | ||||||||
| qSSL5b | 5 | 105.11–115.85 | 2.71 | 6.2 | −1.047 | 2.73 | 2.9 | −0.632 | ||
| unnamed | 5 | Dang et al. 2014 | ||||||||
| qSSL6a | 6 | 37.72–38.10 | 2.72 | 2.9 | −0.747 | |||||
| qSSL6b | 6 | 81.13–89.41 | 2.87 | 2.6 | −0.574 | |||||
| qSSL7 | 7 | 40.99–47.01 | 2.55 | 4.3 | 0.732 | |||||
| unnamed | 7 | Anandan et al. 2016 | ||||||||
| qSSL9 | 9 | 0.377–7.92 | 2.75 | 3.1 | 0.605 | |||||
| qSSL10a | 10 | 0.97–9.24 | 3.47 | 7.7 | 1.221 | 2.96 | 7.8 | 1.038 | ||
| qSSL10b | 10 | 42.30–61.11 | 2.82 | 2.5 | 0.637 | 3.05 | 5.2 | 0.790 | ||
| qSSL10c | 10 | 66.41–72.15 | 3.43 | 9.7 | 1.298 | |||||
| qSSL11a | 11 | 26.05–30.35 | 2.90 | 5.7 | 0.848 | |||||
| qSSL11b | 11 | 60.81–74.18 | 2.70 | 9.2 | −1.272 | |||||
| qSL11 | Cao et al. 2002 | |||||||||
| qSSL12 | 12 | 101.84–107.21 | 3.70 | 3.7 | −0.804 | 2.94 | 7.4 | −0.844 | ||
| qCSH12 | 12 | Han et al. 2007 | ||||||||
|
| ||||||||||
| SRL | qSRL2 | 2 | 120.50–127.40 | 3.18 | 6.4 | −0.377 | ||||
| unnamed | 2 | http://archive.gramene.org/db/qtl/ | ||||||||
| qSRL3 | 3 | 38.35–41.66 | 2.91 | 4.4 | 0.025 | |||||
| qSRL5 | 5 | 27.92–31.30 | 3.72 | 6.1 | 0.376 | 3.54 | 5.4 | 0.356 | ||
| unnamed | 5 | http://archive.gramene.org/db/qtl/ | ||||||||
| qSRL7 | 7 | 25.17–26.13 | 3.02 | 3.9 | −0.252 | |||||
| unnamed | 7 | http://archive.gramene.org/db/qtl/ | ||||||||
| qSRL8a | 8 | 28.49–45.77 | 2.50 | 6.6 | 0.411 | |||||
| qSRL8b | 8 | 56.05–68.96 | 3.36 | 4.6 | 0.255 | 2.53 | 3.8 | 0.303 | ||
| qSRL10 | 10 | 51.46–64.12 | 3.59 | 11.7 | 0.569 | |||||
|
| ||||||||||
| SWW | qSWW1a | 1 | 21.83–28.08 | 3.20 | 7.2 | 0.154 | ||||
| unnamed | 1 | http://archive.gramene.org/db/qtl/ | ||||||||
| qSWW1b | 1 | 54.35–72.27 | 2.87 | 6.6 | 0.112 | |||||
| unnamed | 1 | http://archive.gramene.org/db/qtl/ | ||||||||
| qSWW1c | 1 | 117.12–123.82 | 4.72 | 5.5 | −0.138 | 3.77 | 3.7 | −0.079 | ||
| unnamed | 1 | http://archive.gramene.org/db/qtl/ | ||||||||
| qSWW2a | 2 | 43.21–58.17 | 4.42 | 1.8 | 0.075 | 2.59 | 1.2 | 0.046 | ||
| qSWW2b | 2 | 95.70–108.16 | 4.46 | 2.6 | −0.093 | 2.82 | 1.9 | −0.058 | ||
| unnamed | 2 | http://archive.gramene.org/db/qtl/ | ||||||||
| qSWW4 | 4 | 111.58–114.27 | 3.08 | 4.8 | 0.091 | |||||
| unnamed | 4 | http://archive.gramene.org/db/qtl/ | ||||||||
| qSWW5 | 5 | 31.49–34.98 | 5.73 | 1.5 | −0.020 | 2.89 | 3.1 | −0.033 | ||
| qSWW6a | 6 | 37.72–38.68 | 4.48 | 3.2 | −0.109 | |||||
| qSWW6b | 6 | 53.36–54.32 | 2.95 | 4.1 | −0.127 | 3.14 | 3.5 | −0.034 | ||
| unnamed | 6 | http://archive.gramene.org/db/qtl/ | ||||||||
| qSWW7 | 7 | 40.68–47.11 | 2.74 | 3.8 | 0.082 | |||||
| unnamed | 7 | http://archive.gramene.org/db/qtl/ | ||||||||
| qSWW8 | 8 | 63.20–67.80 | 2.55 | 3.6 | 0.086 | |||||
| qSWW10a | 10 | 35.77–44.47 | 4.41 | 4.3 | 0.076 | 2.91 | 4.9 | 0.030 | ||
| qSWW10b | 10 | 66.41–72.15 | 4.33 | 13.8 | 0.218 | |||||
| qSWW12 | 12 | 101.84–106.21 | 3.23 | 6.9 | −0.061 | 3.12 | 5.3 | −0.101 | ||
|
| ||||||||||
| SDW | qSDW1a | 1 | 52.62–72.27 | 3.12 | 7.7 | 0.015 | ||||
| unnamed | 1 | http://archive.gramene.org/db/qtl/ | ||||||||
| qSDW1b | 1 | 92.28–95.76 | 2.66 | 6.2 | −0.013 | |||||
| qSDW1c | 1 | 117.12–131.89 | 5.27 | 7.5 | −0.018 | 4.13 | 8.1 | −0.011 | ||
| qSDW2a | 2 | 47.89–59.89 | 5.56 | 1.2 | 0.008 | 3.04 | 1.3 | 0.006 | ||
| qSDW2b | 2 | 95.70–111.45 | 5.81 | 2.7 | −0.013 | 5,22 | 2.4 | −0.011 | ||
| unnamed | 2 | http://archive.gramene.org/db/qtl/ | ||||||||
| qSDW3 | 3 | 33.89–51.21 | 4.49 | 2.3 | 0.012 | |||||
| qSDW4 | 4 | 123.31–127.72 | 5.35 | 3.5 | 0.005 | 4.15 | 3.4 | 0.009 | ||
| qSDW5a | 5 | 27.92–34.00 | 6.08 | 5.1 | −0.001 | 3.85 | 10.2 | −0.004 | ||
| unnamed | 5 | Huang et al. 2004 | ||||||||
| qSDW5b | 5 | 112.96–115.27 | 2.98 | 5.9 | −0.019 | |||||
| qSDW6a | 6 | 37.91–38.68 | 5.81 | 3.3 | −0.015 | |||||
| qSDW6b | 6 | 53.36–54.32 | 3.70 | 3.1 | −0.015 | 3.31 | 2.7 | −0.004 | ||
| qTDW6-2 | 6 | Cui et al. 2002 | ||||||||
| qSDW8a | 8 | 22.53–27.34 | 6.33 | 1.7 | 0.001 | 2.50 | 2.2 | 0.001 | ||
| qSDW8b | 8 | 63.96–64.93 | 3.30 | 4.1 | 0.011 | |||||
| qSDW9 | 9 | 3.65–11.39 | 5.30 | 2.8 | 0.002 | 2.59 | 3.2 | 0.009 | ||
| qSDW10a | 10 | 44.47–57.33 | 5.79 | 1.9 | 0.010 | 2.83 | 1.9 | 0.007 | ||
| qSDW10b | 10 | 66.03–72.15 | 3.75 | 11.9 | 0.027 | |||||
| qSDW12a | 12 | 64.46–72.76 | 3.99 | 10.2 | −0.017 | |||||
| qSDW12b | 12 | 101.84–106.21 | 4.20 | 2.0 | −0.011 | 3.38 | 6.4 | −0.013 | ||
|
| ||||||||||
| TSW | qTSW1 | 1 | 16.98–26.32 | 3.11 | 6.9 | 0.633 | ||||
| gw1a | 1 | Hua et al. 2002 | ||||||||
| qTSW2 | 2 | 134.00–148.00 | 3.61 | 12.9 | 0.896 | 3.52 | 12.9 | 0.972 | ||
| qTGWT-2-2 | 2 | Zhuang et al. 2002 | ||||||||
| qTSW3 | 3 | 41.85–42.63 | 3.41 | 4.8 | 0.597 | |||||
| qTSW4a | 4 | 27.25–39.00 | 2.80 | 5.3 | 0.611 | |||||
| qTSW4b | 4 | 118.10–134.12 | 2.84 | 13.1 | 0.884 | |||||
| tgwt4 | 4 | Lin et al. 1996 | ||||||||
| qTSW8 | 8 | 55.47–60.87 | 2.62 | 3.1 | 0.435 | |||||
| qTSW9 | 9 | 49.00–51.82 | 2.97 | 2.2 | −0.405 | |||||
| qTSW11 | 11 | 3.69–16.92 | 3.23 | 8.8 | 0.788 | |||||
SSL: seedling shoot length, SRL: seedling root length, SDW: seedling dry weight, SWW: seedling wet weight, TSW: 1000-seed weight.
Fig. 4.
Location of QTLs for SSL, SRL, SWW, SDW and TSW from Lingshui and Hangzhou on the genetic map. SSL: seedling shoot length, SRL: seedling root length, SDW: seedling dry weight, SWW: seedling wet weight, TSW: 1000-seed weight.
Eighteen QTLs were responsible for SSL, and nine of them were detected from two sites. The phenotypic variance explained by a single QTL ranged from 1.1% to 13.6%. One major QTL qSSL1b had highest LOD (5.48) from Lingshui and relative higher LOD (3.77) from Hangzhou, with R2 of 10.5% and 11.2%, respectively. The additive effect of qSSL1b showed negative, indicating that the positive allele from PA64s contributed to the length of seedling shoot. Genetic distance of qSSL1b was mapped between 117.12 and 133.43 cM, the same locus with qCSH1, a previously reported major QTL for seedling height (Han et al. 2007).
Seven QTLs associated with SRL were detected on chromosomes 2, 3, 5, 7, 8 and 10, respectively. Most of the positive alleles of QTLs were from 93-11. As seedling root length of seeds from Hangzhou was much longer than that from Lingshui (Table 1), only two QTLs were identified from two sites, indicating seedling vigor affected by seeds harvested in different environments.
SWW and SDW are closely related traits, and correlation coefficient parameter of these two traits reached more than 90% (Table 2). As a result, though 14 QTLs for SWW and 18 QTLs for SDW were identified, most of them shared the same locus. Cluster of qSWW1c and qSDW1c detected from two sites also contained the major QTL qSSL1b.
Detection of QTLs for 1000-seed weight
A total of 8 QTLs were detected for 1000-seed weight in Lingshui and/or Hangzhou, distributing on chromosomes 1, 2, 3, 4, 8, 9 and 11 (Table 3, Fig. 4). Only one QTL qTSW2 was identified in both sites, which revealed the trait was environment dependent. The QTL qTSW1 was mapped to the locus overlapped with qSWW1a, consistent with significant correlation between 1000-seed weight and SWW.
Fine mapping of qSSL1b
We produced a CSSL carrying the major QTL qSSL1b from PA64s in 93-11 background by a repeated backcrossing to 93-11. Then phenotypic character was measured in F2 population including 1,233 individuals derived from a CSSL-qSSL1b BC3F1 line exhibiting heterozygous across the entire qSSL1b region screened with markers SNP1-202 and SNP1-257. By comparing the sequences of the parents, four InDel markers were developed (Table 4). Combining the genotype and phenotype of homozygous individuals, the QTL was fine mapped between two InDel markers IND1-3 and IND1-4 within around 80.5-kb region of the long arm of chromosome 1 (Fig. 5), where 16 annotated genes were identified by Rice Genome Annotation Project (http://rice.plantbiology.msu.edu/cgi-bin/gbrowse/rice/), including one encoding retrotransposon and four coding for expressed protein (Table 5).
Fig. 5.
Graphical genotypes of BC3F2 generation and location of qSSL1b. White and black boxes indicate homozygous regions for 93-11 and PA64s, respectively. Mean seedling shoot length (SSL) and standard deviation values were included. ** indicate significant difference (P < 0.01) between 93-11 and PA64s or BC3F2 individual by t test.
Table 5.
Annotated genes included in the 80.5 kb region for qSSL1b
| Gene ID | Annotation |
|---|---|
| LOC_Os01g65870 | expressed protein |
| LOC_Os01g65880 | nodulin MtN3 family protein, putative, expressed |
| LOC_Os01g65890 | DNA repair metallo-beta-lactamase, putative, expressed |
| LOC_Os01g65900 | chitin-inducible gibberellin-responsive protein, putative, expressed |
| LOC_Os01g65902 | apocytochrome f precursor, putative, expressed |
| LOC_Os01g65904 | expressed protein |
| LOC_Os01g65920 | F-box/LRR-repeat protein 2, putative, expressed |
| LOC_Os01g65940 | expressed protein |
| LOC_Os01g65950 | thioesterase family protein, putative, expressed |
| LOC_Os01g65970 | transcription factor HBP-1b, putative, expressed |
| LOC_Os01g65980 | retrotransposon protein, putative, Ty3-gypsy subclass, expressed |
| LOC_Os01g65986 | DUF803 domain containing, putative, expressed |
| LOC_Os01g65992 | expressed protein |
| LOC_Os01g66000 | NADH dehydrogenase I subunit N, putative, expressed |
| LOC_Os01g66010 | amino acid transporter, putative, expressed |
| LOC_Os01g66020 | protein kinase family protein, putative, expressed |
Discussion
Seedling vigor is mainly determined by genetic inheritance, conditions of seed storage and environments of germination and early seedlings growth stage (Sun et al. 2007, Yang et al. 2015). In this study, seeds of the 132 RILs from Lingshui and Hangzhou were stored at 4°C before germination, and seedlings were grown in a nutrient solution in greenhouse. By these measurements, the influence of non-genetic factors on QTL detection for seedling vigor can be greatly reduced. McKenzie et al. (1980) reported that seedling traits measured under controlled laboratory conditions were correlated with those under field conditions, demonstrating that QTLs identified for seedling vigor in laboratory could be applied in the field. In addition, experiments conducted in laboratory have its advantages: environment factors can be easily controlled and experiments can be conducted at any time. However, owing to different environmental conditions in Lingshui and Hangzhou during filling stage, only 28 of 57 QTLs for seedling vigor were shared by the seeds harvested from two locations.
It was proposed that seedling vigor is greatly influenced by seed weight or seed size (Milosevic et al. 2010). Therefore, correlation coefficients between 1000-seed weight and the four indices were analyzed and significant correlations were found between 1000-seed weight and SWW or SDW, suggesting seed weight may contribute to seedling vigor at seedling stage (Table 2). And we found 15 QTL clusters for seedling vigor and TSW, revealing they may be controlled by the same locus.
Seedling vigor is defined as seed properties which determine the potential for rapid, uniform emergence and development of normal seedlings (McDonald 1993). Therefore, the most crucial step in QTL mapping for seedling vigor is the evaluation and screening of the quantitative traits (Wang et al. 2010). We only measured SSL, SRL, SWW and SDW during early seedling growth stage because they were identified as representative indicators for seedling vigor (Redoña and Mackill 1996, Regan et al. 1992). Previous study also found that germination rate (speed) and early seedling growth were correlated in rice (Cui et al. 2002).
Based on a high quality genetic map from sequencing of the RIL genomes (Gao et al. 2013), a total of 57 QTLs were identified for seedling vigor during early seedling growth stage. So far, other researchers have identified several major QTLs for seedling vigor authentically (Dang et al. 2014, Huang et al. 2004, Liu et al. 2014, Wang et al. 2010) and some QTLs were fine mapped to a narrow region and even cloned (Abe et al. 2012, Fujino et al. 2008, 2011, Xie et al. 2014, Yano et al. 2012). Actually, 24 QTLs detected in our study shared the same loci with previously identified QTLs. For example, qSSL4, a QTL detected for seedling shoot length was consistent with qPHS4, a QTL for height of seedlings mapped between markers RM3534 and RM349 in Abe’s study (Abe et al. 2012). The QTL for seedling dry weight on chromosome 6, named as qSDW6b, shared the same locus with qTDW6-2 reported by Cui et al. (2002). In addition, 33 QTLs were first detected here for seedling vigor, such as loci for SWW and SDW on chromosome 12. Owing to high density of SNP markers in our genetic map, most QTLs were detected in a relatively narrower region compared with previous studies (1.93 Mb vs 8.80 Mb on average). However, we did not detect a QTL covering OsGA20ox1, a gene reported to increase plant height and leaf sheath length at the initial growth stage (Yano et al. 2012).
In the study, we identified and delimited a major QTL for seedling shoot length, qSSL1b within 80.5 kb region on chromosome 1, where 16 genes were annotated (Table 5), including the candidate gene for ph1, a major locus affecting plant height in rice encoding a chitin-inducible gibberellin-responsive protein (Kovi et al. 2011). Certainly, a larger genetic population is required for further fine mapping and cloning of the QTL. Meanwhile, series of InDel markers developed here will help improve rice seedling vigor by marker assisted selection (MAS) in future.
Acknowledgments
This work was supported by Grants from the National Natural Science Foundation of China (Grant No 31471167).
Literature Cite
- Abe, A., Takagi, H., Fujibe, T., Aya, K., Kojima, M., Sakakibara, H., Uemura, A., Matsuoka, M. and Terauchi, R. (2012) OsGA20ox1, a candidate gene for a major QTL controlling seedling vigor in rice. Theor. Appl. Genet. 125: 647–657. [DOI] [PubMed] [Google Scholar]
- Anandan, A., Anumalla, M., Pradhan, S.K. and Ali, J. (2016) Population structure, diversity and trait association analysis in rice (Oryza sativa L.) germplasm for early seedling vigor (ESV) using trait linked SSR markers. PLoS ONE 11: e0152406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, L.Y., Zhu, J., Ren, L.F., Zhao, S.T. and Yan, Q.C. (2002) Mapping QTLs and epistasis for seeding vigor in rice (Oryza sativa L.). Acta Agronomica Sinica 28: 809–815. [Google Scholar]
- Cui, K.H., Peng, S.B., Xing, Y.Z., Xu, C.G., Yu, S.B. and Zhang, Q. (2002) Molecular dissection of seedling-vigor and associated physiological traits in rice. Theor. Appl. Genet. 105: 745–753. [DOI] [PubMed] [Google Scholar]
- Dang, X., Thi, T.G., Dong, G., Wang, H., Edzesi, W.M. and Hong, D. (2014) Genetic diversity and association mapping of seed vigor in rice (Oryza sativa L.). Planta 239: 1309–1319. [DOI] [PubMed] [Google Scholar]
- Fujino, K., Sekiguchi, H., Matsuda, Y., Sugimoto, K., Ono, K. and Yano, M. (2008) Molecular identification of a major quantitative trait locus, qLTG3-1, controlling low-temperature germinability in rice. Proc. Natl. Acad. Sci. USA 105: 12623–12628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujino, K. and Sekiguchi, H. (2011) Origins of functional nucleotide polymorphisms in a major quantitative trait locus, qLTG3-1, controlling low-temperature germinability in rice. Plant Mol. Biol. 75: 1–10. [DOI] [PubMed] [Google Scholar]
- Gao, Z.Y., Zhao, S.C., He, W.M., Guo, L.B., Peng, Y.L., Wang, J.J., Guo, X.S., Zhang, X.M., Rao, Y.C., Zhang, C.et al. (2013) Dissecting yield-associated loci in super hybrid rice by resequencing recombinant inbred lines and improving parental genome sequences. Proc. Natl. Acad. Sci. USA 110: 14492–14497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han, L., Qiao, Y., Zhang, S., Zhang, Y., Cao, G., Kim, J., Lee, K. and Koh, H. (2007) Identification of quantitative trait loci for cold response of seedling vigor traits in rice. J. Genet. Genomics 34: 239–246. [DOI] [PubMed] [Google Scholar]
- Han, L.Z., Zhang, Y.Y., Qiao, Y.L., Cao, G.L., Zhang, S.Y., Kim, J.H. and Koh, H.J. (2006) Genetic and QTL analysis for low-temperature vigor of germination in rice. Yi Chuan Xue Bao 33: 998–1006. [DOI] [PubMed] [Google Scholar]
- Hua, J.P., Xing, Y.Z., Xu, C.G., Sun, X.L., Yu, S.B. and Zhang, Q. (2002) Genetic dissection of an elite rice hybrid revealed that heterozygotes are not always advantageous for performance. Genetics 162: 1885–1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, X.Z., Qian, Q., Liu, Z.B., Sun, H.Y., He, S.Y., Luo, D., Xia, G.G., Chu, C.C., Li, J.Y. and Fu, X.D. (2009) Natural variation at the DEP1 locus enhances grain yield in rice. Nat. Genet. 41: 494–497. [DOI] [PubMed] [Google Scholar]
- Huang, Z., Yu, T., Su, L., Yu, S.B., Zhang, Z.H. and Zhu, Y.G. (2004) Identification of chromosome regions associated with seedling vigor in rice. Yi Chuan Xue Bao 31: 596–603. [PubMed] [Google Scholar]
- Kovi, M.R., Zhang, Y., Yu, S., Yang, G., Yan, W. and Xing, Y. (2011) Candidacy of a chitin-inducible gibberellin-responsive gene for a major locus affecting plant height in rice that is closely linked to Green Revolution gene sd1. Theor. Appl. Genet. 123: 705–714. [DOI] [PubMed] [Google Scholar]
- Lin, H.X., Qian, H.R., Zhuang, J.Y., Lu, J., Min, S.K., Xiong, Z.M., Huang, N. and Zheng, K.L. (1996) RFLP mapping of QTLs for yield and related characters in rice (Oryza sativa L.). Theor. Appl. Genet. 92: 920–927. [DOI] [PubMed] [Google Scholar]
- Liu, L.F., Lai, Y.Y., Cheng, J.P., Wang, L., Du, W.L., Wang, Z.F. and Zhang, H.S. (2014) Dynamic quantitative trait locus analysis of seed vigor at three maturity stages in rice. PLoS ONE 9: e115732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, J., Tang, S.Q., Hu, P.S., Aleman, L., Jiao, G.A. and Tang, J. (2007) Analysis on factors affecting seedling establishment in rice. Rice Science 14: 27–32. [Google Scholar]
- McCouch, S.R. (2008) Gene nomenclature system for rice. Rice 1: 72–84. [Google Scholar]
- McDonald, M.B. (1993) The history of seed vigor testing. J. Seed Technol. 17: 93–101. [Google Scholar]
- McKenzie, K.S., Rutger, J.N. and Peterson, M.L. (1980) Relation of seedling vigor to semidwarfism, early maturity, and pubescence in closely related rice lines. Crop Sci. 20: 169–172. [Google Scholar]
- Milosevic, M., Vujakovic, M. and Karagic, D. (2010) Vigour tests as indicators of seed viability. Genetika 42: 103–118. [Google Scholar]
- Rao, A.N., Johnson, D.E., Sivaprasad, B., Ladha, J.K. and Mortimer, A.M. (2007) Weed management in direct-seeded rice. Adv. Agron. 93: 153–255. [Google Scholar]
- Redoña, E.D. and Mackill, D.J. (1996) Mapping quantitative trait loci for seedling vigor in rice using RFLPs. Theor. Appl. Genet. 92: 395–402. [DOI] [PubMed] [Google Scholar]
- Regan, K.L., Siddique, K.H.M., Turner, N.C. and Whan, B.R. (1992) Potential for increasing early vigour and total biomass in spring wheat. II. characteristics associated with early vigour. Aust. J. Agric. Res. 43: 541–553. [Google Scholar]
- Song, X.J., Huang, W., Shi, M., Zhu, M.Z. and Lin, H.X. (2007) A QTL for rice grain width and weight encodes a previously unknown RING-type E3 ubiquitin ligase. Nat. Genet. 39: 623–630. [DOI] [PubMed] [Google Scholar]
- Sun, Q., Wang, J.H. and Sun, B.Q. (2007) Advances on seed vigor physiological and genetic mechanisms. Agric. Sci. China 6: 1060–1066. [Google Scholar]
- Wang, Y.X., Xiong, G.S., Hu, J., Jiang, L., Yu, H., Xu, J., Fang, Y.X., Zeng, L.J., Xu, E.B., Xu, J.et al. (2015) Copy number variation at the GL7 locus contributes to grain size diversity in rice. Nat. Genet. 47: 944–948. [DOI] [PubMed] [Google Scholar]
- Wang, Z.F., Wang, J.F., Bao, Y.M., Wang, F.H. and Zhang, H.S. (2010) Quantitative trait loci analysis for rice seed vigor during the germination stage. J. Zhejiang Univ. Sci. B11: 958–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, L., Tan, Z., Zhou, Y., Xu, R., Feng, L., Xing, Y. and Qi, X. (2014) Identification and fine mapping of quantitative trait loci for seed vigor in germination and seedling establishment in rice. J. Integr. Plant Biol. 56: 749–759. [DOI] [PubMed] [Google Scholar]
- Yang, P., Chen, C.L., Zou, G.X., Peng, Z.Q., Wu, Y.S., Huang, Y.P., Xiong, Y.H. and Yin, J.H. (2015) Research progress in relevant theories of increasing breeding level of direct-seeding rice. Acta Agriculturae Jiangxi 27: 33–35. [Google Scholar]
- Yano, K., Takashi, T., Nagamatsu, S., Kojima, M., Sakakibara, H., Kitano, H., Matsuoka, M. and Aya, K. (2012) Efficacy of microarray profiling data combined with QTL mapping for the identification of a QTL gene controlling the initial growth rate in rice. Plant Cell Physiol. 53: 729–739. [DOI] [PubMed] [Google Scholar]
- Zhuang, J.Y., Fan, Y.Y., Rao, Z.M., Wu, J.L., Xia, Y.W. and Zheng, K.L. (2002) Analysis on additive effects and additive-by-additive epistatic effects of QTLs for yield traits in a recombinant inbred line population of rice. Theor. Appl. Genet. 105: 1137–1145. [DOI] [PubMed] [Google Scholar]