Abstract
Radish (Raphanus sativus L. var. sativus), a widely cultivated root vegetable crop, possesses a large sink organ (the root), implying that photosynthetic activity in radish can be enhanced by altering both the source and sink capacity of the plant. However, since radish is a self-incompatible plant, improved mutation-breeding strategies are needed for this crop. TILLING (Targeting Induced Local Lesions IN Genomes) is a powerful method used for reverse genetics. In this study, we developed a new TILLING strategy involving a two-step mutant selection process for mutagenized radish plants: the first selection is performed to identify a BC1M1 line, that is, progenies of M1 plants crossed with wild-type, and the second step is performed to identify BC1M1 individuals with mutations. We focused on Rubisco as a target, since Rubisco is the most abundant plant protein and a key photosynthetic enzyme. We found that the radish genome contains six RBCS genes and one pseudogene encoding small Rubisco subunits. We screened 955 EMS-induced BC1M1 lines using our newly developed TILLING strategy and obtained six mutant lines for the six RsRBCS genes, encoding proteins with four different types of amino acid substitutions. Finally, we selected a homozygous mutant and subjected it to physiological measurements.
Keywords: reverse genetics, TILLING, radish, Raphanus sativus, self-incompatibility, sink capacity, Rubisco small subunit
Introduction
The leaf photosynthetic rate is an important potential breeding target for improving crop yields (Long et al. 2006, Raines 2011). There are large variations in photosynthetic capacity among plant genotypes and even within a single species (Driever et al. 2014, Flood et al. 2011), implying that photosynthetic productivity can be manipulated using genetic and molecular biology techniques. Photosynthetic capacity is potentially limited by various physical and biochemical parameters. First, upregulation of the amount and activity of photosynthetic proteins, such as electron transport components and Calvin cycle enzymes, may increase photosynthetic capacity (Farquhar et al. 1980). Second, limiting CO2 diffusion by altering various parameters, such as the size and density of stomata and mesophyll conductance, is an important way to increase photosynthetic capacity because the photosynthetic rate strongly depends on the CO2 concentration at the carboxylation site (Farquhar and Sharkey 1982). Third, altering carbon utilization in sink organs (sink strength) also influences the photosynthetic rate; the accumulation of carbohydrates such as sugars imposes negative feedback regulation on photosynthesis (Krapp and Stitt 1995, Sheen 1994). Such downregulation of photosynthesis is often observed in plants grown under elevated CO2 levels (Ainsworth and Rogers 2007).
Radish (Raphanus sativus L. var. sativus) is a widely recognized medicinal root vegetable crop that was first domesticated in Europe (Lugasi et al. 2005). Radish is an ideal target for improving photosynthetic productivity because genome information for this crop is currently available (Kitashiba et al. 2014a). Furthermore, there are many radish cultivars with different sink strengths (sink sizes). For example, ‘Sakurajima’ radish is the largest variety worldwide, achieving a weight of 6 kg, whereas some leafy varieties such as ‘Kosena’ do not have enlarged hypocotyls or roots. Radish can be used as a model plant to study the effects of sink-source balance on photosynthesis and growth (Sugiura et al. 2015, Usuda and Shimogawara 1998). Establishing a reverse genetics method to produce mutant radish plants would facilitate investigations of the molecular mechanisms underlying source-sink balance and the breeding of high-yielding plants.
Artificial genetic modification is a useful strategy for breeding plants with improved traits. The recently developed genome editing tools CRISPR/Cas9 and TALEN are powerful methods that can be used in reverse genetics studies in any organism (Mao et al. 2017, Nemudryi et al. 2014, Rani et al. 2016, Zhu et al. 2017). However, the altered genomes resulting from the use of these tools are not completely welcome in vegetables and other crops grown for human use. Furthermore, genetic transformation techniques have not yet been established for radish. TILLING (Targeting Induced Local Lesions IN Genomes) is a powerful tool for reverse genetics to detect mismatched sequences induced via point mutations (with EMS) by the specific nuclease, CEL 1, which does not require the use of genetic transformation (Wang et al. 2012). TILLING is a particularly efficient tool for mutation breeding in plants. The use of this method has dramatically expanded since its initial use in the model plant Arabidopsis thaliana (McCallum et al. 2000a, 2000b). Over the past 15 years, TILLING has been adapted for use in major model plants, such as rice, wheat, and barley (Acevedo-Garcia et al. 2017, Hwang et al. 2016, Lai et al. 2012), as well as major vegetable crops including tomato, soybean, pumpkin, canola, cucumber, melon, and oilseed rape (Anai 2012, Boualem et al. 2014, Okabe et al. 2011, Rashid et al. 2011, Stephenson et al. 2010). Although this method continues to be developed (Colbert et al. 2001), to date, a TILLING platform has not been established for the self-incompatible plant radish.
In the present study, we developed a TILLING strategy for use with the self-incompatible plant radish. We chose ribulose 1,5-bisphosphate carboxylase/oxygenase (Rubisco) as the primary target because it is the most abundant plant protein and the most important enzyme, catalyzing the first step of CO2 fixation (carboxylation) during photosynthesis at the chloroplast stroma (Evans 1989). Rubisco is composed of two types of subunits, i.e., large and small octamers formed by 55 kDa and 15 kDa proteins, respectively, in all photosynthetic organisms (Spreitzer 2003). The small subunit (RBCS) is encoded by multiple genes in the nuclear genome and is post-translationally imported and processed by the chloroplast. In Arabidopsis thaliana, a major model Brassica plant, the RBCS gene family consists of four members divided into two subfamilies based on sequence similarity. Type B includes three genes (RBCS 1B, 2B, and 3B) that are tandemly arranged at a single locus on chromosome 5. Type A includes the fourth gene (RBCS 1A) on chromosome 1, which is shorter than type B genes (Krebbers et al. 1988). The expression of RBCS genes is differentially regulated by light quality and condition (Dedonder et al. 1993). The five RBCS genes in tomato are also differentially expressed in each tissue (Wanner and Gruissem 1991). Brassica rapa, which is closely related to radish, contains six RBCS genes, including four and two on chromosomes 2 and 4, respectively, among the 10 chromosomes of this species (Anisimov et al. 2007). While RBCS genes have been analyzed in some plant species, radish RBCS (RsRBCS) genes remain to be identified.
To obtain a more accurate estimate of mutation density from TILLING, the target genes should occupy as large a genomic region as possible. The multiple RBCS genes provide an optimum target for testing the quality of TILLING methods. Here, we identified RBCS genes in radish and developed a TILLING strategy to obtain homozygous mutant plants with amino acid substitutions in RBCS proteins.
Materials and Methods
Plant materials and EMS mutagenesis
Open pollinated radish (Raphanus sativus L. var. sativus cv. ‘Comet’) seeds were purchased from Takii Seed Co., Ltd. (Kyoto, Japan). The seeds were vernalized by soaking in water at 4°C in the dark for 2 weeks prior to EMS treatment. The seeds were incubated in solutions containing various concentrations of EMS (0.015–1.0%) for 15 h in a 100 mL volume (~330 seeds) at room temperature, followed by washing in a detergent solution comprising 0.1% Triton X-100. After rinsing the seeds with H2O, the treated seeds were sown in pots containing commercial soil with nutrients (Nippi Engei Baido 1; Nihon Hiryo Co., Tokyo, Japan). The treatments were independently performed four times (total of 4300 seeds) in the spring and fall for a 2-year period. EMS-treated seeds (M1) were sown in a square container (56 cm × 17 cm × 17 cm) and transferred to a greenhouse. After 1 month of growth, the plants were thinned to eight plants per container.
DNA extraction and pooling
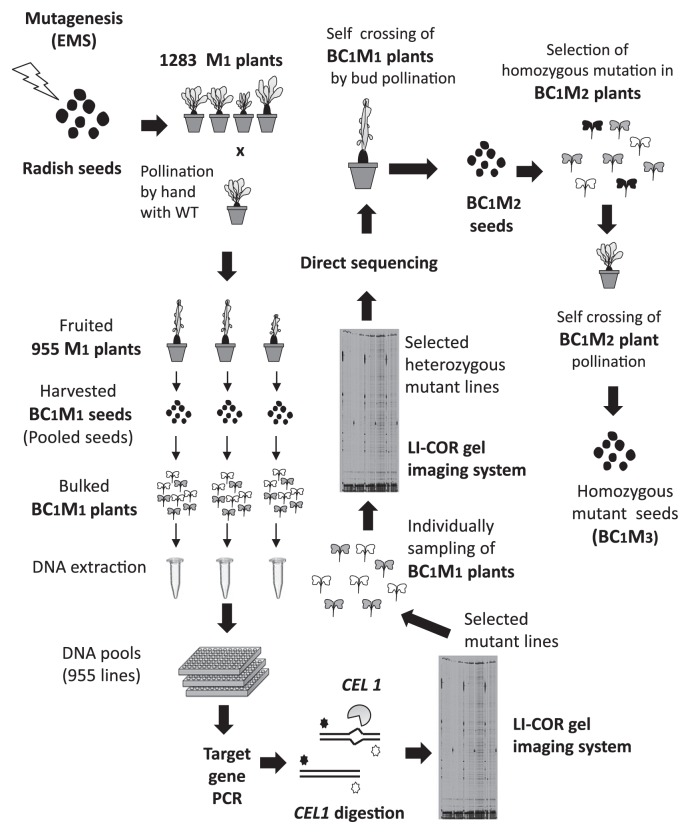
The mutagenized 1283 M1 plants were grown and open-pollinated with wild-type (‘Comet’) pollen by hand at the mature stage. Seeds were successfully harvested from 955 M1 individuals (Fig. 3). These seeds, which were the progenies of M1 plants (♀) backcrossed with wild-type radish (‘Comet’) plants (♂), were termed BC1M1 in this study. To obtain bulked DNA for the TILLING assay, eight plants were grown for each BC1M1 line, and genomic DNA was extracted from the bulked leaf samples using a Chloropure Kit (Beckman Coulter, CA, USA). The bulked DNA samples from the 955 BC1M1 individuals were stored at −20°C.
Fig. 3.
The Radish-TILLING platform. In this TILLING system, radish seeds were mutagenized with the chemical mutagen EMS to produce M1 plants. The M1 plants were crossed with wild-type plants because radish is self-incompatible, producing 955 BC1M1 lines. DNA was extracted from eight bulked BC1M1 plants per line for analysis. The target DNA was amplified using a forward primer labeled with 700 nm dye and a reverse primer labeled with 800 nm dye. The amplified PCR products were digested with CEL 1, and the resulting DNA fragments were separated and detected in the 700 and 800 dye channels of a LI-COR DNA Analyzer. Since the BC1M1 population (in bulk) was predicted to exhibit a mutation ratio of 1:3, a peak sequence signal corresponding to a mutation would have one-fourth the signal intensity of wild type. Therefore, it would be difficult to detect the mutated nucleotide by sequencing DNA from the BC1M1 population. To facilitate mutant identification, an individual BC1M1 plant was grown and analyzed again using TILLING gel analysis. A peak sequence signal corresponding to a mutation in the selected BC1M1 individual should have the same intensity as that of wild type. Selected BC1M1 individuals with heterozygous mutations were self-crossed by bud pollination. The homozygous mutant was selected based on the presence of a single peak in the sequencing signal representing a mutation. The population was then harvested as BC1M2 seeds from BC1M2 plants subjected to bud pollination.
Two-step TILLING, PCR amplification, and detection of mismatched DNA
High-throughput screening was performed via gel-based screening of heteroduplex PCR products as described in Till et al. (2003). Genomic DNA samples were independently extracted from eight plants per BC1M1 line. In most cases, two BC1M1 DNA samples were combined and used as template DNA for PCR. To screen the 955 BC1M1 lines, approximately 477 combined DNA samples were subjected to TILLING analysis. Forward primer labeled with 700 nm infrared fluorescent dye (IRD) label and reverse primer labeled with 800 nm IRD label attached to their 5′ ends were designed using the Tm Calculator program (Applied Biosystems; http://www6.appliedbiosystems.com/support/techtools/calc/) to identify the best amplicons for TILLING, with the aim of obtaining a predicted primer with Tm of 70°C and >30 bp, with PCR products 1000–1300 bp long. After the target genes were amplified using labeled and unlabeled primers, nucleotide mismatches of these PCR products were specifically digested with CEL 1 prepared as described in Till et al. (2003). The reactions were stopped by adding 0.15 M EDTA, pH 8.0. The CEL 1-digested PCR products were purified using a Fast Gene Gel/PCR Extraction Kit (Nippon Genetics Co., Ltd., Tokyo, Japan), loaded onto a 6.5% acrylamide gel with 7 M urea, separated by electrophoresis, and detected using a LI-COR DNA Analyzer (CA, USA). For size standards, 806 bp and 379 bp PCR products amplified from wild-type genomic DNA using primer sets IRDye700RsRBCS3A-22F (AGAGAACGAAGAAGAATTAGTC)/IRDye800RsRBCS3A784R (CGATGAAACTGATGCACTGC) and IRDye700RsRBCS4A313F (ATTCACCAACTGGAAATGCG)/IRDye800RsRBCS4A691R (ACTTCCTTCAACACTTGAGC), respectively, were used. When the PCR products were digested with CEL 1, each genomic DNA sample was independently amplified and digested with CEL 1 to identify the BC1M1 population with a mutation. The position of the mutation was confirmed by direct sequencing using a Genome Lab DTCS Quick Start Kit and Genome Lab GeXP (Beckman Coulter, CA, USA). A second TILLING was performed to isolate individual BC1M1 plants from the heterozygous populations to obtain more precise TILLING results.
Confirmation of polymorphisms in wild-type (‘Comet’)
A total of 180 wild-type radish (‘Comet’) seeds were grown in soil in small pots, and genomic DNA was extracted from the bulked four plants as described above. Each genomic DNA sample was mixed with control wild-type (‘Comet’) genomic DNA and used as template DNA to amplify the PCR products of six target DNAs with specific primers (Supplemental Table 1). After digesting the PCR products with CEL 1, DNA fragments were visualized using a LI-COR DNA Analyzer.
Measurement of photosynthetic parameters
The plants were grown in a greenhouse in an experimental garden at Tohoku University. BC1M2 seeds, i.e., the progenies of self-crossing of BC1M1 plants using the bud pollination technique, were sown in a 4-L pot filled with commercial soil with nutrients (Nippi Engei Baido 1). Homozygous mutants were selected based on the presence of single mutated sequences in individual BC1M2 lines. At 6 weeks after germination, the photosynthetic parameters of the leaves were determined using a LI-6400 portable gas exchange system (LI-COR Inc., NE, USA). The CO2-dependent photosynthesis rates (A) were measured at 35°C, saturating light of 2000 μmol photons m−2 s−1 under CO2 concentrations (Ca) of 100, 200, 300, 400, 700, 1000, and 1500 μmol mol−1. The net photosynthesis rate (A) was plotted against intercellular CO2 concentration (Ci), and RuBP carboxylation capacity was evaluated based on the initial slope of the A-Ci curve (IS) obtained with the rates measured at Ca of 100, 200, and 300 μmol mol−1 CO2.
Results
Radish plants and bud pollination
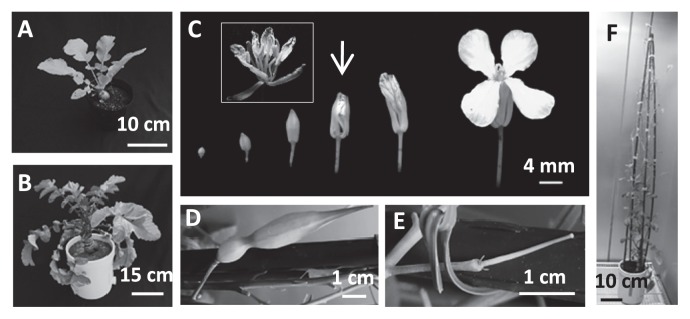
Plants from EMS-mutagenized radish seeds that were grown in soil in the greenhouse exhibited swollen bulbs at 5 weeks, started bolting at 3 months, and reached the flowering stage at 4 months after sowing (Fig. 1A, 1B, 1F). In general, M1 plants obtained from mutagenized seeds were self-crossed to obtain homozygous mutants (as M2 plants). However, it is difficult to obtain M2 radish plants by self-crossing because radish is a self-incompatible plant. Therefore, the M1 plants were crossed with wild-type (‘Comet’) pollen by open pollination (by hand). Among the 1283 M1 plants that were backcrossed with the wild type (‘Comet’), 955 M1 plants successfully produced BC1M1 seeds.
Fig. 1.
Radish plant morphology. Five-week-old plant (A) and 3-month-old plant that had initiated bolting (B). Flower development is shown in (C). A flower used for bud pollination is indicated by an arrow, and a flower in which the carpels were artificially opened is shown in the inset photograph. A seedpod that was successfully produced by self-crossing via bud pollination (D) and a pistil that failed to be pollinated (E) are shown. A 4-month-old radish plant that flowered is shown in (F).
The fertility of the M1 plants depended on the concentration of EMS used for mutagenesis (Supplemental Fig. 1). At concentrations greater than 0.75%, the M1 plants became increasingly less fertile. Fig. 1D and 1E show a fruit that was successfully produced and a pistil that failed to be fertilized, respectively. To obtain BC1M2 seeds, flowers with petals that slightly protruded from sepals were used for bud pollination (Yamaji and Ohsawa 2015). Petals, sepals, and stamens were removed from the flowers with tweezers, and stamens with pollen from the same mutated flower were attached to carpels to produce self-crossed BC1M2 plants (Fig. 1C arrow and inset). After TILLING selection, it took 1–1.5 years to obtain BC1M3 because the lifecycle of radish is long compared to the lifecycles of model plants such as A. thaliana and efficient fruit production depends on the season.
Identification of RsRBCS genes
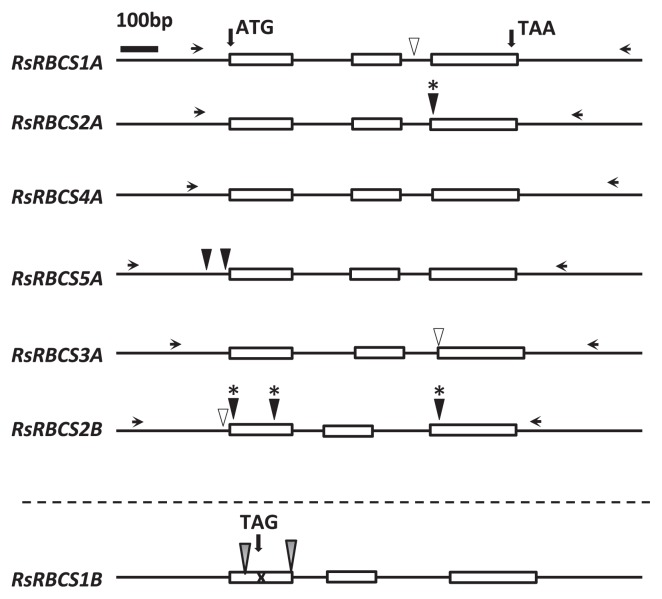
The RBCS gene family consists of two subfamilies, type A and B, in A. thaliana. Six RBCS genes have been found in Brassica rapa, which correspond to type B genes in A. thaliana, whereas type A genes like those found in A. thaliana are absent in B. rapa. To identify radish RBCS (RsRBCS) genes, we used the sequences of four AtRBCS genes in A. thaliana (At5G38410, At5G38420, At5G38430, and At1G67090) from TAIR (The Arabidopsis Information Resource; https://www.arabidopsis.org/) as queries to search for homologous regions in the two available radish genomes (Michigan State University (MSU), http://radish.plantbiology.msu.edu; and Kazusa DNA Research Institute (Kazusa), http://radish.kazusa.or.jp/) with TBLASTN software (e-value < 1e−10 and coverage > 70%) (Boratyn et al. 2013) (Supplemental Table 1). We manually removed redundant contigs in the two radish genomes and identified 10 regions homologous to RsRBCS genes (Supplemental Table 1). We designed 10 sets of primer pairs for the homologous regions to identify RsRBCS genes in ‘Comet’ (Supplemental Table 1). Only seven RsRBCS genes were amplified by PCR among the 10 RBCS genes predicted in the databases, indicating that R. sativus has seven RsRBCS genes. Supplemental Table 2 shows the putative chromosome positions of the RsRBCS genes on the nine radish chromosomes. Since genome analysis in radish has not yet been completed, we estimated the locations of these genes on the radish chromosomes based on high-resolution linkage maps between B. rapa and R. sativus ‘Daikon’ (Kitashiba et al. 2014a, 2014b). Seven RsRBCS genes were predicted to be distributed on LG1/LG4 and LG3. These genes contain three exons and two introns, a highly conserved structure throughout photosynthetic organisms (Fig. 2). The genome and amino acid sequences are also highly conserved among RsRBCS genes (Supplemental Figs. 2, 3). Furthermore, these genes were categorized into two types, RsRBCS1A–5A and RsRBCS1B/2B, based on differences in intronic sequences.
Fig. 2.
Genomic structures of radish Rubisco small subunit (RsRBCS) genes. The three exons are indicated by white boxes. Forward and reverse primers labeled with dye (indicated by black arrows) were designed to amplify approximately 1000–1300 bp PCR products for TILLING analysis. The locations of the detected mutations induced by EMS treatment are indicated by black arrowheads. Asterisks above the black arrowheads indicate effective missense mutations inducing amino acid substitutions. White arrowheads indicate nucleotide insertions or deletions, which are likely natural mutations. Gray arrowheads indicate problematic conserved mutations in the radish (‘Comet’) and R. sativus family, e.g., ‘Daikon’. RsRBCS1B is a pseudogene with a specific frame-shift site and stop codon.
Six loci are likely to be functional RBCS genes, but one gene, RsRBCS1B, was predicted to be a pseudogene because a conserved donor site in the first intron was mutated and a nonsense mutation was also found in the first exon (Fig. 2, Supplemental Fig. 2). We confirmed this prediction by analyzing the expression patterns of the genes using gel electrophoresis analysis of RT-PCR products and quantitative RT-PCR of RsRBCS1B (Supplemental Fig. 4). The seven RsRBCS genes resemble each other (Supplemental Fig. 2), with RsRBCS1B and RsRBCS2B sharing the closest homology (87%). We compared the expression level of RsRBCS1B in the wild type (‘Comet’) with that of RsRBCS2B using specific primers (Supplemental Fig. 4). As shown in Supplemental Fig. 4C and 4D, RsRBCS1B was not expressed, whereas RsRBCS2B was expressed, indicating that RsRBCS1B is indeed a pseudogene. We therefore subjected the six standard RsRBCS genes to TILLING.
Mutant screening by two-step TILLING targeting the key photosynthetic factor RBCS
We pollinated EMS-treated radish plants with wild-type radish (‘Comet’) pollen because it was difficult to self-cross M1 plants, even at the bud stage (Fig. 3). Of these M1 plants, 955 successfully produced BC1M1 seeds. Eight individual BC1M1 seedlings, with an expected wild type: heterozygous mutant ratio of 1:1, were harvested for DNA extraction. In general, approximately 1200 bp PCR products were amplified using the specific primer set IRD 700 (labeled forward primer) and IRD 800 (labeled reverse primer) for TILLING (Supplemental Table 1). Since we used DNA samples extracted from eight bulked BC1M2 seedlings, the bulked DNA samples contained wild-type sequences: mutated sequences at a ratio of 3:1. Therefore, we did not need to add non-mutated (wild-type) DNA in order to form hetero-double-stranded DNA between wild-type sequences and mutated sequences, which should be digested by CEL 1 endonuclease. After CEL 1 digestion, fluorescently-labeled DNA fragments were detected using a LI-COR DNA Analyzer (Till et al. 2003) (Supplemental Fig. 5). The putative position of the mutation was confirmed by direct sequencing, but the results were unclear because the ratio of the mutated gene to the genes examined (the mutated gene: wild-type gene) in the eight bulked BC1 M1 samples was 1:3 or less. To obtain more accurate results, a second TILLING was performed to isolate the heterozygous mutant from the BC1M1 populations.
We attempted to isolate radish mutants with nucleotide transitions in all six RsRBCS genes, but not the pseudogene (RsRBCS1B), using our TILLING strategy. Supplemental Fig. 5 shows an image of a gel in which genomic DNA from RsRBCS2B was amplified by PCR, digested with CEL 1, and separated by gel electrophoresis. The results indicate that the RsRBCS2B DNA fragments from six BC1M1 lines (lines 21, 27, 37, 50, and 52) were digested by CEL 1. In the second TILLING step, BC1M1 seeds from six TILLING-positive lines were re-sown and individually subjected to CEL 1 digestion. The mutations were confirmed by direct sequencing, resulting in the identification of mutations in lines 27, 37, and 52 from EMS-treated seeds, with a GC to AT nucleotide transition (Supplemental Table 3). A mutation was not found in line 43, and lines 21 and 50 had identical single base-pair deletions (Fig. 2).
Point mutations induced by EMS mutagenesis were detected in the five other RsRBCS genes using the same TILLING strategy, resulting in the identification of four missense mutants and two intronic mutants for the six RsRBCS genes out of the 955 BC1M1 lines screened (Table 1). Thirteen TILLING-positive PCR products were digested with CEL 1 from among the 6237 kb of DNA screened, not including the primer region. However, only six PCR products contained nucleotide transitions induced by EMS, i.e., the frequency of nucleotide transitions in TILLING-positive lines was 46%. Based on the six target genes screened, the average mutation density in the Radish-TILLING collection was estimated to be 1/1039 kb−1 (Table 1). In this mutagenesis, six mutants were successfully obtained from BC1M1 lines derived from M1 plants treated with 0.25% or 0.5% EMS (Supplemental Table 3). We estimated the mutation density obtained for each EMS concentration, finding that the efficiency of 0.25% and 0.5% EMS was 1/483 kb−1 and 1/588 kb−1, respectively (Table 1).
Table 1.
Mutation frequencies for six RBCS genes in the radish mutant population revealed by two-step TILLING
| Gene ID (EMS concentration) | PCR product screened (bp) | Primer length (bp) | No. of screened lines | Total screened length (kb) | No. of TILLING positive | No. of mutations obtained | Mutation density (kb−1) | ||
|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||
| Missense | Intronica | Other | |||||||
| RsRBCS1A | 1224 | 57 | 955 | 1114 | 1 | 0 | 0 | 1 (deletion) | 0 |
| RsRBCS2A | 1082 | 62 | 955 | 974 | 1 | 1 | 0 | 0 | 1/974 |
| RsRBCS4A | 1180 | 62 | 955 | 1067 | 0 | 0 | 0 | 0 | 0 |
| RsRBCS5A | 1166 | 70 | 955 | 1046 | 2 | 0 | 2 | 0 | 1/523 |
| RsRBCS3A | 1172 | 72 | 955 | 1050 | 4 | 0 | 0 | 4 (natural variation) | 0 |
| RsRBCS2B | 1105 | 72 | 955 | 986 | 5 | 3 | 0 | 2 (deletion) | 1/328 |
|
| |||||||||
| (0.25% EMS) | 6929 | 395 | 222 | 1450 | – | 1 | 2 | – | 1/483 |
|
| |||||||||
| (0.50% EMS) | 6929 | 395 | 270 | 1764 | – | 3 | 0 | – | 1/588 |
|
| |||||||||
| Total | 6929 | 395 | 5730 | 6239 | 13 | 4 | 2 | 7 | 1/1039 |
The mutation frequency was calculated as [total number of identified mutations]/[(PCR product size screened – total primer size) × (total number of screened lines)] (Lai et al. 2012). The average mutation frequency was estimated to be one mutation per 1039 kb.
A nucleotide was substituted out of an exon region.
In this TILLING assay, 13 TILLING-positive lines were obtained, including four missense mutations and two intronic mutations (Table 1). Among the seven remaining TILLING-positive lines, four and two lines had an identical mutation in RsRBCS3A and RsRBCS2B, respectively. Since it is unlikely that these identical mutations arose independently, we attempted to identify a polymorphism in six RsRBCS genes from the wild type (‘Comet’). First, we independently isolated genomic DNA from 20 wild-type (‘Comet’) plants, mixed the DNA with reference wild-type genomic DNA, amplified DNA fragments from six RsRBCS genes, and treated this DNA with CEL 1. CEL 1-digested-DNA fragments were not detected. Furthermore, we examined a polymorphism in RsRBCS3A and RsRBCS2B using 160 additional wild-type radish plants. One polymorphism in RsRBCS3A (G580A, R101Q in the third exon) was found in one of the 160 plants, suggesting that G580A (R101Q) in RsRBCS3A is a natural variation. The same nucleotide mutation was found in four BC1M1 lines via TILLING analysis. However, the same deletion in RsRBCS2B was not detected in the 160 wild-type radish plants.
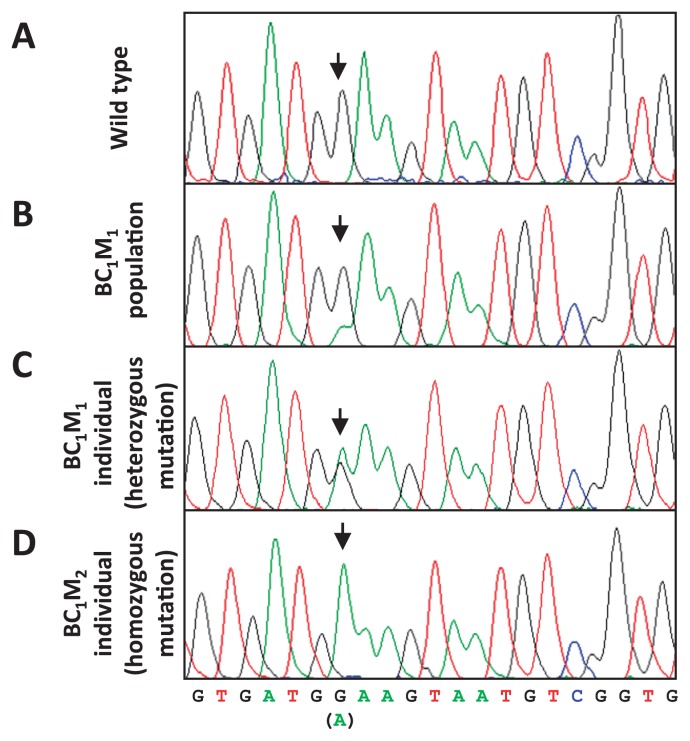
In this study, we developed an effective TILLING strategy for radish (Fig. 3). It was possible to perform direct sequencing after the first gel analysis, but the sequencing signal of the mutation harboring a nucleotide transition was not clear because the bulked sample from BC1M1 plants had a wild-type: mutated sequence ratio of 3:1 (or less) (Fig. 4A, 4B). By contrast, it is easier to detect the mutation in an individual heterozygous mutant, because the mutation can theoretically be present at a 1:1 ratio. BC1M1 seeds from the candidate lines selected after performing the first TILLING step were separately planted and subjected to a second TILLING step, yielding heterozygous mutants: wild type at a ratio of 1:1 (or less). The peak sequencing signal of the heterozygous mutated nucleotides was similar in intensity to that of the wild type (Fig. 4C). The homozygous mutant lines were subsequently segregated from BC1M2 individual plants by sequencing after self-crossing by bud pollination (Fig. 3).
Fig. 4.
Comparison of DNA sequencing chromatograms of RsRBCS2B during two-step TILLING. In the BC1M1 population (line 27), a G-to-A point mutation was detected at nucleotide 134 from ATG, which causes the amino acid substitution S45F. Direct sequencing chromatograms using DNA from wild type (A), a bulk BC1M1 population with a wild-type: mutated sequences ratio of 3:1 (or less) (B), a heterozygous mutant individual BC1M1 plant (C), and a selected homozygous mutant from a BC1M2 population (D) are shown.
In this TILLING, mutants with four different amino acid substitutions were obtained: i.e., RsRBCS2AG104R, RsRBCS2BS4F, RsRBCS2BS45F, and RsRBCS2BD118N (Supplemental Table 3). We estimated the photosynthetic activity of one of these mutants, the homozygous mutant RsRBCS2BS45F, by performing A-Ci curve analysis (A; CO2 assimilation, Ci; internal CO2) of plants grown in the field (Supplemental Fig. 6). The homozygous RsRBCS2BS45F and non-mutated control plants (homozygous RsRBCS2BWT) were segregated in the BC1M2 plant population and identified by sequencing. The each values for the A-Ci curve were collected at 100, 200, 300, 400, 500, 700, 1000, and 1500 μmol mol−1 CO2 concentrations under a light intensity of 2000 μmol photons m−2 s−1. The maximum CO2 assimilation rate under 1000 μmol mol−1 CO2 in the control and the homozygous mutant was 39.83 ± 2.60 and 40.70 ± 5.46, respectively (Supplemental Fig. 6A). We calculated the RuBP carboxylation rates based on the initial slopes of the A-Ci curve (under 100, 200, and 300 μmol mol−1 CO2), finding that the photosynthetic activity of RsRBCS2BS45F was similar to that of the control (Supplemental Fig. 6B), suggesting that one amino acid substitution in this highly redundant protein has no effect on photosynthetic parameters.
Discussion
In the present study, we established a new two-step TILLING platform for radish (Radish-TILLING). Since radish is a self-incompatible plant, we used an unconventional technique for pollination to obtain progeny. Fortunately, the crossing problem in this plant family can be overcome using bud pollination, but the fertility ratio is extremely low. Instead of self-crossing the mutagenized M1 plants, they were backcrossed with wild-type plants (‘Comet’), as we needed to collect as many populations of the next generation and as many seeds as possible for TILLING. As a result, the BC1M1 populations included wild-type and heterozygous-mutant plants at a ratio of approximately 1:1. Meanwhile, the use of backcrossing makes it difficult to identify mutations via direct sequencing of bulked samples of BC1M1 DNA, which include wild-type and mutated sequences at a ratio of 3:1 (Fig. 4). If the mutated chromosome migrates to all megaspore mother cells, half of the egg cells would contain the mutated chromosome. However, the actual ratio of mutation: wild-type is thought to be 1:3 or less because the germline in M1 plants is a chimera of wild type and heterozygous mutants and because not all female megaspore mother cells have mutations. This estimation is consistent with our results, as heterozygous mutants were usually found in 10–50% of BC1M1 plants in our study. This mutation rate in the BC1M1 population reduces the strength of the sequencing signal from the mutated nucleotide when using DNA from this population. To facilitate the identification of a mutation by sequencing, we used an improved TILLING strategy in which the BC1M1 pool was individually re-planted and individually subjected once again to gel electrophoresis (second TILLING step) to identify a heterozygous BC1M1 mutant individual. As a result, the intensity of the sequencing-signal peak of the mutated nucleotide in DNA from the heterozygous BC1M1 mutant individual became similar to that of the wild type.
In this study, we obtained 13 TILLING-positive lines from a total of 6239 bp of DNA, with mutations in six target genes, including six nucleotide transitions, four natural variations, and three deletions (Table 1). Analysis of polymorphisms using 180 wild-type plants revealed that the nucleotide transition (G580A) in RsRBCS3A found in four BC1M1 lines is also present in wild-type radish (‘Comet’), suggesting that it represents a natural variation. By contrast, a deletion of the nucleotide A at position −13 from ATG in RsRBCS2B that was found in two BC1M1 lines, but not in the 180 wild-type (‘Comet’) plants, implying a possibility that this deletion does not represent a polymorphism in wild-type plants. EMS predominantly induces nucleotide transitions from GC to AT. However, Shirasawa et al. (2016) showed that among mutations detected in EMS-treated tomato plants, 1.3% were INDELS (insertions/deletions) of nucleotides whereas 98.7% were single nucleotide transitions. Therefore, the possibility that these INDELS detected in the current study were induced by EMS mutagenesis cannot be ruled out. On the other hand, the six nucleotide transitions were likely induced by EMS, as they were GC/AT transitions and we were not able to find two or more BC1M1 lines that had the same GC/AT nucleotide transition in our screening. Since the mutation density in TILLING analysis is generally estimated by counting only GC/AT transitions, we calculated this rate in our radish-TILLING using six nucleotide transitions. As a result, the mutation density under 0.25% and 0.5% EMS treatment in this study was estimated to be 1/483 kb and 1/588 kb, respectively (Table 1).
According to a recent report, tetraploid plants are more tolerant to EMS than diploid plants, suggesting that polyploidy affects the mutation efficiency of EMS treatment (Tsai et al. 2013). In previous TILLING analyses of various crops, the mutant densities of diploid plants were usually estimated to be between 1/100 kb and 1/700 kb (Rashid et al. 2011). The mutation density estimated in the current study is similar to that obtained in diploids using other TILLING approaches because radish is a diploid plant.
Four amino acid substitution mutants and two mutants with no effect (in introns) were selected from approximately 1000 BC1M2 populations, but no nonsense mutants were obtained. RBCS genes are highly redundant, which explains why an amino acid substitution in a single protein had no effect on photosynthesis activity (Supplemental Fig. 6). There are at least two possible explanations for this: perhaps the mutation had no effect on Rubisco activity, or perhaps other redundant RBCS proteins complement the activity of the mutant enzyme. Indeed, single knock-out T-DNA mutants of RBCS1A or RBCS3B in A. thaliana have the same plant mass and maximum quantum yield in Photosystem II as the wild type, whereas the double mutant has drastic phenotypes (Izumi et al. 2012). To reduce the amounts and activities of a highly redundant component, multiple mutants would be required. It might be possible to produce double mutants from some of our RsRBCS mutants because RsRBCS2A and RsRBCS1A are likely located on chromosomes LG3 and LG1/LG4, respectively (Supplemental Table 2).
Radish can be used as a model plant to investigate the effects of source-sink balance on photosynthesis and growth (Sugiura et al. 2015, Usuda and Shimogawara 1998). Rubisco, which contains 20–40% of total leaf nitrogen, functions in a rate-limiting step of photosynthesis at ambient and lower CO2 concentrations under high light (von Caemmerer and Farquhar 1981), but this protein is present in excess under elevated CO2 conditions because RuBP regeneration and triose phosphate utilization limit photosynthesis (Sage 1990). Reducing Rubisco contents may improve nitrogen use efficiency during plant growth at elevated CO2 levels (Hikosaka and Hirose 1998, Medlyn 1996, Sage 1994). The accumulation of carbohydrates (e.g., sugar or starch) in the chloroplast also affects photosynthetic activity and plant growth (Abramson et al. 2016). Forward genetics or genome analyses in model plants such as A. thaliana should increase our understanding of the factors that limit photosynthesis. The Radish-TILLING platform developed in this study could facilitate such analyses, as well as plant breeding research.
Supplementary Information
Acknowledgments
We are grateful to Prof. Hideyuki Takahashi (Tohoku University) for stimulating discussions, and Dr. Qing-Wei Wang and Mr. Tomoyuki Kawasaki (Tohoku University) for assistance with radish cultivation in the greenhouse. We also thank Dr. Yoshinobu Takada, Prof. Kinya Toriyama, and Prof. Masao Watanabe for advice on how to describe the generation of plants. This work was supported by JST CREST Grant Number JPMJCR11B3, Japan.
Literature Cited
- Abramson, B.W., Kachel, B., Kramer, D.M. and Ducat, D.C. (2016) Increased photochemical efficiency in Cyanobacteria via an engineered sucrose sink. Plant Cell Physiol. 57: 2451–2460. [DOI] [PubMed] [Google Scholar]
- Acevedo-Garcia, J., Spencer, D., Thieron, H., Reinstadler, A., Hammond-Kosack, K., Phillips, A.L. and Panstruga, R. (2017) mlo-based powdery mildew resistance in hexaploid bread wheat generated by a non-transgenic TILLING approach. Plant Biotechnol. J. 15: 367–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ainsworth, E.A. and Rogers, A. (2007) The response of photosynthesis and stomatal conductance to rising [CO2]: mechanisms and environmental interactions. Plant Cell Environ. 30: 258–270. [DOI] [PubMed] [Google Scholar]
- Anai, T. (2012) Potential of a mutant-based reverse genetic approach for functional genomics and molecular breeding in soybean. Breed. Sci. 61: 462–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anisimov, A., Koivu, K., Kanerva, A., Kaijalainen, S., Juntunen, K. and Kuvshinov, V. (2007) Cloning of new rubisco promoters from Brassica rapa and determination of their activity in stably transformed Brassica napus and Nicotiana tabacum plants. Mol. Breed. 19: 241–253. [Google Scholar]
- Boratyn, G.M., Camacho, C., Cooper, P.S., Coulouris, G., Fong, A., Ma, N., Madden, T.L., Matten, W.T., McGinnis, S.D., Merezhuk, Y.et al. (2013) BLAST: a more efficient report with usability improvements. Nucleic Acids Res. 41: W29–W33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boualem, A., Fleurier, S., Troadec, C., Audigier, P., Kumar, A.P., Chatterjee, M., Alsadon, A.A., Sadder, M.T., Wahb-Allah, M.A., Al-Doss, A.A.et al. (2014) Development of a Cucumis sativus TILLinG platform for forward and reverse genetics. PLoS ONE 9: e97963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colbert, T., Till, B.J., Tompa, R., Reynolds, S., Steine, M.N., Yeung, A.T., McCallum, C.M., Comai, L. and Henikoff, S. (2001) High-throughput screening for induced point mutations. Plant Physiol. 126: 480–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dedonder, A., Rethy, R., Fredericq, H., Van Montagu, M. and Krebbers, E. (1993) Arabidopsis rbcS genes are differentially regulated by light. Plant Physiol. 101: 801–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driever, S.M., Lawson, T., Andralojc, P.J., Raines, C.A. and Parry, M.A. (2014) Natural variation in photosynthetic capacity, growth, and yield in 64 field-grown wheat genotypes. J. Exp. Bot. 65: 4959–4973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans, J.R. (1989) Photosynthesis and nitrogen relationships in leaves of C3 plants. Oecologia 78: 9–19. [DOI] [PubMed] [Google Scholar]
- Farquhar, G.D., von Caemmerer, S. and Berry, J.A. (1980) A biochemical model of photosynthetic CO2 assimilation in leaves of C3 species. Planta 149: 78–90. [DOI] [PubMed] [Google Scholar]
- Farquhar, G.D. and Sharkey, T.D. (1982) Stomatal conductance and photosynthesis. Annu. Rev. Plant Physiol. 33: 317–345. [Google Scholar]
- Flood, P.J., Harbinson, J. and Aarts, M.G. (2011) Natural genetic variation in plant photosynthesis. Trends Plant Sci. 16: 327–335. [DOI] [PubMed] [Google Scholar]
- Hikosaka, K. and Hirose, T. (1998) Leaf and canopy photosynthesis of C3 plants at elevated CO2 in relation to optimal partitioning of nitrogen among photosynthetic components: theoretical prediction. Ecol. Modell. 106: 247–259. [Google Scholar]
- Hwang, J.E., Jang, D.S., Lee, K.J., Ahn, J.W., Kim, S.H., Kang, S.Y., Kim, D.S. and Kim, J.B. (2016) Identification of gamma ray irradiation-induced mutations in membrane transport genes in a rice population by TILLING. Genes Genet. Syst. 91: 245–256. [DOI] [PubMed] [Google Scholar]
- Izumi, M., Tsunoda, H., Suzuki, Y., Makino, A. and Ishida, H. (2012) RBCS1A and RBCS3B, two major members within the Arabidopsis RBCS multigene family, function to yield sufficient Rubisco content for leaf photosynthetic capacity. J. Exp. Bot. 63: 2159–2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitashiba, H., Li, F., Hirakawa, H., Kawanabe, T., Zou, Z., Hasegawa, Y., Tonosaki, K., Shirasawa, S., Fukushima, A., Yokoi, S.et al. (2014a) Draft sequences of the radish (Raphanus sativus L.) genome. DNA Res. 21: 481–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitashiba, H. and Nasrallah, J.B. (2014b) Self-incompatibility in Brassicaceae crops: lessons for interspecific incompatibility. Breed. Sci. 64: 23–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krapp, A. and Stitt, M. (1995) An evaluation of direct and indirect mechanisms for the ‘sink-regulation’ of photosynthesis in spinach: changes in gas exchange, carbohydrates, metabolites, enzyme activities and steady-state transcript levels after cold-girdling source leaves. Planta 195: 313–323. [Google Scholar]
- Krebbers, E., Seurinck, J., Herdies, L., Cashmore, A.R. and Timko, M.P. (1988) Four genes in two diverged subfamilies encode the ribulose-1,5-bisphosphate carboxylase small subunit polypeptides of Arabidopsis thaliana. Plant Mol. Biol. 11: 745–759. [DOI] [PubMed] [Google Scholar]
- Lai, K.S., Kaothien-Nakayama, P., Iwano, M. and Takayama, S. (2012) A TILLING resource for functional genomics in Arabidopsis thaliana accession C24. Genes Genet. Syst. 87: 291–297. [DOI] [PubMed] [Google Scholar]
- Long, S.P., Zhu, X.G., Naidu, S.L. and Ort, D.R. (2006) Can improvement in photosynthesis increase crop yields? Plant Cell Environ. 29: 315–330. [DOI] [PubMed] [Google Scholar]
- Lugasi, A., Blazovics, A., Hagymasi, K., Kocsis, I. and Kery, A. (2005) Antioxidant effect of squeezed juice from black radish (Raphanus sativus L. var niger) in alimentary hyperlipidaemia in rats. Phytother. Res. 19: 587–591. [DOI] [PubMed] [Google Scholar]
- Mao, Y., Botella, J.R. and Zhu, J.K. (2017) Heritability of targeted gene modifications induced by plant-optimized CRISPR systems. Cell. Mol. Life Sci. 74: 1075–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCallum, C.M., Comai, L., Greene, E.A. and Henikoff, S. (2000a) Targeted screening for induced mutations. Nat. Biotechnol. 18: 455–457. [DOI] [PubMed] [Google Scholar]
- McCallum, C.M., Comai, L., Greene, E.A. and Henikoff, S. (2000b) Targeting induced local lesions IN genomes (TILLING) for plant functional genomics. Plant Physiol. 123: 439–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medlyn, B.E. (1996) Interactive effects of atmospheric carbon dioxide and leaf nitrogen concentration on canopy light use efficiency: a modeling analysis. Tree Physiol. 16: 201–209. [DOI] [PubMed] [Google Scholar]
- Nemudryi, A.A., Valetdinova, K.R., Medvedev, S.P. and Zakian, S.M. (2014) TALEN and CRISPR/Cas genome editing systems: tools of discovery. Acta Naturae 6: 19–40. [PMC free article] [PubMed] [Google Scholar]
- Okabe, Y., Asamizu, E., Saito, T., Matsukura, C., Ariizumi, T., Bres, C., Rothan, C., Mizoguchi, T. and Ezura, H. (2011) Tomato TILLING technology: development of a reverse genetics tool for the efficient isolation of mutants from Micro-Tom mutant libraries. Plant Cell Physiol. 52: 1994–2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raines, C.A. (2011) Increasing photosynthetic carbon assimilation in C3 plants to improve crop yield: current and future strategies. Plant Physiol. 155: 36–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rani, R., Yadav, P., Barbadikar, K.M., Baliyan, N., Malhotra, E.V., Singh, B.K., Kumar, A. and Singh, D. (2016) CRISPR/Cas9: a promising way to exploit genetic variation in plants. Biotechnol. Lett. 38: 1991–2006. [DOI] [PubMed] [Google Scholar]
- Rashid, M., He, G., Guanxiao, Y. and Khurram, Z. (2011) Relevance of tilling in plant genomics. Aust. J. Crop Sci. 5: 411–420. [Google Scholar]
- Sage, R.F. (1990) A model describing the regulation of ribulose-1,5-bisphosphate carboxylase, electron transport, and triose phosphate use in response to light intensity and CO2 in C3 plants. Plant Physiol. 94: 1728–1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sage, R.F. (1994) Acclimation of photosynthesis to increasing atmospheric CO2: The gas exchange perspective. Photosyn. Res. 39: 351–368. [DOI] [PubMed] [Google Scholar]
- Sheen, J. (1994) Feedback control of gene expression. Photosyn. Res. 39: 427–438. [DOI] [PubMed] [Google Scholar]
- Shirasawa, K., Hirakawa, H., Nunome, T., Tabata, S. and Isobe, S. (2016) Genome-wide survey of artificial mutations induced by ethyl methanesulfonate and gamma rays in tomato. Plant Biotechnol. J. 14: 51–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spreitzer, R.J. (2003) Role of the small subunit in ribulose-1,5-bisphosphate carboxylase/oxygenase. Arch. Biochem. Biophys. 414: 141–149. [DOI] [PubMed] [Google Scholar]
- Stephenson, P., Baker, D., Girin, T., Perez, A., Amoah, S., King, G.J. and Østergaard, L. (2010) A rich TILLING resource for studying gene function in Brassica rapa. BMC Plant Biol. 10: 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiura, D., Betsuyaku, E. and Terashima, I. (2015) Manipulation of the hypocotyl sink activity by reciprocal grafting of two Raphanus sativus varieties: its effects on morphological and physiological traits of source leaves and whole-plant growth. Plant Cell Environ. 38: 2629–2640. [DOI] [PubMed] [Google Scholar]
- Till, B.J., Colbert, T., Tompa, R., Enns, L.C., Codomo, C.A., Johnson, J.E., Reynolds, S.H., Henikoff, J.G., Greene, E.A., Steine, M.N.et al. (2003) High-throughput TILLING for functional genomics. Methods Mol. Biol. 236: 205–220. [DOI] [PubMed] [Google Scholar]
- Tsai, H., Missirian, V., Ngo, K.J., Tran, R.K., Chan, S.R., Sundaresan, V. and Comai, L. (2013) Production of a high-efficiency TILLING population through polyploidization. Plant Physiol. 161: 1604–1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usuda, H. and Shimogawara, K. (1998) The effects of increased atmospheric carbon dioxide on growth, carbohydrates, and photosynthesis in radish, Raphanus sativus. Plant Cell Physiol. 39: 1–7. [Google Scholar]
- von Caemmerer, S. and Farquhar, G.D. (1981) Some relationships between the biochemistry of photosynthesis and the gas exchange of leaves. Planta 153: 376–387. [DOI] [PubMed] [Google Scholar]
- Wang, T.L., Uauy, C., Robson, F. and Till, B. (2012) TILLING in extremis. Plant Biotechnol. J. 10: 761–772. [DOI] [PubMed] [Google Scholar]
- Wanner, L.A. and Gruissem, W. (1991) Expression dynamics of the tomato rbcS gene family during development. Plant Cell 3: 1289–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaji, F. and Ohsawa, T.A. (2015) Breaking-bud pollination: a new pollination process in partially opened flowers by small bees. J. Plant Res. 128: 803–811. [DOI] [PubMed] [Google Scholar]
- Zhu, C., Bortesi, L., Baysal, C., Twyman, R.M., Fischer, R., Capell, T., Schillberg, S. and Christou, P. (2017) Characteristics of genome editing mutations in cereal crops. Trends Plant Sci. 22: 38–52. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.