Abstract
DNA-encoded combinatorial libraries are increasingly being used as tools for the discovery of small organic binding molecules to proteins of biological or pharmaceutical interest. In the majority of cases, synthetic procedures for the formation of DNA-encoded combinatorial libraries incorporate at least one step of amide bond formation between amino modified DNA and a carboxylic acid. We investigated reaction conditions and established a methodology by using 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide, 1-hydroxy-7-azabenzotriazole and N,N′-diisopropylethylamine (EDC/HOAt/DIPEA) in combination, which provided conversions greater than 75% for 423/543 (78%) of the carboxylic acids tested. These reaction conditions were efficient with a variety of primary and secondary amines, as well as with various types of amino-modified oligonucleotides. The reaction conditions, which also worked efficiently over a broad range of DNA concentrations and reaction scales, should facilitate the synthesis of novel DNA-encoded combinatorial libraries.
Keywords: DNA-encoded chemical libraries, amide bond formation, synthesis, bioconjugation
DNA-encoded combinatorial libraries (DECLs) are collections of organic compounds, individually coupled to distinct DNA fragments, serving as identification barcodes.1–4 Since DNA can be efficiently amplified by the polymerase chain reaction (PCR) and read by high-throughput DNA sequencing methods, the encoding of combinatorial libraries with DNA barcodes allows both the facile identification of specific ligands to protein targets immobilized on a solid support and the convenient handling of the libraries as mixtures of compounds. Compared to High-Throughput Screening, which relies on expensive compound collections and a sophisticated management system, DECLs provide an avenue for researchers from academia or small companies to identify hits that bind to targets of pharmaceutical interest. Numerous binders have been discovered from recent DECL selections, like ADAMTS-5 inhibitors,5 tankyrase 1 inhibitors,6 XIPA inhibitors 7 and integrin lymphocyte function-associated antigen 1 (LFA-1) antagonists,8 just to name a few.9
Various chemical strategies have been developed for the generation of large combinatorial DECLs, including DNA-templated synthesis,10,11 DNA-encoded routing,12 hybridization-based chemical assembling 13–17and DNA-encoded solid-phase synthesis.18 In addition, the iterative assembly of sets of building blocks in a “split-and-pool” methodology, accompanied by the stepwise addition of DNA barcodes (which unambiguously identify the chemical identity of the nascent molecules), is the most generally used strategy for DECLs construction. This strategy facilitates the preparation of DECLs containing millions or even billions of compounds, starting from just few hundred building blocks and oligonucleotides.19,20,21
In the majority of DECLs disclosed so far, at least one synthesis step involved the formation of an amide bond.4 More than 20’000 carboxylic acids can be purchased from commercial sources at moderate costs (less than $150/g).22 The acylation of DNA-attached amines with carboxylic acids can be performed on protected DNA structures linked to Controlled Pore Glass (CPG) supports 6 or in solution, 23 followed by deprotection and HPLC purification of the individual conjugates at early stages of library construction. Additionally, for unprotected DNA-attached amines, acylation can be performed either in solution or on “pseudo-solid phase” (i.e., on amino-modified oligonucleotide derivatives non-covalently immobilized on solid supports, facilitating the use of large molar excess of reagents and repeated washing steps for purification). 24,25 While reaction conditions with DNA on solid phase can work very efficiently, they typically consume several milligrams of each carboxylic acid for each coupling, which makes synthesis expensive and may limit the use of precious building blocks for the construction of different chemical libraries. By contrast, the acylation in solution may allow the synthesis of large DECLs using only minute amounts of building blocks.
Amide bond-forming reactions, performed in solution with unprotected oligonucleotides, have been described using EDC/sulfo-NHS26 or DMT-MM as coupling reagent for DECL synthesis purposes.19,20 Recently, Satz and colleagues have explored the scope and applicability of organic reactions in aqueous solution for the construction of DECLs, reporting also a sequential acylation method with DMT-MM for multistep synthesis.27 In this letter, we report an alternative a new method (featuring EDC/HOAt/DIPEA as coupling combination), which afforded >75% conversions for the majority of the tested building blocks (423 out of 543, corresponding to 78%).
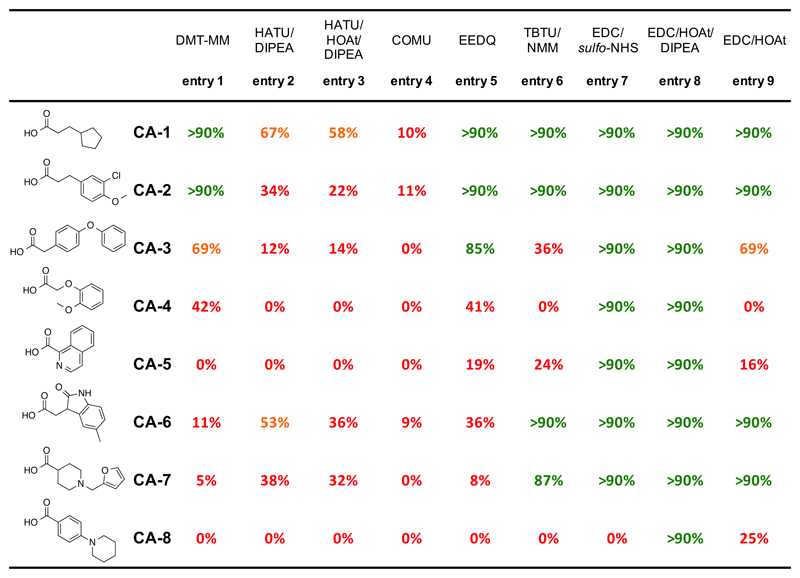
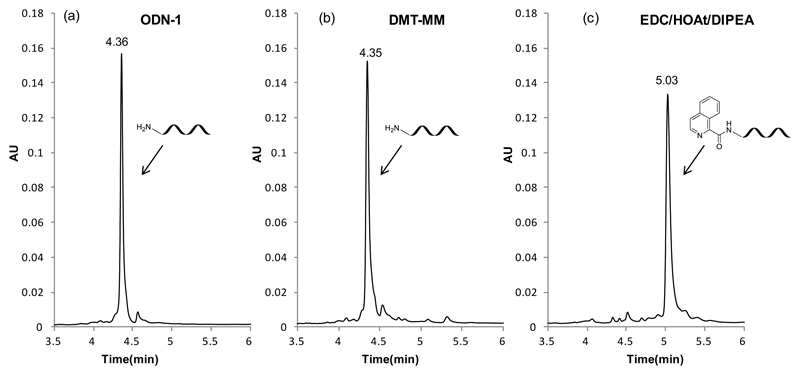
Motivated by previous experience of our laboratory in the synthesis of DECLs, which involved carboxylic acids as building blocks, we characterized the reaction of eight representative compounds, leading to very different conversion rates with DMT-MM as coupling reagent [Figure 1, entry 1]. While two carboxylic acids (CA-1 and CA-2) formed amides with 5’-aminomodified DNA (ODN-1) with excellent conversion rates (>90%; determined by UV absorbance trace at 260 nm of UPLC), the remaining six structures exhibited suboptimal conversion (ranging between 0 and 11% for 4 out of 8 compounds). A systematic investigation for alternative coupling methods is summarized in Figure 1 and different peptide coupling reagents were tested to enhance the yield of acylation. Coupling reagents like HATU [Figure 1, entries 2 and 3] and COMU [Figure 1, entry 4] consistently provided poor conversion rates for all the reactions tested. EEDQ [Figure 1, entry 5] and TBTU [Figure 1, entry 6] afforded better conversion only for some acids, while the coupling efficacy remained very low for the other substrates. Promising results were obtained by using EDC/sulfo-NHS [Figure 1, entry 7] as coupling combination, a well established and widely used method. This led to a complete conversion for 7 out of 8 compounds, while for one carboxylic acid (CA-8) no product could be observed. Further optimization revealed a novel method, featuring the combination of EDC/HOAt/DIPEA [Figure 1, entry 8], which yielded >90% conversion for all the tested compounds. For four tested acids, the coupling efficiency decreased dramatically by using only EDC/HOAt [Figure 1, entry 9], highlighting the importance of DIPEA for proper activation. Figure 2 and Supplementary Figure S3 show representative LC and MS profiles for CA-5, one carboxylic acid that did not couple at all using DMT-MM, which however provided excellent conversion (>90%) with the EDC/HOAt/DIPEA methodology.
Figure 1.
Summary of coupling conversions by using different coupling reagents with eight representative carboxylic acids. In all cases, 72 μL ODN-1 (MOPS buffer pH 8.0, 0.5 nmol), 45 μL carboxylic acid (60 mM in DMSO) and coupling reagent were reacted for 16h at RT, followed by an additional coupling step with the same activated carboxylic acid for 6h at RT. Coupling reagents: entry 1: 4 μL DMT-MM (300 mM in water); entry 2: 4 μL HATU (300 mM in DMSO), 4 μL DIPEA (300 mM in DMSO); entry 3: 4 μL HATU (300 mM in DMSO), 4 μL HATU (60 mM in DMSO), 4 μL DIPEA (300 mM in DMSO); entry 4: 4 μL COMU (300 mM in DMSO); entry 5: 4 μL EEDQ (300 mM in DMSO); entry 6: 4 μL TBTU (300 mM in DMSO), 4 μL NMM (300 mM in DMSO); entry 7: 4 μL EDC (300 mM in DMSO), 4 μL sulfo-NHS (60 mM in DMSO:water = 2:1); entry 8: 4 μL EDC (300 mM in DMSO), 4 μL HOAt (60 mM in DMSO), 4 μL DIPEA (300 mM in DMSO); entry 9: 4 μL EDC (300 mM in DMSO), 4 μL HOAt (60 mM in DMSO).
Figure 2.
UPLC profiles of (a) 5’-C12-amino oligonucleotide (ODN-1). (b) crude reaction mixture after coupling with CA-5 using DMT-MM. (c) crude reaction mixture after coupling with CA-5 using EDC/HOAt/DIPEA. Detection at 260 nm.
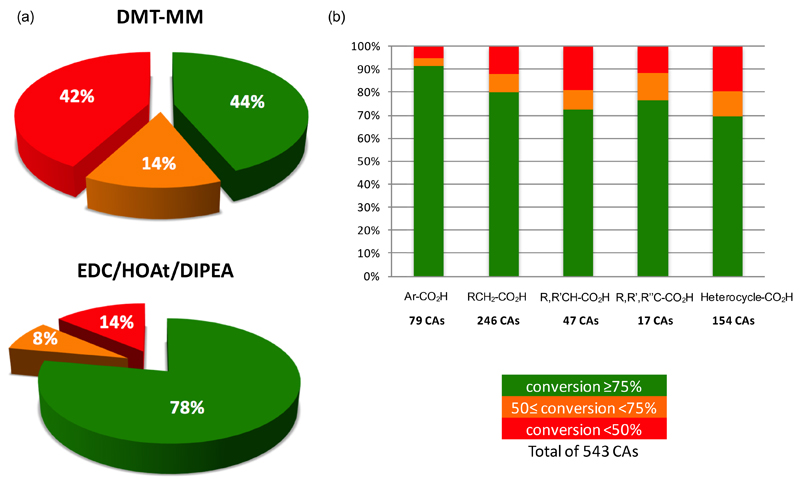
In order to compare the performance of the DMT-MM and EDC/HOAt/DIPEA methods over a broader range of substrates, we performed reactions using ODN-1 and a structurally diverse set of 543 carboxylic acids. Using an optimized DMT-MM protocol, 42% of the compounds exhibited a conversion lower than 50%, while 14% had a conversion between 50% and 75%, as assessed by UPLC analysis and MS confirmation [Figure 3a]. Conversely, the EDC/HOAt/DIPEA method exhibited a conversion rate of >75% for 78% of the 543 carboxylic acids. However, no clear correlation between structural features and conversion rate was found. For instance, different types of carboxylic acids (i.e., benzoic acids; primary, secondary, and tertiary carboxylic acids; aromatic or heterocyclic acids) provided similar coupling efficiencies [Figure 3b]. The experimental findings underline the importance of screening all building blocks in model reactions before DECL construction, rather than studying coupling conditions on a small set of test compounds.
Figure 3.
(a) Pie chart analysis of the coupling yields of ODN-1 with 543 carboxylic acids using DMT-MM and EDC/HOAt/DIPEA-mediated coupling, according to the conditions described in Figure 1, entries 1 and 8. (b) Histogram analysis of the coupling yields of structurally diverse carboxylic acids to ODN-1 with EDC/HOAt/DIPEA method. Benzoic (79 acids), primary (246 acids), secondary (47 acids), and tertiary carboxylic acids (17 acids), heterocyclic acids (154 acids).
In a previous report, it was described that adduct formation of EDC and G- and T-nucleobases could occur during the formation of the acylation product by using EDC as coupling reagent on “pseudo-solid phase”.25 While the impact of such possible base modifications on the performance of PCR amplification is so far unexplored, it may also compromise the integrity of the DNA-barcodes.28 We systematically checked the mass spectra of all obtained conjugates from the solution phase EDC/HOAt/DIPEA method and found no detectable EDC adducts associated with the conjugates (i.e., <5%). Supplementary Figure S4 shows an exemplary MS profile for CA-9 (trans-4-hydroxycyclohexane-1-carboxylic acid)-DNA conjugate, one of the carboxylic acids that exhibited significant EDC adduct formation with the “pseudo-solid phase” method. No adduct was detected with the EDC/HOAt/DIPEA methodology.
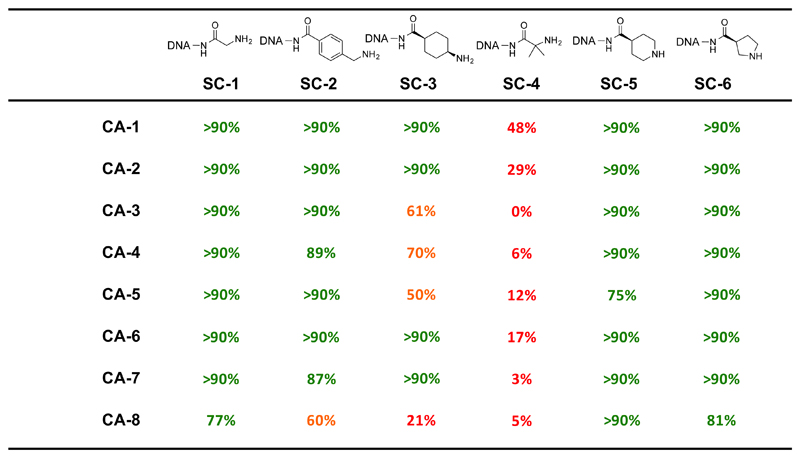
We further tested the coupling efficiency between eight representative carboxylic acids and six DNA scaffolds, containing amines with different chemical features [Figure 4]. The DNA-conjugate of glycine (SC-1) and the DNA-conjugate linked to 4-(aminomethyl)benzoic acid (SC-2) provided fair to good conversion yields for all the eight tested acids. Good conversions were also achieved with two cyclic secondary amines SC-5 and SC-6 respectively.
Figure 4.
Coupling yields of six different DNA-Scaffold Conjugates with eight representative carboxylic acids, according to the condition described in Figure 1, entry 8.
Using our EDC/HOAt/DIPEA coupling protocol, conversions of amide bond forming reactions were distinctly lower for sterically hindered substrates (SC-3). More steric bulk at the α-carbon further decreased the coupling efficiency extensively. For example, poor coupling results were obtained with a DNA-conjugate of 2-aminoisobutyric acid (SC-4). With the exception of CA-1, all the remaining seven carboxylic acids had conversion yields <30% under the optimized conditions.
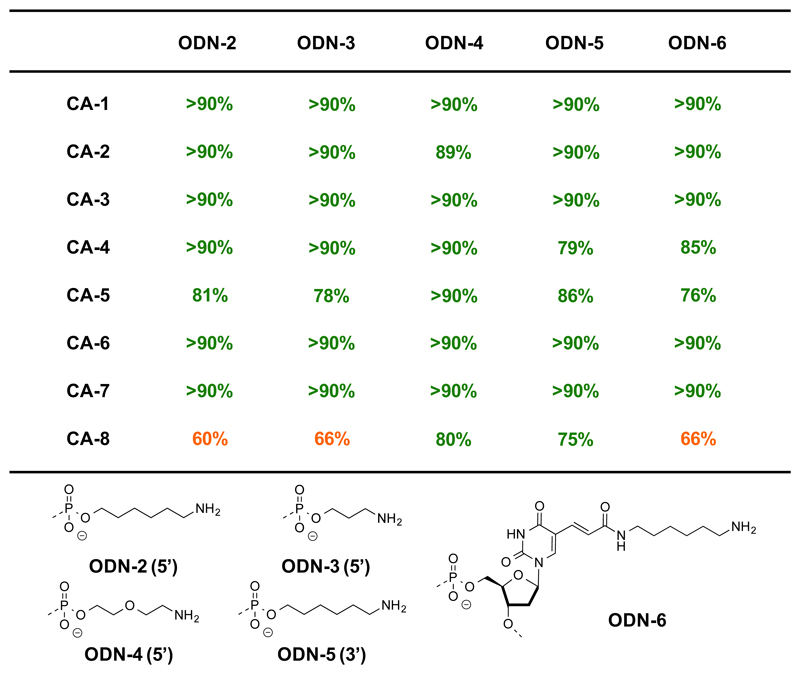
In order to further investigate the dependence of reaction yields on the nature of amines on DNA, five additional amino-modified oligonucleotides bearing different tethers were used. Such reactions are particularly useful for the construction of encoded self-assembling chemical libraries.17 Compared to ODN-1, which featured a long C-12 tether, amino-modified oligonucleotides (i.e., ODN-2, ODN-3 and ODN-4) with various short linkers at the 5’ end were examined. Good conversions were observed in 22/24 reactions and, in all cases, a conversion >60% was achieved [Figure 5]. ODN-5, carrying an aminohexyl-tether at the 3’ end, was also efficiently converted into the corresponding acylated conjugates (>75%). As the chemical modification of internal sites within the DNA structure may be used for the construction of DECLs using the Yoctoliter reactor method, 29 the conversion of a primary amine on the side chain of a thymine modified at the C-6 position in ODN-6 was investigated and led to good yields for 7 out of 8 reactions, while only one carboxylic acid (CA-8) yielded a slightly lower conversion (66%).
Figure 5.
Coupling yields of five different amino-modified oligonucleotides with eight representative carboxylic acids, according to the condition described in Figure 1, entry 8.
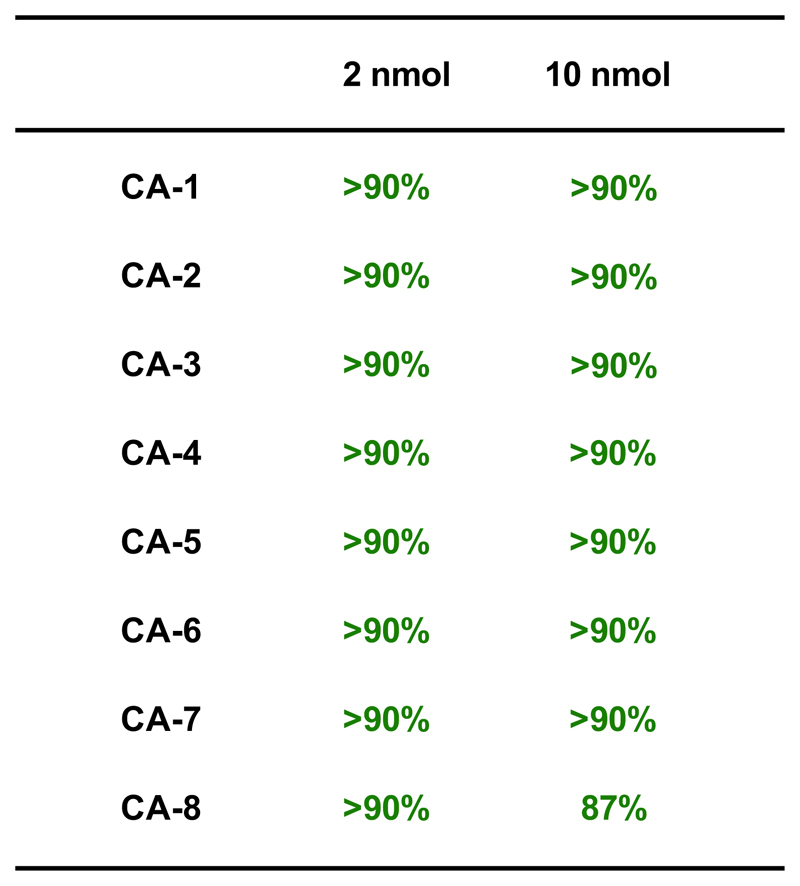
Importantly, the EDC/HOAt/DIPEA method worked well also at different scales of amino-modified DNA (ranging between 2 and 10 nmol), which are customarily utilized for library construction [Figure 6].
Figure 6.
Coupling yields of ODN-1 at 2 nmol and 10 nmol scale with eight representative carboxylic acids, according to the condition described in Figure 1, entry 8.
The execution of synthetic steps with high yields and good purity is particularly important for the construction of DNA-encoded combinatorial libraries that require multiple steps of assembly (e.g., those based on split-and-pool methodologies). While individual DNA conjugates can be purified and analyzed after the first step of library construction, the processing of synthetic intermediates at later stages becomes more complicated. In order to minimize the presence of incomplete reaction products (e.g., residual unreacted amines) in the library, methods have been developed for the modification and removal of such truncated products.30 While these methods can be useful and may lead to improved library purity, they inevitably lead to an uneven distribution of library members, thus complicating decoding procedures. The method described in this letter should facilitate library synthesis without the need for “cap-and-catch” strategies.
In summary, we have systematically investigated different experimental conditions for amide bond formation on DNA in solution phase and established a novel efficient methodology (EDC/HOAt/DIPEA as coupling combination). We tested 543 structurally diverse carboxylic acids as building blocks and found that the method provides good conversion yields for the majority of the compounds. The reaction works well with various types of amino-modified oligonucleotides and at different reaction scales. While the EDC/HOAt/DIPEA method worked well in most experimental conditions, additional research efforts will be needed for certain substrates (e.g., amines featuring a substantial steric hindrance), as chemical purity greatly contributes to the performance of DECL technology.
Supplementary Material
Experimental Procedures
Detailed experimental prcedures and oligonucleotide sequences are described in the Supporting Information.
Acknowledgements
The authors thank Melanie Buehler and Davor Bajic for technical support.
Funding
Financial contribution from ETH Zürich, Swiss National Science Foundation (Grants 310030B_163479/1 and CRSII2_160699/1), ERC Advanced Grant “Zauberkugel” and Philochem AG are gratefully acknowledged.
Abbreviations
- CA
carboxylic acid
- COMU
(1-cyano-2-ethoxy-2-oxoethylidenaminooxy)-dimethylamino-morpholino-carbenium hexafluorophosphate
- DIPEA
N,N′-diisopropylethylamine
- MSO
dimethyl sulfoxide
- DMT-MM
4-(4, 6-dimethoxy-1, 3, 5-triazin-2-yl)-4-methylmorpholinium chloride
- EDC
1-ethyl-3- (3-dimethylaminopropyl) carbodiimide
- EEDQ
2-Ethoxy-1-ethoxycarbonyl-1,2-dihydroquinoline
- HATU
1-[Bis(dimethylamino)methylene]-1H-1,2,3-triazolo[4,5-b]pyridinium 3-oxid hexafluorophosphate
- HOAt
1-hydroxy-7-azabenzotriazole
- UPLC
Ultra-performance liquid chromatography
- LC-MS
liquid chromatography mass spectrometry
- NMM
N-methylmorpholine
- ODN
oligonucleotide
- SC
scaffold conjugate
- sulfo-NHS
N-hydroxysulfosuccinimide
- TBTU
O-(Benzotriazol-1-yl)-N,N,N',N'-tetramethyluronium tetrafluoroborate
- TEAA
triethylammonium acetate
- TEA
triethylamine
Footnotes
The authors declare no competing financial interest.
Submitted to ACS Combinatorial Science as a Letter
References
- (1).Clark MA. Selecting chemicals: the emerging utility of DNA-encoded libraries. Curr Opin Chem Biol. 2010;14:396–403. doi: 10.1016/j.cbpa.2010.02.017. [DOI] [PubMed] [Google Scholar]
- (2).Mannocci L, Leimbacher M, Wichert M, Scheuermann J, Neri D. 20 years of DNA-encoded chemical libraries. Chem Commun. 2011;47:12747–12753. doi: 10.1039/c1cc15634a. [DOI] [PubMed] [Google Scholar]
- (3).Franzini RM, Neri D, Scheuermann J. DNA-Encoded Chemical Libraries: Advancing beyond Conventional Small-Molecule Libraries. Acc Chem Res. 2014;47:1247–1255. doi: 10.1021/ar400284t. [DOI] [PubMed] [Google Scholar]
- (4).Franzini RM, Randolph C. Chemical Space of DNA-Encoded Libraries. J Med Chem. 2016 Feb 25; doi: 10.1021/acs.jmedchem.5b01874. published online. [DOI] [PubMed] [Google Scholar]
- (5).Deng H, O’Keefe H, Davie CP, Lind KE, Acharya RA, Franklin GJ, Larkin J, Matico R, Neeb M, Thompson MM, Lohr T, et al. Discovery of Highly Potent and Selective Small Molecule ADAMTS-5 Inhibitors That Inhibit Human Cartilage Degradation via Encoded Library Technology (ELT) J Med Chem. 2012;55:7061–7079. doi: 10.1021/jm300449x. [DOI] [PubMed] [Google Scholar]
- (6).Franzini RM, Ekblad T, Zhong N, Wichert M, Decurtins W, Nauer A, Zimmermann M, Samain F, Scheuermann J, Brown PJ, Hall J, et al. Identification of structure-activity relationships from screening a structurally compact DNA-encoded chemical library. Angew Chem Int Ed Engl. 2015;54:3927–3931. doi: 10.1002/anie.201410736. [DOI] [PubMed] [Google Scholar]
- (7).Seigal BA, Connors WH, Fraley A, Borzilleri RM, Carter PH, Emanuel SL, Fargnoli J, Kim K, Lei M, Naglich JG, Pokross ME, et al. The Discovery of Macrocyclic XIAP Antagonists from a DNA-Programmed Chemistry Library, and Their Optimization To Give Lead Compounds with in Vivo Antitumor Activity. J Med Chem. 2015;58:2855–2861. doi: 10.1021/jm501892g. [DOI] [PubMed] [Google Scholar]
- (8).Kollmann CS, Bai X, Tsai C-H, Yang H, Lind KE, Skinner SR, Zhu Z, Israel DI, Cuozzo JW, Morgan BA, Yuki K, et al. Application of encoded library technology (ELT) to a protein–protein interaction target: Discovery of a potent class of integrin lymphocyte function-associated antigen 1 (LFA-1) antagonists. Bioorg Med Chem. 2014;22:2353–2365. doi: 10.1016/j.bmc.2014.01.050. [DOI] [PubMed] [Google Scholar]
- (9).Salamon H, Klika Škopić M, Jung K, Bugain O, Brunschweiger A. Chemical Biology Probes from Advanced DNA-encoded Libraries. ACS Chem Biol. 2016;11:296–307. doi: 10.1021/acschembio.5b00981. [DOI] [PubMed] [Google Scholar]
- (10).Gartner ZJ, Tse BN, Grubina R, Doyon JB, Snyder TM, Liu DR. DNA-Templated Organic Synthesis and Selection of a Library of Macrocycles. Science. 2004;305:1601–1605. doi: 10.1126/science.1102629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Li Y, Zhao P, Zhang M, Zhao X, Li X. Multistep DNA-Templated Synthesis Using a Universal Template. J Am Chem Soc. 2013;135:17727–17730. doi: 10.1021/ja409936r. [DOI] [PubMed] [Google Scholar]
- (12).Wrenn SJ, Weisinger RM, Halpin DR, Harbury PB. Synthetic Ligands Discovered by in Vitro Selection. J Am Chem Soc. 2007;129:13137–13143. doi: 10.1021/ja073993a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Melkko S, Scheuermann J, Dumelin CE, Neri D. Encoded self-assembling chemical libraries. Nat Biotech. 2004;22:568–574. doi: 10.1038/nbt961. [DOI] [PubMed] [Google Scholar]
- (14).Daguer JP, Ciobanu M, Alvarez S, Barluenga S, Winssinger N. DNA-templated combinatorial assembly of small molecule fragments amenable to selection/amplification cycles. Chem Sci. 2011;2:625–632. [Google Scholar]
- (15).Reddavide FV, Lin W, Lehnert S, Zhang Y. DNA-Encoded Dynamic Combinatorial Chemical Libraries. Angew Chem Int Ed. 2015;54:7924–7928. doi: 10.1002/anie.201501775. [DOI] [PubMed] [Google Scholar]
- (16).Li G, Zheng W, Chen Z, Zhou Y, Liu Y, Yang J, Huang Y, Li X. Design, preparation, and selection of DNA-encoded dynamic libraries. Chem Sci. 2015;6:7097–7104. doi: 10.1039/c5sc02467f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Wichert M, Krall N, Decurtins W, Franzini RM, Pretto F, Schneider P, Neri D, Scheuermann J. Dual-display of small molecules enables the discovery of ligand pairs and facilitates affinity maturation. Nat Chem. 2015;7:241–249. doi: 10.1038/nchem.2158. [DOI] [PubMed] [Google Scholar]
- (18).MacConnell AB, McEnaney PJ, Cavett VJ, Paegel BM. DNA-Encoded Solid-Phase Synthesis: Encoding Language Design and Complex Oligomer Library Synthesis. ACS Comb Sci. 2015;17:518–534. doi: 10.1021/acscombsci.5b00106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Clark MA, Acharya RA, Arico-Muendel CC, Belyanskaya SL, Benjamin DR, Carlson NR, Centrella PA, Chiu CH, Creaser SP, Cuozzo JW, Davie CP, et al. Design, synthesis and selection of DNA-encoded small-molecule libraries. Nat Chem Biol. 2009;5:647–654. doi: 10.1038/nchembio.211. [DOI] [PubMed] [Google Scholar]
- (20).Litovchick A, Dumelin C, Habeshian S, Gikunju D, GuiÈ M, Centrella P, Zhang Y, Sigel E, Cuozzo J, Keefe A, Clark M. Encoded Library Synthesis Using Chemical Ligation and the Discovery of sEH Inhibitors from a 334-Million Member Library. Sci Rep. 2015;5:10916. doi: 10.1038/srep10916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Buller F, Steiner M, Frey K, Mircsof D, Scheuermann J, Kalisch M, Bühlmann P, Supuran CT, Neri D. Selection of Carbonic Anhydrase IX Inhibitors from One Million DNA-Encoded Compounds. ACS Chem Biol. 2011;6:336–344. doi: 10.1021/cb1003477. [DOI] [PubMed] [Google Scholar]
- (22).Kalliokoski T. Price-Focused Analysis of Commercially Available Building Blocks for Combinatorial Library Synthesis. ACS Comb Sci. 2015;17:600–607. doi: 10.1021/acscombsci.5b00063. [DOI] [PubMed] [Google Scholar]
- (23).McMinn DL, Greenberg MM. Postsynthetic Conjugation of Protected Oligonucleotides Containing 3‘-Alkylamines. J Am Chem Soc. 1998;120:3289–3294. [Google Scholar]
- (24).Halpin DR, Lee JA, Wrenn SJ, Harbury PB. DNA Display III. Solid-Phase Organic Synthesis on Unprotected DNA. PLoS Biol. 2004;2:e175. doi: 10.1371/journal.pbio.0020175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Franzini RM, Samain F, Abd Elrahman M, Mikutis G, Nauer A, Zimmermann M, Scheuermann J, Hall J, Neri D. Systematic Evaluation and Optimization of Modification Reactions of Oligonucleotides with Amines and Carboxylic Acids for the Synthesis of DNA-Encoded Chemical Libraries. Bioconjug Chem. 2014;25:1453–1461. doi: 10.1021/bc500212n. [DOI] [PubMed] [Google Scholar]
- (26).Dumelin CE, Scheuermann J, Melkko S, Neri D. Selection of Streptavidin Binders from a DNA-Encoded Chemical Library. Bioconjug Chem. 2006;17:366–370. doi: 10.1021/bc050282y. [DOI] [PubMed] [Google Scholar]
- (27).Satz AL, Cai J, Chen Y, Goodnow R, Gruber F, Kowalczyk A, Petersen A, Naderi-Oboodi G, Orzechowski L, Strebel Q. DNA Compatible Multistep Synthesis and Applications to DNA Encoded Libraries. Bioconjug Chem. 2015;26:1623–1632. doi: 10.1021/acs.bioconjchem.5b00239. [DOI] [PubMed] [Google Scholar]
- (28).Malone ML, Paegel BM. What is a “DNA-Compatible” Reaction? ACS Comb Sci. 2016;18:182–187. doi: 10.1021/acscombsci.5b00198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Hansen MH, Blakskjær P, Petersen LK, Hansen TH, Højfeldt JW, Gothelf KV, Hansen NJV. A Yoctoliter-Scale DNA Reactor for Small-Molecule Evolution. J Am Chem Soc. 2009;131:1322–1327. doi: 10.1021/ja808558a. [DOI] [PubMed] [Google Scholar]
- (30).Franzini RM, Biendl S, Mikutis G, Samain F, Scheuermann J, Neri D. “Cap-and-Catch” Purification for Enhancing the Quality of Libraries of DNA Conjugates. ACS Comb Sci. 2015;17:393–398. doi: 10.1021/acscombsci.5b00072. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.