Summary
Host defense to RNA viruses depends on rapid intracellular recognition of viral RNA by two cytoplasmic RNA helicases, RIG-I and MDA5. RNA transfection experiments indicate that RIG-I responds to naked double-stranded (ds) RNAs with a triphosphorylated 5′ (5′ppp) terminus. However, identity of the RIG-I stimulating viral structures in an authentic infection context remains unresolved. We show that incoming viral nucleocapsids containing a 5′ppp dsRNA “panhandle” structure trigger antiviral signaling that commences with RIG-I, is mediated through the adaptor protein MAVS, and terminates with transcription factor IRF-3. Independent of mammalian cofactors or viral polymerase activity, RIG-I bound to viral nucleocapsids, underwent a conformational switch, and homo-oligomerized. Enzymatic probing and super-resolution microscopy suggest that RIG-I interacts with the panhandle structure of the viral nucleocapsids. These results define cytoplasmic entry of nucleocapsids as the proximal RIG-I-sensitive step during infection, and establish viral nucleocapsids with a 5′ppp dsRNA panhandle as a RIG-I activator.
Keywords: RIG-I, RNA virus, nucleocapsids, 5′ triphosphate dsRNA, panhandle
Introduction
Host defenses to RNA viruses are dependent on a rapid detection by pathogen recognition receptors (PRRs). Intracellular recognition of virus infection is mediated by two cytoplasmic RNA helicases, RIG-I and MDA5 (termed RIG-like receptors, RLRs) (Kato et al., 2011). The binding of an RNA ligand to these PRRs leads to phosphorylation and dimerization of interferon regulated factor 3 (IRF-3), which subsequently activates genes for type I interferons (IFN-α/β) (Hiscott, 2007). These cytokines trigger the expression of IFN-stimulated gene (ISG) products that have antiviral and immunomodulatory activities (Randall and Goodbourn, 2008).
The infection cycle of RNA viruses consists of the phases attachment, entry, mRNA transcription, genome replication, assembly, and exit. Negative-strand RNA viruses carry an RNA-dependent RNA polymerase (RdRp) within their particles and immediately start transcribing their genome after entering the cell. The protein products of this so-called primary transcription then drive the replication of the genome via a positive-sense intermediate. Positive-strand RNA viruses do not carry an RdRp in their particles and directly translate their genome after entry. The newly synthesized RdRp then produces a negative-sense intermediate, mRNA, and progeny genome. Usually, the genome of both negative- and positive-strand RNA viruses is packaged by a viral nucleocapsid protein (often called N). Most studies on activation of RLRs were either based on infection with RNA viruses undergoing a full replication cycle, or on transfection of cells with naked viral or synthetic RNAs. The infection experiments established that RIG-I and MDA5 recognize mostly non-overlapping subsets of viruses (McCartney and Colonna, 2009). The RNA transfection experiments revealed that RIG-I responds to long double-stranded (ds) RNA molecules, short dsRNAs with a triphosphorylated 5′ (5′ppp) terminus, and poly U/UC-rich sequences, whereas MDA5 activation is more dependent on branched dsRNA structures (Binder et al., 2011; Hornung et al., 2006; Kato et al., 2008; Pichlmair et al., 2006; Pichlmair et al., 2009; Saito et al., 2008; Schlee et al., 2009; Schmidt et al., 2009). Despite these achievements, the question of the natural RLR ligands, i.e. which viral structures are stimulating the RLRs in the authentic infection context, is largely unsolved. Two recent RNA fractionation studies showed that full-length and shortened RNAs arising during genome replication of 5′ppp-RNA viruses are natural stimulators of RIG-I (Baum et al., 2010; Rehwinkel et al., 2010). However, it remained open whether these RIG-I ligands were naked RNA side-products of viral genome replication, or whether RIG-I could also recognize nucleoprotein-encapsidated RNA, the main viral structure in the infected cell. Our study presented here addresses this problem and indicates that RIG-I is capable of reacting to incoming, encapsidated RNA virus genomes. This immediate early IFN response requires the viral 5′ppp dsRNA “panhandle” structure but is independent of viral RNA synthesis. RIG-I thereby directly interacts with the panhandle on the viral nucleocapsids, switches conformation, oligomerizes, and triggers the activation of IRF-3.
Results
Our first aim was to identify the earliest infection step that triggers IFN induction. In the hope of drawing conclusions on the responsible viral determinant, we employed a set of RNA viruses with different genomic features. Dependent on the particular virus, a block of viral mRNA translation by cycloheximide (CHX) has different effects on primary transcription and genome replication (Table S1).
Entry of negative-sense RNA viruses can activate IFN induction in the absence of replication
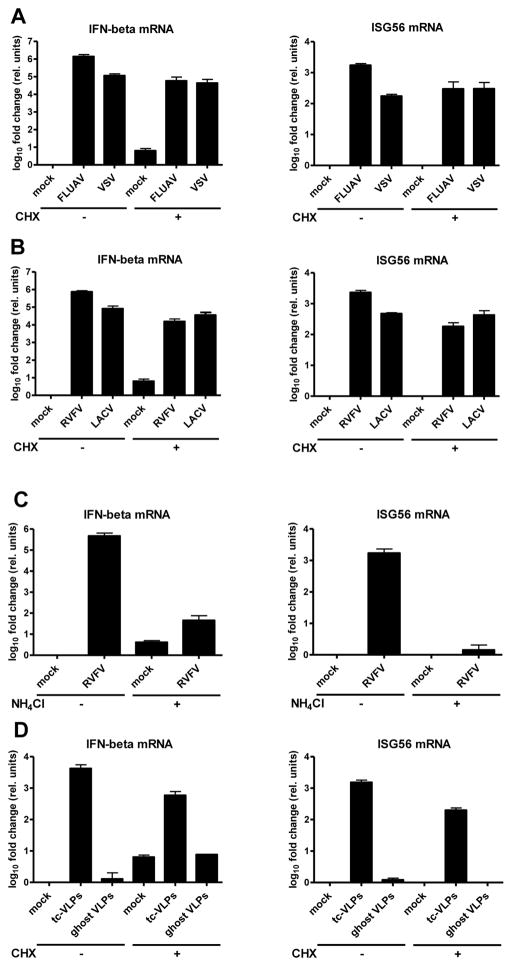
Influenza A virus (FLUAV) and vesicular stomatitis virus (VSV) are negative-strand RNA viruses. Application of CHX allows particle attachment, entry of nucleocapsids, and primary transcription, but not genome replication (Fig. S1). We measured the virus-inducible genes for IFN-β and ISG56 in human A549 cells by real-time RT-PCR. Figure 1A shows that both FLUAV and VSV are strongly activating these genes, even when CHX was applied. Thus, an IFN response can occur before the viruses start replicating their genome.
Fig. 1. IFN response to incoming negative-strand RNA viruses.
Cells were treated with inhibitors, infected, incubated for 24 h, and then assayed for mRNA levels of IFN-β (left panels) and ISG56 (right panels) using real-time RT-PCR. Here and in all following figures, mean values and standard deviations from three independent experiments are shown. (A and B) IFN response in the absence of viral genome replication. Cells were treated for 1 h with 0 or 50 μg/ml CHX, and then infected either with FLUAVΔNS1 (FLUAV) and VSV (A), or with RVFVΔNSs::GFP (RVFV) and LACVdelNSs (LACV) (B). (C and D) Requirement for virus entry. (C) Cells were treated for 1 h with 0 or 50 mM NH4Cl, and then infected with RVFVΔNSs::GFP. (D) Infection with different types of VLPs. Cells were CHX treated and infected with tc-VLPs or the equivalent amount of ghost VLPs. See also Figs. S1 to S7.
To narrow down the IFN-relevant infection step, we repeated the experiment using Rift Valley fever virus (RVFV) and La Crosse virus (LACV). These bunyaviruses are peculiar in that their transcription depends on concurrent translation (Raju et al., 1989). CHX treatment arrests viral primary transcription at a very early step, but had only a minor influence on IFN-β and ISG56 induction (Fig. 1B). Thus, apparently, not even a fully operating primary transcription is required for triggering an IFN response.
Recent reports suggested that particle attachment or membrane fusion can activate an antiviral response (Holm et al., 2012; Noyce et al., 2011), although other groups did not observe this in their systems (Handke et al., 2009; Spiropoulou et al., 2007; Stoltz and Klingstrom, 2010). We employed several methods to study the involvement of particle attachment. Firstly, we pretreated cells with NH4Cl, an agent that inhibits bunyaviral entry into the cytoplasm (Filone et al., 2006), but does not impede the IFN response (Fig. S2). As shown in figure 1C, NH4Cl almost entirely abrogated the host response to RVFV. Similarly, three other bunyavirus entry inhibitors (de Boer et al., 2012) (Fig. S3), as well as virus inactivation by β-propiolactone (Fig. S4) or UV irradiation (Fig. S5) all led to a reduction of IFN induction. In a complementary approach, we employed two types of virus-like particles (VLPs) of RVFV. Complete VLPs contain nucleocapsids with a reporter minigenome, whereas “ghost” VLPs have just the N protein inside (Fig. S6). The complete VLPs, also called transcriptionally competent (tc-) VLPs (Hoenen et al., 2011), are capable of primary transcription, but not of replication (Habjan et al., 2009a). tc-VLPs triggered IFN-β and ISG56 gene expression, even under CHX (Fig. 1D). Ghost VLPs, however, did not elicit any IFN response, although they bind equally well to the cells (Fig. S7). These data indicate that, in our system, virus entry or primary transcription are necessary and sufficient for IFN induction.
The immediate early IFN response requires viral 5′triphosphate RNA and RIG-I
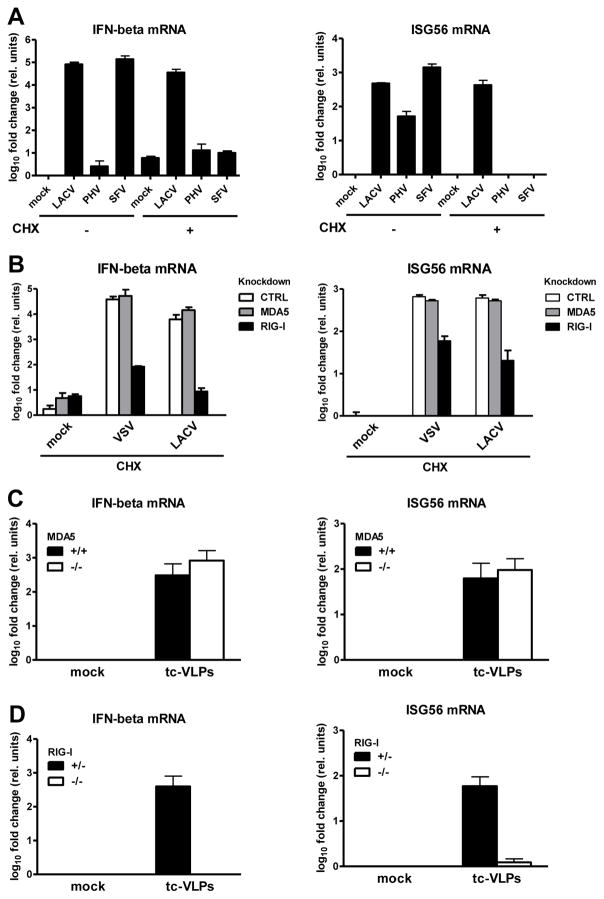
All viruses used so far contained the RIG-I-activating 5′ppp group on their RNA genome (Habjan et al., 2008a; Hornung et al., 2006; Pichlmair et al., 2006). To clarify the contribution of the genome end, we employed two RNA viruses that lack this feature (see table S1). Prospect Hill virus (PHV; family Bunyaviridae) has a monophosphate at the 5′ end (Garcin et al., 1995; Habjan et al., 2008a). Semliki Forest virus (SFV; family Togaviridae) has a 5′ cap structure (McInerney et al., 2005). CHX inhibits PHV in the same way as the related RVFV or LACV (Fig. S1). SFV is a positive-strand RNA virus, i.e. its genome is directly translated after entry. In the absence of CHX, PHV minimally activated the IFN-β promoter and moderately activated ISG56 transcription, as expected (Prescott et al., 2005), whereas the IFN response to SFV was similar as to LACV (Fig. 2A). Strikingly, application of CHX completely abrogated the IFN response to both PHV and SFV. In line with this, the transcription factor IRF-3 was activated under CHX-restricted infection with VSV or LACV, but not PHV or SFV (Fig. S8). Thus, IFN responses to incoming RNA virus nucleocapsids occur only if the genome contains a 5′ ppp.
Fig. 2. Influence of the viral 5′ppp group and of RIG-I.
(A) Cells were CHX treated, infected with LACVdelNSs, PHV or SFV, and assayed as described for Fig. 1. (B) Innate response to VSV or LACVdelNSs by cells treated with CHX and with siRNAs targeting MDA5 or RIG-I. (C) MEFs lacking MDA5 or (D) RIG-I, and the corresponding wt MEFs, were tested for their innate response to tc-VLPs. See also Figs. S8 to S10.
To pin down the responsible RLR, we evaluated the influence of specific knock-downs. Cells were transfected with siRNAs for MDA5, RIG-I, or a control (Fig. S9), treated with CHX, and infected with VSV or LACV. Knockdown of RIG-I, but not of MDA5, impaired IFN induction by VSV and LACV nucleocapsids (Fig. 2B). To bolster these findings, we utilized mouse embryo fibroblasts (MEFs) deficient in RIG-I or MDA5. As these cells were extremely senstitive to CHX (data not shown), we employed tc-VLPs as a virus system halted at the stage of primary transcription. MEFs lacking MDA5 displayed similar host responses to tc-VLPs as wt MEFs (Fig. 2C). MEFs lacking RIG-I, by contrast, completely lost their ability to respond (Fig. 2D). Knockout of MAVS (Seth et al., 2005), the adaptor common to MDA5 and RIG-I, had a similar impact (Fig. S10). Taken together, these data suggest that nucleocapsids of 5′ppp RNA viruses activate an IFN response via the RIG-I-MAVS signaling cascade.
Activation of RIG-I
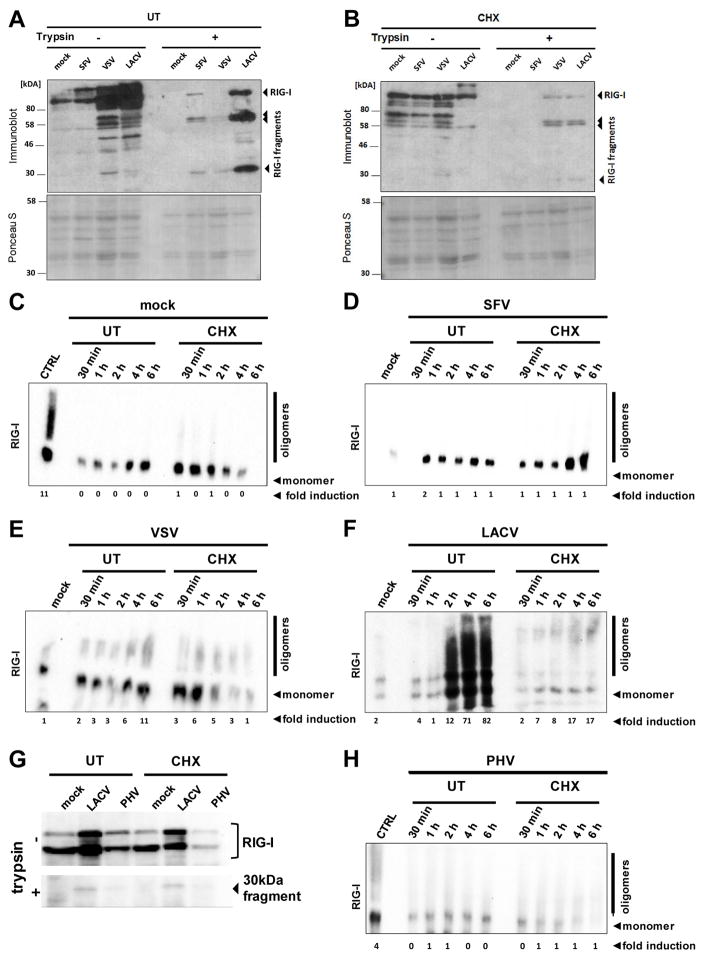
Binding of a target RNA to RIG-I triggers a conformational switch (Saito et al., 2007; Takahasi et al., 2008) and oligomerization (Binder et al., 2011; Saito et al., 2007). The conformational switch is indicated by partial resistance to trypsin digestion. We observed that the full infection cycle of SFV, VSV and LACV leads to the emergence of trypsin-resistant fragments of RIG-I (Fig. 3A). When cells had been pretreated with CHX, VSV and LACV still triggered the conformational switch of RIG-I, whereas SFV infection had no effect (Fig. 3B). The oligomerization of RIG-I was assayed by native PAGE. In uninfected cells, only monomers of RIG-I were detected (Fig. 3C). Curiously, also in SFV-infected cells only monomeric RIG-I is present (Fig. 3D), although RIG-I switches conformation (see Fig. 3A). For the 5′ppp RNA viruses VSV and LACV both the full and the CHX-aborted replication cycle resulted in RIG-I oligomerization (Fig. 3E and F). The 5′monophosphorylated PHV, by contrast, only weakly activated the conformational switch (Fig. 3G), and not the oligomerization (Fig. 3H). Taken together, these results suggest that RIG-I is rapidly and strongly activated by viral nucleocapsids in the absence of genome replication, provided the genome is carrying a 5′ppp group.
Fig. 3. Activation of RIG-I.
(A and B) Conformational switch. A549 cells were treated with 0 (A) or 50 μg/ml CHX (B), infected with the indicated viruses, and lysed 6 h later. Cell extracts were subjected to limited trypsin digestion (right panels), or left untreated (left panels). Upper panels show Western Blot analysis with an anti-RIG-I antibody, lower panels show Ponceau S staining as loading control. (C to F) Oligomerization. A549 cells were left untreated (UT) or treated with 50 μg/ml CHX. Then, cells were either mock infected (C) or infected with SFV (D), VSV (E), or LACVdelNSs (F). At the indicated time points, RIG-I was analyzed by native PAGE and Western blotting. As positive and negative controls, VSV-infected cells at 6 h post-infection (CTRL) and mock infected cells, respectively, were used. (G and H) PHV and RIG-I. A549 cells infected with LACVdelNSs or PHV were monitored for the RIG-I conformation at 6 h p.i. (G), or for RIG-I oligomerization over a time course (H). The ratio of oligomers to monomers (normalised to the actin signal and in relation to mock cells) is indicated below the blots (fold induction).
Interaction between RIG-I and incoming nucleocapsids
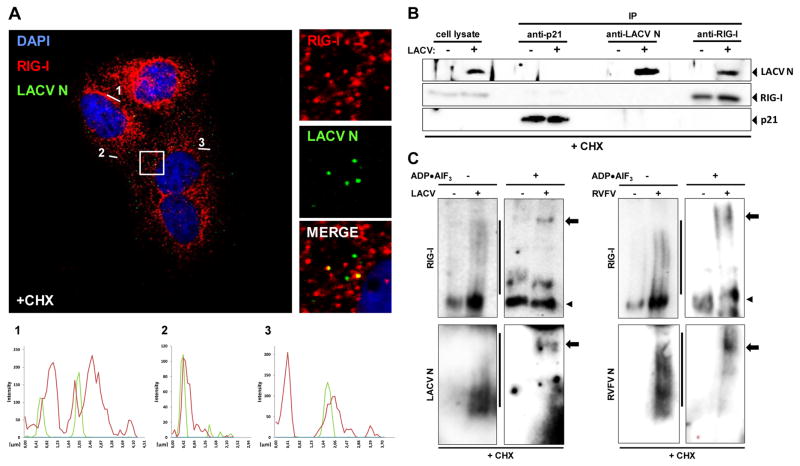
We wondered whether RIG-I could form a complex with viral 5′ppp RNA nucleocapsids. As experimental system, we chose the set-up closest to the immediate early phase (arrested primary transcription), namely infection with bunyaviruses such as LACV under CHX treatment. The N protein thereby served as a marker for viral nucleocapsids. Using confocal immunofluorescence microscopy, we detected the nucleocapsids of incoming LACV particles as individual dots in the cytoplasm (Fig. 4A). These LACV N dots co-localize with RIG-I (Fig. S11), as indicated by overlay pictures and intensity profiles of the fluorescence signals. Quantitative analysis revealed that LACV nucleocapsids co-localize with RIG-I in 51% of the cases, but with the cytoplasmic control protein glyceraldehyde-3-phosphate dehydrogenase (GAPDH) in 33 % of the cases (data not shown). N/RIG-I co-localization was also observed in cells undergoing a full replication cycle (data not shown), in agreement with observations on FLUAV (Onomoto et al., 2012). The incoming LACV nucleocapsids also co-localized with peroxisomes (Fig. S12), cytoplasmic organelles involved in immediate early activation of RIG-I (Dixit et al., 2010). Co-immunoprecipitation demonstrated that RIG-I was capable of binding the incoming LACV nucleocapsids (Fig. 4B). The same RIG-I/LACV nucleocapsid interaction was observed when a full replication cycle was allowed, whereas MDA5 did not interact under any condition (data not shown). Overexpressed N protein, by contrast, could not be precipitated via RIG-I (Fig. S13). We performed similar experiments with RVFV. Again, incoming nucleocapsids triggered the conformational switch and oligomerization of RIG-I, and activated IRF-3 (Fig. S14). Moreover, the nucleocapsids of RVFV co-localized (Fig. S15) and co-precipitated with RIG-I (Fig. S16). Strikingly, incoming nucleocapsids could be forced into a high molecular weight RIG-I complex. The ATP analogue ADP•AlF3 traps RIG-I in an RNA-bound, closed conformation unable to cycle between the different states (Kowalinski et al., 2011). When CHX-treated and infected cells were incubated with ADP•AlF3, the oligomers of RIG-I shifted from a smear into a single, high molecular weight band (Fig. 4C, upper panels). Probing of the lysates for the viral N protein revealed a similar shift from an oligomeric state to a high molecular weight complex (Fig. 4C, lower panels). This was true for LACV as well as for RVFV, indicating a general phenomenon. Together these findings demonstrate that RIG-I is able to associate with nucleocapsids of 5′ppp RNA viruses directly after entry into the cell, leading to RIG-I activation and innate immune signaling.
Fig. 4. Interaction of RIG-I with LACV nucleocapsids.
(A) Co-localization analysis. CHX-treated A549 cells were infected with LACVdelNSs and analyzed 5 h later by double immunofluorescence using antisera against LACV N (green channel) or RIG-I (red channel). Cell nuclei were counterstained with DAPI (blue channel). The square area of the inset is digitally magnified on the right hand side. Three fluorescence intensity profiles are shown on the bottom. (B) Co-immunoprecipitation. CHX-treated A549 cells were infected with LACVdelNSs (MOI 10), lysed 5 h later, and subjected to immunoprecipitation (IP) and Western blot analysis using antibodies against p21 (negative control), LACV N, and RIG-I. As input control, 10% of the cell lysate were analyzed in parallel (left lanes). (C) ADP–aluminium fluoride trapping. CHX-treated A549 cells were infected with LACVdelNSs (left panels) or RVFVΔNSs::REN (right panels). At 5 h post-infection lysates were incubated with ADP•AlF3 and analyzed by native PAGE and Western blot using antibodies against RIG-I (upper panels), or viral N (lower panels). Lines indicate oligomers, arrowheads monomers, and arrows point towards high molecular-weight complexes. See also Figs. S11 to S16.
Nucleocapsid binding and activation of RIG-I are independent of mammalian cofactors
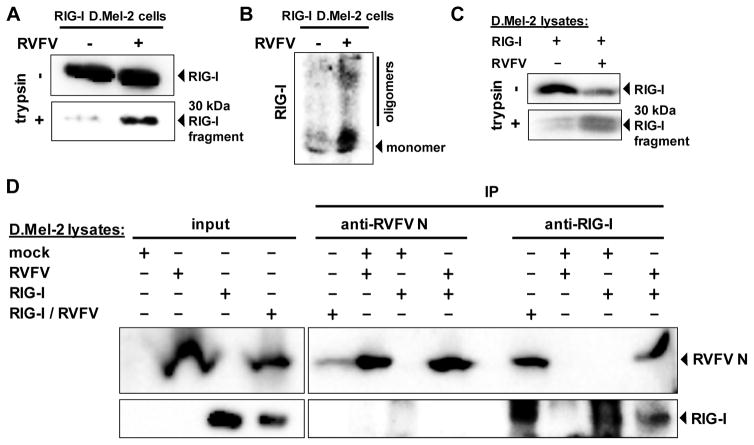
The RIG-I/nucleocapsid interaction could either be direct, or mediated by one of the cellular cofactors of RIG-I (Kato et al., 2011; Kok et al., 2011; Miyashita et al., 2011). To distinguish between these possibilities, we employed an insect cell system. Drosophila melanogaster cells are a useful tool to reconstitute mammalian signaling complexes in a background-free setting (Yang and Reth, 2012), and are infectable with RVFV (Kortekaas et al., 2011). When human RIG-I was expressed in Drosophila D.mel-2 cells, conformational switching and oligomerization were observed after infection with RVFV (Fig. 5A and B). RIG-I was also activated in vitro, by mixing of lysates from RIG-I-transfected D.mel-2 cells with lysates of infected D.mel-2 cells (Fig. 5C). Co-immunoprecipitation experiments demonstrated binding of RIG-I to viral nucleocapsids in the Drosophila system (Fig. 5D). Again, this nucleocapsid interaction was independent of whether RIG-I contacted RVFV during authentic virus infection, or whether lysates containing either RIG-I or RVFV nucleocapids were mixed in vitro. Thus, activation and nucleocapsid interaction of RIG-I occur in a direct manner and without the contribution of a mammalian cofactor.
Fig. 5. Activation of RIG-I and binding to viral nucleocapsids in insect cells.
D.Mel-2 cells expressing human RIG-I were infected for 72 h with RVFVΔNSs::REN (RVFV) and then tested for RIG-I conformation (A) and oligomerization (B). (C) In vitro activation of RIG-I. Lysates of RIG-I-expressing D.Mel-2 cells were mixed with lysates of naïve or RVFVΔNSs::REN-infected D.Mel-2 cells, and assayed for RIG-I conformation. (D) Co-immunoprecipitation. D.Mel-2 cells were either left naïve (mock), or only infected with RVFVΔNSs::REN (RVFV), only expressing RIG-I (RIG-I), or were both expressing RIG-I and superinfected with RVFVΔNSs::REN (RIG-I/RVFV). Combinations of lysates were subjected to IP and Western blot analysis using antibodies against RVFV N or RIG-I. As input control, 10% of the cell lysate were analyzed in parallel (left lanes).
Nucleocapsid-borne 5′ ppp-dsRNA is necessary and sufficient for RIG-I activation
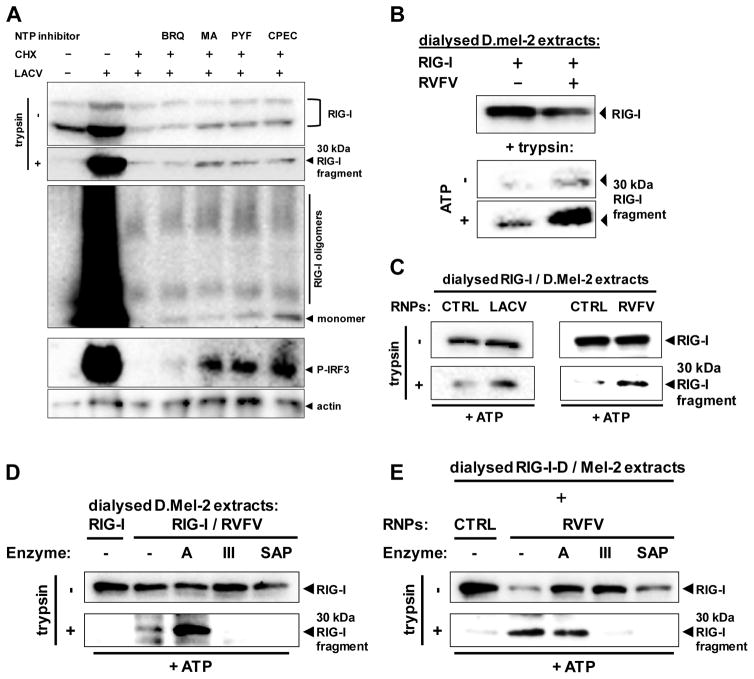
Our experiments with bunyaviruses and CHX implicated that the transcriptional activity of nucleocapsids may not be relevant for RIG-I activation. However, CHX still allows some abortive transcription (Raju et al., 1989). To clarify whether RIG-I requires this residual RNA synthesis, we performed several experiments. Firstly, we depleted the cellular NTP pool with the compounds Brequinar (BRQ; inhibits pyrimidine synthesis), mycophenolic acid (MA; reduces GTP levels), pyrazofurin (PYF; reduces CTP and UTP levels), or cyclopentenylcytosine (CPEC; depletes the CTP pool) (Linke et al., 1996; Qing et al., 2010). Each of these inhibitors affected viral RNA synthesis (Fig. S17). Nonetheless, even when combined with CHX, neither inhibitor diminished RIG-I activation by LACV (Fig. 6A). In fact, some of the compounds slightly increased RIG-I activation, and enhanced the activation of IRF-3 (Fig. 6A). Secondly, we depleted Drosophila D.Mel-2 cell lysates containing either RIG-I or nucleocapsids from NTPs by dialysis. When these dialysed lysates were mixed with each other, the conformational switch of RIG-I still occurred, and was enhanced by adding back ATP (Fig. 6B). Thirdly, we employed nucleocapsids that had been isolated from purified virus particles. Dialysed, RIG-I-containing D.Mel-2 extracts were incubated with nucleocapsids of RVFV or LACV. The nucleocapsids clearly induced the conformational switch of RIG-I in vitro (Fig. 6C). We conclude from these experiments that viral transcription is not necessary for triggering RIG-I. As outlined above, the genomic RNA of bunyaviruses forms a “panhandle” structure which has remarkable similarity with the optimal 5′ppp dsRNA ligand identified by transfection and in vitro binding experiments (Fig. S18). Indeed, destroying the dsRNA structures with RNase III or cleaving the triphosphates with a phosphatase abolished the ability of viral nucleocapsids to activate RIG-I (Fig. 6D and 6E). Treatment with the ssRNA-specific RNase A, by contrast, had no effect. Thus, a 5′ppp dsRNA structure is indeed necessary for the activation of RIG-I by nucleocapsids.
Fig. 6. Activation of RIG-I solely depends on the nucleocapsid-borne panhandle structure.
(A) Effect of NTP withdrawal on activation of RIG-I and IRF-3. Pretreated A549 cells were infected with LACVdelNSs for 5 h. Pretreatment with CHX (50 μg/ml) was for 1h, and with BRQ (10 μM, stocks dissolved in DMSO), MA (10 μM, stocks dissolved in methanol), PYF (10 μM, stocks dissolved in DMSO), or CPEC (5 μM, stocks dissolved in DMSO) for 24 h. RIG-I conformation (upper two panels), RIG-I oligomerization (upper middle panel), and IRF-3 phosphorylation (lower middle panel) were monitored. Immunoblot for actin served as loading control. (B) RIG-I activation in dialysed samples. Lysates from D.Mel-2 cells expressing RIG-I (RIG-I) or infected with RVFVΔNSs::REN (RVFV) were dialysed against PBS, mixed with each other, and incubated with or without ATP. After 1 h incubation, mixes were subjected to the RIG-I conformational switch assay. (C) RIG-I activation by purified viral nucleocapsids (RNPs). Lysates from D.Mel-2 cells expressing RIG-I were dialysed against PBS and mixed with purified nucleocapsids from particles of LACV (left panels) or RVFV (right panels). Incubation with ATP and conformational switch assay were performed as described for (B). Equivalent fractions of gradient-purified supernatants from mock-infected cells were used as negative control (CTRL). (D and E) Structural requirements for viral nucleocapsids to activate RIG-I. Lysates from RVFV-infected D.Mel-2 cells (D) or purified RVFV nucleocapsids (E) were incubated with ATP and one of the indicated enzymes, namely RNase A (A), RNase III (III), or Shrimp Alkaline Phosphatase (SAP). After mixing and incubation with dialysed lysates from RIG-I-expressing D.Mel-2 cells, the RIG-I conformational switch assay was performed. Negative controls (CTRL) were performed as described for (C). See also Figs. S17 and S18.
A single site on the nucleocapsids is contacted by RIG-I
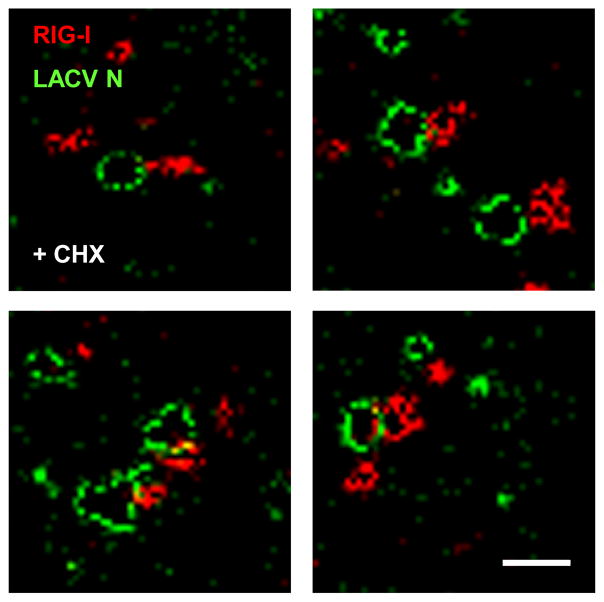
We employed GSD (Ground State Depletion) microscopy to visualize the RIG-I/LACV nucleocapsid complex at 20 nm resolution. This technique allows to display individual nucleocapsids in their characteristic pseudocircular shape (Fig. 7, green channel), which is caused by the dsRNA panhandle formation between the 5′ and 3′ genome ends (Obijeski et al., 1976). Strikingly, accumulations of RIG-I are contacting nucleocapsids at a single site, lending the co-complexes a “diamond ring”-like appearance (Fig. 7, red channel). The negative control GAPDH, by contrast, appeared to be more distant and not accumulated at the nucleocapsids (Fig. S19). These observations add additional weight to our hypothesis that RIG-I binds the nucleocapsids via the terminal 5′ppp dsRNA panhandle.
Fig. 7. Super-resolution immunofluorescence microscopy of RIG-I/LACV nucleocapsid complexes.
CHX-treated A549 cells were infected with LACVdelNSs and analyzed 5 h later by GSDIM double immunofluorescence using antisera against LACV N (green channel) or RIG-I (red channel). Four example areas with nucleocapsids are shown. Scale bar 200 nm. See also Fig. S19.
Discussion
The aim of our study was to clarify whether RIG-I is capable of recognizing the RNA contained within incoming viral nucleocapsids. Our results indicate that this is indeed the case. RIG-I was rapidly activated by viruses with the prototypical 5′ppp dsRNA panhandle structure, even when viral RNA synthesis was abolished. Moreover, we detected a direct, single-site interaction between RIG-I and nucleocapsids which was dependent on 5′ppp dsRNA. Thus, viral nucleocapsids containing a 5′ppp panhandle represent a pathogen-associated molecular pattern (PAMP) for RIG-I. These findings advance the proximal RIG-I-sensitive step of the viral infection cycle from the late stage of genome replication (Baum et al., 2010; Rehwinkel et al., 2010) to the immediate early step of nucleocapsids entering the cytoplasm.
Erroneous, non-encapsidated replication products appear during infection with FLUAV (Vreede et al., 2004). It is quite likely that such naked replication products (Rehwinkel et al., 2010; Vreede et al., 2004), along with defective interfering RNAs (Baum et al., 2010; Killip et al., 2011; Strahle et al., 2006) and newly formed nucleocapsids (this study) are responsible for RIG-I induction under a full viral multiplication cycle. Previous reports already demonstrated that genome replication is not an absolute requirement for IFN induction (Killip et al., 2012; Marcus and Sekellick, 1980), and that transfected nucleocapsids of the 5′ppp viruses measles and VSV can activate IRF-3, even if they are transcriptionally inactive (tenOever et al., 2002; tenOever et al., 2004). None of these studies, however, had addressed the involvement of RIG-I or the requirement for a specific 5′ genome end. Our work extends their conclusions by showing that incoming viral nucleocapsids can directly activate RIG-I, thus triggering a rapid innate immune response.
It remains to be shown how RIG-I manages to access the encapsidated viral RNA. Although the dsRNA-panhandle structure is covered by the viral polymerase (Resa-Infante et al., 2011), some cytoplasmic exposure is necessary e.g. to initiate mRNA transcription. The global presence of RIG-I in the cytoplasm and its high affinity for 5′ppp dsRNA may allow to rapidly entrap a panhandle, even if it is only briefly exposed. Our observation of an early and substantial formation of RIG-I oligomers in response to incoming nucleocapsids is supporting this hypothesis.
The standard model of RIG-I activation implies that ligand binding induces a conformational change followed by oligomerization (Kowalinski et al., 2011; Saito et al., 2007; Takahasi et al., 2008). Interestingly, oligomerization seems not to be a strict consequence of the conformational switch. While infection with the negative-stranded 5′ppp-RNA viruses VSV, LACV and RVFV triggers both the conformational switch and the oligomerization, infection with the positive-strand 5′capped RNA virus SFV triggers only the conformational switch. It is known that innate immune recognition of SFV occurs mainly through MDA5, with a modest contribution from RIG-I (Schulz et al., 2010). VSV, LACV and RVFV are mainly recognized by RIG-I (Habjan et al., 2008a; Kato et al., 2006; Verbruggen et al., 2011). Thus, a weak RIG-I trigger like SFV may only cause the conformational switch, whereas stronger RIG-I triggers continue to the subsequent oligomerization.
We have introduced Drosophila cells as a tool to establish that RIG-I activation occurs independent of mammalian cofactors. In mammalian cells, RIG-I is fine-tuned by inhibitors and cofactors. The Drosphila cells, which can co-express up to 12 different recombinant proteins (Yang and Reth, 2012), hold promise as a system to reconstitute the RIG-I signaling complex for studying the details of its regulation.
The classical studies on PRRs and PAMPs involved the usage of purified RNA ligands. However, as we and others (Baum and Garcia-Sastre, 2011; Kato et al., 2011) have pointed out, viral RNAs in their physiological context are complexed with host cell or viral proteins. Recent reports on the PRRs PKR and TLR3 provide increasing evidence that protein-bound RNA ligands are comparable, or even more powerful PRR ligands than naked RNAs are (Dauber et al., 2009; Lai et al.). It would be interesting to test other PRRs for activation by physiological nucleic acid-protein complexes.
In summary, our results indicate that RIG-I, the major intracellular PRR for viral pathogens, is capable of recognizing the 5′ppp-dsRNA of viral nucleocapsids. This enables a cytoplasmic response to incoming negative-strand RNA viruses at the earliest possible time point of infection.
Experimental Procedures
Cells, viruses, plasmids, and reagents
A549, 293T, and MEFs deficient in RIG-I, MDA5 (Kato et al., 2006) or MAVS (Seth et al., 2005) were cultivated in DMEM supplemented with 10% FCS at 37°C and 5% CO2. D.Mel-2 cells (Gibco) were cultivated in Spodopan (Pan Biotech) at 28°C with no additional CO2. VSV, SFV, FLUAVΔNS1 (Garcia-Sastre et al., 1998), RVFVΔNSs::GFP (Habjan et al., 2008b; Kuri et al., 2010), RVFVΔNSs::REN (Habjan et al., 2008b; Kuri et al., 2010), and LACVdelNSs (Blakqori et al., 2007) were propagated on Vero cells. Plasmids pI.18_RVFV_N, pI.18_RVFV_L, pI.18_RVFV_M, and pHH21_RVFV_vMGFP were described previously (Habjan et al., 2008b; Habjan et al., 2009a). Plasmid pRmHA3-RIG-I was constructed by cloning a PCR-generated RIG-I cDNA fragment into pRmHA3 (Bunch et al., 1988), using engineered 5′ KpnI and 3′ SalI restriction sites. Primer sequences are available upon request. CHX, NH4Cl, CuSO4, BRQ and MA were from Sigma. PYF (NSC-143095) and CPEC (NSC-375575) were kindly obtained from the Drug Synthesis and Chemistry Branch of the National Cancer Institute.
Production of VLPs
RVFV VLPs were generated as described (Habjan et al., 2009a). Briefly, subconfluent monolayers of 293T cells in 90 mm dishes were transfected with 3 μg each of pI.18_RVFV_N, pI.18_RVFV_L, pI.18_RVFV_M, and pHH21_RVFV_vMGFP (tc-VLPs), or pI.18_RVFV_N and pI.18_RVFV_M only (ghost VLPs), using Nanofectin (PAA Laboratories). At 5 h post-transfection, medium was changed and 48 h later supernatants collected and clarified from cell debris by centrifugation (6000×g, 10 min at 4°C). VLPs were concentrated with Amicon Ultra-15 (Millipore) Ultracel-100kDa filter devices.
Infection of cells
Cells grown to 90% confluency were inoculated for 1 h with viruses or VLPs dissolved in 200 μl OptiMEM (Invitrogen; for mammalian cells) or Spodopan (for insect cells) at an MOI of 5. FLUAVΔNS1 was washed off with PBS, whereas for all other infections the inoculum was directly replaced with DMEM 5% FCS (mammalian cells) or Spodopan (insect cells). If required, cells were pretreated with inhibitors dissolved in complete medium for 1 h (CHX, NH4Cl) or 24 h (BRQ, MA, PYF, CPEC). The inhibitors were also added to the virus inoculum and the incubation medium.
Real-time RT-PCR
Cellular RNA was isolated with the NucleoSpin RNA II kit (Macherey-Nagel). Six hundred ng were used for cDNA synthesis and PCR, employing the QuantiTect SYBR Green RT-PCR Kit (Qiagen) and a LightCycler II (Roche). mRNAs of human and murine IFN-β and ISG56 were detected with specific QuantiTect primers (see Supplemental Information) and normalized against γ-actin (human cells) or GAPDH (murine cells) using the ddCT method (Livak and Schmittgen, 2001). Up-regulation of inducible genes is depicted in relation to non-stimulated, non-infected (mock) cells.
siRNA knockdown
Knockdown of gene expression was achieved by two-fold reverse transfection of siRNAs. siRNAs (25 nM each; see Supplemental Information) were diluted in 100 μl DMEM, mixed with 3 μl HiPerFect (Qiagen), incubated for 10 min at room temperature, and dropped onto a 12-well plate. Then, 1.5×10E5 cells in DMEM 10% FCS were seeded on top. After 48 h at 37°C, cells were harvested, counted, and 1.5×10E5 cells were again reverse transfected as described.
Activation state of RIG-I
To assay the conformation of RIG-I, cells were lysed in PBS/0.5% Triton X-100, and incubated on ice for 10 min. Then, samples were sonified in a Branson 3200 Ultrasonic cleaner at 4°C for 10 min, and centrifuged at 4°C for 10 min at 10,000×g. An aliquot of 25 μg of total protein in 10 μl PBS was digested for 25 min with 0.2 μg/μl L-1-tosylamido-2-phenylethyl chloromethyl ketone-treated trypsin (Sigma-Aldrich) at 37°C. Reaction was stopped by adding 5× sample buffer (250 mM Tris-HCl pH 6.8, 10% SDS, 50% glycerol, 25% β-Mercaptoethanol (β-ME), 0.5% Bromphenol Blue) and heating at 95°C for 5 min. Samples were subjected to 12% SDS-PAGE and Western Blot analysis using mouse monoclonal anti-RIG-I antibody (ALME-1; Enzo Life Sciences) at 1:1000. Staining of the blot with 0.1% Ponceau S in 5% acetic acid served as a loading control.
To investigate the oligomerization of RIG-I, 50 μg of sonified cell lysate in native loading buffer (50 mM Tris-HCl pH 6.8, 10% glycerol, 0.1% Bromphenol Blue) were loaded onto a nondenaturing 8% polyacrylamide gel. Proteins were separated by electrophoresis with 50 mM Tris-NaOH pH 9.0, 384 mM glycin as anode buffer and 50 mM Tris pH 8.3, 384 mM glycin, 1% sodium deoxycholate as cathode buffer. Western blot analysis was performed as outlined above.
Immunofluorescense assays
Cells were grown on coverslips to 30–50% confluency, infected, and incubated for the indicated time. Cells were fixed with 3% paraformaldehyde, permeabilized with 0.5% Triton X-100 in PBS, and washed three times with PBS. Primary antibodies were diluted in PBS/1% FCS. Primary antibodies were either rabbit polyclonal anti-LACV N (1:1000) (Blakqori et al., 2007) or rabbit polyclonal anti-RVFV N (1:1000) (Lorenzo et al., 2008), combined with mouse polyclonal anti-human RIG-I (1:200; (Baum et al., 2010). After 1 h incubation at room temperature, coverslips were washed 3× in PBS then treated with goat anti-rabbit Cy2 and goat anti-mouse Cy3 at a dilution of 1:200. After 3× washing in PBS, coverslips were mounted with Fluorsave solution (Calbiochem) and examined using a Leica SP5 confocal microscope.
For the super-resolution microscopy, a Leica SR GSD (ground state depletion) microscope was used. Samples were prepared as described for confocal microscopy except that goat anti-rabbit Alexa Fluor 488 (LACV N) and goat anti-mouse Alexa Fluor 647 (RIG-I) were employed as secondary antibody antibodies. Samples were embedded in freshly prepared 100 mM β-Mercaptoethylamine (MEA) in PBS (pH 7.4) directly before imaging.
Co-immunoprecipitation assays
Cells grown in two T175 flasks were scraped off in 10 ml PBS, centrifuged at low speed, and the pellets lysed in 1050 μl RIPA buffer (prepared with DEPC-treated H2O) containing protease inhibitors, incubated on ice for 30 min and centrifuged at 4°C for 10 min at 10,000×g. Supernatants were transferred to fresh tubes and 10% kept as input control. The remaining 90% of the lysates were subjected to immunoprecipitation (IP) using Dynabeads (Invitrogen). In parallel, 1.5 mg beads per IP were coupled with the appropriate antibodies using the Dynabeads antibody coupling kit (Invitrogen). For the anti-p21 and anti-RIG-I IPs, beads were coupled with rabbit polyclonal anti-mouse antibody (DAKO), for the anti-LACV N IP beads were coupled with rabbit polyclonal anti-LACV N (each at a 1:200 dilution per IP), and for the anti-RVFV N IP beads were coupled with mouse polyclonal anti-N serum (Habjan et al., 2009b) for 20 h at 37°C. After antibody coupling, beads were washed 2× with RIPA buffer, and the anti-mouse antibody beads were further incubated with mouse monoclonal anti-p21 (Santa Cruz Biotechnology) or mouse monoclonal anti-RIG-I antibody ALME-1 at a 1:200 dilution in RIPA buffer for 2 h at 4°C. The beads were then incubated with the lysates for 2 h at 4°C, the immunoprecipitates washed 3× with RIPA buffer, and then eluted with sample buffer (50 mM Tris-HCl pH 6.8, 2% SDS, 10% glycerol, 5% β-ME, 0.1% Bromphenol Blue) for 5 min at 95°C. Eluates were analyzed by Western blotting using rabbit polyclonal anti-LACV N or anti-RVFV N antisera (1:1000), mouse monoclonal anti-RIG-I antibody ALME-1 (1:1000), or mouse monoclonal anti-p21 (1:500). Protein A-HRP conjugate (Millipore; 1:10 000) was used for detection.
ADP-aluminium fluoride trapping
ADP•AlF3 trapping was performed essentially as described elsewhere (Chaney et al., 2001). Briefly, 50 μg cell protein were prepared in STA buffer (25 mM Tris-Acetate pH 8.0, 8 mM Mg-actetate, 10 mM KCl, 3.5% w/v PEG 6000, 1 mM DTT) with 0.2 mM ADP and 10mM NaF, and incubated at 37°C for 5 min. After addition of 0.4 mM AlCl3 the reactions were incubated for further 10 min, and then supplemented with native loading buffer (50 mM Tris-HCl pH 6.8, 10% glycerol, 0.1% bromophenol blue). Native gel electrophoresis and Western blot analysis were performed as described above.
Expression of RIG-I in Drosophila cells
D.Mel-2 cells (1× 10E7) resuspended in 10 ml Spodopan were transfected with 16 μg of pRmHa3-RIG-I plasmid mixed with 48 μl Cellfectin (Invitrogen), and incubated at 28°C in T75 flasks. RIG-I expression was induced 48 h later with 1 mM CuSO4, and 24 h later cells were scraped off in 10 ml PBS. Cells were pelleted by 5 min centrifugation at 100×g, and resuspended in 800 μl RIPA buffer (for IPs) or in 150 μl 0.5% Triton X-100 in PBS (for RIG-I assays). The suspensions were incubated for 10 min at 4°C, centrifuged at 10000×g, and the supernantants kept at 4°C.
Dialysis and ATP supplementation of cell lysates
Visking dialysis tubing 27/32 with a diameter of 21 mm (Serva Electrophoresis) were used to deplete cell lysates from low-molecular weight compounds. The membrane tubes were activated by submerging in H2O and boiling for 1 min in the microwave oven, followed by a transfer into H2O at room temperature. For cell lysates, 100 μl were dialysed at 4°C against PBS in 1 ml Eppendorf tubes covered with the dialysis membrane. For nucleocapsids, 10 μl were dialysed using 200 μl PCR tubes. The buffer was exchanged after 1 h, 12 h, and then again 1 h. To test the contribution of ATP, lysates or nucleocapsids were first mixed and then incubated with or without 1 mM ATP for 1 h at 37°C. The RIG-I conformational switch assay was performed as indicated above.
Purification of viral nucleocapsids
BHK cells seeded in 10 T175 flasks were infected with virus at an MOI of 0.01. Cell supernatants were harvested at 3 days later and virions purified by centrifugation through a 30% glycerol cushion at 25.000 rpm for 1.5 h at 4°C in a SW-32 rotor. Pellets were resuspended in hypotonic lysis buffer (10 mM Tris/HCl pH 7.8, 150 mM NaCl, 1 mM EDTA, 1% NP40) in the presence of Complete protease inhibitors (Roche). Nucleocapsids were purified in a CsCl gradient as described elsewere (Mavrakis et al., 2002), with minor modifications. Briefly, the cleared lysate was loaded on top of a continuous 20 to 40% CsCl gradient in 20 mM Tris/HCl pH 7.9, 200 mM NaCl, centrifuged at 52.000 rpm for 2 h at 12°C in a SW60 rotor, and the recovered fraction pelleted at 45.000 rpm for 1 h at 4°C in a TLA45 rotor. Nucleocapsids were resuspended in PBS and dialysed against PBS to remove residual CsCl. Nucleocapsid-containing fractions were identified by SDS-PAGE with Coomassie Blue staining and Western blot analysis.
Enzymatic treatment of lysates
Dialysed lysates of RVFV-infected D.Mel-2 cells (50 μg protein in 10 μl) or nucleocapsids were supplemented with 1 mM ATP and incubated either with 5 μg RNase A, 1 U RNase III, or 2 U Shrimp Alkaline Phosphatase (SAP) for 1 h at 37°C. Samples were then mixed 1:1 with dialysed lysates from RIG-I expressing D.Mel-2 cells (50 μg protein in 10 μl) and incubated for 1 h at 37°C. The RIG-I conformation assay was performed with half of the sample, whereas the other half was kept as input control.
Supplementary Material
Highlights.
RIG-I is activated by the incoming RNA virus nucleocapsids during infection
RIG-I activation requires a 5′triphosphate dsRNA structure on the nucleocapsids
Viral nucleocapsids trigger conformational switching and oligomerization of RIG-I
RIG-I directly binds to viral nucleocapsids containing a 5′triphosphate dsRNA structure
Acknowledgments
We thank Valentina Wagner and Jörg Schmidt for excellent technical assistance, Michael Reth for drawing our attention to Drosophila cells, and Stefan Bauer and Marco Binder for critically reading the manuscript. We also thank the antibody facility at MSSM NY, and Shizuo Akira, Zhijian J. Chen, Gema Lorenzo, Alejandro Brun, Markus Schnare, and the NCI for reagents. FW is supported by the DFG grants We2616/2-3 and We2616/5-2, grant 47/2012MR of the Forschungsförderung (§2 Abs. 3) Kooperationsvertrag UKGM, and the Leibniz Graduate School for Emerging viral diseases (EIDIS), GK by DFG grant Ko1579/52, and AGS by the NIAID grants R01AI046954, U19AI083025 and the CEIRS program of NIAID under contract HHSN266200700010C.
References
- Baum A, Garcia-Sastre A. Differential recognition of viral RNA by RIG-I. Virulence. 2011;2:166–169. doi: 10.4161/viru.2.2.15481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum A, Sachidanandam R, Garcia-Sastre A. Preference of RIG-I for short viral RNA molecules in infected cells revealed by next-generation sequencing. Proc Natl Acad Sci U S A. 2010;107:16303–16308. doi: 10.1073/pnas.1005077107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binder M, Eberle F, Seitz S, Mucke N, Huber CM, Kiani N, Kaderali L, Lohmann V, Dalpke A, Bartenschlager R. Molecular Mechanism of Signal Perception and Integration by the Innate Immune Sensor Retinoic Acid-inducible Gene-I (RIG-I) J Biol Chem. 2011;286:27278–27287. doi: 10.1074/jbc.M111.256974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blakqori G, Delhaye S, Habjan M, Blair CD, Sanchez-Vargas I, Olson KE, Attarzadeh-Yazdi G, Fragkoudis R, Kohl A, Kalinke U, et al. La Crosse bunyavirus nonstructural protein NSs serves to suppress the type I interferon system of mammalian hosts. J Virol. 2007;81:4991–4999. doi: 10.1128/JVI.01933-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunch TA, Grinblat Y, Goldstein LS. Characterization and use of the Drosophila metallothionein promoter in cultured Drosophila melanogaster cells. Nucleic Acids Res. 1988;16:1043–1061. doi: 10.1093/nar/16.3.1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaney M, Grande R, Wigneshweraraj SR, Cannon W, Casaz P, Gallegos MT, Schumacher J, Jones S, Elderkin S, Dago AE, et al. Binding of transcriptional activators to sigma 54 in the presence of the transition state analog ADP-aluminum fluoride: insights into activator mechanochemical action. Genes Dev. 2001;15:2282–2294. doi: 10.1101/gad.205501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dauber B, Martinez-Sobrido L, Schneider J, Hai R, Waibler Z, Kalinke U, Garcia-Sastre A, Wolff T. Influenza B virus ribonucleoprotein is a potent activator of the antiviral kinase PKR. PLoS Pathog. 2009;5:e1000473. doi: 10.1371/journal.ppat.1000473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Boer SM, Kortekaas J, Spel L, Rottier PJ, Moormann RJ, Bosch BJ. Acid-activated structural reorganization of the rift valley Fever virus gc fusion protein. J Virol. 2012;86:13642–13652. doi: 10.1128/JVI.01973-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixit E, Boulant S, Zhang Y, Lee AS, Odendall C, Shum B, Hacohen N, Chen ZJ, Whelan SP, Fransen M, et al. Peroxisomes are signaling platforms for antiviral innate immunity. Cell. 2010;141:668–681. doi: 10.1016/j.cell.2010.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filone CM, Heise M, Doms RW, Bertolotti-Ciarlet A. Development and characterization of a Rift Valley fever virus cell-cell fusion assay using alphavirus replicon vectors. Virology. 2006;356:155–164. doi: 10.1016/j.virol.2006.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Sastre A, Egorov A, Matassov D, Brandt S, Levy DE, Durbin JE, Palese P, Muster T. Influenza A virus lacking the NS1 gene replicates in interferon-deficient systems. Virology. 1998;252:324–330. doi: 10.1006/viro.1998.9508. [DOI] [PubMed] [Google Scholar]
- Garcin D, Lezzi M, Dobbs M, Elliott RM, Schmaljohn C, Kang CY, Kolakofsky D. The 5′ ends of Hantaan virus (Bunyaviridae) RNAs suggest a prime-and-realign mechanism for the initiation of RNA synthesis. J Virol. 1995;69:5754–5762. doi: 10.1128/jvi.69.9.5754-5762.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habjan M, Andersson I, Klingstrom J, Schumann M, Martin A, Zimmermann P, Wagner V, Pichlmair A, Schneider U, Muhlberger E, et al. Processing of genome 5′ termini as a strategy of negative-strand RNA viruses to avoid RIG-I-dependent interferon induction. PLoS One. 2008a;3:e2032. doi: 10.1371/journal.pone.0002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habjan M, Penski N, Spiegel M, Weber F. T7 RNA polymerase-dependent and -independent systems for cDNA-based rescue of Rift Valley fever virus. J Gen Virol. 2008b;89:2157–2166. doi: 10.1099/vir.0.2008/002097-0. [DOI] [PubMed] [Google Scholar]
- Habjan M, Penski N, Wagner V, Spiegel M, Overby AK, Kochs G, Huiskonen JT, Weber F. Efficient production of Rift Valley fever virus-like particles: The antiviral protein MxA can inhibit primary transcription of bunyaviruses. Virology. 2009a;385:400–408. doi: 10.1016/j.virol.2008.12.011. [DOI] [PubMed] [Google Scholar]
- Habjan M, Pichlmair A, Elliott RM, Overby AK, Glatter T, Gstaiger M, Superti-Furga G, Unger H, Weber F. NSs protein of rift valley fever virus induces the specific degradation of the double-stranded RNA-dependent protein kinase. J Virol. 2009b;83:4365–4375. doi: 10.1128/JVI.02148-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handke W, Oelschlegel R, Franke R, Kruger DH, Rang A. Hantaan virus triggers TLR3-dependent innate immune responses. J Immunol. 2009;182:2849–2858. doi: 10.4049/jimmunol.0802893. [DOI] [PubMed] [Google Scholar]
- Hiscott J. Triggering the innate antiviral response through IRF-3 activation. J Biol Chem. 2007;282:15325–15329. doi: 10.1074/jbc.R700002200. [DOI] [PubMed] [Google Scholar]
- Hoenen T, Groseth A, de Kok-Mercado F, Kuhn JH, Wahl-Jensen V. Minigenomes, transcription and replication competent virus-like particles and beyond: Reverse genetics systems for filoviruses and other negative stranded hemorrhagic fever viruses. Antiviral Res. 2011;91:195–208. doi: 10.1016/j.antiviral.2011.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm CK, Jensen SB, Jakobsen MR, Cheshenko N, Horan KA, Moeller HB, Gonzalez-Dosal R, Rasmussen SB, Christensen MH, Yarovinsky TO, et al. Virus-cell fusion as a trigger of innate immunity dependent on the adaptor STING. Nat Immunol. 2012 doi: 10.1038/ni.2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornung V, Ellegast J, Kim S, Brzozka K, Jung A, Kato H, Poeck H, Akira S, Conzelmann KK, Schlee M, et al. 5′-Triphosphate RNA Is the Ligand for RIG-I. Science. 2006;314 doi: 10.1126/science.1132505. [DOI] [PubMed] [Google Scholar]
- Kato H, Takahasi K, Fujita T. RIG-I-like receptors: cytoplasmic sensors for non-self RNA. Immunol Rev. 2011;243:91–98. doi: 10.1111/j.1600-065X.2011.01052.x. [DOI] [PubMed] [Google Scholar]
- Kato H, Takeuchi O, Mikamo-Satoh E, Hirai R, Kawai T, Matsushita K, Hiiragi A, Dermody TS, Fujita T, Akira S. Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid-inducible gene-I and melanoma differentiation-associated gene 5. J Exp Med. 2008;205:1601–1610. doi: 10.1084/jem.20080091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato H, Takeuchi O, Sato S, Yoneyama M, Yamamoto M, Matsui K, Uematsu S, Jung A, Kawai T, Ishii KJ, et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441:101–105. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- Killip MJ, Young DF, Precious BL, Goodbourn S, Randall RE. Activation of the beta interferon promoter by paramyxoviruses in the absence of virus protein synthesis. J Gen Virol. 2012;93:299–307. doi: 10.1099/vir.0.037531-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Killip MJ, Young DF, Ross CS, Chen S, Goodbourn S, Randall RE. Failure to activate the IFN-beta promoter by a paramyxovirus lacking an interferon antagonist. Virology. 2011;415:39–46. doi: 10.1016/j.virol.2011.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kok KH, Lui PY, Ng MH, Siu KL, Au SW, Jin DY. The double-stranded RNA-binding protein PACT functions as a cellular activator of RIG-I to facilitate innate antiviral response. Cell Host Microbe. 2011;9:299–309. doi: 10.1016/j.chom.2011.03.007. [DOI] [PubMed] [Google Scholar]
- Kortekaas J, Oreshkova N, Cobos-Jimenez V, Vloet RP, Potgieter CA, Moormann RJ. Creation of a nonspreading Rift Valley fever virus. J Virol. 2011;85:12622–12630. doi: 10.1128/JVI.00841-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalinski E, Lunardi T, McCarthy AA, Louber J, Brunel J, Grigorov B, Gerlier D, Cusack S. Structural Basis for the Activation of Innate Immune Pattern-Recognition Receptor RIG-I by Viral RNA. Cell. 2011;147:423–435. doi: 10.1016/j.cell.2011.09.039. [DOI] [PubMed] [Google Scholar]
- Kuri T, Habjan M, Penski N, Weber F. Species-independent bioassay for sensitive quantification of antiviral type I interferons. Virol J. 2010;7:50. doi: 10.1186/1743-422X-7-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai Y, Yi G, Chen A, Bhardwaj K, Tragesser BJ, Rodrigo AV, Zlotnick A, Mukhopadhyay S, Ranjith-Kumar CT, Kao CC. Viral Double-Strand RNA-Binding Proteins Can Enhance Innate Immune Signaling by Toll-Like Receptor 3. PLoS One. 2011;6:e25837. doi: 10.1371/journal.pone.0025837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linke SP, Clarkin KC, Di Leonardo A, Tsou A, Wahl GM. A reversible, p53-dependent G0/G1 cell cycle arrest induced by ribonucleotide depletion in the absence of detectable DNA damage. Genes Dev. 1996;10:934–947. doi: 10.1101/gad.10.8.934. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lorenzo G, Martin-Folgar R, Rodriguez F, Brun A. Priming with DNA plasmids encoding the nucleocapsid protein and glycoprotein precursors from Rift Valley fever virus accelerates the immune responses induced by an attenuated vaccine in sheep. Vaccine. 2008;26:5255–5262. doi: 10.1016/j.vaccine.2008.07.042. [DOI] [PubMed] [Google Scholar]
- Marcus PI, Sekellick MJ. Interferon induction by viruses. III. Vesicular stomatitis virus: interferon-inducing particle activity requires partial transcription of gene N. J Gen Virol. 1980;47:89–96. doi: 10.1099/0022-1317-47-1-89. [DOI] [PubMed] [Google Scholar]
- Mavrakis M, Kolesnikova L, Schoehn G, Becker S, Ruigrok RW. Morphology of Marburg virus NP-RNA. Virology. 2002;296:300–307. doi: 10.1006/viro.2002.1433. [DOI] [PubMed] [Google Scholar]
- McCartney SA, Colonna M. Viral sensors: diversity in pathogen recognition. Immunol Rev. 2009;227:87–94. doi: 10.1111/j.1600-065X.2008.00726.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McInerney GM, Kedersha NL, Kaufman RJ, Anderson P, Liljestrom P. Importance of eIF2alpha phosphorylation and stress granule assembly in alphavirus translation regulation. Mol Biol Cell. 2005;16:3753–3763. doi: 10.1091/mbc.E05-02-0124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyashita M, Oshiumi H, Matsumoto M, Seya T. DDX60, a DEXD/H box helicase, is a novel antiviral factor promoting RIG-I-like receptor-mediated signaling. Mol Cell Biol. 2011;31:3802–3819. doi: 10.1128/MCB.01368-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noyce RS, Taylor K, Ciechonska M, Collins SE, Duncan R, Mossman KL. Membrane Perturbation Elicits an IRF3-Dependent, Interferon-Independent Antiviral Response. J Virol. 2011;85:10926–10931. doi: 10.1128/JVI.00862-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obijeski JF, Bishop DH, Palmer EL, Murphy FA. Segmented genome and nucleocapsid of La Crosse virus. J Virol. 1976;20:664–675. doi: 10.1128/jvi.20.3.664-675.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onomoto K, Jogi M, Yoo JS, Narita R, Morimoto S, Takemura A, Sambhara S, Kawaguchi A, Osari S, Nagata K, et al. Critical Role of an Antiviral Stress Granule Containing RIG-I and PKR in Viral Detection and Innate Immunity. PLoS ONE. 2012;7:e43031. doi: 10.1371/journal.pone.0043031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichlmair A, Schulz O, Tan CP, Naslund TI, Liljestrom P, Weber F, Reis ESC. RIG-I-Mediated Antiviral Responses to Single-Stranded RNA Bearing 5′ Phosphates. Science. 2006;314:997–1001. doi: 10.1126/science.1132998. [DOI] [PubMed] [Google Scholar]
- Pichlmair A, Schulz O, Tan CP, Rehwinkel J, Kato H, Takeuchi O, Akira S, Way M, Schiavo G, Reis ESC. Activation of MDA5 requires higher order RNA structures generated during virus infection. J Virol. 2009 doi: 10.1128/JVI.00770-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prescott J, Ye C, Sen G, Hjelle B. Induction of innate immune response genes by Sin Nombre hantavirus does not require viral replication. J Virol. 2005;79:15007–15015. doi: 10.1128/JVI.79.24.15007-15015.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qing M, Zou G, Wang QY, Xu HY, Dong H, Yuan Z, Shi PY. Characterization of dengue virus resistance to brequinar in cell culture. Antimicrobial agents and chemotherapy. 2010;54:3686–3695. doi: 10.1128/AAC.00561-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raju R, Raju L, Kolakofsky D. The translational requirement for complete La Crosse virus mRNA synthesis is cell-type dependent. J Virol. 1989;63:5159–5165. doi: 10.1128/jvi.63.12.5159-5165.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randall RE, Goodbourn S. Interferons and viruses: an interplay between induction, signalling, antiviral responses and virus countermeasures. J Gen Virol. 2008;89:1–47. doi: 10.1099/vir.0.83391-0. [DOI] [PubMed] [Google Scholar]
- Rehwinkel J, Tan CP, Goubau D, Schulz O, Pichlmair A, Bier K, Robb N, Vreede F, Barclay W, Fodor E, et al. RIG-I Detects Viral Genomic RNA during Negative-Strand RNA Virus Infection. Cell. 2010;140:397–408. doi: 10.1016/j.cell.2010.01.020. [DOI] [PubMed] [Google Scholar]
- Resa-Infante P, Jorba N, Coloma R, Ortin J. The influenza virus RNA synthesis machine: advances in its structure and function. RNA biology. 2011;8:207–215. doi: 10.4161/rna.8.2.14513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito T, Hirai R, Loo YM, Owen D, Johnson CL, Sinha SC, Akira S, Fujita T, Gale M., Jr Regulation of innate antiviral defenses through a shared repressor domain in RIG-I and LGP2. Proc Natl Acad Sci U S A. 2007;104:582–587. doi: 10.1073/pnas.0606699104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito T, Owen DM, Jiang F, Marcotrigiano J, Gale M., Jr Innate immunity induced by composition-dependent RIG-I recognition of hepatitis C virus RNA. Nature. 2008;454:523–527. doi: 10.1038/nature07106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlee M, Roth A, Hornung V, Hagmann CA, Wimmenauer V, Barchet W, Coch C, Janke M, Mihailovic A, Wardle G, et al. Recognition of 5′ triphosphate by RIG-I helicase requires short blunt double-stranded RNA as contained in panhandle of negative-strand virus. Immunity. 2009;31:25–34. doi: 10.1016/j.immuni.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt A, Schwerd T, Hamm W, Hellmuth JC, Cui S, Wenzel M, Hoffmann FS, Michallet MC, Besch R, Hopfner KP, et al. 5′-triphosphate RNA requires base-paired structures to activate antiviral signaling via RIG-I. Proc Natl Acad Sci U S A. 2009;106:12067–12072. doi: 10.1073/pnas.0900971106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz O, Pichlmair A, Rehwinkel J, Rogers NC, Scheuner D, Kato H, Takeuchi O, Akira S, Kaufman RJ, Reis e Sousa C. Protein kinase R contributes to immunity against specific viruses by regulating interferon mRNA integrity. Cell Host Microbe. 2010;7:354–361. doi: 10.1016/j.chom.2010.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seth RB, Sun L, Ea CK, Chen ZJ. Identification and Characterization of MAVS, a Mitochondrial Antiviral Signaling Protein that Activates NF-kappaB and IRF3. Cell. 2005;122:669–682. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- Spiropoulou CF, Albarino CG, Ksiazek TG, Rollin PE. Andes and Prospect Hill Hantaviruses Differ in Early Induction of Interferon although Both Can Downregulate Interferon Signaling. J Virol. 2007;81:2769–2776. doi: 10.1128/JVI.02402-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoltz M, Klingstrom J. Alpha/beta interferon (IFN-alpha/beta)-independent induction of IFN-lambda1 (interleukin-29) in response to Hantaan virus infection. J Virol. 2010;84:9140–9148. doi: 10.1128/JVI.00717-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahle L, Garcin D, Kolakofsky D. Sendai virus defective-interfering genomes and the activation of interferon-beta. Virology. 2006;351:101–111. doi: 10.1016/j.virol.2006.03.022. [DOI] [PubMed] [Google Scholar]
- Takahasi K, Yoneyama M, Nishihori T, Hirai R, Kumeta H, Narita R, Gale M, Jr, Inagaki F, Fujita T. Nonself RNA-sensing mechanism of RIG-I helicase and activation of antiviral immune responses. Mol Cell. 2008;29:428–440. doi: 10.1016/j.molcel.2007.11.028. [DOI] [PubMed] [Google Scholar]
- tenOever BR, Servant MJ, Grandvaux N, Lin R, Hiscott J. Recognition of the measles virus nucleocapsid as a mechanism of IRF-3 activation. J Virol. 2002;76:3659–3669. doi: 10.1128/JVI.76.8.3659-3669.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- tenOever BR, Sharma S, Zou W, Sun Q, Grandvaux N, Julkunen I, Hemmi H, Yamamoto M, Akira S, Yeh WC, et al. Activation of TBK1 and IKK epsilon kinases by vesicular stomatitis virus infection and the role of viral ribonucleoprotein in the development of interferon antiviral immunity. J Virol. 2004;78:10636–10649. doi: 10.1128/JVI.78.19.10636-10649.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verbruggen P, Ruf M, Blakqori G, Overby AK, Heidemann M, Eick D, Weber F. Interferon antagonist NSs of La Crosse virus triggers a DNA damage response-like degradation of transcribing RNA polymerase II. J Biol Chem. 2011;286:3681–3692. doi: 10.1074/jbc.M110.154799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vreede FT, Jung TE, Brownlee GG. Model suggesting that replication of influenza virus is regulated by stabilization of replicative intermediates. J Virol. 2004;78:9568–9572. doi: 10.1128/JVI.78.17.9568-9572.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Reth M. Drosophila S2 Schneider cells: a useful tool for rebuilding and redesigning approaches in synthetic biology. Methods Mol Biol. 2012;813:331–341. doi: 10.1007/978-1-61779-412-4_20. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.