Abstract
Clinically, combined therapy of cisplatin (CDDP) and metformin is an effective treatment for non-small cell lung cancer (NSCLC). The success is attributed to synergistic effects between the two drugs. Therefore, we hypothesize that co-encapsulation of CDDP and metformin will avoid the prominent toxicity of CDDP while maintaining the synergy between the regimens. CDDP was first conjugated to polyglutamic acid (PGA) to form anionic PGA-CDDP which was electrostatically complexed with the cationic polymeric metformin (polymet). The nano-sized complex was then stabilized with cationic liposomes composed of DOTAP (2, 3-Dioleoyloxy-propyl)-trimethylammonium/Cholesterol/DSPE-PEG-anisamide aminoethyl. Both in vitro and in vivo experiments confirmed the synergy between polymet and CDDP. CDDP delivered with nanoparticles (NPs) exhibited significantly increased tumor accumulation over free CDDP and suppressed tumor growth through apoptosis in NSCLC H460 tumor-bearing mice without nephrotoxicity. The synergistic effect of polymet alongside CDDP demonstrates that polymet-CDDP NPs can activate the AMP-activated protein kinase α (AMPKα) pathway and inhibit mammalian target rapamycin (mTOR) activity to enhance growth suppression. In all, this platform is the first to successfully co-load polymet, a polymeric metformin, and CDDP into the same nanoparticle for successful treatment of NSCLC.
Keywords: Cisplatin, Polymeric metformin, Core-membrane nanoparticle, Non-small cell lung cancer
1. Introduction
Lung cancer remains the leading cause of cancer-related deaths worldwide, and about 85% of all lung cancers are non-small cell lung cancer (NSCLC) [1,2]. The overall 5-year survival rate for patients with NSCLC is 13% in Europe and 16% in the United States [3,4]. Thus, the efficacy and success rate of treatment needs improvement. Cisplatin (CDDP) remains the leading therapy for advanced NSCLC [5]. For patients with advanced (stage III and IV) NSCLC, CDDP was often used in combination with paclitaxel, gemcitabine, docetaxel, vinorelbine or irinotecan, in concurrence with radiotherapy [6].
Recent studies have shown that in addition to the therapies listed above, metformin, a common antidiabetic drug (N′,N′-dimethylbiguanide), displayed significant growth-inhibition and proapoptotic effects in several cancers, including NSCLC [7,8]. Metformin activates AMP-activated protein kinase (AMPK), inhibits the mammalian target of rapamycin (mTOR) and down-regulates excision repair cross-complementation group 1 (ERCC1) [9,10]. Consequently, metformin may be used in combination with CDDP to treat NSCLC [11,12].
While preclinical studies of free CDDP and metformin in combination exhibited promising outcomes, the clinical application of this combination is severely restricted by a collection of issues. Firstly, CDDP and metformin are administered through different routes, i.e. oral for metformin and intravenous (IV) injection for CDDP, which challenges patient compliance. The difference in administrative routes leads to a subsequent discrepancy in the pharmacokinetic (PK) profiles. Second, proper evaluation of the synergy between these two drugs is further complicated by the different physicochemical properties of the drugs, which leads to subpar anti-tumor efficacies. Furthermore, well-known toxicities such as nephro and neurotoxicity limit the use of CDDP. Thus, formulating these two drugs into a single nanoparticle will be a promising strategy to treat NSCLC and overcome the aforementioned limitations.
Formulating CDDP into nanoparticles (NPs), such as liposomal or polymeric formulations, significantly reduces the adverse side effects of CDDP while maintaining its anti-tumor efficacy [13,14]. This advantage can be partially attributed to the fact that NPS can be modified to avoid undesired uptake by the reticuloendothelial system (RES), leading to prolonged blood circulation and increased tumor accumulation. Due to promising preclinical studies, several NPs incorporating CDDP have already been approved for clinical trials, such as Lipoplatin [15] and Nanoplatin [16]. However, unlike the co-delivery of CDDP with other hydrophobic drugs, which works by modifying CDDP into hydrophobic platinum IV prodrugs, co-delivery of CDDP with a hydrophilic drug (e.g. metformin) has rarely been reported [17]. Herein, we report a novel strategy for the co-encapsulation of CDDP and metformin into a single self-assembled core-membrane NPs. We took advantage of recently discovered polymeric metformin (polymet) which exhibits similar anticancer efficacy as metformin and acts through the same mechanistic pathway [18]. Namely, polymet both activates the AMPK pathway, and reduces mTOR activation. It is appropriate to consider polymet as a substitute for metformin. CDDP was chemically conjugated to polyglutamic acid (PGA) through the displacement of chlorine atoms by hydrogen of carboxyl groups on PGA side-chains to form anionic PGA-CDDP as reported before [14,19]. From here, cationic polymet was then subject to an electrostatic interaction with an anionic PGA-CDDP conjugates to produce a negatively charged core which was then coated with PEGylated cationic liposomes to form the final core-membrane structure. The dissociation of polymeric ion pairs then controls the release of both therapeutic moieties.
Herein we report the synergistic anti-cancer activity of NPs containing both polymet and PGA-CDDP. The experiments were carried out in H460 human lung cancer xenograft as a model for human NSCLC.
2. Materials and methods
2.1. Chemicals and materials
CDDP was purchased from Sigma-Aldrich (Dorset, UK). Linear polyethylenimine (PEI) hydrochloride with average molecular weight 8000, Poly-L-glutamic acid sodium salt (molecular weight 3000–15,000), cholesterol, dicyandiamide, p-Anisic acid, Ethylene dichloride (EDC), N-Hydroxysuccinimide (NHS), 3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide (MTT), N,N-Diisopropylethylamine (DIPEA), Radio-Immunoprecipitation Assay (RIPA), dichloromethane and silver nitrate were obtained from Sigma-Aldrich (St Louis, MO) without further purification. (2, 3-Dioleoyloxy-propyl)-trimethylammonium (DOTAP) and 1, 2-distearoryl-sn-glycero-3-phosphoethanolamine-N [methoxy (polyethyleneglycol-2000)] (ammonium salt) (DSPE-PEG2000) were purchased from Avanti Polar Lipids, Inc. (Alabaster, AL). DSPE-PEG-aminoethyl anisamide (DSPE-PEG-AA) was synthesized in our lab as described previously [20]. TdT-dependent dUTP-biotin nick end labeling (TUNEL) assay kits was obtained from Promega (Madison, WI). 4′,6-diamidino-2-phenylindole (DAPI) was obtained from Vector laboratories (Burlingame, CA). Bicinchoninine acid (BCA) protein assay reagent kit was purchased from Thermo Fisher Scientific Inc. Rabbit monoclonal antibodies: Phospho-AMPKα (Thr172) (40H9), AMPKα (D5A2), Phospho-mTOR (Ser2448) (D9C2) XP®, mTOR (7C10), Phospho-p70 S6 Kinase (Ser371), Phospho-4E-BP1 (Thr37/46), and horseradish peroxidase (HRP)-linked Antibody were purchased from Cell signaling. Reduced glyceraldehyde-phosphate dehydrogenase (GAPDH) (14C10), mouse monoclonal antibodies: ERCC1 (3H11), anti-rabbit IgG, xeroderma pigmentosum complementation group A (XPA) (12F5), poly ADP-ribose polymerase-1 (PARP-1) (F-2) and goat antimouse-IgG2b, HRP-linked Antibody were purchased from Santa Cruz Biotechnology, Inc.
2.2. Cell lines and experimental animals
H460 human NSCLC cells was obtained from American Type Culture Collection (ATCC) and was cultured in RPMI 1640 Media (Sigma Aldrich, UK) supplemented with 10% fetal bovine serum (Life Technologies, Carlsbad, California), 100 U/mL penicillin, and 100 μg/mL streptomycin (Invitrogen). Cells were cultivated in a humidified incubator at 37 °C with 5% CO2 and harvested with 0.05% trypsin-EDTA (ethylene diamine tetraacetic acid) before subculture.
Female nude mice of 6–8 weeks old were purchased from National Cancer Institute (Bethesda, MD) and bred by the Division of Laboratory Animal Medicine (DLAM) at University of North Carolina at Chapel Hill. To establish the xenograft models, 5 × 106 H460 cells in 100 μL of phosphate-buffered saline (PBS) were inoculated subcutaneously into the right flank of each mouse. All procedures involving experimental animals were performed in accordance with the protocols approved by the University of North Carolina Institutional Animal Care and Use Committee and conformed to the Guide for the Care and Use of Laboratory Animals (NIH publication No. 86-23, revised 1985).
2.3. Preparation of polymet
Polymet (ca. molecular weight is 4300 Da) was synthesized as previous reported [18]. Briefly, 0.2 g of linear PEI and 2 g of dicyandiamide were mixed in 10 mL deionized water. Two milliliters HCl were added into the solution (Fig. 1A). The compounds reacted in a 100 °C oil bath for 4 h. Polymet was then purified through an ultrafiltration tube with a cutoff of 3000 Da, washed with deionized water for two times and lyophilized.
Fig. 1.
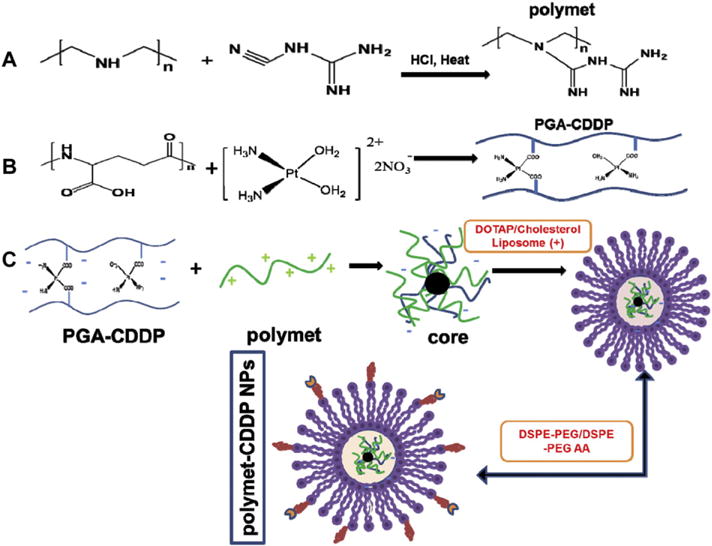
Preparation and characteristic for polymet-CDDP NPs. (A) Synthesis scheme of polymet. (B) Synthesis scheme of PGA-CDDP. (C) Preparation of polymet-CDDP NPs.
2.4. Preparation and characterization of polyglutamic acid-CDDP conjugation (PGA-CDDP)
The preparation of PGA-CDDP was synthesized as previous reported [19]. First the cis-[Pt(NH3)2(H2O)2] (NO3)2 precursor was as previously described [21]. Briefly, AgNO3 (66.2 mg or 0.39 mmol) was added to a suspension of CDDP (60 mg or 0.2 mmol) in 1 mL water. The mixture was heated at 60 °C for 3 h and then stirred overnight in a flask protected from light with aluminum foil. The mixture was centrifuged at 16,000 rpm for 15 min to precipitate AgCl. The supernatant was then filtered using a 0.22 μm syringe filter to afford the desired product (CDDP precursor). PGA was solved in deionized water (25 mM) and then added to the solution of CDDP precursor (6.25 mM) at the same volume. The mixture was then put in dark at room temperature for 72 h with gently stirring to form the PGA-CDDP conjugation (Fig. 1B). The formation of the conjugation was certified by the 1H NMR nuclear magnetic resonance (1H NMR) (Fig. S1).
2.5. Preparation and characterization of the NPs
The formulation of polymet-CDDP NPs involves two key steps after the production of PGA-CDDP. Firstly, an anionic core must be formed from a complex of polymet and PGA-CDDP. The anionic core is then combined with cationic liposomes composed of DOTAP and cholesterol to produce the outer membrane of the core-membrane structure (Fig. 1C).
2.5.1. Preparation of cores
In optimizing polymet-CDDP NPs, the core should be composed of a ratio of polymet to PGA-CDDP to yield the smallest anionic NPs. So, 200 μL of PGA-CDDP (3.1 mM CDDP) and various volume of polymet solution (12.5 mM) were mixed at different ratios (1∶0.6, 1∶0.8, 1∶1, 1∶1.2) by pipetting up and down and allowed to stand at room temperature for 10 min before the size and zeta potential are determined. The optimal ratio of the complex was determined by the particle size and zeta potential results determined by dynamic light scattering (DLS) using a Malvern ZetaSizer Nano series (Westborough, MA). After the optimal ratio of the core was chosen, the core was mixed with cationic DOTAP/cholesterol liposomes and incubated for another 10 min for lipid coating.
2.5.2. Preparation of the lipid membrane
To prepare the lipid membrane, the DOTAP (20 mM, 1 mL) and cholesterol (20 mM, 1 mL) were dissolved (1∶1 mol/mol) in chloroform and the solvent was removed under reduced pressure. The lipid film was hydrated overnight with 1 mL distilled water to form cationic liposomes (20 mM), which were sequentially extruded through polycarbonate membranes (200 nm × 20 times and 50 nm × 20 times) (Millipore, Bil-lerica, MA).
2.5.3. Preparation of the NPs
To determine the optimal amount of DOTAP/cholesterol liposomes for the core, different ratios (8∶1, 10∶1, 12∶1) of liposome to PGA-CDDP conjugates were designed. The optimal NPs were defined as the ratio that yielded the smallest particle size, smallest polydispersity index (PDI) and appropriate cationic charge. Furthermore, previous studies have shown that H460 cells exhibited over expression of sigma receptors [22]. Therefore, aminoethyl anisamide, was specifically employed as targeting moiety to sigma receptors to direct polymet-CDDP NPs to the H460 tumors. The lipid-coated NPs were PEGylated using a post-insertional approach by adding 15 μL DSPE-PEG (50 mg/mL) and 15 μL DSPE-PEG-AA (50 mg/mL) and incubated with NPs at 50 °C for 15 min. The ratio of DOTAP∶DSPE-PEG∶DSPE-PEG-AA was maintained as (1∶0.12∶0.11).
The preparation of PEI-CDDP NPs followed the same procedure as polymet-CDDP NPs except polymet was replaced with PEI (12.5 mM). Also polymet NPs was prepared following the same procedure as polymet-CDDP NPs except replacing PGA-CDDP with PGA.
2.5.4. Characterization of the NPs
The size distribution and the zeta potential of particles were deter mined using a Malvern ZetaSizer Nano series (Westborough, MA). Transmission electron microscope (TEM) images of polymet-CDDP NPs were acquired through the use of JEOL 100CX II TEM (Tokyo, Japan). Briefly, freshly prepared polymet-CDDP NPs (20 μL) were carefully dropped onto a 300-mesh carbon-coated copper grid (Ted Pella, Inc., Redding, CA) and allowed to stand at room temperature for 15 min and then removed the assess liquid using a dry filter paper. Images were acquired at an accelerating voltage of 100 kV.
2.6. Cellular uptake study in H460 cell lines
H460 cells were seeded into a 12-well plate (1.5 × 105 cells/well) containing 1 ml of media. Twenty hours later, 1 ml of the free CDDP, PGA-CDDP, polymet-CDDP NPs, PEI-CDDP NPs at a concentration of 10 μM CDDP were incubated with cells in a serum-free medium. Four hours later, cells were treated with 0.05% trypsin-EDTA (Sigma-Aldrich) and the cell lysate was centrifuged at 14,000 rpm for 20 min at 4 °C to collect cells and then the cells were digested with HNO3. The concentration of CDDP was measured using inductively coupled plasma mass spectrometry (ICP-MS).
2.7. In vitro release of CDDP
The dialysis technique was employed to study the in vitro release of CDDP from free CDDP, PGA-CDDP, polymet-CDDP NPs, PEI-CDDP NPs in phosphate buffered saline (PBS) (pH 7.4 with 100 mM NaCl) at 37 °C. Five hundred microliters free CDDP, PGA-CDDP, polymet-CDDP NPs, PEI-CDDP NPs, with 180 μg/mL CDDP separately were added into the dialysis tube with a molecular weight cut off of 3000 Da and dialyzed against 15 mL PBS in a thermo-controlled shaker with a stirring speed of 100 rpm at 37 °C for up to 144 h. At each predetermined time point, 300 μL samples were withdrawn and replaced with fresh media. The CDDP concentration was then determined by ICP-MS at specified times. All experiments were performed in triplicate and the data were reported as the mean ± SEM of 3 individual experiments.
2.8. In vitro cell viability on H460 cells and analysis of synergistic drug combinations
An MTT assay was conducted to evaluate in vitro viability of free CDDP, PGA-CDDP, polymet-CDDP NPs, polymet NPs and PEI-CDDP NPs in which the molar ratio of CDDP to polymet or PEI was 1 to 1. In brief, H460 cells were seeded in 96-well plates at a density of 2000 cells per well 24 h prior to drug treatment. Subsequently, cells were treated with free drugs and the drug combination with various molar ratios at a series of dilutions in full medium. Following 48 h of treatment, MTT (20 μL, 5 mg/mL) reagent was added for an additional 4 h incubation period at 37 °C. The medium was discarded, the formed formazan salt was dissolved in 100 μL of DMSO and absorbance was read at 570 nm using a multidetection microplate reader (Plate CHAMELEON ™ V-Hidex). Cell survival rates were calculated as normalized to untreated control wells. Each concentration was tested in 5 wells and data presented as mean ± SEM. The mean drug concentration required for 50% growth inhibition (IC50) was determined with CompuSyn software (Version 1.0, Combo-Syn Inc., U.S.) using the median effect equation: Fa = [1 + (IC50 / D)m]−1, where Fa is the fraction of affected cells, D is drug concentration and m is the Hill slope.
Combination Index (CI) Analysis of free CDDP combined with polymet based on the Chou and Talalay method [23] was conducted using CompuSyn software. Briefly, for each level of Fa, the CI values for CDDP and polymet combinations were calculated according to the following equation: CI = (D)1 / (Dx)1 + (D)2 / (Dx)2, where (D)1 and (D)2 are the concentrations of each drug in the combination resulting in Fa × 100% growth inhibition, and (Dx)1 and (Dx)2 are the concentrations of the drugs alone resulting in Fa × 100% growth inhibition. CI values for drug combinations were plotted as a function of Fa. CI values less than 1 or more than 1 demonstrate synergism or antagonism of drug combinations, respectively. Notably, CI values between Fa 0.2 to 0.8 are considered validate [24].
2.9. Biodistribution of CDDP in tumors and other major organs in H460 tumor–bearing mice
H460 tumor–bearing mice were administered a single dose of free CDDP, polymet-CDDP NPs and PEI-CDDP NPs at the dose of 2.5 mg/kg CDDP. Each group contained 4 mice, which were sacrificed 24 h following IV administration. About 30 mg tumor and tissue samples (heart, liver, spleen, lung, and kidney) were digested with 300 μL 60% nitric acid (Acros Organic) at 60 °C overnight and proceeded to quantify CDDP by ICP-MS. The uptake of CDDP was expressed as the percentage of the injected dose per gram tumor or tissue.
2.10. Anti-tumor efficacy in H460 human NSCLC xenografts
Human NSCLC xenografts were used as previously described [18,22]. H460 cells (5.0 × 106) were subcutaneously injected into the right flanks of female athymic nu/nu mice. On day 8 after tumor implantation (tumor size 60–80 mm2), mice were randomized into 5 groups (n = 4–6) as follows: untreated control (PBS), free CDDP, polymet NPs, polymet-CDDP NPs and PEI-CDDP NPs. IV injections were performed every second day for a total of 5 injections with CDDP dose of 2.5 mg/kg and/or the corresponding amount of polymet as determined by the ratio in the ultimately optimizing formulation. Animal body weight and tumor volumes were measured each day. The tumor length (L) and width (W) were used to calculate volume (V) by the equation: V = 1/2 × L × W2. Mice were sacrificed the second day after the last injection by CO2 asphyxiation and tumors were excised.
2.11. TUNEL assay
Slides were deparaffinized through xylene and a graded alcohol series and prefixed with 4% formaldehyde. Apoptosis in situ was then detected by a TUNEL assay using an apoptosis detection kit according to the manufacturer’s instructions. Slides were then rinsed with PBS and mounted to a cover slip using Vectashield. All staining was evaluated and digital images were acquired by Eclipse Ti-U inverted microscope (Nikon Corp., Tokyo, Japan) at 20× magnification. Five randomly selected microscopic fields were quantitatively analyzed on Image J.
2.12. Serum biochemical value analysis and hematology assay
After 5 injections, the whole blood was collected and centrifuged at 4000 rpm for 5 min to obtain the serum. Blood urea nitrogen (BUN), creatinine, serum aspartate aminotransferase (AST) and alanine aminotransferase (ALT) levels were assayed as indicators of renal and hepatic function. Whole blood was collected for the detection of myelosuppression by counting the Red blood cells (RBC), white blood cells (WBC), platelets (PLT), hemoglobin (HGB) and hematocrits (HCT). Organs (heart, liver, spleen, lung, and kidney) were fixed with 4% PFA and sectioned for H&E staining to evaluate organ-specific toxicity.
2.13. Western-blot analysis
2.13.1. Western-blot analysis in tumor of H460 bearing mice
Twenty-four hours after 5 times IV injections, H460 bearing mice were sacrificed and tumor lysates were prepared as previously described [25]. Briefly, tumor samples of each group were lysed with RIPA buffer supplemented with 1% protease inhibitor cocktail (PIC). Protein concentration in the tumor or cell lysate was determined using a BCA protein assay according to the manufacturer’s instruction (Invitrogen). Then 40 μg of protein per lane was separated by 4–12% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE electrophoresis) (Invitrogen) before being transferred to polyvinylidene difluoride (PVDF) membranes (Bio-Rad). The membranes were blocked for 1 h with 5% Bovine Serum Albumin (BSA) at room temperature and then incubated overnight at 4 °C with and then incubated with primary antibodies overnight at 4 °C. The membranes were washed 3 times and then incubated with a secondary antibody (1∶4000 dilution) at room temperature for 1 h. Finally, the membranes were washed 4 times and developed using a chemiluminescence system according to the manufacturer’s instructions (Thermo Scientific). GAPDH antibody (1∶2000 dilution) was probed as the loading control. The membranes were washed 3 times and then incubated with a Rabbit anti-mouse secondary antibody (1∶4000 dilution) or an Anti-rabbit IgG, HRP-linked Antibody (1∶4000 dilution) at room temperature for 1 h. Finally, the membranes were washed 4 times and developed by an enhanced chemiluminescence system according to the manufacturer’s instructions (Thermo Scientific). The relative protein expression level was quantified with Image J software (National Institutes of Health, Bethesda, MD).
2.13.2. Western-blot analysis in vitro
H460 cells were seeded into a 6-well plate (5 × 105 cells/well) containing 2 ml of media. Twenty hours later, 2 ml of free CDDP (1 μM, 10 μM), polymet (1 μM, 10 μM), and their combo at the molar ratio of 1∶1 were incubated with cells in a RPMI 1640 medium. Twenty-four hours later, cells were treated with RIPA buffer and the cell lysates were quantified for the protein concentration by BCA. Forty micrograms of protein per lane was separated by 4–12% SDS-PAGE electrophoresis before being transferred to PVDF membranes. The subsequent procedure was as indicated above.
2.14. Statistical analysis
Quantitative data were expressed as mean ± SEM. The analysis of variance was completed using a one-way Analysis of Variance (ANOVA). P < 0.05 was considered statistically significant. All statistical analyses were carried out using GraphPad Prism Software (Version 6.0, GrapPad Software, San Diego, CA).
3. Results and discussions
3.1. Preparation of polymet-CDDP NPs
3.1.1. Preparation of polymet
Polymet was synthesized successfully as previous reported [18] (Fig. 1A). The matrix-assisted laser desorption/ionization time of flight mass spectrometry (MALDI-TOF-MS) results indicated the modification of PEI by dicyandiamide. Additionally, polymet displayed a significantly lower cytotoxicity measured using a MTT assay with a 3-fold increase in maximum tolerable dose (MTD) compared to PEI on H460 cells. These results indirectly indicated the conversion of the secondary amine of PEI to polymet [18].
3.1.2. Preparation of PGA-CDDP
CDDP was chemically conjugated PGA through the substitution of hydrogen atoms on the carboxyl groups of PGA side-chains (Fig. 1B). The free CDDP and water-soluble conjugate, PGA-CDDP, were both quantified via ICP-MS. To ensure reaction completion, the product was subject to ultrafiltration with a 3000 Da cutoff membrane. The supernatant was analyzed for any traces of unreacted free CDDP. Findings confirmed a ~100% reaction yield after 72 h of stirring (Fig. S1) and 1H NMR results further confirmed the successful manufacture of PGA-CDDP (Fig. S1).
3.1.3. Preparation and optimization of polymet-CDDP NPs
As Fig. 2A demonstrated, 1∶1 polymet∶CDDP produced the smallest cores (132 ± 2 nm) with a zeta potential of (−36 ± 1 mV). The anionic inner core composed of a 1∶1 polymet∶PGA-CDDP was then subject to electrostatic interactions with cationic DOTAP/cholesterol liposomes. Considering the parameters of defining the nanoparticle mentioned in Section 2.5.3, 10∶1 DOTAP/cholesterol liposome to CDDP molar ratio was chosen as the optimal and yielded NPs with sizes of 145 ± 1 nm, zeta potentials of 49 ± 1 mV (Fig. 2B) and PDI of 0.2. After introducing DSPE-PEG and DSPE-PEG-AA into the NPs, the final NPs were formed. The size, PDI and the zeta potential of both cores and the final NPs were summarized in the Table 1 and final NPs had an approximate diameter of 150 nm and positive surface charge of about 50 mV.
Fig. 2.
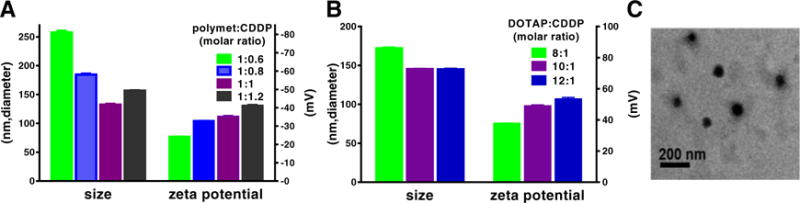
The optimization of polymet-CDDP core and NPs. (A) Effects of different molar ratio of polymet (unit of metformin) to CDDP on the size and zeta potential of polymet-CDDP core. Ratios were calculated according to the metformin unit of polymet. (B) Effects of different molar ratio of DOTAP to CDDP on the size and zeta potential of polymet-CDDP NPs. (C) Representative TEM image of polymet-CDDP NPs.
Table 1.
Sizes, PDI and zeta potentials of different cores and final nanoparticles (n = 3).
| Diameter (nm) | PDI | Zeta potential (mv) | |
|---|---|---|---|
| polymet-CDDP Corea | 132 ± 2 | 0.2 ± 0.0 | −35 ± 1 |
| polymet-CDDP NPsb | 151 ± 1 | 0.2 ± 0.0 | 49 ± 1 |
| PEI-CDDP corec | 150 ± 3 | 0.38 ± 0.1 | −30 ± 0 |
| PEI-CDDP NPsd | 189 ± 2 | 0.2 ± 0.0 | 48 ± 1 |
Molar ratio of CDDP to PGA = 1∶4.
Molar ratio of CDDP to polymet = 1∶1.
Molar ratio of DOTAP to CDDP = 10∶1.
Molar ratio of DOTAP∶Cholesterol∶DSPE-PEG∶DSPE-PEG-AA = 1∶1∶0.12∶0.11.
Despite a cationic surface charge, Fig. S4 and S5 demonstrated that NPs induced no significant toxicities on healthy tissues of the heart, liver, spleen, lung and kidney tested in vivo. Furthermore, no significant deviations in hematological parameters and serum biochemistry were found in NP treated groups. These findings confirmed that NPs had no observable toxicities on normal tissues despite their cationic charge, which can most likely be attributed to the PEG shielding effect of the PEGylated NPs. Additionally, a recently published manuscript found that PEGylated DOTAP particles with similar surface charges and NP structure, had low toxicity on normal lung fibroblasts MRC-5, further confirming the minimal toxicity properties of the blank NPs [26].
Polymet and PGA-CDDP were condensed into the solid core due to electrostatic interaction. The zeta potential of the core is negative, so, it is reasonable to assume the encapsulation efficiency of the cationic polymet is approximately 100%. The negative core was further coated with an excess of liposomes, to produce a final cationic charge. Similarly, the encapsulation of the anionic PGA-CDDP should be around 100%. Our lab has previously investigated that in lipid-polycation nanoparticles, the core was coated by two cationic lipid bilayers which were the inner bilayer stabilized by charge–charge interaction and the outer layer. When 10.6 mol% of DSPE-PEG2000 presented on the particle surface, approximately 60% of the total cationic lipids which located in the outer bilayer, were stripped off during the PEGylation [27]. So, we calculated the loading capacity of both polymet and CDDP with their actual weights according to their ratio in the NPs excluding the excess lipids. The loading ratio of CDDP and polymet were about 2.5% and 1.1% respectively.
Imaging by TEM revealed polymet-CDDP cores as mono-dispersed spheroids with a diameter of approximately 50 nm (Fig. 2C), which was smaller than the value measured by DLS (average 130 nm). This significant discrepancy is to be expected, as the size determined through DLS regards the hydrodynamic size, which includes both solvent (hydro) and shape (dynamic) effects of the NPs, while TEM just reveals the size of the core [28,29] (Fig. S2). Due to the water solubility and ionic complex of PGA-CDDP to polymet, the hydrodynamic diameter is expected to be larger than the dehydrated diameter determined by TEM (Table 1).
3.2. In vitro Cellular uptake of NPs in H460 tumor cells
In vitro cellular uptake studies showed that polymet-CDDP NPs and PEI-CDDP NPs shared comparable NP uptake profiles when measuring Pt content in H460 cells (Fig. 3A). However, the uptake of both aforementioned NP platforms was significantly higher than the uptake of either free CDDP or PGA-CDDP on H460, and can be attributed to the cationic lipid coating along with the active targeting capabilities of aminoethyl anisamide. The results from these preliminary studies serve to confirm the efficient cellular uptake of polymet-CDDP NPs.
Fig. 3.
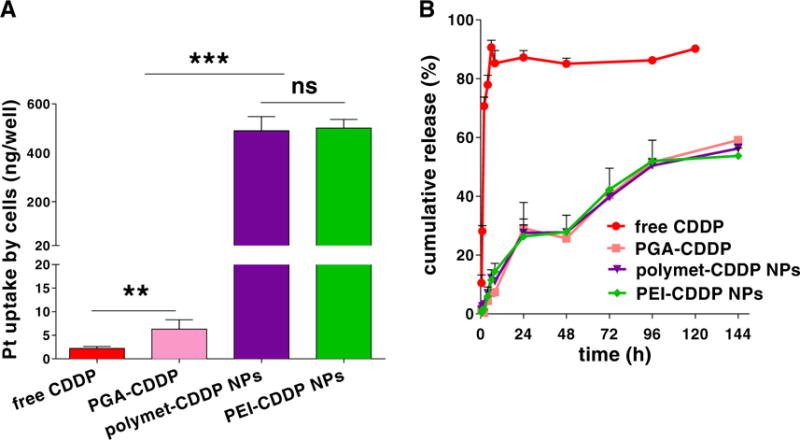
Cellular uptake and release of CDDP (based on Pt) from polymet-CDDP NPs. (A) In vitro uptake of CDDP in free CDDP, PGA-CDDP, polymet-CDDP NPs, and PEI-CDDP NPs at 37 °C for 4 h in H460 cells. (B) Cumulative in vitro release kinetics of CDDP, PGA-CDDP, polymet-CDDP NPs, PEI-CDDP NPs in PBS (pH 7.4, Cl− concentration: 100 mM) at 37 °C. ns: no significance; **P < 0.01, ***P < 0.001 (n = 3).
3.3. In vitro CDDP release from NPs
The release of CDDP was determined by dialyzing the NPs and PGA-CDDP conjugate against a buffer containing 10 mM phosphate (pH 7.4) and 100 mM NaCl at 37 °C through a membrane with a cutoff size of 3000 Da (Fig. 3B). This particular size was selected as membrane pores are large enough to allow diffusion of free CDDP but not NPs or the PGA-CDDP conjugate. The mechanism of CDDP release occurred through an exchange reaction between the chloride and carboxylate groups of PGA in the conjugate [30] and released CDDP was quantified via ICP-MS. Results demonstrated similar release patterns (40% cumulative release) for PGA-CDDP conjugate, polymet-CDDP NPs and PEI-CDDP NPs over 72 h. Significant deviations in release did not occur until after 144 h. However, molecular CDDP was quickly cleared through the membrane after just 6 h. These results indicate that the PGA conjugated to CDDP was the main mechanism responsible for prolonged drug release.
In addition to their low toxicity to normal tissues, lipid coated polyplexes were shown to be stable both in vitro and in vivo [18,31]. Using 11.5 mol% of DSPE-PEG coating, the majority of particles showed RES evasion; subsequently promoting delivery of the therapeutic agent to neoplasms. In vitro release of platinum from the NPs revealed a sustained release profile of CDDP, and no burst release was observed. Overall, NPs demonstrated high stability in physiological conditions (Fig. 3B).
3.4. Analysis of synergistic drug combination effects of and in vitro cell viability on H460 cells
In vitro studies of the synergy between free CDDP and polymet using the Chou-Talalay method [23] indicated that the combination exhibited a strong synergy at a wide range of CDDP/polymet ratios. This range spans from 2∶1 to 1∶6 with respect to the CDDP∶polymet molar ratio calculated according to the number of metformin units in polymet. Throughout this range, the CI at the predetermined IC50 for each drug was below 0.5, indicating that any ratio within this range is suitable for synergy (Fig. 4A).
Fig. 4.
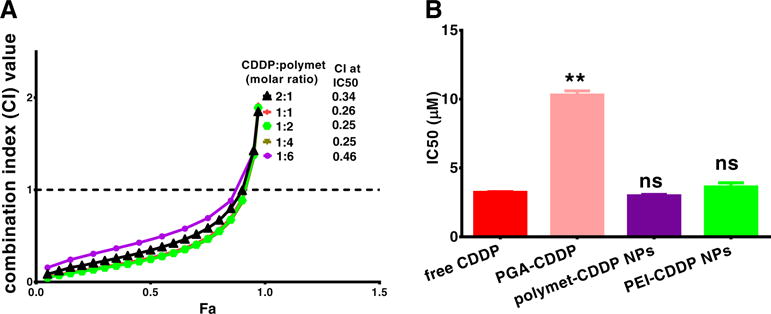
Synergy between CDDP and polymet on H460 cells. The corresponding combination index (CI) vs Fa plots of polymet-CDDP NPs and free CDDP were shown (A). IC50 of free CDDP, PGA-CDDP, polymet-CDDP NPs (molar ratio of CDDP and polymet at 1∶1), polymet NPs and PEI-CDDP NPs (molar ratio of CDDP and PEI at 1∶1) were shown (B). Ratios were calculated according to the unit of polymet or PEI. ns: no significance, **P < 0.01 (n = 3).
The synergistic effects of CDDP and polymet combinations could be further confirmed by the analyzing the combination’s IC50 on H460. Although the release of CDDP from PGA-CDDP is expected to cause a subsequent increase IC50 over molecular CDDP, any increase can most likely be attributed to low uptake and slow CDDP release from the conjugate. However, in vitro viability studies indicated that integration of PGA-CDDP into NPs significantly reduced the IC50 to values comparable of free CDDP (Fig. 4B). These results could be attributed to the previous cell uptake studies which confirmed that polymet-CDDP NPs and PEI-CDDP NPs exhibited significantly increased cellular uptake over free CDDP and PGA-CDDP. Therefore, the increased uptake could compensate for the slow release of CDDP from the NPs.
The IC50 values of polymet and polymet NPs were 373 ± 3 μM and 35 ± 1 μM, respectively, which offered no significant reduction compared to free CDDP and further confirms the synergy between PGA-CDDP and polymet.
3.5. Polymet-CDDP NPs inhibited tumor growth in vivo on H460 lung cancer xenograft model and triggered significant tumor cell apoptosis
Athymic nude mice bearing subcutaneous H460 xenografts were treated with 5 times IV injections of the selected treatment. Treatments include PBS, PEI-CDDP NPs, polymet NPs, free CDDP, or polymet-CDDP NPs (n = 4–6 per group). The dose for mice receiving treatment was 2.5 mg/kg with respect to CDDP and/or the corresponding amount of polymet as determined by the 1∶1 ratio mentioned in Section 3.1.3. As shown in Fig. 5A, polymet-CDDP NPs, free CDDP, polymet NPs and PEI-CDDP NPs exhibited significant tumor growth inhibition effects when compared with PBS control group. There was no significant difference of antitumor efficacy between polymet NPs and PEI-CDDP NPs. However, polymet-CDDP NPs exhibited comparable efficacy with free CDDP and significantly improved anti-cancer efficacy over PBS, PEI-CDDP NPs and polymet-NPs. Tumor volumes at the end of the study comparable between the polymet-CDDP NPs and the free CDDP group (Fig. 5B). This result demonstrates that polymet-CDDP NPs were able to induce similar degrees of efficacy compared to free CDDP. Tumor cell apoptosis was quantified via a TUNEL assay after treatment as shown in Fig. 5C. Free CDDP and polymet-CDDP NPs treated cells showed a comparable number of apoptotic cells, which are significantly more than the PBS and other control groups. This finding correlates with the tumor inhibition data shown in Fig. 5A. While there was not a significant difference in cell apoptosis between polymet NPs and PEI-CDDP NPs, both groups showed significantly more apoptotic cells than the PBS group. This suggests that polymet alone acts as a competent anticancer agent and plays a critical role in the overall synergy. While polymet-CDDP NPs exhibited comparable efficacy as free CDDP, they did not induce adverse effects on other tissues as in the case of free CDDP.
Fig. 5.
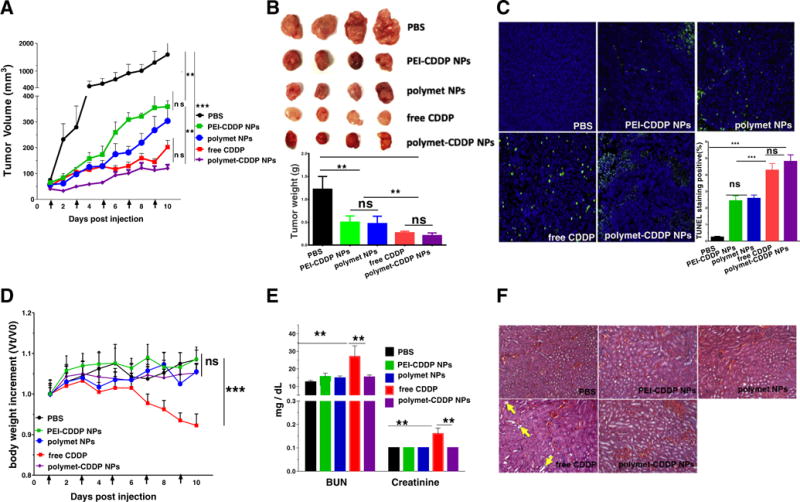
Tumor inhibition effects and systemic toxicity of free CDDP, polymet-CDDP NPs, polymet NPs and PEI-CDDP NPs on H460 tumor–bearing mice. (A) H460 tumor–bearing mice were injected intravenously Q.O.D 5 times at a dose of 2.5 mg/kg CDDP or the corresponding polymet quantity in all the treatment groups. Arrows indicate time of injection. The tumor volumes were measured every day. (B) Visual observations of the H460 tumor sizes in each treatment group at the end time point. Tumor weights were measured 24 h after the final injection. (C) TUNEL assay on H460 tumor cells after treatment with different formulations in vivo. The percentage denotes the average percentage of TUNEL positive cells (green). Five randomly selected microscopic fields were quantitatively analyzed using Image J. (D) Effect of free CDDP, polymet-CDDP NPs, polymet NPs and PEI-CDDP NPs on body weight of H460 tumor– bearing mice. The body weights were measured every day. Data represents the mean ± SEM (n = 4–6 per group). **P < 0.01, ***P < 0.001. ns: no significance. (E) Kidney functional parameters, blood urea nitrogen (BUN) and creatinine. Data represents the mean ± SEM (n = 4–6 per group). **P < 0.01, ***P < 0.001. ns: no significance. (F) H&E staining of kidney tissue from H460 tumor–bearing mice that received 5 doses of treatment. Arrows indicate tubular cell atrophy.
Fig. 5D, E and F indicated that mice treated with polymet-CDDP NPs exhibited significantly reduced toxicities as illustrated by BUN and creatinine levels, body weight, and hematoxylin-eosin (H&E) staining of kidney tissues. Primarily, polymet-CDDP NPs successfully mitigated the well-documented nephrotoxicity that currently limits the clinical use of CDDP while still maintaining an anti-cancer efficacy comparable to CDDP. These results indicated that while the CDDP released from polymet-CDDP NPs exhibited slow release, the synergy between CDDP and polymet promoted enhanced antitumor efficacy over PEI-CDDP NPs.
3.6. In vivo polymet-CDDP NPs accumulation in tumors and other major organs of H460 tumor–bearing mice
It was postulated that polymet-CDDP NPs were able to achieve comparable antitumor efficacy as molecular CDDP despite its slow release for two reasons. Firstly, the synergistic effect between polymet and CDDP compensated for slow release with enhanced potency. Secondly, polymet-CDDP NPs were able to deliver CDDP into the tumor more efficiency than free CDDP. This second advantage is two tiered. Data in Fig. S3 firstly indicated that tumors treated with polymet-CDDP NPs and PEI-CDDP NPs exhibited significantly higher levels of platinum than tumors treated with free CDDP. This can most likely be attributed to the enhanced permeability and retention (EPR) effect of the tumor vasculature [32]. In addition, previously conducted cell uptake studies indicated that both polymet-CDDP NPs and PEI-CDDP NPs exhibited greater cellular uptake, most likely attributed to the employment of sigma receptor mediated targeting. Therefore, polymet-CDDP NPs and PEI-CDDP NPs were able to promote greater platinum uptake into the tumor, and further into tumor cells specifically. Because polymet-CDDP NPs and PEI-CDDP NPs showed no significant difference in tumor accumulation, it is likely that the improved anticancer efficacy of the polymet CDDP NPs can be attributed to synergistic effects between polymet and CDDP on H460 NSCLC. As for the accumulation of CDDP in other organs, similar to other nano-platforms, the CDDP loaded NPs distributed more in the liver and spleen (approximately 20% ID/g) than free CDDP while not significantly different in heart, kidney, and lung tissues 24 h post injection (Fig. S3). Due to similar particle size and zeta potential, polymet-CDDP NPs and PEI-CDDP NPs presented almost identical biodistribution behaviors in vivo.
3.7. Evaluation of systemic toxicity of polymet-CDDP NPs
The clinical use of CDDP is severely limited by systemic toxicities, and many studies are ongoing to mitigate the toxicities of CDDP while preserving the anticancer efficacy and dose [13,15]. However, polymet-CDDP NPs demonstrated the capacity to induce efficacy comparable to CDDP while significantly reducing systemic toxicities. The in vivo toxicity herein was evaluated by monitoring the body weight. After 5 total doses at 2.5 mg/kg CDDP administered every other day, the free CDDP group showed a significant body weight reduction of up to 10% (Fig. 5D). However, animals treated with polymet-CDDP NPs or PEI-CDDP NPs exhibited no statistically significant changes in weight to the PBS control.
Hematological parameters and histopathology of major organs (heart, liver, spleen, lung, and kidney) were measured 24 h after the last treatment of either PBS, PEI-CDDP NPs, polymet NPs, free CDDP or polymet-CDDP NPs to investigate the toxicity. Hematological parameters AST, ALT, BUN and creatinine were all within the normal range for PEI-CDDP NPs, polymet NPs, polymet-CDDP NPs groups (Fig. 5D, Fig. S5). Additionally, no noticeable histological changes were seen in H&E-stained tissue sections of the liver, kidney and spleen in these groups (Figs. 5F, S4). Noticeable histological changes were observed in H&E-stained kidney tissue sections of free CDDP treated mice, indicating severe nephrotoxicity (Fig. 5F). These findings were coincided with the elevated BUN and creatinine levels (Fig. 5E). Since the chemotherapy always cause the change of blood cells, the count of RBC, WBC, PLT and HGB were checked. The results showed that all groups including the free CDDP group were within the normal range (Fig. S5). These studies demonstrated that polymet-CDDP NPs have successfully mitigated the prominent toxicities of CDDP-based treatment while maintaining significant synergistic therapeutic efficacy. Therefore, polymet-CDDP NPs is a promising outlook for enhancing the therapy of NSCLC while reducing systemic toxicities of current treatments.
3.8. Mechanism of synergistic effect of polymet and CDDP combo NPs
It is well known that conjugation of PGA to CDDP decreases the anticancer efficacy of CDDP, primarily due to its slow release from PGA [19, 33]. Therefore, previous reports [14,30] have indicated that treatment with PGA-CDDP alone requires a much higher dose to achieve the same anticancer efficacy as that of free CDDP. This result was mirrored in the current study, as the tumor accumulation of PEI-CDDP NPs was significantly higher than free CDDP, but the anticancer efficacy of PEI-CDDP NPs was significantly lower over free CDDP. This observation provides further evidence that successful anticancer efficacy of polymet-CDDP NPs can be attributed to the synergistic effects between CDDP and polymet. Herein, the dose of CDDP in polymet-CDDP NPs was only 2.5 mg/kg, 37% lower than 4 mg/kg required to induce significant anticancer efficacy with PGA-CDDP alone [19,33]. Despite this lower dose, results in Fig. 5 indicated polymet-CDDP NPs maintain the best an-titumor efficiency. Thus, it is hypothesized that the synergy between polymet with CDDP is responsible for the successful efficacy even at a lower dose.
Emerging evidence indicates that AMPK is a promising metabolic tumor suppressor and target for cancer prevention and therapy [34, 35]. Recent studies have demonstrated the antidiabetic drug metformin could act as an AMPK activator and mTOR inhibitor for use as a cancer therapeutic [36,37]. Thus, the activity of polymet was examined with respect to its role in these two pathways.
It was found that treatment with polymet or polymet-CDDP NPs increased the phosphorylation levels of AMPKα in the H460 tumor compared with the PBS group. This finding was not observed in PEI-CDDP NPs group (Fig. 6A and B), suggesting that polymet is the key factor for activation of the AMPKα pathway. Polymet’s ability to activate the AMPKα pathway while simultaneously inhibiting mTOR suggests its mechanism of synergy with CDDP. The AMPK pathway is known to intersect with the congogenic Ras/Phosphatidylinositol 3-kinase (PI3K)/mTOR and extra cellular signal regulated kinase (ERK) pathways at multiple points [38,39]. Collectively, it is known that AMPK responds to a decrease in cellular ATP by phosphorylating effectors that inhibit mTOR activity. Therefore, AMPK activation leads to a subsequent decrease in mTOR activity [40]. In vivo, S6K1 or 4E-BP1 phosphorylation is primarily used as readout for mTOR activity. Therefore, the effect of polymet and polymet-CDDP NPs on the activity of mTOR and its downstream P-p70-s6K, P-4E-BP1 was investigated in this respect (Fig. 6A and B). Analysis indicated a significant inhibition of mTOR activity as well as inhibition of its downstream P-p70-s6K and P-4E-BP1 compared to the PBS group and PEI-CDDP NPs group. From the western blot analysis, it could be concluded that polymet was loaded in polymet NPs and co-loaded with CDDP in polymet-CDDP NPs.
Fig. 6.
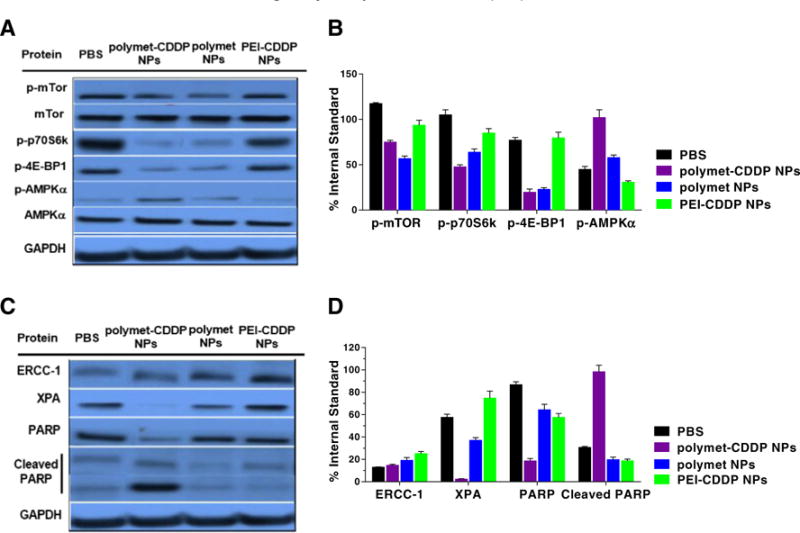
Western blot analysis of H460 tumor protein levels after five injections of polymet-CDDP NPs, polymet NPs and PEI-CDDP NPs. (A) p-mTOR, p-p70S6k, p-4E-BP1, p-AMPKα protein levels were evaluated. (B) Quantification of the protein level of p-mTOR, P-p70-s6, P-4E-BP1 and p-AMPKα which were concerned with the polymet pathway. Both polymet NPs and polymet-CDDP NPs could decrease p-mTOR, p-p70S6k, and p-4E-BP1 as well as increase the p-AMPKα. (C) ERCC-1, XPA, PARP and Cleaved PARP protein levels were evaluated. Polymet combined with CDDP into NPs will decrease the XPA and ERCC-1 and increased the cleaved PARP. (D) Quantification of the protein level of ERCC-1, XPA, PARP and Cleaved PARP. Comparing PEI-CDDP NPs, polymet-CDDP NPs could decrease ERCC-1, XPA and increase Cleaved PARP more efficiently. All quantifications were done by using Image J. Data are mean ± S.D. (n = 3).
Cleaved PARP acts as a reliable marker for the suppression of DNA repair and induction of apoptosis [22,41]. During apoptosis, intact PARP is cleaved into a small and large subunit. The level of cleaved PARP was further investigated to determine the underlying basis of synergy. Fig. 6C and D indicates that the level of cleaved PARP increased in tumors treated with polymet-CDDP NPs and was accompanied by a decrease in the level of uncleaved PARP. Conclusively, polymet-CDDP NPs exhibited greater efficacy in inhibiting DNA repair, leading to intensified apo-ptosis compared to CDDP and polymet in separate NPs.
Separately, high ERCC1 and XPA levels have been implicated to produce resistance to platinum based chemotherapy due to their ability to repair DNA damage done by platinum adducts [42,43]. Therefore, lower levels of ERCC1 and XPA offer a good prognosis for tumor growth inhibition. The effect of combining CDDP and polymet on ERCC1 and XPA was examined via western blot analysis. Results indicated that tumors treated with polymet-CDDP NPs expressed significantly decreased expression of ERCC1 and XPA in comparison to tumors treated with only PEI-CDDP NPs or polymet NPs (Fig. 6C and D). The lowered levels of ERCC1 and XPA permitted the use of multiple (5) doses of CDDP without incurring significant drug resistance. The ability for sequential dosing without inducing resistance showcases yet another advantage to utilizing the combination of CDDP with poly-metformin.
As indicated in Fig. 7A and B, treatment with polymet alone produces only negligible differences in the expression of ERCC1 and XPA. Yet, cells treated with CDDP caused a subsequent increase in both ERCC1 and XPA. On the other hand, a combination of CDDP and polymet results in the ablation of both ERCC1 and XPA expression to levels lower than the PBS control. Furthermore, the level of cleaved PARP displayed a simultaneous increase with using the combination. These results serve to reinforce work done in vivo, which describes the role combining CDDP and polymet on regulating ERCC1, XPA, and cleaved PARP levels. Treatments consisting of both CDDP and polymet delivered simultaneously decrease the expression of key DNA repair proteins, inhibiting successful repair of platinum DNA adducts and result in the subsequent apoptosis of affected cells. Apoptosis was confirmed both by TUNEL and by the increase in cleaved PARP expression, a reliable marker for cell apoptosis.
Fig. 7.
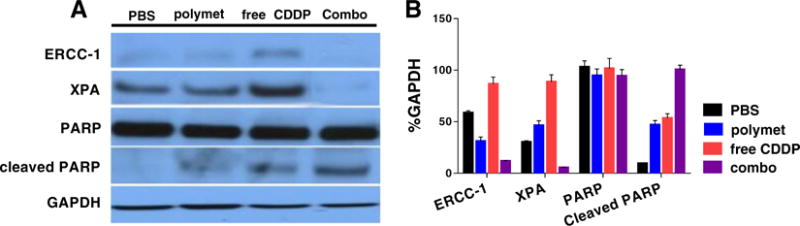
Western blot analysis of protein levels after treating 24 h with polymet, CDDP and their combination (combo) on H460 tumor cells in vitro. (A) ERCC-1, XPA, PARP and cleaved PARP protein levels were evaluated. In vitro test certified that polymet combined with CDDP will decrease the XPA and ERCC-1 and increased the Cleaved PARP. The concentration of polymet and free CDDP were 1 μM and combo including both 1 μM polymet (calculated according to the metformin unit of polymet) and 1 μM free CDDP. (B) Quantification of the protein level using image J. Data are mean ± S.D. (n = 3).
In summary, the combination of CDDP and polymet allows the downregulation of ERCC1 and XPA, key DNA repair proteins that prevent platinum adduct-based apoptosis. Downregulation of ERCC1 and XPA are followed by increased cell apoptosis as determined by the increase in level of cleaved PARP and decrease of uncleaved PARP. Simultaneously, activity in the AMPK tumor suppressor pathway is significantly augmented by the presence of polymet, which working in conjunction with the downregulation of ERCC1 and XPA, inhibits tumor proliferation. In addition, further examination into downstream targets indicates that the upregulation of AMPK is followed by the decrease of mTOR activity. Collectively, these results indicate the mechanism of the synergy between CDDP and polymet.
4. Conclusions
In summary, the development of core-membrane NPs capable of co-encapsulating two drugs of different physicochemical properties have permitted the creation of a novel anti-cancer regimen composed of CDDP and polymeric metformin. By employing the prominent synergy between the two drugs, polymet-CDDP NPs have displayed significantly enhanced anticancer efficacy in comparison to PBS, polymet NPs and PEI-CDDP NPs. Furthermore, the efficacy of polymet-CDDP NPs was statistically equivalent to that of free CDDP. However, polymet-CDDP NPs have successfully mitigated the prominent toxicities that hamper the clinical application of CDDP, namely nephrotoxicity and significant weight loss. Investigations into the mechanisms of the successful anticancer efficacy of polymet-CDDP NPs have pointed to a host of reasons for its achievement. High tumor and cellular uptake of polymet-CDDP NPs indicate the formulation’s ability to deliver large amounts of drug into the tumor. Moreover, active targeting through sigma receptors allows tumor specific delivery on the cellular level. After successful delivery, the synergy between CDDP and polymet ensures the limited drug release due to conjugation of CDDP with PGA during formulation does not hamper the efficacy of the treatment. Rather, polymet-CDDP NPs displayed the greatest anticancer efficacy among all treatment groups. Additionally, in vitro examination into the anticancer ability of polymet-CDDP NPs is in accordance with all in vivo studies, ensuring the mechanistic basis behind the successful efficacy is accurate. Relative to free CDDP and even a PBS control, polymet-CDDP NPs decreased the expression of XPA and ERCC-1. Ablating the mechanism of cisplatin induced drug resistance allowed polymet-CDDP NPs to be delivered in many doses without incurring drug tolerance, a feat that is not possible with CDDP alone. Overall, the polymet-CDDP NP platform serves an influential role for cancer drug delivery in its ability to co-encapsulate two drugs of differing properties for synergistic effects. Furthermore, the platform successfully alleviates the prominent toxicities that are the current clinical barrier to further implementation of CDDP as a front line anti-cancer therapeutic while maintaining a level of efficacy comparable to CDDP.
Supplementary Material
Acknowledgments
This work was supported by NIH grants CA151652, CA149363, CA198999 and DK100664. And also supported by project of the National Natural Science Foundation of China (No. 81473434, No. 81202926).
Abbreviations
- NSCLC
non-small cell lung cancer
- CDDP
cisplatin
- polymet
polymeric metformin
- AMPK
AMP-activated protein kinase
- mTOR
mammalian target of rapamycin
- ERCC1
excision repair cross-complementation group 1
- IV
intravenous
- PK
pharmacokinetic
- NPs
nanoparticles
- RES
reticuloendothelial system
- PGA
polyglutamic acid
- PEI
polyethylenimine
- EDC
ethylene dichloride
- NHS
N-hydroxysuccinimide
- MTT
3-[4.5-dimethylthiazol-2-yl]-2.5-diphenyltetrazolium bromide
- DAPI
4′,6-diamidino-2-phenylindole
- DIPEA
N,N-Diisopropylethylamine
- DOTAP
2,3-dioleoyloxy-propyl)-trimethylammonium
- RIPA
Radio-Immunoprecipitation Assay
- BCA
Bicinchoninine acid
- HRP
horseradish peroxidase
- DSPE-PEG2000
1,2-distearoryl-sn-glycero-3-phosphoethanolamine-N[methoxy (polyethyleneglycol-2000)](ammonium salt)
- DSPE-PEG-AA
DSPE-PEG-aminoethyl anisamide
- TUNEL
TdT-dependent dUTP-biotin nick end labeling
- GAPDH
glyceraldehyde-phosphate dehydrogenase
- XPA
xeroderma pigmentosum complementation group A
- PARP-1
poly ADP-ribose polymerase-1
- PBS
phosphate-buffered saline
- 1H NMR
nuclear magnetic resonance
- DLS
dynamic light scattering
- PDI
polydispersity index
- TEM
transmission electron microscope
- ICP-MS
inductively coupled plasma mass spectrometry
- BUN
blood urea nitrogen
- AST
aspartate aminotransferase
- ALT
alanine aminotransferase
- RBC
red blood cells
- WBC
white blood cells
- PLT
platelets
- HGB
hemoglobin
- HCT
hematocrits
- PIC
protease inhibitor cocktail
- SDS-PAGE electrophoresis
sodium dodecyl sulfate polyacrylamide gel electrophoresis
- PVDF
polyvinylidene difluoride
- BSA
Bovine Serum Albumin
- MALDI-TOF-MS
matrix-assisted laser desorption/ionization time of flight mass spectrometry
- MTD
maximum tolerable dose
- PI3K
phosphatidyl inositol 3 kinase
- ERK
extracellular signal regulated kinase
Appendix A. Supplementary data
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.jconrel.2016.11.005.
References
- 1.http://www.wcrf.org/cancerstatistics/worldcancerstatistics.php.2012; Available at: http://www.crf.org/cancerstatistics/worldcancerstatistics.php. (Accessed March 2014)
- 2.Spiro SG, Tanner NT, Silvestri GA, Janes SM, Lim E, Vansteenkiste JF, Pirker R. Lung cancer: progress in diagnosis, staging and therapy. Respirology. 2010;15:44–50. doi: 10.1111/j.1440-1843.2009.01674.x. [DOI] [PubMed] [Google Scholar]
- 3.Surveillance, Epidemiology, Sur and End Results [SEER] incidence and National Center for HealthStatistics [NHCS] mortality statistics. Available at: http://www.cancer.gov/cancertopics/types/lung/cancer-survival-prognosis. (Last accessed 4 August 2014)
- 4.De Angelis R, Sant M, Coleman MP, Francisci S, Baili P, Pierannunzio D, Trama A, Visser O, Brenner H, Ardanaz E, Bielska-Lasota M, Engholm G, Nennecke A, Siesling S, Berrino F, Capocaccia R. Cancer survival in Europe 1999–2007 by country and age: results of EUROCARE-5—a population-based study. Lancet Oncol. 2014;15:23–34. doi: 10.1016/S1470-2045(13)70546-1. [DOI] [PubMed] [Google Scholar]
- 5.Yin JY, Huang Q, Zhao YC, Zhou HH, Liu ZQ. Meta-analysis on pharmacogenetics of platinum-based chemotherapy in non small cell lung cancer (NSCLC) patients. PLoS One. 2012;7:e38150. doi: 10.1371/journal.pone.0038150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.N.C. Institute. Non-small cell lung cancer treatment. 2014 http://www.cancer.gov/cancertopics/pdq/treatment/non-small-cell-lung/healthprofessional/page10.
- 7.Dingemans AM, Mellema WW, Groen HJ, van Wijk A, Burgers SA, Kunst PW, Thunnissen E, Heideman DA, Smit EF. A phase II study of sorafenib in patients with platinum-pretreated, advanced (stage IIIb or IV) non-small cell lung cancer with a KRAS mutation. Clin Cancer Res. 2013;19:743–751. doi: 10.1158/1078-0432.CCR-12-1779. [DOI] [PubMed] [Google Scholar]
- 8.Morgillo F, Della CMC, Festino L, Manzo A, Martinelli E, Troiani T, Capuano A, Ciardiello F. Metformin in lung cancer rationale for a combination therapy. Expert Opin Investig Drugs. 2013;22:1401–1409. doi: 10.1517/13543784.2013.828691. [DOI] [PubMed] [Google Scholar]
- 9.Zhou G, Myers R, Li Y, Chen Y, Shen X, Fenyk-Melody J, Wu M, Ventre J, Doebber T, Fujii N, Musi N, Hirshman MF, Goodyear LJ, Moller DE. Role of AMP-activated protein kinase in mechanism of metformin action. J Clin Invest. 2001;108:1167–1174. doi: 10.1172/JCI13505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dowling RJ, Zakikhani M, Fantus IG, Pollak M, Sonenberg N. Metformin inhibits mammalian target of rapamycin-dependent translation initiation in breast cancer cells. Cancer Res. 2007;67:10804–10812. doi: 10.1158/0008-5472.CAN-07-2310. [DOI] [PubMed] [Google Scholar]
- 11.Lin CC, Yeh HH, Huang WL, Yan JJ, Lai WW, Su WP, Chen HH, Su WC. Metformin enhances cisplatin cytotoxicity by suppressing signal transducer and activator of transcription-3 activity independently of the liver kinase B1-AMP-activated protein kinase pathway. Am J Respir Cell Mol Biol. 2013;49:241–250. doi: 10.1165/rcmb.2012-0244OC. [DOI] [PubMed] [Google Scholar]
- 12.Teixeira SF, Madeira KP, Daltoé RD, Silva IV, Rangel LB. Metformin synergistically enhances antiproliferative effects of cisplatin and etoposide in NCI-H460 human lung cancer cells. J Bras Pneumol. 2013;39(6):644–649. doi: 10.1590/S1806-37132013000600002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guo S, Wang Y, Miao L, Xu Z, Lin CM, Zhang Y, Huang L. Lipid-coated cisplatin nanoparticles induce neighboring effect and exhibit enhanced anticancer efficacy. ACS Nano. 2013;7:9896–9904. doi: 10.1021/nn403606m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nishiyama N, Okazaki S, Cabral H, Miyamoto M, Kato Y, Sugiyama Y, Nishio K, K K, Matsumura Y. Novel cisplatin-incorporated polymeric micelles can eradicate solid tumors can eradicate solid tumors in mice. Cancer Res. 2003;63:8977–8983. [PubMed] [Google Scholar]
- 15.Boulikas T. Clinical overview on Lipoplatin- a successful liposomal formulation of cisplatin. Expert Opin Investig Drugs. 2009;18:1197–1218. doi: 10.1517/13543780903114168. [DOI] [PubMed] [Google Scholar]
- 16.Plummer R, Wilson RH, Calvert H, Boddy AV, Griffin M, Sludden J, Tilby MJ, Eatock M, Pearson DG, Ottley CJ, Matsumura Y, Kataoka K, Nishiya T. A phase I clinical study of cisplatin-incorporated polymeric micelles (NC-6004) in patients with solid tumours. Br J Cancer. 2011;104:593–598. doi: 10.1038/bjc.2011.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miao L, Guo S, Zhang J, Kim WY, Huang L. Nanoparticles with precise ratiometric co-loading and co-delivery of gemcitabine monophosphate and cisplatin for treatment of bladder cancer. Adv Funct Mater. 2014;24:6601–6611. doi: 10.1002/adfm.201401076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao Y, Wang W, Guo S, Wang Y, Miao L, Xiong Y, Huang L. Polymetformin combines carrier and anticancer activities for in vivo siRNA delivery. Nat Commun. 2016;7:11822. doi: 10.1038/ncomms11822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ye H, Jin L, Hu R, Yi Z, Li J, Wu Y, Xi X, Wu Z. Poly(gamma, L-glutamic acid)-cisplatin conjugate effectively inhibits human breast tumor xenografted in nude mice. Biomaterials. 2006;27:5958–5965. doi: 10.1016/j.biomaterials.2006.08.016. [DOI] [PubMed] [Google Scholar]
- 20.Banerjee R, Tyagi P, Li S, Huang L. Anisamide-targeted stealth liposomes: a potent carrier for targeting doxorubicin to human prostate cancer cells. Int J Cancer. 2004;112:693–700. doi: 10.1002/ijc.20452. [DOI] [PubMed] [Google Scholar]
- 21.Guo S, Miao L, Wang Y, Huang L. Unmodified drug used as a material to construct nanoparticles: delivery of cisplatin for enhanced anti-cancer therapy. J Control Release. 2014;174:137–142. doi: 10.1016/j.jconrel.2013.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang Y, Kim WY, Huang L. Systemic delivery of gemcitabine triphosphate via LCP nanoparticles for NSCLC and pancreatic cancer therapy. Biomaterials. 2013;34:3447–3458. doi: 10.1016/j.biomaterials.2013.01.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chou TC, Talalay P. Quantitative analysis of dose-effect relationships: the combined effects of multiple drugs or enzyme inhibitors. Adv Enzym Regul. 1984;22:27–55. doi: 10.1016/0065-2571(84)90007-4. [DOI] [PubMed] [Google Scholar]
- 24.Han Y, He Z, Schulz A, Bronich TK, Jordan R, Luxenhofer R, Kabanov AV. Synergistic combinations of multiple chemotherapeutic agents in high capacity poly(2-oxazoline) micelles. Mol Pharm. 2012;9:2302–2313. doi: 10.1021/mp300159u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miao L, Wang Y, Lin CM, Xiong Y, Chen N, Zhang L, Kim WY, Huang L. Nanoparticle modulation of the tumor microenvironment enhances therapeutic efficacy of cisplatin. J Control Release. 2015;217:27–41. doi: 10.1016/j.jconrel.2015.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Luo C, Miao L, Zhao Y, Musetti S, Wang Y, Shi K, Huang L. A novel cationic lipid with intrinsic antitumor activity to facilitate gene therapy of TRAIL DNA. Biomaterials. 2016;102:239–248. doi: 10.1016/j.biomaterials.2016.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li SD, Huang L. Stealth nanoparticles: high density but sheddable PEG is a key for tumor targeting. J Control Release. 2010;145:178–181. doi: 10.1016/j.jconrel.2010.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.De Palma R, Peeters S, Van Bael MJ, Van den Rul H, Bonroy K, Laureyn W, Mullens J, Borghs G, Maes G. Silane ligand exchange to make hydrophobic superparamagnetic nanoparticles water-dispersible. Chem Mater. 2007;19:1821–1831. [Google Scholar]
- 29.Jenkins SI, Pickard MR, Furness DN, Yiu HH, Chari DM. Differences in magnetic particle uptake by CNS neuroglial subclasses: implications for neural tissue engineering. Nanomedicine. 2012;8:951–968. doi: 10.2217/nnm.12.145. [DOI] [PubMed] [Google Scholar]
- 30.Xiong Y, Jiang W, Shen Y, Li H, Sun C, Ouahab A, Tu J. A poly(gamma, L-glutamic acid)-citric acid based nanoconjugate for cisplatin delivery. Biomaterials. 2012;33:7182–7193. doi: 10.1016/j.biomaterials.2012.06.071. [DOI] [PubMed] [Google Scholar]
- 31.Wang Y, Miao L, Satterlee A, Huang L. Delivery of oligonucleotides with lipid nanoparticles. Adv Drug Deliv Rev. 2015;87:68–80. doi: 10.1016/j.addr.2015.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vasey PA, Kaye SB, Morrison R, Twelves C, Wilson P, Duncan R, Thomson AH, Murray LS, Hilditch TE, Murray T, Burtles S, Fraier D, Frigerio E, Cassidy J. Phase I clinical and pharmacokinetic study of PK1 [N-(2Hydroxypropyl) methacrylamide copolymer doxorubicin]: first member of a new class of chemotherapeutic agents—drug-polymer conjugates. Clin Cancer Res. 1999;5:83–94. [PubMed] [Google Scholar]
- 33.Mochida Y, Cabral H, Miura Y, Albertini F, Fukushima S, Osada K, Nishiyama N, Kataoka K. Bundled assembly of helical nanostructures in polymeric micelles loaded with platinum drugs enhancing therapeutic efficiency against pancreatic tumor. ACS Nano. 2015;8:6724–6738. doi: 10.1021/nn500498t. [DOI] [PubMed] [Google Scholar]
- 34.Song P, Kwon Y, Yea K, Moon HY, Yoon JH, Ghim J, Hyun H, Kim D, Koh A, Berggren PO, Suh PG, Ryu SH. Apolipoprotein a1 increases mitochondrial biogenesis through AMP-activated protein kinase. Cell Signal. 2015;27:1873–1881. doi: 10.1016/j.cellsig.2015.05.003. [DOI] [PubMed] [Google Scholar]
- 35.LeRoith D. Insulin-like Growth Factors and Cancer: From Basic Biology to Therapeutics. Humana Press; New York: 2012. [Google Scholar]
- 36.Vallianou NG, Evangelopoulos A, Kazazis C. Metformin and cancer. Rev Diabet Stud. 2013;10:228–235. doi: 10.1900/RDS.2013.10.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Faubert B, Vincent EE, Poffenberger MC, Jones RG. The AMP-activated protein kinase (AMPK) and cancer: many faces of a metabolic regulator. Cancer Lett. 2015;356:165–170. doi: 10.1016/j.canlet.2014.01.018. [DOI] [PubMed] [Google Scholar]
- 38.Lopez-Cotarelo P, Escribano-Diaz C, Gonzalez-Bethencourt IL, Gomez-Moreira C, Deguiz ML, Torres-Bacete J, Gomez-Cabanas L, Fernandez-Barrera J, Delgado-Martin C, Mellado M, Regueiro JR, Miranda-Carus ME, Rodriguez-Fernandez JL. A novel MEK-ERK-AMPK signaling axis controls chemokine receptor CCR7-dependent survival in human mature dendritic cells. J Biol Chem. 2015;290:827–840. doi: 10.1074/jbc.M114.596551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mihaylova MM, Shaw RJ. The AMPK signalling pathway coordinates cell growth, autophagy and metabolism. Nat Cell Biol. 2011;13:1016–1023. doi: 10.1038/ncb2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Naoki K, Chiharu T, Sushila D, Christine R, Yoshino K, Kenta H, Bruce EK, Lee AW, Osamu M, Kazuyoshi Y. A possible linkage between AMP-activated protein kinase (AMPK) and mammalian target of rapamycin (mTOR) signalling pathway. Genes Cells. 2003;8:65–79. doi: 10.1046/j.1365-2443.2003.00615.x. [DOI] [PubMed] [Google Scholar]
- 41.Zhang Y, Peng L, Mumper RJ, Huang L. Combinational delivery of c-myc siRNA and nucleoside analogs in a single, synthetic nanocarrier for targeted cancer therapy. Biomaterials. 2013;34:8459–8468. doi: 10.1016/j.biomaterials.2013.07.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mazzoni F, Cecere FL, Meoni G, Giuliani C, Boni L, Camerini A, Lucchesi S, Martella F, Amoroso D, Lucherini E, Torricelli F, Di Costanzo F. Phase II trial of customized first line chemotherapy according to ERCC1 and RRM1 SNPs in patients with advanced non-small-cell lung cancer. Lung Cancer. 2013;82:288–293. doi: 10.1016/j.lungcan.2013.08.018. [DOI] [PubMed] [Google Scholar]
- 43.Liu Y, Bernauer AM, Yingling CM, Belinsky SA. HIF1α regulated expression of XPA contributes to cisplatin resistance in lung cancer. Carcinogenesis. 2012;33:1187–1192. doi: 10.1093/carcin/bgs142. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


