Abstract
For many years, only a small fraction of the human genome was believed to regulate cell function and development. This protein-coding portion composed only 1% to 2% of 3 billion human DNA base pairs— the remaining sequence was classified as junk DNA. Subsequent research has revealed that most of the genome is transcribed into a broad array of noncoding RNAs, ranging in size from microRNA (20–23 nucleotides) to long noncoding RNA (lncRNA, more than 200 nucleotides). These noncoding RNA classes have been shown to use diverse molecular mechanisms to control gene expression and organ system development. As anticipated, alterations in this large control system can contribute to disease pathogenesis and carcinogenesis. We review the involvement of noncoding RNAs—lncRNAs in particular—in development of Barrett's esophagus and esophageal carcinoma.
Barrett's esophagus (BE) is a premalignant condition in which the lining of the esophagus transitions from normal stratified squamous epithelium to a metaplastic specialized columnar epithelium1. Chronic gastroesophageal reflux disease (GERD) causes BE, which increases risk of esophageal adenocarcinoma (EAC)2,3. The major histologic types of esophageal cancer are esophageal squamous cell carcinoma (ESCC) and EAC. A recent study by The Cancer Genome Atlas Research Network used a broad array of genomic research methods to show that ESCCs are similar to squamous cell cancers of the head and neck, whereas EACs resemble certain gastric adenocarcinomas4. We review the potential roles of a poorly understood but abundant class of RNAs, known as long noncoding RNAs (lncRNAs), in development and/or neoplastic progression of BE.
Coding and Non-coding RNAs
RNA polymerase II interacts with transcription factors at specific gene promoter DNA sequences to initiate synthesis of RNA. Some RNAs (mRNAs) are then processed, transported to the cytoplasm,5,6 and translated into proteins. RNA polymerase I synthesizes RNAs ribosomal RNAs (rRNAs)—the first known noncoding RNAs (ncRNAs), which are required for protein synthesis. RNA polymerases II and III synthesize other ncRNAs, which account for most of the RNA in a cell and do not encode proteins. Nonetheless, ncRNAs have a wide range of functions that affect all aspects of homeostasis and development 7,8. Over the past few decades, a vast and diverse universe of ncRNAs has been discovered and explored (see Table 1).
Table 1. Human Noncoding RNAs.
| Symbol | Name | Length (nt) | Function |
|---|---|---|---|
| tRNA | transfer RNA | 73–93 | Amino acid incorporation into protein. |
| rRNA | ribosomal RNA | 121–5070 | Ribosome production |
| snRNA | small nuclear RNA | 100-300 | Removal of introns from spliceosome |
| snoRNA | small nuclear RNA | 70 | Modification of rRNAs |
| TERC | telomerase RNA | 451 | Template for telomere synthesis |
| miRNA | microRNA | 20–23 | Regulation of gene expression |
| piRNA | piwi-interacting RNA | 25–33 | Silencing of transposons |
| T-UCR | transcribed ultra-conserved region | > 350 | Potential regulation of RNA levels |
| lncRNA | long noncoding RNA | > 200 | Control of mRNA & miRNA expression |
During the 1980's, small nuclear RNAs (snRNAs, 100–300 nucleotides) were identified and found to participate in removal of introns from heterogenous RNAs, which are transcribed in the nucleus9 (Fig. 1a). Cells contain up to 1 million snRNAs, which combine with certain proteins to form spliceosomes to facilitate removal of introns10. Another class of RNAs, small nucleolar ribonucleoproteins (snoRNAs), are small in size (70 nucleotides) and participate in the processing of pre-rRNA molecules into mature rRNAs in ribosomes11. Finally, the piwi-interacting RNAs (piRNAs) are small RNAs (25–33 nucleotides) that interact with piwi proteins to suppress gene expression via transposable elements.
Figure 1. Cell functions controlled by lncRNAs.
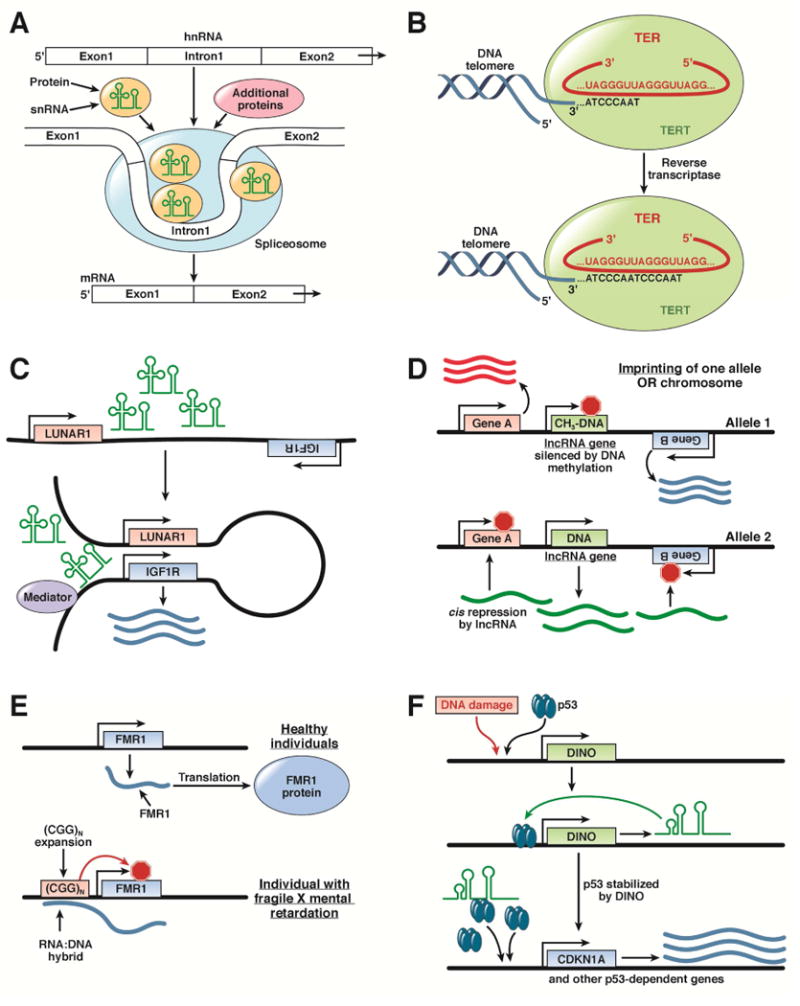
A. snRNAs form part of a nuclear spliceosome, which precisely removes introns from heterogeneous RNA to yield mature mRNAs consisting only of exons. B. Telomerase comprises the TERT protein subunit and a noncoding telomerse RNA (TER), which provides the template for synthesis and extension of telomeres. C. The lncRNA LUNAR causes chromosomal looping, resulting in placement of a transcriptional activating complex named mediator to act as an enhancer near the promoter of the IGF1R gene. D. During imprinting, 1 allele of a gene (such as an inhibitory lncRNA) is silenced by DNA methylation, permitting expression of neighboring genes. The other allele has reduced or absent methylation, resulting in lncRNA expression and cis repression of neighboring genes. E. The FMR1 mRNA encodes the FMR1 protein; the 5′ untranslated region of the FMR1 mRNA binds an expanded CGG triplet sequences at the gene promoter. This DNA–RNA hybrid prevents transcription of the FMR1 gene, leading to fragile X mental retardation. F. In response to DNA damage, cell levels of the lncRNA DINO greatly increase. DINO binds to and stabilizes TP53, allowing TP53 to regulate hundreds of gene targets.
A new class of small ncRNAs (19–23 nucleotides) that affect protein production was discovered in Caenorhabditis elegans in 199312,13 and in humans in 200114. These microRNAs (miRNAs) regulate levels of most mRNAs by targeting them for degradation or translational inhibition. Mammalian cells have approximately 2000 unique miRNAs, each of which usually has multiple mRNA targets15. In fact, some miRNAs are actually derived from lncRNAs, such as miRNA-675-3p and miRNA-675-5p, which come from the lncRNA H1916,17. The involvement of miRNAs in the progression of BE to EAC has been well studied and is not covered here (for more information, see refs 18,19,20).
Telomerase is a ribonucleoprotein that maintains the ends of the linear human chromosomes (telomeres). The number of these telomeric repeats essentially regulates how old a cell is and can also determine its level of proliferation. Telomerase comprises a coding telomerase reverse transcriptase (TERT) subunit and a ncRNA (TER or TERC) subunit; this ncRNA provides a template for synthesis of part of the telomeric DNA repeats21 (Fig. 1b).
LncRNAs
ncRNAs longer than 200 nucleotides are called lncRNAs. These molecules control gene expression by many different mechanisms. Within the past 10 years, the ENCODE project has catalogued over 9600 lncRNAs, most of which were originally thought to represent transcriptional noise, having been transcribed from what was previously termed junk DNA.
How are lncRNAs identified? They are transcribed from areas of the genome that are not annotated as having open reading frames that produce proteins22. Transcribed RNAs are detected by a variety of methods, each with its own strengths and limitations. Tiling assays use microarray slides containing overlapping oligonucleotides covering the whole genome, which are hybridized to cDNA made from all cellular RNAs. Serial analysis of gene expression was an early method to identify and quantify cellular transcripts using only small parts (tags) of each transcript.
Tremendous improvements in DNA sequencing techniques have led to efficient total RNA sequencing, wherein RNA is converted to cDNA and all sequences are determined at a very high level of coverage. Computer analysis then determines which sequences are protein-coding RNAs and which are not protein-coding, and this method is both qualitative and quantitative. New lncRNA studies are now being published daily, and this field is expanding so rapidly that only a fraction of notable examples of lncRNAs' involvement in gene regulation can be discussed here.
Enhancer RNAs
Transcriptional enhancers, which have been studied for several decades, comprise distinct DNA elements that facilitate increased transcription at a specific gene promoters23. Some lncRNAs act as enhancer RNAs, such as LUNAR1, associated with T-cell acute lymphoblastic leukemia. LUNAR1 mediates the positioning of an enhancer element, named mediator, near the promoter of the insulin-like growth factor 1 receptor, resulting in increased transcription of this gene24 (Fig. 1c).
At imprinted genes and chromosomes
Epigenetic modification of genes via a process known as imprinting can silence 1 allele of a gene. This process occurs on autosomes as well as sex chromosomes. The most widely known example of this phenomenon occurs on the X chromosome in female human cells—1 of the X chromosomes is transcribed while the other is silenced. More than 20 years ago, the lncRNA H19 was found to be epigenetically controlled and expressed only from the maternally inherited allele25.
Cells have 2 alleles for each gene but 1 one is expressed; this imprinting process is controlled in cis 26 (Fig. 1d). It is now widely believed that lncRNAs are needed for the allele-specific regulation of gene expression that occurs at imprinted loci27. In general, methylation of a lncRNA gene prevents its transcription, which in turn allows the expression of nearby mRNA-encoding genes28. Conversely, the absence of methylation of a lncRNA gene is thought to allow its expression, which in turn blocks expression of nearby mRNA genes, in cis 29.
Autoregulation
Many protein-coding genes autoregulate their own expression, in that production of the final protein product reduces further production of the same product. Some lncRNAs also appear to autoregulate. For example, a transcript of the fragile X mental retardation gene (FMR1) can act as either an mRNA that is translated into the FMR1 protein or as an lncRNA that forms a DNA–RNA hybrid at CGG repeats within the FMR1 gene promoter30 (Fig. 1e). This hybrid then represses transcription; individuals with expanded CGG repeats at this promoter region have fragile X mental retardation31.
Antisense lncRNAs block sense transcription
Many protein-coding genes have an antisense lncRNA cognate gene partner present on the opposite strand. When both such genes are actively transcribed, the respective RNA polymerase II synthesis complexes are blocked from proceeding, and transcription of both genes is prevented. An example of this phenomenon is repression of the sense insulin-like growth factor 2 receptor (IGF2R) gene by the overlapping antisense lncRNA AIR gene, in which each gene blocks transcription of its counterpart32.
LncRNA DINO stabilizes TP53
Schmitt et al. investigated the role of the lncRNA damage induced noncoding (DINO) in the cell's response to DNA damage mediated by TP53 expression33 (Fig. 1f). TP53 regulates expression of hundreds of genes, including the cyclin-dependent kinase inhibitor 1A (CDKN1A). The relatively short half-life of TP53 affects expression of these genes. DINO, which is greatly upregulated upon DNA damage, binds to and stabilizes TP53, allowing it to regulate its gene targets.
The surprising stabilization of p53 by DINO is another example of an interesting function of lncRNA. This is 1 of the many areas of lncRNA research that is likely to expand. Other important areas of study will include the regulation of lncRNA expression. Researchers have investigated mechanisms of transcriptional regulation of protein-coding genes vs long intervening ncRNAs (lincRNAs), a subgroup of lncRNA. They found different patterns of RNA polymerase II phosphorylation, depending on which type of RNA was produced; these patterns were also dependent on progression of transcription varying from initiation to termination34.
LncRNAs in BE and EAC
It has been 20 years since Hibi et al first reported altered expression of a lncRNA (H19) in human esophageal cancer cells. Although these researchers did not specify whether they were studying ESCCs or EAC, they did describe loss of H19 imprinting in esophageal tumors35. Normally, the paternally inherited H19 allele is methylated and the maternally inherited allele is hypomethylated or unmethylated, which in turn permits transcription of the H19 gene and the production of a 2.3-kilobase RNA molecule36. Expression levels of H19 and IGF2, each located on chromosome 11, are closely linked in an inversely controlled manner—increased expression of lncRNA H19 reduces expression of IGF2 protein, whereas decreased expression of H19 increases expression of IGF2 37. The same research group found a marked loss of imprinting in esophageal cancers, but not in colorectal cancers. Loss of imprinting in tumor cells can result in methylation of the maternal H19 allele, reducing its expression and thereby increasing IGF2 expression.
There have been several reports of involvement of lncRNAs in esophageal cancer, but most of these investigations focused on ESCC and therefore fall outside the scope of this review38,39,40,41,42,43,44. One recent comprehensive study of lncRNA expression evaluated 5037 human tumor specimens, of 13 distinct cancer types45. The goal of this study was to identify and catalog lncRNAs that promote carcinogenesis and predict their functions. However, esophageal cancer was not included in this study.
Another recent study analyzed global methylation patterns in EAC or BE samples vs matched normal esophageal tissues 46. This work identified a lncRNA, AFAP1-AS1 (6810 bases in length), that is transcribed from the antisense or noncoding DNA strand at the locus where the coding DNA strand produces actin filament-associated protein 1 (AFAP1). AFAP1 interacts with the proto-oncogene SRC and was proposed to modulate the integrity of actin filaments47,48. This lncRNA provides another example of a sense–antisense relationship, wherein RNA transcription complexes at these sites can physically hamper transcription. AFAP1-AS1 expression levels were measured by quantitative reverse-transcriptase PCR, which showed increased expression of this lncRNA in BE vs matched normal tissue. AFAP1-AS1 expression was also markedly increased in 3 EAC cell lines (OE33, SKGT4, Flo-1) compared with normal primary esophageal epithelial cells (HEEpic).
Transfection of OE33 and SKGT4 cells with small interfering RNAs (siRNAs) against AFPA1-AS1 significantly reduced cell migration and invasion, but did not significantly reduce expression of AFAP1. This finding suggests that the phenotypic changes induced by siRNA transfection were mediated directly by AFAP1-AS1, rather than indirectly, via AFAP1. The authors propose that aberrant increases in expression of AFAP1-AS1 could contribute to development of EAC. Subsequently, an independent investigation of 65 patients with ESCC found increased levels of AFAP1-AS1 in their tumor tissues 38. Silencing of this lncRNA in ESCC cells inhibited proliferation and reduced colony-forming capacity, so AFAP1-AS1 appears to function as an oncogene that promotes ESCC development or progression.
Levels of the lncRNA metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) were reported to be higher in esophageal tumor specimens (n=132) than in normal esophageal tissues49. However, it is not clear whether this study analyzed ESCC or EAC samples, although levels of MALAT1 correlated with lymphatic invasion, distant metastasis, and tumor differentiation. In another report, RNA sequence analyses revealed that the lncRNA CASC9 was upregulated in ESCC50. Knockdown of CASC9 in ESCC cells reduced their migration and invasion. In an investigation of gastric adenocarcinomas, levels of lncRNA KRT7-AS were significantly higher in tumors than in matched normal adjacent gastric tissues51. This lncRNA contains regions that are antisense to the protein-coding keratin 7 gene (KRT7); surprisingly, RNA hybrids formed between these transcripts increase expression of KRT7 mRNA and protein.
Yang et al investigated lncRNAs expressed during the progression of normal esophageal tissue to EAC by performing RNA-seq analyses of matched normal, BE, and EAC tissues. They identified 6216 lncRNAs expressed in EAC 52. Of these, 61 unique lncRNAs were substantially upregulated in EAC compared with normal esophageal tissue, with an average increase of more than 8-fold. One of the lncRNAs chosen for further study, HNF1A-AS1, is a 2455-nucleotide molecule located on the antisense DNA strand, whereas its cognate sense counterpart, the hepatic nuclear factor 1 alpha gene (HNF1A), is on the sense strand. HNF1A-AS1 was overexpressed in 4 EAC cell lines (Flo-1, OE3, JHU-EsoAd1, and SKGT4) compared with normal primary esophageal cells (HEEpic). siRNA-mediated knockdown of HNF1A-AS1 reduced EAC cell proliferation and decreased anchorage-independent growth, measured by colony-formation assays, compared with a scrambled siRNA control. These experiments also showed reduced cell migration and invasion in EAC cell lines after siRNA knockdown of HNF1A-AS1. Surprisingly, HNF1A-AS1 did not appear to directly regulate HNF1A mRNA or protein—both were significantly upregulated in EACs compared with normal esophageal tissues. Interestingly, lncRNA H19 was the gene most markedly dysregulated by knockdown of HNF1A-AS1; the correlation between these lncRNAs was verified in primary EACs. The lncRNA HNF1A-AS1 might therefore serve as a marker of EAC aggressiveness.
A meta-analysis examined 7 sets of RNA expression data from normal esophagus, BE, EAC and ESCC tissues. This study evaluated differentially expressed protein-coding genes and their relationship to noncoding lncRNAs53. The salient finding from this study was that the number of lncRNA and protein-coding gene pairs varied greatly. The number of such pairs specific to BE and normal esophagus was 2690, whereas 29 pairs were specific to EAC and BE, 2000 were specific to EAC and normal esophagus, and 19,815 were specific to ESCC and normal esophagus. It is important to remember that the authors' conclusions were based on several different experimental datasets from various research groups. The authors noted a large difference in described lncRNA and protein-coding gene pairs between EAC/BE and ESCC/normal esophagus. This difference could be due to the distinct etiologies of these 2 cancer subtypes, which might involve differences in lncRNA expression and function. Other studies have examined circulating lncRNAs as cancer biomarkers. In the circulation, lncRNAs are protected from degradation, due to their encasement in exosomes. Two studies have described circulating lncRNAs as possible markers for ESCC54,55.
Ultra-conserved ncRNAs (UCRs) are another important class of ncRNAs; these were discovered in 2004 by bioinformatic analyses of mouse, rat, and human genomes56,57,58. There are at least 481 genomic UCRs, ranging in size from 200 to 779 bases, with 100% identity between mouse, rat, and human sequences59,60. Over 90% of these UCRs are transcribed (T-UCRs) in certain normal tissues, resulting in tissue-specific expression patterns, but T-UCR functions are in general poorly understood61,62. Fassan et al measured levels of 481 human T-UCRs in patients with BE and EAC matched with normal esophagus63. A signature of 9 T-UCRs was associated with BE vs normal esophagus (6 upregulated T-UCRs and 3 downregulated T-UCRs). When the authors studied this profile in mouse and rat models of BE 64, they found 4 of these T-UCRs to have similar expression patterns. Progression from normal esophagus to BE to EAC correlated with increased expression of 4 T-UCRs and reduced expression of a single T-UCR. This T-UCR expression pattern of EACs did not resemble those reported in other tumor types (leukemia, colon cancer, liver cancer, prostate cancer, or neuroblastoma)65,66,67. In addition, there was chromosomal clustering of the dysregulated T-TCRs (7p15.3, 9q33.3, 11p13) in areas that had previously been reported as deleted or amplified in BE; genomic instability might therefore be involved in regulation of T-UCR expression68,69,70.
Telomerase is a ribozyme comprising the enzymatic subunit TERT and a lncRNA (TER or TERC), which serves as a template for the synthesis of telomeres at the termini of chromosomes. A study that measured TERC levels in paraffin-embedded esophageal biopsies correlated the transition from low-grade to high-grade dysplasia with a significant increase in TERC, and showed that EAC samples contained high levels of TERC 71. A subsequent study reported a strong, sequential increase in TERT expression during the progression of normal esophagus to BE and EAC 72. Souza et al directly measured telomere lengths in biopsy specimens from individuals with vs without GERD, and found shorter telomeres in patients with GERD, although TERT levels seemed to be similar between groups73. In addition, TERT activity is significantly higher in esophageal tissues from patients with BE with dysplasia or BE with EAC than from patients with BE without dysplasia74.
Most basic medical research over the past several decades has focused on the tiny fraction of the human genome that is transcribed into RNAs (mRNAs) that are translated into proteins. However, researchers are becoming more interested in the remainder of the genome, which is transcribed into the vast and complex universe of ncRNAs. Increasing our knowledge and understanding of these molecules could provide exciting new insights into mechanisms of cell function, development, and disease pathogenesis.
Table 2. LncRNAs Dysregulated in BE or Esophageal Cancer.
| Name | Direction | Length (nt) | Chromosome | Reference |
|---|---|---|---|---|
| H19 | Increased in Esophageal Carcinoma | 2300 | 11p15 | 35 |
| AFAP-AS1 | Increased in BE, EAC | 6,810 | 4p16.1 | 46 |
| AFAP-AS1 | Increased in ESCC | 6,810 | 4p16.1 | 38 |
| MALAT1 | Increased in Esophageal Carcinoma | 8,000 | 11q13.1 | 49 |
| MALAT1 | Increased in ESCC | 8,000 | 11q13.1 | 39 |
| CASC9 | Increased in ESCC | 2,400 | 8q21.13 | 50 |
| KRT7-AS | Increased in Gastric Cancer | unknown | 12q13.13 | 51 |
| HNF1A-AS1 | Increased in EAC | 2,455 | 12q24.31 | 52 |
Acknowledgments
Funding Sources: NCI CA190040, American Cancer Society Clinical Professorship (SJM)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Badillo R, Francis D. Diagnosis and treatment of gastroesophageal reflux disease. World J Gastrointest Pharmacol Ther. 2014;5:105–12. doi: 10.4292/wjgpt.v5.i3.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sappati Biyyani RS, Chak A. Barrett's esophagus: review of diagnosis and treatment. Gastroenterol Rep (Oxf) 2013;1:9–18. doi: 10.1093/gastro/got015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Modiano N, Gerson LB. Barrett's esophagus: Incidence, etiology, pathophysiology, prevention and treatment. Ther Clin Risk Manag. 2007;3:1035–145. [PMC free article] [PubMed] [Google Scholar]
- 4.Cancer Genome Atlas Research N, Analysis Working Group: Asan U, Agency BCC et al. Integrated genomic characterization of oesophageal carcinoma. Nature. 2017;541:169–175. doi: 10.1038/nature20805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Choi YS, Patena W, Leavitt AD, et al. Widespread RNA 3′-end oligouridylation in mammals. RNA. 2012;18:394–401. doi: 10.1261/rna.029306.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Quaresma AJ, Sievert R, Nickerson JA. Regulation of mRNA export by the PI3 kinase/AKT signal transduction pathway. Mol Biol Cell. 2013;24:1208–21. doi: 10.1091/mbc.E12-06-0450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cech TR, Steitz JA. The noncoding RNA revolution-trashing old rules to forge new ones. Cell. 2014;157:77–94. doi: 10.1016/j.cell.2014.03.008. [DOI] [PubMed] [Google Scholar]
- 8.Quinn JJ, Chang HY. Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet. 2016;17:47–62. doi: 10.1038/nrg.2015.10. [DOI] [PubMed] [Google Scholar]
- 9.Busch H, Reddy R, Rothblum L, et al. SnRNAs, SnRNPs, and RNA processing. Annu Rev Biochem. 1982;51:617–54. doi: 10.1146/annurev.bi.51.070182.003153. [DOI] [PubMed] [Google Scholar]
- 10.Wahl MC, Will CL, Luhrmann R. The spliceosome: design principles of a dynamic RNP machine. Cell. 2009;136:701–18. doi: 10.1016/j.cell.2009.02.009. [DOI] [PubMed] [Google Scholar]
- 11.Liu J, Maxwell ES. Mouse U14 snRNA is encoded in an intron of the mouse cognate hsc70 heat shock gene. Nucleic Acids Res. 1990;18:6565–71. doi: 10.1093/nar/18.22.6565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–54. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 13.Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–62. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- 14.Lagos-Quintana M, Rauhut R, Lendeckel W, et al. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–8. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- 15.Gurtan AM, Sharp PA. The role of miRNAs in regulating gene expression networks. J Mol Biol. 2013;425:3582–600. doi: 10.1016/j.jmb.2013.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Keniry A, Oxley D, Monnier P, et al. The H19 lincRNA is a developmental reservoir of miR-675 that suppresses growth and Igf1r. Nat Cell Biol. 2012;14:659–65. doi: 10.1038/ncb2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dey BK, Pfeifer K, Dutta A. The H19 long noncoding RNA gives rise to microRNAs miR-675-3p and miR-675-5p to promote skeletal muscle differentiation and regeneration. Genes Dev. 2014;28:491–501. doi: 10.1101/gad.234419.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bansal A, Gupta V, Wang K. MicroRNA Expression Signatures During Malignant Progression From Barrett's Esophagus. J Cell Biochem. 2016;117:1288–95. doi: 10.1002/jcb.25497. [DOI] [PubMed] [Google Scholar]
- 19.Mallick R, Patnaik SK, Wani S, et al. A Systematic Review of Esophageal MicroRNA Markers for Diagnosis and Monitoring of Barrett's Esophagus. Dig Dis Sci. 2016;61:1039–50. doi: 10.1007/s10620-015-3959-3. [DOI] [PubMed] [Google Scholar]
- 20.Bus P, Kestens C, Ten Kate FJ, et al. Profiling of circulating microRNAs in patients with Barrett's esophagus and esophageal adenocarcinoma. J Gastroenterol. 2016;51:560–70. doi: 10.1007/s00535-015-1133-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Greider CW, Blackburn EH. A telomeric sequence in the RNA of Tetrahymena telomerase required for telomere repeat synthesis. Nature. 1989;337:331–7. doi: 10.1038/337331a0. [DOI] [PubMed] [Google Scholar]
- 22.Fatica A, Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development. Nat Rev Genet. 2014;15:7–21. doi: 10.1038/nrg3606. [DOI] [PubMed] [Google Scholar]
- 23.Shlyueva D, Stampfel G, Stark A. Transcriptional enhancers: from properties to genome-wide predictions. Nat Rev Genet. 2014;15:272–86. doi: 10.1038/nrg3682. [DOI] [PubMed] [Google Scholar]
- 24.Trimarchi T, Bilal E, Ntziachristos P, et al. Genome-wide mapping and characterization of Notch-regulated long noncoding RNAs in acute leukemia. Cell. 2014;158:593–606. doi: 10.1016/j.cell.2014.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang Y, Tycko B. Monoallelic expression of the human H19 gene. Nat Genet. 1992;1:40–4. doi: 10.1038/ng0492-40. [DOI] [PubMed] [Google Scholar]
- 26.Adalsteinsson BT, Ferguson-Smith AC. Epigenetic control of the genome-lessons from genomic imprinting. Genes (Basel) 2014;5:635–55. doi: 10.3390/genes5030635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Royo H, Cavaille J. Non-coding RNAs in imprinted gene clusters. Biol Cell. 2008;100:149–66. doi: 10.1042/BC20070126. [DOI] [PubMed] [Google Scholar]
- 28.Mancini-Dinardo D, Steele SJ, Levorse JM, et al. Elongation of the Kcnq1ot1 transcript is required for genomic imprinting of neighboring genes. Genes Dev. 2006;20:1268–82. doi: 10.1101/gad.1416906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee JT. Epigenetic regulation by long noncoding RNAs. Science. 2012;338:1435–9. doi: 10.1126/science.1231776. [DOI] [PubMed] [Google Scholar]
- 30.Guil S, Esteller M. Cis-acting noncoding RNAs: friends and foes. Nat Struct Mol Biol. 2012;19:1068–75. doi: 10.1038/nsmb.2428. [DOI] [PubMed] [Google Scholar]
- 31.Oberle I, Rousseau F, Heitz D, et al. Instability of a 550-base pair DNA segment and abnormal methylation in fragile X syndrome. Science. 1991;252:1097–102. doi: 10.1126/science.252.5009.1097. [DOI] [PubMed] [Google Scholar]
- 32.Latos PA, Pauler FM, Koerner MV, et al. Airn transcriptional overlap, but not its lncRNA products, induces imprinted Igf2r silencing. Science. 2012;338:1469–72. doi: 10.1126/science.1228110. [DOI] [PubMed] [Google Scholar]
- 33.Schmitt AM, Garcia JT, Hung T, et al. An inducible long noncoding RNA amplifies DNA damage signaling. Nat Genet. 2016;48:1370–1376. doi: 10.1038/ng.3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schlackow M, Nojima T, Gomes T, et al. Distinctive Patterns of Transcription and RNA Processing for Human lincRNAs. Mol Cell. 2017;65:25–38. doi: 10.1016/j.molcel.2016.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hibi K, Nakamura H, Hirai A, et al. Loss of H19 imprinting in esophageal cancer. Cancer Res. 1996;56:480–2. [PubMed] [Google Scholar]
- 36.Moore T, Haig D. Genomic imprinting in mammalian development: a parental tug-of-war. Trends Genet. 1991;7:45–9. doi: 10.1016/0168-9525(91)90230-N. [DOI] [PubMed] [Google Scholar]
- 37.Leighton PA, Saam JR, Ingram RS, et al. An enhancer deletion affects both H19 and Igf2 expression. Genes Dev. 1995;9:2079–89. doi: 10.1101/gad.9.17.2079. [DOI] [PubMed] [Google Scholar]
- 38.Luo HL, Huang MD, Guo JN, et al. AFAP1-AS1 is upregulated and promotes esophageal squamous cell carcinoma cell proliferation and inhibits cell apoptosis. Cancer Med. 2016;5:2879–2885. doi: 10.1002/cam4.848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yao W, Bai Y, Li Y, et al. Upregulation of MALAT-1 and its association with survival rate and the effect on cell cycle and migration in patients with esophageal squamous cell carcinoma. Tumour Biol. 2016;37:4305–12. doi: 10.1007/s13277-015-4223-3. [DOI] [PubMed] [Google Scholar]
- 40.Zhang X, Xu Y, He C, et al. Elevated expression of CCAT2 is associated with poor prognosis in esophageal squamous cell carcinoma. J Surg Oncol. 2015;111:834–9. doi: 10.1002/jso.23888. [DOI] [PubMed] [Google Scholar]
- 41.Hu L, Wu Y, Tan D, et al. Up-regulation of long noncoding RNA MALAT1 contributes to proliferation and metastasis in esophageal squamous cell carcinoma. J Exp Clin Cancer Res. 2015;34:7. doi: 10.1186/s13046-015-0123-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wei G, Luo H, Sun Y, et al. Transcriptome profiling of esophageal squamous cell carcinoma reveals a long noncoding RNA acting as a tumor suppressor. Oncotarget. 2015;6:17065–80. doi: 10.18632/oncotarget.4185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang X, Li M, Wang Z, et al. Silencing of long noncoding RNA MALAT1 by miR-101 and miR-217 inhibits proliferation, migration, and invasion of esophageal squamous cell carcinoma cells. J Biol Chem. 2015;290:3925–35. doi: 10.1074/jbc.M114.596866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen FJ, Sun M, Li SQ, et al. Upregulation of the long non-coding RNA HOTAIR promotes esophageal squamous cell carcinoma metastasis and poor prognosis. Mol Carcinog. 2013;52:908–15. doi: 10.1002/mc.21944. [DOI] [PubMed] [Google Scholar]
- 45.Yan X, Hu Z, Feng Y, et al. Comprehensive Genomic Characterization of Long Non-coding RNAs across Human Cancers. Cancer Cell. 2015;28:529–40. doi: 10.1016/j.ccell.2015.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu W, Bhagat TD, Yang X, et al. Hypomethylation of noncoding DNA regions and overexpression of the long noncoding RNA, AFAP1-AS1, in Barrett's esophagus and esophageal adenocarcinoma. Gastroenterology. 2013;144:956–966 e4. doi: 10.1053/j.gastro.2013.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Qian Y, Gatesman AS, Baisden JM, et al. Analysis of the role of the leucine zipper motif in regulating the ability of AFAP-110 to alter actin filament integrity. J Cell Biochem. 2004;91:602–20. doi: 10.1002/jcb.10725. [DOI] [PubMed] [Google Scholar]
- 48.Baisden JM, Gatesman AS, Cherezova L, et al. The intrinsic ability of AFAP-110 to alter actin filament integrity is linked with its ability to also activate cellular tyrosine kinases. Oncogene. 2001;20:6607–16. doi: 10.1038/sj.onc.1204802. [DOI] [PubMed] [Google Scholar]
- 49.Huang C, Yu Z, Yang H, et al. Increased MALAT1 expression predicts poor prognosis in esophageal cancer patients. Biomed Pharmacother. 2016;83:8–13. doi: 10.1016/j.biopha.2016.05.044. [DOI] [PubMed] [Google Scholar]
- 50.Pan Z, Mao W, Bao Y, et al. The long noncoding RNA CASC9 regulates migration and invasion in esophageal cancer. Cancer Med. 2016;5:2442–7. doi: 10.1002/cam4.770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Huang B, Song JH, Cheng Y, et al. Long non-coding antisense RNA KRT7-AS is activated in gastric cancers and supports cancer cell progression by increasing KRT7 expression. Oncogene. 2016;35:4927–36. doi: 10.1038/onc.2016.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yang X, Song JH, Cheng Y, et al. Long non-coding RNA HNF1A-AS1 regulates proliferation and migration in oesophageal adenocarcinoma cells. Gut. 2014;63:881–90. doi: 10.1136/gutjnl-2013-305266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li S, Xu Y, Sun Z, et al. Identification of a lncRNA involved functional module for esophageal cancer subtypes. Mol Biosyst. 2016;12:3312–3323. doi: 10.1039/c6mb00101g. [DOI] [PubMed] [Google Scholar]
- 54.Tong YS, Wang XW, Zhou XL, et al. Identification of the long non-coding RNA POU3F3 in plasma as a novel biomarker for diagnosis of esophageal squamous cell carcinoma. Mol Cancer. 2015;14:3. doi: 10.1186/1476-4598-14-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hu HB, Jie HY, Zheng XX. Three Circulating LncRNA Predict Early Progress of Esophageal Squamous Cell Carcinoma. Cell Physiol Biochem. 2016;40:117–125. doi: 10.1159/000452529. [DOI] [PubMed] [Google Scholar]
- 56.Bejerano G, Pheasant M, Makunin I, et al. Ultraconserved elements in the human genome. Science. 2004;304:1321–5. doi: 10.1126/science.1098119. [DOI] [PubMed] [Google Scholar]
- 57.Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. 2011;12:861–74. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- 58.Baira E, Greshock J, Coukos G, et al. Ultraconserved elements: genomics, function and disease. RNA Biol. 2008;5:132–4. doi: 10.4161/rna.5.3.6673. [DOI] [PubMed] [Google Scholar]
- 59.Calin GA, Liu CG, Ferracin M, et al. Ultraconserved regions encoding ncRNAs are altered in human leukemias and carcinomas. Cancer Cell. 2007;12:215–29. doi: 10.1016/j.ccr.2007.07.027. [DOI] [PubMed] [Google Scholar]
- 60.Wojcik SE, Rossi S, Shimizu M, et al. Non-codingRNA sequence variations in human chronic lymphocytic leukemia and colorectal cancer. Carcinogenesis. 2010;31:208–15. doi: 10.1093/carcin/bgp209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shen H, Lu C, Jiang Y, et al. Genetic variants in ultraconserved elements and risk of breast cancer in Chinese population. Breast Cancer Res Treat. 2011;128:855–61. doi: 10.1007/s10549-011-1395-4. [DOI] [PubMed] [Google Scholar]
- 62.Yang R, Frank B, Hemminki K, et al. SNPs in ultraconserved elements and familial breast cancer risk. Carcinogenesis. 2008;29:351–5. doi: 10.1093/carcin/bgm290. [DOI] [PubMed] [Google Scholar]
- 63.Fassan M, Dall'Olmo L, Galasso M, et al. Transcribed ultraconserved noncoding RNAs (T-UCR) are involved in Barrett's esophagus carcinogenesis. Oncotarget. 2014;5:7162–71. doi: 10.18632/oncotarget.2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ingravallo G, Dall'Olmo L, Segat D, et al. CDX2 hox gene product in a rat model of esophageal cancer. J Exp Clin Cancer Res. 2009;28:108. doi: 10.1186/1756-9966-28-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Braconi C, Valeri N, Kogure T, et al. Expression and functional role of a transcribed noncoding RNA with an ultraconserved element in hepatocellular carcinoma. Proc Natl Acad Sci U S A. 2011;108:786–91. doi: 10.1073/pnas.1011098108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lujambio A, Portela A, Liz J, et al. CpG island hypermethylation-associated silencing of non-coding RNAs transcribed from ultraconserved regions in human cancer. Oncogene. 2010;29:6390–401. doi: 10.1038/onc.2010.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Paparidis Z, Abbasi AA, Malik S, et al. Ultraconserved non-coding sequence element controls a subset of spatiotemporal GLI3 expression. Dev Growth Differ. 2007;49:543–53. doi: 10.1111/j.1440-169X.2007.00954.x. [DOI] [PubMed] [Google Scholar]
- 68.Jin Y, Jin C, Law S, et al. Cytogenetic and fluorescence in situ hybridization characterization of clonal chromosomal aberrations and CCND1 amplification in esophageal carcinomas. Cancer Genet Cytogenet. 2004;148:21–8. doi: 10.1016/s0165-4608(03)00213-9. [DOI] [PubMed] [Google Scholar]
- 69.Paulson TG, Maley CC, Li X, et al. Chromosomal instability and copy number alterations in Barrett's esophagus and esophageal adenocarcinoma. Clin Cancer Res. 2009;15:3305–14. doi: 10.1158/1078-0432.CCR-08-2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zaninotto G, Minnei F, Guirroli E, et al. The Veneto Region's Barrett's Oesophagus Registry: aims, methods, preliminary results. Dig Liver Dis. 2007;39:18–25. doi: 10.1016/j.dld.2006.09.021. [DOI] [PubMed] [Google Scholar]
- 71.Morales CP, Lee EL, Shay JW. In situ hybridization for the detection of telomerase RNA in the progression from Barrett's esophagus to esophageal adenocarcinoma. Cancer. 1998;83:652–9. [PubMed] [Google Scholar]
- 72.Lord RV, Salonga D, Danenberg KD, et al. Telomerase reverse transcriptase expression is increased early in the Barrett's metaplasia, dysplasia, adenocarcinoma sequence. J Gastrointest Surg. 2000;4:135–42. doi: 10.1016/s1091-255x(00)80049-9. [DOI] [PubMed] [Google Scholar]
- 73.Souza RF, Lunsford T, Ramirez RD, et al. GERD is associated with shortened telomeres in the squamous epithelium of the distal esophagus. Am J Physiol Gastrointest Liver Physiol. 2007;293:G19–24. doi: 10.1152/ajpgi.00055.2007. [DOI] [PubMed] [Google Scholar]
- 74.Merchant NB, Dutta SK, Girotra M, et al. Evidence for enhanced telomerase activity in Barrett's esophagus with dysplasia and adenocarcinoma. Asian Pac J Cancer Prev. 2013;14:679–83. doi: 10.7314/apjcp.2013.14.2.679. [DOI] [PubMed] [Google Scholar]


