Abstract
Although muscle atrophy is a serious problem during spaceflight, little is known about the sequence of molecular events leading to atrophy in response to microgravity. We carried out a spaceflight experiment using Caenorhabditis elegans onboard the Japanese Experiment Module of the International Space Station. Worms were synchronously cultured in liquid media with bacterial food for 4 days under microgravity or on a 1-G centrifuge. Worms were visually observed for health and movement and then frozen. Upon return, we analyzed global gene and protein expression using DNA microarrays and mass spectrometry. Body length and fat accumulation were also analyzed. We found that in worms grown from the L1 larval stage to adulthood under microgravity, both gene and protein expression levels for muscular thick filaments, cytoskeletal elements, and mitochondrial metabolic enzymes decreased relative to parallel cultures on the 1-G centrifuge (95% confidence interval (P⩽0.05)). In addition, altered movement and decreased body length and fat accumulation were observed in the microgravity-cultured worms relative to the 1-G cultured worms. These results suggest protein expression changes that may account for the progressive muscular atrophy observed in astronauts.
Introduction
Spaceflight-induced muscle atrophy poses a significant risk for astronaut health and mission performance. Accordingly, it is a major obstacle for manned deep-space exploration. In contrast to observations on the Earth, exercise countermeasures employed during spaceflight have proven ineffective for maintaining skeletal muscle mass.1,2 Although the optimal exercise and nutritional strategies might simply have not yet been developed, there is growing evidence that muscle may be intrinsically sensitive to the effects of spaceflight per se. For example, vascular and cardiac muscles undergo atrophy, without reported muscle unloading,3 and cultured embryonic muscle synthesizes less protein as a direct consequence of spaceflight.4 However, the molecular events regulating these changes remain unclear.
Here we aimed to identify the mechanisms that are triggered in response to microgravity using the nematode Caenorhabditis elegans. In this space experiment (CERISE; C. elegans RNA Interference in Space Experiment), we synchronously cultured L1 larvae-stage animals to adulthood in liquid media for 4 days either in microgravity or 1-G (gravitational acceleration unit) centrifuge conditions onboard the Japanese Experiment Module of the International Space Station (ISS).5,6 The nematode larvae were launched to the ISS onboard the Space Shuttle Atlantis STS-129 on 16 November 2009, cultures were initiated on 20 November 2009, and the post-cultivation frozen samples were returned by the Space Shuttle Endeavour STS-130 on 21 February 2010.
C. elegans is an ideal model organism for studies of the physiological effects of space environments because of its small size, short life cycle, and because of its common use on Earth as a model organism for human medical pathologies. The first examinations of the effects of the space environment, including cosmic radiation, on C. elegans were performed using essentially standard laboratory culturing techniques with agar plates. C. elegans can mate, reproduce, and undergo embryonic development during spaceflight, and radiation-induced mutations can be monitored during spaceflight experiments.7–10 During the Dutch Soyuz mission DELTA to the ISS in April 2004, an international collaboration of laboratories carried out the ‘‘the first International C. elegans Experiment in Space’’ (ICE-First). In the ICE-First experiment, we found that muscle-, metabolic-, and aging-related genes showed 10–20% lower expression levels in response to spaceflight.11–14 These studies were carried out in a chemically defined medium, which allowed study of the effects of surface tension in flight but which significantly alters C. elegans life history in a manor akin to dietary restriction.15
In this study, we wished to confirm that gene and protein expression changes observed in ICE-FIRST were not unique to the operational issues associated with that spaceflight, were not owing to the use of a non-standard laboratory diet, and were owing to exposure to microgravity. To achieve these goals we flew worms on a different launch system, cultured them in a different segment of the ISS, froze them on orbit, used the standard nutritionally rich bacterial diet, and cultured them both in microgravity and on a 1-G centrifuge onboard the ISS. In our previous study ICE-First, worms exposed to microgravity showed ~20% lower expression of thick filament components such as myosin heavy chain and paramyosin upon return to Earth. In the current study CERISE, changes in the expression of these same genes were found in the worms grown in microgravity versus in those grown at 1 G (changes were 53–71%). These results confirm that the past reported gene and protein expression changes for C. elegans during spaceflight are not unique to specific operational constrains or diet, and are owing to microgravity. In addition to alterations in cytoskeletal genes we have observed reproducible changes in metabolic genes including sirtuin, which is also known to influence ageing. Consistent with the gene and protein expression changes we also found lower fat accumulation and shorter body length in the microgravity-exposed worms. Last, alterations in the movement of the microgravity worms were observed. These results demonstrate a reproducible set of C. elegans gene and protein expression changes in response to microgravity and provide the basis for future mechanistic studies of how microgravity impacts muscle alteration and metabolism in C. elegans and possibly astronauts.
Results
Proteomic and transcriptomic analysis of space-flown C. elegans
To identify proteins that were differentially expressed in the nematodes during spaceflight, we quantitatively analyzed the expression levels of 475 proteins by matrix-assisted laser desorption/ionization time-of-flight tandem mass spectrometry. This analysis revealed significantly increased expression of 16 proteins and significantly decreased expression of 43 proteins in microgravity (P⩽0.05). Of the 16 proteins with increased expression, the expression of ASP-1 (aspartic protease) was increased 1.89-fold (log2[μG/1 G]=0.92) with the remaining proteins only slightly increased; these include proteins involved in protein synthesis such as ribosomal proteins and translation elongation factors (Figure 1, Supplementary Table S1). As expected, cytoskeletal proteins, including those thought as muscle proteins, were among the significantly decreased proteins. These include the structural proteins UNC-54 (myosin heavy chain, 0.87-fold (log2[μG/1 G]=−0.20)) and UNC-15 (paramyosin, 0.91-fold (log2[μG/1 G]=−0.14)), and MLC-3 (myosin light chain, 0.81-fold (log2[μG/1 G]=−0.30); Figure 1, Supplementary Table S1). UNC-87 and DIM-1, which are essential for the maintenance of muscle integrity and function,16,17 were also decreased by the same extent in microgravity (UNC-87: −0.79-fold (log2[μG/1 G]=−0.34), DIM-1: −0.89-fold (log2[μG/1 G]=−0.17). Gene expression analysis confirmed the downregulation of muscle-related proteins such as the myosin heavy chains, troponins, and intermediate filaments in the microgravity-cultured worms (Figure 1, Supplementary Table S2). Importantly, the pattern of downregulation of expression of muscle genes such as myosin heavy-chain genes myo-3 (log2[μG/1 G]=−0.89) and unc-54 (log2[μG/1 G]=−0.49) as well as the paramyosin gene unc-15 (log2[μG/1 G]=−0.62) were reproducibly observed in both the current experiment and the previous flight experiment (Table 1).
Figure 1.
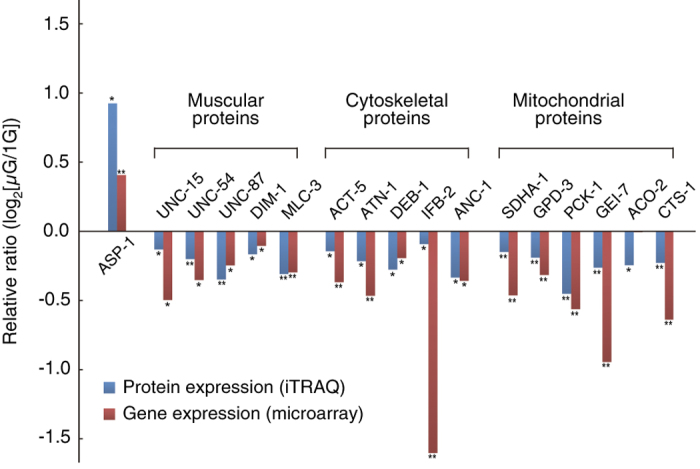
Quantitative analysis of protein (iTRAQ) and gene expression (microarray) levels in space-flown worms. The levels of certain muscular, cytoskeletal, and mitochondrial proteins and an aspartic protease (ASP-1) were significantly altered in worms cultured under microgravity conditions. *P⩽0.05, **P⩽0.01, Student’s t-test for analyses (details are shown in Supplementary Tables S1 and S2).
Table 1. Reproducible gene expression changes of myosin, paramyosin, unc-87, and dim-1 are induced by spaceflight.
| Gene name | Description |
CERISE (current experiment)
|
ICE-First (previous experiment)
|
||
|---|---|---|---|---|---|
| Expression change log2[μG/1 G] | t-test P-value | Expression change log2 [μG/1 G] | t-test P-value | ||
| myo-3 | Myosin heavy chain A | −0.89 | 0.00 | −0.34 | 0.00 |
| unc-54 | Myosin heavy chain B | −0.49 | 0.01 | −0.45 | 0.01 |
| unc-15 | Paramyosin | −0.62 | 0.01 | −0.33 | 0.04 |
| unc-87 | Maintain the myofilament structure | −0.25 | 0.05 | −0.34 | 0.00 |
| dim-1 | Required for muscle assembly | −0.11 | 0.03 | −0.53 | 0.00 |
Abbreviations: CERISE, C. elegans RNA Interference in Space Experiment; ICE-First, the first International C. elegans Experiment in Space.
CERISE and ICE-First experiments were carried out in 2009 and 2004, respectively. The worms were synchronously cultivated in a bacterial diet for 4 days from L1 larvae in CERISE. Adult worms frozen on ISS were used for the microarray analysis. In ICE-First, the worms were non-synchronously cultivated in a chemically defined diet for 10 days. Mixed stage worms frozen within 30 min of Soyuz landing were used for the analysis. Microarray analysis was performed in triplicate (n=3).
Other cytoskeletal components also displayed decreased expression during culture in microgravity, for example ACT-5 (actin), ATN-1 (α-actinin), DEB-1 (vinculin), IFB-2 (intermediate filament), and ANC-1 (nuclear and mitochondrial anchorage protein; Figure 1, Supplementary Table S1). In addition, significantly reduced levels of metabolic proteins were observed during culture in microgravity, for example, components involved in glycolysis (GPD-3), gluconeogenesis (PCK-1: phosphoenolpyruvate carboxykinase), and the glyoxylate cycle, a variation of the tricarboxylic acid (TCA) cycle (GEI-7) (Figure 1, Supplementary Table S1). Moreover, the levels of components of the electron transport chain (SDHA-1) and the TCA cycle (ACO-2 and CTS-1) also significantly decreased during spaceflight (Figure 1, Supplementary Table S1). DNA microarray analysis confirmed these decreased protein expression levels in response to microgravity (Figure 1) and suggested other energy metabolism gene expression level changes in microgravity (Supplementary Table S3). For example, genes encoding NADH dehydrogenase (complex I) and succinate dehydrogenase (complex II) displayed lower gene expression levels in the worms cultured in microgravity versus those cultured on the 1-G centrifuge (complex I: log2[μG/1 G]=−0.18 to −1.03, complex II: log2[μG/1 G]=−0.56 to −0.76). Similarly, ATP synthase expression was down (log2[μG/1 G]=−0.38 to −0.76).
Whereas DNA microarray analysis suggests that major muscle components, cytoskeletal elements, metabolic genes, and mitochondrial electron transport genes largely are downregulated in microgravity, some genes in these broad classes appear to be upregulated (Supplementary Table S2 and S3). Notably, a sirtuin gene, sir-2.1 encoding SIRT-1 protein, was upregulated. Sirtuin proteins deacetylate histones and several transcriptional factors, resulting in the activation or repression of target genes, and the sirtuin family may act as a global regulator of tissue health in response to diet and other stimuli.18–20 It was reported that sir-2.1 is involved in longevity and anti-aging of C. elegans,21 and that abu-6, abu-7, and pqn-5 are downstream targets that are negatively regulated by sir-2.1.22 Our microarray data indicate not only that the expression level of sir-2.1 significantly increases in microgravity but also that the downstream target gene expression levels significantly decrease under microgravity condition (Table 2); notably, significant changes in the same directions of expression alteration were also noted on our previous flight. Real-time quantitative PCR (qPCR) analysis confirmed the increased expression of sir-2.1 and the decreased expression of downstream genes, which are negatively regulated by sir-2.1, including abu-6, abu-7, and pqn-5 (Figure 2).
Table 2. Reproducible gene expression changes of sirtuin and a subset of its downstream target genes.
| Gene name | Description |
CERISE (current experiment)
|
ICE-First (previous experiment)
|
||
|---|---|---|---|---|---|
| Expression change log2 [μG/1 G] | t-test P-value | Expression change log2 [μG/1 G] | t-test P-value | ||
| sir-2.1 | Yeast SIR-related family member | 0.53 | 0.00 | 0.03 | 0.67 |
| abu-7 | Activated in blocked unfolded protein response | −1.24 | 0.00 | −0.63 | 0.00 |
| abu-6 | −2.18 | 0.00 | −0.78 | 0.00 | |
| pqn-78 | Prion-like-(Q/N-rich)-domain-bearing protein | −3.08 | 0.00 | −0.80 | 0.00 |
| pqn-5 | −4.18 | 0.00 | −0.90 | 0.00 | |
| fat-7 | Fatty acid desaturase | −0.38 | 0.01 | ND | — |
| lbp-6 | Lipid-binding protein | −0.47 | 0.00 | −0.61 | 0.00 |
Abbreviations: CERISE, C. elegans RNA Interference in Space Experiment; ICE-First, the first International C. elegans Experiment in Space; ND, not determined.
CERISE and ICE-First experiments were carried out in 2009 and 2004, respectively. The worms were synchronously cultivated in a bacterial diet for 4 days from L1 larvae in CERISE. Adult worms frozen on ISS were used for the microarray analysis. In ICE-First, the worms were non-synchronously cultivated in a chemically defined diet for 10 days. Mixed stage worms frozen within 30 min of Soyuz landing were used for the analysis. A sirtuin gene, sir-2.1, was significantly upregulated under microgravity. As expected, negatively regulated downstream target genes, abu and pqn families displayed complementary decreases in expression. Microarray analysis was performed in triplicate (n=3).
Figure 2.
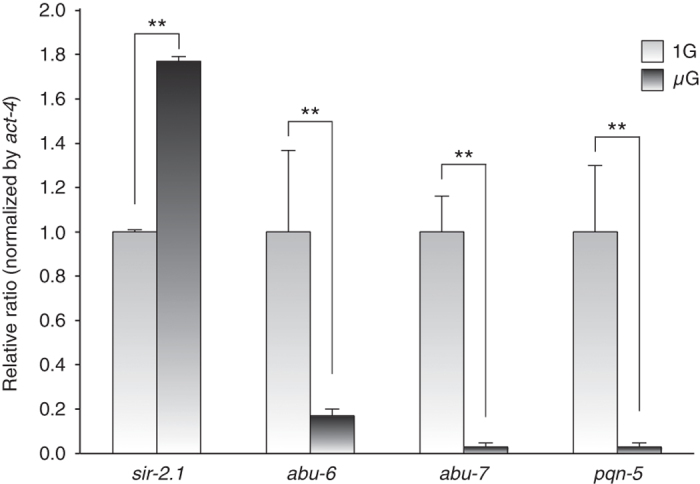
Altered gene expression of the sirtuin gene transcript (sir-2.1) and downstream target gene transcripts assessed by quantitative real-time PCR. The expression of sir-2.1 increases in the worms grown in microgravity and SIR-2.1 downstream target genes, abu-6, abu-7, and pqn-5, decrease as expected. Signals were normalized to act-4, which encodes an actin isoform, as a housekeeping gene. **P⩽0.01, Student’s t-test.
Body length and fat accumulation of nematodes cultured in microgravity
As proteomic and microarray analyses demonstrated decreased muscle and metabolic gene expression in microgravity, body lengths of a small subset of worms not used for omic analyses were microscopically measured. The body length of worms cultured in microgravity were slightly but significantly decreased by ~5.5% versus worms cultured on the centrifuge (μG: 1.37±0.053 mm, 1 G: 1.45±0.094 mm, n=15 worms per group, P⩽0.05; Figure 3). The data from the omics analyses suggest that the space-flown worms had lower energy metabolism capacity. Several molecules involved in fat metabolism are downstream targets of sirtuin signaling.23 Our DNA microarray analyses indicated that microgravity induced the decreased expression of fat genes encoding fatty acid desaturases24 and lbp genes that are involved in lipid binding activity (Figure 4c). The sir-2.1 downstream target genes fat-7 and lbp-6 were among the downregulated transcripts (log2[μG/1 G]=−0.38 (P=0.01) and −0.47 (P=0.00), respectively). Therefore, we assessed lipid stores in a small subset of worms not used for omic analysis. Sudan Black staining indicated that the accumulation of fat in microgravity-cultured nematodes was significantly reduced compared with 1-G cultured worms (Figure 4a,b).
Figure 3.
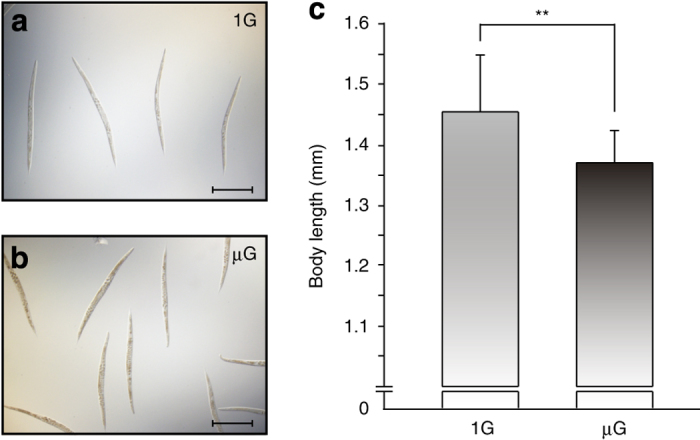
Alteration of body length of space-flown C. elegans. (a) Microscopic image of worms grown on the 1 G centrifuge for 4 days. (b) Microscopic image of worms grown in microgravity for 4 days. The worms in both conditions had normal levels of eggs in their bodies. (c) The mean of body length of the worms grown at 1 G was 1.45±0.09 mm, and for microgravity was 1.37±0.05 mm, respectively (n=15, means±s.d.). **P⩽0.01, Welch’s t-test. Scale bars indicate 0.5 mm.
Figure 4.
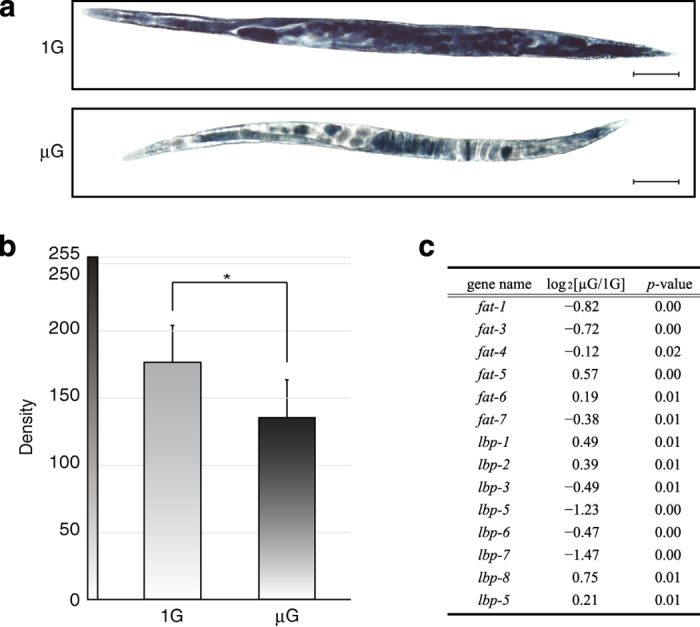
Altered fat storage in microgravity-cultured C. elegans. (a) Images of Sudan Black staining of accumulated fat in C. elegans. Worms grown onboard the 1 G centrifuge (upper image) and in microgravity (lower image) are displayed. Scale bars indicate 100 μm. There was less fat accumulation in microgravity-cultured worms compared with 1 G controls. (b) Density measurements of Sudan Black stained worms (as described in Methods). The mean densities were 135.36±28.12 in microgravity-cultured worms and 176.55±27.61 in 1-G cultured worms (means±s.d.). *P⩽0.05, Student’s t-test. (c) Alteration of fat-related gene expression determined by microarray analysis. P-values as indicated in the panel, Student’s t-test.
Effect of microgravity on swimming behavior of C. elegans
To examine the swimming behavior changes of C. elegans during spaceflight, we analyzed the swimming motion of worms grown under microgravity or 1 G on the ISS for 4 days. As shown in Table 3, wavelength showed no difference between microgravity and 1-G cultured worms. However, frequency of swimming was significantly different, and this likely caused a significant difference in wave velocity. Amplitude was measured at the point of 0.1 L, and no significant difference was observed. These results suggest that microgravity induced lower beating frequency and slower wave velocity without changing the shape feature of movement, i.e., the movement of the worm became slower under microgravity.
Table 3. Characteristics of moving behaviors of space-flown nematodes.
| Culture condition | μG (n=6) | 1 G (n=3) | P-value a |
|---|---|---|---|
| Wavelength (Lb) | 2.29±0.71 | 2.07±0.30 | >0.1 |
| Frequency (Hz) | 1.53±0.64 | 2.61±0.12 | <0.01 |
| Wave velocity (L/s) | 3.19±0.76 | 5.39±0.64 | <0.01 |
| Amplitudec (rad/L) | 4.09±0.76 | 4.75±0.40 | >0.1 |
Statistical test was an unequal sample sizes and unequal variance t-test. Wavelength and amplitude showed no significant difference but frequency and wave velocity showed significant difference with P⩽0.01.
‘L’ indicates the distance from the tip of the head to the tail end of individual worm.
Amplitude is measured at the point of distance s=0.1 L.
Discussion
It is well known that microgravity exerts considerable effects on physiological processes. Muscle atrophy is one of the main concerns for astronauts during spaceflight. To counter atrophy, astronauts are forced to perform several exercises such as using an ergometer. In 2004, we had the opportunity to conduct an experiment using the nematode C. elegans on the ISS. Nematodes flown in microgravity for ten days exhibited reduced gene expression of muscle-related molecules.11,25 Both gene and protein expression levels of myosin heavy chain and paramyosin, main components of thick filaments in invertebrates, significantly decreased in space-flown worms. In the present study, we also observed lower expression levels of both the major components for myofilament assembly such as myosin heavy chain and paramyosin. This repeated observation suggests that these changes are caused by spaceflight and not operational, technical, or dietary differences between these two spaceflight experiments.
In our previous study, ICE-First, we found small (10–20%) decreases in expression of muscle-, metabolic-, and aging-related genes in response to ten day spaceflight.11–14 In the present study the magnitude of change in the majority of statistically significant gene expression changes is also small. Although the reproducibility of gene and protein expression changes across these two flights increases confidence in these changes being due to spaceflight, it does raise the question of whether such small changes in gene expression are physiologically relevant. People display a loss of roughly 3–10% of muscle cross sectional area in response to a week of spaceflight or bedrest.26 Thus, one would anticipate relatively small-scale changes in gene and protein expression. Data from rodent studies suggest that this is likely true. In hindlimb-unloaded rats displaying a 60% reduction muscle wet weight, the majority of significantly altered transcripts displayed less than a 50% alteration.27 Conversely, in rats displaying a 6% increase in muscle fiber cross sectional area in response to focal adhesion kinase overexpression, a 9% increase in MHC1 was observed.28 In both rodent studies, the small-scale changes in gene expression appear to be physiologically relevant as they have the potential to explain the known fiber-type switching and altered metabolism observed in response to “unloading” atrophy and “loading” hypertrophy. While on a small scale, our observations of body size, fat content, and locomotion are all consistent with the predicted physiologic impact(s) of the altered gene and protein expression we have observed in microgravity. These phenotypes, and others such as strength, ATP production, and mitochondrial networking in muscle seem logical and necessary measurements to make in future mechanistic/interventional flight experiments. Thus, while the presently observed gene expression changes are of a small magnitude, the fact they translate to reduced expression of their encoded protein, and as we observe alterations in the physiological processes they would be predicted to regulate, suggest these expression differences are physiologically meaningful. Clearly, however, future interventional studies looking at both omic and physiologic changes, preferably in the same animals, are required in order to make firm mechanistic links.
Another difference between our previous study and the current study is the successful use of an onboard 1-G centrifuge. We have compared our microgravity data with the onboard 1-G control in order to isolate the effects of microgravity alone. This comparison allows us to confirm that reproducible changes between ICE-First and the present study are due primarily to the removal of mechanical factors during microgravity exposure and not to other environmental differences (for example, oxygen or radiation). Although we were primarily interested in attempting to demonstrate microgravity effects, it is entirely possible that others may be interested in comparing our flight data directly to the ground control, we simply have not done that here in order to make the presentation easier to follow. Similarly, it is important to note that because our observations suggest that altered mechanical loading of muscle is responsible for the gene expression changes we detect by microarray analysis and because altered mechanical loading of muscle is well documented to alter cytoskeletal and metabolic gene expression in human and rodent muscle, use of common normalization genes/proteins such as actin and GAPDH that decrease in expression in response to spaceflight (Supplementary Tables S2 and S3) may result in under estimation of decreased expression changes and over estimation of increased expression change in other genes/proteins when used in PCR and/or western analysis of space-flown samples. It is precisely for this reason that microarray and proteomic data are not normalized to any single gene/protein in this study. We have, however, used actin for normalization in the confirmatory qPCR studies fully aware of this limitation. This choice was made as actin is a well validated normalization that provides consistently reproducible results and as it shows less of a decrease by microarray analysis than GAPDH.
Muscle cells in C. elegans are mechanically linked to the cuticle through transepidermal attachment, and intermediate filaments, which display decreased expression in response to microgravity (Figure 1, Supplementary Table S1) are a component of these attachments.29,30 Transepidermal attachments are specialized cell–matrix attachments that span epidermal cells, connecting muscles via basement membrane to the apical cuticle; they are restricted to epidermal regions that overlie muscle cells, and their formation occurs concurrently with muscle development.29 The reduced gene expression of intermediate filaments could reasonably account for decreased mechanical loading of muscle and subsequent gene expression changes. This is consistent with the previous speculation that changes in expression of integrin attachment complexes at the muscle surface, which connect to the transepidermal attachments, underlie the gene expressions changes and movement alterations in response to microgravity.31,32 Interestingly, these same proteins are believed to be associated with muscle hypertrophy and atrophy in man32 and therefore might be targets for investigation and/or manipulation in astronauts. However, in the present study, we do not find reproducibly decreased the levels of all components of the attachment complexes (for example, pat-2 α-integrin expression decreases as expected but both ina-1 and pat-3 α- and β-integrin expression increase contrary to expectation (Supplementary Table S2)). Thus, as with most omics-based work further interventional studies are required to make mechanistic conclusions (for example, pharmacologically or genetically manipulating these complexes in flight).
Other cytoskeletal components including ATN-1, DEB-1, ACT-5, and ANC-1 also display altered expression (Figure 1, Supplementary Table S1) and this agrees with the previous observation that microgravity causes disorganization of the cytoskeletal network in cultured cells.33 This suggests that the positions of nuclei and mitochondria in all cells may be supported by the cytoskeletal network to counteract the force of gravity. Interestingly, the mammalian ANC-1 homologs SYNE-1/Nesprin-1 and -2 were shown to be involved in the pathogenesis of Emery Dreifuss muscular dystrophy.34 Thus, loss of mechanical sensation by muscle in flight may be via intramuscular mechanisms such as ANC-1, speculation that again calls for future interventional studies in flight.
In addition to reproducible changes in muscle gene expression we have found reproducible changes, in the expression of sirtuin and downstream targets (Table 2), and in several major enzymes in the TCA cycle (Figure 5 and the microarray analyses of the ICE-First experiment (GSE71771)).
Figure 5.
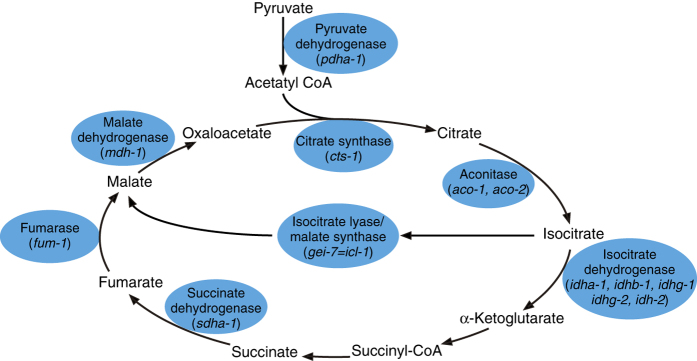
Schematic of the TCA cycle and the expression changes under microgravity. The enzymes indicated by blue symbols exhibited lower gene and/or protein expression in space-flown worms. Almost all enzymes (citrate synthase, aconitase, isocitrate dehydrogenase, succinate dehydrogenase, fumarase, malate dehydrogenase) participated in the TCA cycle showed lower expression.
As with the muscle gene expression changes the alteration of sirtuin and downstream targets as well as TCA cycle components appears to be owing to microgravity and not specific to idiosyncrasies of an individual spaceflight or owing to diet. It has been reported that the mammalian SIRT-1 protein may have novel role in muscle mitochondrial turnover and biogenesis.35,36 Similar to our observations with space-flown C. elegans, a previous study found that SIRT-1 activity is increased, whereas mitochondrial content is decreased in hindlimb muscle following denervation.37 In addition, a negative relationship between SIRT-1 protein content and oxidative capacity across diverse fiber types in rodent muscles has been reported.38 Thus, it is possible that muscle gene expression changes in response to microgravity are driven by SIRT-1-based remodeling of muscle. We suspect that the relatively small expression changes in sirtuin versus large downstream effects in target genes occur because sirtuin acts as a master regulatory gene and is therefore expected to be under tight control and that small changes would result in larger amplified effects in downstream-regulated genes. However, while abu/pqn families are located downstream of sir-2.1, they also interact with other pathways. For example, abu-6 is negatively regulated by unc-95,39 and abu-7 and pqn-5 are negatively regulated by eri-1 and arr-1, which interestingly were upregulated under microgravity (eri-1: log2[μG/1 G]=0.43 (P=0.00), arr-1: log2[μG/1 G]=0.70 (P=0.00)). Thus, future flight studies with sir-2.1 mutants seem required provide mechanistic insight.
As we found that most of the enzymes involved in the TCA cycle were downregulated in microgravity, it may be that turnover of the cycle is lower during space flight (Table 4). Rahman et al.40 reported that downregulation of the TCA cycle lead to a G2-like arrest of the one-cell stage embryo by preventing the activation of cyclin B-cyclin-dependent kinase, Cdk1. However, we did not find growth differences between μG and 1 G. This might be explained by our observation that the cyb family encoding cyclin B was upregulated 1.6- to 1.9-fold and cdk-1 1.4-fold in microgravity. Thus, upregulation of cyclin and cdk-1 may counter the effects of TCA cycle downregulation in microgravity. Edwards et al.41 reported that the TCA cycle is a key determinant of longevity, and malate-mediated lifespan extension requires SIR-2.1. Honda et al.13 reported that the downregulation of genes involved in longevity-induced slower ageing, and the upregulation of sir-2.1 was observed in this study. These observations may suggest increased longevity as the result of lower turnover of the TCA cycle during space flight. Future experiments directly measuring lifespan and metabolism of C. elegans in microgravity are required to address this hypothesis.
Table 4. Downregulated gene expression of the TCA cycle components.
| Gene name | Coding protein | Expression change log2[μG/1 G] | t-test P-value |
|---|---|---|---|
| pdha-1 | Pyruvate dehydrogenase (Lipoamide) α1 | −0.45 | 0.00 |
| C04C3.3 | Pyruvate dehydrogenase complex E1 β subunit | −0.36 | 0.00 |
| dld-1 | Dihydrolipoamide dehydrogenase | −0.54 | 0.00 |
| cts-1 | Citrate synthase | −0.76 | 0.00 |
| aco-1 | Aconitase | −0.32 | 0.00 |
| aco-2 | Aconitase | −0.14 | 0.01 |
| idha-1 | NAD+-dependent mitochondrial isocitrate dehydrogenase α subunit | −0.64 | 0.00 |
| idhb-1 | NAD+-dependent mitochondrial isocitrate dehydrogenase β subunit | −0.38 | 0.00 |
| idhg-1 | NAD+-dependent mitochondrial isocitrate dehydrogenase γ subunit | −0.30 | 0.00 |
| idhg-2 | NAD+-dependent mitochondrial isocitrate dehydrogenase γ subunit | −0.56 | 0.00 |
| idh-2 | Mitochondrial isocitrate dehydrogenase | −0.30 | 0.00 |
| gei-7 | Isocitrate lyase/malate synthase | −1.09 | 0.00 |
| dlst-1 | Dihydrolipoamide S-succinyltransferase | −0.43 | 0.01 |
| dld-1 | Dihydrolipoamide dehydrogenase | −0.54 | 0.00 |
| sdha-1 | Succinate dehydrogenase complex, subunit A | −0.67 | 0.01 |
| sdhb-1 | Succinate dehydrogenase complex, subunit B | −0.56 | 0.00 |
| mev-1 | Succinate dehydrogenase complex, subunit C | −0.76 | 0.00 |
| sdhd-1 | Succinate dehydrogenase complex, subunit D | −0.60 | 0.00 |
| fum-1 | Fumarase | −0.56 | 0.00 |
| mdh-1 | Malate dehydrogenase | −0.38 | 0.00 |
Last, the TCA cycle and electron transport chain are localized to mitochondria. Several previous studies have reported and reviewed the critical influence of the cellular mitochondrial distribution and the cytoskeletal network on mitochondrial function and movement.42,43 Here we have observed that microgravity weakens the cytoskeletal network, including major muscle proteins, and switches mitochondrial metabolism to an energy-saving mode. In mammals, this would lead to specific atrophy of type-1 muscle fibers, and indeed type-1 slow oxidative muscle fibers experience the greatest level of atrophy in humans in space.1 A key open question is do cytoskeletal alterations drive mitochondrial alterations, do mitochondrial alterations drive cytoskeletal alterations, or is there a coordinated co-alteration of these two sets of proteins that underlies C. elegans’ muscular response to spaceflight? Answering this question is currently technically feasible by flying various mutants and, once answered, would allow testing of whether this same mechanism is acting in astronauts.
Materials and methods
Spaceflight experiment
To synchronize the growth of the worms, an egg preparation was made in a laboratory at the Kennedy Space Center. The N2 Bristol (wild-type) strain was used. Eggs were prepared using the alkaline bleach method with 0.5 M KOH and 1.0% NaClO and incubated overnight in M9 buffer containing 5 mg/l cholesterol (M9C) at 20 °C. The hatched L1 larvae were then prepared to a concentration of 9,000 per 2 ml of M9C and loaded into one end of a polyethylene bag. Volume of 12 ml of S-basal media suspended Escherichia coli (OD600=5) were separately packed into the other end of the polyethylene bag with separation of the two ends of the bag achieved via a U-pin (Supplementary Figure S1). The prepared worms were launched by STS-129 (16 November 2009). A full description of the experimental procedures and flight hardware, including illustrations, employed in this experiment can be found in previous work.5,6 A summary illustration appears in Supplementary Figure S1. Cultures were exposed to one of two experimental conditions onboard the ISS: microgravity or 1-G centrifugation. All L1 larvae were maintained in stasis within M9C until the initiation of the experiment. The experiment was activated once onboard the ISS by reintroducing the worms to bacterial food. All experiments were carried out for 4 days at 20 °C in the Cell Biology Experiment Facility within the ISS. Nutrition and therefore maintained growth synchronization were improved compared with those in our previous spaceflight experiments, which had used a chemically defined medium.11–14,44 This was confirmed by observing the worms via light microscopy, installed on the Clean Bench on the ISS, immediately prior to termination of culturing. The 4-day cultures were confirmed to have grown to synchronous adulthood in both the microgravity and onboard 1-G centrifuge cultures. Movies of the microscopic observation were filmed and downlinked from the ISS to the JAXA Tsukuba control center. The cultivation was terminated by freezing with subsequent storage at −80 °C in the freezer installed in the ISS (MELFI: the −80 °C laboratory freezer for ISS). All frozen samples were returned by STS-130 (21 February 2010).
Proteomic analyses
Frozen worms were thawed on ice, 300 adult worms were selected under a microscope with a cooled stage and were washed with ice-cold M9 buffer to exclude larvae and the bacterial food. Total proteins were extracted from 100 adult hermaphrodites in 20 μl 5 M urea, 2 M thiourea, and 0.5% CHAPS with sonication at five 5-s intervals on ice. The proteins extracted from 100 worms grown in microgravity were 3.32±0.11 μg, and 3.05±0.23 μg for the worms grown onboard the 1 G centrifuge. The proteins were normalized by bias correction on ABI 5800 LC-MALDI-TOF MS/MS system (AB Sciex, Framingham, MA, USA). The crude extracts were denatured, reduced, and digested with trypsin overnight at 37 °C. The digested peptides were labeled with iTRAQ Reagents (AB Sciex) according to the manufacturer’s instructions. Each peptide was quantitatively analyzed using the ABI 5800 system. The experiments were independently performed in triplicate with statistical significance (P) determined using a Student’s t-test.
Extraction of total RNA
Frozen worms were thawed on ice, 1,000 adult worms were selected under a microscope with a cooled stage and were washed with ice-cold M9 buffer to exclude larvae and the bacterial food. Washed worms were homogenized with four rounds of 20-s beating at 5,000 r.p.m. in a Micro Smash MS-100 bead beater (Tomy Seiko, Tokyo, Japan) with 3 min of cooling on ice between rounds. Total RNA was isolated using an Isogen Reagent (Nippon Gene, Tokyo, Japan) following the manufacturer’s protocol. The residual DNA was eliminated by treating with DNase I (Takara Bio, Shiga, Japan) during an isolation process of total RNA. The isolated total RNA was subjected to DNA microarray and real-time qPCR.
Microarray
The isolated RNA was analyzed on the C. elegans Oligo Microarray 44 k version 2.0 (Agilent Technologies, Santa Clara, CA, USA). Isolated total RNA was amplified using the Low Input Quick Amp Labeling Kit, one-color (Agilent Technologies). Cy3 labeled cRNA was prepared according to the manufacturer’s protocol. The labeled cRNA (1.65 μg/sample) was hybridized to a microarray using the Agilent Gene Expression Hybridization kit at 65 °C for 17 h. The hybridized microarrays were scanned with an Agilent high-resolution scanner G2505C and processed with Agilent Feature Extraction Software version 10.7.3.1. Biological triplicates were used for the microarray experiments. Microarray data were analyzed using Gene Spring software (Agilent Technologies). Normalized data were produced by “Normalize to 75 percentile shift protocol” and “baseline to median of control sample protocol”. Arrays were filtered on expression 20–100th percentile in raw data. All statistical analyses were carried out by a Student’s t-test with significance set at P⩽0.05. All global gene expression data are MIAME compliant and were deposited into the gene expression omnibus (GEO) database (accession numbers GSE71770).
Real-time quantitative PCR
Real-time qPCR was performed to measure the expression differences of genes. The isolated RNA was reverse-transcribed with the ExScript RT reagent kit (Takara Bio) and an iCycler thermal cycler (Bio-Rad Laboratories, Hercules, CA, USA). The reaction mixture for measuring the complementary DNA quantity was performed using Premix EX Taq (Takara Bio) according to the manufacturer’s instruction. Reaction and fluorescent monitoring were performed using Applied Biosystems 7500 Real-Time PCR System (Thermo Fischer Scientific, Waltham, MA, USA). The following primer pairs were used for PCR amplification: sir-2.1 (forward: 5′-TGGGAATGAGATTCGTGAGGAA-3′, reverse: 5′-ACTCCTTCGCAACGTTTGCAAT-3′), abu-6 (forward: 5′-AATACGACACCACCACTGAAGCT-3′, reverse: 5′-ATTGGGATGATTGAGCAACAGA-3′), abu-7 (forward: 5′-CCATTGCCTTCCTTGCATGT-3′, reverse: 5′-CCACACTGTCTCTTTTCACGGATT-3′), pqn-5 (forward: 5′-ACTTTCTATTGCGTTCCTTGCAT-3′, reverse: 5′-TCTCTTTTCACGGATTGCACTTC-3′), act-4 (forward: 5′-GACAGGTCATCACCGTTGCAA-3′, reverse: 5′-CGGCAGACTCCATACCCAAGA-3′). The qPCR data were normalized to act-4, which encodes an actin isoform, as a housekeeping gene. The analyses were performed in triplicate, and statistical significance was determined using a Student’s t-test.
Body-length measurement of space-flown worms
The body length of space-flown worms was measured quantitatively using an Olympus BX51 microscope system with a DP73 digital camera (Olympus, Tokyo, Japan). The observation was immediately performed after thawing the frozen samples. Fifteen worms were randomly selected from each condition (n=15). Significant differences were determined by a Welch’s t-test.
Sudan Black staining and microscopic observation
Sudan Black (Sigma-Aldrich, St Louis, MO, USA) staining of stored fat was performed as previously described45,46 with slight modification. A piece of frozen culture containing space-flown C. elegans was slowly thawed in an equal amount of 2% paraformaldehyde in M9 buffer, and then fixed for 30 min at room temperature. The fixed worms were then subjected to three freeze–thaw cycles, and then sequentially dehydrated through washes with 25, 50, and 70% ethanol. Staining was performed overnight in a saturated Sudan Black solution in 70% ethanol. At the end of the staining procedure, seven worms cultured in microgravity and five worms cultured onboard the 1-G centrifuge were recovered and analyzed. Bright-field images were obtained using an Olympus BX51 microscope system with a DP73 digital camera (Olympus, Tokyo, Japan). The obtained images were converted to 8-bit gray-scale images and the extent of staining was quantitatively measured using ImageJ software version 1.49 (National Institutes of Health, Bethesda, MD, USA). The distribution of density across the nematode was converted to histograms using the “Histogram” function on the software.
Analysis of movement in space-flown C. elegans
Moving behavior of C. elegans on the ISS was observed using the microscope installed on the Clean Bench on the ISS Japanese Experiment Module (http://iss.jaxa.jp/en/kiboexp/pm/cb/). To measure moving behavior, downlinked video images of swimming C. elegans were converted to dark-field images, and the positions of individuals were determined using Bohboh tracking software (Bohboh Soft, Tokyo, Japan).47 The means of frequency, phase, and amplitude were calculated from changes in distance at specific time intervals (30 f.p.s.). Considering the individual differences between worm body lengths (L), the wavelength of motion was calculated as a function of L. Curves were plotted as cosine functions of time fitted to measurements by the least squares method.38 Six frames were analyzed for microgravity culture (n=6) and three frames for 1-G culture (n=3). Statistical significance was determined by an unequal sample size and unequal variance t-test.
Acknowledgments
We are grateful to the entire crew of the CERISE for their work on STS-129, STS-130, and the ISS. The CERISE was organized with the support of the JAXA. This experiment was supported by the Cell Biology Experiment Project conducted by the Institute of Space and Astronautical Science in JAXA, and was funded in part by JSPS KAKENHI Grant Numbers 26506029, 15H05937, the Medical Research Council UK (G0801271), and National Institutes of Health (NIH NIAMS ARO54342).
Footnotes
The authors declare no conflict of interest.
References
- Fitts, R. H. , Riley, D. R. & Widrick, J. J. Functional and structural adaptations of skeletal muscle to microgravity. J. Exp. Biol. 204, 3201–3208 (2001). [DOI] [PubMed] [Google Scholar]
- Fitts, R. H. et al. Prolonged space flight-induced alterations in the structure and function of human skeletal muscle fibres. J. Physiol. 588, 3567–3592 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, L. F. , Yu, Z. B. , Ma, J. & Mao, Q. W. Peripheral effector mechanism hypothesis of postflight cardiovascular dysfunction. Aviat. Space Environ. Med. 72, 567–575 (2001). [PubMed] [Google Scholar]
- Vandenburgh, H. , Chromiak, J. , Shansky, J. , Del Tatto, M. & Lemaire, J. Space travel directly induces skeletal muscle atrophy. FASEB J. 13, 1031–1038 (1999). [DOI] [PubMed] [Google Scholar]
- Etheridge, T. et al. The effectiveness of RNAi in Caenorhabditis elegans is maintained during spaceflight. PLoS ONE 6, e20459 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashitani, A. et al. C. elegans RNAi space experiment (CERISE) in Japanese Experiment Module KIBO. Biol. Sci. Space 23, 183–187 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman, P. S. et al. A comparison of mutations induced by accelerated iron particles versus those induced by low earth orbit space radiation in the FEM-3 gene of Caenorhabditis elegans. Mutat. Res. 474, 47–55 (2001). [DOI] [PubMed] [Google Scholar]
- Johnson, T. E. & Nelson, G. A. Caenorhabditis elegans: a model system for space biology studies. Exp. Gerontol. 26, 299–309 (1991). [DOI] [PubMed] [Google Scholar]
- Nelson, G. A. , Schubert, W. W. , Kazarians, G. A. & Richards, G. F. Development and chromosome mechanics in nematodes: results from IML-1. Adv. Space Res. 14, 209–214 (1994). [DOI] [PubMed] [Google Scholar]
- Nelson, G. A. et al. Radiation effects in nematodes: results from IML-1 experiments. Adv. Space Res. 14, 87–91 (1994). [DOI] [PubMed] [Google Scholar]
- Higashibata, A. et al. Decreased expression of myogenic transcription factors and myosin heavy chains in Caenorhabditis elegans muscles developed during spaceflight. J. Exp. Biol. 209, 3209–3218 (2006). [DOI] [PubMed] [Google Scholar]
- Higashitani, A. et al. Checkpoint and physiological apoptosis in germ cells proceeds normally in spaceflown Caenorhabditis elegans. Apoptosis 10, 949–954 (2005). [DOI] [PubMed] [Google Scholar]
- Honda, Y. et al. Genes down-regulated in spaceflight are involved in the control of longevity in Caenorhabditis elegans. Sci. Rep. 2, 487 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selch, F. et al. Genomic response of the nematode Caenorhabditis elegans to spaceflight. Adv. Space Res. 41, 807–815 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szewczyk, N. J. et al. Delayed development and lifespan extension as features of metabolic lifestyle alteration in C. elegans under dietary restriction. J. Exp. Biol. 209, 4129–4139 (2006). [DOI] [PubMed] [Google Scholar]
- Kranewitter, W. J. , Ylanne, J. & Gimona, M. UNC-87 is an actin-bundling protein. J. Biol. Chem. 276, 6306–6312 (2001). [DOI] [PubMed] [Google Scholar]
- Rogalski, T. M. , Gilbert, M. M. , Devenport, D. , Norman, K. R. & Moerman, D. G. DIM-1, a novel immunoglobulin superfamily protein in Caenorhabditis elegans, is necessary for maintaining bodywall muscle integrity. Genetics 163, 905–915 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, D. et al. Tissue-specific regulation of SIRT1 by calorie restriction. Genes Dev. 22, 1753–1757 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, D. & Guarente, L. SIR2: a potential target for calorie restriction mimetics. Trends Mol. Med. 13, 64–71 (2007). [DOI] [PubMed] [Google Scholar]
- Jadiya, P. et al. Sir-2.1 modulates 'calorie-restriction-mediated' prevention of neurodegeneration in Caenorhabditis elegans: implications for Parkinson's disease. Biochem. Biophys. Res. Commun. 413, 306–310 (2011). [DOI] [PubMed] [Google Scholar]
- Tissenbaum, H. A. & Guarente, L. Increased dosage of a sir-2 gene extends lifespan in Caenorhabditis elegans. Nature 410, 227–230 (2001). [DOI] [PubMed] [Google Scholar]
- Viswanathan, M. , Kim, S. K. , Berdichevsky, A. & Guarente, L. A role for SIR-2.1 regulation of ER stress response genes in determining C. elegans life span. Dev. Cell 9, 605–615 (2005). [DOI] [PubMed] [Google Scholar]
- Walker, A. K. et al. Conserved role of SIRT1 orthologs in fasting-dependent inhibition of the lipid/cholesterol regulator SREBP. Genes Dev. 24, 1403–1417 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts, J. L. & Browse, J. A palmitoyl-CoA-specific Δ9 fatty acid desaturase from Caenorhabditis elegans. Biochem. Biophys. Res. Commun. 272, 263–269 (2000). [DOI] [PubMed] [Google Scholar]
- Higashibata, A. et al. Space flight induces reduction of paramyosin and troponin T: Proteomic analysis of space-flown Caenorhabditis elegans. Curr. Biotechnol. 2, 262–271 (2013). [Google Scholar]
- Adams, G. R. , Caiozzo, V. J. & Baldwin, K. M. Skeletal muscle unweighting: spaceflight and ground-based models. J. Appl. Physiol. (1985) 95, 2185–2201 (2003). [DOI] [PubMed] [Google Scholar]
- Wittwer, M. et al. Prolonged unloading of rat soleus muscle causes distinct adaptations of the gene profile. FASEB J. 16, 884–886 (2002). [DOI] [PubMed] [Google Scholar]
- Durieux, A. C. et al. Focal adhesion kinase is a load-dependent governor of the slow contractile and oxidative muscle phenotype. J. Physiol. 587, 3703–3717 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding, M. , Woo, W. M. & Chisholm, A. D. The cytoskeleton and epidermal morphogenesis in C. elegans. Exp. Cell Res. 301, 84–90 (2004). [DOI] [PubMed] [Google Scholar]
- Francis, R. & Waterston, R. H. Muscle cell attachment in Caenorhabditis elegans. J Cell Biol 114, 465–479 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etheridge, T. et al. Calpains mediate integrin attachment complex maintenance of adult muscle in Caenorhabditis elegans. PLoS Genet. 8, e1002471 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etheridge, T. et al. The integrin-adhesome is required to maintain muscle structure, mitochondrial ATP production, and movement forces in Caenorhabditis elegans. FASEB J. 29, 1235–1246 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schatten, H. , Lewis, M. L. & Chakrabarti, A. Spaceflight and clinorotation cause cytoskeleton and mitochondria changes and increases in apoptosis in cultured cells. Acta Astronaut. 49, 399–418 (2001). [DOI] [PubMed] [Google Scholar]
- Zhang, Q. et al. Nesprin-1 and -2 are involved in the pathogenesis of Emery Dreifuss muscular dystrophy and are critical for nuclear envelope integrity. Hum. Mol. Genet. 16, 2816–2833 (2007). [DOI] [PubMed] [Google Scholar]
- Gurd, B. J. , Little, J. P. & Perry, C. G. Does SIRT1 determine exercise-induced skeletal muscle mitochondrial biogenesis: differences between in vitro and in vivo experiments? J. Appl. Physiol. 112, 926–928 (2012). [DOI] [PubMed] [Google Scholar]
- Menzies, K. J. & Hood, D. A. The role of SirT1 in muscle mitochondrial turnover. Mitochondrion 12, 5–13 (2012). [DOI] [PubMed] [Google Scholar]
- Chabi, B. , Adhihetty, P. J. , O'Leary, M. F. , Menzies, K. J. & Hood, D. A. Relationship between Sirt1 expression and mitochondrial proteins during conditions of chronic muscle use and disuse. J. Appl. Physiol. 107, 1730–1735 (2009). [DOI] [PubMed] [Google Scholar]
- Gurd, B. J. , Yoshida, Y. , Lally, J. , Holloway, G. P. & Bonen, A. The deacetylase enzyme SIRT1 is not associated with oxidative capacity in rat heart and skeletal muscle and its overexpression reduces mitochondrial biogenesis. J. Physiol. 587, 1817–1828 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broday, L. , Hauser, C. A. , Kolotuev, I. & Ronai, Z. Muscle-epidermis interactions affect exoskeleton patterning in Caenorhabditis elegans. Dev. Dyn. 236, 3129–3136 (2007). [DOI] [PubMed] [Google Scholar]
- Rahman, M. M. , Rosu, S. , Joseph-Strauss, D. & Cohen-Fix, O. Down-regulation of tricarboxylic acid (TCA) cycle genes blocks progression through the first mitotic division in Caenorhabditis elegans embryos. Proc. Natl Acad. Sci. USA 111, 2602–2607 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards, C. B. , Copes, N. , Brito, A. G. , Canfield, J. & Bradshaw, P. C. Malate and fumarate extend lifespan in Caenorhabditis elegans. PLoS One 8, e58345 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anesti, V. & Scorrano, L. The relationship between mitochondrial shape and function and the cytoskeleton. Biochim. Biophys. Acta 1757, 692–699 (2006). [DOI] [PubMed] [Google Scholar]
- Boldogh, I. R. & Pon, L. A. Mitochondria on the move. Trends Cell Biol. 17, 502–510 (2007). [DOI] [PubMed] [Google Scholar]
- Szewczyk, N. J. , Kozak, E. & Conley, C. A. Chemically defined medium and Caenorhabditis elegans. BMC Biotechnol. 3, 19 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogg, S. & Ruvkun, G. The C. elegans PTEN homolog, DAF-18, acts in the insulin receptor-like metabolic signaling pathway. Mol. Cell 2, 887–893 (1998). [DOI] [PubMed] [Google Scholar]
- Kimura, K. D. , Tissenbaum, H. A. , Liu, Y. & Ruvkun, G. daf-2, an insulin receptor-like gene that regulates longevity and diapause in Caenorhabditis elegans. Science 277, 942–946 (1997). [DOI] [PubMed] [Google Scholar]
- Ohmuro, J. , Mogami, Y. & Baba, S. A. Progression of flagellar stages during artificially delayed motility initiation in sea urchin sperm. Zool. Sci. 21, 1099–1108 (2004). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.