SUMMARY
Increasing evidence suggests that diverse RNA binding proteins (RBPs) interact with regulatory RNAs to regulate transcription. RBFox2 is a well-characterized pre-mRNA splicing regulator, but we now encounter an unexpected paradigm where depletion of this RBP induces widespread increase in nascent RNA production in diverse cell types. Chromatin immunoprecipitation sequencing (ChIP-seq) reveals extensive interaction of RBFox2 with chromatin in a nascent RNA-dependent manner. Bayesian network analysis connects RBFox2 to Polycomb complex 2 (PRC2) and H3K27me3, and biochemical experiments demonstrate the ability of RBFox2 to directly interact with PRC2. Strikingly, RBFox2 inactivation eradicates PRC2 targeting on the majority of bivalent gene promoters and leads to transcriptional de-repression. Together, these findings uncover a mechanism underlying the enigmatic association of PRC2 with numerous active genes, highlight the importance of gene body sequences to gauge transcriptional output, and suggest nascent RNAs as critical signals for transcriptional feedback control to maintain homeostatic gene expression in mammalian genomes.
Graphical Abstract
INTRODUCTION
RNA binding proteins (RBPs) and ribonucleoprotein particles (RNPs) have been increasingly appreciated for their involvement in multiple layers of regulated gene expression beyond their traditional roles in RNA processing, which likely act in conjunction with diverse regulatory RNAs expressed in mammalian genomes (Fu, 2014; Rinn and Chang, 2012). For instance, the splicing commitment factor SRSF2 is now known to play a key role in facilitating RNA Polymerase II (Pol II) pause release from gene promoters (Ji et al., 2013) and modulating transcriptional elongation in gene bodies (Lin et al., 2008). The small nuclear ribonucleoprotein particle U1 small nuclear RNP (snRNP), besides its role in 5′ splice site definition, helps prevent cryptic transcriptional termination and premature polyadenylation (Kaida et al., 2010). The regulatory functions of RBPs via specific non-coding RNAs (ncRNAs) in transcriptional control are also exemplified by the interaction of FUS/TLS with a cyclin D1 promoter-associated ncRNA to repress transcription in response to DNA damage (Wang et al., 2008) and by the ability of both hnRNP U and hnRNP K to bind the Xist ncRNA to mediate transcriptional repression (Chu et al., 2015). These findings mark an emerging trend for wide participation of RBPs in transcriptional control, likely via numerous ncRNAs expressed in mammalian genomes (ENCODE Project Consortium, 2012).
The mechanism underlying ncRNA-mediated epigenetic control of gene expression is perhaps best illustrated with Polycomb complex 2 (PRC2), which catalyzes H3K27 trimethylation on chromatin (Cao et al., 2002) and is critical for X-inactivation in the mouse (Lee and Bartolomei, 2013) and responsible for establishing bivalent promoters in different cell lineages (Voigt et al., 2013). Interestingly, at least two components of PRC2 (EZH2 and SUZ12) appear to have the capacity to directly bind RNA, but whether they bind with a degree of specificity has been a subject of debate (Davidovich et al., 2013; Kaneko et al., 2013; Kanhere et al., 2010; Zhao et al., 2010). A more recent biochemical comparison suggests that PRC2 does have a degree of preference for different RNAs despite its capacity to bind diverse RNAs with high affinity, suggesting that promiscuous and specific RNA-binding activities of PRC2 do not have to be mutually exclusive (Davidovich et al., 2015).
Genome-wide analysis of PRC2-RNA interactions reveals that RNAs from modestly expressed protein-coding genes constitute the vast majority of the PRC2 interactome (Kaneko et al., 2013; Zhao et al., 2010). Because PRC2 does not bind all RNAs, especially from highly expressed genes, this has been part of ongoing debate on direct or indirect mechanisms for PRC2 recruitment in mammalian genomes, leading to speculation for the involvement of certain RBPs as adaptors (Bonasio and Shiekhattar, 2014). The observation that bivalency is established during the transition from naive to primed embryonic stem (ES) cells suggests that PRC2 targeting is a highly regulated process (Marks et al., 2012; Theunissen et al., 2014). Consistently, a recent study reveals a key role of the DNA/RBP ATRX in Xist loading/spreading on X chromosome and in facilitating PRC2 targeting to many other genomic loci (Sarma et al., 2014).
The RBFox family of RBPs has been characterized as splicing regulators (Kuroyanagi, 2009) by recognizing the evolutionarily conserved GCAUG motif (Jin et al., 2003; Zhang et al., 2008) to induce alternative splicing in a position-dependent manner (Jangi et al., 2014; Yeo et al., 2009). RBFox1 (a.k.a. A2BP1) is expressed in muscles and brain; RBFox2 (a.k.a. RMB9 or Fxh) is more ubiquitously expressed with higher abundance in most cell types; and RBFox3 (a.k.a. NeuN) is predominantly detected in mature neurons (Underwood et al., 2005). It is also important to bear in mind that RBFox1 was first discovered as a X chromosome dosage-compensation regulator in C. elegans (Hodgkin et al., 1994; Nicoll et al., 1997); RBFox2 was found earlier as a co-transcriptional repressor in nuclear-receptor-regulated gene expression (Norris et al., 2002); and RBFox3/NeuN was recently shown to modulate microRNA (miRNA) processing (Kim et al., 2014). This literature information suggests that RBFox proteins may regulate gene expression at different levels and via multiple mechanisms.
We recently dissected the RBFox2-regulated splicing program during pressure overloading-induced heart failure (Wei et al., 2015). However, many specific cardiac phenotypes caused by RBFox2 ablation could not readily be attributed to altered splicing (unpublished data), suggesting the need to investigate the function of RBFox2 in regulated gene expression without being confined to the traditional framework. Here, we present evidence that RBFox2 broadly interacts with nascent RNA on chromatin to mediate transcriptional repression through its functional interplay with PRC2. Strikingly, RBFox2 ablation causes genome-wide eradication of PRC2-chromatin interactions and dynamic H3K27me3 deposition on gene promoters in diverse cell types. This reveals RBFox2 as a global regulator of PRC2 targeting in mammalian genomes, which also supports the concept that nascent RNAs from both non-coding and protein-coding genes act as cis-acting signals for dynamic control of mammalian gene expression.
RESULTS
Widespread Transcriptional Induction in Response to RBFox2 Knockout
As part of our ongoing efforts to understand severe cardiomyopathy triggered by RBFox2 knockout in the mouse heart (Wei et al., 2015), we noted transcriptional induction of multiple mature miRNAs, which is coincident with the development of cardiac phenotype (Figure S1A). Unlike the reported role of RBFox3/NeuN in modulating post-transcriptional miRNA processing (Kim et al., 2014), however, we found miRNA induction at the pri-miRNA level without significant changes at the pre-miRNA level, which applies to all induced and at least a subset of repressed miRNAs we detected at mature miRNA levels (Figures S1A–S1C). This finding indicates a previously unrecognized function of RBFox2 in transcriptional control. Although we were able to link some induced miRNAs to a specific cardiac defect (unpublished data), we suspected an even greater altered transcription program that may underlie the overall disease phenotype in our animal model. We therefore decided to first pursue this intriguing and unexpected paradigm for RBFox2-regulated gene expression.
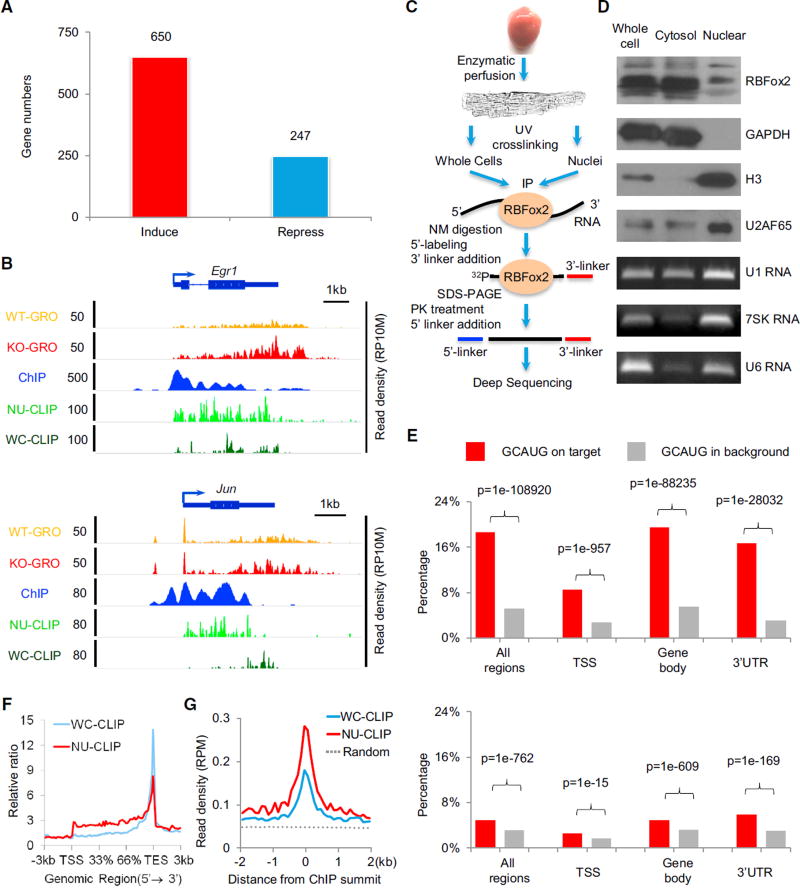
We isolated primary cardiomyocytes from WT and RBFox2 knockout heart by enzymatic perfusion and performed global nuclear run-on coupled with deep sequencing (GRO-seq) to detect nascent RNA production (Table S1A). We found that RBFox2 ablation caused a >1.5-fold change in nascent RNA production among 650 upregulated genes and 247 downregulated genes, indicating a dominant role (directly or indirectly at this point) of RBFox2 in transcriptional repression (Figure 1A). This includes a set of miRNAs we examined by real-time RT-PCR (Figure S1A), which corroborates with increased GRO-seq signals (Figure S1D), both at the primary miRNA level. Importantly, we also detected elevated transcription from a large number of protein-coding genes, as illustrated for Egr1 and Jun (Figure 1B). These data provide initial evidence for a critical role of RBFox2 in transcriptional control, which we decided to rigorously pursue in both primary cardiomyocytes and more experimentally manipulable cellular models.
Figure 1. RBFox2 Knockout-Induced Transcription in Primary Cardiomyocytes.
(A) Significantly induced and repressed genes in RBFox2 knockout cardiomyocytes. The data are based on a >1.5-fold change and p value < 0.1 from GRO-seq analysis. See also Figures S1A–S1C.
(B) RBFox2 knockout-induced gene expression detected by GRO-seq in comparison with RBFox2-RNA interactions detected by CLIP-seq and RBFox2-DNA interactions detected by ChIP-seq on two representative protein-coding genes, Egr1 and Jun. All experiments were on isolated cardiomyocytes from 9-week-old mice. The scale on they axis indicates the read density per 10 million of total normalized reads (RP10M). See also Figure S1D and Table S1.
(C) Diagram showing key steps in CLIP-seq analysis on whole-cell or nuclear-enriched cardiomyocytes.
(D) Characterization of fractionated cardiomyocytes by western blotting using both cytoplasmic (GAPDH) and nuclear proteins (histone H3 and U2AF65) and RNAs (U1, 7SK, and U6) as markers.
(E) Distribution of the GCAUG motif in RBFox2 WC CLIP-seq reads (top) or NU CLIP-seq reads (bottom) in different regions of RefSeq coding genes (red bars) relative to background (gray bars). TSS, transcription stare site to +/−1 kb downstream regions; gene body, regions in RefSeq coding genes without a TSS and 3′ UTR. See also Figures S1E and S1F.
(F) Meta-gene analysis of whole-cell (WC; blue) and nuclear (NU; red) RBFox2 CLIP-seq signals on all RefSeq protein-coding genes by the ngsplot program.
(G) Alignment of the WC (blue) and NU (red) RBFox2 CLIP-seq signals on the center of RBFox2 ChIP-seq signals. Randomized signals (dashed line) served as control.
RBFox2 Interaction with RNA and DNA
Because RBFox2 is an RBP that has been implicated in regulated RNA metabolism in both the nucleus and the cytoplasm, we hypothesized that RBFox2 might interact with RNA on chromatin during co-transcriptional pre-mRNA splicing to influence transcription. We therefore mapped its interaction with RNA by cross-linking immunoprecipitation followed by deep sequencing (CLIP-seq) on fractionated primary cardiomyocytes from WT mouse heart (Figure 1C). We optimized the cell fractionation protocol for cardiomyocytes, which involves a more elaborated procedure to disrupt multi-nuclei muscle cells (see Experimental Procedures), and showed the expected distribution of cytoplasmic and nuclear proteins (e.g., GAPDH and histone 3) and nuclear RNAs (e.g., 7SK and U6), despite a degree of leakage of nuclear components into the cytosolic fraction (i.e., U2AF65 and U1), which is common in preparing nuclear extracts even from cultured cells (Figure 1D). Such leakage would in fact enrich for RBFox2-RNA interactions on chromatin.
From 67 and 22 million initially mapped reads, we obtained 14.9 million whole-cell (WC) CLIP-seq reads and 3.2 million nuclear (NU) CLIP-seq reads that were uniquely mapped to the mouse genome after removing PCR duplicates (Table S1B). In line with the prediction by Ray et al. (2013), a large fraction of the WC RBFox2 CLIP-seq reads were mapped in 3′ UTRs, which becomes further evident after size normalization (Figure S1E). The WC RBFox2 CLIP-seq reads are enriched with the expected GCAUG motif (Figures 1E and S1F), similar to our early report on ES cells (Yeo et al., 2009). Interestingly, RBFox2 appears to interact with RNA near gene promoters (from the transcription start site [TSS] to +/−1 kb downstream sequences) with much reduced specificity, as indicated by less frequent association of the consensus motif with its binding events near the TSS relative to gene body or 3′ UTR (Figure 1E, top). In contrast, the NU RBFox2 CLIP-seq signals showed a general underrepresentation of the GCAUG motif underlying RBFox2 binding events (Figure 1E, bottom), which also exhibited a 5′ shift toward promoter and gene body from 3′ UTR (Figure 1F). These observations hint on a more relaxed binding mode for RBFox2 to interact with chromatin-associated RNA.
We next hypothesized that chromatin-associated RNA might mediate RBFox2 association with chromatin and thus performed RBFox2 chromatin immunoprecipitation sequencing (ChIP-seq) on isolated cardiomyocytes, obtaining 26 million uniquely mapped reads (Table S2). As anticipated, we detected extensive RBFox2 interaction with genomic DNA near the TSS as well as within the gene body, which was correlated with induced nascent RNA production, as illustrated on representative Egr1 and Jun genes (Figure 1B). By relating RBFox2 CLIP-seq peaks to its ChIP-seq signals, we observed that RBFox2-DNA interactions were more related to its RNA binding activities enriched in the nucleus (Figure 1G). This suggests that RBFox2 may become associated with chromatin via nascent RNA, which we later demonstrate to be the case both on representative genes and at a global level (see below).
RBFox2 Binding in Relationship with Key Chromatin Features
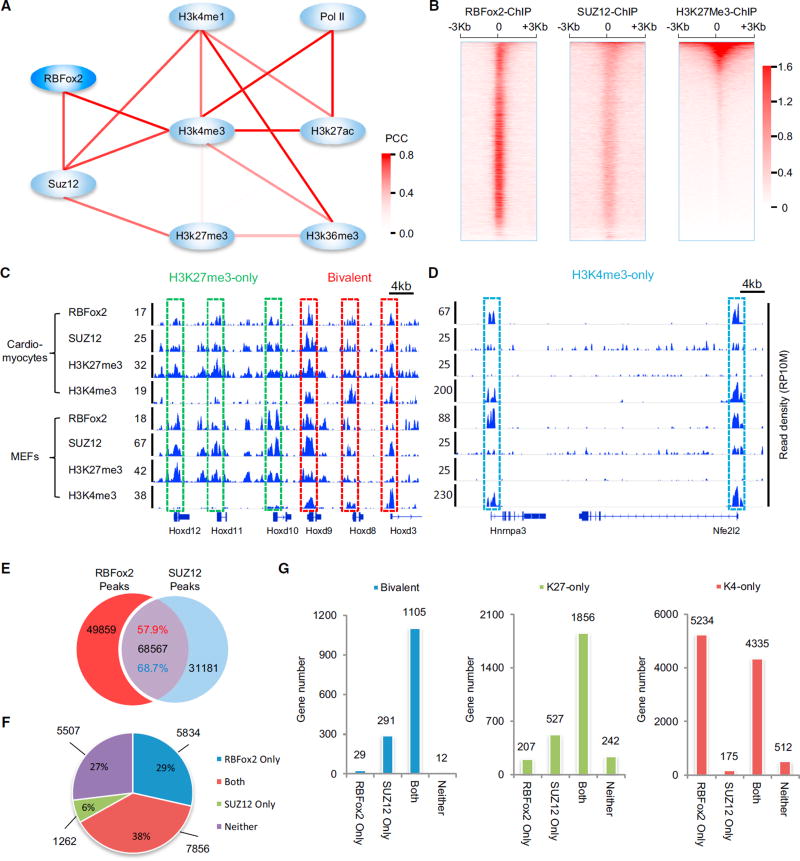
To uncover potential mechanism(s) underlying RBFox2-mediated transcriptional repression, we performed Bayesian network analysis, a bioinformatics approach for inferring global relationships among detected binding events (Liu et al., 2013), by relating RBFox2 ChIP-seq signals to public ChIP-seq data from mouse heart near the TSS region (see Experimental Procedures). The resulting network suggests a connection of RBFox2 to SUZ12, which is linked to the repressive H3K27me3 mark on chromatin (Figure 2A). This was further confirmed by meta-gene analysis near the TSS with RBFox2 and SUZ12 ChIP-seq signals generated on the same cardiomyocytes (Table S2; Figure 2B). These relationships therefore predicted a functional interplay between RBFox2 binding and PRC2 targeting, raising an intriguing possibility that RBFox2 might mediate transcription repression via PRC2. We seized this opportunity to understand the enigmatic association with PRC2 with numerous active genes in mammalian genomes.
Figure 2. Connecting RBFox2-DNA Interactions to PRC2.
(A) Bayesian network analysis of the interactions in gene promoter regions (TSS ± 1kb) among RBFox2, SUZ12, PolII and key histone modifications by SeqSpider program. The color intensity of an edge indicates the Pearson correlation coefficient (PCC) between paired nodes based on the total reads counts in their promoter regions.
(B) Heatmaps of RBFox2 ChIP-seq signals in comparison with SUZ12 and H3K27me3 ChIP-seq signals ±3 kb from the TSS. The RBFox2 and SUZ12 ChIP-seq data were generated on isolated cardiomyocytes from 9-week-oldwild-type (WT) mice. The H3K27me3 ChIP-seq dataset was extracted from the GEO database (GEO: GSM1000131). All heatmaps were normalized by total reads, input signals subtracted, and sorted by mean H3K27me3 density. The numbers beside the color bar indicate the read density per 10 million total reads.
(C and D) RBFox2, SUZ12, H3K27me3 and H3K4me3 ChIP-seq signals on representative genes. Bivalent genes (red in C) are highlighted in comparison with genes exclusively marked by H3K27me3 (green in C) and with genes exclusively marked with H3K4me3 (blue in D).
(E) Overlap between RBFox2 and SUZ12 ChIP-seq peaks. Shown are both the number and the relative percentage of overlapped peaks on each class of genes. Hypergeometric test shows the overlapping is extremely significant (p < 1e–10).
(F) Distribution of protein-coding genes with or without RBFox2 or/and SUZ12 binding at TSS regions. Hypergeometric test shows the overlapping is extremely significant (p < 1e–10).
(G) RBFox2 and/or SUZ12 ChIP binding genes on different groups of genes classified by the presence of H3K4me3, H3K27me3, or both (bivalent) on their promoter regions. See also Figure S2 and Table S2.
Because PRC2-regulated gene expression applies to all cell types in mammals (Davidovich et al., 2013), we next extended the analysis to mouse embryonic fibroblasts (MEFs), which would minimize potential compound effects in knockout animals and allow efficient experimental manipulations. As on cardiomyocytes, we fractionated MEFs to obtain nuclear enriched RBFox2, noting that RBFox2 appears to be predominantly associated with the nuclear fraction in MEFs (Figure S2A). To begin to investigate the potential contribution of nascent RNA to RBFox2’s association with chromatin, we briefly treated MEFs for 3 hr with the transcription inhibitor 5,6-dichloro-1-β-D-ribofuranosylbenzimidazole (DRB), which targets the active site in the kinase p-TEFb required for Pol II pause release (Adelman and Lis, 2012). While the treatment did not affect RBFox2 at the protein level (Figure S2B, bottom), we noted that RBFox2 immunofluorescence signals became greatly dispersed (Figure S2B, top). Furthermore, RNase A and DNase I treatment on Triton X-100-permeabilized MEFs essentially eliminated RBFox2 staining signals (Figure S2B, top). These data indicate the requirement of RNA for RBFox2 to interact with DNA in the nucleus.
We next performed both WC and NU CLIP-seq for RBFox2 in MEFs (Table S3A). Duplicated experiments based on 2 to 3 million uniquely mapped reads in the mouse genome demonstrated the reproducibility of RBFox2 WC and NUCLIP-seq analyses (Figures S2C and S2D). As indicated by biochemical fractionation, RBFox2 appears to be tightly retained in the nucleus of MEFs (Figure S2A), and consequently, both datasets showed the enriched GCAUG motif, but with much reduced representation underlying RBFox2 binding events across all genic regions (Figures S2E and S2F), which is similar to the trend observed with RBFox2 NU CLIP-seq on primary cardiomyocytes (see Figure 1E, bottom).
In mammalian genomes, H3K4me3 and H3K27me3 generally mark active and repressed genes, respectively (Rivera and Ren, 2013).However, many are associated with both chromatin marks, known as bivalent genes, and such bivalent genes are thought to be particularly sensitive to regulatory signals during development (Bernstein et al., 2006; Mikkelsen et al., 2007). To link RBFox2 and SUZ12 binding to specific histone modification events under uniform conditions, we generated an elaborated set of ChIP-seq data with 10 to 25 million uniquely mapped reads in each (Table S3B). In both cardiomyocytes and MEFs, we noted concordant ChIP-seq signals for RBFox2, SUZ12, and H3K27me3 on both repressed (marked exclusively with H3K27me3) and bivalent genes (Figure 2C), while RBFox2 ChIP-seq signals were also prevalent on active genes marked exclusively with H3K4me3 (Figure 2D). These reflect a global trend: RBFox2 and SUZ12 ChIP-seq signals overlap on more than half of their peaks (Figure 2E), which is particularly significant (38%) on protein-coding genes (Figure 2F). RBFox2 and SUZ12 ChIP-seq peaks are largely co-incident on both bivalent (both H3K4me3 and H3K27me3) and largely repressed genes (H3K27me3-only), as well as on approximately half of active genes (H3K4me3-only) (Figure 2G). These observations indicate extensive functional interplay between RBFox2 and PRC2 in the mouse genome.
RBFox2 Mediates Global PRC2 Targeting Near Gene Promoters via Nascent RNA
To determine potential function of RBFox2 in PRC2 targeting, we performed functional perturbation in response to small interfering RNA (siRNA)-mediated knockdown of RBFox2 and SUZ12 in MEFs. In comparison with MEFs treated with nonspecific control siRNA (NC), two independent siRBFox2 reduced RBFox2 protein, and two independent siSUZ12 reduced not only SUZ12 but also EZH2 (likely due to destabilized PRC2 in the cell) (Figure S3A). Interestingly, in comparison with significant reduction of H3K27me3 levels in siSUZ12-treated MEFs, we detected little effect on this histone modification at the global level in siRBFox2-treated MEFs (Figure S3A), which is consistent with unaltered expression of all key PRC2 components at both the mRNA and protein levels (data not shown).
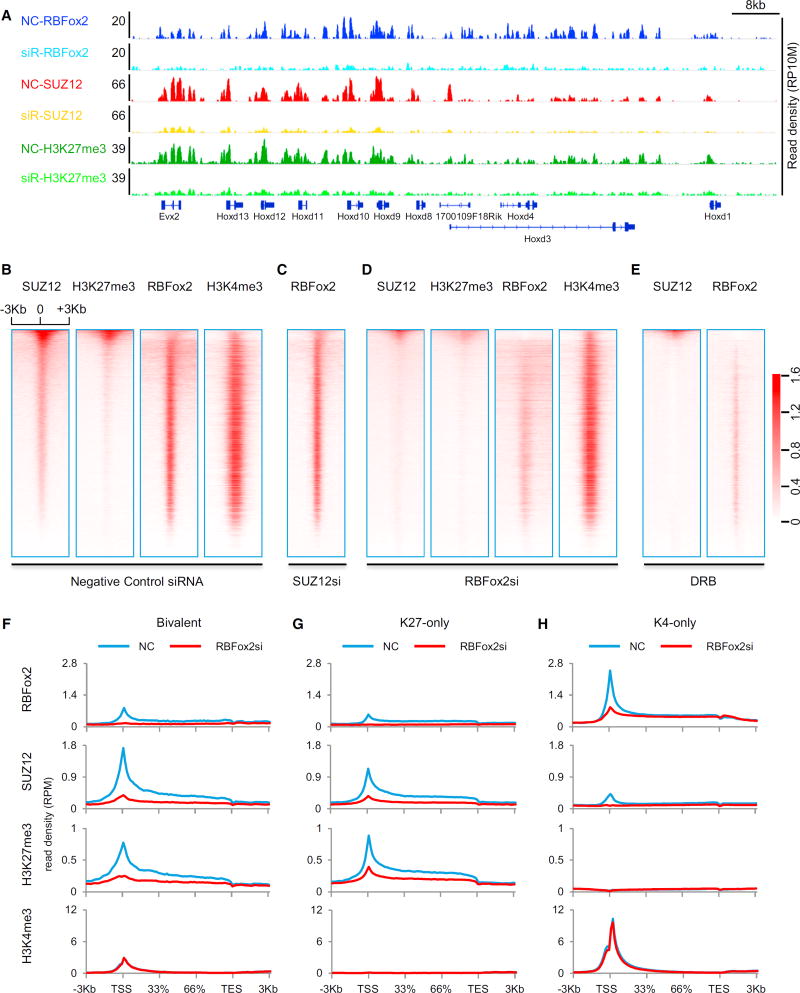
We next performed ChIP-seq on SUZ12, H3K27me3, and H3K4me3 in response to siRBFox2 in MEFs (Figure S3B; Table S3B). Strikingly, in comparison with control siRNA-treated MEFs, siRBFox2 not only abolished RBFox2 ChIP-seq signals, as expected (this also helps validate the specificity of anti-RBFox2 antibody in ChIP-seq analysis), but also significantly reduced ChIP-seq signals for both SUZ12 and H3K27me3, as shown on a representative genomic segment in the HoxD locus (Figure 3A). Focusing on TSS-associated ChIP-seq signals, we found that RBFox2 ChIP signals were unaffected by siSUZ12, indicating that PRC2 is not required for RBFox2 to interact with chromatin (Figures 3B and 3C). By contrast, siRBFox2 not only diminished global RBFox2 binding but also abolished SUZ12 and H3K27me3 ChIP-seq signals around the TSS, without measurable impact on H3K4me3 ChIP-seq signals (Figure 3D).
Figure 3. Functional Requirement for RBFox2 to Mediate Global PRC2 Targeting.
(A) ChIP-seq signals on a representative chromosomal segment for RBFox2, SUZ12, and H3K27me3 before and after siRNA-mediated RBFox2 knockdown (siR) in MEFs. The y axis indicates the read density per 10 million total reads (RP10M).
(B–E) Sorted ChIP-seq signals for SUZ12, H3K27me3, RBFox2, and H3K4me3 at TSS regions in control siRNA-treated (B), SUZ12 siRNA-treated (C), RBFox2 siRNA-treated (D), and DRB-treated (E) MEFs. The data were sorted according to mean SUZ12 ChIP-seq signals in control siRNA-treated MEFs. All ChIP-seq data were normalized by total reads after subtracting input signals. The numbers beside the color bar indicate the read density per million. See also Figure S3 and Table S3.
(F–H) Responses of the indicated ChIP-seq signals to RBFox2 knockdown on different gene classes (bivalent, H3K27me3 only, or H3K4me3 only) in MEFs.
See also Figure S4 and Table S4 on related functional data from C2C12 cells.
Because siRBFox2 had little effect on the global level of H3K27me3 (Figure S3A), we determined whether siRBFox2 caused redistribution of PRC2 in the mouse genome, as reported in comparing between mouse naive and primed ES cells (Marks et al., 2012). We computed the genomic distribution of H3K27me3 peaks by binning H3K27me3-enriched regions to tag density per peak (based on normalization of total tags to 10 million), as described earlier (Marks et al., 2012), and found that, in control siRNA-treated MEFs (Figure S3C, left), more than half of H3K27me3 peaks with high tag density were associated with gene promoters (blue), but in response to RBFox2 knockdown (Figure S3C, right), those peaks were significantly reduced near the TSS with accompanying increases in inter-genic regions (green). Together, these data suggest that RBFox2 may not be essential for general PRC2 activities but are critical for PRC2 targeting to dynamically regulated gene promoters.
Given RBFox2 is a well-established RBP, we next asked whether the coordinated events for factor binding and histone modification were RNA dependent. By treating MEFs with the transcription inhibitor DRB for 2 hr to block nascent RNA production in the gene body, we observed a major reduction of both RBFox2 and SUZ12 ChIP-seq signals (Figure 3E), which we further validated on specific genes by ChIP-qPCR (Figure S3D). While we cannot rule out other potential indirect effects of DRB, the data strongly suggest the requirement for nascent RNAs to mediate the association of RBFox2 with active genes, which in turn recruits PRC2.
Because RBFox2 frequently interacts with PRC2 on bivalent genes, we separately analyzed the functional impact of siRB-Fox2 on different gene classes. We found that RBFox2 knockdown in MEFs greatly reduced SUZ12 and H3K27me3 ChIP-seq signals on bivalent genes as well as on largely repressed genes that were exclusively marked by H3K27me3 (Figures 3F and 3G), but without detectable effect on H3K4me3 signals on most active genes that are exclusively decorated with this chromatin mark (Figure 3H).
To generalize the function of RBFox2 in global PRC2 targeting to gene promoters, we extended the analysis to C2C12 cells (Table S4). RBFox2 mainly bound bivalent and active genes in C2C12 cells (Figure S4A), and in response to siRBFox2, global H3K27me3 (Figure S4B), but not H3K4me3 (Figure S4C), went down. Interestingly, the Pol II ChIP-seq signals were also modestly increased, consistent with a role of RBFox2 in global transcriptional repression in C2C12 cells (Figure S4D). These data suggest a general requirement for RBFox2 to mediate global PRC2 targeting in diverse cell types.
Functional Consequence of RBFox2-Regulated PRC2 Targeting
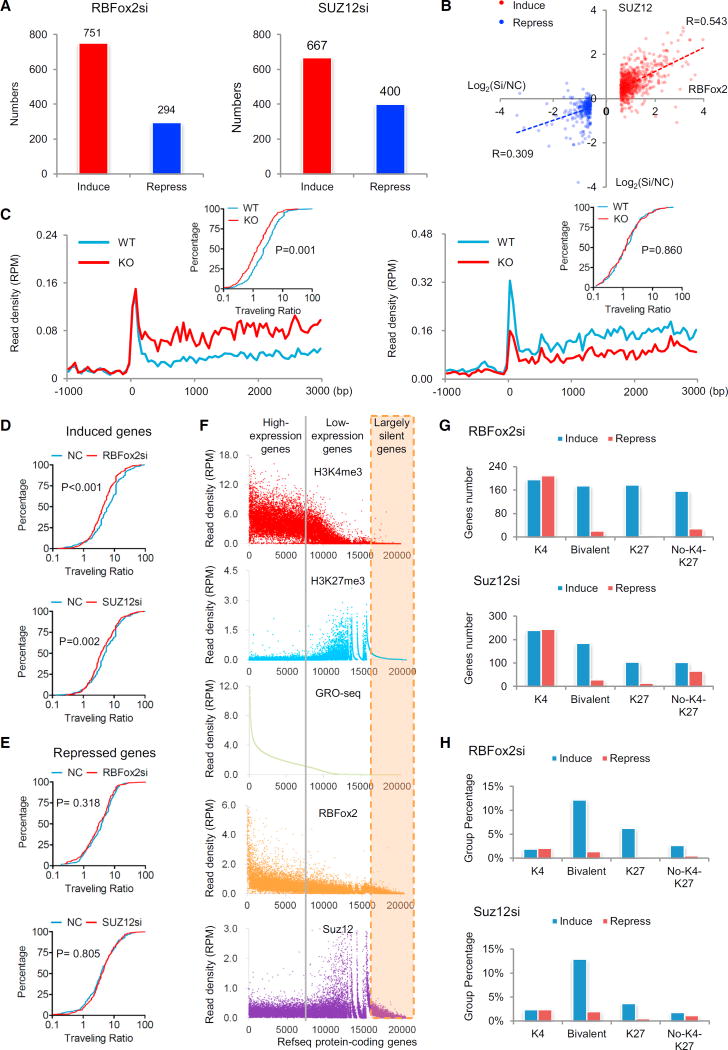
A previous study recorded little functional consequence in response to PRC2 knockdown based on analysis of steady-state mRNA by RNA-sequencing, which led to a “junk mail” model for PRC2 targeting, emphasizing additional requirements in conjunction with PRC2 to have measurable functional consequences on a rather restricted subset of PRC2-targeted genes (Davidovich et al., 2013). We revisited this critical issue by using more sensitive GRO-seq to measure transcriptional response at the levels of nascent RNA production, as we could clearly detect transcriptional response in RBFox2 knockout cardiomyocytes (see Figures 1A and 1B), and more importantly, we wished to relate RBFox2-mediated PRC2 targeting to the functional impact on gene expression, at least on a subset of PRC2-targeted genes. In MEFs with 6–7 million unique GRO-seq reads under individual conditions (Table S5), we detected both up- and down-regulated nascent RNA production in response to siRBFox2 and siSUZ12 treatment, and as observed on cardiomyocytes, de-repressed genes outnumbered repressed genes in both cases (Figure 4A). By comparing the responsive genes with DNA binding evidence for both RBFox2 and SUZ12, we found that siRB-Fox2 and siSUZ12 had similar effects on induced genes (R = 0.543), but much less so on repressed genes (R = 0.309) (Figure 4B). These results support a more related function of RBFox2 and SUZ12 in transcriptional repression than activation, the latter of which likely results from various indirect effects.
Figure 4. Transcription Repression Coordinately Regulated by RBFox2 and PRC2.
(A) Induced and repressed genes according to a >1.5-fold change and p value < 0.1 in GRO-seq in response to knockdown of RBFox2 (left) or SUZ12 (right) relative to control siRNA-treated MEFs. See also Table S5.
(B) Comparison between induced and repressed transcription (in log2) in response to knockdown of RBFox2 and SUZ12, showing a significant concordance between RBFox2 and SUZ12 siRNA treatments among induced, but not repressed, genes.
(C) Meta-gene analysis of GRO-seq signals and the distribution of the traveling ratio (TR) of induced (left) or repressed genes (right) in RBFox2 knockout car-diomyocytes. A total of 209 and 138 significantly induced and repressed genes were selected for the analysis. The × axis indicates the distance from the TSS and the y axis shows the read density per million (RPM). The inserts show changes in TRs plotted in the accumulative fashion. Statistical significance was determined using the Kolmogorov-Smirnov test.
(D and E) Calculated TR changes in response to knockdown of RBFox2 and SUZ12 in MEFs based on significantly induced genes (476 genes from the siRBFox2 group and 435 genes from the siSUZ12 group) (D) and repressed genes (196 genes from the siRBFox2 group and 222 genes from the siSUZ12 group) (E). (F) Sorting of protein-coding genes according to GRO-seq levels at TSS regions (±3 kb) to segregate highly active (left from the gray line), intermediately expressed (between the gray line and orange-shaded regions), and largely silent genes (orange shaded), relative to their association with levels of H3K4me3, H3K27me3, SUZ12, and RBFox2.
(G and H) Association (G) and percentage (H) of induced and repressed genes in response to knockdown of RBFox2 or SUZ12 with active and/or repressive chromatin marks.
Based on the GRO-seq signals from both cardiomyocytes (Table S1A) and MEFs (Table S5), which reflect transcriptionally engaged Pol II (Ji et al., 2013), we calculated the Pol II traveling ratio [TR, defined by the read density at the TSS divided by the read density in the gene body, see (Rahl et al., 2010)] for both induced and repressed genes and plotted the data in the accumulative fashion (inserts in Figure 4C). In RBFox2 knockout cardiomyocytes, we linked decreased TR to induced gene expression (Figure 4C, left) but detected little difference with repressed genes (Figure 4C, right). We made similar observations with MFFs treated with siRBFox2 or siSUZ12 (Figures 4D and 4E). Note that the differences are not as striking as on cardiomyocytes, but still statistically significant, which was likely due to incomplete knockdown versus knockout in combination with ∼5-fold fewer GRO-seq reads from MEFs (Table S5) relative to cardiomyocytes (Table S1A). Nonetheless, these findings are consistent with one another, together suggesting that RBFox2 and PRC2 may act in a coordinated fashion to induce Pol II pausing near the promoters of a set of responsive genes.
Sorting the GRO-seq signals in MEFs further revealed a positive correlation of gene expression with H3K4me3 and a negative correlation with H3K27me3 and SUZ12 binding, and largely silent genes (orange-shaded) typically lacked either mark (Figure 4F). While higher on highly expressed genes, the RBFox2 ChIP-seq signals were also associated with bivalent genes, but much less or none on largely silent genes (Figure 4F). We further observed that, while siRBFox2 and siSUZ12 caused induction and repression of approximately equal numbers of genes among those exclusively marked with H3K4me3, perhaps indicative of both direct and indirect effects, both treatments preferentially de-repressed bivalent genes as well as a significant number of largely repressed genes exclusively marked by H3K27me3 and genes that lacked detectable H3K4me3 and H3K27me3 (Figure 4G). The preferential impact on bivalent genes became more evident when comparing the relative percentages of induced versus repressed genes in different gene classes (Figure 4H). Together, these results strongly suggest that RBFox2 is directly involved in the dynamic regulation of transcriptional repression via PRC2.
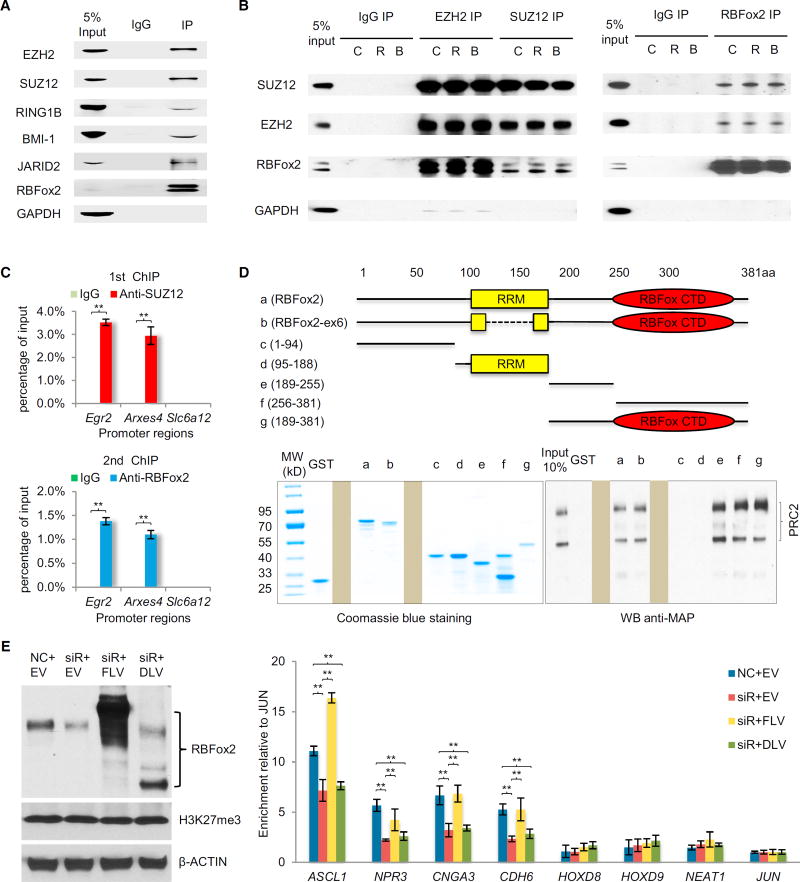
Interactions between RBFox2 and PRC2 In Vivo and In Vitro
Finally, to understand the molecular basis for RBFox2-dependent recruitment of PRC2, we performed co-immunoprecipitation (co-IP) on MEFs. We detected the association of RBFox2 with multiple components of PRC2 as well as several key components of PRC1 we examined (Figure 5A). To determine whether RNA is required to mediate and/or stabilize the interaction, we treated the cell lysate with RNase A (R) or Benzonase (B, which degrades both DNA and RNA) and found that neither treatment affected reciprocal immunoprecipitation between SUZ12 and RBFox2 (Figure 5B). In these immunoprecipitation experiments, GAPDH (Figures 5A and 5B) as well as abundant nuclear protein SRSF1 and histone H3 (data not shown) served as negative controls, suggesting that RBFox2 is part of larger Polycomb complexes in the cell.
Figure 5. Interaction between RBFox2 and PRC2.
(A) Co-IP of RBFox2 with key components of PRC1 and PRC2 in MEFs.
(B) Reciprocal co-IP of RBFox2 with PRC2 in a RNA-independent manner. Before immunoprecipitation, whole-cell extracts were treated with either RNase A (R) or benzonase (B) for 30 min at room temperature. GAPDH served as negative control for immunoprecipitation.
(C) Re-ChIP confirms co-binding of SUZ12 and RBFox2 on the TSS regions of two bivalent genes (Egr2 and Arxes4), but not on Slc6a12 without H3K4me3 or H3K27me3 signals. ChIP signals are presented as percentage of input. Data are compared to immunoglobulin G control and are presented as mean ± SEM. **p < 0.01.
(D) The RBFox2 CTD responsible for interactions with PRC2. Full-length and various deletion constructs of RBFox2 are depicted (top). Individual proteins were expressed and purified from bacteria (bottom left), which were used to conduct GST pull-down assays with baculovirus expressed PRC2 complex (bottom right). Two irrelevant lanes in each gel were blocked.
(E) Western blotting analysis of endogenous RBFox2 (middle band), overexpressed full-length RBFox2 containing N-terminally tagged FLAG and Strep (top band), and similarly tagged C-terminal deletion mutant (bottom band), H3K27me3, and actin in transfected 293T cells (left). NC, negative siRNA control; EV, empty pcDNA3.0 vector; siR, RBFox2 RNAi; FLV, full-length RBFox2 vector; DLV, C-terminal deletion RBFox2 vector. Right panel shows ChIP-qPCR analysis of H3K27me3 levels on the TSS regions of four positive and four negative genes in 293T cells. Results was calculated as fold-changes normalized to JUN and are presented as mean ± SEM. **p < 0.01. See also Figure S5.
Given efficient co-IP between RBFox2 and PRC2, we explored a possibility that RBFox2 might directly interact with PRC2. We expressed core PRC2 components individually as well as together to form the PRC2 complex in baculovirus-infected Sf9 cells, and purified these proteins to nearly homogeneity via the maltose-binding protein (MBP) tag (Figure S5A) according to the established conditions (Davidovich et al., 2013). We also prepared the PRC2 complex after removing the MBP tags from individual components. We then used these pure proteins for glutathione S-transferase (GST) pull-down assays with bacterially expressed GST-RBFox2, finding that GST-RBFox2, but not GST alone, efficiently pulled down the PRC2 complex with or without the MBP tag (Figure S5B). Interestingly, under the same conditions, we detected little binding with individual PRC2 components (data not shown), which might be due to induced conformation in both EZH2 and SUZ12 that only occurs within the fully assembled PRC2 complex (Jiao and Liu, 2015). Given their direct interaction, we performed sequential ChIP on two SUZ12-positive loci (Egr2 and Arxes4) and one negative locus (Slc6a12), finding the ability of RBFox2 and SUZ12 to simultaneously interact with both positive loci (Figure 5C).
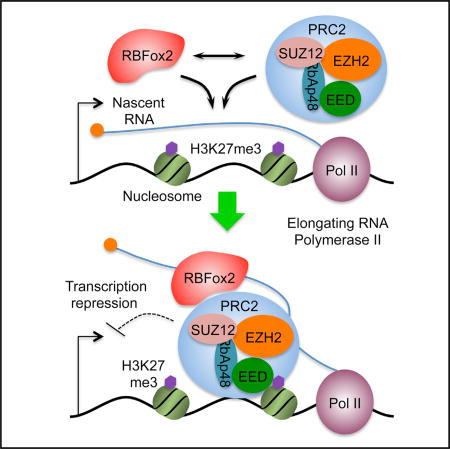
We further mapped the domain in RBFox2 responsible for PRC2 binding and found that its RNA binding domain, which is conserved among all RBFox family members, is not required, and the C-terminal domain (CTD), which is unique in RBFox2, is sufficient to pull down PRC2 (Figure 5D). Interestingly, we found that two segments in the RBFox2 CTD were able to interact with PRC2, indicating multiple contacts between RBFox2 and PRC2 (Figure 5D). Finally, we performed the functional rescue experiments with full-length FLAG- and Strep-tagged RBFox2 (FLV) and its C-terminal deletion mutant (DLV) in transfected 293T cells (Figure 5E, left), finding that full-length RBFox2, but not the mutant, could rescue RBFox2-dependent H3K27me3 deposition on four SUZ12-positive loci, but not four SUZ12-negative loci, as previously reported (Gao et al., 2012) (Figure 5E, right). Together, these findings provide a biochemical basis for RBFox2-dependent PRC2 targeting in mammalian cells. Combining the current observations with the existing information in literature, we propose a model for dynamic control of gene expression through nascent RNA-mediated and context-dependent functional interplays between RBFox2 and PRC2 (Figure 6; see Discussion).
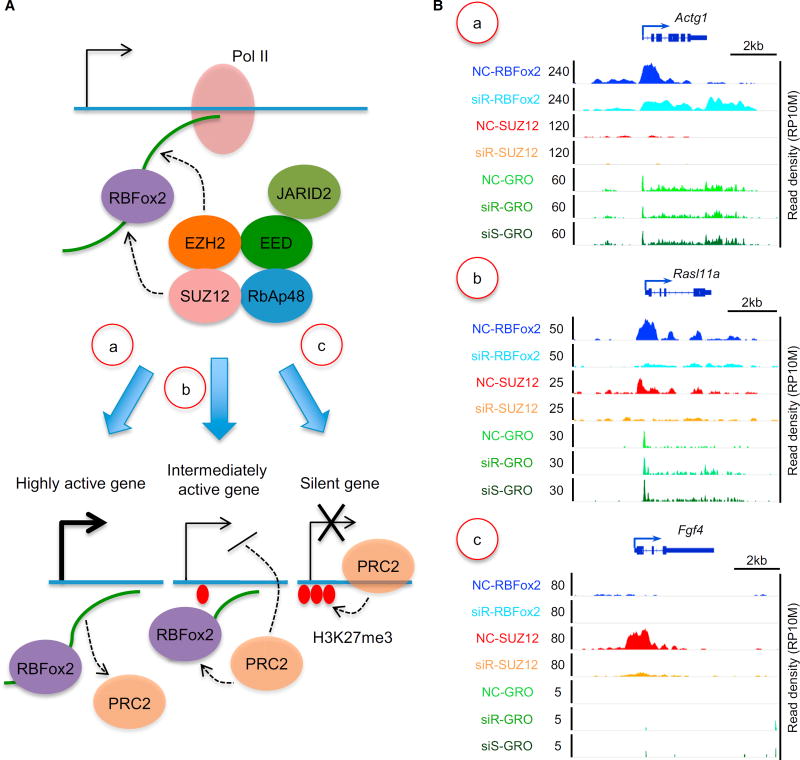
Figure 6. Proposed Model for Functional Interplays of RBFox2, PRC2, and H3K27me3 that Result in Different Gene Classes.
(A) Model for nascent RNA-mediated, RBFox2-dependent recruitment of PRC2. (a) On highly expressed genes, nascent RNA may modulate the RNA sensor function of PRC2 to cause PRC2 repulsion. (b) On modestly expressed genes, functional interplay between RBFox2 and PRC2 may dynamically regulate homeostatic gene expression, as a modest increase or decrease in nascent RNA production may cause increased or decreased PRC2 recruitment to induce feedback controls. A level of pre-deposited H3K27me3 may help stabilize recruited PRC2. (c) On nearly silent genes, interaction of PRC2 with chromatin is switched from RNA-dependent to EED-mediated interactions, leading to the amplification of the repressive signal to eventually shut down the genes. It is important to also emphasize that functional outcomes on individual genes likely depend on the contribution of transcription regulators in conjunction with specific nucleosome states.
(B) Representative genes indifferent classes based on the genomic data generated in the current study: NC, siR, and siS represent ChIP-seq or GRO-seq signals in nonspecific control siRNA-, siRBFox2-, and siSUZ12-treated MEFs, respectively. (a–c) Examples of specific gene in each class described in (A, a–c), showing RBFox2 and SUZ12 binding before and after RBFox2 knockdown, as well as nascent RNA production before and after RBFox2 or SUZ12 knockdown. NC, nonspecific siRNA; siR, siRBFox2; siS, siSUZ12.
DISCUSSION
Multifaceted Functions of RBPs in Mammalian Cells
Through pursuing altered transcription programs in RBFox2 knockout heart and extending the analysis to additional model cell lines, we discovered a general role of RBFox2 as a transcription repressor. A conservative estimate based on RBPs cross-linked to oligo-dT captured mRNA suggests that ∼1,000 RBPs are expressed in the human genome (Castello et al., 2012). However, many RBPs have been studied in their traditional frameworks, which are largely confined to their originally elucidated functions in RNA metabolism. Our current findings, together with earlier work demonstrating the splicing-independent role of U1 snRNP in genome protection (Kaida et al., 2010) and the involvement of SRSF2 in transcription pause release (Ji et al., 2013), suggest the direct participation of RBPs in gene expression before engaging in co-transcriptional RNA processing. Given the pervasive transcription of mammalian genomes, various forms of ncRNAs must enlist specific RBPs to execute their regulatory functions in the genome (Cech and Steitz, 2014; Fu, 2014).
Distinct RBFox2 Binding Modes with Different RNA Populations
RBPs execute biological functions by binding to their target RNAs. In isolation, each RBP shows distinct binding affinity and specificity, and related RNA binding specificities have been observed with RBPs that bear similar RNA recognition motifs (Ray et al., 2013). However, even with purified RBPs, each RBP appears to bind diverse motifs (Lambert et al., 2014). This is also the case with RBFox2, which appears to be capable of binding multiple RNA motifs that can fit into its RNA binding pocket (Auweter et al., 2006; Lambert et al., 2014). Inside cells, individual RBPs may have even more complex binding profiles, as indicated by numerous published CLIP-seq studies. In some extreme cases, such as PTB and U2AF, most of their in vivo binding events are associated with their in vitro binding consensus – pyrimidine-rich sequences (Shao et al., 2014; Xue et al., 2009), but the CLIP-seq data for many other RBPs show diverse motifs underlying their binding peaks with the best consensus representing only a fraction of their binding events in the cell. An extreme case is exemplified by RBFox3/NeuN, which seems to bind RNA with much relaxed specificity in vivo (Kim et al., 2014). While these variable modes of RNA binding may be affected by the quality of CLIP-seq data generated in individual studies, such variations may also reflect the influence of RNA secondary structure and the involvement of more than one RBP, as well as the interaction with different populations of RNAs in different cellular compartments.
Here, we found that even within the same cell type, RBFox2 binds RNA from different genic regions with different specificities. The general trend is that RBFox2 binds chromatin-associated RNA, especially those near gene promoters, with much reduced specificity during transcription relative to its interaction with late RNA segments during co- and post-transcriptional RNA processing. The reduced RNA binding specificity is likely due to the action of RBFox2 as part of larger protein complexes, including PRC2. Because PRC2 has intrinsic RNA binding capability, but with much less specificity (Davidovich et al., 2015; Kaneko et al., 2013), RBFox2 and PRC2 (perhaps other RBPs) in the complex may undertake some coordinated interactions with RNA to regulate a broad spectrum of genes in mammalian cells.
Regulated Targeting of Polycomb Complexes
The most striking discovery here is the unexpected function of RBFox2 in directing PRC2 targeting in the genome. Polycomb complexes are well known to play vital roles in developmental control of gene expression (Schuettengruber et al., 2007). Polycomb complexes have two separate cores, PRC1 and PRC2, acting either separately or in a concerted fashion to induce gene silencing (Simon and Kingston, 2013). PRC2 contains four major subunits (Cao et al., 2002), with EZH2/SUZ12 for catalyzing H3K27me2/3, EED for amplifying repressive signals via its affinity for H3K27me3, and Rbap48 capable of interacting with histone H4 (Voigt et al., 2013).
EZH2/SUZ12 may target PRC2 to specific genomic loci via RNA, but because PRC2 does not bind RNAs proportional to their abundance in the cell, this process appears to be regulated. A recent study demonstrates a role of the DNA/RBP ATRX in Xist loading and spreading on X chromosome as well as in global regulation of PRC2 targeting; however, it remains unclear whether such regulatory function is mediated by its DNA and/or RNA binding activity (Sarma et al., 2014). A recent proteomic analysis of Xist-containing RNP reveals a large number of proteins associated with this long ncRNA, and we noted that RBFox2 is actually part of this large RNP (Chu et al., 2015). PRC2 has also been proposed to act as an RNA sensor because RNA can effectively block its enzymatic activity both in vitro and inside cells (Kaneko et al., 2013). These findings suggest that Polycomb Complex targeting is likely regulated by various RNAs and RBPs in mammalian cells.
Mechanisms for Sorting Genes in Different Functional States
As depicted in Figure 6A and exemplified on representative genes in Figure 6B, our data in combination with literature information suggest that RBFox2 may initially interact with nascent RNA on chromatin and then recruit PRC2 via protein-protein interactions. In this process, the level of nascent RNA may provide a key sensory function: On highly active genes, as proposed earlier (Kaneko et al., 2013), high RNA levels coupled with relatively open chromatin may prevent stable PRC2 binding on chromatin, even initially recruited (Figures 6A and 6Ba). On modestly expressed genes, nascent RNA together with JARID2 (da Rocha et al., 2014; Kaneko et al., 2014a) may help initial recruitment of PRC2, which may become stabilized by relatively compacted chromatin (Yuan et al., 2012) and a degree of predisposed H3K27me3 for EED to bind (Margueron et al., 2009). As a result, increased RNA production would elevate RNA-mediated recruitment of PRC2, leading to increased chromatin compaction, whereas stabilized PRC2 would then limit nascent RNA production, thereby damping further PRC2 recruitment (Figures 6A and 6Bb). This mechanism may help maintain the bivalent nature of those genes for homeostatic expression at intermediate levels. Finally, on a subset of these initially bivalent genes, diminished nascent RNA production may cause a switch from RNA-dependent to H3K27me3-mediated PRC2 recruitment, likely due to excessive chromatin compaction, predisposed H3K27me3, and the action of EED, among other mechanisms, as suggested by a recent study (Kaneko et al., 2014b), together leading to eventual silencing of those genes (Figures 6A and 6Bc).
Last, but not least, our findings emphasize the regulatory function of RNA, even from protein-coding genes. Interestingly, nascent RNA appears to use multiple strategies to regulate the expression of their host genes. A recent study showed that nascent RNA base pairs with a virus-encoded long ncRNA to facilitate the recruitment of the transcription factor PAX5 to regulate the expression of neighbor genes during replication of Epstein-Barr virus (Lee et al., 2015). Our findings now suggest that nascent RNAs produced from active protein-coding genes are not only products of transcription but also critical signals for the regulation of gene expression. Therefore, RNAs are also multitasking in mammalian cells regardless of whether they are ncRNAs or part of RNAs from protein-coding genes.
EXPERIMENTAL PROCEDURES
RBFox2 Knockout, miRNA Profiling, Immunostaining, Western Blotting, Co-IP, GST Pull-Down, and RNAi
Handling of mice was approved by the IRB of University of California, San Diego. Conditional RBFox2 knockout mice were previously described (Wei et al., 2015). Immunostaining, western blotting, co-IP, GST pull-down, and RNAi assays were conducted with various antibodies listed in Supplemental Experimental Procedures. The siRNA sequences used in the current study and primer sequences for RT-PCR are listed in Table S6.
CLIP-Seq, ChIP-Seq, and GRO-Seq
CLIP-seq was performed as described previously (Xue et al., 2009). Isolation of nuclei from cardiomyocytes was performed according to Widnell et al. (1967). To break up myofilaments and release nuclei from cardiomyocytes, cells were homogenized with a homogenizer (Kinematica, POLYTRON PT 2100) at maximum speed for 30 s and ice cooled for 30 s for three rounds. To perform CLIP-seq, WCs or isolated nuclei were digested with 0.1 U/µl RQ1 DNase and 2 U/µl RNaseOUT in lysis buffer (1xPBS buffer with 0.3% SDS, 0.5% deoxycholate sodium, and 0.5% NP-40) for 5 min at 37°C. Each lysate was processed for CLIP-seq with rabbit polyclonal anti-RBFox2 antibody (Bethyl, A300-864A). ChIP-seq was performed as described previously (Ji et al., 2013) using anti-RBFox2 (Bethyl, A300-864A), anti-SUZ12 (Cell Signaling, D39F6), anti-H3K27me3 (Active Motif, 61017), anti-H3K4me3 (Active Motif, 39159) or anti-Pol II (Santa Cruz Biotechnology, sc-899). GRO-seq was performed as described previously (Wang et al., 2011).
Analysis of Genomic Data
Bayesian network construction was performed according to Liu et al. (2013) by using the RBFox2 and SUZ12 ChIP-seq data generated on isolated cardiomyocytes from 9-week-old mice and the following publically available datasets generated from mouse heart: GSM769017 (for H3K4me3), GSM769025 (for H3K4me1), GSM1000130 (for H3K36me3), GSM1000131 (for H3K27me3), GSM1000093 (for H3K27ac), and GSM918723 (for total Pol II). SeqSpider software (Liu et al., 2013) was used to generate binding matrix and construct the network. Other details on data analysis can be found in Supplemental Experimental Procedures.
Highlights.
RBFox2 has a direct role in transcriptional control besides splicing regulation
Nascent RNAs are required for RBFox2 interaction with chromatin
RBFox2 directly interacts with PRC2 to mediate PRC2 targeting genome-wide
Gene-body-associated nascent RNA serves as feedback sensor for transcription output
In Brief.
PRC2 is known to interact with RNA, but it has been unclear whether this process requires additional regulators. Wei et al. report that the RNA binding protein RBFox2 is a global regulator of PRC2 recruitment to active genes, particularly those with bivalent histone modifications, to mediate dynamic transcriptional control.
Acknowledgments
The authors are grateful to members of the X.-D.F lab for cooperation, reagent sharing, and stimulating discussion during the course of this investigation. We are grateful to Dr. Tom Cech for providing the expression plasmids for individual PRC2 subunits and to Drs. Yi Zhang and John Lis for critical comments on the manuscript. H.C. was supported by China Scholarship Council. This work was supported by National Institutes of Health grants GM049369 and HG004659 (to X-D.F.).
Footnotes
ACCESSION NUMBERS
The accession number for the CLIP-seq, ChIP-seq, and GRO-seq data reported in this paper and summarized in Tables S1–S5 is GEO: GSE57926.
SUPPLEMENTAL INFORMATION
Supplemental Information includes Supplemental Experimental Procedures, five figures, and six tables and can be found with this article online at http://dx.doi.org/10.1016/j.molcel.2016.04.013.
AUTHOR CONTRIBUTIONS
C.W. and X.-D.F. designed the study; R.X., C.W., L.C., and H.C. generated most data; Y.X. and J.H. performed CLIP-seq and cell fractionation; T.T., J.Q., and H.L. contributed to expression of recombinant proteins and deep sequencing; Y.Z. and B.Z. analyzed genomic data; L.T guided H.C. in initial study; and C.W. and X.-D.F. wrote the paper.
References
- Adelman K, Lis JT. Promoter-proximal pausing of RNA polymerase II: emerging roles in metazoans. Nat. Rev. Genet. 2012;13:720–731. doi: 10.1038/nrg3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auweter SD, Fasan R, Reymond L, Underwood JG, Black DL, Pitsch S, Allain FH. Molecular basis of RNA recognition by the human alternative splicing factor Fox-1. EMBO J. 2006;25:163–173. doi: 10.1038/sj.emboj.7600918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Bonasio R, Shiekhattar R. Regulation of transcription by long noncoding RNAs. Annu. Rev. Genet. 2014;48:433–455. doi: 10.1146/annurev-genet-120213-092323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298:1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- Castello A, Fischer B, Eichelbaum K, Horos R, Beckmann BM, Strein C, Davey NE, Humphreys DT, Preiss T, Steinmetz LM, et al. Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell. 2012;149:1393–1406. doi: 10.1016/j.cell.2012.04.031. [DOI] [PubMed] [Google Scholar]
- Cech TR, Steitz JA. The noncoding RNA revolution-trashing old rules to forge new ones. Cell. 2014;157:77–94. doi: 10.1016/j.cell.2014.03.008. [DOI] [PubMed] [Google Scholar]
- Chu C, Zhang QC, da Rocha ST, Flynn RA, Bharadwaj M, Calabrese JM, Magnuson T, Heard E, Chang HY. Systematic discovery of Xist RNA binding proteins. Cell. 2015;161:404–416. doi: 10.1016/j.cell.2015.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Rocha ST, Boeva V, Escamilla-Del-Arenal M, Ancelin K, Granier C, Matias NR, Sanulli S, Chow J, Schulz E, Picard C, et al. Jarid2 Is implicated in the initial Xist-induced targeting of PRC2 to the inactive X chromosome. Mol. Cell. 2014;53:301–316. doi: 10.1016/j.molcel.2014.01.002. [DOI] [PubMed] [Google Scholar]
- Davidovich C, Zheng L, Goodrich KJ, Cech TR. Promiscuous RNA binding by Polycomb repressive complex 2. Nat. Struct. Mol. Biol. 2013;20:1250–1257. doi: 10.1038/nsmb.2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidovich C, Wang X, Cifuentes-Rojas C, Goodrich KJ, Gooding AR, Lee JT, Cech TR. Toward a consensus on the binding specificity and promiscuity of PRC2 for RNA. Mol. Cell. 2015;57:552–558. doi: 10.1016/j.molcel.2014.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu XD. Non-coding RNA: a new frontier in regulatory biology. Natl. Sci. Rev. 2014;1:190–204. doi: 10.1093/nsr/nwu008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Z, Zhang J, Bonasio R, Strino F, Sawai A, Parisi F, Kluger Y, Reinberg D. PCGF homologs, CBX proteins, and RYBP define functionally distinct PRC1 family complexes. Mol. Cell. 2012;45:344–356. doi: 10.1016/j.molcel.2012.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgkin J, Zellan JD, Albertson DG. Identification of a candidate primary sex determination locus, fox-1, on the X chromosome of Caenorhabditis elegans. Development. 1994;120:3681–3689. doi: 10.1242/dev.120.12.3681. [DOI] [PubMed] [Google Scholar]
- Jangi M, Boutz PL, Paul P, Sharp PA. Rbfox2 controls autoregulation in RNA-binding protein networks. Genes Dev. 2014;28:637–651. doi: 10.1101/gad.235770.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji X, Zhou Y, Pandit S, Huang J, Li H, Lin CY, Xiao R, Burge CB, Fu XD. SR proteins collaborate with 7SK and promoter-associated nascent RNA to release paused polymerase. Cell. 2013;153:855–868. doi: 10.1016/j.cell.2013.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao L, Liu X. Structural basis of histone H3K27 trimethylation by an active polycomb repressive complex 2. Science. 2015;350 doi: 10.1126/science.aac4383. aac4383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y, Suzuki H, Maegawa S, Endo H, Sugano S, Hashimoto K, Yasuda K, Inoue K. A vertebrate RNA-binding protein Fox-1 regulates tissue-specific splicing via the pentanucleotide GCAUG. EMBO J. 2003;22:905–912. doi: 10.1093/emboj/cdg089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaida D, Berg MG, Younis I, Kasim M, Singh LN, Wan L, Dreyfuss G. U1 snRNP protects pre-mRNAs from premature cleavage and polyadenylation. Nature. 2010;468:664–668. doi: 10.1038/nature09479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko S, Son J, Shen SS, Reinberg D, Bonasio R. PRC2 binds active promoters and contacts nascent RNAs in embryonic stem cells. Nat. Struct. Mol. Biol. 2013;20:1258–1264. doi: 10.1038/nsmb.2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko S, Bonasio R, Saldaña-Meyer R, Yoshida T, Son J, Nishino K, Umezawa A, Reinberg D. Interactions between JARID2 and noncoding RNAs regulate PRC2 recruitment to chromatin. Mol. Cell. 2014a;53:290–300. doi: 10.1016/j.molcel.2013.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko S, Son J, Bonasio R, Shen SS, Reinberg D. Nascent RNA interaction keeps PRC2 activity poised and in check. Genes Dev. 2014b;28:1983–1988. doi: 10.1101/gad.247940.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanhere A, Viiri K, Araújo CC, Rasaiyaah J, Bouwman RD, Whyte WA, Pereira CF, Brookes E, Walker K, Bell GW, et al. Short RNAs are transcribed from repressed polycomb target genes and interact with polycomb repressive complex-2. Mol. Cell. 2010;38:675–688. doi: 10.1016/j.molcel.2010.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KK, Yang Y, Zhu J, Adelstein RS, Kawamoto S. Rbfox3 controls the biogenesis of a subset of microRNAs. Nat. Struct. Mol. Biol. 2014;21:901–910. doi: 10.1038/nsmb.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroyanagi H. Fox-1 family of RNA-binding proteins. Cell. Mol. Life Sci. 2009;66:3895–3907. doi: 10.1007/s00018-009-0120-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert N, Robertson A, Jangi M, McGeary S, Sharp PA, Burge CB. RNA Bind-n-Seq: quantitative assessment of the sequence and structural binding specificity of RNA binding proteins. Mol. Cell. 2014;54:887–900. doi: 10.1016/j.molcel.2014.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JT, Bartolomei MS. X-inactivation, imprinting, and long noncoding RNAs in health and disease. Cell. 2013;152:1308–1323. doi: 10.1016/j.cell.2013.02.016. [DOI] [PubMed] [Google Scholar]
- Lee N, Moss WN, Yario TA, Steitz JA. EBV noncoding RNA binds nascent RNA to drive host PAX5 to viral DNA. Cell. 2015;160:607–618. doi: 10.1016/j.cell.2015.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S, Coutinho-Mansfield G, Wang D, Pandit S, Fu XD. The splicing factor SC35 has an active role in transcriptional elongation. Nat. Struct. Mol. Biol. 2008;15:819–826. doi: 10.1038/nsmb.1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Qiao N, Zhu S, Su M, Sun N, Boyd-Kirkup J, Han JD. A novel Bayesian network inference algorithm for integrative analysis of heterogeneous deep sequencing data. Cell Res. 2013;23:440–443. doi: 10.1038/cr.2013.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margueron R, Justin N, Ohno K, Sharpe ML, Son J, Drury WJ, 3rd, Voigt P, Martin SR, Taylor WR, De Marco V, et al. Role of the polycomb protein EED in the propagation of repressive histone marks. Nature. 2009;461:762–767. doi: 10.1038/nature08398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks H, Kalkan T, Menafra R, Denissov S, Jones K, Hofemeister H, Nichols J, Kranz A, Stewart AF, Smith A, Stunnenberg HG. The transcriptional and epigenomic foundations of ground state pluripotency. Cell. 2012;149:590–604. doi: 10.1016/j.cell.2012.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicoll M, Akerib CC, Meyer BJ. X-chromosome-counting mechanisms that determine nematode sex. Nature. 1997;388:200–204. doi: 10.1038/40669. [DOI] [PubMed] [Google Scholar]
- Norris JD, Fan D, Sherk A, McDonnell DP. A negative coregulator for the human ER. Mol. Endocrinol. 2002;16:459–468. doi: 10.1210/mend.16.3.0787. [DOI] [PubMed] [Google Scholar]
- Rahl PB, Lin CY, Seila AC, Flynn RA, McCuine S, Burge CB, Sharp PA, Young RA. c-Myc regulates transcriptional pause release. Cell. 2010;141:432–445. doi: 10.1016/j.cell.2010.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray D, Kazan H, Cook KB, Weirauch MT, Najafabadi HS, Li X, Gueroussov S, Albu M, Zheng H, Yang A, et al. A compendium of RNA-binding motifs for decoding gene regulation. Nature. 2013;499:172–177. doi: 10.1038/nature12311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu. Rev. Biochem. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera CM, Ren B. Mapping human epigenomes. Cell. 2013;155:39–55. doi: 10.1016/j.cell.2013.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarma K, Cifuentes-Rojas C, Ergun A, Del Rosario A, Jeon Y, White F, Sadreyev R, Lee JT. ATRX directs binding of PRC2 to Xist RNA and Polycomb targets. Cell. 2014;159:869–883. doi: 10.1016/j.cell.2014.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuettengruber B, Chourrout D, Vervoort M, Leblanc B, Cavalli G. Genome regulation by polycomb and trithorax proteins. Cell. 2007;128:735–745. doi: 10.1016/j.cell.2007.02.009. [DOI] [PubMed] [Google Scholar]
- Shao C, Yang B, Wu T, Huang J, Tang P, Zhou Y, Zhou J, Qiu J, Jiang L, Li H, et al. Mechanisms for U2AF to define 3′ splice sites and regulate alternative splicing in the human genome. Nat. Struct. Mol. Biol. 2014;21:997–1005. doi: 10.1038/nsmb.2906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon JA, Kingston RE. Occupying chromatin: Polycomb mechanisms for getting to genomic targets, stopping transcriptional traffic, and staying put. Mol. Cell. 2013;49:808–824. doi: 10.1016/j.molcel.2013.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theunissen TW, Powell BE, Wang H, Mitalipova M, Faddah DA, Reddy J, Fan ZP, Maetzel D, Ganz K, Shi L, et al. Systematic identification of culture conditions for induction and maintenance of naive human pluripotency. Cell Stem Cell. 2014;15:471–487. doi: 10.1016/j.stem.2014.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Underwood JG, Boutz PL, Dougherty JD, Stoilov P, Black DL. Homologues of the Caenorhabditis elegans Fox-1 protein are neuronal splicing regulators in mammals. Mol. Cell. Biol. 2005;25:10005–10016. doi: 10.1128/MCB.25.22.10005-10016.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voigt P, Tee WW, Reinberg D. A double take on bivalent promoters. Genes Dev. 2013;27:1318–1338. doi: 10.1101/gad.219626.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Arai S, Song X, Reichart D, Du K, Pascual G, Tempst P, Rosenfeld MG, Glass CK, Kurokawa R. Induced ncRNAs allosterically modify RNA-binding proteins in cis to inhibit transcription. Nature. 2008;454:126–130. doi: 10.1038/nature06992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Garcia-Bassets I, Benner C, Li W, Su X, Zhou Y, Qiu J, Liu W, Kaikkonen MU, Ohgi KA, et al. Reprogramming transcription by distinct classes of enhancers functionally defined by eRNA. Nature. 2011;474:390–394. doi: 10.1038/nature10006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei C, Qiu J, Zhou Y, Xue Y, Hu J, Ouyang K, Banerjee I, Zhang C, Chen B, Li H, et al. Repression of the central splicing regulator RBFox2 is functionally linked to pressure overload-induced heart failure. Cell Rep. 2015;10:1521–1533. doi: 10.1016/j.celrep.2015.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widnell CC, Hamilton TH, Tata JR. The isolation of enzymically active nuclei from the rat heart and uterus. J. Cell Biol. 1967;32:766–770. doi: 10.1083/jcb.32.3.766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y, Zhou Y, Wu T, Zhu T, Ji X, Kwon YS, Zhang C, Yeo G, Black DL, Sun H, et al. Genome-wide analysis of PTB-RNA interactions reveals a strategy used by the general splicing repressor to modulate exon inclusion or skipping. Mol. Cell. 2009;36:996–1006. doi: 10.1016/j.molcel.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo GW, Coufal NG, Liang TY, Peng GE, Fu XD, Gage FH. An RNA code for the FOX2 splicing regulator revealed by mapping RNA-protein interactions in stem cells. Nat. Struct. Mol. Biol. 2009;16:130–137. doi: 10.1038/nsmb.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan W, Wu T, Fu H, Dai C, Wu H, Liu N, Li X, Xu M, Zhang Z, Niu T, et al. Dense chromatin activates Polycomb repressive complex 2 to regulate H3 lysine 27 methylation. Science. 2012;337:971–975. doi: 10.1126/science.1225237. [DOI] [PubMed] [Google Scholar]
- Zhang C, Zhang Z, Castle J, Sun S, Johnson J, Krainer AR, Zhang MQ. Defining the regulatory network of the tissue-specific splicing factors Fox-1 and Fox-2. Genes Dev. 2008;22:2550–2563. doi: 10.1101/gad.1703108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Ohsumi TK, Kung JT, Ogawa Y, Grau DJ, Sarma K, Song JJ, Kingston RE, Borowsky M, Lee JT. Genome-wide identification of polycomb-associated RNAs by RIP-seq. Mol. Cell. 2010;40:939–953. doi: 10.1016/j.molcel.2010.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]