Abstract
BACKGROUND
Excessive reactive oxygen species generated in mitochondria has been implicated as a causal event in hypertensive cardiomyopathy. Multiple recent studies suggest that microRNAs (miRNAs) are able to translocate to mitochondria to modulate mitochondrial activities, but the medical significance of such a new miRNA function has remained unclear. Here, we characterized spontaneous hypertensive rats (SHRs) in comparison with Wistar rats, finding that micro RNA-21 (miR-21) was dramatically induced in SHRs relative to Wistar rats. We designed a series of experiments to determine whether miR-21 is involved in regulating reactive oxygen species generation in mitochondria, and if so, how induced miR-21 may either contribute to hypertensive cardiomyopathy or represent a compensatory response.
METHODS
Western blotting was used to compare the expression of key nuclear genome (nDNA)–encoded and mitochondrial genome (mtDNA)–encoded genes involved in reactive oxygen species production in SHRs and Wistar rats. Bioinformatics was used to predict miRNA targets followed by biochemical validation using quantitative real-time polymerase chain reaction and Ago2 immunoprecipitation. The direct role of miRNA in mitochondria was determined by GW182 dependence, which is required for miRNA to function in the cytoplasm, but not in mitochondria. Recombinant adeno-associated virus (type 9) was used to deliver miRNA mimic to rats via tail vein, and blood pressure was monitored with a photoelectric tail-cuff system. Cardiac structure and functions were assessed by echocardiography and catheter manometer system.
RESULTS
We observed a marked reduction of mtDNA-encoded cytochrome b (mt-Cytb) in the heart of SHRs. Downregulation of mt-Cytb by small interfering RNA in mitochondria recapitulates some key disease features, including elevated reactive oxygen species production. Computational prediction coupled with biochemical analysis revealed that miR-21 directly targeted mt-Cytb to positively modulate mt-Cytb translation in mitochondria. Circulating miR-21 levels in hypertensive patients were significantly higher than those in controls, showing a positive correlation between miR-21 expression and blood pressure. Remarkably, recombinant adeno-associated virus–mediated delivery of miR-21 was sufficient to reduce blood pressure and attenuate cardiac hypertrophy in SHRs.
CONCLUSIONS
Our findings reveal a positive function of miR-21 in mitochondrial translation, which is sufficient to reduce blood pressure and alleviate cardiac hypertrophy in SHRs. This observation indicates that induced miR-21 is part of the compensatory program and suggests a novel theoretical ground for developing miRNA-based therapeutics against hypertension.
Keywords: cytochrome b; MIR-21 microRNA, human; mitochondria; reactive oxygen species
Hypertension has remained a major public health burden, because it is associated with considerable morbidity and mortality.1 Patients with high blood pressure frequently show abnormalities in cardiac structure and function, including left ventricular hypertrophy, systolic/diastolic dysfunction, and, in many cases, heart failure.2 Despite advances in developing antihypertensive therapies, the number of patients with uncontrolled hypertension continues to rise, thus stressing the importance of understanding the mechanisms underlying hypertension and hypertensive cardiomyopathy to develop new intervention strategies.
Recent studies have linked mutations in mitochondrial DNA (mtDNA) to hypertension,3,4 which has been attributed, at least in part, to increased reactive oxygen species (ROS) production.5 In fact, elevated ROS has been implicated in diverse pathologies, such as heart failure, atherosclerosis, diabetes mellitus, chronic kidney disease, and cancer. However, most clinical trials of antioxidants (eg, vitamin E and vitamin C) on hypertension have met limited success.6 The conundrum might result from inaccessibility of orally administered antioxidants to sources of free radicals, in particular, those generated by the mitochondria.7 Recent results have also attributed angiotensin II–induced hypertension to increased mt-ROS.8 Importantly, although inhibition of mt-ROS by MitoQ showed a measureable degree of benefit in reducing blood pressure and alleviating cardiac hypertrophy in spontaneous hypertensive rats (SHRs),9 mitochondria-targeted antioxidants seem relatively inefficient in scavenging mt-ROS.6
MicroRNAs (miRNAs) are a class of small (≈22 nt) non-coding RNAs that negatively regulate gene expression at posttranscriptional levels.10 Interestingly, recent data suggest that miRNAs also may regulate gene expression in a positive fashion.11,12 Early studies have linked altered miRNA expression to hypertension, atherosclerosis, arrhythmia, and fibrosis.13 A recent study indicates that stress-induced nuclear miR-181c compromises mitochondrial translation, leading to defective mitochondrial electron transport chain (ETC) function and increased mt-ROS.14 However, the functional consequences have not yet been linked directly to the action of this miRNA within the mitochondria. Interestingly, another recent report demonstrates that miR-1, a miRNA specifically induced during myogenesis, is not only efficiently targeted to mitochondria, but is also able to enhance mitochondrial translation directly.15 These studies have thus raised a new regulatory paradigm for miRNAs to function in mitochondria.
The ETC complexes are well known to be the major source of mt-ROS.16 Previous studies have focused on nuclear-encoded subunits of the ETC and their contributions to mt-ROS production in the SHR model.17,18 Because mutations in mtDNA have been linked to essential hypertension, we hypothesized that altered expression of mtDNA-encoded proteins might be causal to initiation and progression of hypertension. In the present study, we identified downregulation of mtDNA-encoded cytochrome b (mt-Cytb) in SHRs, which seems to contribute directly to increased mt-ROS. We found that miRNA-21 (miR-21), a key miRNA induced in SHRs, is able to translocate into mitochondria to counteract mt-Cytb down-regulation, and strikingly, we showed that exogenous miR-21 delivered by recombinant adeno-associated virus (rAAV) was sufficient to lower blood pressure in the SHR model, suggesting a new therapeutic strategy against hypertension.
METHODS
All animal studies were conducted with the approval of the Animal Research Committee of Tongji Medical College, and in accordance with the NIH Guide for the Care and Use of Laboratory Animals. Patients and controls were enrolled in accordance with the Declaration of Helsinki and approved by the Ethics Committee of Tongji Hospital. Blood samples were collected via venous puncture after obtaining written informed consent. Expanded versions of Methods, including list of antibodies (online-only Data Supplement Table I) and a list of polymerase chain reaction (PCR) primers (online-only Data Supplement Table II), are presented in the online-only Data Supplement.
Mitochondria Isolation
Mitochondria were isolated by using the MACS Mitochondria Extraction Kit from Miltenyi Biotec GmbH as described.19 In brief, cells were lysed, and mitochondria were magnetically captured with anti-TOM22 antibody-coated beads, and eluted mitochondria were collected by centrifugation at 13 000g for 2 minutes at 4°C.
Quantifcation of ROS Production
The oxidative fluorescent dye, dihydroethidium (DHE; Invitrogen) was applied to frozen, 7-µm sections from various organs at 40 µmol for 30 minutes. Fluorescence intensity was measured under a Nikon DXM1200 fluorescence microscope and images were analyzed with the Image-Pro software (Media Cybernetics).
mt-ROS was measured with MitoSOX Red (Invitrogen) on live cells, as described.20 In brief, cells were washed 3 times with phosphate-buffered saline. MitoSOX Red (diluted to a final concentration of 5 µmol/L) was added to the media and incubated for 30 minutes at 37°C in the dark. After incubation, cells were trypsinized and washed with ice-cold phosphate-buffered saline 3 times. mt-ROS was quantified by flow cytometry (BD Biosciences) with 510 nm excitation/580 nm emission filters. Total ROS was quantified by using 2,7-dichlorodihydrofluorescein diacetate (Invitrogen), as described.21
Quantifcation of the Copy Number of miR-21 and Cytb in the Mitochondria
To quantify the levels of Cytb mRNA in the mitochondria, total mitochondrial RNA was reverse transcribed to cDNA. A standard curve was generated by quantitative PCR using serially diluted Cytb DNA, which was used to determine the absolute levels of Cytb. miR-21 was similarly quantified in the mitochondria based on a standard curve generated with serially diluted miR-21 (Riobio Co., Ltd). Quantitative analysis of miR-21 in purified mitochondria from cells and tissues was according to the published procedure.15,22
Polysome Analysis
Mitochondrial polysome profiling on sucrose gradient was performed as described previously.15,23 In brief, mitochondria purified from H9c2 cells were suspended in the lysis buffer on ice for 20 minutes. The lysate was centrifuged at 9000g for 30 minutes to remove particles, loaded on a 10% to 30% sucrose gradient, and centrifuged at 180 000g for 260 minutes in Beckman SW41-Ti rotor. After centrifugation, 13 fractions were collected for RNA and protein analysis. Purified mitochondria were free of cytoplasmic ribosomes, as indicated by the lack of representative cytoplasmic ribosomal proteins (RPS3 and RPL4) in Western blots. Representative mitochondrial ribosomal proteins (MRPS27 and MRPL45) on individual gradient fractions were detected by Western blotting and specific rRNA, mRNA, and miRNA transcripts were quantified by real-time PCR. The assignment of small and large ribosomal subunits, monosomes, and putative polysomes was based on the distribution of both rRNAs and ribosomal proteins and comparison with published mitochondrial polysome profiles. To characterize putative polysomes, the lysate was treated with 5 U/mL RNase I for 40 minutes at 25°C to convert polysome to monosome. The relative abundance of individual transcripts in each fraction was presented as the percentage of the total fraction.
Application of rAAV to Animals
rAAVs (type 9) containing miR-21, mut-miR-21, anti-miR-21, mut-anti-miR-21, or green fluorescent protein (GFP) were prepared by triple plasmid cotransfection in HEK293T cells, as previously described.24 Detail information is provided in the online-only Data Supplement Expanded Methods.
Statistical Analysis
Data were presented as mean±standard error of the mean (n noted in specific figure legends). Student t tests and analysis of variance with Bonferroni post hoc analysis were performed to determine statistical significance between different treatment groups. The data of blood pressure in rats over the time course were statistically analyzed by repeated measures analysis of variance. To assess the significance of the correlations, Spearman rank correlation coefficient was calculated. In all cases, statistical significance was defined as P<0.05.
RESULTS
Downregulation of Mitochondrial Cytb Linked to Increased ROS Production in SHR Heart
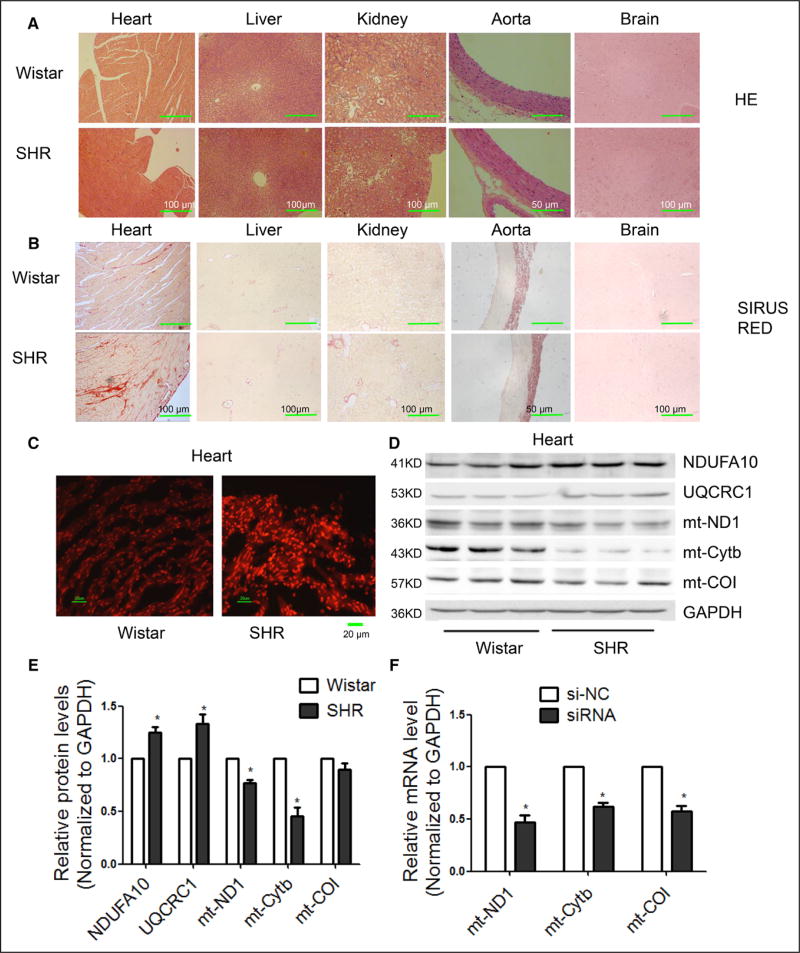
We performed hematoxylin and eosin staining to detect gross morphological changes and Sirius red staining to detect fibrosis in various organs of SHRs at the age of 9 months. In comparison with other organs, we observed significant hypertrophy and fibrosis in the heart of SHRs (Figure 1A and 1B). We therefore focused on cardiac damage in SHRs.
Figure 1. Downregulated mitochondrial Cytb linked to increased ROS production in the heart of SHRs.
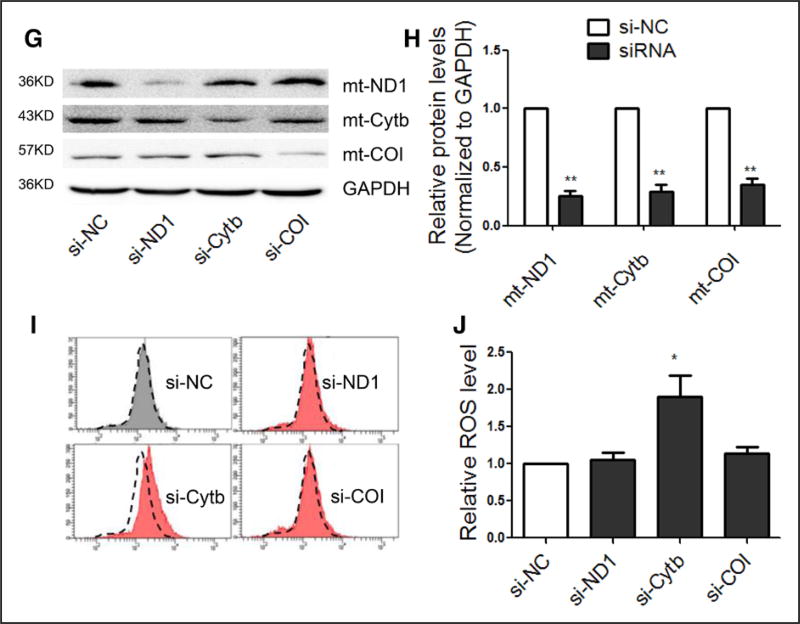
A, Representative images of HE staining in various organs of SHRs in comparison with Wistar rats. B, Representative images of Sirius red staining in various organs of SHRs in comparison with Wistar rats. C, Representative images of ROS detected by DHE probe in frozen heart sections of SHRs in comparison with Wistar rats. D and E, Western blot analysis of the ETC subunits in the heart of SHRs. n=3, results from Wistar rats were set to 1, *P<0.05 in comparison with Wistar rats. F, mRNA levels of mitochondrial subunits in H9c2 cells transfected with the indicated siRNAs. n=3, results from si-NC were set to 1, *P<0.05 relative to si-NC. G and H, Protein levels of mitochondrial subunits in H9c2 cells transfected with the indicated siRNAs. I and J, ROS levels in H9c2 cells transfected with si-ND1, si-Cytb or si-COI. n=3, results from si-NC were set to 1. *P<0.05. **P<0.01 related to si-NC. DHE indicates dihydroethidium; ETC, electron transport chain; HE, hematoxylin and eosin; ROS, reactive oxygen species; SHR, spontaneous hypertensive rat; and si, small interfering.
We detected total superoxide and substantially increased ROS in frozen heart sections of SHRs in comparison with Wistar controls (Figure 1C). We next measured the expression of several representative proteins of the ETC by Western blotting, and consistent with earlier reports,17,18 we found significant upregulation of nuclear genome (nDNA)-encoded NDUFA10 (subunit of ETC I) and UQCRC1 (subunit of ETC III) (Figure 1D and 1E). Interestingly, we also detected a significant decrease of mtDNA-encoded ND1 (subunit of ETC I) and Cytb (subunit of ETC III) in SHR hearts relative to controls, whereas COI (subunit of ETC IV) remained unaltered (Figure 1D and 1E). These data suggest a compromised coordination of nDNA- and mtDNA-encoded ETC components in the heart of SHRs.
By measuring ROS production in other organs, we also found increased ROS in liver, kidney, and aorta of SHRs than in Wistar controls (online-only Data Supplement Figure IA). We also detected decreased expression of Cytb in both liver and kidney in SHRs relative to Wistar controls (online-only Data Supplement Figure IB through II).
Based on the demonstrated ability of exogenous small interfering RNAs (siRNAs) to target mtDNA-encoded transcripts in the mitochondria (X. Zhang and X.-D. Fu, unpublished results, 2016), we explored the use of specific siRNAs against mtDNA-encoded Cytb, ND1, and COI to determine their contributions to ROS production in H9c2 cells, a rat cell line derived from embryonic heart tissue. We found that each siRNA treatment led to specific downregulation of their mitochondrial targets at both the mRNA (Figure 1F) and protein (Figure 1G and 1H) levels. Interestingly, siCytb specifically caused elevated ROS in comparison with siND1 and siCOI (Figure 1I), which is evident from quantified results (Figure 1J and online-only Data Supplement Figure IJ and IK). These data indicate a critical role of Cytb in ROS generation.
Identification of miR-21 Involved in the Regulation of Cytb Expression
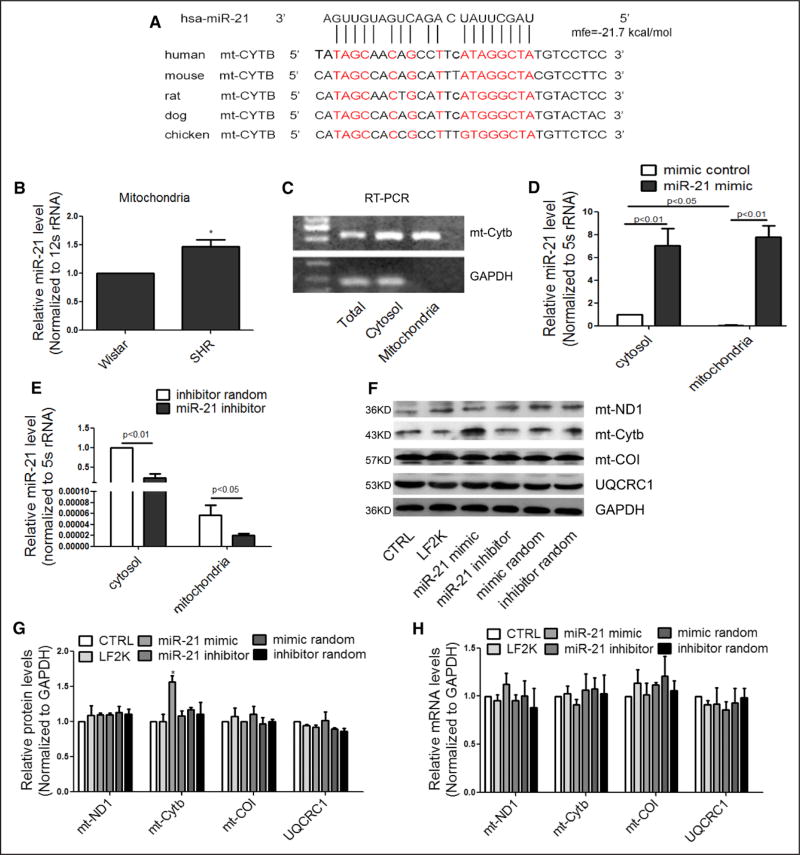
Given the recently reported roles of miRNAs in mitochondria, we tested the hypothesis that Cytb downregulation might be mediated by a specific miRNA in SHRs. We thus used RNAhybrid software25 to search for potential miRNAs that may specifically target the Cytb transcript. This led to the identification of miR-21, which showed the potential to target Cytb mRNA in a highly conserved region among human, rat, and mouse (Figure 2A). More importantly, we found that miR-21 was overexpressed in the mitochondria of SHR hearts (Figure 2B and online-only Data Supplement Figure IIA), which might be responsible for causing Cytb downregulation.
Figure 2. miR-21 enhanced translation of Cytb in the mitochondria.
A, Sequence alignment of miR-21 on the Cytb mRNA from different organisms. Extensive base pairings are evident both at the 5′ seed and the 3′ region of miR-21. B, miR-21 levels in purified mitochondria for SHR and Wistar hearts. n=3, results from Wistar rats were set to 1, *P<0.05. C, Real-time PCR analysis of mitochondrial Cytb mRNA and cytoplasmic GAPDH mRNA in purified mitochondria. D and E, miR-21 levels in the cytosol and mitochondria of transfected H9c2 cells. n=3, results from control cytosol were set to 1. F and G, Effects of transfected miR-21 on mitochondrial subunits at the protein levels. n=3, results from control were set to 1, *P<0.05. H, Effects of miR-21 on mitochondrial subunits at the mRNA levels. n=3, results from control were set to 1. mt indicates mitochondrial; RT-PCR, real-time polymerase chain reaction; and SHR, spontaneous hypertensive rat.
To determine the potential effect of miR-21 on Cytb expression, we transfected a miR-21 mimic into H9c2 cells. To demonstrate that transfected miR-21 mimic entered the mitochondria, we used anti-TOM22–coated microbeads to purify the mitochondria from transfected cells, showing no measurable contamination of cytoplasmic GAPDH mRNA in our preparation (Figure 2C). We next quantified miR-21 in both cytosol and purified mitochondria from transfected cells, detecting transfected miR-21 in the mitochondria (Figure 2D). By transfecting a miR-21 inhibitor into the cell, we detected decreased miR-21 in both cytosol and purified mitochondria (Figure 2E). We made similar observations on both HK2 and HEK293 cells (online-only Data Supplement Figure IIB through IIE).
Contrary to our expectation, we detected a specific increase in the Cytb protein level in miR-21 mimic-transfected H9c2 cells (Figure 2F). Quantitative analysis based on triplicated experiments demonstrated increased Cytb protein without change at the mRNA level (Figure 2G and 2H). The effect was specific, because other subunits of the ETC were unaffected (Figure 2F through 2H). We did not observe the converse effect with the miR-21 inhibitor on the expression of the Cytb protein in transfected H9c2 cells (Figure 2F and 2G), indicating the limited abundance of endogenous miR-21 in the mitochondria of H9c2 cells, which we confirmed by measuring the ratio of miR-21 molecules per Cytb transcript in the mitochondria, showing insignificant amounts of miR-21 in untransfected cells (Table 1). We made similar observations on multiple other cell types, including HK2, HEK293, and HUVEC cells (online-only Data Supplement Figure IIF through IIK). These data are reminiscent of the positive role of miR-1 in mitochondrial translation,15 and suggest a protective role of induced miR-21 as part of the compensatory program in SHRs.
Table 1.
The Ratio of miR-21 Molecules per Cytb Transcript in Mitochondria of Cultured Cells
| Cell Types | Negative Control |
miR-21 Mimic | miR-21 Inhibitor |
|---|---|---|---|
| H9c2 | 0.110 | 97* | 0.066* |
| HK2 | 0.073 | 47* | 0.028* |
| HEK293 | 0.052 | 40* | 0.029* |
Cytb indicates cytochrome b; and miR-21, microRNA-21.
P<0.05 versus negative control, n=3.
MiR-21 Directly Enhanced Cytb mRNA Translation in the Mitochondria
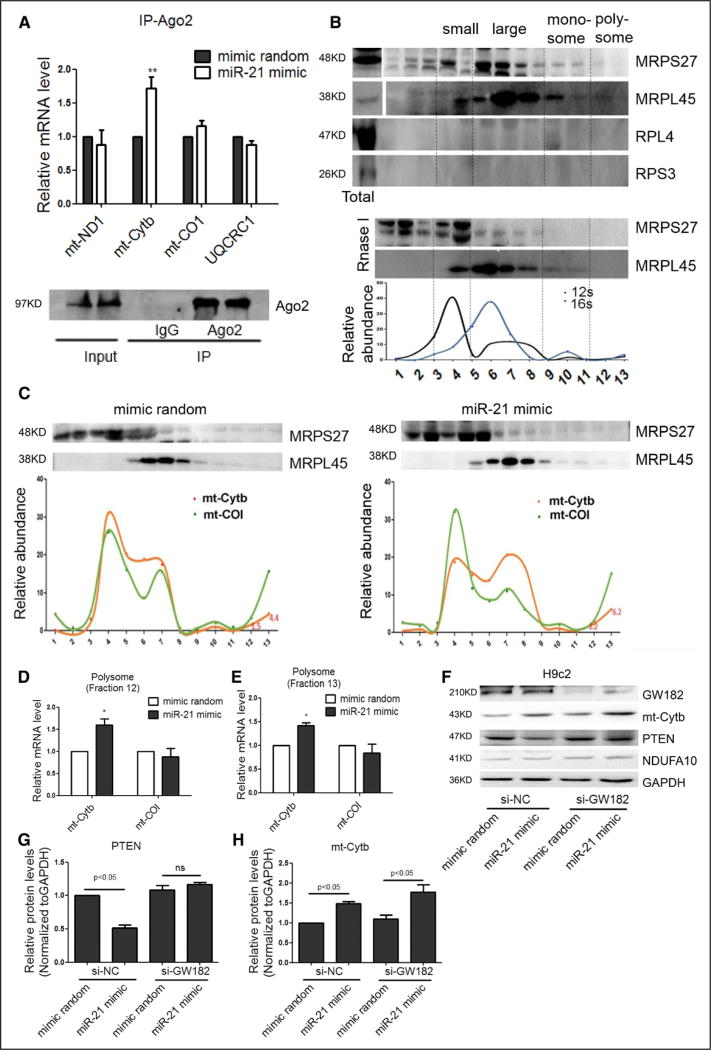
To pursue the mechanism underlying miR-21 enhanced mitochondrial translation, we performed RNA coimmunoprecipitation with anti-Ago2, which has been previously shown to have a fraction localized in the mitochondria.15 We found that Ago2 showed increased association with the Cytb mRNA after miR-21 transfection (Figure 3A), consistent with miR-21-guided targeting of the microRNA machinery to Cytb.
Figure 3. miR-21-stimulated interaction of the Cytb mRNA with the mitochondrial translation machinery.
A, Real-time PCR analysis of mRNA in association with Ago2 in H9c2 cells. n=3, results from control were set to 1, **P<0.01. B, Mitochondrial polysome profiling. Isolated mitochondria free of markers of cytoplasmic ribosomes (RPS3 and RPL4) were fractionated on a sucrose gradient. The assignment of small and large ribosomal subunits, monosomes, and putative polysomes was based on the distribution of both 12/16S RNAs and mitochondrial ribosomal proteins (MRPS27 and MRPL45). The putative polysome fractions near the bottom of the gradient (fractions 12 and 13) could be converted to monosomes by RNase I treatment. The relative abundance of individual transcripts in each fraction was presented as the percentage of the total fraction. C through E, The association of the Cytb mRNA with putative polysome fractions (fractions 12 and 13) in response to miR-21 mimic treatment. n=3, results from control were set to 1, *P<0.05. F through H, Effects of miR-21 on Cytb and PTEN protein expression with the miRNA machinery selectively inactivated by knocking down GW182 in the cytoplasm, n=3, results from si-NC control were set to 1. miRNA indicates microRNA; mt, mitochondrial; PCR, polymerase chain reaction; and si, small interfering.
To determine how miR-21 might enhance the expression of Cytb at the protein level, we performed polysome analysis, a gold standard for studying translation. We characterized the 10% to 30% sucrose gradient fraction by showing the distribution of representative small and large mitochondrial ribosomal proteins (MRPS27 and MRPL45) and 12S and 16S rRNAs, as well, in the expected fractions (Figure 3B), as reported in the literature.15,23 We detected no contamination of cytoplasmic ribosomes based on immunoblotting for 18S and 28S ribosomal proteins RPL4 and RPS3, respectively, in our gradient fractions. We further showed that the putative polysome fractions near the bottom of the gradient (fractions 12 and 13) could be converted to monosome by RNase I treatment (Figure 3B). Significantly, by real-time quantitative PCR, we detected increased Cytb mRNA in the putative polysome fractions (fractions 12 and 13) in response to transfected miR-21 mimic (Figure 3C) in comparison with the internal CO1 mRNA control (Figure 3D and 3E). These data suggest that miR-21 directly enhanced Cytb mRNA translation in mitochondria.
To rigorously rule out indirect effects of the miR-21 mimic via cytoplasmic targets, we performed a diagnostic analysis on the differential requirement for the Ago2 cofactor GW182, which is required for miRNA effects in the cytoplasm,26 but not in mitochondria.18 As expected, GW182 knockdown prevented miR-21–mediated translational repression of its cytoplasmic target PTEN (Figure 3F through 3H).27 Under these conditions, GW182 RNAi-treated H9c2 cells continued to show enhanced Cytb translation by the miR-21 mimic (Figure 3F through 3H). The same results also were obtained in transfected HK2 and HEK293 cells (online-only Data Supplement Figure IIIA through IIIF). Together, these data strongly suggest a direct role of miR-21 in enhancing Cytb translation in mitochondria.
Ability of Transfected miR-21 Mimic to Quench ROS
To determine the functional consequence of the transfected miR-21 mimic, we used MitoSOX Red to monitor mitochondrial ROS production. This reagent rapidly and selectively targets mitochondria in live cells, which can be oxidized by superoxide but not by other ROS- or reactive nitrogen species–generating systems. We found that the transfected miR-21 mimic had no significant impact on mt-ROS in H9c2 cells (Figure 4A), nor in HK2 and HEK293 cells (online-only Data Supplement Figure IVA and IVB). mt-ROS could be induced with either siCytb or antimycin A (inhibitor of electron transfer at ETC III) (Figure 4B) and siCytb was unable to further elevate mt-ROS in cells treated with antimycin A (Figure 4B). Under these conditions, we found that the transfected miR-21 mimic was able to suppress siCytb-induced, but not antimycin A–elevated, mt-ROS (Figure 4C and 4D).
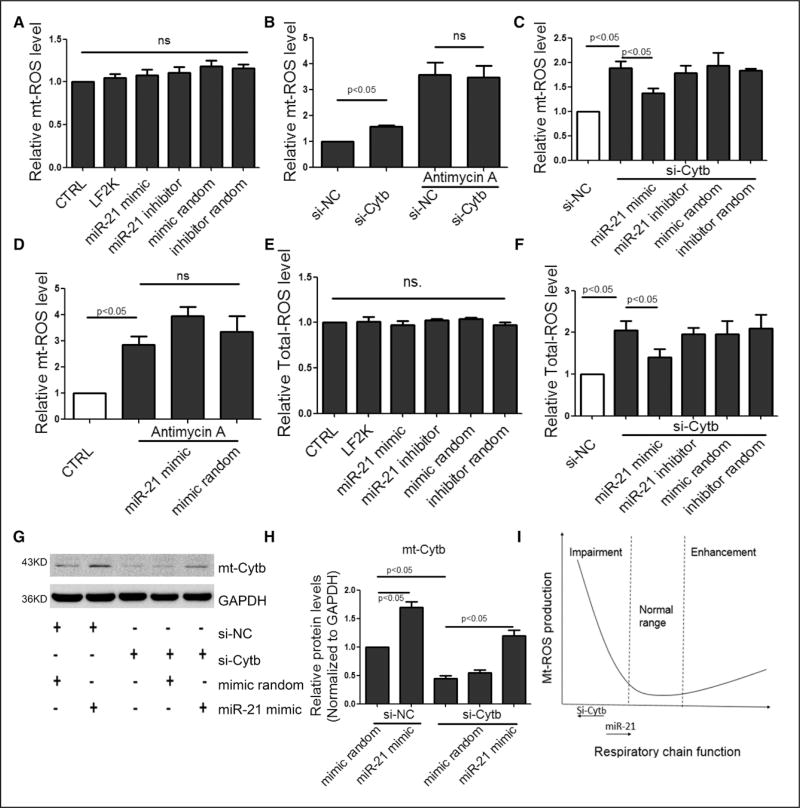
Figure 4. Transfected miR-21 mimic to quench ROS.
A, Effect of miR-21 on mt-ROS in transfected H9c2 cells. B, Effects of si-Cytb and Antimycin A on mt-ROS. C, Effect of miR-21 on mt-ROS in si-Cytb–treated cells. D, Effect of miR-21 on mt-ROS in Antimycin A–treated cells. E, Effect of miR-21 on total-ROS. F, Effect of miR-21 on total-ROS in si-Cytb–treated cells. G and H, Effect of miR-21 on the Cytb protein level, results from si-NC control were set to 1. I, A model to illustrate the effects of si-Cytb and miR-21 on mt-ROS production. All data were presented as mean±SEM, n=3. mt indicates mitochondrial; ROS, reactive oxygen species; SEM, standard error of the mean; and si, small interfering.
These data indicated that the protective effect conferred by miR-21 via increased Cytb was dependent on electron transfer at ETC III, suggesting that the unbalanced composition in the ETC III complex interfered with its normal function, leading to increased mt-ROS. Total ROS detected by 2,7-dichlorodihydrofluorescein diacetate showed a similar trend as mt-ROS under similar conditions (Figure 4E and 4F). We obtained the same results in HK2 and HEK293 cells (online-only Data Supplement Figure IVC through IVF). We further confirmed that miR-21 was able to counteract siCytb-induced mt-ROS by up-regulating Cytb at the protein level (Figure 4G and 4H). Together, these data support a model in which increased mt-ROS is induced by siCytb and suppressed by miR-21 (Figure 4I).
Blood Pressure Reduction by Short-Term rAAV-miR-21 Treatment
To investigate the functional relevance of our molecular findings, we characterized clinical characteristics among patients with hypertension in comparison with control subjects (online-only Data Supplement Table III). With the exception of blood pressure, there were no significant differences in age, sex, body mass index, levels of fasting glucose and lipids, and other biochemical parameters among the 2 comparison groups. However, hypertensive patients showed significantly higher levels of circulating miR-21 than healthy controls (Figure 5A). We thus further investigated the relationship between plasma miR-21 concentrations and blood pressure levels among all participants, finding that the plasma levels of miR-21 were correlated positively with the levels of either systolic or diastolic blood pressure (Figure 5B and 5C). These findings suggest a potential role of increased miR-21 as part of the compensatory program in hypertensive patients in light of our findings in SHRs.
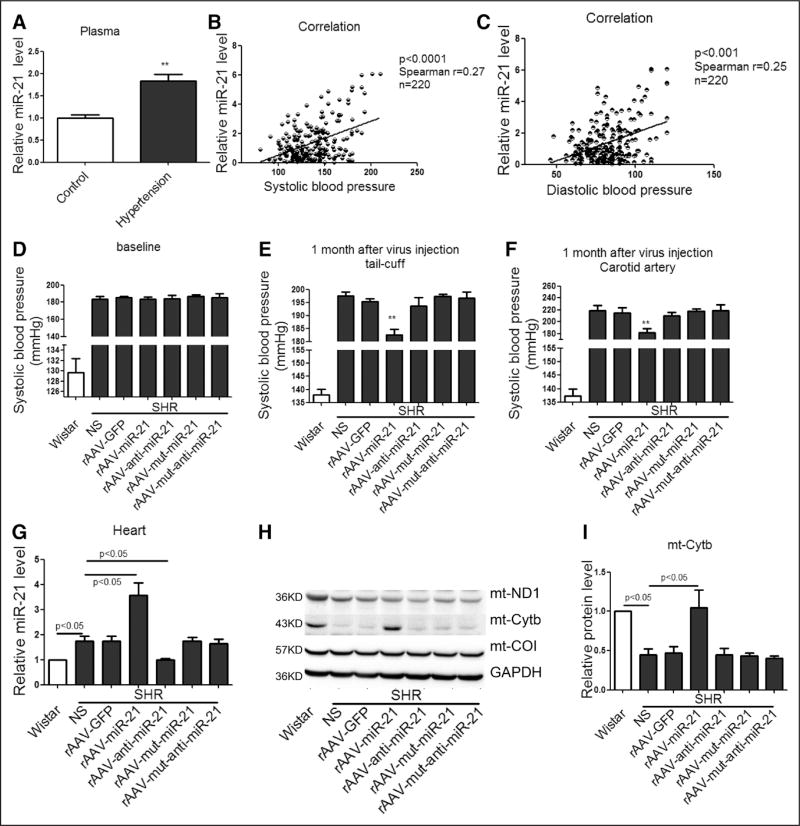
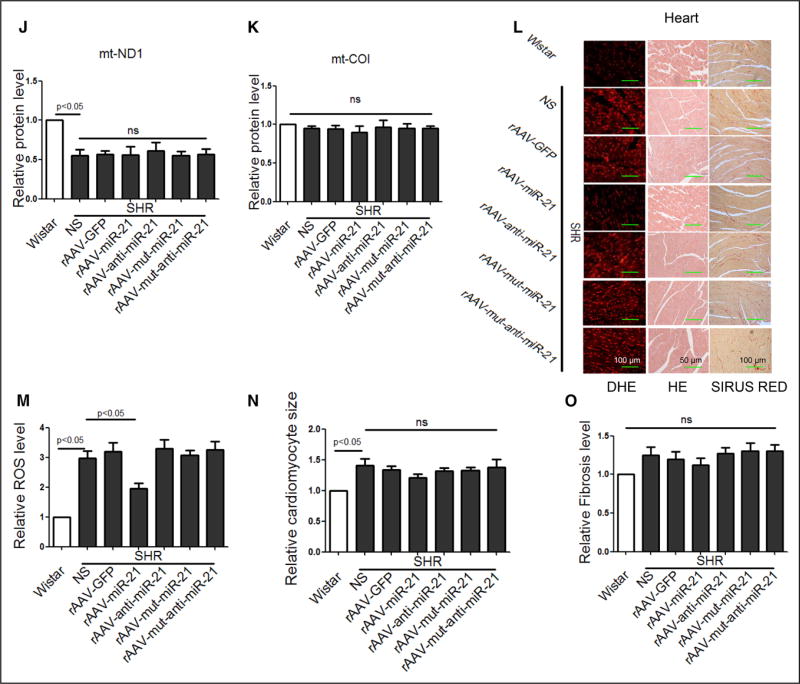
Figure 5. Short-term treatment of rAAV-delivered miR-21 reduced blood pressure in SHRs.
A, The levels of circulating miR-21 in healthy control and hypertensive patients. *P<0.01. B, The correlation between circulating miR-21 levels and systolic pressure. C, The correlation between circulating miR-21 levels and diastolic pressure. D and E, Systolic blood pressure in SHRs measured by tail-cuff method. F, Maximum pressure of carotid artery monitored by Millar Pressure-Volume System. G, miR-21 levels in heart of various groups, results from Wistar rats were set to 1. H through K, Western blot analysis of Cytb protein in the heart of rAAV-miR-21–treated SHRs, results from Wistar rats were set to 1. L through O, HE, Sirius red, and DHE staining of myocardial tissue in SHRs. All data were presented as mean±SEM, n=10. *P<0.05. **P<0.01. DHE indicates dihydroethidium; GFP, green fluorescent protein; HE, hematoxylin and eosin; mt, mitochondrial; rAAV, recombinant adeno-associated virus; ROS, reactive oxygen species; SEM, standard error of the mean; and SHR, spontaneous hypertensive rat.
We next used the SHR model to determine whether miR-21 could reduce blood pressure and alleviate cardiac hypertrophy by using rAAV to deliver miR-21 into the animal. We first monitored potential short-term effects of miR-21 treatment on blood pressure and cardiac hypertrophy. Eight-week-old male SHRs were divided into 6 groups (n=10 in each), each treated with NS (control saline), rAAV-GFP, rAAV-miR-21, rAAV-mut-miR-21, rAAV-anti-miR-21, or rAAV-mut-anti-miR-21 for 4 weeks. The blood pressure among various groups of SHRs showed no significant differences before rAAV delivery (Figure 5D). After 1 month treatment, rAAV-miR-21 significantly reduced systolic blood pressure measured either by tail-cuff or hemodynamic detection in SHRs, and, in contrast, rAAV-anti-miR-21 showed no obvious effect (Figure 5E and 5F).
We confirmed increased miR-21 in hearts of rAAV-miR-21–treated SHRs by real-time PCR (Figure 5G), and upregulated Cytb by Western blot (Figure 5H through 5K). Importantly, DHE detected decreased ROS only in rAAV-miR-21–treated SHRs (Figure 5L and 5M). We observed no significant difference in cardiac function and morphology among various groups of SHRs (Table 2 and Figure 5N and 5O).
Table 2.
Comparison of Echocardiographic and Hemodynamic Measurements in Short-Term rAAV-miR-21–Treated SHRs
| Wistar | SHRs | ||||||
|---|---|---|---|---|---|---|---|
| NS | rAAV-GFP | rAAV-miR-21 | rAAV-anti- miR-21 |
rAAV-mut- miR-21 |
rAAV-mut- anti-miR-21 |
||
| HR, bpm | 396±3.9 | 407±9.3 | 429±9.6 | 416±10.3 | 413±5.3 | 433±7.3 | 425±6.7 |
| Pes, mm Hg | 140±1 | 219±3* | 214±9 | 181±6† | 210±6 | 218±3 | 219±9 |
| dp/dtmax | 13 558±1015 | 15 245±1130 | 15 927±1154 | 14 238±775 | 15 421±732 | 15 968±833 | 16 965±1106 |
| dp/dtmin | −10 645±1566 | −13 561±593 | −12 860±1226 | −11 797±766 | −13 745±1178 | −13 629±1248 | −13 038±1269 |
| EF, % | 79.8±3.1 | 88.1±2.3* | 88.0±2.2 | 86.8±2.9 | 84.5±1.7 | 85.4±3.0 | 86.5±3.1 |
| IVSTd, mm | 2.02±0.07 | 2.24±0.12 | 2.20±0.10 | 2.09±0.06 | 2.21±0.14 | 2.20±0.09 | 2.26±0.11 |
| IVSTs, mm | 3.30±0.13 | 3.25±0.09 | 3.43±0.12 | 3.37±0.17 | 3.33±0.15 | 3.27±0.14 | 3.35±0.13 |
Data were presented as mean±SEM, n=10. dp/dtmax indicates peak instantaneous rate of left ventricular pressure increase; dp/dtmin, peak instantaneous rate of left ventricular pressure increase decline; EF, ejection fraction of left ventricular; GFP, green fluorescent protein; HR, heart rate; IVSTd, interventricular septum thickness in end-diastolic of left ventricle; IVSTs, interventricular septum thickness in end-systolic of left ventricular; miR-21, microRNA-21; mut, mutation; NS, control saline; Pes, end systolic pressure; rAAV, recombinant adeno-associated virus; SEM, standard error of the mean; and SHR, spontaneous hypertensive rat.
P<0.05 versus Wistar control.
P<0.05 versus SHRs treated with NS.
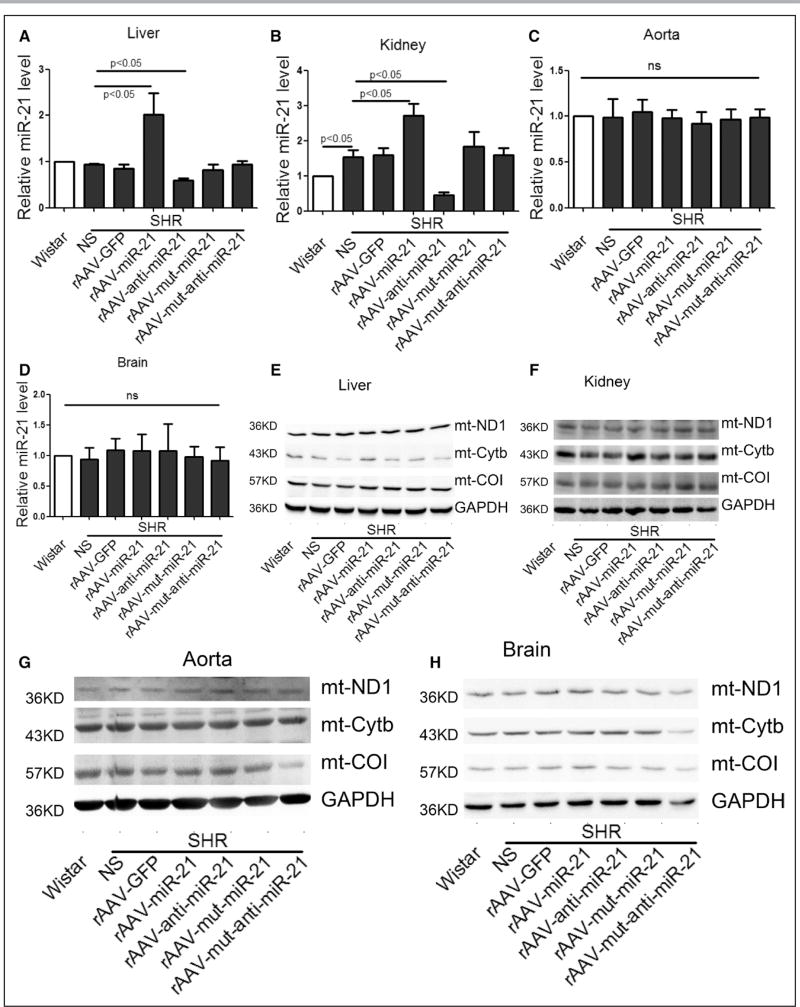
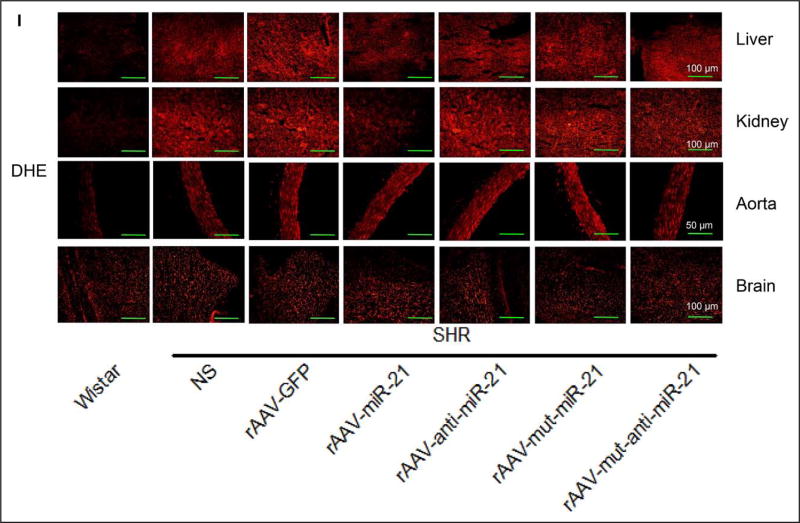
To search for potential organs that may contribute to elevated blood pressure, we examined several organs, particularly liver and kidney, the latter of which has recently been linked to hypertension.28,29 Although lung is known to contribute to pulmonary hypertension, we did not detect any differences in morphology, miR-21 levels, and expression of representative nDNA- and mtDNA-encoded mitochondrial proteins between Wistar and SHR (online-only Data Supplement Figure V). We thus excluded lung from further analysis. Strikingly, we detected increased miR-21, elevated Cytb protein, and decreased ROS in liver and kidney, but not aorta, in rAAV-miR-21–treated SHRs (Figure 6A through 6I). Somewhat unexpectedly, we did not detect the effect of miR-21 inhibitor in increasing the blood pressure in SHRs as anticipated by inhibiting the compensatory program, despite the ability of rAAV-anti-miR-21 to decrease the expression of miR-21 in various organs (Figure 6A and 6B).
Figure 6. Effects of short-term rAAV-miR-21 treatments in various organs of SHRs.
A through D, miR-21 levels in liver, kidney, aorta, and brain of various groups, results from Wistar rats were set to 1. E through H, Western blot analysis of Cytb protein in liver, kidney, aorta, and brain of various groups. I, ROS detected by DHE probe in liver, kidney, aorta, and brain of various groups. All data were presented as mean±SEM, n=5. *P<0.05. **P<0.01. DHE indicates dihydroethidium; GFP, green fluorescent protein; mut, mutation; rAAV, recombinant adeno-associated virus; ROS, reactive oxygen species; SEM, standard error of the mean; and SHR, spontaneous hypertensive rat.
Together, our data indicate that exogenous miR-21 is able to counteract the disease state, but reduction of endogenous miR-21 by antagomir is insufficient to alter the phenotype in the established disease state, at least in the treatment window we studied (see further in Discussion).
Cardiac Hypertrophy Alleviated by Long-Term rAAV-miR-21 Treatment
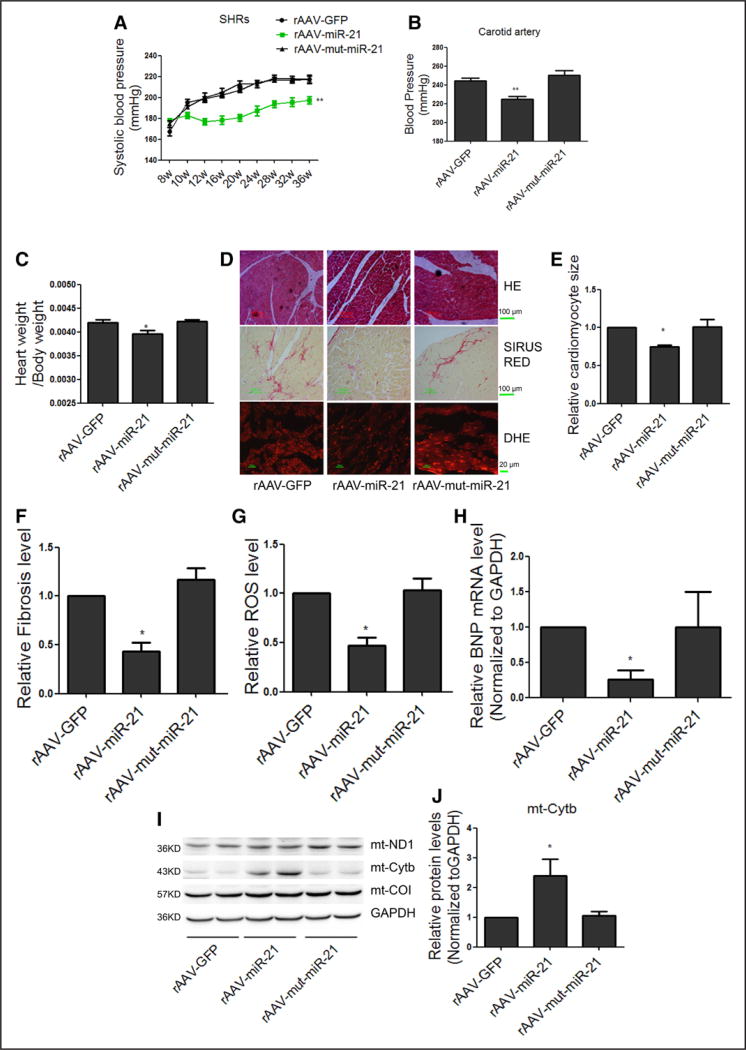
We next measured the effects of chronic miR-21 treatment on blood pressure and cardiac hypertrophy. Eight-week-old male SHRs were divided into 3 groups (n=5 in each), each treated with rAAV-GFP, rAAV-miR-21, and rAAV-mut-miR-21 for 28 weeks. We found that rAAV-miR-21, but not rAAV-GFP or rAAV-mut-miR-21, could indeed significantly lower systolic blood pressure in SHRs (Figure 7A). We achieved ≈20 mm Hg reduction in blood pressure after 4 weeks of rAAV-miR-21 treatment, which was maintained throughout the remaining study period (Figure 7A).
Figure 7. Long-term treatment of rAAV delivered miR-21-reduced blood pressure and inhibited cardiac hypertrophy in SHRs.
A, Systolic blood pressure in SHRs measured by tail-cuff method. B, Maximum pressure of carotid artery monitored by Millar Pressure-Volume System. C, Cardiac mass index in SHRs treated with rAAV-miR-21. D, Representative images of HE staining, Sirius red staining and DHE of myocardial tissue in SHRs. E through G, Quantification of cardiomyocyte size, fibrosis, and ROS under different treatment conditions. H, Real-time PCR analysis of BNP mRNA levels in SHR hearts, results from rAAV-GFP were set to 1. I and J, Western blot analysis of Cytb protein in the heart of rAAV-miR-21–treated SHRs, results from rAAV-GFP were set to 1. All data were presented as mean±SEM, curves were compared using repeated measures ANOVA, n=5. *P<0.05. **P<0.01. ANOVA indicates analysis of variance; BNP, brain natriuretic peptide; DHE, dihydroethidium; GFP, green fluorescent protein; HE, hematoxylin and eosin; mt, mitochondrial; mut, mutation; PCR, polymerase chain reaction; rAAV, recombinant adeno-associated virus; ROS, reactive oxygen species; SEM, standard error of the mean; and SHR, spontaneous hypertensive rat.
By week 36, hemodynamic variables of SHRs were measured by a catheter tip manometer advanced from the right carotid artery into the left ventricle. We found that rAAV-miR-21 was still able to reduce peak systolic blood pressure significantly in SHRs (Figure 7B). We further characterized hemodynamic and echocardiographic variables in the SHR model. Although we found no difference in cardiac performance (dp/dtmax and dp/dtmin) among 3 rAAV treatment groups, the interventricular septum thickness in end-diastolic and end-systolic of the left ventricle decreased significantly in SHRs treated with rAAV-miR-21, but not rAAV-mut-miR-21 (Table 3). Neither body weight (data not shown) nor heart rate was affected, but cardiac mass index was clearly reduced in SHRs treated with rAAV-miR-21 (Figure 7C).
Table 3.
Comparison of Echocardiographic and Hemodynamic Measurements in Long-Term Treated SHRs
| rAAV-GFP | rAAV-miR-21 | rAAV-mut-miR-21 | P Value | |
|---|---|---|---|---|
| HR, bpm | 395±7 | 384±16 | 398±12 | 0.59 |
| Pes, mm Hg | 244±3 | 224±7† | 250±5 | 0.002 |
| dp/dtmax | 14 474±438 | 14 162±1163 | 14 125±383 | 0.826 |
| dp/dtmin | −14 003±620 | −12 619±889 | −14 250±738 | 0.266 |
| EF, % | 85.2±0.6 | 81.0±1.1* | 86.2±1.1 | 0.016 |
| FS, % | 33.9±2.8 | 30.7±2.9 | 33.6±2.5 | 0.450 |
| IVSTd, mm | 2.92±0.07 | 2.66±0.04* | 2.88±0.12 | 0.014 |
| IVSTs, mm | 3.76±0.14 | 3.30±0.10* | 3.76±0.09 | 0.030 |
Data are presented as mean±SEM, n=5. dp/dtmax indicates peak instantaneous rate of left ventricular pressure increase; dp/dtmin, peak instantaneous rate of left ventricular pressure increase decline; EF, ejection fraction of left ventricular; FS, fractional shortening; GFP, green fluorescent protein; HR, heart rate; IVSTd, interventricular septum thickness in end-diastolic of left ventricle; IVSTs, interventricular septum thickness in end-systolic of left ventricular; miR-21, microRNA-21; mut, mutation; Pes, end systolic pressure; rAAV, recombinant adeno-associated virus; SEM, standard error of the mean; and SHR, spontaneous hypertensive rat.
P<0.05 versus rAAV-GFP.
P<0.01 versus rAAV-GFP.
Hematoxylin and eosin staining of myocardial tissue sections showed that rAAV-miR-21 treatment significantly reduced the size of the cardiac myocytes in SHRs (Figure 7D and 7E). Furthermore, microscopic analysis after Sirius red staining to detect myocardial fibrosis and matrix proliferation showed that the red-stained area of cardiac tissues was significantly decreased in rAAV-miR-21–treated animals in comparison with other groups (Figure 7D and 7F). We also performed DHE staining and Western blotting, observing decreased levels of ROS (Figure 7D and 7G). Consistently, brain natriuretic peptide, an indicator for cardiac hypertrophy, was significantly reduced in SHRs treated with rAAV-miR-21 (Figure 7H). Real-time PCR revealed increased miR-21 both in the cytosol and mitochondria (online-only Data Supplement Figure VIA and VIB), and Western blot showed increased Cytb protein in the heart of rAAV-miR-21–treated SHRs (Figure 7I and 7J). Among other organs, we found that the expression of miR-21 and Cytb were also increased only in liver and kidney, which was accompanied with decreased ROS (online-only Data Supplement Figure VIC through VIO). Together, these data demonstrated significant benefits of in vivo delivered miR-21 in the improvement of pathophysiology of this hypertensive animal model.
DISCUSSION
In the present study, we observed downregulation of the mitochondrial genome–encoded Cytb in SHRs, which appears to directly contribute to increased mt-ROS. We found that miR-21 was able to translocate into the mitochondria to counteract mt-Cytb downregulation, likely as part of the compensatory program to hypertension. Strikingly, we showed that delivery of exogenous miR-21 by rAAV9 was sufficient to lower blood pressure in the SHR model, suggesting a new therapeutic strategy against hypertension.
In the cardiovascular system, complex I and complex III of the ETC are the major sites for ROS production.30,31 In fact, multiple studies suggest that complex III is more important than complex I in generating mt-ROS in the heart.31–33 Consistent with these reports, we found that siRNA-mediated silencing of Cytb, which is the only mtDNA-encoded subunit of complex III, caused elevated ROS. In contrast, siRNAs against ND1 (complex I) and COI (complex IV) did not elevate ROS. Given that previous studies failed to induce ROS by downregulating nuclear genome-encoded subunits of complex III,34,35 our data suggest a more crucial role for the mtDNA-encoded subunit in dictating the functional state of ETC III in mitochondria.
It has been widely accepted that miRNAs negatively regulate gene expression at the posttranscriptional levels in the cytoplasm.36 Considering the fact that miR-21 was upregulated, we initially thought that such induced miR-21 might positively contribute to hypertension in the SHR model. Unexpectedly, our data suggest that miR-21 is part of the compensatory program to counteract Cytb downregulation by enhancing Cytb translation in mitochondria. Notably, these data are consistent with the recent report by Zhang et al,15 showing miR-1 enhanced mitochondrial translation during C2C12 differentiation. This study also established the requirement for GW182 in the negative effects of microRNA in the cytoplasm, but not for their positive effects in mitochondria, which indicates direct microRNA actions in mitochondria. Using this criterion, we showed that miR-21 directly acts on Cytb to enhance its translation in mitochondria.
It has been reported previously that both damaged and enhanced respiratory chain activities are able to induce considerable amounts of ROS.37,38 Similarly, we showed that only when the respiratory chain is in its normal state, ROS production is set at its lowest point (maintained at physiological level). Hypertension decreased Cytb expression, impaired ETC III complex function, and increased electron leakage, which eventually led to elevated ROS production. In accordance, miR-21 restored Cytb to normal levels, thus decreasing ROS. Consistent with our findings, it has been reported in human coronary arterioles cells that both catalase and gp-91ds-tat inhibited bradykinin-induced ROS, but showed no effect on ROS under normal conditions.39 Overall, various compensatory mechanisms may help maintain ROS under normal physiological conditions, but, under certain pathophysiologic conditions, such a homeostatic program may be compromised, leading to elevated ROS production.
Strikingly, we found that exogenous miR-21 delivered by rAAV was sufficient to lower blood pressure in the SHR model. We note a previous report by Kaeppel et al40 suggesting the ability of rAAV to integrate into mtDNA directly. Whether rAAV was inserted into the mitochondrial genome along with miR-21 in our system remains to be investigated. A concern regarding viral vectors is their potential toxicity. To address this, we measured liver function, kidney function, and the morphology of the liver and kidney at the end of the experiment (online-only Data Supplement Figure VII). Our results showed no evidence for liver and kidney toxicity, which implies that the rAAV9 was nontoxic, although we could not completely rule out potential off-target effects of rAAV in the current study.
miR-21 has been extensively studied in cardiovascular diseases. Consistent with our findings, miR-21 has been reported to have a protective role in ischemia/reperfusion by reducing cardiomyocyte apoptosis through targeting the PDCD4 mRNA.41 Furthermore, in vivo silencing of miR-21 using a specific antagomir has been found to reduce cardiac fibrosis and cardiac dysfunction in pressure-overloaded heart.42 Our present work demonstrated that miR-21 delivered by rAAV9, which is predominantly expressed in cardiomyocytes rather than cardiac fibroblasts,43,44 not only reduced hypertension, but also improved hypertensive hypertrophy and cardiac fibrosis in SHRs. It is important to point out that, besides cardiomyocytes, rAAV-delivered miR-21 also targets other organs, such as kidney and liver, which may collectively contribute to the observed reduction of blood pressure, consistent with a recent report linking kidney to hypertension.28,29 Importantly, our data showed that rAAV did not infect the aorta, implying that the inhibition of ROS production in the aorta may be a consequence, but not a cause of lowered blood pressure. These possibilities are intriguing subjects for future studies to understand complex and coordinated regulation of hypertension in mammals.
Another interesting issue is that a miR-21 mimic could induce Cytb, but miR-21 inhibitor showed no effect both in vitro and in vivo. This is likely because of the low abundance of miR-21 in the mitochondria under normal physiological conditions, as observed earlier.45 Even under pathologic conditions, we did not record the effect of miR-21 inhibitor on exacerbating the hypertensive phenotype. We reason that induced miR-21 is likely part of the compensatory program, but the endogenous miR-21, although clearly induced in SHRs, may not have reached a sufficient threshold to antagonize an already established disease state in the animal model. Consequently, the miR-21 inhibitor showed no measurable effect in worsening the disease phenotype. In addition, lowering miR-21 may require extended periods of time to alter the disease state to a measurable degree. It also is possible that induced miR-21 is only part of the proposed compensatory program in SHRs such that inhibition of miR-21 alone was insufficient to alter the program.
In summary, our findings reveal a positive function of miR-21 in mitochondrial translation, and its systemic delivery to animals is sufficient to reduce blood pressure and suppress cardiac hypertrophy in SHRs. These observations provide a theoretical basis for developing miRNA-based therapeutics against hypertension and associated diseases.
Supplementary Material
Clinical Perspective.
What Is New?
Mitochondrial DNA-encoded cytochrome b is down-regulated in the spontaneous hypertensive rat model
Cytochrome b downregulation contributes to elevated mitochondrial reactive oxygen species
MicroRNA-21 (miR-21) directly targets cytochrome b to enhance its translation in mitochondria
Induced miR-21 positively correlates to high blood pressure in human patients
Recombinant adeno-associated virus (type 9)–delivered miR-21 mimic is able to lower blood pressure and relieve cardiac hypertrophy in animal model
What Are the Clinical Implications?
The induction of miR-21 is part of the compensatory program, not cause, in the genetic animal model for hypertensive cardiomyopathy
Delivery of exogenous miR-21 mimic may be used to treat patients who have high blood pressure
Acknowledgments
We thank colleagues in Dr Wang’s group for various technical help and stimulating discussion during the course of this investigation.
SOURCES OF FUNDING
This work was supported by grants from the National Natural Science Foundation of China (Nos. 91439203, 91440102 and 31571197) and the 973 program of the Ministry of Science and Technology of China (No. 2012CB517800).
Footnotes
DISCLOSURES
None.
The online-only Data Supplement is available with this article at http://circ.ahajournals.org/lookup/suppl/doi:10.1161/CIRCULATIONAHA.116.023926/-/DC1.
References
- 1.Drazner MH. The progression of hypertensive heart disease. Circulation. 2011;123:327–334. doi: 10.1161/CIRCULATIONAHA.108.845792. [DOI] [PubMed] [Google Scholar]
- 2.Lip GY, Felmeden DC, Li-Saw-Hee FL, Beevers DG. Hypertensive heart disease. A complex syndrome or a hypertensive ‘cardiomyopathy’? Eur Heart J. 2000;21:1653–1665. doi: 10.1053/euhj.2000.2339. [DOI] [PubMed] [Google Scholar]
- 3.Teng L, Zheng J, Leng J, Ding Y. Clinical and molecular characterization of a Han Chinese family with high penetrance of essential hypertension. Mitochondrial DNA. 2012;23:461–465. doi: 10.3109/19401736.2012.710205. [DOI] [PubMed] [Google Scholar]
- 4.Nikitin AG, Lavrikova EY, Chistiakov DA. The heteroplasmic 15059G>A mutation in the mitochondrial cytochrome b gene and essential hypertension in type 2 diabetes. Diabetes Metab Syndr. 2012;6:150–156. doi: 10.1016/j.dsx.2012.09.005. [DOI] [PubMed] [Google Scholar]
- 5.Ding Y, Xia B, Yu J, Leng J, Huang J. Mitochondrial DNA mutations and essential hypertension (Review) Int J Mol Med. 2013;32:768–774. doi: 10.3892/ijmm.2013.1459. [DOI] [PubMed] [Google Scholar]
- 6.Touyz RM. Reactive oxygen species, vascular oxidative stress, and redox signaling in hypertension: what is the clinical significance? Hypertension. 2004;44:248–252. doi: 10.1161/01.HYP.0000138070.47616.9d. [DOI] [PubMed] [Google Scholar]
- 7.Li X, Fang P, Mai J, Choi ET, Wang H, Yang XF. Targeting mitochondrial reactive oxygen species as novel therapy for inflammatory diseases and cancers. J Hematol Oncol. 2013;6:19. doi: 10.1186/1756-8722-6-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dikalova AE, Bikineyeva AT, Budzyn K, Nazarewicz RR, McCann L, Lewis W, Harrison DG, Dikalov SI. Therapeutic targeting of mitochondrial superoxide in hypertension. Circ Res. 2010;107:106–116. doi: 10.1161/CIRCRESAHA.109.214601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Graham D, Huynh NN, Hamilton CA, Beattie E, Smith RA, Cochemé HM, Murphy MP, Dominiczak AF. Mitochondria-targeted antioxidant MitoQ10 improves endothelial function and attenuates cardiac hypertrophy. Hypertension. 2009;54:322–328. doi: 10.1161/HYPERTENSIONAHA.109.130351. [DOI] [PubMed] [Google Scholar]
- 10.Thomson DW, Bracken CP, Goodall GJ. Experimental strategies for microRNA target identification. Nucleic Acids Res. 2011;39:6845–6853. doi: 10.1093/nar/gkr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ørom UA, Nielsen FC, Lund AH. MicroRNA-10a binds the 5’UTR of ribosomal protein mRNAs and enhances their translation. Mol Cell. 2008;30:460–471. doi: 10.1016/j.molcel.2008.05.001. [DOI] [PubMed] [Google Scholar]
- 12.Vasudevan S, Steitz JA. AU-rich-element-mediated upregulation of translation by FXR1 and Argonaute 2. Cell. 2007;128:1105–1118. doi: 10.1016/j.cell.2007.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Small EM, Frost RJ, Olson EN. MicroRNAs add a new dimension to cardiovascular disease. Circulation. 2010;121:1022–1032. doi: 10.1161/CIRCULATIONAHA.109.889048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Das S, Ferlito M, Kent OA, Fox-Talbot K, Wang R, Liu D, Raghavachari N, Yang Y, Wheelan SJ, Murphy E, Steenbergen C. Nuclear miRNA regulates the mitochondrial genome in the heart. Circ Res. 2012;110:1596–1603. doi: 10.1161/CIRCRESAHA.112.267732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang X, Zuo X, Yang B, Li Z, Xue Y, Zhou Y, Huang J, Zhao X, Zhou J, Yan Y, Zhang H, Guo P, Sun H, Guo L, Zhang Y, Fu XD. MicroRNA directly enhances mitochondrial translation during muscle differentiation. Cell. 2014;158:607–619. doi: 10.1016/j.cell.2014.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang DX, Gutterman DD. Mitochondrial reactive oxygen species-mediated signaling in endothelial cells. Am J Physiol Heart Circ Physiol. 2007;292:H2023–H2031. doi: 10.1152/ajpheart.01283.2006. [DOI] [PubMed] [Google Scholar]
- 17.Meng C, Jin X, Xia L, Shen SM, Wang XL, Cai J, Chen GQ, Wang LS, Fang NY. Alterations of mitochondrial enzymes contribute to cardiac hypertrophy before hypertension development in spontaneously hypertensive rats. J Proteome Res. 2009;8:2463–2475. doi: 10.1021/pr801059u. [DOI] [PubMed] [Google Scholar]
- 18.Jüllig M, Hickey AJ, Chai CC, Skea GL, Middleditch MJ, Costa S, Choong SY, Philips AR, Cooper GJ. Is the failing heart out of fuel or a worn engine running rich? A study of mitochondria in old spontaneously hypertensive rats. Proteomics. 2008;8:2556–2572. doi: 10.1002/pmic.200700977. [DOI] [PubMed] [Google Scholar]
- 19.Barrey E, Saint-Auret G, Bonnamy B, Damas D, Boyer O, Gidrol X. Pre-microRNA and mature microRNA in human mitochondria. PLoS One. 2011;6:e20220. doi: 10.1371/journal.pone.0020220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mukhopadhyay P, Rajesh M, Haskó G, Hawkins BJ, Madesh M, Pacher P. Simultaneous detection of apoptosis and mitochondrial superoxide production in live cells by flow cytometry and confocal microscopy. Nat Protoc. 2007;2:2295–2301. doi: 10.1038/nprot.2007.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang S, Chen C, Wang H, Rao X, Wang F, Duan Q, Chen F, Long G, Gong W, Zou MH, Wang DW. Protective effects of acyl-coA thioesterase 1 on diabetic heart via PPARα/PGC1α signaling. PLoS One. 2012;7:e50376. doi: 10.1371/journal.pone.0050376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lu J, Tsourkas A. Imaging individual microRNAs in single mammalian cells in situ. Nucleic Acids Res. 2009;37:e100. doi: 10.1093/nar/gkp482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Antonicka H, Sasarman F, Nishimura T, Paupe V, Shoubridge EA. The mitochondrial RNA-binding protein GRSF1 localizes to RNA granules and is required for posttranscriptional mitochondrial gene expression. Cell Metab. 2013;17:386–398. doi: 10.1016/j.cmet.2013.02.006. [DOI] [PubMed] [Google Scholar]
- 24.Xie J, Xie Q, Zhang H, Ameres SL, Hung JH, Su Q, He R, Mu X, Seher Ahmed S, Park S, Kato H, Li C, Mueller C, Mello CC, Weng Z, Flotte TR, Zamore PD, Gao G. MicroRNA-regulated, systemically delivered rAAV9: a step closer to CNS-restricted transgene expression. Mol Ther. 2011;19:526–535. doi: 10.1038/mt.2010.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rehmsmeier M, Steffen P, Hoechsmann M, Giegerich R. Fast and effective prediction of microRNA/target duplexes. [Accessed March 3, 2014];RNA. 2004 10:1507–1517. doi: 10.1261/rna.5248604. http://bibiserv.techfak.uni-bielefeld.de/rnahybrid/submission.html. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eulalio A, Huntzinger E, Izaurralde E. GW182 interaction with Argonaute is essential for miRNA-mediated translational repression and mRNA decay. Nat Struct Mol Biol. 2008;15:346–353. doi: 10.1038/nsmb.1405. [DOI] [PubMed] [Google Scholar]
- 27.Wu XT, Li XL. miR-21 contributes to cardiac aging by targeting PTEN. J Am Coll Cardiol. 2015;66:C4–C4. [Google Scholar]
- 28.Cuevas S, Zhang Y, Yang Y, Escano C, Asico L, Jones JE, Armando I, Jose PA. Role of renal DJ-1 in the pathogenesis of hypertension associated with increased reactive oxygen species production. Hypertension. 2012;59:446–452. doi: 10.1161/HYPERTENSIONAHA.111.185744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brezniceanu ML, Liu F, Wei CC, Chénier I, Godin N, Zhang SL, Filep JG, Ingelfinger JR, Chan JS. Attenuation of interstitial fibrosis and tubular apoptosis in db/db transgenic mice overexpressing catalase in renal proximal tubular cells. Diabetes. 2008;57:451–459. doi: 10.2337/db07-0013. [DOI] [PubMed] [Google Scholar]
- 30.Murphy E, Steenbergen C. Preconditioning: the mitochondrial connection. Annu Rev Physiol. 2007;69:51–67. doi: 10.1146/annurev.physiol.69.031905.163645. [DOI] [PubMed] [Google Scholar]
- 31.Chen Q, Vazquez EJ, Moghaddas S, Hoppel CL, Lesnefsky EJ. Production of reactive oxygen species by mitochondria: central role of complex III. J Biol Chem. 2003;278:36027–36031. doi: 10.1074/jbc.M304854200. [DOI] [PubMed] [Google Scholar]
- 32.St-Pierre J, Buckingham JA, Roebuck SJ, Brand MD. Topology of superoxide production from different sites in the mitochondrial electron transport chain. J Biol Chem. 2002;277:44784–44790. doi: 10.1074/jbc.M207217200. [DOI] [PubMed] [Google Scholar]
- 33.Demin OV, Kholodenko BN, Skulachev VP. A model of O2.-generation in the complex III of the electron transport chain. Mol Cell Biochem. 1998;184:21–33. [PubMed] [Google Scholar]
- 34.Aguilera-Aguirre L, Bacsi A, Saavedra-Molina A, Kurosky A, Sur S, Boldogh I. Mitochondrial dysfunction increases allergic airway inflammation. J Immunol. 2009;183:5379–5387. doi: 10.4049/jimmunol.0900228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Korde AS, Yadav VR, Zheng YM, Wang YX. Primary role of mitochondrial Rieske iron-sulfur protein in hypoxic ROS production in pulmonary artery myocytes. Free Radic Biol Med. 2011;50:945–952. doi: 10.1016/j.freeradbiomed.2011.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: microRNAs can up-regulate translation. Science. 2007;318:1931–1934. doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]
- 37.Tsutsui H, Kinugawa S, Matsushima S. Mitochondrial oxidative stress and dysfunction in myocardial remodelling. Cardiovasc Res. 2009;81:449–456. doi: 10.1093/cvr/cvn280. [DOI] [PubMed] [Google Scholar]
- 38.Finkel T. Cell biology: a clean energy programme. Nature. 2006;444:151–152. doi: 10.1038/444151a. [DOI] [PubMed] [Google Scholar]
- 39.Larsen BT, Bubolz AH, Mendoza SA, Pritchard KA, Jr, Gutterman DD. Bradykinin-induced dilation of human coronary arterioles requires NADPH oxidase-derived reactive oxygen species. Arterioscler Thromb Vasc Biol. 2009;29:739–745. doi: 10.1161/ATVBAHA.108.169367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kaeppel C, Beattie SG, Fronza R, van Logtenstein R, Salmon F, Schmidt S, Wolf S, Nowrouzi A, Glimm H, von Kalle C, Petry H, Gaudet D, Schmidt M. A largely random AAV integration profile after LPLD gene therapy. Nat Med. 2013;19:889–891. doi: 10.1038/nm.3230. [DOI] [PubMed] [Google Scholar]
- 41.Qin Y, Yu Y, Dong H, Bian X, Guo X, Dong S. MicroRNA 21 inhibits left ventricular remodeling in the early phase of rat model with ischemia-reperfusion injury by suppressing cell apoptosis. Int J Med Sci. 2012;9:413–423. doi: 10.7150/ijms.4514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thum T, Gross C, Fiedler J, Fischer T, Kissler S, Bussen M, Galuppo P, Just S, Rottbauer W, Frantz S, Castoldi M, Soutschek J, Koteliansky V, Rosenwald A, Basson MA, Licht JD, Pena JT, Rouhanifard SH, Muckenthaler MU, Tuschl T, Martin GR, Bauersachs J, Engelhardt S. MicroRNA-21 contributes to myocardial disease by stimulating MAP kinase signalling in fibroblasts. Nature. 2008;456:980–984. doi: 10.1038/nature07511. [DOI] [PubMed] [Google Scholar]
- 43.Kho C, Lee A, Jeong D, Oh JG, Chaanine AH, Kizana E, Park WJ, Hajjar RJ. SUMO1-dependent modulation of SERCA2a in heart failure. Nature. 2011;477:601–605. doi: 10.1038/nature10407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Aoyama Y, Kobayashi K, Morishita Y, Maeda K, Murohara T. Wnt11 gene therapy with adeno-associated virus 9 improves the survival of mice with myocarditis induced by coxsackievirus B3 through the suppression of the inflammatory reaction. J Mol Cell Cardiol. 2015;84:45–51. doi: 10.1016/j.yjmcc.2015.04.009. [DOI] [PubMed] [Google Scholar]
- 45.Ricci EP, Limousin T, Soto-Rifo R, Allison R, Pöyry T, Decimo D, Jackson RJ, Ohlmann T. Activation of a microRNA response in trans reveals a new role for poly(A) in translational repression. Nucleic Acids Res. 2011;39:5215–5231. doi: 10.1093/nar/gkr086. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.