Abstract
Orthologous genes that maintain a single-copy status in a broad range of species may indicate a selection against gene duplication. If this is the case, then duplicates of such genes that do survive may have escaped the dosage control by rapid and sizable changes in their function. To test this hypothesis and to develop a strategy for the identification of novel gene functions, we have analyzed 22 primate-specific intrachromosomal duplications of genes with a single-copy ortholog in all other completely sequenced metazoans. When comparing this set to genes not exposed to the single-copy status constraint, we observed a higher tendency of the former to modify their gene structure, often through complex genomic rearrangements. The analysis of the most dramatic of these duplications, affecting ∼10% of human Chromosome 2, enabled a detailed reconstruction of the events leading to the appearance of a novel gene family. The eight members of this family originated from the highly conserved nucleoporin RanBP2 by several genetic rearrangements such as segmental duplications, inversions, translocations, exon loss, and domain accretion. We have experimentally verified that at least one of the newly formed proteins has a cellular localization different from RanBP2's, and we show that positive selection did act on specific domains during evolution.
Gene duplication is known to play a leading role in evolution for the creation of novel gene function (Ohno 1970; Kimura 1983). The fate of duplicated genes is dependent on the selective advantage they bring to the organism, and the vast majority of them are deleted or degrade into pseudogenes (Nadeau and Sankoff 1997; Li et al. 2001). More rarely, the new copies acquire novel functions under the driving force of adaptive evolution, which exposes them to different selective constraints from those of the parental genes (Walsh 1995; Lynch and Conery 2000). Many gene functions have evolved through such a mechanism (Prince and Pickett 2002), and some of them involve primate-specific gene families (Courseaux and Nahon 2001; Johnson et al. 2001; Maston and Ruvolo 2002; Zhang et al. 2002; Paulding et al. 2003). Gene duplicability, defined as the tendency to retain duplicates, increases with organismal complexity, probably because of their increased adaptability (Yang et al. 2003). Yet, even in higher eukaryotes duplicability varies among gene families. There are classes of genes whose duplication is disadvantageous and consequently suppressed during evolution. Such a selection against duplication has been reported mostly for housekeeping genes (Hooper and Berg 2003) and for genes coding for complex subunits (Papp et al. 2003; Yang et al. 2003). The most likely explanation is that, as duplication directly affects gene dosage, it sometimes prevents the proper function of the gene product. Indeed, an inappropriate gene dosage balance is involved in the etiology of genetic disorders such as the Charcot-Marie-Tooth disease type 1A, which is caused by the duplication of the PMP22 gene (Lupski et al. 1992).
The maintenance of a single-copy status across diverse metazoans is likely to be an indicator for such a selection against duplication, considering the high frequency of duplications in eukaryotes (Lynch and Conery 2000; Yang et al. 2003). It is thus likely that lineage-specific duplications of otherwise single-copy orthologs have acquired a novel function. Although functional distinctiveness is difficult to quantify, here we mean functional divergence beyond differences in tissue expression or substrate specificity. Identification of such lineage-specific duplications would imply a simple strategy to identify recently evolved functions.
In order to apply this strategy to human, we searched for primate-specific intrachromosomal duplications of genes with single orthologs in all other metazoans. The detailed study of the most dramatic of such duplications revealed that the break from a well-conserved genetic stability is, indeed, linked to the birth of new gene function and is caused by complex genetic mechanisms. Using comparative genomics, we could reconstruct the genomic events leading to the emergence of a novel human gene function.
Results
Detection of primate-specific gene duplications
We systematically searched for single-copy orthologs in all completely sequenced metazoan genomes except human, and then identified their multicopy orthologs located in the same human chromosome (inparalogs) (Table 1). We restricted the analysis to intrachromosomal duplications because these events are expected to be more recent when compared to interchromosomal translocations (D. Torrents, M. Suyama, and P. Bork, unpubl.), and hence more likely to have occurred after the rodent-primate split. Furthermore, the inclusion of orthologs located in different chromosomes would have affected the specificity of our analysis, owing to the possible inclusion of processed pseudogenes. In total we detected 22 metazoan single-copy orthologs with at least one additional paralog in human. Of those we excluded four identical copies that might represent copy number polymorphisms (Sebat et al. 2004). Of the duplicates, 55% are localized in the subtelomeric and pericentromeric regions of the human chromosomes, highlighting the genetic dynamism of those regions (Guy et al. 2003; She et al. 2004). At least 82% of the detected duplications are clearly part of larger segments that have undergone recent duplication (Bailey et al. 2002). This confirms the primary role of large segmental duplications particularly in the appearance of primate-specific traits (Eichler 2001; Samonte and Eichler 2002).
Table 1.
Primate-specific duplications of metazoan single-copy genes
| Chrom. band | Copy numbera | Mouse pairwise score | Gene name | Protein AC | Description |
|---|---|---|---|---|---|
| 1p36.33 | 2 | L | ATAD3B | NP_114127 | AAA-ATPase TOB3 |
| 1q23.2 | 2 | H | VANGL1 | NP_620409 | Vang-like protein |
| 1p13.1 | |||||
| 2p23.1 | 2 | H | XDHA | NP_000370 | Xanthine oxidoreductase |
| 2q33.1 | |||||
| 2q12.3 | 6 | H | RANBP2 | NP_006258 | RanBP2 |
| 3p12.3 | 3 | L | SB153 | BAC086025 | SB153 protein, isoform 1 |
| 3q22.1 | |||||
| 3q21.1 | 2 | H | EAF2 | NP_060926 | ELL associated factor 2 |
| 3p25.1 | |||||
| 5q13.2 | 3 | L* | SMN1 | NP_000335 | Survival motor neuron 1 |
| 5q13.2 | 3 | L* | GTF2H2 | NP_001506 | Transcription factor IIH |
| 5q35.2 | 2 | L | THOC3 | NP_115737 | THO complex subunit 3 |
| 5q35.3 | |||||
| 6p12.3 | 3 | H | TFAP2A | NP_003211 | Transcription factor AP-2 α |
| 6p24.3 | |||||
| 7p22.1 | 2 | L* | C7orf28A | NP_056437 | CGI-43 protein |
| 7q22.1 | 4 | L | POLR2J2 | NP_116580 | RNA polymerase II subunit 11 |
| 7p13 | |||||
| 7q11.3 | 3 | H | WBSCR20A | NP_060514 | Williams Beuren syndrome-associated gene |
| 8p21.2 | 2 | H | DPYS | NP_001376 | Dihydropyrimidinase |
| 8p22.3 | |||||
| 12p11.1 | 2 | L | ALG10 | NP_116223 | Glucosyltransferase |
| 12p12 | |||||
| 12q24.13 | 2 | H | PTPN11 | NP_002825 | Protein tyrosine phosphatase |
| 12p13.31 | |||||
| 15q24.1 | 3 | H | COMMD4 | NP_060298 | COMM domain containing 4 |
| 15q24.2 | |||||
| 15q24.3 | |||||
| 16p12.1 | 2 | L* | EIF3S8 | NP_003743 | Translation initiation factor 3 |
| 16p13.11 | 3 | L | PM5 | NP_055102 | pM5 protein |
| 16p13.3 | |||||
| 16q22.3 | 2 | H | PDRD | NP_060460 | Pyruvate dehydrogenase phosphatase |
| 17q23.2 | 2 | H | USP32 | NP_115971 | Ubiquitin-specific hydrolase 32 |
| 17p13.2 | |||||
| 17q23.3 | 2
|
H
|
TLK2
|
NP_006843
|
Tousled-like kinase 2
|
| 17q11.2 |
The database accession number of the longest of the human paralogs is reported. The phylogenetic relationship between the human copies has been assessed on the basis of the global score of their pairwise alignment (see text). H indicates that the global alignment of one of the human copies was scoring better with the mouse ortholog than with the other paralogs; L indicates the opposite, L* indicates that the human copies were almost identical (>99% sequence identity). Note that, because of the specific constraints used, this list reports only a subset of primate-specific genes, namely, the ones with single-copy orthologs in other metazoans and with duplications in primates. For a full list of primate-specific genes, see Long et al. (2003) and references therein.
This refers to the number of gene copies annotated in Ensembl at the time of analysis (see Methods), In the case of RanBP2, six genes were annotated in Ensembl, but we collected expression evidence for nine genes (RanBP2 and eight related genes). Moreover, five out of the six Ensembl genes appear to be fragments (the encoded predicted proteins are 46, 156, and 905 residues long).
In order to refine the duplication events and to predict the putative functional role of the duplicated genes, we reconstructed the functional relationships between the human copies and their orthologs in the other species. We thus compared the score of the global alignment within the human paralogs and between them and the corresponding mouse ortholog. Higher scores between one of the human copies and the mouse ortholog than within the human paralogs should reveal divergent paralogs in terms of rearrangements in the gene structure. Thus, depending on this score, we defined two categories for the classification of the observed duplications. When the pairwise alignment score is higher within the human paralogs, it is likely that the duplications occurred recently and were not affected by genomic rearrangements. These gene copies might not have been subjected to functional selection, might differ only in their expression patterns (Gu et al. 2004), or might have other relatively small functional differences that are difficult to measure.
Of the 18 human paralogous families recorded here, six (33%) fall into this group, while in the remaining 12 duplications (67%) one of the human genes aligns globally better with the mouse ortholog than with the other paralogs. To test whether our data set is, indeed, enriched in genes with a modified structure, we performed a comparable analysis on a reference set obtained by relaxing the constraint of a single-copy status across metazoa. For this purpose, we required a single ortholog in rodents but allowed complete freedom elsewhere, meaning gene absence or presence of multiple paralogs in other metazoans. In this case we detected 215 duplications, and in 55% of them the human copies align better with the mouse ortholog than with the other paralogs. It should be noted that the reference set is likely to include ancient duplications that either underwent rodent-specific loss or whose rodent paralogs were overlooked in the database. This might inflate the final count by increasing the number of human paralogs more similar to the mouse orthologs than to the other copies. Despite this possible bias, we found an enrichment in divergent human paralogs in the data set of genes that have originated from metazoan single-copy orthologs. This supports our hypothesis that there are considerable functional differences among this specific set of novel genes.
The duplicated human genes with a modified structure are all candidates for the rapid evolution of new functions through rapid genomic rearrangements. As a case study to test the validity of our strategy, we analyzed in detail the most dramatic of these duplications, interspersed in more than 10% of human Chromosome 2.
A novel gene family descended from RanBP2 and with a domain contribution from GCC2
The most striking of the primate-specific gene families identified in our screen originated from RanBP2, the largest nucleoporin characterized so far (Wu et al. 1995; Yokoyama et al. 1995). The duplicated copies also acquired an additional domain from the recently described trans-Golgi protein GCC2 (GRIP and coiled-coil domain-containing protein 2) (Luke et al. 2003).
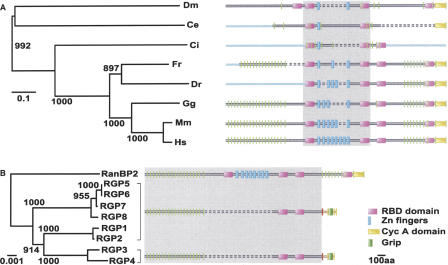
Single-copy orthologs of RanBP2 are unambiguously detectable in all fully sequenced animal genomes, but not in other eukaryotes such as plants and fungi (Fig. 1A). Although there are slight differences in the domain architectures among the species, the overall domain organization of the protein is well conserved (Fig. 1A). The N-terminal leucine-rich region is followed by RanGTP-binding domains (RanBP1 homologous domains or RBD) related to the one present in RanBP1, a variable number of zinc-finger motifs, and a C terminus with homology to cyclophilin A (Fig. 1A).
Figure 1.
Conservation of the RanBP2 gene during metazoan evolution and its expansion in human. For each protein, the corresponding domain architecture is reported. The proteins are depicted reproducing the sequence alignments, the dashed bars representing the gaps in the alignments. The regions used to build the trees are highlighted in gray. (A) Phylogenetic tree of the RanBP2 orthologs in representatives of fully sequenced metazoan genomes. The light-blue bars represent protein regions that were not predictable because of gaps in the corresponding genomes. The exon-intron boundaries of the encoding genes are reported as vertical green bars. (Ce) Caenorhabditis elegans; (Ci) Ciona intestinalis; (Dm) Drosophila melanogaster; (Dr) Danio rerio; (Fr) Fugu rubripes; (Gg) Gallus gallus; (Hs) Homo sapiens; (Mm) Mus musculus.(B) Family tree of RanBP2 and RGP genes. The exon-intron boundaries of RanBP2-derived region (exons 1-20) are shown in green, those of GCC2-derived part (exons p-r) are in yellow. The RGP-specific intron, bearing the fusion between the RanBP2- and GCC2-derived regions, is depicted in red. The RGP regions encoded by the RanBP2-derived DNA are shown in black, the ones encoded by the GCC2-derived DNA in brown.
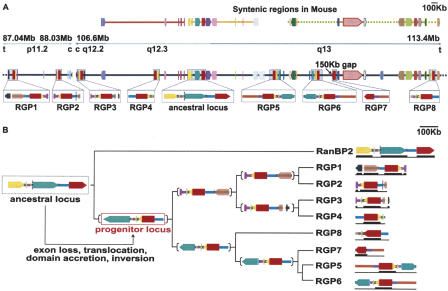
In addition to the human RanBP2 ortholog, we detected eight partial copies located in regions that have arisen from intrachromosomal segmental duplications (Figs. 1B, 2A). Despite the fact that human Chromosome 2 appears to be relatively poor in segmental duplications (Bailey et al. 2002), the eight segments containing the RanBP2-related genes added up to 1.5 Mb of DNA interspersed in 26 Mb of the chromosome sequence. Two of the gene copies are located in the pericentromeric region of the short arm (band 2p11.2) and the other seven on the long arm (bands 2q12.3 and 2q13) (Fig. 2A). The entire region of duplication is proximal to the site of fusion between the two ancestral ape chromosomes that originated the human Chromosome 2 (Yunis and Prakash 1982). This area is known to be prone to rearrangements occurring before and after the fusion (Fan et al. 2002a,b). It is worth noting that only five of the eight RanBP2-related genes were identified by automated gene prediction pipelines (Table 1). Only a manual analysis of this large, complex chromosomal region uncovered the entire duplication event. Although all of the eight additional copies maintain more than 95% local sequence similarity to RanBP2, the overall gene structure is significantly modified. The new gene copies lack part of RanBP2 exons 20 and 21 and all of exons 22-29. As a consequence, the predicted encoded proteins lack the whole zinc-finger region, the first and the last RBD, and the cyclophilin A homologous domain (Fig. 1B). In addition, they acquire the 3′-terminal exons p-r from the GCC2 gene, coding for a GRIP (Golgin-97, RanBP2α, Imh1p, and p230/golgin-245) domain (Munro and Nichols 1999), which has been reported to be implicated in Golgi localization (McConville et al. 2002; see the legend to Fig. 1 for the exon nomenclature). Because of its novel domain architecture, we named the gene family RGP (RanBP2-like, GRIP domain containing proteins). The predicted length for each of the eight encoded proteins exceeds 1700 amino acids. Several lines of evidence suggest that all eight gene copies are expressed. Firstly, a full-length copy aligning with 99.6% sequence identity both to RGP5 and RGP6 has been reported as a testis-specific protein BS-63 (RANBP2L1 isoform 1, RefSeq accession no. NP_005045) (Cai et al. 2002). Secondly, ubiquitous expression has been assessed for a short variant encoding the first 18 exons of RGP5, 6, or 7 (they are 100% identical in this part of their sequence; RANBP2L1 isoform 2, RefSeq accession no. NP_115636) (Nothwang et al. 1998). Thirdly, ESTs unambiguously matching all of the copies are detectable. These ESTs derive from different human tissues, including fetal, adult, normal, and tumoral cells. Finally, by RT-PCR we could amplify DNA fragments specifically corresponding to RGP1, 3, 4, 7, 8 and to RGP1 or 2 and to RGP5 or 6 from both testis and HeLa cell cDNA libraries (Fig. 3).
Figure 2.
Evolutionary mechanism for the origin of the new gene family. The genes are shown as arrows with different colors associated with different genes. (Red) RanBP2; (yellow) GCC2; the RGP paralogs are shown in red for the RanBP2-derived region and in yellow for the GCC2-derived domain. The color scheme of the other genes as well as a larger version of the figure are given in Supplemental Figures S4 and S5, respectively. (A) Gene composition of the regions on human Chromosome 2 where the duplicated fragments containing the RGP paralogs are interspersed. The duplicated segments are highlighted in gray and enlarged in the lower boxes, in which similar intergenic sequences are also reported (see Methods for more details). The vertical dashed bars in both the segments containing RGP2 and RanBP2 indicate intergenic regions with no detectable intrachromosomal matches. The 3′-end of the RGP7 copy could not be assessed, as the human genome build 34 has a 150-kb gap in that region. The chromosomal bands, the region borders, and the direction to the centromere (c) and telomeres (t) are shown in the upper bar. The syntenic regions in mouse Chromosomes 10 (yellow), 17 (orange), and 2 (green) are also shown. (B) Family tree of the RGP and the RanBP2 paralogs, and putative mechanism for the formation of the RGP progenitor locus. At each branching point, the genomic structure of the putative progenitor is depicted. The ancestral locus, which contains RanBP2 and GCC2 and is syntenic in mouse, underwent several genetic rearrangements leading to the formation of the progenitor locus. The rearrangements included an inversion of the entire region, a loss of the 3′-exons from RanBP2, a partial deletion of the RanBP2 exon 20, and a translocation that places the 3′ noncoding region just downstream of the last four exons of the GCC2 duplicated gene. This event leads to the accretion of the GRIP domain. We assume that the progenitor locus already contained the newly formed RGP gene, as all the RGP duplicates contain the GRIP domain and a shorter version of RanBP2-derived exon 20. The bars reported under each of the duplicated segments indicate the presence of unambiguous expression data (ESTs and cDNAs) for the corresponding gene.
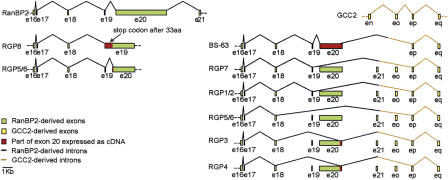
Figure 3.
Gene structure and expression evidences of the RGP gene copies. The gene structure of the regions of the RGP genes amplified by RT-PCR is shown. For comparison, the corresponding regions of the RanBP2 (exons 16-19) and GCC (exons n-q) genes are reported. By comparing the expression data to the genomic sequences, it is possible to predict the existence of different splice variants for each copy. Indeed, some of the cDNAs detect exon-skipping.
Genomic rearrangements led to the formation of the new gene
Although the current coverage and quality of the chimp genome do not allow a final assessment, we were able to detect at least four partial genes of the RGP family in addition to the complete RanBP2 ortholog. This is an indication that at least some of the RGP duplications occurred before the human-chimp split. A further confirmation derives from the recent publication of hominoid-specific gene duplications detected by cDNA-array-based comparative genomic hybridization, in which the cDNA corresponding to RanBP2L1 isoform 2 cohybridizes with the genomic DNA of several hominoids (Fortna et al. 2004). The conservation in synteny of the RanBP2 and GCC2 surrounding regions between mouse and human allowed us to unambiguously identify the starting segment (ancestor locus) for the following duplications. Through the analysis of gene order and the detection of significant sequence similarity in the noncoding regions, we were able to extend the borders of the duplicated segment and reconstruct the most parsimonious evolutionary scenario leading to the progenitor locus (Fig. 2B). This region of ∼200 kb already contains the gene structure of the newly formed RGP family, which evolved from RanBP2 and GCC2 by successive rearrangements. Although it is not possible to assess the temporal order of the events, the ancestor locus underwent duplication, inversion, partial deletion of the long RanBP2 exon 20, and acquisition of the 3′-end of the GCC2 gene coding for the GRIP domain (see Fig. 2B legend for more details). The resulting progenitor locus contains the core coding and noncoding regions common to all the duplicated copies. We performed a similar comparative analysis using the surrounding genomic regions of the eight RGP copies to reconstruct the entire duplication scenario and to assess the parental relationships between them (Fig. 2B). The reliability of this reconstruction is confirmed by the agreement in topology between the trees obtained by using both the gene order approach (Fig. 2B) and the cDNA sequences of RanBP2 and RGPs (Fig. 1B).
The region surrounding the junction site of each of the duplicated segments is enriched in DNA repeats (55%) when compared to a random control (39%) (Supplemental Fig. S1a; Supplemental Table S1; see the legend to the figure for the procedure used to detect the content in DNA repeats). The repeats that mainly contribute to the enrichment are the Alu elements with a peak localized at the junction site (Supplemental Fig. S1b). These results are in agreement with the proposed mechanism for segmental duplications involving Alu-mediated recombinations (Babcock et al. 2003; Bailey et al. 2003).
Alternative splicing of the RGP genes
The presence in the protein databases of short versions of the RGP genes, such as the variant bearing only the first 18 exons (RANBP2L1 isoform 2) (Nothwang et al. 1998), points to the possibility of alternative splicing events involving the RGP genes. Additional evidence for alternative splicing was provided by the results of the RGP amplification by RT-PCR (Fig. 3). We observed a variable presence of exon 20 in several clones as well as an invariable lack of the exon o, which is present in the genomic sequences of all of the RGP genes (Fig. 3; see legend to Fig. 1 for the exon nomenclature).
Positive selection drives the evolution of the RGP family
Although in each of the eight large duplicated segments there are other potential coding regions, the RGP genes are the only ones for which specific ESTs can always be detected (Fig. 2B). This indicates that the RGP gene copies represent the evolutionarily active parts of each duplicated segment. We then searched specifically for particular regions within the RGP genes jthat are under selective pressure, measuring the Ka/Ks ratios for each of the RGP exons. We used the branch-site models of the ML method, which is particularly useful for studying the evolution of gene families (Yang and Nielsen 2002; see Supplemental Fig. S2 for details). We observed that positive selection is acting on the first 20 exons of the RGP genes, for which the Ka/Ks ratio is always higher than 1. The remaining part of the gene (exons p-q) is under purifying selection, showing a Ka/Ks ratio lower than 1 (Table 2). All the RGP genes contain the mutated residues, at least at the level of their full-length transcripts.
Table 2.
Positive selection of different exons of the RGP genes
| Exon | Ka/Ks | 2Δℓ | p | Sites under positive selection |
|---|---|---|---|---|
| (Ka/Ks)0 = 0.74798 | ||||
| 1-19 | (Ka/Ks)1 = 0.12120 | 21.47 | 2.210−5 | D77E; S305N; N834K (P > 0.7) |
| (Ka/Ks)2 = 4.21470 | ||||
| (Ka/Ks)0 = 0.05678 | S2523R; R2039G;Q2121R (0.85 > P > 0.8) | |||
| 20 | (Ka/Ks)1 = 0.16790 | 28.50 | 6.510−7 | L1950V; D2031G;K2338E;T2391S (P > 0.85) |
| (Ka/Ks)2 = 3.99336 | ||||
| (Ka/Ks)0 = 0.12345 | ||||
| p-q | (Ka/Ks)1 = 0.43336 | 7.47 | 2.410−2 | |
| (Ka/Ks)2 = 0.25333 |
(Ka/Ks)0 and (Ka/Ks)1 indicate the Ka/Ks ratios associated to the background lineages, while (Ka/Ks)2 refers to Ka/Ks of the foreground branch (see Methods and Supplemental Fig. S2). The parameters indicating positive selection are represented in bold. 2Δℓ is twice the difference between log likelihood scores (ℓ) obtained under models B and M3 (k=2). The resulting values were then compared against a χ2 distribution with degrees of freedom equal to 2 (it is the difference in the number of parameters estimated in models B, five, and in M3, three). The sites under positive selection located in the RDB domains are underlined. The numbering is relative to the human RanBP2. p represents the probability associated to each prediction.
Five of the seven residues of exon 20 that are potentially under positive selection belong to the RBD domains of the RGP proteins (underlined in Table 2). Four of them correspond to residues directly involved in the binding of RanBP2 to Ran (Vetter et al. 1999; Supplemental Fig. S3a). In particular, the two mutations R2039G and K2338E modify the two RBD domains in a similar way, both affecting the same region of contact to the C terminus of Ran (Supplemental Fig. S3b,c).
Distinct cellular localization of the RGP proteins
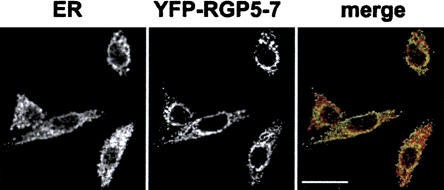
The different domain architecture together with the substitution of key sites for the binding to Ran suggest a novel function for the RGP family. As a proof of such a functional divergence of the RGP proteins from RanBP2, we decided to study the subcellular localization of the short splice variant named RANBP2L1 isoform 2, which is ubiquitously expressed (Nothwang et al. 1998). RANBP2L1 isoform 2 is 100% identical to the first 18 exons of RGP5, 6, or 7 and can be readily amplified by PCR from both HeLa and testis cDNA libraries. The latter suggests that the corresponding mRNA is expressed at relatively high levels. RANBP2L1 isoform 2 was detected in discrete cytoplasmic regions (Fig. 4), in sharp contrast to the reported localization of RanBP2, which is almost exclusively found at the nuclear envelope at steady state (Wu et al. 1995; Yokoyama et al. 1995).
Figure 4.
Subcellular localization of RanBP2L1 isoform 2 (RGP5-7). Confocal images of fixed HeLa cells expressing a GFP-fusion of RANBPL1 isoform 2. Cells were stained with a polyclonal anti-calnexin antibody. In the merged image the calnexin signal is shown in red and the GFP signal is shown in green. Scale bar, 20 μm.
Discussion
In this study we report primate-specific duplications of genes that have maintained a single-copy status in all other metazoans sequenced so far. Our hypothesis is that the escape from an evolutionarily conserved single-copy status in one specific lineage is connected to the acquisition of novel molecular functions. In addition, we expect that this functional divergence is acquired through massive genetic rearrangements over a relatively short period of time so as to avoid negative dosage effects. Indeed, in the set of 22 recent duplications with a single-copy ortholog in all other metazoans, we observe an enrichment of genes with a modified structure, compared to human duplications not exposed to the same evolutionary constraints. Further confirming the validity of our strategy, we were also able to detect previously reported acquisitions of primate-specific functions by gene duplication. An example is the TRE2 gene (Table 1), which derived from the chimeric fusion of the duplicates of two parental genes (USP32 and TBC1D3) and was recently described as a novel hominoid-specific gene (Paulding et al. 2003). Other duplications within the set of 22 recently duplicated human genes have been reported as parts of bigger segments that underwent primate-specific segmental duplications (Samonte and Eichler 2002). Examples are the genes EIF3S8 and PM5, which are contained in the 12-Mb duplicated region of human Chromosome 16 (Loftus et al. 1999).
Lineage-specific duplications with novel gene structures not only hint at new function, but also allow the tracing of the evolutionary events leading to the actual genomic arrangements. This is particularly true for primate-specific duplications as they are recent and more likely to be still contained in larger segments. Thus, the comparison of the paralogous and orthologous regions permits the identification of the ancestor locus and allows a detailed reconstruction of the evolutionary mechanism.
Both the discovery of new gene function and the reconstruction of the underlying mechanism were applicable to the most dramatic of the primate-specific duplications reported here, which involves the nucleoporin RanBP2. Diverse genetic events were apparently required over a short time period to allow the emergence of the RGP gene family (Figs. 1, 2). Moreover, within the duplicated segments, RGPs represent the vast majority of the surviving coding regions, indicating that they drive the evolution of the entire region. Finally, the cytoplasmic localization of one of the RGP variants, originally termed RanBP2L1 isoform 2 (Nothwang et al. 1998), is clearly distinct from that of RanBP2 and implies a distinct function of the RGP family. Although precise functional assignment requires more investigation, possible hints about the RGP function can be derived from the analysis of their molecular features. The fact that the sites most likely to be under positive selection reside in the RDB domains and mediate the binding of RanBP2 to Ran (Supplemental Fig. S3) suggests a change in the specificity of this binding, and hence in its function. These sites are changed in all of the RGP genes, suggesting that the mutations happened early in the evolution of the RGP family and were suddenly fixed in the population. They also took place in comparable regions in the two RBD domains (Supplemental Fig. S3b). Some of the possible splice variants of the RGP genes gain a GRIP domain at their C termini. In addition to its function in targeting proteins to the Golgi apparatus (Munro and Nichols 1999; McConville et al. 2002), the GRIP domain has been shown to form homodimers that interact with the GTP-bound form of the ARF/Arl GTPase family (Wu et al. 2004). Some of the GRIP-domain-containing proteins have been implicated in the maintenance of trans-Golgi network integrity and in the vesicular transport of proteins to the plasma membrane (Yoshino et al. 2003). The acquisition of the GRIP domain points toward a possible role of the RGP genes in intracellular trafficking. Domain accretion is a recurrent mechanism for the formation of new genes with distinct functions (Long et al. 2003 and references therein). It has been shown, for example, that the acquisition of the Kua domain determines the localization of the Kua-UEV1 protein to the cytoplasm rather to the nucleus (Thomson et al. 2000).
Why did RGP appear in primates and nowhere else? There are two possible answers to this question depending on the evolutionary scenario considered. Under the more neutral one, the high number of complex rearrangements required to escape the dosage imbalance renders this event so unlikely that it would only happen in a single lineage. The alternative, and more selective scenario, requires that the positive contribution of the newly formed genes to lineage-specific traits outweighs the negative impact of the dosage imbalance. In this case, the new genes that we detected are likely to have evolved in primates because of their specific genomic and functional context. As is usual in evolutionary biology, it is difficult to determine which one of these scenarios is correct; however, if the selective one is true, a thorough functional characterization of the new genes reported here would reveal the functionally most relevant areas for primate evolution.
Methods
Detection of primate-specific gene duplications of single-copy genes
The complete protein sets of Anopheles gambiae (BDGP build 2a), Caenorhabditis briggsae (Cb25.agp8), Caenorhabditis elegans (Wormbase build 102), Drosophila melanogaster (BDGP build 3a), Fugu rubripes (Fugu build 2.0), Homo sapiens (NCBI build 34), Mus musculus (NCBI build 30), and Rattus norvegicus (BGDP build 3.1) were downloaded from the Ensembl Web site (http://www.ensembl.org, Dec 20th 2003), and reduced to one translation at each locus (longest transcript). All-against-all Smith-Waterman searches and a previously described algorithm (Tatusov et al. 1997; Zdobnov et al. 2002) were then used to search for orthologous groups containing at least one protein from each of the analyzed species. Only the groups that had a single representative in each of the nonhuman species, but at least two representatives in human, were extracted. The resulting list was extended by searching for additional cases possibly overlooked because of artifacts in the gene-prediction procedure (partial/fragmentary prediction or accidental fusion with surrounding genes). All orthologous groups were searched for the presence of additional paralogs in all the genomes (with reduced stringency). Only those human paralogs which resided on the same chromosome and with markedly better similarity scores than paralogs in any of the other genomes were included to the list (the similarity of paralogs was expressed in bit scores normalized by self-hit bit scores; this value had to be above 0.4 for putative paralogs in the human lineage to be included, all other paralogs occurring elsewhere had to be below 0.3). In order to exclude processed pseudogenes and events predating the human/mouse split, only the intrachromosomal duplications were kept. To avoid unprocessed pseudogenes, ESTs from dbEST (Feb. 5th 2004) were aligned against the DNA region corresponding to each of the human proteins using stand-alone BLAT (Kent 2002). Of the resulting alignments, only the best alignment with a percent identity higher than 96% and a length greater than 100 bases was kept. In cases where the best alignment could not be unambiguously assigned, the EST was discarded. For comparison, a background set of unrestricted gene duplications in the human lineage was derived. This set included all detectable duplications in the human lineage located on the same chromosome and with single-copy orthologs in rodents. In all other metazoans duplications as well as absences/losses were allowed.
Calculation of the global alignment score and analysis of the primate-specific duplications
For both data sets, pairwise Smith-Waterman alignments between the paralogous human genes and their orthologs in mouse were built in order to assess whether any of the duplicates had undergone structural changes (truncations, indels, and rearrangements). Absolute bit scores were assessed using the BLOSUM62 matrix, no low complexity filters, and gap costs of 11 for gap opening and 1 for gap extension. Any human gene copy was classified as structurally altered when it had a lower bit score to its parental human gene as did the corresponding full-length mouse ortholog.
RanBP2 orthology assignment, sequence analysis, and gene structure definition
The sequence of the human protein RanBP2 (NP_006258) was used as a query for stand-alone TBLASTN (Altschul et al. 1997) in representatives of fully sequenced metazoan genomes. Once the corresponding gene regions were detected, the gene structure was assigned using BLAST2GENE (Suyama et al. 2004) and GeneWise (Birney et al. 1996). The protein domain architectures were retrieved using SMART (http://smart.embl-heidelberg.de; Letunic et al. 2004).
Analysis of the genomic DNA sequence and detection of the duplicated fragments
A nonredundant collection of known genes (SWISS-PROT, TrEMBL, TrEMBL-NEW, and mRNA from GenBank) from Ensembl (http://www.ensembl.org/) and UCSC (http://genome.ucsc.edu/) in the regions of RanBP2 duplication (87.040-88.030 Mb and 106.600-113.400 Mb) was used for stand-alone BLAT (Kent 2002) and BLAST2seq (Tatusova and Madden 1999) against human Chromosome 2 to search for total or partial duplications. Each gene was considered fully duplicated if the copy covered at least 70% of its length. For retrieving intrachromosomal duplications in the noncoding regions, each intergenic DNA fragment was used as a query for BLAT against Chromosome 2.
Evolutionary analysis
Phylogenetic trees were derived by MEGA2 (http://www.megasoftware.net/; Kumar et al. 2001) and CLUSTALX (Thompson et al. 1997), using the neighbor-joining method with 1000 bootstrap replications. For the tree of the RanBP2 orthologs and the cDNA regions in between the first and second RBD domains were used. For deriving the tree of RanBP2 and the RGP copies, a multiple alignment of the corresponding first 20 exons were used.
The Ka/Ks ratios were measured using the ML method implemented in PAML (http://abacus.gene.ucl.ac.uk/software/paml.html/; Yang 1997). The DNA alignments for exons 1-19, the conserved part of exon 20, and exons p-q were used to assess variation of the RGP genes in respect to the corresponding exons in the RanBP2 and GCC2 orthologs across the species. The exon r was excluded as it is not present in all duplicates. The branch-site model (Yang and Nielsen 2002) was applied, defining as a foreground branch the RGP gene branch, and all the others as background branches (see Supplemental Fig. S2). Two models of evolution were used. In model A (neutral), positive selection is only allowed in the foreground branch, while it is fixed at 0 in the background branches (Nielsen and Yang 1998). In model B (discrete), Ka/Ks ratios are considered free in all branches and estimated directly from the data (Yang et al. 2000). As comparable results were obtained, only the results using model B are shown in Table 2. The likelihood ratio test (LRT) was applied to measure the statistical significance of models A and B when compared to the corresponding neutral models (M1 and M3, k =2) (Nielsen and Yang 1998; Yang and Nielsen 2002).
Amplification of cDNAs corresponding to the RGP gene family
To confirm the expression of RGP genes, cDNA fragments were amplified by PCR using human testis Marathon-Ready cDNA library (Clontech) or a oligo(dT)-primed HeLa cell cDNA as template and primers introducing unique restriction sites. The 5′-primer (5′-GTAAAAGTTACAAGTATTCTCCC) corresponds to the RanBP2 exon 16-17 boundary, predicted to be present in all RGP genes. Two different 3′-primers were used: the first (5′-GTGGATCAAGAAAATTCACCTTC) corresponding to the region of RanBP2 exon 20 present in RGPs and the second (5′-GCAACATCGTATTTATAACAGG) corresponding to the C-terminal part of the GRIP domain. All PCR reactions were performed with the Expand high-fidelity PCR system (Roche). The amplified cDNAs were cloned into a derivative of pBS-SK and sequenced.
Localization of RGP proteins
The RANBPL1 isoform 2 cDNA was amplified with primers containing appropriate restriction sites, using a (dT)15-primed HeLa cDNA library or a human testis Marathon-Ready cDNA library (Clontech) as templates. The amplified cDNAs were then cloned into the vector pECFP-C1 (Clontech) for localization experiments.
HeLa cells were grown on coverslips in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum, 100 U/mL penicillin, and 100 μg/mL streptomycin. Transfections were performed in six-well plates with Lipofectamine Plus reagent (Invitrogen), according to the manufacturer's instructions. Then, 24 h after transfection, cells were washed in PBS, fixed for 10 min in 3.7% paraformaldehyde in PBS, washed again in PBS, and stained with a polyclonal anti-calnexin antibody (Santa Cruz). Cells were then mounted in Fluoromount G (Southern Biotechnology). Images were acquired using a Zeiss LSM510 FCS confocal microscope.
Acknowledgments
We thank Elena Conti for the RBD structure analysis, Sean Hooper for his help with the Ka/Ks calculations, and David Torrents for helpful discussions.
Footnotes
[Supplemental material is available online at www.genome.org. and at http://www.bork.embl.de/~ciccarel/RGP_add_data.html.]
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.3266405. Article published online ahead of print in February 2005.
References
- Altschul, S.F., Madden, T.L., Schaffer, A.A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D.J. 1997. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25: 3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babcock, M., Pavlicek, A., Spiteri, E., Kashork, C.D., Ioshikhes, I., Shaffer, L.G., Jurka, J., and Morrow, B.E. 2003. Shuffling of genes within low-copy repeats on 22q11 (LCR22) by Alu-mediated recombination events during evolution. Genome Res. 13: 2519-2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey, J.A., Gu, Z., Clark, R.A., Reinert, K., Samonte, R.V., Schwartz, S., Adams, M.D., Myers, E.W., Li, P.W., and Eichler, E.E. 2002. Recent segmental duplications in the human genome. Science 297: 1003-1007. [DOI] [PubMed] [Google Scholar]
- Bailey, J.A., Liu, G., and Eichler, E.E. 2003. An Alu transposition model for the origin and expansion of human segmental duplications. Am. J. Hum. Genet. 73: 823-834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birney, E., Thompson, J.D., and Gibson, T.J. 1996. PairWise and SearchWise: Finding the optimal alignment in a simultaneous comparison of a protein profile against all DNA translation frames. Nucleic Acids Res. 24: 2730-2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai, Y., Gao, Y., Sheng, Q., Miao, S., Cui, X., Wang, L., Zong, S., and Koide, S.S. 2002. Characterization and potential function of a novel testis-specific nucleoporin BS-63. Mol. Reprod. Dev. 61: 126-134. [DOI] [PubMed] [Google Scholar]
- Courseaux, A. and Nahon, J.L. 2001. Birth of two chimeric genes in the Hominidae lineage. Science 291: 1293-1297. [DOI] [PubMed] [Google Scholar]
- Eichler, E.E. 2001. Recent duplication, domain accretion and the dynamic mutation of the human genome. Trends Genet. 17: 661-669. [DOI] [PubMed] [Google Scholar]
- Fan, Y., Linardopoulou, E., Friedman, C., Williams, E., and Trask, B.J. 2002a. Genomic structure and evolution of the ancestral chromosome fusion site in 2q13-2q14.1 and paralogous regions on other human chromosomes. Genome Res. 12: 1651-1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan, Y., Newman, T., Linardopoulou, E., and Trask, B.J. 2002b. Gene content and function of the ancestral chromosome fusion site in human chromosome 2q13-2q14.1 and paralogous regions. Genome Res. 12: 1663-1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortna, A., Kim, Y., MacLaren, E., Marshall, K., Hahn, G., Meltesen, L., Brenton, M., Hink, R., Burgers, S., Hernandez-Boussard, T., et al. 2004. Lineage-specific gene duplication and loss in human and great ape evolution. PLoS Biol. 2: E207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu, Z., Rifkin, S.A., White, K.P., and Li, W.H. 2004. Duplicate genes increase gene expression diversity within and between species. Nat. Genet. 36: 577-579. [DOI] [PubMed] [Google Scholar]
- Guy, J., Hearn, T., Crosier, M., Mudge, J., Viggiano, L., Koczan, D., Thiesen, H.J., Bailey, J.A., Horvath, J.E., Eichler, E.E., et al. 2003. Genomic sequence and transcriptional profile of the boundary between pericentromeric satellites and genes on human chromosome arm 10p. Genome Res. 13: 159-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper, S.D. and Berg, O.G. 2003. On the nature of gene innovation: Duplication patterns in microbial genomes. Mol. Biol. Evol. 20: 945-954. [DOI] [PubMed] [Google Scholar]
- Johnson, M.E., Viggiano, L., Bailey, J.A., Abdul-Rauf, M., Goodwin, G., Rocchi, M., and Eichler, E.E. 2001. Positive selection of a gene family during the emergence of humans and African apes. Nature 413: 514-519. [DOI] [PubMed] [Google Scholar]
- Kent, W.J. 2002. BLAT—The BLAST-like alignment tool. Genome Res. 12: 656-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura. 1983. The neutral theory of molecular evolution. Cambridge University Press, Cambridge, UK.
- Kumar, S., Tamura, K., Jakobsen, I.B., and Nei, M. 2001. MEGA2: Molecular evolutionary genetics analysis software. Bioinformatics 17: 1244-1245. [DOI] [PubMed] [Google Scholar]
- Letunic, I., Copley, R.R., Schmidt, S., Ciccarelli, F.D., Doerks, T., Schultz, J., Ponting, C.P., and Bork, P. 2004. SMART 4.0: Towards genomic data integration. Nucleic Acids Res. 32: D142-D144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, W.H., Gu, Z., Wang, H., and Nekrutenko, A. 2001. Evolutionary analyses of the human genome. Nature 409: 847-849. [DOI] [PubMed] [Google Scholar]
- Loftus, B.J., Kim, U.J., Sneddon, V.P., Kalush, F., Brandon, R., Fuhrmann, J., Mason, T., Crosby, M.L., Barnstead, M., Cronin, L., et al. 1999. Genome duplications and other features in 12 Mb of DNA sequence from human chromosome 16p and 16q. Genomics 60: 295-308. [DOI] [PubMed] [Google Scholar]
- Long, M., Betran, E., Thornton, K., and Wang, W. 2003. The origin of new genes: Glimpses from the young and old. Nat. Rev. Genet. 4: 865-875. [DOI] [PubMed] [Google Scholar]
- Luke, M.R., Kjer-Nielsen, L., Brown, D.L., Stow, J.L., and Gleeson, P.A. 2003. GRIP domain-mediated targeting of two new coiled-coil proteins, GCC88 and GCC185, to subcompartments of the trans-Golgi network. J. Biol. Chem. 278: 4216-4226. [DOI] [PubMed] [Google Scholar]
- Lupski, J.R., Wise, C.A., Kuwano, A., Pentao, L., Parke, J.T., Glaze, D.G., Ledbetter, D.H., Greenberg, F., and Patel, P.I. 1992. Gene dosage is a mechanism for Charcot-Marie-Tooth disease type 1A. Nat. Genet. 1: 29-33. [DOI] [PubMed] [Google Scholar]
- Lynch, M. and Conery, J.S. 2000. The evolutionary fate and consequences of duplicate genes. Science 290: 1151-1155. [DOI] [PubMed] [Google Scholar]
- Maston, G.A. and Ruvolo, M. 2002. Chorionic gonadotropin has a recent origin within primates and an evolutionary history of selection. Mol. Biol. Evol. 19: 320-335. [DOI] [PubMed] [Google Scholar]
- McConville, M.J., Ilgoutz, S.C., Teasdale, R.D., Foth, B.J., Matthews, A., Mullin, K.A., and Gleeson, P.A. 2002. Targeting of the GRIP domain to the trans-Golgi network is conserved from protists to animals. Eur. J. Cell Biol. 81: 485-495. [DOI] [PubMed] [Google Scholar]
- Munro, S. and Nichols, B.J. 1999. The GRIP domain—A novel Golgi-targeting domain found in several coiled-coil proteins. Curr. Biol. 9: 377-380. [DOI] [PubMed] [Google Scholar]
- Nadeau, J.H. and Sankoff, D. 1997. Comparable rates of gene loss and functional divergence after genome duplications early in vertebrate evolution. Genetics 147: 1259-1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen, R. and Yang, Z. 1998. Likelihood models for detecting positively selected amino acid sites and applications to the HIV-1 envelope gene. Genetics 148: 929-936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nothwang, H.G., Rensing, C., Kubler, M., Denich, D., Brandl, B., Stubanus, M., Haaf, T., Kurnit, D., and Hildebrandt, F. 1998. Identification of a novel Ran binding protein 2 related gene (RANBP2L1) and detection of a gene cluster on human chromosome 2q11-q12. Genomics 47: 383-392. [DOI] [PubMed] [Google Scholar]
- Ohno, S. 1970. Evolution by gene duplication. Springer-Verlag, Berlin-Heidelberg-New York.
- Papp, B., Pal, C., and Hurst, L.D. 2003. Dosage sensitivity and the evolution of gene families in yeast. Nature 424: 194-197. [DOI] [PubMed] [Google Scholar]
- Paulding, C.A., Ruvolo, M., and Haber, D.A. 2003. The Tre2 (USP6) oncogene is a hominoid-specific gene. Proc. Natl. Acad. Sci. 100: 2507-2511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prince, V.E. and Pickett, F.B. 2002. Splitting pairs: The diverging fates of duplicated genes. Nat. Rev. Genet. 3: 827-837. [DOI] [PubMed] [Google Scholar]
- Samonte, R.V. and Eichler, E.E. 2002. Segmental duplications and the evolution of the primate genome. Nat. Rev. Genet. 3: 65-72. [DOI] [PubMed] [Google Scholar]
- Sebat, J., Lakshmi, B., Troge, J., Alexander, J., Young, J., Lundin, P., Maner, S., Massa, H., Walker, M., Chi, M., et al. 2004. Large-scale copy number polymorphism in the human genome. Science 305: 525-528. [DOI] [PubMed] [Google Scholar]
- She, X., Jiang, Z., Clark, R.A., Liu, G., Cheng, Z., Tuzun, E., Church, D.M., Sutton, G., Halpern, A.L., and Eichler, E.E. 2004. Shotgun sequence assembly and recent segmental duplications within the human genome. Nature 431: 927-930. [DOI] [PubMed] [Google Scholar]
- Suyama, M., Torrents, D., and Bork, P. 2004. BLAST2GENE: A comprehensive conversion of BLAST output into independent genes and gene fragments. Bioinformatics 20: 1968-1970. [DOI] [PubMed] [Google Scholar]
- Tatusov, R.L., Koonin, E.V., and Lipman, D.J. 1997. A genomic perspective on protein families. Science 278: 631-637. [DOI] [PubMed] [Google Scholar]
- Tatusova, T.A. and Madden, T.L. 1999. BLAST 2 Sequences, a new tool for comparing protein and nucleotide sequences. FEMS Microbiol. Lett. 174: 247-250. [DOI] [PubMed] [Google Scholar]
- Thompson, J., Gibson, T., Plewniak, F., Jeanmougin, F., and Higgins, D. 1997. The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25: 4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson, T.M., Lozano, J.J., Loukili, N., Carrio, R., Serras, F., Cormand, B., Valeri, M., Diaz, V.M., Abril, J., Burset, M., et al. 2000. Fusion of the human gene for the polyubiquitination coeffector UEV1 with Kua, a newly identified gene. Genome Res. 10: 1743-1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vetter, I.R., Nowak, C., Nishimoto, T., Kuhlmann, J., and Wittinghofer, A. 1999. Structure of a Ran-binding domain complexed with Ran bound to a GTP analogue: Implications for nuclear transport. Nature 398: 39-46. [DOI] [PubMed] [Google Scholar]
- Walsh, J.B. 1995. How often do duplicated genes evolve new functions? Genetics 139: 421-428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, J., Matunis, M.J., Kraemer, D., Blobel, G., and Coutavas, E. 1995. Nup358, a cytoplasmically exposed nucleoporin with peptide repeats, Ran-GTP binding sites, zinc fingers, a cyclophilin A homologous domain, and a leucine-rich region. J. Biol. Chem. 270: 14209-14213. [DOI] [PubMed] [Google Scholar]
- Wu, M., Lu, L., Hong, W., and Song, H. 2004. Structural basis for recruitment of GRIP domain golgin-245 by small GTPase Arl1. Nat. Struct. Mol. Biol. 11: 86-94. [DOI] [PubMed] [Google Scholar]
- Yang, Z. 1997. PAML: A program package for phylogenetic analysis by maximum likelihood. Comput. Appl. Biosci. 13: 555-556. [DOI] [PubMed] [Google Scholar]
- Yang, Z. and Nielsen, R. 2002. Codon-substitution models for detecting molecular adaptation at individual sites along specific lineages. Mol. Biol. Evol. 19: 908-917. [DOI] [PubMed] [Google Scholar]
- Yang, Z., Nielsen, R., Goldman, N., and Pedersen, A.M. 2000. Codon-substitution models for heterogeneous selection pressure at amino acid sites. Genetics 155: 431-449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, J., Lusk, R., and Li, W.-H. 2003. Organismal complexity, protein complexity, and gene duplicability. Proc. Natl. Acad. Sci. 100: 15661-15665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama, N., Hayashi, N., Seki, T., Pante, N., Ohba, T., Nishii, K., Kuma, K., Hayashida, T., Miyata, T., Aebi, U., et al. 1995. A giant nucleopore protein that binds Ran/TC4. Nature 376: 184-188. [DOI] [PubMed] [Google Scholar]
- Yoshino, A., Bieler, B.M., Harper, D.C., Cowan, D.A., Sutterwala, S., Gay, D.M., Cole, N.B., McCaffery, J.M., and Marks, M.S. 2003. A role for GRIP domain proteins and/or their ligands in structure and function of the trans Golgi network. J. Cell Sci. 116: 4441-4454. [DOI] [PubMed] [Google Scholar]
- Yunis, J.J. and Prakash, O. 1982. The origin of man: A chromosomal pictorial legacy. Science 215: 1525-1530. [DOI] [PubMed] [Google Scholar]
- Zdobnov, E.M., von Mering, C., Letunic, I., Torrents, D., Suyama, M., Copley, R.R., Christophides, G.K., Thomasova, D., Holt, R.A., Subramanian, G.M., et al. 2002. Comparative genome and proteome analysis of Anopheles gambiae and Drosophila melanogaster. Science 298: 149-159. [DOI] [PubMed] [Google Scholar]
- Zhang, J., Zhang, Y.P., and Rosenberg, H.F. 2002. Adaptive evolution of a duplicated pancreatic ribonuclease gene in a leaf-eating monkey. Nat. Genet. 30: 411-415. [DOI] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- http://genome.ucsc.edu/; University of California San Francisco genome browser.
- http://www.ensembl.org/; Ensembl.
- http://www.bork.embl.de/~ciccarel/RGP_add_data.html; Supplemental material to this paper.
- http://abacus.gene.ucl.ac.uk/software/paml.html/; PAML.
- http://smart.embl-heidelberg.de/; SMART database.
- http://www.megasoftware.net/; MEGA.
- http://www.ncbi.nlm.nih.gov/; National Center for Biotechnology Information.