Abstract
The Drosophila (fruit fly) model system has been instrumental in our current understanding of human biology, development, and diseases. Here, we used a high-throughput yeast two-hybrid (Y2H)-based technology to screen 102 bait proteins from Drosophila melanogaster, most of them orthologous to human cancer-related and/or signaling proteins, against high-complexity fly cDNA libraries. More than 2300 protein-protein interactions (PPI) were identified, of which 710 are of high confidence. The computation of a reliability score for each protein-protein interaction and the systematic identification of the interacting domain combined with a prediction of structural/functional motifs allow the elaboration of known complexes and the identification of new ones. The full data set can be visualized using a graphical Web interface, the PIMRider (http://pim.hybrigenics.com), and is also accessible in the PSI standard Molecular Interaction data format. Our fly Protein Interaction Map (PIM) is surprisingly different from the one recently proposed by Giot et al. with little overlap between the two data sets. Analysis of the differences in data sets and methods suggests alternative strategies to enhance the accuracy and comprehensiveness of the post-genomic generation of broad-scale protein interaction maps.
The availability of an increasing number of fully sequenced genomes demands processes facilitating functional interpretation of the genomic information. Two-hybrid methods can be industrialized and robotized and thus offer some answer to this challenge. The sequencing of the fruit fly Drosophila melanogaster genome (Adams et al. 2000) considerably increased our ability to integrate genetic and biochemical information for functional annotation of Drosophila proteins. Given the importance of Drosophila as a model system, this will profoundly improve our understanding of orthologous protein function in human biology.
Until recently, however, there have been few global studies performed under well-controlled conditions to investigate on a large scale, protein-protein interactions (PPI) in multicellular eukaryotes (Walhout et al. 2000), similar to those performed for Helicobacter pylori (Rain et al. 2001) or Saccharomyces cerevisiae (Uetz et al. 2000; Ito et al. 2001; Gavin et al. 2002). Recently, fly (Giot et al. 2003) and nematode (Li et al. 2004) genome-wide interactome maps have been generated using yeast two-hybrid methods different one from each other, and different from ours.
We have built a high-density protein interaction map of the fly orthologs of a set of human proteins (see Fig. 1). We report here the results obtained for the first 154 yeast two-hybrid analyses performed using 102 Drosophila proteins as baits, screened against two different fruit fly cDNA libraries derived from whole-body fly embryos or adult heads. These screens led to connections of >1700 additional Drosophila proteins. The Y2H strategy and methods are described as well as the bioinformatics tools developed to handle large bodies of data, to compute a confidence score for each protein interaction and to explore the map using a Web-based graphical interface. An analysis of the distribution of a set of Gene Ontology (GO) categories in the map and in the fly genome are presented together with an analysis of the distribution of predicted structural/functional domains among the experimentally defined interacting domains. The exploration of the present map leads to numerous biological hypothesis and expands our knowledge of regulatory protein networks important in human cancer as shown by the biological analysis of a particularly interesting network surrounding the Ras oncogene.
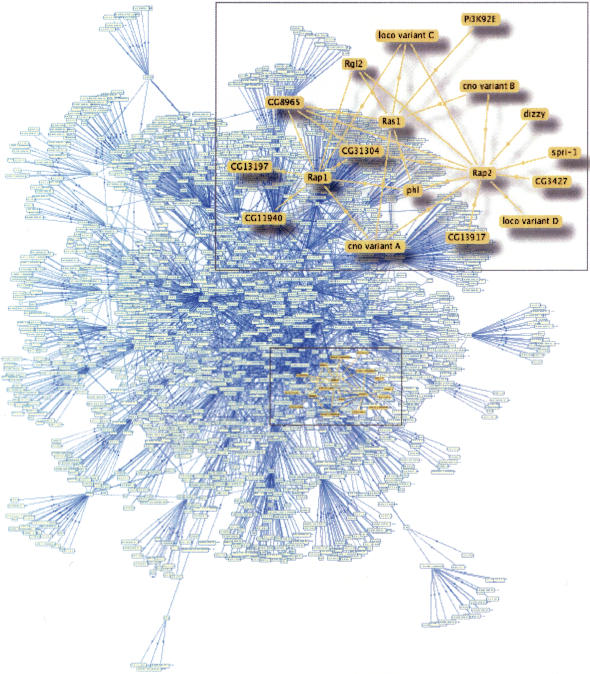
Figure 1.
A global view of the Drosophila embryo interaction map. This view of the Protein Interaction Map (PIM) resulting from our work is generated using the PIM Walker graphical interface, a tool developed for network display. The proteins in the map that bear an RA (Ras Association) or RBD (Raf-like Ras-binding) domain (colored in orange) define a discrete subnetwork around Ras-like GTPases (colored in yellow).
Striking differences were observed when comparing our partial fly interaction map to the genome-wide fly interactome published recently by Giot et al. (2003). These differences define the intrinsic limitations of each approach and highlight important considerations for the production of high-quality PPI maps.
Results
Bait selection and design
The protein interaction map presented here was constructed using as baits Drosophila orthologs of human proteins involved in oncogenesis or in “generic” cellular functions such as signaling, intracellular trafficking, or maintenance of genome integrity. The complete set of 102 bait proteins is listed in Supplemental Table 1.
For each selected protein, a custom bait design was performed to generate the best suited constructs for interaction screening; for example, bona fide or predicted hydrophobic transmembrane domains, signal peptides, and transcriptional transactivation domains were excluded from the bait constructs. Alternatively, specific structural or functional domains located within a larger polypeptide chain were selected as baits. In some cases, dominant mutant alleles with defined biological/biochemical properties that stabilize or promote protein/protein interactions were introduced into appropriate baits; for example, mutations that convert small GTPases to constitutively activated, GTP-bound forms. A total of 154 baits were thus generated from the 102 starting proteins (Supplemental Table 1).
Drosophila screens, interacting domain identification, and computation of interaction reliability scores
We constructed two high-complexity libraries of randomly primed cDNA fragments from poly(A)+ RNA isolated from adult fly head and embryos. Each screen was first performed on a small scale with various concentrations (0.5 to 200 mM) of 3-aminotriazol (3-AT), an inhibitor of imidazole glycerol phosphate dehydratase, the product of the HIS3 reporter gene. This procedure reduced the background generated by baits that activate transcription alone (so-called autoactivating baits). Concentrations of 3-AT for the full-sized screens were chosen in order to obtain up to 380 independent positive clones per screen (Supplemental Table 1). A minimum of 5 × 107 interactions were screened for each bait. All positive clones were then picked, and prey fragments were identified by sequence analysis and comparison with BDGP v3.1 (Adams et al. 2000; Celniker et al. 2002; Misra et al. 2002) and/or GenBank databases using a dedicated integrated laboratory production management system, the PIM-Builder (Rain et al. 2001). Following identification of positive clones, overlapping prey fragments derived from the same gene were clustered into families. The common sequence shared by these fragments defines the Selected Interacting Domain (SID). Thus, a SID presumably contains all the structural determinants required for a given interaction to occur. For each interaction, a Predicted Biological Score (PBS) was computed to assess interaction reliability. This score represents the probability of an interaction being nonspecific. For practical use, the scores were divided into four categories, from A (lowest probability) to D (highest probability). A fifth category, E, was added to specifically tag interactions involving highly connected prey domains. This category represents highly likely two-hybrid artifacts (see Methods).
General features of the Drosophila interaction map and visualization tool
We carried out a total of 154 screens on 102 Drosophila proteins. In all, 18,353 positive colonies were picked and 15,201 prey inserts were successfully sequenced. From these prey fragments, 2338 PPI and 2484 SIDs (mean size: 267 residues) were identified. Of these, 153 interactions fell into the “sticky” category (PBS E). The remaining set defined 2185 interactions connecting 1711 Drosophila proteins, which corresponds to an average connectivity of 2.55 partners per connected proteins and of 21.4 partners per bait (Table 1). The average connectivity for the high-confidence map (PBS A to C) is of seven partners per bait, an estimate in accordance with those proposed in recent studies (Grigoriev 2003; Lehner and Fraser 2004).
Table 1.
General features of the Drosophila protein interaction map
| Value | Comments | |
|---|---|---|
| Screens | 154 | |
| Combinations of bait/prey polypeptide tested | 7.7 × 109 | |
| Preys tested/bait | 5 × 107 | |
| Selected colonies/bait | 0-380 | On average 119 colonies/bait |
| Selected colonies/million pairs tested | 1.7 | |
| Selected clones | 18,353 | |
| Sequenced clones | 16,123 | 88% of selected clones |
| Clones identified | 15,201 | 94.3% of sequenced clones |
| 6.7% not predicted cDNA | ||
| Protein-protein interactions | 710 (PBS A to C) | |
| 2185 (PBS A to D) | ||
| 2338 (PBS A to E) | ||
| Proteins connected | 641 (PBS A to C) | |
| 1711 (PBS A to D) | ||
| 1727 (PBS A to E) | ||
| Mean connectivity | 2.22 (PBS A to C) | 2.9/bait (PBS A) |
| 2.55 (PBS A to D) | 5.8/bait (PBS A, B) | |
| 2.71 (PBS A to E) | 7/bait (PBS A to C) | |
| 21.4/bait (PBS A to D) | ||
| SID identified | 2484 |
For display and analysis of large interaction data sets, we used a previously developed software platform, the PIMRider (Colland et al. 2004). This tool has been considerably enhanced to visualize multiple PIMs from different libraries and includes several viewers (Supplemental Fig. 1). Access to all data through the PIMRider software is freely available following registration at http://pim.hybrigenics.com.
Drosophila protein interaction map analysis
Global analysis of Gene Ontology (GO) distribution in the map and in the fly genome
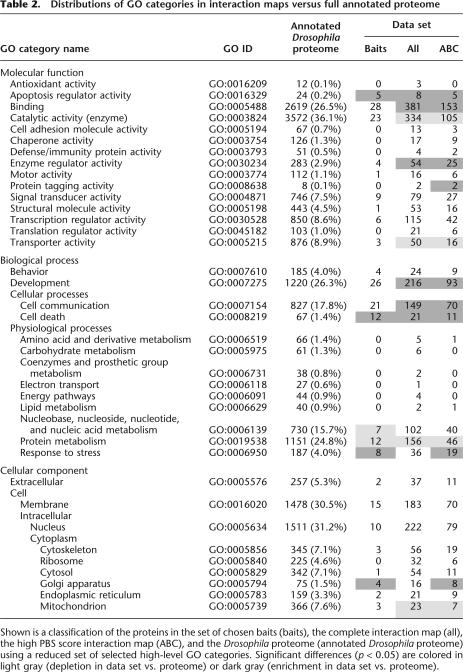
The complete interaction map and the high-confidence score map were classified according to a reduced set of selected GO categories (Table 2). This distribution was compared to the one of the annotated Drosophila proteome. A few classes are enriched or depleted because of a primary bias in the distribution of the bait proteins (e.g., apoptosis regulator, cell death, or protein metabolism). An expected enrichment in the binding function class is observed. Interestingly, there is no significant depletion in membrane proteins. This suggests that the Y2H strategy used in our study (specific bait design, use of prey fragment libraries) overcomes the frequent bias against these cellular components reported in other studies (Giot et al. 2003). The ability of our Y2H assay to efficiently detect interactions involving membrane proteins is convincingly exemplified in a recent study (Boeda et al. 2002). Conversely, we did not observe an enrichment in nuclear proteins as recently reported (Giot et al. 2003).
Table 2.
Among the other classes, proteins with catalytic activity are underrepresented. The observed depletion may reflect the recognized difficulty to detect transient enzyme-substrate interactions even using a rather sensitive assay. Alternatively, exogenous catalytic activities may, in some circumstances, be toxic to yeast.
Global analysis of SID and predicted structural/functional domains
The identification of the interacting domains (SID) and the systematic prediction of InterPro domains, transmembrane segments (TM), and signal peptide (SP) in the whole interaction map and in the Drosophila proteome allowed us to compare the distribution of these experimental and predicted domains.
As shown in Table 3, the predicted transmembrane domains are underrepresented in the SIDs, and proteins with a predicted signal peptide are significantly depleted from the map. Surprisingly, the percentage of proteins in the map with at least one predicted InterPro domain is somewhat lower than those in the fly genome. A likely explanation for this observation is that several proteins in the map contain yet uncharacterized structural/functional domains. For example, the SID on the tumor-suppressor gene CG33193 (Sav) mediating its interaction with CG11228 (Hpo) maps to its last 90 C-terminal residues (amino acids 518 to 608). This domain precisely encompasses a novel protein interaction motif, termed SARAH, mapped from amino acids 552 to 602 of CG33193 (Scheel and Hofmann 2003). Thus, our PIM can predict and define the boundaries of novel domains. We are currently conducting a global search to identify novel interaction motifs in our map.
Table 3.
Distribution of domains in the fly proteome, interacting proteins, and SID
| Drosophila proteome | Proteins in data set | SID in data set | |||
|---|---|---|---|---|---|
| With TM | 20% | 205 | 12%a | 105 | 6%a,b |
| With SP | 21% | 247 | 14%a | 85 | 5%a,b |
| With InterPro domain | 78% | 1258 | 73%a | 1024 | 62%a,b |
| Total | 1727 | 1651 | |||
Transmembrane (TM) segments (Krogh et al. 2001), signal peptide (SP) (Nielsen et al. 1997), and InterPro domains (Ashburner and Lewis 2002) were predicted from the primary sequences of the proteins in the present data set and the complete Drosophila proteome. Mapped domains were then compared with the SID.
Significantly different (p < 0.05) from representation in Drosophila proteome.
Significantly different (p < 0.05) from representation in interacting proteins.
To further explore the relationship between SIDs and predicted structural domains, we identified among the different InterPro domains those known to mediate protein/protein interactions using an existing list as starting point (http://www.mshri.on.ca/pawson/domains.html). We then analyzed the frequency of appearance of these binding domains among SIDs versus the fly genome. This analysis revealed that SIDs were significantly enriched in known binding domains (Table 4). The most frequently encountered domains are: PDZ, PH, ANK, SH3, RING, and WD40, with the most enriched being PDZ, RA, ANK, 14-3-3, FERM, and LIM. Interestingly, some of the enriched domains are thought to require their specific target motifs to be post-translationally modified in order to recognize and bind them. This is the case for the 14-3-3, PH, and FHA domains, which recognize phosphorylated serine or threonine residues, or for the Bromo domain, which binds acetylated lysines. Enrichment of these domains in the map suggests that many bait proteins are modified by endogenous yeast enzymes.
Table 4.
Distribution of known binding motlfs in interacting proteins, in the SID, and in the fly proteome
| IPR ID | Domains | No. in map ORF | No. in map SID | No. in Drosophila proteome |
|---|---|---|---|---|
| IPR001478 | PDZ | 39a | 28a | 110 |
| IPR000159 | RA | 11a | 7a | 21 |
| IPR002110 | ANK | 27a | 18a | 136 |
| IPR000308 | 14_3_3 | 3 | 3a | 9 |
| IPR000299 | Band4.1/FERM | 12a | 7a | 38 |
| IPR001781 | LIM | 13 | 11a | 78 |
| IPR001487 | Bromo | 11a | 5a | 28 |
| IPR000225 | ARM | 6a | 4a | 21 |
| IPR003169 | GYF | 1 | 1a | 2 |
| IPR003116 | Ras BD | 4a | 2a | 7 |
| IPR000253 | FHA | 7a | 4a | 27 |
| IPR001849 | PH | 29a | 10a | 110 |
| IPR001440 | TPR | 17 | 10a | 114 |
| IPR000157 | TIR | 2 | 2a | 12 |
| IPR001092 | bHLH | 18 | 9 | 109 |
| IPR001715 | CH | 13a | 5 | 53 |
| IPR001452 | SH3 | 26a | 9 | 124 |
| IPR008251 | Chromo Shadow | 1 | 1 | 6 |
| IPR000727 | t-SNARE | 5 | 1 | 18 |
| IPR000306 | FYVE | 6a | 1 | 20 |
| IPR000980 | SH2 | 8 | 3 | 64 |
| IPR000569 | HECT | 7a | 1 | 22 |
| IPR000953 | Chromo | 6a | 1 | 22 |
| IPR000449 | UBA | 6 | 2 | 50 |
| IPR001680 | WD-40 | 24 | 11 | 269 |
| IPR001841 | RING | 27 | 7 | 175 |
| IPR000270 | PB1 | 1 | 0 | 4 |
| IPR002014 | VHS | 4a | 0 | 6 |
| IPR001026 | ENTH | 1 | 0 | 8 |
| IPR001660 | SAM | 8 | 1 | 43 |
| IPR001960 | EVH1 | 3 | 0 | 10 |
| IPR000488 | Death Domain | 1 | 0 | 10 |
| IPR001810 | F-Box | 4 | 1 | 44 |
| IPR000313 | PWWP | 3 | 0 | 11 |
| IPR000342 | RGS | 2 | 0 | 13 |
| IPR001496 | SOCS | 3 | 0 | 19 |
Shown is a list of InterPro domains known to be involved in protein-protein interactions and their occurrence in the connected proteins (No. in map ORFs; total 1727), in the SID (No. in map SID; total 2484), and in the Drosophila proteome (No. in Drosophila proteome; total 20,532). Their distribution in the connected proteins and in the SIDs are compared with those in the Drosophila proteome.
Significantly enriched (p < 0.05) from representation in the Drosophila proteome.
Some domains were not represented in SIDs. These include WW, MH2, and BH1_2_3_4. The exclusion of WW and BH domains from the SIDs is due to the bias in the bait protein distribution (data not shown) combined with the low occurrence of these domains in the annotated fly proteome.
Comparison with known fly interactions and with other large-scale projects
We compared our data with the genome-wide exploration of the fly interactome recently published by Giot et al. (2003). This study includes both the screening of a collection of 10,306 full-length prey proteins and of two commercially available cDNA libraries. The resulting draft map includes 7048 proteins and 20,405 interactions. The study also presents a refined, high-confidence map of 4679 proteins and 4780 interactions.
We examined the number of known interactions recapitulated in each data set. We restricted both experimental sets to the PPI between two named genes (CG were excluded) in order to maximize overlap with the literature-derived set, and compared them with 945 Drosophila PPI curated from the literature (A. Baudot, P. Mouren, B. Jacq, and C. Brun, in prep.). This restricted our set to 885 interactions and the one of Giot et al. (2003) to 2787 interactions. Also, 51 interactions (1.8%) from the Giot data set and 20 interactions (2.3%) from the present study were previously known to interact or to occur in the same complex. Taken together with a recent Y2H screen concerned with the cell cycle (Stanyon et al. 2004), all three large-scale screens should be sources of novel information.
To examine how the Giot data set overlapped with ours, we restricted both maps to the PPI involving common bait proteins. Out of the 102 proteins analyzed in the present study, only 30 were found as baits in the genome-wide interaction map. These 30 common bait proteins are implicated in 216 PPI in the Giot map and in 662 PPI in ours. Unexpectedly, the overlap was rather small as the data sets share only 24 interactions (Supplemental Table 2).
We also analyzed the overlap between both data sets and the 4829 genetic interactions gathered in FlyBase. The present map and that of Giot et al. (2003) recapitulated 2.15% (19/885 PPI) and 1% (29/2787 PPI) of the genetic interactions, respectively (Supplemental Table 3). This small overlap indicates that the two approaches provide complementary tools to decipher biological processes.
Benchmarking of the PBS confidence scoring system
Among the 20 interactions already described in the literature, 17 fall into the PBS A category, two into the PBS B category, and the last one into the D category (Supplemental Table 4). On average, 13.1% of all the PBS A interactions (17/130) were found in the literature set, confirming that the PBS correlates with the biological significance of the interaction. In addition, 36 interactions from the present map or the related data set obtained with human orthologs were experimentally tested using independent methodologies (pull-down or coimmunoprecipitation). In all, 25 (69%) interactions were reconfirmed. These figures increase to 23/29 (79%) for the high-confidence (ABC) data set, while two out of seven (29%) PBS D interactions were independently confirmed, demonstrating that biologically relevant interactions can be found in the low-confidence set (Supplemental Table 5). Manuscripts describing in detail these interactions and their biological relevance are in preparation. These observations are consistent with several published studies by us and by others. These use the biological approach and scoring method described here and show that on average at least 60% of the high-confidence interactions are biologically relevant (Rain et al. 2001; Wojcik et al. 2002; Colland et al. 2004; Terradot et al. 2004).
The distribution of PBS categories in the 19 shared “genetic” interactions (10 PBS A, 1 PBS B, and 8 PBS D) is strikingly different from that in the literature set (Supplemental Table 3). The higher proportion of PBS D in the “genetic” data set overlap may indicate that the PBS D subset contains a significant proportion of bona fide interactions that have been refractory to identification using classical protein interaction mapping approaches as suggested by their significant depletion in the “literature” set.
A closer look at the local pathway around the Ras oncogene
Normal and pathological Ras signaling has been largely deciphered using Drosophila genetics (Rubin et al. 1997). However, PPI around Ras have been more thoroughly investigated in vertebrates. Ras has been shown to act via direct interactions with at least three types of effectors, the serine/threonine kinases Raf, the PI3kinases, and GEFs of the RalGDS family. AF6 is another partner of Ras although it might be a biological effector of Rap. The tumor-suppressor gene RASSF1 has been proposed to be a Ras effector regulating its pro-apoptotic function (Vos et al. 2000; Khokhlatchev et al. 2002) and whether RIN proteins, which are Rab5 GEFs, are bona fide Ras effectors still awaits further investigation (Wang et al. 2002).
Our Y2H screens used an activated allele of Ras1 (Ras1G12V) deprived of its C-terminal CAAX motif to avoid prenylation that would target Ras to the plasma membrane and impair Y2H analysis. These screens identified the orthologs of the three main effectors of human Ras, confirming previously described genetic and/or physical interactions: Phl (pole hole), p110, and Rgl2 are the fly orthologs of Raf proteins, the p110 subunit of PI3K, and the RalGDS proteins, respectively (Supplemental Fig. 1A). Cno (canoe, AF6 in mammals) and Loco (orthologous to RGS12 and RGS14) were also identified in this screen. Of significance, the SIDs on these different effectors map exactly to the RA (Ras Association) or RBD (Raf-like Ras Binding Domain) domains previously proposed to mediate Ras binding (Supplemental Fig. 1C).
There is no fly protein containing a C2 domain (Protein Kinase C Conserved Region 2) and a C-terminal RA domain like RASSF1. In vertebrates, RASSF1 interacts with the STE20-like kinases MST1/2 (Khokhlatchev et al. 2002). CG4656 was identified in our screen performed with the fly ortholog of MST1/2 (CG11228/hippo). CG4656 bears an N-terminal LIM (Lin-11 Isl-1 Mec-3) domain instead of the C2 domain found in RASSF1, and a RA domain. The fact that Ras and CG4656 were not found as partners in screens with Ras or with CG4656 as baits while the CG4656-CG11228 interaction was identified suggests that (1) the CG11228-CG4656 interaction is the orthologous interaction of the vertebrate MST1/2-RASSF1 interaction and (2) the Ras-RASSF1 interaction is either specific to vertebrates or, because it is not conserved, questionable.
Prd (paired) and qua (quail) are proteins identified as single interacting fragments (PBS D) in our Ras screens. The Prd transcription factor is orthologous to vertebrate Pax3/Pax7. Qua is the fly ortholog of advillin. Neither prd/pax nor qua/advillin is a known Ras partner, and neither protein contains a domain predicted to mediate binding to Ras. Confidence in a partnership identified once, with no conserved interaction in mammals and no predicted binding domain, should be considered as low.
RIN proteins are vertebrate partners of Ras, although to what extent they are bona fide effectors of Ras remains uncertain. However, the RA-containing fly ortholog CG33175 was not identified in Ras screens, despite its presence in our cDNA library.
Reciprocally, CG8965 was identified as a good Ras effector candidate in flies (PBS A). It contains two RA domains, but only the first one is present in the SID derived from more than 20 independent prey fragments, suggesting that the second RA domain might not be functional regarding Ras binding (Supplemental Fig. 1B). We find an ortholog of CG8965 in Anophaele gambia but not in vertebrates. CG8965 might represent the first case of a Ras effector transducing Ras signals only in insects.
Giot et al. (2003) have screened a wild-type, full-length Ras. The single partner identified, encoded by CG3428, contains no predicted domain (including RA domain) other than an N-terminal F-box. F-box proteins are components of multisubunit E3 ligases, like the SCF complexes. F-box-only proteins like FBX4 might well be the orthologs of CG3428. The absence of an RA domain and the fact that in our screens performed with activated Ras we did not find CG3428 suggest that CG3428 interacts with an unloaded or GDP-bound form of Ras. Protein degradation by the proteasome has been recently demonstrated to control the stability of and the signaling by some GTPases (Wang et al. 2003). The data by Giot et al. (2003) suggest that Ras might join this group, with Ras ubiquitination and subsequent degradation being dependent on CG3428/FBX4.
Discussion
Large-scale two hybrid analysis can be industrialized and robotized and is thus one of the methods of choice for functional annotation by interaction mapping in the post-genomic era. While it has so far been mostly applied to unicellular organisms, recent projects have tackled the genome-wide analysis of multicellular interactomes (i.e., Caenorhabditis elegans and Drosophila melanogaster). These two exhaustive studies have provided the scientific community with functional predictions for a large number of unannotated proteins from these model organisms. In the present study, we report the identification of 2300 interactions connecting 1727 Drosophila proteins. For each interaction, a reliability score is computed, and the interacting domain is mapped. The high-confidence set comprises 710 interactions and connects 641 proteins. Our data set largely complements a recently published genome-wide map of the Drosophila interactome (Giot et al. 2003), as the two projects poorly overlap.
This striking difference points to some of the limitations of large-scale two-hybrid analyses, and several explanations can be proposed. First, full-length proteins could, in several cases, be inappropriate for interaction screening. This is exemplified by plasma membrane proteins whose transmembrane domains hinder Y2H analysis. A rational bait design combined with the screening of short fragmented preys' libraries significantly decrease the rate of false negatives within this protein class as shown in the present study. Although more difficult to adapt to large-scale studies, such customized bait design should improve the quality of yeast two-hybrid screens. In addition, analyzing PPI using multiple truncated forms of bait and prey polypeptides maximize the chance of obtaining properly folded, stable domains and thus successful interactions. This has been substantiated in yeast, where screening a domain library rather than full-length proteins has considerably enriched the yeast protein interaction database (Fromont-Racine et al. 1997). Finally, interacting domains are, in a number of full-length proteins, masked by intramolecular interactions and are therefore not accessible to their ligands unless being exposed following a specific activation signal. Conversely, an interaction can be missed because of a discontinuous interaction domain and would be more likely detected using full-length proteins. It is clear also that full-length prey collections have the advantage of being normalized and fully representative of a predicted genome.
Second, the depth of the screening procedure dramatically impacts the false-negative rate. In the present study, exhaustive screening and analysis of up to 380 clones per screen (on average 119 preys/bait) were performed. The resulting coverage is close to comprehensiveness, as suggested by a reproducibility rate close to 90% (Supplemental Table 6).
Third and finally, additional methodological differences may explain the small overlap between the Drosophila studies. Each project used unique yeast expression vectors with distinct fusion systems that might impact the folding and stability of the hybrid proteins. The stringency of the selection was also different; in the present study, the selective pressure is adapted to the intrinsic properties of each bait and a single reporter gene is used, while in the genome-wide analysis, a unique selection condition and two reporter genes are used. When applied to the present screening procedure, a double-reporter system gave an insignificant attrition rate and increased the false-negative rate (data not shown). A single-reporter strategy was therefore adopted.
A common theme in the increasing number of large-scale Y2H studies is the occurrence of false-positive interactions that need to be filtered out. The high reproducibility of the screening strategy described here has allowed the development of a systematic statistical approach to assign a confidence score, the PBS, to each interaction. The PBS has been shown here and in other independent studies (Rain et al. 2001; Wojcik et al. 2002; Colland et al. 2004; Terradot et al. 2004) to positively correlate with the biological significance of interactions. Previous attempts at confidence scoring used only the redundancy in prey fragments (Ito et al. 2001) or identified some of the parameters used in PBS calculation as global predictors in a trained linear model (Giot et al. 2003). Ideally, the establishment of experimental standards to uniformly evaluate false positives would be needed to compare or integrate data from different sources. The rate of false positives in the two Drosophila studies can be broadly estimated by comparing the percentage of low-confidence interactions in both data sets. It appears that 77% (15,625/20,405) of Giot et al. (2003) interactions have been classified as low confidence versus 70% (1628/2338) in the present study. This rather high prevalence of low-confidence interactions may partly explain the small overlap between the two studies.
Conversely, large-scale two-hybrid data sets are largely incomplete because of a significant rate of false negatives. This is inherent in the two-hybrid technology as it cannot apply to all PPI and is probably biased toward specific interacting domain pairs (see above). In addition, different approaches using distinct types of bait and prey constructs, expression vectors, or selective pressure will favor different segments of a given interactome. This has been substantiated in a recent study comparing two Drosophila maps generated using different bait and prey constructs (Stanyon et al. 2004).
Clearly, genome-wide and more focused studies have different limitations. The former allows a cost- and time-effective exploration of complex interactomes but results in maps that have a much lower coverage than the latter. Conversely, the focused approach provides a more comprehensive map but is less adaptable to genome-wide exploration mainly because of cost, time, and managerial considerations. Cost refers mainly to the screening cost and, in particular, the prey sequencing up to 380 clones/bait; time refers mainly to the custom bait design as opposed to full-length, wild-type cDNA cloning. Finally, managerial consideration refers to the requirement of experts in each targeted field for proper custom bait design. The original goal of the strategy described here was to find a compromise between throughput and quality (i.e., reproducibility of the screens, density of the map, interaction scoring). Currently, the union of processed data obtained through different approaches, such as those discussed here, should complement each other and considerably improve our knowledge of protein interactomes.
Ultimately, the integration of multiple, independent sets of genomic and proteomic data, as proposed for yeast and C. elegans (Ge et al. 2001; Li et al. 2004), proves to be even more powerful in assigning unannotated proteins to biological pathways. However, the recent correlation between some of these data sets from C. elegans appears to be poorer than those observed in yeast, possibly reflecting the overall higher complexity of metazoan biological processes. The controlled, multiple integration of genomic and proteomic data sets generated in different uni- and pluricellular organisms may overcome some of these limitations. But efforts in ortholog identification through phylogenetic analyses need to be accomplished.
Reliable exploration tools are urgently needed to visualize, explore, and analyze these large proteomic data sets. To this end, our dedicated Web-based graphical interface, the PIMRider is freely accessible at http://pim.hybrigenics.com to explore the present map. Our Drosophila protein interactions data are also available in the standard data model recently developed by the Proteomics Standards Initiative (PSI) and can be visualized using the PIMWalker tool (http://pim.hybrigenics.com/pimwalker) which graphically displays interaction networks described in this format (Hermjakob et al. 2004). Finally, we have deposited these interactions with FlyBase (http://flybase.bio.indiana.edu/) and with BIND (http://bind.ca), DIP (http://dip.doe-mbi.ucla.edu/), IntAct (http://www.ebi.ac.uk/intact/index.html), MINT (http://mint.bio.uniroma2.it/mint/), and MIPS (http://mips.gsf.de/) via the IMEX (International Molecular Interaction Exchange) consortium (http://imex.mbi.ucla.edu:60606/imex/IMEX.jsp).
Methods
Bait cloning
The tim, per, cry, and cyc bait constructs were provided by Michael Rosbash (Brandeis University, MA, USA). Baits were either directly subcloned in the pB27 plasmid or were first PCR-amplified and then transferred. The pB27 plasmid is derived from the original pBTM116 (Vojtek and Hollenberg 1995) in which the AmpR gene has been replaced by a TetR gene. In addition, the multiple cloning site has been modified to GAATTCGGGGCCGGACGGGCCGCGGCCGCACTAGTGGGGATCCTTAATTAAGGGCCACTGGGGCCCCTCGACCTGCAG. All bait constructs were checked by full insert sequencing. Bait plasmids were transformed in the L40ΔGAL4 yeast strain (Fromont-Racine et al. 1997).
Library construction
Random-primed cDNA libraries from Drosophila (Canton strain) adult heads (provided by Michael Rosbash) and embryos (provided by Peter Maroy [University of Szeged, Hungary]) poly(A)+ RNA were constructed in the pP6 plasmid. The embryo library is an equimolar pool of two cDNA libraries prepared from 0-12 h (zygotic + maternal mRNA) and 12-24-h embryo mRNA. The pP6 plasmid is derived from the original pGADGH (Clontech) in which the multiple cloning site has been modified to CCATGGCCGCAGGGGCCGCGGCCGCACTAGTGGGGATCCTTAATTAAGGGCCACTGGGGCCCCTCGAGTAGCTAGTGTCTAGA. The cDNA fragments shorter than 400 bp were removed by gel filtration chromatography. In all, 90% of the plasmids contained a cDNA insert with an average size of 700 bp. After amplification in Escherichia coli (50 million independent clones), the cDNA libraries were transformed into the Y187 yeast strain. Ten million independent yeast colonies were collected, pooled, and stored at -80°C as equivalent aliquot fractions of the same library.
Screening procedure and identification of interacting fragments
The screens were performed using a mating method previously described (Fromont-Racine et al. 2002). Each screen was first performed on a small scale to adapt the selective pressure to the intrinsic properties of the bait. Then, the full-size screen was performed to ensure a minimum of 50 million interactions tested (five times the primary complexity of the yeast-transformed cDNA libraries). Positive clones were selected on medium lacking leucine, tryptophane, and histidine. Up to 380 positive clones per independent screen were picked, and the corresponding prey fragments were amplified by PCR and sequenced at their 5′ and 3′ junctions. 5′ and 3′ sequences were then filtered by using PHRED (Ewing and Green 1998; Ewing et al. 1998) and masking ALU repeats. Sequence contigs were built using CAP3 (Huang and Madan 1999) and compared to the recent release 3.1 of BDGP using BLASTN (Altschul et al. 1997). In the few cases where no matching transcripts were identified, the contigs were compared to the latest release of the GenBank database using the same BLASTN procedure. The issue of the splice variants was addressed as follows: When the sequences of the prey fragment(s) allow the identification of a specific splice variant, the identification procedure assigns this prey (or the prey family) to this specific variant. When the prey fragment sequences do not allow the distinction between different isoforms, the process picks the best annotated variant and assigns the prey fragment(s) to this isoform.
Interaction scoring
The method for calculating the Predicted Biological Score (PBS), previously described for genomic libraries (Rain et al. 2001), has been adapted for randomly primed cDNA libraries. The PBS takes into account the redundancy and independency of prey fragments (number of times an interaction was observed with one given bait fragment), the distributions of reading frames and stop codons in overlapping fragments, and the local topology of the interaction network: highly connected prey regions, interactions confirmed by two independent screens in the bait/prey and prey/bait orientations, and cycles and cliques. The PBS E-value ranges from 0 to 1 and has been classified in five distinct categories: A to E. Intercategory thresholds were chosen manually with respect to a training data set containing known true-positive and false-positive interactions (data not shown): A < 1e-10 < B < 1e-5 < C < 1e-2.5 < D < 1. The E category corresponds to prey domains nonspecifically selected by baits (Rain et al. 2001) for which the corresponding PBS has been set to 1, and then represents most probably false positives. Categories A, B, and C represent probable true-positive interactions at different levels of confidence. In between, the D category gathers protein interactions detected by only one prey fragment for a given bait: It may represent false positives (prey fragment selected nonspecifically by the bait) or interesting rare events (interaction with a protein encoded by a rare mRNA or interaction difficult to detect in classical two-hybrid assays). Those categories have been shown to be positively correlated to the biological significance of interactions (Rain et al. 2001; Wojcik et al. 2002). More detailed explanations about PBS calculation can be found as Supplemental material.
In addition, proteins connected in the PIM were automatically annotated in terms of structural and functional domains by use of bioinformatics algorithms: TMHMM (Krogh et al. 2001) and SignalP (Nielsen et al. 1997) for the detection of transmembrane helices and signal peptides, respectively, and IpScan for the prediction of InterPro domains (Apweiler et al. 2001). Proteins are also linked to functional categories defined in the Gene Ontology (GO) classification (Ashburner and Lewis 2002).
Acknowledgments
This work was supported by a GenHomme Network Grant (02490-6088) to Hybrigenics and Institut Curie as well as by the Robert E. Welch Foundation (I-1414) and the Association de Recherche sur le Cancer (5440). We thank Spyros Artavanis, Hervé Chneiweiss, Jean-Louis Couderc, Laurence Del Maestro, Christelle Dousset, Sylvaine Fouix, Bill Gelbart, Don Gilbert, Claudie Lamour-Isnard, Séverine Martin-Lannerée, Mélanie Paces-Fessy, Matthieu Sanial, Stephanie Vogeling, and all Hybrigenics staff for their contribution. Special thanks to the Hybrigenics Department of Industrial Operation headed by Gerlinde Lenzen, Emmanuelle Vanmerris, and Catherine Borg-Capra. We are very indebted to Donny Strosberg, Daniel Louvard, and Jean-Christophe Rain for stimulating discussions. We are very grateful to Michael Rosbash for critical reading of the manuscript and helpful suggestions. S.A. is a recipient of a Marie Curie Industry Host Fellowship (contract # QLK5-CT-2001-60042).
Footnotes
[Supplemental material is available online at www.genome.org. The interaction data described in this study have been submitted to FlyBase, BIND (accession numbers: 146576-146804 and 146805-148829 for the Drosophila head and embryo interactions, respectively), and the IMEX (International Molecular Interaction Exchange) consortium (accession numbers: IMEX0000001 and IMEX0000002 for the Drosophila embryo and head interactions, respectively). The following individuals kindly provided reagents, samples, or unpublished information as indicated in the paper: M. Rosbash and P. Maroy.]
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.2659105. Article published online ahead of print in February 2004.
References
- Adams, M.D., Celniker, S.E., Holt, R.A., Evans, C.A., Gocayne, J.D., Amanatides, P.G., Scherer, S.E., Li, P.W., Hoskins, R.A., Galle, R.F., et al. 2000. The genome sequence of Drosophila melanogaster. Science 287: 2185-2195. [DOI] [PubMed] [Google Scholar]
- Altschul, S.F., Madden, T.L., Schaffer, A.A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D.J. 1997. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25: 3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apweiler, R., Attwood, T.K., Bairoch, A., Bateman, A., Birney, E., Biswas, M., Bucher, P., Cerutti, L., Corpet, F., Croning, M.D., et al. 2001. The InterPro database, an integrated documentation resource for protein families, domains and functional sites. Nucleic Acids Res. 29: 37-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner, M. and Lewis, S. 2002. On ontologies for biologists: The Gene Ontology—untangling the web. Novartis Found. Symp. 247: 66-80; discussion 80-63, 84-90, 244-252. [PubMed] [Google Scholar]
- Boeda, B., El-Amraoui, A., Bahloul, A., Goodyear, R., Daviet, L., Blanchard, S., Perfettini, I., Fath, K.R., Shorte, S., Reiners, J., et al. 2002. Myosin VIIa, harmonin and cadherin 23, three Usher I gene products that cooperate to shape the sensory hair cell bundle. EMBO J. 21: 6689-6699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celniker, S.E., Wheeler, D.A., Kronmiller, B., Carlson, J.W., Halpern, A., Patel, S., Adams, M., Champe, M., Dugan, S.P., Frise, E., et al. 2002. Finishing a whole-genome shotgun: Release 3 of the Drosophila melanogaster euchromatic genome sequence. Genome Biol. 3: RESEARCH0079. [DOI] [PMC free article] [PubMed]
- Colland, F., Jacq, X., Trouplin, V., Mougin, C., Groizeleau, C., Hamburger, A., Meil, A., Wojcik, J., Legrain, P., and Gauthier, J.M. 2004. Functional proteomics mapping of a human signaling pathway. Genome Res. 14: 1324-1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewing, B. and Green, P. 1998. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 8: 186-194. [PubMed] [Google Scholar]
- Ewing, B., Hillier, L., Wendl, M.C., and Green, P. 1998. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 8: 175-185. [DOI] [PubMed] [Google Scholar]
- Fromont-Racine, M., Rain, J.C., and Legrain, P. 1997. Toward a functional analysis of the yeast genome through exhaustive two-hybrid screens. Nat. Genet. 16: 277-282. [DOI] [PubMed] [Google Scholar]
- ____. 2002. Building protein-protein networks by two-hybrid mating strategy. Methods Enzymol. 350: 513-524. [DOI] [PubMed] [Google Scholar]
- Gavin, A.C., Bosche, M., Krause, R., Grandi, P., Marzioch, M., Bauer, A., Schultz, J., Rick, J.M., Michon, A.M., Cruciat, C.M., et al. 2002. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415: 141-147. [DOI] [PubMed] [Google Scholar]
- Ge, H., Liu, Z., Church, G.M., and Vidal, M. 2001. Correlation between transcriptome and interactome mapping data from Saccharomyces cerevisiae. Nat. Genet. 29: 482-486. [DOI] [PubMed] [Google Scholar]
- Giot, L., Bader, J.S., Brouwer, C., Chaudhuri, A., Kuang, B., Li, Y., Hao, Y.L., Ooi, C.E., Godwin, B., Vitols, E., et al. 2003. A protein interaction map of Drosophila melanogaster. Science 302: 1727-1736. [DOI] [PubMed] [Google Scholar]
- Grigoriev, A. 2003. On the number of protein-protein interactions in the yeast proteome. Nucleic Acids Res. 31: 4157-4161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermjakob, H., Montecchi-Palazzi, L., Bader, G., Wojcik, J., Salwinski, L., Ceol, A., Moore, S., Orchard, S., Sarkans, U., Von Mering, C., et al. 2004. The HUPO PSI's Molecular Interaction format—A community standard for the representation of protein interaction data. Nat. Biotechnol. 22: 177-183. [DOI] [PubMed] [Google Scholar]
- Huang, X. and Madan, A. 1999. CAP3: A DNA sequence assembly program. Genome Res. 9: 868-877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito, T., Chiba, T., Ozawa, R., Yoshida, M., Hattori, M., and Sakaki, Y. 2001. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl. Acad. Sci. 98: 4569-4574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khokhlatchev, A., Rabizadeh, S., Xavier, R., Nedwidek, M., Chen, T., Zhang, X.F., Seed, B., and Avruch, J. 2002. Identification of a novel Ras-regulated proapoptotic pathway. Curr. Biol. 12: 253-265. [DOI] [PubMed] [Google Scholar]
- Krogh, A., Larsson, B., von Heijne, G., and Sonnhammer, E.L. 2001. Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J. Mol. Biol. 305: 567-580. [DOI] [PubMed] [Google Scholar]
- Lehner, B. and Fraser, A.G. 2004. A first-draft human protein-interaction map. Genome Biol. 5: R63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, S., Armstrong, C.M., Bertin, N., Ge, H., Milstein, S., Boxem, M., Vidalain, P.O., Han, J.D., Chesneau, A., Hao, T., et al. 2004. A map of the interactome network of the metazoan C. elegans. Science 303: 540-543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misra, S., Crosby, M.A., Mungall, C.J., Matthews, B.B., Campbell, K.S., Hradecky, P., Huang, Y., Kaminker, J.S., Millburn, G.H., Prochnik, S.E., et al. 2002. Annotation of the Drosophila melanogaster euchromatic genome: A systematic review. Genome Biol. 3: RESEARCH0083. [DOI] [PMC free article] [PubMed]
- Nielsen, H., Engelbrecht, J., Brunak, S., and von Heijne, G. 1997. A neural network method for identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Int. J. Neural Syst. 8: 581-599. [DOI] [PubMed] [Google Scholar]
- Rain, J.C., Selig, L., De Reuse, H., Battaglia, V., Reverdy, C., Simon, S., Lenzen, G., Petel, F., Wojcik, J., Schachter, V., et al. 2001. The protein-protein interaction map of Helicobacter pylori. Nature 409: 211-215. [DOI] [PubMed] [Google Scholar]
- Rubin, G.M., Chang, H.C., Karim, F., Laverty, T., Michaud, N.R., Morrison, D.K., Rebay, I., Tang, A., Therrien, M., and Wassarman, D.A. 1997. Signal transduction downstream from Ras in Drosophila. Cold Spring Harb. Symp. Quant. Biol. 62: 347-352. [PubMed] [Google Scholar]
- Scheel, H. and Hofmann, K. 2003. A novel interaction motif, SARAH, connects three classes of tumor suppressor. Curr. Biol. 13: R899-R900. [DOI] [PubMed] [Google Scholar]
- Stanyon, C.A., Liu, G., Mangiola, B.A., Patel, N., Giot, L., Kuang, B., Zhang, H., Zhong, J., and Finley Jr., R.L. 2004. A Drosophila protein-interaction map centered on cell-cycle regulators. Genome Biol. 5: R96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terradot, L., Durnell, N., Li, M., Ory, J., Labigne, A., Legrain, P., Colland, F., and Waksman, G. 2004. Biochemical characterization of protein complexes from the Helicobacter pylori protein interaction map: Strategies for complex formation and evidence for novel interactions within type IV secretion systems. Mol. Cell Proteomics 3: 809-819. [DOI] [PubMed] [Google Scholar]
- Uetz, P., Giot, L., Cagney, G., Mansfield, T.A., Judson, R.S., Knight, J.R., Lockshon, D., Narayan, V., Srinivasan, M., Pochart, P., et al. 2000. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature 403: 623-627. [DOI] [PubMed] [Google Scholar]
- Vojtek, A.B. and Hollenberg, S.M. 1995. Ras-Raf interaction: Two-hybrid analysis. Methods Enzymol. 255: 331-342. [DOI] [PubMed] [Google Scholar]
- Vos, M.D., Ellis, C.A., Bell, A., Birrer, M.J., and Clark, G.J. 2000. Ras uses the novel tumor suppressor RASSF1 as an effector to mediate apoptosis. J. Biol. Chem. 275: 35669-35672. [DOI] [PubMed] [Google Scholar]
- Walhout, A.J., Sordella, R., Lu, X., Hartley, J.L., Temple, G.F., Brasch, M.A., Thierry-Mieg, N., and Vidal, M. 2000. Protein interaction mapping in C. elegans using proteins involved in vulval development. Science 287: 116-122. [DOI] [PubMed] [Google Scholar]
- Wang, Y., Waldron, R.T., Dhaka, A., Patel, A., Riley, M.M., Rozengurt, E., and Colicelli, J. 2002. The RAS effector RIN1 directly competes with RAF and is regulated by 14-3-3 proteins. Mol. Cell. Biol. 22: 916-926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, H.R., Zhang, Y., Ozdamar, B., Ogunjimi, A.A., Alexandrova, E., Thomsen, G.H., and Wrana, J.L. 2003. Regulation of cell polarity and protrusion formation by targeting RhoA for degradation. Science 302: 1775-1779. [DOI] [PubMed] [Google Scholar]
- Wojcik, J., Boneca, I.G., and Legrain, P. 2002. Prediction, assessment and validation of protein interaction maps in bacteria. J. Mol. Biol. 323: 763-770. [DOI] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- http://bind.ca; BIND.
- http://dip.doe-mbi.ucla.edu/; DIP.
- http://flybase.bio.indiana.edu/; FlyBase.
- http://imex.mbi.ucla.edu:60606/imex/IMEX.jsp; IMEX.
- http://mint.bio.uniroma2.it/mint/; MINT.
- http://mips.gsf.de/; MIPS.
- http://pim.hybrigenics.com; Drosophila PIMRider.
- http://pim.hybrigenics.com/pimwalker/; PIMWalker.
- http://www.ebi.ac.uk/intact/index.html; IntAct.
- http://www.mshri.on.ca/pawson/domains.html; Pawson Lab domains' page.