Abstract
Objective Sinonasal teratocarcinosarcomas are rare, aggressive tumors of the skull base. Treatment options are limited and outcomes are poor. Little is known in regard to the genetic factors regulating these tumors. Characterization of actionable molecular alterations in these tumors could provide potentially successful therapeutic options.
Methods We performed targeted exome sequencing on an index sinonasal teratocarcinosarcoma specimen to identify potential driver mutations. We performed immunohistochemical stains for β-catenin on paraffin-embedded tissue on the index tumor and a subsequent teratocarcinosarcoma. Online databases of cancer mutations (Catalogue of Somatic Mutations in Cancer and The Cancer Genome Atlas) were accessed.
Results We identified an activating p.S45F mutation in β-catenin in our index sinonasal teratocarcinosarcoma. This mutation results in constitutive signaling in the Wnt/β-catenin pathway. We confirmed β-catenin overexpression and nuclear localization via immunohistochemistry in the index tumor and a second patient. The p.S45F activating mutation was found in a variety of solid tumors, and accounts for 3.3 to 10.4% of all known β-catenin mutations.
Conclusion We identified a potential driver mutation in β-catenin in a sinonasal teratocarcinosarcoma, resulting in β-catenin overexpression. These findings suggest a role for the Wnt/β-catenin pathway in sinonasal teratocarcinosarcoma tumorigenesis and a role for anti-β-catenin targeted therapy.
Keywords: CTNNB1, Wnt, pathogenetics, sinonasal teratocarcinosarcoma, teratocarcinosarcoma, β-catenin
Introduction
Teratocarcinosarcomas are aggressive tumors arising primarily in the sinonasal region and anterior cranial base. They are extremely rare, with less than 100 cases ever reported in the literature. 1 These tumors are heterogeneous, and comprised epithelial, mesenchymal, and neural tissues. 2 Sinonasal teratocarcinosarcomas are highly difficult to treat, in part due to their location in the anterior skull base (with difficulty to achieve adequate margins) and in part due to their inherent aggressiveness. Current standard of care includes aggressive surgical resection followed by radiation therapy. 3 There are no established surgical guidelines, but surgical options historically reported include open anterior craniofacial resection, maxillectomy, and lateral rhinotomy. 1 Currently open, endoscopic, or a combination thereof have been used to approach sinonasal teratocarcinosarcomas to achieve complete resection.
Unfortunately, disease recurrence is frequent (31.4%), with a high rate for metastasis (17.6%) and intracranial extension. 1 Most frequently, the lungs are the site of distant metastasis. Overall survival historically was dismal, and although improving (56% overall survival at 45 months after surgery and radiation), 1 patients still have a guarded prognosis.
Little is known biologically or genetically about teratocarcinosarcomas. Although previous immunohistochemical staining has confirmed the presence of epithelial, mesenchymal, and neural markers, 2 4 there has been no insight into the pathogenesis of these tumors. The genetic pathways governing teratocarcinosarcomas have not been studied, and no reported disruptive genomic events driving these tumors have been described. Identification of the underlying genetic drivers of sinonasal teratocarcinosarcomas could provide potentially successful adjuvant or alternative precision medicine treatment options for these aggressive tumors. 5 6 7 8 Here, we describe a case of a patient with a sinonasal teratocarcinosarcoma and perform targeted next-generation sequencing to identify critical genomic drivers of the disease.
Methods
Patient Data
Patients were identified from the University of Michigan Cranial Base Program. Institutional Review Board (IRB) approval was obtained from the University of Michigan IRB. Patient data and images were collected from clinical records retrospectively.
Targeted Exomic Sequencing
Using targeted, amplicon-based sequencing with the Oncomine Cancer Panel, we assessed the mutation status of common oncologic therapeutic targets as described. 9 10 Briefly, amplicon-based DNA sequencing was performed using 40 ng of DNA isolated with a RNA/DNA formalin-fixed paraffin-embedded isolation kit (Qiagen) on the Ion Torrent Personal Genome Machine, utilizing the AmpliSeq Oncomine Cancer Panel. 11 Nucleotide variants and indels were identified using the Torrent Variant Caller plugin, annotated using Annovar. 12 Sequencing the tumor library on a single 318 chip, we generated 5,254,571 mapped reads with a mean depth of 315 x and 96% of all targeted bases covered by at least 20 reads. Alignment and variant calling were performed using standard plugins in the Torrent Server Suite (v3.6). We identified 1,209 variants, which were then filtered through a standardized pipeline to remove low confidence calls, sequencing errors, germline variants and common polymorphisms, and results were compared with known mutations in Catalogue of Somatic Mutations in Cancer (COSMIC), the Oncomine Powertools Database (containing pan-cancer The Cancer Genome Atlas [TCGA] data), and our internal database of cancer sequencing results.
Sanger Sequencing
Confirmatory Sanger sequencing of the index tumor, and sequencing of additional index patient tissue was performed to confirm the mutation was a somatic variant. Forward and reverse primers were designed to amplify the region of the c.134C>T mutation ( Supplementary Table 1 ). Standard polymerized chain reaction conditions were used to amplify the DNA product. Sanger sequencing was performed by the University of Michigan Sequencing Core. Data analysis was performed with Sequence Scanner Software 1.0 from Applied Biosystems, Inc..
Immunohistochemical Staining of β-catenin
Immunohistochemical staining for β-catenin overexpression was performed on patient tumors. Briefly, the formalin-fixed paraffin-embedded sections from the primary tumor were heated, and underwent peroxidase blocking. A β-catenin mouse monoclonal antibody (224M; Cell Marque) was applied at a 1:50 dilution. A Mouse EnVision + HRP Kit (Dako) was used for staining. The slides were then counterstained with Harris's hematoxylin.
Analysis of Data from the Cancer Genome Atlas and COSMIC
The COSMIC is an online database cataloguing mutations in a wide array of cancers. 13 TCGA is an online repository of mutational findings collected from next-generation sequencing on a variety of cancers. 14 Mutational reports from both datasets are publicly available and were accessed to identify mutations in β-catenin in other tumors as previously described. 15 16
Results
Patient Clinical Courses
The index patient was a 48-year-old man who initially presented with a left-sided nasal mass. His symptoms included a 2-month history of left-sided nasal obstruction, hyposmia, and recurrent left-sided epistaxis. On clinical exam, he had a large friable left-sided nasal mass filling the nasal cavity and extending into the nasopharynx. Biopsy in clinic demonstrated poorly differentiated sarcomatoid adenocarcinoma. Imaging demonstrated a T2 hyperintense, T1 hypointense left nasal cavity mass extending to the left ethmoid sinuses and nasopharynx. It also involved the lamina papyracea, abutting the left orbit and extending up to the cribriform plate ( Fig. 1A ). Positron emission tomography imaging did not reveal any metastatic disease, and he was staged as T2N0M0.
Fig. 1.
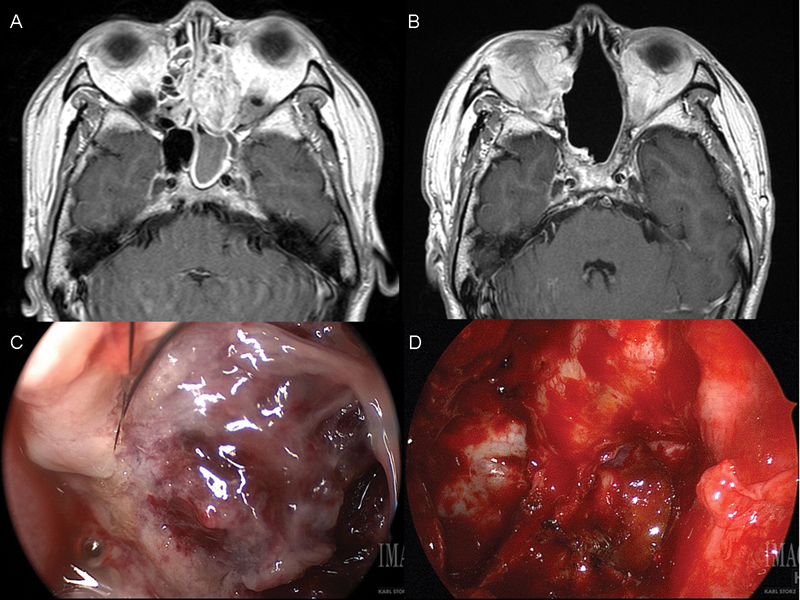
( A ) Preoperative and ( B ) 24-month postoperative axial T1-weighted MRI with contrast of the index teratocarcinosarcoma. Intraoperative appearance of sinonasal teratocarcinosarcoma ( C ) before and ( D ) after resection. MRI, magnetic resonance imaging.
The patient underwent a margin-negative endoscopic endonasal approach for resection of his nasal mass, including cribriform resection and nasoseptal vascularized pedicled flap reconstruction ( Fig. 1C, D ). Final pathology was confirmed as a sinonasal teratocarcinosarcoma, with negative margins. He underwent 60 gray of postoperative radiation to the left neck and bilateral retropharyngeal nodal basins. He recovered well postoperatively, and is currently 43 months status posttreatment with no evidence of disease recurrence ( Fig. 1B ).
The second patient was a 50-year-old man who had slowly progressive headaches, nausea, and emesis for 2 to 3 years. He had worsening headaches, nausea, and emesis, along with behavioral changes, dysosmias, a recurrent right-sided nasal obstruction and epistaxis. Magnetic resonance imaging of the brain revealed a large mass in the right inferior frontal lobe ( Fig. 2A, B ). Biopsies were consistent with sinonasal teratocarcinosarcoma. The tumor was not surgically resectable; thus, the patient underwent chemoradiation. While he initially tolerated chemoradiation, his neurologic status deteriorated. Subsequent imaging revealed progressive disease and hemorrhagic infarctions ( Fig. 2C, D ). Given his worsening clinical status, he was made comfort care and died ∼1 month after presentation.
Fig. 2.
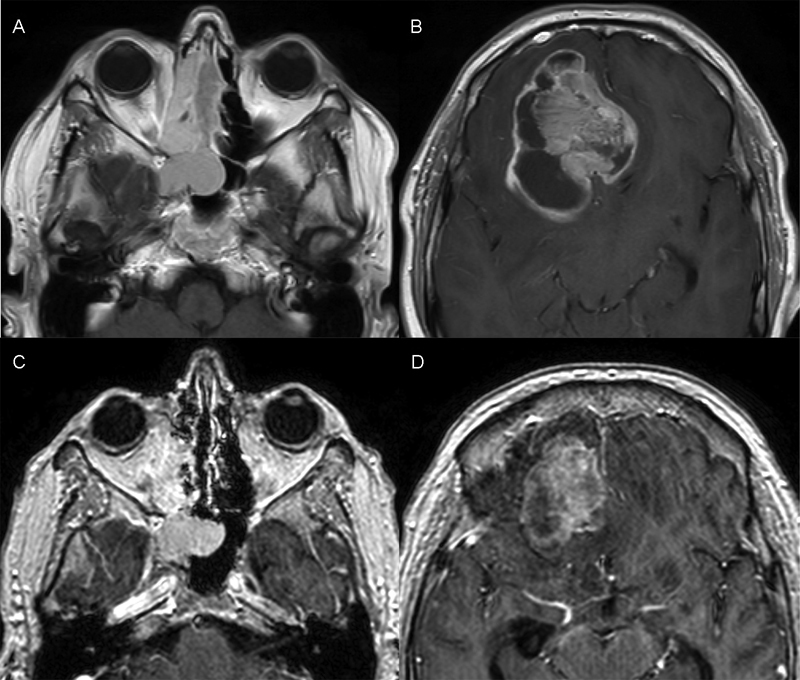
( A, B ) Initial and ( C, D ) follow-up axial T1-weighted MRI of the second teratocarcinosarcoma. The follow-up MRI showed new hemorrhagic infarcts over the course of chemoradiation treatment. MRI, magnetic resonance imaging.
Targeted Exome Sequencing of the Sinonasal Teratocarcinosarcoma
Given the limited understanding of the biology behind sinonasal teratocarcinosarcomas, we screened for commonly mutated cancer drivers, as determined by the Oncomine Cancer Panel. This analysis pipeline prioritized a high confidence mutation in the CTNNB1 (β-catenin) gene, resulting in a p.S45F codon change. To confirm our finding of the p.S45F mutation in β-catenin, we performed Sanger sequencing. We confirmed the c.134C>T point mutation in one copy of β-catenin ( Fig. 3A ), resulting in the p.S45F activating mutation. Sequencing of surrounding tissue did not reveal this mutation ( Fig. 3B ), verifying this is a somatic, not germline, mutation.
Fig. 3.
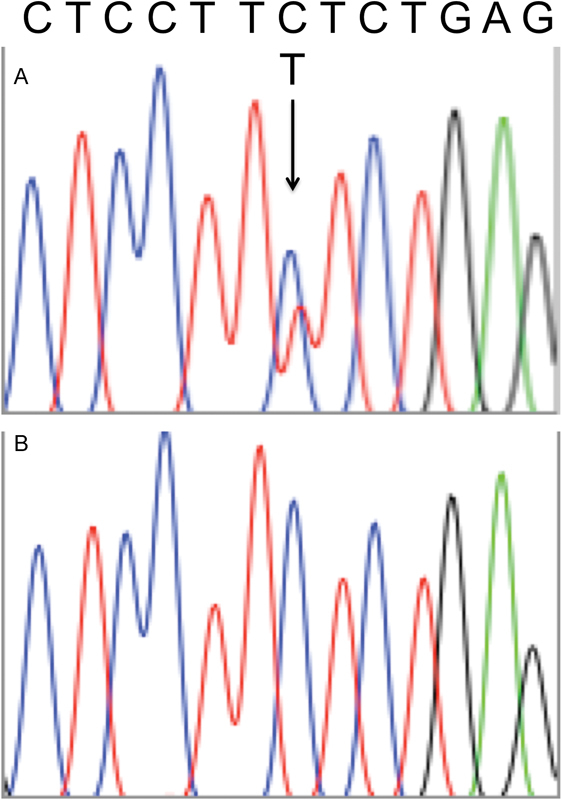
( A ) Chromatogram of c.134C>T mutation in β-catenin, resulting in p.S45F. Arrow pointing to the mutation. ( B ) No mutation seen in surrounding tissue, confirming this is a somatic mutation.
Immunohistochemical Staining of β-catenin
We performed immunohistochemical staining to identify β-catenin protein expression patterns in our index tumor and the second tumor. In the tumors, the carcinoma component was adenocarcinoma. The teratoma component contained benign respiratory, benign squamous, and primitive mesenchymal components. Both tumors demonstrated significant overexpression of β-catenin throughout the tumor, when compared with surrounding nontumor tissue ( Fig. 4 ). Specifically, β-catenin staining was predominantly nuclear in the mesenchymal component and in a subset of the epithelial component. The majority of the epithelial component demonstrated membranous β-catenin staining.
Fig. 4.
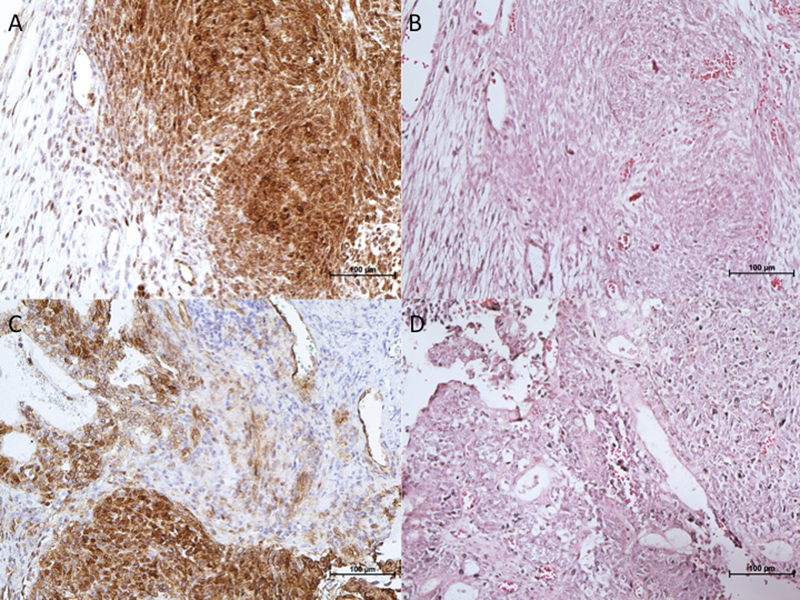
( A, C ) Immunohistochemistry demonstrating overexpression of β-catenin in index and second teratocarcinosarcoma tumor tissues, with surrounding normal tissue with no overexpression of β-catenin. ( B, D ) Corresponding hematoxylin and eosin stains were captured. ×20 magnification.
β-catenin S45F Mutations Identified in Mutation Databases
We next searched for p.S45F (c.134C>T) β-catenin mutations in publically available cancer databases using Oncomine. Overall, 454 mutations in β-catenin were identified in 10,194 tumor samples (4.5% of all tumors). Of the 454 mutations, 44 were missense mutations in S45 (9.7% of all β-catenin mutations), and 15 were specifically p.S45F mutations (3.3% of all β-catenin mutations). The tumors containing this specific mutation were most frequently hepatocellular carcinomas ( Table 1 ).
Table 1. Known p.S45F β-catenin mutations in other tumors.
| Database | % Tumors with β-catenin mutations | % β-catenin mutations p.S45F | p.S45F tumor types |
|---|---|---|---|
| COSMIC | 10.1% (4,974/49,121) | 10.4% (518/4,974) | 65% Desmoid fibromatosis (337/518) 12% Colorectal (61/518) 11% Hepatic (59/518) 4% Wilm's tumor (21/518) 3% Endometrial (13/518) 2% Melanoma (8/518) 1% Biliary (5/518) < 1% Adrenal (3/518) < 1% Ovarian (3/518) < 1% CNS (2/518) < 1% Cervical (1/518) < 1% Gastric (1/518) < 1% Lung (1/518) < 1% Lymphoma (1/518) < 1% Pancreatic (1/518) < 1% Urethral (1/518) |
| TCGA | 4.5% (454/10,194) | 3.3% (15/454) | 60% Hepatic (9/15) 13% Endometrial (2/15) 7% Adrenal (1/15) 7% Lung (1/15) 7% Colorectal (1/15) 7% Melanoma (1/15) |
Abbreviations: CNS, central nervous system; COSMIC, Catalogue of Somatic Mutations in Cancer; TCGA, The Cancer Genome Atlas.
We then queried the p.S45F (c.134C>T) mutation in the COSMIC database. The p.S45F mutation was identified in more than 500 combined soft tissue tumors (including adrenal, biliary tract, central nervous system, endometrial, kidney, and liver systems, Table 1 ). The most common tumor harboring the mutation is desmoid fibromatosis.
Discussion
The Wnt//β-catenin pathway is a well-studied cell proliferation pathway. Dysregulation in Wnt//β-catenin signaling has been implicated in several cancers. 17 18 19 Using next-generation sequencing techniques, we have identified an actionable p.S45F mutation in β-catenin in a sinonasal teratocarcinosarcoma. This mutation is of great interest as it leads to decreased degradation of β-catenin, resulting in increased signaling for cell growth and division. 20 21 We have confirmed that this mutation results in an overexpression and nuclear translocation of β-catenin based on immunohistochemical stains. Given the resulting constitutive Wnt/β-catenin signaling, we postulate this mutation is a potential genetic driver mutation and an alluring prospect for treatment using inhibitors of the Wnt/β-catenin pathway. 22
Interestingly, the p.S45F mutation is identified in tumors with a significant mesenchymal component (particularly desmoid fibromatosis of soft tissue). This is consistent with the histology in this sinonasal teratocarcinosarcoma, which contains a significant amount of mesenchymal tissue. Notably, this mutation has been demonstrated to predict increased recurrence and poor response to meloxicam therapy in desmoid tumors. 23 24 The presence of the p.S45F mutation in multiple soft tissue tumors, and identification of a worse prognosis in certain tumors with this mutation, suggests a potential beneficial role for inhibitors targeting β-catenin.
Precision medicine trials are increasingly being employed to identify and apply targeted therapy options in cancers refractory to current treatment paradigms. 25 26 Currently, multiple Wnt/β-catenin pathway agents are in development, including agents targeted specifically against β-catenin. 27 28 Future trials using these inhibitors in patients with p.S45F mutations will be informative in their applicability. In patients with this specific mutation, targeted regulators of the Wnt pathway upstream of β-catenin may prove to be ineffective given the likely dominant overexpression of the β-catenin protein. Rather, specific inhibitors of β-catenin and downstream targets may prove to be more successful.
Given the current limitations in care and poor prognosis of patients with sinonasal teratocarcinosarcoma, new treatment paradigms need to be identified. Next-generation sequencing techniques provide valuable data for potential targeted therapy options. Specifically in this instance, we have identified a p.S45F mutation in β-catenin in a patient with a sinonasal teratocarcinosarcoma, providing a targetable treatment option if this index patient were to suffer disease recurrence. This is the first study to identify a targetable genetic mutation and possible tumor driver in a sinonasal teratocarcinosarcoma. In the future, it will be important to screen for p.S45F mutations and other mutations/aberrations in β-catenin in other teratocarcinosarcoma specimens to identify if this is a common driver mutation in these tumors. If so, application of Wnt/β-catenin pathway inhibitors may provide a successful adjuvant or alternative treatment option in a historically difficult-to-treat cancer. Further sequencing of teratocarcinosarcomas and other rare skull base tumors will be useful to increase our knowledge of potential genetic drivers of disease.
Acknowledgments
A.C.B. and S.J.B. received funding from NIH Grant T32-DC005356. J.C.B. received funding from NIH Grants T32-DC005356, U01-DE025184, P30-CA046592, and R01-CA194536. This article was presented at the 26th Annual North American Skull Base Society Meeting, February 12–14, 2016, Scottsdale, Arizona, United States.
Footnotes
These authors contributed equally to this work.
These are cosenior authors.
Supplementary Material
References
- 1.Misra P, Husain Q, Svider P F, Sanghvi S, Liu J K, Eloy J A. Management of sinonasal teratocarcinosarcoma: a systematic review. Am J Otolaryngol. 2014;35(01):5–11. doi: 10.1016/j.amjoto.2013.04.010. [DOI] [PubMed] [Google Scholar]
- 2.Fatima S S, Minhas K, Din N U, Fatima S, Ahmed A, Ahmad Z. Sinonasal teratocarcinosarcoma: a clinicopathologic and immunohistochemical study of 6 cases. Ann Diagn Pathol. 2013;17(04):313–318. doi: 10.1016/j.anndiagpath.2013.01.003. [DOI] [PubMed] [Google Scholar]
- 3.Wei S, Carroll W, Lazenby A, Bell W, Lopez R, Said-Al-Naief N. Sinonasal teratocarcinosarcoma: report of a case with review of literature and treatment outcome. Ann Diagn Pathol. 2008;12(06):415–425. doi: 10.1016/j.anndiagpath.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 4.Su Y Y, Friedman M, Huang C C, Wilson M, Lin H C. Sinonasal teratocarcinosarcoma. Am J Otolaryngol. 2010;31(04):300–303. doi: 10.1016/j.amjoto.2009.02.022. [DOI] [PubMed] [Google Scholar]
- 5.Birkeland A C, Brenner J C. Personalizing medicine in head and neck squamous cell carcinoma: The rationale for combination therapies. Med Res Arch. 2015;2015(03) doi: 10.18103/mra.v0i3.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Birkeland A C, Ludwig M L, Meraj T S, Brenner J C, Prince M E. The tip of the iceberg: clinical implications of genomic sequencing projects in head and neck cancer. Cancers (Basel) 2015;7(04):2094–2109. doi: 10.3390/cancers7040879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Birkeland A C, Uhlmann W R, Brenner J C, Shuman A G. Getting personal: head and neck cancer management in the era of genomic medicine. Head Neck. 2016;38 01:E2250–E2258. doi: 10.1002/hed.24132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Giefing M, Wierzbicka M, Szyfter K et al. Moving towards personalised therapy in head and neck squamous cell carcinoma through analysis of next generation sequencing data. Eur J Cancer. 2016;55:147–157. doi: 10.1016/j.ejca.2015.10.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Birkeland A C, Yanik M, Tillman B N et al. Identification of targetable ERBB2 aberrations in head and neck squamous cell carcinoma. JAMA Otolaryngol Head Neck Surg. 2016;142(06):559–567. doi: 10.1001/jamaoto.2016.0335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tillman B N, Yanik M, Birkeland A C et al. Fibroblast growth factor family aberrations as a putative driver of head and neck squamous cell carcinoma in an epidemiologically low-risk patient as defined by targeted sequencing. Head Neck. 2016;38 01:E1646–E1652. doi: 10.1002/hed.24292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Warrick J I, Hovelson D H, Amin A et al. Tumor evolution and progression in multifocal and paired non-invasive/invasive urothelial carcinoma. Virchows Arch. 2015;466(03):297–311. doi: 10.1007/s00428-014-1699-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38(16):e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Forbes S A, Beare D, Gunasekaran Pet al. COSMIC: exploring the world's knowledge of somatic mutations in human cancer Nucleic Acids Res 201543(Database issue):D805–D811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weinstein J N, Collisson E A, Mills G B et al. The Cancer Genome Atlas Pan-Cancer analysis project. Nat Genet. 2013;45(10):1113–1120. doi: 10.1038/ng.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ludwig M L, Birkeland A C, Hoesli R, Swiecicki P, Spector M E, Brenner J C. Changing the paradigm: the potential for targeted therapy in laryngeal squamous cell carcinoma. Cancer Biol Med. 2016;13(01):87–100. doi: 10.28092/j.issn.2095-3941.2016.0010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Michmerhuizen N L, Birkeland A C, Bradford C R, Brenner J C.Genetic determinants in head and neck squamous cell carcinoma and their influence on global personalized medicine Genes Cancer 20167(5-6):182–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pez F, Lopez A, Kim M, Wands J R, Caron de Fromentel C, Merle P. Wnt signaling and hepatocarcinogenesis: molecular targets for the development of innovative anticancer drugs. J Hepatol. 2013;59(05):1107–1117. doi: 10.1016/j.jhep.2013.07.001. [DOI] [PubMed] [Google Scholar]
- 18.Arend R C, Londoño-Joshi A I, Straughn J M, Jr, Buchsbaum D J. The Wnt/b-catenin pathway in ovarian cancer: a review. Gynecol Oncol. 2013;131(03):772–779. doi: 10.1016/j.ygyno.2013.09.034. [DOI] [PubMed] [Google Scholar]
- 19.Stewart D J. Wnt signaling pathway in non-small cell lung cancer. J Natl Cancer Inst. 2014;106(01):djt356. doi: 10.1093/jnci/djt356. [DOI] [PubMed] [Google Scholar]
- 20.Rubinfeld B, Robbins P, El-Gamil M, Albert I, Porfiri E, Polakis P.Stabilization of beta-catenin by genetic defects in melanoma cell lines Science 1997275(5307):1790–1792. [DOI] [PubMed] [Google Scholar]
- 21.Morin P J, Sparks A B, Korinek Vet al. Activation of beta-catenin-Tcf signaling in colon cancer by mutations in beta-catenin or APC Science 1997275(5307):1787–1790. [DOI] [PubMed] [Google Scholar]
- 22.Liu J, Pan S, Hsieh M H et al. Targeting Wnt-driven cancer through the inhibition of Porcupine by LGK974. Proc Natl Acad Sci U S A. 2013;110(50):20224–20229. doi: 10.1073/pnas.1314239110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hamada S, Futamura N, Ikuta K et al. CTNNB1 S45F mutation predicts poor efficacy of meloxicam treatment for desmoid tumors: a pilot study. PLoS One. 2014;9(05):e96391. doi: 10.1371/journal.pone.0096391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Colombo C, Miceli R, Lazar A J et al. CTNNB1 45F mutation is a molecular prognosticator of increased postoperative primary desmoid tumor recurrence: an independent, multicenter validation study. Cancer. 2013;119(20):3696–3702. doi: 10.1002/cncr.28271. [DOI] [PubMed] [Google Scholar]
- 25.Conley B A, Doroshow J H. Molecular analysis for therapy choice: NCI MATCH. Semin Oncol. 2014;41(03):297–299. doi: 10.1053/j.seminoncol.2014.05.002. [DOI] [PubMed] [Google Scholar]
- 26.Tsimberidou A M, Wen S, Hong D S et al. Personalized medicine for patients with advanced cancer in the phase I program at MD Anderson: validation and landmark analyses. Clin Cancer Res. 2014;20(18):4827–4836. doi: 10.1158/1078-0432.CCR-14-0603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Delgado E R, Yang J, So J et al. Identification and characterization of a novel small-molecule inhibitor of b-catenin signaling. Am J Pathol. 2014;184(07):2111–2122. doi: 10.1016/j.ajpath.2014.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lenz H J, Kahn M. Safely targeting cancer stem cells via selective catenin coactivator antagonism. Cancer Sci. 2014;105(09):1087–1092. doi: 10.1111/cas.12471. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


