Abstract
This study analyzed the origin and evolution of snake venom proteome by means of phylogenetic analysis of the amino acid sequences of the toxins and related nonvenom proteins. The snake toxins were shown to have arisen from recruitment events of genes from within the following protein families: acetylcholinesterase, ADAM (disintegrin/metalloproteinase), AVIT, complement C3, crotasin/β defensin, cystatin, endothelin, factor V, factor X, kallikrein, kunitz-type proteinase inhibitor, LYNX/SLUR, L-amino oxidase, lectin, natriuretic peptide, βnerve growth factor, phospholipase A2, SPla/Ryanodine, vascular endothelial growth factor, and whey acidic protein/secretory leukoproteinase inhibitor. Toxin recruitment events were found to have occurred at least 24 times in the evolution of snake venom. Two of these toxin derivations (CRISP and kallikrein toxins) appear to have been actually the result of modifications of existing salivary proteins rather than gene recruitment events. One snake toxin type, the waglerin peptides from Tropidolaemus wagleri (Wagler's Viper), did not have a match with known proteins and may be derived from a uniquely reptilian peptide. All of the snake toxin types still possess the bioactivity of the ancestral proteins in at least some of the toxin isoforms. However, this study revealed that the toxin types, where the ancestral protein was extensively cysteine cross-linked, were the ones that flourished into functionally diverse, novel toxin multigene families.
Venomous snakes possess one of the most sophisticated integrated weapons systems in the natural world. The advanced snakes (superfamily Colubroidea) make up >80% of the ∼2900 species of snake currently described, and contain all of the known venomous forms (Greene 1997; Vidal 2002). Snake venom glands evolved a single time, at the base of the colubroid radiation, 60-80 million years ago, with extensive subsequent “evolutionary tinkering” (Vidal and Hedges 2002; Fry and Wüster 2004). Evidence comes from comparative morphology, embryology, and developmental biology, as well as the demonstrated homology of venom-secreting glands of different colubroid families (Kochva 1963, 1965, 1978; Underwood and Kochva 1993; Underwood 1997; Jackson 2003), as well as the distribution of these glands across the full spectrum of “colubrid” families (Vidal 2002) in addition to phylogenetic analyses of toxin sequences (Fry et al. 2003a,b; Fry and Wüster 2004). As maxillary fangs and a venom gland are a colubroid synapomorphy, the distinction between the “Duvernoy's gland” and the atractaspidid/elapid/viperid venom glands has been abandoned (Fry et al. 2003c).
It has been previously postulated that some of the snake toxin types (such as three-finger toxins) evolved from a single ribonuclease ancestor (Strydom 1973). It has also been hypothesized that the snake venom gland itself evolved in the mouth region as a consequence of an evolutionary change in the pancreatic trait, and consequently, some of the toxins should show strong affinities to pancreatic proteins (Kochva 1987). Therefore, a fundamental question that has remained unanswered is what gene types were recruited for use in the snake venom proteome and what were the tissue locations from which these genes were harvested? Another major remaining unanswered question is what biochemical characteristics do these ancestral proteins share?
The purpose of this study was to use phylogenetic analyses of toxin and related body proteins to reconstruct the evolutionary history of snake venom proteome in order to provide a frame-work for use in answering these questions. Examined in this study were the following snake toxin types: 3FTx (three-finger toxin), acetylcholinesterase, ADAM (disintegrin/metalloproteinase), CVF/C3 (cobra venom factor/complement C3), crotamine, cystatin, factor V, factor X, kallikrein, kunitoxins, L-amino oxidase, lectins (C-type and galactose binding), MIT (mamba intestinal toxin), natriuretic peptide, NGF (nerve growth factor), PLA2 (phospholipase A2), sarafotoxin, SPRY (SPla/Ryanodine), VEGF (vascular endothelial growth factor), wagerlin, and waprin (Table 1).
Table 1.
Data sets analyzed
| Toxin type and related body proteins | # of sequences | # of characters | # parsimony informative |
|---|---|---|---|
| 3FTx/LYNX/SLUR | 378 | 483 | 272 |
| Acetylcholinesterase | 97 | 1259 | 773 |
| ADAM | 141 | 750 | 673 |
| Cobra venom factor/C3 | 100 | 2403 | 1932 |
| CRISP | 96 | 826 | 681 |
| Crotamine/Crotasin/β Defensin | 20 | 72 | 21 |
| Cystatin | 90 | 200 | 179 |
| Factor V | 34 | 2594 | 2131 |
| Factor X | 60 | 750 | 508 |
| Kallikrein | 249 | 476 | 352 |
| Kunitz | 250 | 236 | 201 |
| L-Amino oxidase | 48 | 1005 | 648 |
| Lectin | 145 | 518 | 382 |
| Mamba intestinal peptide/Prokineticin | 22 | 135 | 79 |
| Natriuretic peptide | 82 | 329 | 183 |
| Nerve growth factor | 95 | 440 | 256 |
| Phospholipase A2 | 327 | 250 | 206 |
| Sarafotoxin/endothelin | 37 | 557 | 217 |
| SPRY | 141 | 300 | 134 |
| VEGF | 93 | 594 | 411 |
| Waprin/Whey acidic protein | 124 | 288 | 197 |
Results
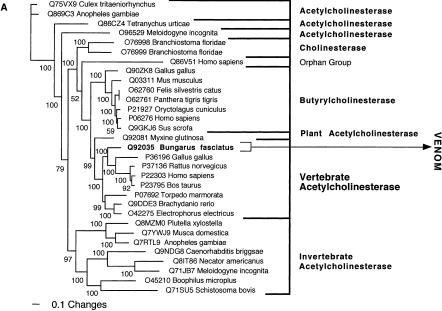
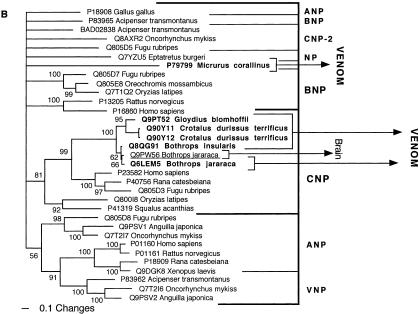
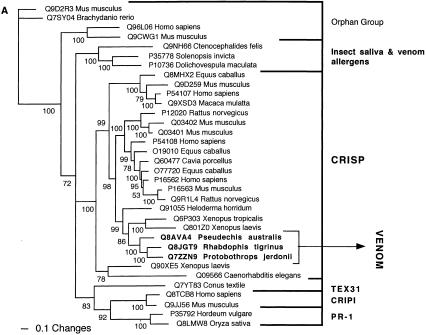
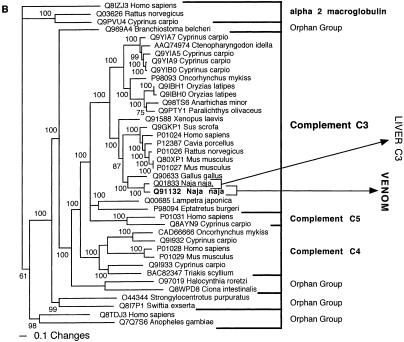
The conventionally recognized, major classes of a particular protein type, characterized by activity type and specific functional motifs, formed monophyletic groups. These groupings were congruent whether by Bayesian analysis (Figs. 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11) or maximum parsimony (data not shown) and supported by high posterior probabilities. Of the 24 snake toxin types examined, 23 had matches with known protein types (Table 2). In 11 data sets (acetylcholinesterase, CNP-BPP, CRISP, CVF, crotamine, factor V, factor X, L-amino oxidase, type IB PLA2, and type IIA PLA2) the toxin sequences were nested within a nontoxin subclade, with high posterior probability support, thus allowing for clear inference of gene origin (Figs. 1, 2, 3, 4, 5). In seven data sets (ADAM, cystatin, MIT, kallikrein, lectin, sarafotoxin, and SPRY), the toxin sequences formed sister groups to body protein types with high bootstrap value support, allowing for inference of ancestral nontoxin gene, but definitive assignment to a particular subclade not being possible (Figs. 6, 7, 8, 9A). In five data sets (3FTx, BNP, kunitz type proteinase inhibitor, VEGF, and waprin), high levels of saturation, combined with short sequence length, produced polytomies that did not allow for assignment of the toxins to specific clade within the larger protein family (Figs. 1B, 9B, 10, 11). The waglerin toxins from Tropidolaemus wagleri (Wagler's Viper) (Aiken et al. 1992) did not have a match with any known protein types, and genome mining did not reveal any high confidence matches within protein-coding regions.
Figure 1.
Bayesian analysis of representative (A) acetylcholinesterase and (B) natriuretic related sequences. In order to minimize confusion, all sequences are referred to by their SWISS-PROT accession numbers (http://www.expasy.org/cgi-bin/sprot-search-ful). Snake venom sequences are in bold. Snake body proteins are underlined.
Figure 2.
Bayesian analysis of representative (A) CRISP and (B) complement-related sequences. In order to minimize confusion, all sequences are referred to by their SWISS-PROT accession numbers (http://www.expasy.org/cgi-bin/sprot-search-ful). Snake venom sequences are in bold. Snake body proteins are underlined.
Figure 3.
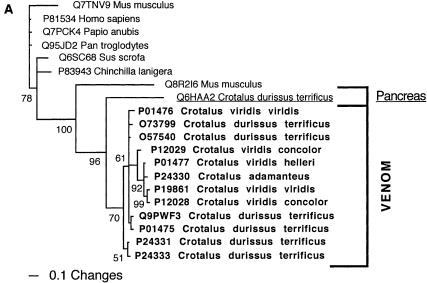
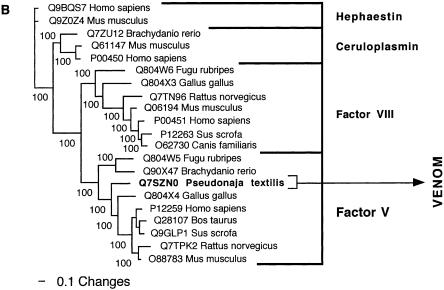
Bayesian analysis of representative (A) crotamine/crotasin/β defensin and (B) factor V related sequences. In order to minimize confusion, all sequences are referred to by their SWISS-PROT accession numbers (http://www.expasy.org/cgi-bin/sprot-search-ful). Snake venom sequences are in bold. Snake body proteins are underlined.
Figure 4.
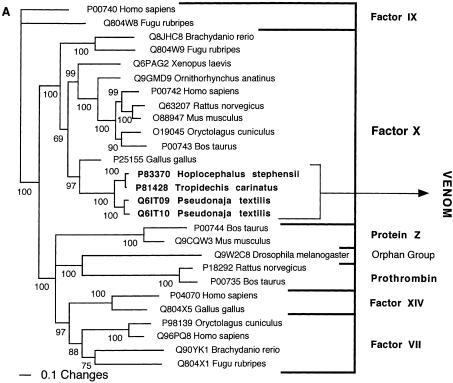
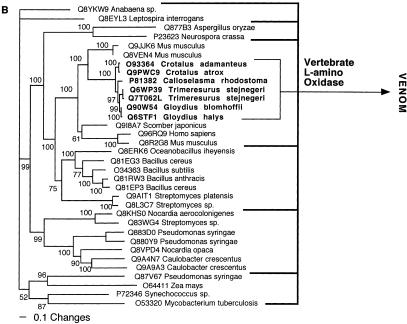
Bayesian analysis of representative (A) factor X and (B) L-amino oxidase-related sequences. In order to minimize confusion, all sequences are referred to by their SWISS-PROT accession numbers (http://www.expasy.org/cgi-bin/sprot-search-ful). Snake venom sequences are in bold.
Figure 5.
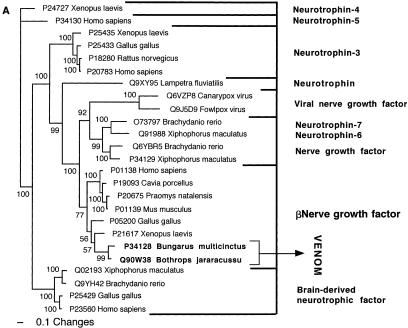
Bayesian analysis of representative (A) nerve growth factor and (B) phospholipase A2-related sequences. In order to minimize confusion, all sequences are referred to by their SWISS-PROT accession numbers (http://www.expasy.org/cgi-bin/sprot-search-ful). Snake venom sequences are in bold. Snake body proteins are underlined.
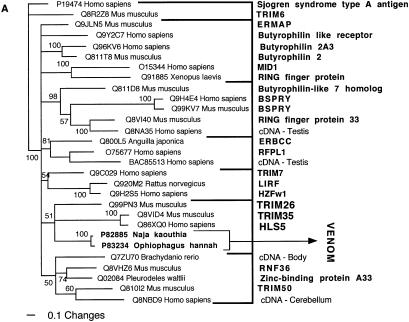
Figure 6.
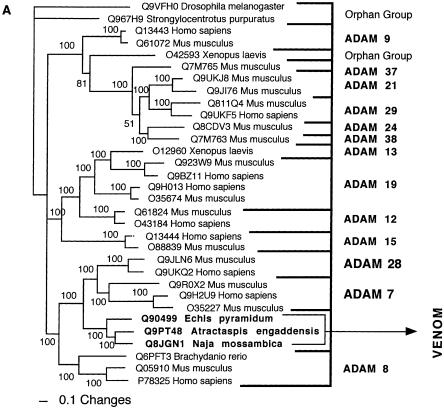
Bayesian analysis of representative (A) ADAM and (B) AVIT-related sequences. In order to minimize confusion, all sequences are referred to by their SWISS-PROT accession numbers (http://www.expasy.org/cgi-bin/sprot-search-ful). Snake venom sequences are in bold.
Figure 7.
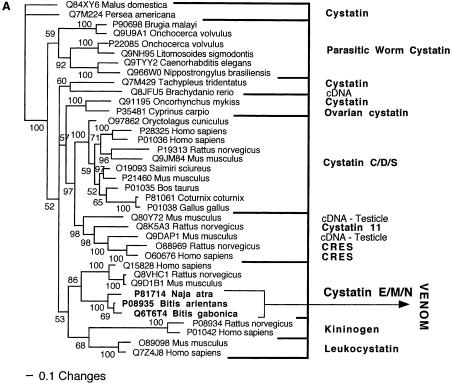
Bayesian analysis of representative (A) cystatin and (B) sarafotoxin/endothelin-related sequences. In order to minimize confusion, all sequences are referred to by their SWISS-PROT accession numbers (http://www.expasy.org/cgi-bin/sprot-search-ful). Snake venom sequences are in bold.
Figure 8.
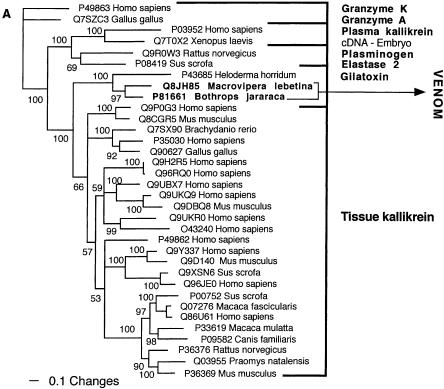
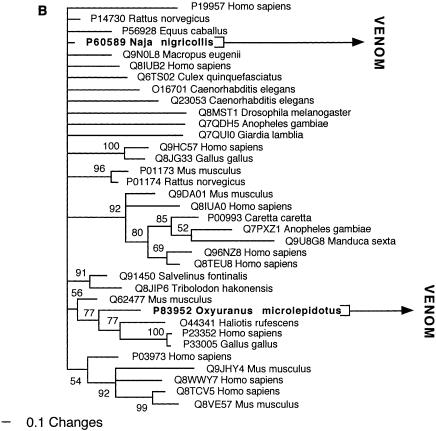
Bayesian analysis of representative (A) kallikrein and (B) lectin-related sequences. In order to minimize confusion, all sequences are referred to by their SWISS-PROT accession numbers (http://www.expasy.org/cgi-bin/sprot-search-ful). Snake venom sequences are in bold.
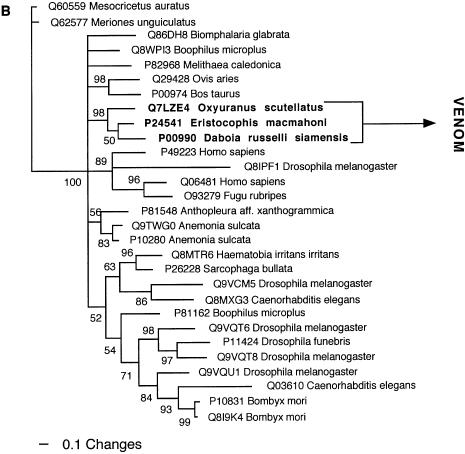
Figure 9.
Bayesian analysis of representative (A) SPRY and (B) kunitz-type proteinase inhibitor-related sequences. In order to minimize confusion, all sequences are referred to by their SWISS-PROT accession numbers (http://www.expasy.org/cgi-bin/sprot-search-ful). Snake venom sequences are in bold.
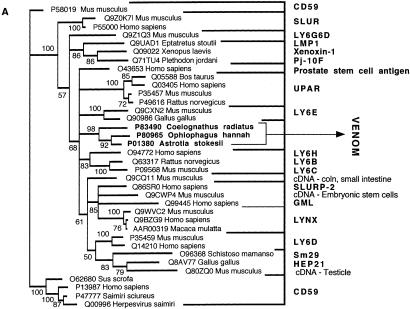
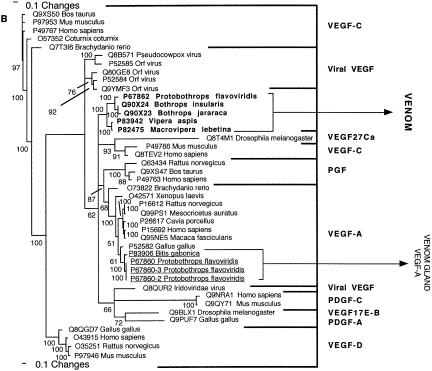
Figure 10.
Bayesian analysis of representative (A) 3FTx/LYNX/SLUR and (B) VEGF-related sequences. In order to minimize confusion, all sequences are referred to by their SWISS-PROT accession numbers (http://www.expasy.org/cgi-bin/sprot-search-ful). Snake venom sequences are in bold. Snake body proteins are underlined.
Figure 11.
Molecular phylogenetic analysis of whey acidic proteins/secretory leukoproteinase inhibiting peptides-related sequences (A) Unrooted maximum parsimony network and (B) Outgroup rooted Bayesian analysis. Representative toxin and nontoxin sequences are labeled. In order to minimize confusion, all proteins sequences are referred to by their SWISS-PROT accession numbers (http://www.expasy.org/cgi-bin/sprot-search-ful). Snake venom sequences are in bold.
Table 2.
Summary of toxin relationships with ancestral proteins
| Toxin type | Nearest ancestral protein match |
|---|---|
| 3FTx | Part of a polytomy that includes the nicotinic acetylcholine receptor binding LYNX (e.g., Q9BZG9 from Homo sapiens [Human]) and SLUR peptides (e.g., P55000 from Homo sapiens). |
| Acetylcholinesterase | Vertebrate acetylcholinesterase (e.g., P36196 from Gallus gallus). |
| ADAM | Sister group to the ADAM 7 and ADAM 28 clades. |
| BNP | Related to the variable BNP forms. |
| Cobra venom factor | Complement C3 sequence Q01833 from Naja naja (Spectacled cobra). |
| CNP-BPP | CNP-BPP Q9PW56 from Bothrops jararaca (Jararaca). |
| CRISP | CRISP protein subclade that includes Q91055 (helothermine) from Heloderma horridum (Beaded lizard). |
| Crotamine | Crotasin (Q6HAA2) from Crotalus durissus terrificus (Neotropical Rattlesnake). |
| Cystatin | Sister group to the E/M/N type cystatins. |
| Factor V | Factor V (Q804X4) from Gallus gallus (Chicken). |
| Factor X | Factor X (P25155) from Gallus gallus (Chicken). |
| Kallikrein | Nearest neighbor is P43685 (gilatoxin) from Heloderma horridum (Beaded lizard) and this clade is then sister group to the tissue (glandular) kallikrein clade. |
| Kunitz | Overall protein family a polytomy. |
| L-amino oxidase | Vertebrate L-amino oxidase. |
| Lectin | Unresolved but within main lectin clade. |
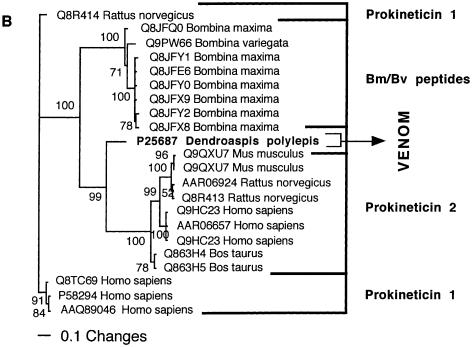
| Mamba intestinal toxin | Sister group to the prokineticin 2 clade. |
| Nerve growth factor | β NGF. |
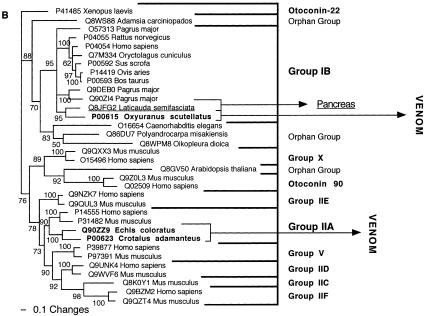
| PLA2 (type IB) | Endogenous type IB PLA2 (e.g., Q8JFG2 from Laticauda semifasciata). |
| PLA2 (type IIA) | Endogenous type IIA PLA2. |
| Sarafotoxin | Sister group to endothelin-3. |
| SPRY | Weakly related to proteins of hemopoietic origin. |
| VEGF | Relationships unresolved. |
| Wagerlin | No endogenous protein match currently known. |
| Waprin | Relationships unresolved. |
Discussion
Past hypotheses suggested that proteins of pancreatic origin should be the dominant snake toxins (Strydom 1973; Kochva 1987). However, the results of this study reveal a diverse array of tissues from which toxin encoding genes were harvested and the bioactivities equally diverse (Tables 3, 4). While there was no pattern with regard to specific tissue type, with many of the ancestral gene types expressed in several tissues, all coded for secretory proteins. Comparison of toxins and ancestral body proteins allowed for a determination of ancestral versus derived activities (Tables 5, 6). For example, the phylogenetic association of the 3FTx with the nicotinic acetylcholine receptor-binding LYNX and SLUR peptides (e.g., Miwa et al. 1999; Ibanez-Tallon et al. 2002; Chimienti et al. 2003) is consistent with the basal α-neurotoxic activity of the 3FTx (Fry et al. 2003a). This is in contrast to previous hypotheses of an ancestral digestive action for this toxin type (Strydom 1973).
Table 3.
Tissue specificities of the ancestral body proteins
| Ancestral protein | Tissue location |
|---|---|
| Acetylcholinesterase | Muscle |
| ADAM 7 and ADAM 28 | ADAM 7 - epididymis as well as colon; ADAM 28 - variety of tissues including epididymis, lung, lymph node and thymus. |
| BNP | Heart |
| C3 | Liver |
| CNP-BPP | Brain |
| CRISP | Myriad of exocrine tissues including salivary. |
| Crotasin | Expressed in high levels in the pancreas, and lower but still significant levels expressed in the heart, liver, brain, and kidneys. |
| E/M/N Cystatin | Restricted to the stratum granulosum of normal skin, the stratum granulosum/spinosum of psoriatic skin, the secretory coils of exocrine sweat glands with low expression levels also found in the nasal cavity. |
| Endothelin-3 | Endothelium |
| Factor V | Liver |
| Factor X | Liver |
| Kallikrein (glandular/tissue) | Variety of exocrine tissues including pancreas as well as the salivary glands. |
| Kunitz | Wide variety of tissues, including brain, conceptus membrane, lung, ovary, placenta, and uterus. |
| L-amino oxidase | Variety of exocrine and immune tissues. |
| Lectin | Myriad |
| LYNX/SLUR | LYNX - large projection neurons in the hippocampus, cortex and cerebellum; SLUR - granulocytes, expressed in the skin. |
| β Nerve growth factor | Wide variety of tissues including the brain, eye, prostate and salivary glands. |
| PLA2 (type IB) | Pancreas. |
| PLA2 (type IIA) | Synovial fluid and elsewhere. |
| Prokineticin 2 | Expressed at high levels in testis and at lower levels in brain, lung, ovary, spleen, thymus, and uterus. |
| SPRY | Hemopoietic lineages. |
| VEGF | Various tissues ranging from the brain to the heart. |
| Whey acidic peptide | Variety of tissues including lactating mammary gland, lung, ovary, and testis. |
Table 4.
Bioactivities of the ancestral proteins
| Ancestral protein | Activity |
|---|---|
| Acetylcholinesterase | Rapidly hydrolyses choline released into the synapse, resulting in less neurotransmitter available for neuromuscular control. |
| ADAM | ADAM 7 - noncatalytic metalloproteinase-like proteins; ADAM 28 - enzymatic cleavage of the extracellular matrix. |
| BNP | Lowering of blood pressure mediated by the relaxation of vascular smooth muscle subsequent by binding the GC-A receptor and stimulating the intracellular production of cGMP. |
| C3 | Central to both classical and alternative complement pathways. |
| CNP-BPP | BPP - indirectly cause hyperpermeability of blood vessels and hypotension by inhibiting the activity of the angiotensin-converting enzyme and enhances the action of bradykinin by inhibiting the kinases that inactivate; CNP - Lowers blood pressure mediated by the relaxation of vascular smooth muscle by binding the GC-B receptor and stimulating the intracellular production of cGMP. |
| CRISP | Specific actions largely uncharacterized. |
| Crotasin | Unknown; related β defensin peptides have an antimicrobial activity. |
| E/M/N Cystatin | Inhibit cysteine proteases such as the cathepsins B, L, and S. |
| Endothelin-3 | Potently vasoconstrictive, modulating the contraction of cardiac and smooth muscle. |
| Factor V | Blood cofactors that participate with factor Xa to activate prothrombin to thrombin. |
| Factor X | Vitamin K-dependent glycoproteins that convert prothrombin to thrombin in the presence of factor Va, calcium, and phospholipid during blood clotting. |
| Kallikrein (glandular/tissue) | Release kinins from circulatory kininogen. |
| Kunitz | Inhibit a diverse array of serine proteinases. |
| L-amino oxidase | Induce apoptosis in cells by two distinct mechanisms; one rapid and mediated by H2O2, the other delayed and mediated by deprivation of L-lysine. |
| Lectin | Hemagglutination activity. |
| LYNX/SLUR | Bind to the α 7 nicotinic acetylcholine receptor. |
| β Nerve growth factors | Stimulate division and differentiation of sympathetic and embryonic sensory neurons. |
| PLA2 (type IB) | Release arachidonic acid from the sn-2 position of the plasma membrane phospholipids. |
| PLA2 (type IIA) | Release arachidonic acid from the sn-2 position of the plasma membrane phospholipids, involved in inflammatory processes and diseases, such as rheumatoid arthritis and asthma. |
| Prokineticin 2 | Potent constriction of intestinal smooth muscle and induction hyperalgesia. |
| SPRY | Largely uncharacterized. |
| VEGF | Increase the permeability of the vascular bed. |
| Whey acidic peptide | Inhibit leukoproteinases. |
Table 5.
Basal bioactivities of each toxin type
| Toxin type | Basal toxic activity |
|---|---|
| 3FTx | α-neurotoxicity, antagonistically binding to the nicotinic acetylcholine receptor. |
| Acetylcholinesterase | Rapid hydrolysis of choline released into the synapse, resulting in less neurotransmitter available for neuromuscular control. |
| ADAM | Tissue necrosis. |
| BNP | Potent induction of hypotension leading to loss of consciousness. |
| Cobra venom factor | Unregulated activation of the complement cascade, causing rapid and significant problems such as anaphylactic-type problems and/or tissue damage via hemolysis/cytolysis. |
| CNP-BPP | Potent induction of hypotension leading to loss of consciousness. |
| CRISP | Paralysis of peripheral smooth muscle and induction of hypothermia. |
| Crotamine | Myonecrosis. |
| Cystatin | Inhibition of body defensive enzymes. |
| Factor V | In Oxyuranus (Taipan) and Pseudonaja (Brown snake) venoms, combines with toxic form of factor X to potently convert prothrombin to thrombin. |
| Factor X | Potent conversion of prothrombin to thrombin in the presence of factor V, valcium and phospholipid. |
| Kallikrein | Increase of vascular permeability and production of hypotension in addition to stimulation of inflammation. |
| Kunitz | Inhibition of circulating serine proteinases. |
| L-amino oxidase | Apoptosis. |
| Lectin | Platelet aggregation mediated by galactose binding. |
| MIT | Potent constriction of intestinal smooth muscle, resulting in painful cramping, and induction of hyperalgesia. |
| NGF | Unknown. |
| PLA2 (IB) | Lipase activity resulting in inflammation and tissue destruction. |
| PLA2 (IIA) | Lipase activity resulting in inflammation and tissue destruction. |
| Sarafotoxin | Potent vasoconstriction resulting in acute hypertension. |
| SPRY | Induces hypolocomotion and hyperalgesia. Unknown which, if either, is basal activity. |
| VEGF | Increase of the permeability of the vascular bed and binding of heparin. Results in hypotension and shock. |
| Wagerlin | Presynaptically modulates γ-aminobutyric acid activated currents. Postsynaptically antagonistically binds to the ε form of the muscle nicotinic acetylcholine receptors. Presynaptic effect more potent than postsynaptic. Unknown which, if either, is basal activity. |
| Waprin | Bioactivities uncharacterized. |
Table 6.
Structural and functional derivations of each toxin type
| Toxin type | Derivations |
|---|---|
| 3FTx | Basal activity of α-neurotoxicity greatly potentiated by the deletion of the C2 and C3 ancestral cysteines. Functional derivations include binding to the postsynaptic muscarinic acetylcholine receptors, presynaptic neurotoxic action upon the L-type calcium channels, cytotoxic interactions, acetylcholinesterase inhibition, and others. |
| Acetylcholinesterase | None currently documented. |
| ADAM | Prothrombin activation a basal derivation. In Viperidae venoms, proteolytic cleavage of C-terminal domains resulted in myriad of other activities including direct-acting fibrinolytic activity. Liberated disintegrin domain inhibits platelets via GP IIb/IIIa integrin receptor. |
| BNP | Cardiovascular effects independent of the GC-A receptor. Antiplatelet activities evolved for domains upstream of the natriuretic peptide domain. |
| CVF | None currently documented. |
| CNP-BPP | None currently documented. |
| CRISP | Blockage of cyclic nucleotide gated calcium channels. |
| Crotamine | Significant neurotoxic activity, modifying voltage-sensitive Na+ channels, resulting in a potent analgesic effect. |
| Cystatin | None currently documented. |
| Factor V | None currently documented. |
| Factor X | None currently documented. |
| Kallikrein | Derivations affect the blood, particularly targeting fibrinogen. |
| Kunitz | Derivations include inhibition of plasmin and thrombin and the blockage of L-type calcium channels. Structural derivatives form part of neurotoxic complexes with PLA2 molecules. |
| L-amino oxidase | Derivations include hemorrhagic effects, not only by affecting platelet aggregation, but also inhibiting blood factor IX. |
| Lectin | Derivations include stimulation of platelet aggregation (binding GPVI, GPIb, GPIa/IIa or VWF), platelet aggregation inhibition (binding GPIb or GPIa/IIa) or anticoagulant actions by binding blood factors IX, X. |
| MIT | None currently documented. |
| NGF | Immunomodulatory effects mediated through histamine release and plasma extravasation. |
| PLA2 (type IB) | Deletion of pancreatic loop facilitated the derivation of a multiplicity of novel, nonenzymatic activities, including antiplatelet and presynaptic neurotoxicity. Some derivatives are parts of neurotoxic complexes. |
| PLA2 (type IIA) | Derivations include neurotoxic and antiplatelet activity. Some derivatives are parts of complexes. |
| Sarafotoxin | None currently documented. |
| SPRY | None currently documented. |
| VEGF | Sustained hypotension. |
| Wagerlin | None currently documented. |
| Waprin | None currently documented. |
Significant changes in the gene structure have not been currently documented for the snake toxins relative to their protein ancestors, with the exception of the sarafotoxins and the ADAM toxins. The gene structure of sarafotoxins is quite distinct from that of the ancestral endothelin peptides, being arranged in a rosary, with several coding regions in tandem (Ducancel et al. 1993). In a highly derived form of the ADAM toxins (e.g., Q6T6T2 from Bitis gabonica), the regions upstream of the C-terminal disintegrin domain have been excised, and the transcripts code now solely for the disintegrin domain. The selection pressure resulting in this change is indicative of the usefulness of this domain to the snake in prey capture. Currently, the gene location for only one toxin type (crotamine) has been determined, with the transcripts located at the chromosomal tips (Rádis-Baptista et al. 2003). The chromosomal location of the endogenous ancestor crotasin (Q6HAA2) is unknown.
Molecular scaffold characteristics
Extensive cysteine cross-linking was revealed to be a beneficial characteristic of proteins recruited for use as toxins (Table 7). Such proteins were more likely to have come from multigene families, as well as the toxin subgroup flourishing into a new multigene family with extensive functional diversification. This bias is due to the stability of the molecular core, and thus, these proteins are amenable to extensive functional diversification. Supporting this trend, the toxin types that have been independently recruited for use in other animal venoms are also extensively cysteine cross-linked (Table 7).
Table 7.
Comparison of toxins with their protein ancestors
| Toxin type | Ancestors secretory proteins? | Ancestral amino acid:cysteine ratio | Ancestors from a multigene family? | Ancestral proteins functionally diverse? | Ancestral activity retained? | Novel toxic activities evolved? | Related proteins used as toxins by other animals? |
|---|---|---|---|---|---|---|---|
| 3FTx | X | 13:1 | X | X | X | X | X |
| Acn | X | 110:1 | − | − | X | − | − |
| ADAM | X | 18:1 | X | X | X | X | − |
| BNP | X | 65:1 | X | X | X | X | − |
| CNP-BPP | X | 133:1 | X | X | X | − | × (CNP) |
| CRISP | X | 15:1 | X | X | X | X | X |
| Crotamine | X | 10:1 | X | X | X | X | − |
| CTL | X | 23:1 | X | X | X | X | − |
| CVF | X | 61:1 | − | − | X | − | − |
| Cystatin | X | 30:1 | X | X | X | X | − |
| Factor V | X | 53:1 | − | − | X | − | − |
| Factor X | X | 19:1 | − | − | X | − | − |
| GBL | X | 19:1 | X | X | X | X | − |
| Kallikrein | X | 21:1 | X | X | X | X | X |
| Kunitz | X | 12:1 | X | X | X | X | X |
| L-amino oxidase | X | 65:1 | − | − | X | X | − |
| Lectin | X | 9:1 | X | X | X | X | X |
| NGF | X | 39:1 | X | X | X | X | X |
| PLA2 Type IB | X | 15:1 | X | X | X | X | X |
| PLA2 Type IIA | X | 10:1 | X | X | X | X | X |
| Sarafotoxin | X | 19:1 | X | X | X | − | − |
| SPRY | X | 33:1 | X | X | N.A. | N.A. | − |
| VEGF | X | 13:1 | X | X | X | X | − |
| Waprin | X | 8:1 | X | X | N.A. | N.A. | − |
| Waglerin | N.A. | N.A. | N.A. | N.A. | N.A. | N.A. | − |
(N.A.) Bioactivities unknown for entire toxin type (waprin), bioactivity of ancestral protein unknown (SPRY) or ancestral protein undiscovered (waglerin).
However, the apparent bias toward mutations in the toxins of residues at the tips of loops (Fry et al. 2003b) and lower rate of mutations of the cysteines and flanking residues (which have structural, rather than functional roles), may, in fact, be an arti-fact. Intracellular housekeeping enzymes are more likely to proteolytically destroy proteins that have had a mutation of a key structural residue, such as cysteine or a flanking amino acid. This may be a consequence of such changes, potentially resulting in the protein being unable to form stable tertiary conformations. Transcriptome studies may reveal more mRNA transcripts with mutations in key structural residues than in proteins secreted as part of the “venome.”
A similar force may be at work to produce the apparent strong correlation between extensive cysteine connectivity and the tendency for formation of functionally diverse multigene families (Table 7). This may be due to the beneficial stability of an extensively cysteine-linked molecular scaffold in comparison to the much more loosely constructed tertiary structure of globular proteins formed by noncovalent interactions. For such globular proteins, a single amino acid change may be enough to decimate the ability of the protein to fold into a stable conformation, and unstable proteins are once again more likely to be destroyed by intracellular housekeeping enzymes. Thus, mutations that may have enabled functional diversification in globular proteins may have also produced fatal structural changes. Similarly, cDNA/EST studies may yet again reveal more mutations in mRNA transcripts coding for globular proteins than in secreted protein libraries.
In this study, the BNP and CNP-BPP natriuretic peptides had a high amino acid:cysteine ratio (Table 7). The very long preproregion of the natriuretic peptides includes a number of noncysteine-linked small peptides (e.g., cardiodilatin-related peptides) that are cleaved off upstream of the circulating form of the natriuretic peptide itself. The final highly processed form of the natriuretic peptides contains the only two cysteines coded for by the full gene, which form the 17 amino acid loop essential for the GC-A/GC-B receptor binding activity (Bovy 1990).
The molecular evolution of the factor X blood proteins also appeared at first glance to be contrary to the pattern displayed by other data sets. The factor X blood proteins are highly structurally and functionally conserved in the body, despite being extensively cysteine cross-linked (Table 7). However, this is a likely consequence of negative selection pressure against variations in such a crucial aspect of hemostasis, with mutations likely to have catastrophic effects resulting in embryonic mortality. No newly evolved activities have been documented for the factor X toxins, possibly due to their recent recruitment in the Australian elapid snake lineage and existing extremely potent and useful action. However, multiple isoforms exist in each of the venoms containing this toxin type. This is indicative of these toxins evolving via the birth-and-death model, as has been previously shown for the 3FTx (Fry et al. 2003b). As a result, the factor X toxins may ultimately acquire novel activities.
Potential salivary origin of two of the snake toxin types
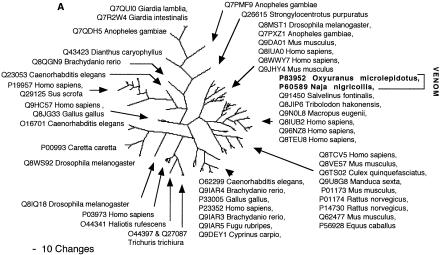
Two of the toxin types present in the snake venome, CRISP and kallikrein, may not actually have been the result of gene recruitment events, but rather modifications of secreted proteins already present in the ancestral salivary tissue that gave rise to the venom gland. Both are phylogenetically extremely close to toxins from the venomous lizards in the Heloderma genus (Beaded Lizards and Gila Monsters) such as Q91055 (helothermine) and P43685 (gilatoxin), respectively (Figs. 2A, 8A). The shared toxins between the snake and Helodermatid lizard venoms represent independent recruitments of proteins for use as toxins, as the venom-secreting structures in advanced snakes and Heloderma lizards are different, nonhomologous structures (glands located in the supralabial and infralabial regions, respectively), and the last common ancestor of Heloderma and advanced snakes would have been a basal varanoid (Forstner et al. 1995; Lee 1997) or even a basal scleroglossan (Rieppel et al. 2003) lizard devoid of a venomous function.
In addition to the close phylogenetic relationships revealed in this study, helothermine and gilatoxin have biochemical actions similar to the ancestral activities of the snake venom equivalents. The snake CRISP toxins induce hypothermia, possibly mediated through their demonstrated relaxation of the peripheral smooth muscle through blockage of K+-induced contraction, potentially by acting upon voltage-gated Ca2+ channels (Yamazaki et al. 2002). Helothermine from the venomous lizard Heloderma horridum (Beaded Lizard) has been reported to have a myriad of activities, including blocking of voltage-gated Ca2+ channels, K+ channels and ryanodine receptors, as well as introduction of hypothermia (Mochca-Morales et al. 1990; Nobile et al. 1994, 1996; Morrissette et al. 1995). The isolation of toxins with the same action (relaxation of smooth muscle and induction of hypothermia) from divergent groups of snakes, as well as the unrelated venomous lizards, indicates that this may be the original activity of the common protein ancestor. Similarly, both the snake and lizard kallikrein toxins are sister groups to the mammalian tissue (glandular) kallikrein, secreted in a wide variety of tissues including saliva glands (Fig. 8A). Gilatoxin also displays significant similarity of action to the snake venom equivalents, releasing bradykinin from kininogen with subsequent lowering of blood pressure (Utaisincharoen et al. 1993).
Clearly, in either case, the snake and lizard toxins were recruited from closely related body proteins, reflective of the wide-spread presence of CRISP and kallikrein proteins in salivary tissue and other exocrine tissue. In light of the strong phylogenetic associations, the similarity of bioactivities and the relationship to known salivary proteins, the most parsimonious explanation is that both toxin types are derivations of previously existing salivary proteins. Sequencing and phylogenetic analysis of both protein types from nontoxin-secreting salivary tissue from venomous and nonvenomous lizards and snakes would be revealing in this context.
Presence of snake toxin types in other animal venoms
In addition to the presence of CRISP and kallikrein in lizard venoms, other protein types have been independently selected for use as toxins by other venomous animals.
Disparate members of the protein superfamily to which the CRISP proteins belong are used in venoms of cone snails, such as Q7YT83 from Conus textile (Cloth-of-gold cone snail) and insects, such as P10736 from Dolichovespula maculata (White-face hornet) and P35778 from Solenopsis invicta (Fire ant). However, the activities of these toxins differ considerably from the reptilian CRISP toxins. The toxin from Conus textile has a proteinase activity, while the insect toxins are the major venom allergens. Interestingly, CRISP-related proteins are also the major allergens in insect saliva, such as Q9NH66 from Ctenocephalides felis (Cat flea), Q7Z0B5 from Stomoxys calcitrans (Stable fly), and Q8T9U1 from Aedes aegypti (Yellowfever mosquito). Intriguingly, CRISP-related proteins are also utilized by plants for defence against fungi (e.g., PR-1 protein P35792 from Hordeum vulgare [Barley]), and other proteins are the hallmark of certain cancer types (e.g., the glioma pathogenesis-related protein P48060). None of these other CRISP-related proteins align near the snake or lizard venom toxins (Fig. 2A).
The PLA2 proteins have also been recruited on numerous occasions, ranging from Q7M4I5 from Apis dorsata (Giant honeybee), Q8WS88 from Adamsia carciniopados (Cloak Sea Anemone), and P80003 from Heloderma suspectum (Gila Monster) venoms, with a wide variety of evolved activities. It is notable that the PLA2 from Heloderma venoms are not biochemically or phylogenetically related to either of the snake PLA2 toxin types, reinforcing the uniqueness of the relationships of the CRISP and kallikrein toxins.
Intriguingly, the kunitz peptides in sea anemone venom have been convergently evolved for the same unique K+ channel-blocking activity as the highly derived Dendroaspis (Mamba) venoms within the Elapidae snake family (Schweitz et al. 1994, 1995). Ornithorhynchus anatinus (Platypus) venom contains high amounts of CNP natriuretic toxins (e.g., de Plater et al. 1998a,b), but of a form that lacks the derived BPP domains found in the snake brain and venom peptides.
Two snake toxin types have been independently recruited for use in amphibian toxic skin secretions. The AVIT peptides on frog skin have a similar toxic action as the snake MIT toxins, causing intestinal cramping and increased sensitivity to pain (Mollay et al. 1999). AVIT-like peptides have also been reported from spider venoms (Szeto et al. 2000), but other than sharing the cysteine arrangement, the arachnid toxins lack all of the invariant residues, as well as the contractive effect upon smooth muscle and the ability to produce hyperalgesia. Toxins related to the snake venom 3FTx are secreted on the skin of Xenopus laevis (African clawed frog) (e.g., Q09022). These toxins are phylogenetically and functionally distinct from the snake venom 3FTx, acting presynaptically by activating dihydropyridine-sensitive Ca2+ channels (Kolbe et al. 1993). These toxins appear to be wide-spread in amphibians, with related peptides (e.g., Q71TU4) having been sequenced from the skin of Plethodon jordani (Jordan's salamander).
Notably, all of the above shared toxin types are extensively cysteine cross-linked, come from functionally diverse multigene families, and all have been developed into toxin multigene families (Table 7). The multiple, independent recruitment of certain protein types for use as toxins also sheds additional light on what sorts of proteins are favored in molecular evolution and what sort of molecular scaffold is beneficial. However, unlike the Heloderma CRISP and kallikrein toxins, they are all phylogenetically very distinct from the snake equivalents. This reinforces the uniqueness of the close relationship between the snake and lizard CRISP and kallikrein toxins and the most parsimonious explanation of these two toxin types arising from modification of existing salivary proteins.
Conclusion
The results of this study suggest that the tissue types from which the toxin recruitment genes were selected were as diverse as the proteins themselves. Two toxin types (CRISP and kallikrein) actually appear to be modifications of proteins already present in the ancestral salivary tissue. While the activity of the ancestral protein has been invariably retained, toxin types originating from extensively cysteine cross-linked proteins have been the ones to flourish into newly derived, functionally diverse, toxin multigene families.
Methods
In order to minimize confusion, all proteins' sequences are referred to by their SWISS-PROT accession numbers (http://www.expasy.org/cgi-bin/sprot-search-ful). Sequences were obtained through BLAST searching using representative toxin sequences (http://www.expasy.org/tools/blast/). Resultant sequence sets were aligned using the program CLUSTAL-X (Thompson et al. 1997), followed by visual inspection for errors. In the cases where peptides have been incorporated as domains into longer preproteins (Kunitz protease inhibitors, whey acidic protein, and SPRY), alignments were trimmed on either side of the domain. Due to the large number of sequences in each data set, phylogenetic analyses were conducted in two steps. For each data set, phylogenetic trees containing all sequenced proteins were initially reconstructed using the maximum parsimony (MP) method, conducted using the program PAUP*4.0b10 (Swofford 2002) and random stepwise taxon addition with TBR branch swapping and the PROTPARS weighting scheme (Felsenstein 2001), which takes into account the number of changes required at the nucleotide level to substitute one amino acid for another. Number of sequences, alignment length (including gaps), and parsimony informative sites are shown in Table 1. In this manner, clades that contained the venom proteins were identified. Once such clades were identified, data sets containing representative venom proteins, all the nearest neighbor nonvenom sequences, and representatives of the full breadth of gene phylogenetic diversity were selected and then analyzed using Bayesian inference implemented on MrBayes, version 3.0b4 (Huelsenbeck and Ronquist 2001; Ronquist and Huelsenbeck 2003). The method uses Markov-chain Monte Carlo methods to generate posterior probabilities for each clade represented in the tree. The analysis was performed by running a minimum of 1*106 generations in four chains, and saving every 100th tree. The log-likelihood score of each saved tree was plotted against the number of generations to establish the point at which the log likelihood scores of the analysis reached their asymptote, and the posterior probabilities for clades established by constructing a majority rule consensus tree for all trees generated after the completion of the burn-in phase. Sequence alignments and unrooted maximum parsimony trees can be obtained by e-mailing Dr. Bryan Grieg Fry (bgf@unimelb.edu.au).
Acknowledgments
I thank the Commonwealth of Australia (Department of Health and Ageing) for the on-going funding of the AVRU, and for the financial assistance of the Australia & Pacific Science Foundation, Australian Research Council (DP0451663), Australian Academy of Science, CASS Foundation, SWISS-PROT/EBI, and the University of Melbourne. I also thank Dr. Heikki Lehvaslaiho for his help in loading the larger data sets to run on the Linux cluster at the European Bioinformatics Institute/SWISS-PROT and Dr. Minna Lehvaslaiho for her kind invitation to visit the EBI/SWISS-PROT.
E-mail bgf@unimelb.edu.au; fax 61-3-93482048.
Footnotes
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.3228405.
[Supplemental material is available online at www.genome.org.]
References
- Aiken, S.P., Sellin, L.C., Schmidt, J.J., Weinstein, S.A., and McArdle, J.J. 1992. A novel peptide toxin from Trimeresurus wagleri acts pre- and post-synaptically to block transmission at the rat neuromuscular junction. Pharmacol. Toxicol. 70: 459-462. [DOI] [PubMed] [Google Scholar]
- Bovy, P.R. 1990. Structure activity in the atrial natriuretic peptide (ANP) family. Med. Res. Rev. 10: 115-142. [DOI] [PubMed] [Google Scholar]
- Chimienti, F., Hogg, R.C., Plantard, L., Lehmann, C., Brakch, N., Fischer, J., Huber, M., Bertrand, D., and Hohl, D. 2003. Identification of SLURP-1 as an epidermal neuromodulator explains the clinical phenotype of Mal de Meleda. Hum. Mol. Genet. 12: 3017-3024. [DOI] [PubMed] [Google Scholar]
- de Plater, G.M., Martin, R.L., and Milburn, P.J. 1998a. The natriuretic peptide (ovCNP-39) from platypus (Ornithorhynchus anatinus) venom relaxes the isolated rat uterus and promotes oedema and mast cell histamine release. Toxicon 36: 847-857. [DOI] [PubMed] [Google Scholar]
- ____. 1998b. A C-type natriuretic peptide from the venom of the platypus (Ornithorhynchus anatinus):structure and pharmacology. Comp. Biochem. Physiol. C Pharmacol. Toxicol. Endocrinol. 120: 99-110. [DOI] [PubMed] [Google Scholar]
- Ducancel, F., Matre, V., Dupont, C., Lajeunesse, E., Wollberg, Z., Bdolah, A., Kochva, E., Boulain, J.C., and Menez, A. 1993. Cloning and sequence analysis of cDNAs encoding precursors of sarafotoxins. Evidence for an unusual “rosary-type” organization. J. Biol. Chem. 268: 3052-3055. [PubMed] [Google Scholar]
- Felsenstein, J. 2001. PHYLIP (Phylogeny Inference Package) version 3.6. Department of Genetics, University of Washington, Seattle, WA. http://evolutiongeneticswashingtonedu/phyliphtml.
- Forstner, M.R.J., Davis, S.K., and Arévalo, E. 1995. Support for the hypothesis of anguimorph ancestry for the suborder Serpentes from phylogenetic analysis of mitochondrial DNA sequences. Mol. Phylogenet. Evol. 4: 93-102. [DOI] [PubMed] [Google Scholar]
- Fry, B.G. and Wüster, W. 2004. Assembling an arsenal: Origin and evolution of the snake venom proteome inferred from phylogenetic analysis of toxin sequences. Mol. Biol. Evol. 21: 870-883. [DOI] [PubMed] [Google Scholar]
- Fry, B.G., Lumsden, N.G., Wüster, W., Wickramaratna, J.C., Hodgson, W.C., and Kini, R.M. 2003a. Isolation of a neurotoxin (α-colubritoxin) from a nonvenomous colubrid: Evidence for early origin of venom in snakes. J. Mol. Evol. 57: 446-452. [DOI] [PubMed] [Google Scholar]
- Fry, B.G., Wüster, W., Kini, R.M., Brusic, V., Khan, A., Venkataraman, D., and Rooney, A.P. 2003b. Molecular evolution and phylogeny of elapid snake venom three-finger toxins. J. Mol. Evol. 57: 110-129. [DOI] [PubMed] [Google Scholar]
- Fry, B.G., Wüster, W., Ramjan, S.F.R., Jackson, T., Martelli, P., and Kini, R.M. 2003c. Analysis of Colubroidea snake venoms by liquid chromatography with mass spectrometry: Evolutionary and toxinological implications. Rapid Commun. Mass Spectrom 17: 2047-2062. [DOI] [PubMed] [Google Scholar]
- Greene, H.W. 1997. Snakes: The evolution of mystery in nature. Univertsity of California Press, Berkeley, CA.
- Huelsenbeck, J.P. and Ronquist, F. 2001. MrBayes—Bayesian inference of phylogeny, Version 30b4 Bioinformatics 17: 754-755. [DOI] [PubMed] [Google Scholar]
- Ibanez-Tallon, I., Miwa, J.M., Wang, H.L., Adams, N.C., Crabtree, G.W., Sine, S.M., and Heintz, N. 2002. Novel modulation of neuronal nicotinic acetylcholine receptors by association with the endogenous prototoxin lynx1. Neuron 33: 893-903. [DOI] [PubMed] [Google Scholar]
- Jackson, K. 2003. The evolution of venom-delivery systems in snakes. Zool. J. Linn. Soc. 137: 337-354. [Google Scholar]
- Kochva, E. 1963. Development of the venom gland and trigeminal muscles in Vipera palaestinae. Acta Anatomica 52: 49-89. [Google Scholar]
- ____. 1965. The development of the venom gland in the opisthoglyph snake Telescopus fallax with remarks on Thamnophis sirtalis (Colubridae, Reptilia). Copeia 147-154.
- ____. 1978. Oral glands of the Reptilia. In: Biology of the Reptilia, (eds. C.K. Gans and A. Gans), Vol. 8, pp. 43-162. Physiology B Academic Press, UK. [Google Scholar]
- Kochva, E. 1987. The origin of snakes and evolution of the venom apparatus. Toxicon 25: 65-106. [DOI] [PubMed] [Google Scholar]
- Kolbe, H.V., Huber, A., Cordier, P., Rasmussen, U.B., Bouchon, B., Jaquinod, M., Vlasak, R., Delot, E.C., and Kreil, G. 1993. Xenoxins, a family of peptides from dorsal gland secretion of Xenopus laevis related to snake venom cytotoxins and neurotoxins. J. Biol. Chem. 268: 16458-16464. [PubMed] [Google Scholar]
- Lee, M.S.Y. 1997. The phylogeny of varanoid lizards and the affinities of snakes. Phil. Trans. Roy. Soc. London Ser. B-Biol. Sci. 352: 53-91. [Google Scholar]
- Miwa, J.M., Ibanez-Tallon, I., Crabtree, G.W., Sanchez, R., Sali, A., Role, L.W., and Heintz, N. 1999. lynx1, an endogenous toxin-like modulator of nicotinic acetylcholine receptors in the mammalian CNS. Neuron 23: 105-114. [DOI] [PubMed] [Google Scholar]
- Mochca-Morales, J., Martin, B.M., and Possani, L.D. 1990. Isolation and characterization of helothermine, a novel toxin from Heloderma horridum horridum (Mexican beaded lizard) venom. Toxicon 28: 299-309. [DOI] [PubMed] [Google Scholar]
- Mollay, C., Wechselberger, C., Mignogna, G., Negri, L., Melchiorri, P., Barra, D., and Kreil, G. 1999. Bv8, a small protein from frog skin and its homologue from snake venom induce hyperalgesia in rats. Eur. J. Pharmacol. 374: 189-196. [DOI] [PubMed] [Google Scholar]
- Morrissette, J., Kratzschmar, J., Haendler, B., el-Hayek, R., Mochca-Morales, J., Martin, B.M., Patel, J.R., Moss, R.L., Schleuning, W.D., Coronado, R., et al. 1995. Primary structure and properties of helothermine, a peptide toxin that blocks ryanodine receptors. Biophys. J. 68: 2280-2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobile, M., Magnelli, V., Lagostena, L., Mochca-Morales, J., Possani, L.D., and Prestipino, G. 1994. The toxin helothermine affects potassium currents in newborn rat cerebellar granule cells. J. Membr. Biol. 139: 49-55. [DOI] [PubMed] [Google Scholar]
- Nobile, M., Noceti, F., Prestipino, G., and Possani, L.D. 1996. Helothermine, a lizard venom toxin, inhibits calcium current in cerebellar granules. Exp. Brain Res. 110: 15-20. [DOI] [PubMed] [Google Scholar]
- Rádis-Baptista, G., Kubo, T., Oguiura, N., Svartman, M., Almeidae, T.M.B., Batistic, R.F., Oliveira, E.B., Vianna-Morgante, A.M., and Yamane, T. 2003. Structure and chromosomal localization of the gene for crotamine, a toxin from the South American rattlesnake, Crotalus durissus terrificus Toxicon 42: 747-752. [DOI] [PubMed] [Google Scholar]
- Rieppel, O., Zaher, H., Tchernov, E., and Polcyn, M.J. 2003. The anatomy and relationships of Haasiophis terrasanctus, a fossil snake with well-developed hind limbs from the Mid-Cretaceous of the Middle East. J. Paleont. 77: 536-558. [Google Scholar]
- Ronquist, F. and Huelsenbeck, J.P. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572-1574. [DOI] [PubMed] [Google Scholar]
- Schweitz, H., Heurteaux, C., Bois, P., Moinier, D., Romey, G., and Lazdunski, M. 1994. Calcicludine, a venom peptide of the Kunitz-type protease inhibitor family, is a potent blocker of high-threshold Ca2+ channels with a high affinity for L-type channels in cerebellar granule neurons. Proc. Natl. Acad. Sci. 91: 878-882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweitz, H., Bruhn, T., Guillemare, E., Moinier, D., Lancelin, J.M., Beress, L., and Lazdunski, M. 1995. Kalicludines and kaliseptine. Two different classes of sea anemone toxins for voltage sensitive K+ channels. J. Biol. Chem. 270: 25121-25126. [DOI] [PubMed] [Google Scholar]
- Strydom, D.J. 1973. Snake venom toxins: The evolution of some of the toxins found in snake venoms. Syst. Zool. 22: 596-608. [Google Scholar]
- Swofford, D.L. 2002. PAUP*—Phylogenetic analysis using parsimony *and other methods beta version 40b10. Sinauer Associates, Sunderland, MA.
- Szeto, T.H., Wang, X.H., Smith, R., Connor, M., Christie, M.J., Nicholson, G.M., and King, G.F. 2000. Isolation of a funnel-web spider polypeptide with homology to mamba intestinal toxin 1 and the embryonic head inducer Dickkopf-1. Toxicon 38: 429-442. [DOI] [PubMed] [Google Scholar]
- Thompson, J.D., Gibson, T.J., Plewniak, F., Jeanmougin, F., and Higgins, D.G. 1997. The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25: 4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Underwood, G. 1997. An overview of venomous snake evolution. In Venomous snakes: Ecology, evolution and snakebite (eds. R.S. Thorpe, W.Wüster, and A. Malhotra), pp. 1-13, Symposia of the Zoological Society of London, No 70, Clarendon Press, Oxford, UK.
- Underwood, G. and Kochva, E. 1993. On the affinities of the burrowing asps Atractaspis (Serpentes: Atractaspididae). Zool. J. Linn. Soc. 107: 3-64. [Google Scholar]
- Utalsincharoen, P., Mackessy, S.P., Miller, R.A., and Tu, A.T. 1993. Complete primary structure and biochemical properties of gilatoxin, a serine protease with kallikrein-like and angiotensin-degrading activities. J. Biol. Chem. 268: 21975-21983. [PubMed] [Google Scholar]
- Vidal, N. 2002. Colubroid systematics: Evidence for an early appearance of the venom apparatus followed by extensive evolutionary tinkering. J. Toxicol. Toxin Rev. 21: 21-41. [Google Scholar]
- Vidal, N. and Hedges, S.B. 2002. Higher-level relationships of caenophidian snakes inferred from four nuclear and mitochondrial genes. C.R. Biol. 325: 987-995. [DOI] [PubMed] [Google Scholar]
- Yamazaki, Y., Koike, H., Sugiyama, Y., Motoyoshi, K., Wada, T., Hishinuma, S., Mita, M., and Morita, T. 2002. Cloning and characterization of novel snake venom proteins that block smooth muscle contraction. Eur. J. Biochem. 269: 2708-2715. [DOI] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- http://www.expasy.org/cgi-bin/sprot-search-ful; SWISS-PROT accessional nos.
- http://www.expasy.org/tools/blast/; BLAST