Abstract
Genome-wide expression profiling of normal tissue may facilitate our understanding of the etiology of diseased organs and augment the development of new targeted therapeutics. Here, we have developed a high-density gene expression database of 18,927 unique genes for 158 normal human samples from 19 different organs of 30 different individuals using DNA microarrays. We report four main findings. First, despite very diverse sample parameters (e.g., age, ethnicity, sex, and postmortem interval), the expression profiles belonging to the same organs cluster together, demonstrating internal stability of the database. Second, the gene expression profiles reflect major organ-specific functions on the molecular level, indicating consistency of our database with known biology. Third, we demonstrate that any small (i.e., n ∼ 100), randomly selected subset of genes can approximately reproduce the hierarchical clustering of the full data set, suggesting that the observed differential expression of >90% of the probed genes is of biological origin. Fourth, we demonstrate a potential application of this database to cancer research by identifying 19 tumor-specific genes in neuroblastoma. The selected genes are relatively underexpressed in all of the organs examined and belong to therapeutically relevant pathways, making them potential novel diagnostic markers and targets for therapy. We expect this database will be of utility for developing rationally designed molecularly targeted therapeutics in diseases such as cancer, as well as for exploring the functions of genes.
At present, it is estimated that the human genome encodes for ∼30,000 genes. However, it has been suggested that only a fraction, perhaps 10,000 genes, are actively transcribed in normal cell processes (Lander et al. 2001; Venter et al. 2001). High-throughput genome-wide expression profiling is a logical approach to decipher the underlying biological processes of normal organ function as well as pathological states such as cancer. The identification of differential transcript levels between a diseased and normal tissue will enhance our understanding of the mechanism of disease and thus may provide clues for the identification of new drug targets as well as facilitate the prediction of potential side effects of therapies.
Previously, several groups have produced oligo-based array transcriptome databases for normal human tissues (Haverty et al. 2002; Shmueli et al. 2003), normal human and normal mouse tissues (Su et al. 2002, 2004), and normal rat tissues (Walker et al. 2004). Here, we have contributed to this field by constructing a gene expression data set of 18,927 genes from 158 normal samples of 19 different organs. To our knowledge, this data set is the largest to date for the analysis of global gene expression profiles within individual organs from multiple patients and is complementary to previously published data sets.
Our database of transcript levels in normal tissues was developed as a reference database that can be compared to data attained from diseased tissues, allowing the detection of disease-related aberrations in transcript levels. In order to produce credible findings from these types of analyses, it is crucial to first validate this reference data set extensively. Therefore, the major focus of this manuscript is on demonstrating the internal consistency of gene expression profiles in the database and verifying that these profiles are biologically meaningful. Additionally, we report that >90% of the measured genes are differentially expressed between the different organs. We show that this finding is not a statistical artifact but does, indeed, reflect biology. Finally, to show the powerful utility of our validated data set, we describe one of many possible applications in which we identify genes that are highly expressed in neuroblastoma, an embryonal cancer of childhood, as compared to any individual organ type profiled in our data set.
Results
Samples and cDNA microarray
A total of 158 samples across 19 different organs were collected from the Brain and Tissue Banks for Developmental Disorders at the University of Maryland (http://medschool.umaryland.edu/btbank/) from 30 individual human donors (Supplemental Table 1). Of the donors, 17 were males and 13 were females. The median age was 20 yr (range 3 mo-39 yr), and the donors came from three ethnic groups (Caucasian, African American, Pacific Ocean Islands). The median postmortem interval was 11 hr (range 4-19 hr). The cause of death varied (see Supplemental Table 1). The median yield of RNA was 0.58 μg/mg (range 2.11 μg/mg for pancreas-0.2 μg/mg for skeletal muscle). Samples were profiled on sequence-verified cDNA libraries containing 42,421 cDNA spots representing 25,933 different UniGene clusters (13,606 known genes and 12,327 ESTs). After quality filtering, 18,927 unique UniGene clusters remained.
Unsupervised analysis using principal component analysis and hierarchical clustering
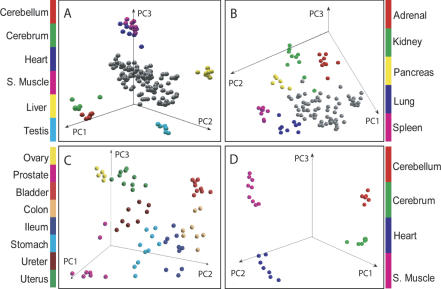
In order to test the internal consistency of our data set, we used principal component analysis (PCA) to check if the individual samples clustered together according to their organ of origin. No gene selection was performed prior to the PCA, except quality filtering. Interestingly, despite the fact that the samples used in this study were from 30 individual donors from different ethnic groups, genders, ages, causes of death, and postmortem intervals (Supplemental Table 1), the PCA plot showed that samples from the same organ clustered together and were distinct from other organs (Fig. 1). The gene expression profiles of cerebellum, cerebrum, heart, skeletal muscle, liver, and testis had relatively higher variance as evidenced by their segregation away from the rest of the organs (Fig. 1A). After removing these organs and recalculating the PCs, it can be seen that adrenal, kidney, pancreas, lung, and spleen also cluster according to organ type and aggregate slightly away from the remaining organs (Fig. 1B). These remaining organs—ileum, colon, stomach, bladder, uterus, ureter, prostate, and ovary—grouped closer together upon recalculation of the PCs (Fig. 1C), but did, in fact, cluster according to organ type. Tissues having similar cellular composition and function, like cerebellum and cerebrum or heart and skeletal muscle, cluster closely together (Fig. 1A) but were clearly separated from each other when examined apart from the rest of the organs (Fig. 1D).
Figure 1.
Principal components analysis. (A) The top three principal components of all 158 samples from all of the 19 organs. Cerebellum, cerebrum, heart, skeletal muscle, liver, and testis clustered separately from the rest of the organs (gray color). (B) After the above six organs in A were removed, PCA was recalculated with the samples from the remaining 13 organs. Adrenal, kidney, pancreas, lung, and spleen clustered separately from the rest of the organs (gray color). (C) After the above five organs in B were removed, PCA was recalculated with the samples from the remaining eight organs (ovary, prostate, bladder, colon, ileum, stomach, ureter, and uterus). (D) Cerebrum, cerebellum, heart, and skeletal muscle were separated completely with recalculated PCA.
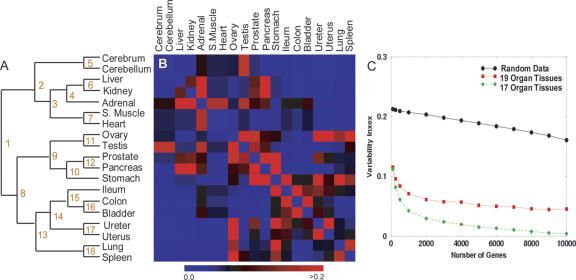
We further investigated how the organs can be classified according to transcript levels using hierarchical clustering (HC) employing the Pearson metric on average expression levels for each organ (Fig. 2A). For many of the organ types, the hierarchical structure reflects the known similarity of biological functions between the organs. For example, cerebrum and cerebellum cluster together, reflecting their physiological similarity. Likewise, the reproductive organs—testis and ovary—and the muscle organs—skeletal muscle and heart—group together. Bladder, colon, ileum, uterus, and ureter, which form one subbranch of the hierarchy, are known to have a similar cellular composition with predominantly smooth muscle as a constituent (Albert et al. 1994).
Figure 2.
Hierarchical clustering structure and stability. (A) Hierarchical clustering of the organs based on mean expression levels across organs for all genes. The branch points are enumerated for reference in the text and Supplemental Table 2. (B) The heat map visualizes the variance of the distance (see Methods) in the HC hierarchy estimated from 1000 HC trees using randomly selected subsets of genes of size 1000. (Black) Variability close to the average for the whole dendrogram; (blue) a lower than average variability; (red) an increased variability. The interpretation of the increased variability indicated by the red squares is that the corresponding organs are close in some of the HC graphs, while they are further away in other HC graphs. (C) The variability index for the whole dendrogram is plotted as a function of the size of the sets of genes drawn randomly from the genes on the microarray. The black line represents the variability index for randomly generated gene expression data (see Methods), the red line for the entire data set (19 organs), and the green when ovary and stomach, which showed the highest variability, are removed from the analysis.
To substantiate the biological relevance of the HC tree, we identified which genes are differentially expressed at the branch points (defined in Fig. 2A) and looked for statistically overrepresented Gene Ontology (GO) terms in these selected genes (Supplemental Table 2). The first branch point distinguished organs with a high level of energy consumption (upper part: cerebrum, cerebellum, liver, kidney, adrenal, skeletal muscle, heart) from organs with lower energy consumption (lower part). Immune response genes were very significant for branch point 8, which divides ileum, colon, bladder, ureter, uterus, lung, and spleen from ovary, testis, prostate, pancreas, and stomach. In branching point 9, excretion-related genes were significant, likely because of the function of prostate, pancreas, and stomach. For the majority of the branch points, the significant GO terms are in logical agreement with known organ functions.
Analysis of the robustness of the hierarchical clustering structure
The hierarchical clustering (HC) of the different tissues reflects well the similarity between the expression profiles of the different organs. In order to explore if this result depends on the specific set of genes, that is, if it reflects specific cell functions, or if it is independent on the specific choice of genes, we analyzed the stability of the HC when small randomly selected subsets of genes were used in this analysis. If the hierarchical structure is unchanged by various gene subset selections, this would indicate that the similarity is present on the whole range of transcriptional regulation and consequently over the majority of biological processes. If, however, changes in gene selection do produce significant changes in the hierarchical structure, this would indicate that the similarities extend only over specific cellular processes. In order to assess the stability of the hierarchical structure to changes in the selected genes, we regenerated 1000 new HCs with randomly chosen subsets of 1000 genes and measured the similarity between the resulting trees where “similarity” was defined geometrically (i.e., the “distance” between any two tissue samples A and B on the dendrogram was defined as the number of branch points visited by a walker traversing the tree from leaf A to leaf B). If these distances are approximately constant for the trees generated with different gene sets, the hierarchical clustering is deemed insensitive to the specific choice of genes. For the details of this procedure, see the Methods section. To quantitate the stability of the hierarchical structure, we calculated the variance of the distance between all pairs of organs within each of the 1000 HC dendrograms. The heat map of Figure 2B visually depicts the interorgan variance. A blue color linking two organs indicates essentially constant distance between these two organs, while a red color indicates a variable distance. Thus if two organs are linked by a red square, it indicates that their relationship depends on the specific choice of genes while a stable (blue) distance suggests that the similarity in the expression of genes is present in (almost) all of the gene sets. For the majority of organs we find that the interorgan similarity is stable, while certain organs cluster with different partners for different gene sets. For example, the red squares for testis in Figure 2B indicate that this sample clusters not only with ovary (Fig. 2A) but with some gene sets clusters with cerebrum, cerebellum, prostate, or adrenal. As a second example, ovary also clusters with ureter, uterus, prostate, or spleen depending on the gene sets used.
We next asked the question, how many genes are necessary to produce essentially the same hierarchical structure obtained for all genes? To do this, we repeated the procedure above with an increasing number of genes starting from 100 and increasing to 10,000. For each subset size, 1000 different random subsets were used to generate HCs. For each of these replicates, R, we estimated the similarity with the dendrogram, D, for the full set of genes. As before, this similarity was quantified by comparing the distances between any two tissues in R and D. A variability index, the average variance of the distances (for details, see Methods), is shown in Figure 2C. It is interesting to note that for subsets as small as 100 randomly selected genes, the dendrogram is significantly more stable than the trees found for randomly generated data (black line), which is shown in the same plot as a reference point for variability. The variability is stabilized in our real data when the size of the subsets approaches ∼1000 out of 18,927. Furthermore, we find that the hierarchical structure is much more stable (lower variability index, green line) when ovary and stomach, which showed the highest variability, were removed from the analysis.
We observed a remarkable heterogeneity of the transcript levels between the different tissues. Using a one-way ANOVA analysis, we found that >90% of the 18,927 unique genes in our study showed differential expression patterns at a 5% significance level after adjustment for multiple comparisons.
Prevalent gene expression patterns
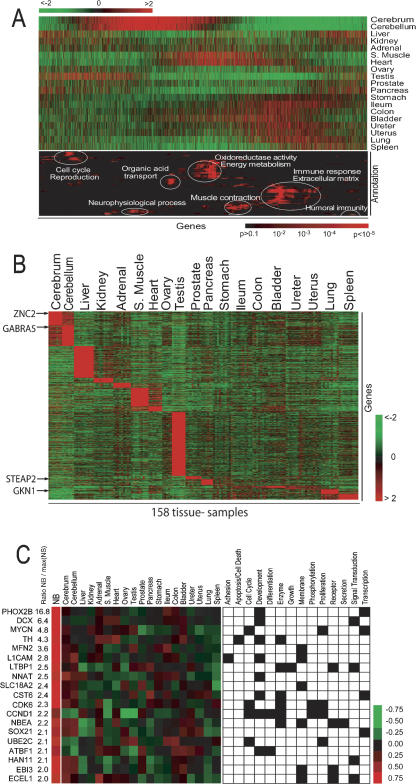
Next, we explored the simultaneous comparison of global patterns of gene expression and their relationship to biological processes by using the Gene Ontology (GO) annotations. We ordered the genes in this database using the reshuffling algorithm (Lund technical report: http://www.thep.lu.se/pub/Preprints/00/lu_tp_00_18.pdf; Cunliffe et al. 2003), which generates a unique order of genes based on the Pearson similarity distance metric. Prior to this step, we removed all genes with low variance in the data set (i.e., <0.25, see Methods) because the current implementation of the reshuffling algorithm cannot effectively deal with the full data set. We analyzed the sorted list of the remaining 7020 genes for regions in which specific GO annotations were overrepresented. To this end, we calculated the probability of a GO term occurring by random chance within a window of 160 adjacent genes (see Methods). The result of this analysis is shown in Figure 3A, which has two parts: a traditional gene-expression heat map (upper part) and a probability heat map (lower part) for gene annotations representing a Gene Ontology term in each row (Supplemental Table 5).
Figure 3.
(A) Gene Ontology densities on reshuffled data. The upper heat map represents the average gene expression in 19 tissues. The genes were ordered by the reshuffling algorithm and analyzed for regions where the density of genes annotated to GO terms was higher than expected by random chance. The results of this analysis are shown in lower heat map, where the probability of finding a GO term by chance represented by a red color according to the scale shown. A red color thus represents a nonrandom density of genes associated with the corresponding GO term around this position (x-direction) in the gene list. The GO terms (y-direction) were selected if at some location the density genes annotated with the corresponding GO terms was considered statistically significantly overrepresented. The selected 98 GO terms were then sorted in the y-direction by HC with respect to the pattern of P values. A detailed list of the GO terms is given in Supplemental Table We have highlighted in white ellipses some of the most predominant terms. (B) Organ-specific gene expression. Heat map of the expression level of all samples for 4291 organ-specific genes identified by performing pairwise t-tests between each organ and all the remaining organs. (C Neuroblastoma-specific gene expression. (Left panel) The heat map the median values for gene expression for the differentially expressed genes in NB and each of the normal organs. The color scale represents the z-scored gene expression ratio (number of standard deviation of median expression ratio from the mean). (Right panel) The GO terms the 19 genes, where a black square in the grid indicates that the gene associated to that GO term. The numbers in the NB/max(NS) column the ratios of the median NB gene expression ratio divided by the maximum median expression ratio of all 19 organs.
An advantage of this analysis is that it demonstrates how different biological processes are shared between organs. For example, the overrepresentation of the GO term “neurophysiologic process” coincides with an overexpression of the corresponding genes in the cerebrum and cerebellum. Likewise, the overrepresented term “muscle contraction” coincides with an increased expression in heart and skeletal muscle and the other smooth muscle organs. Genes associated with the extracellular matrix and immune response are specifically active in bladder, colon, ileum, stomach, ureter, uterus, and lung. Liver and spleen show the strongest expression of genes related to humoral immune response. On the other hand, cerebellum, cerebrum, and testis characterize the lowest expression of genes associated with immune-related GO terms. Testis shows the highest expression level in cell cycle- and reproduction-related genes.
Organ-specific gene expression
We next identified the genes that are specifically overexpressed in one organ in comparison to all other 18 organs, by requiring that the P-value for each pairwise t-test was <0.01. Figure 3B shows the heat map of all the organ-specific genes that passed this selection criterion. We observed that testis, liver, cerebrum, cerebellum, skeletal muscle, and heart had the largest number of specifically expressed genes. For example, we found that sexual reproduction, spermatogenesis, mitosis, and fertilization were the dominating annotations in testis-specific genes. The 639 genes that were specifically expressed in liver were associated with the major physiological functions of the liver including energy process, lipid metabolism, cholesterol metabolism, complement production, detoxification, alcoholic metabolism, and urea cycle. The exact numbers of differentially expressed genes are presented in Supplemental Table 3, where we also list the top 10 genes (lowest P-value) for each organ. We found that in most cases, these genes reflected the organ function, as can be seen from the result of the GO analysis (Supplemental Table 4).
Application of database: Identification of potential therapeutic targets
One potential application of this database is to identify uniquely expressed genes in cancer that may be used as potential diagnostic markers or targets for therapy. We used neuroblastoma (NB) as a model and performed a t-test comparing the gene expression profiles of 100 NB tumors of different stages (1 through 4 including MYCN amplified tumors) with 100 normal organ samples (randomly selected with approximately equal numbers from each organ) from our database. Next, we selected the genes whose (1) NB expression levels were significantly overexpressed (see Methods) compared with the expression ratio of the highest expressed organ for that gene, and (2) GO annotations would suggest good therapeutic targets (see Methods). In all, 19 genes passed this stringent filter (Fig. 3C), of which the top most differentially expressed gene was PHOX2B.
Discussion
We have constructed a gene expression database capturing the mRNA transcriptional levels for 19 different organs from 158 normal human tissues from 30 donors. Previously, three groups (Haverty et al. 2002; Su et al. 2002; Shmueli et al. 2003) established human transcriptomic databases using oligonucleotide microarrays. Our database builds on the published studies by an increased number of samples, an increased number of biological repeats for the same tissue, an increase in the number of detected unique clones, and the different array technology used (cDNA). Biological repeats were hybridized individually, allowing us to estimate intraorgan variation of transcript levels. In our analysis, 18,927 unique genes passed our quality filtering. To our knowledge, our work herein represents the largest normal sample data set profiled in a single study. The second largest data set (Su et al. 2004) contained 79 human samples, and the number of unique clones detected in at least one tissue was only slightly smaller at 16,454 genes. The strength of that work was the use of a custom-made oligonucleotide array, which complemented the commercial array they used in parallel. The custom array focused on expressed sequences predicted by RefSeq, Celera, and Ensembl that were not present on the commercial array. This setup makes that database very valuable for the detection and annotation of novel genes. For our database, the emphasis is to allow researchers to compare their own data to our reference database of normal tissues. Therefore, we only used commercially available and frequently used cDNA libraries for the production of our microarrays to increase the probability that the researcher's genes of interest will be in our data set. Furthermore, our cDNA-based arrays will simplify data analysis for research groups already using cDNA technology and may help them avoid potential cross-platform problems (Park et al. 2004). The main focus of our data analysis was to establish the validity of our database so that it can be used with confidence. In addition, we provide one demonstration of a possible application in the context of identifying cancer-specific genes. For the first part, the statistical analysis of the data therefore aimed at confirming that this database (1) is consistent and (2) contains relevant biological information.
In order to establish consistency, we first analyzed the homogeneity of transcriptional information within samples of the same organ by using PCA. Without prior information, we showed that samples from the same organ but different donors clustered together. This result is remarkable given the potential variability that could have been introduced for each sample with respect to the different source, cause of death, age, sex, postmortem interval, and sampling site. An example of the variability introduced by sampling site was reported by one group, who identified distinctive patterns of gene expression from discrete portions of kidney (Higgins et al. 2004). In our study, despite the fact that there was no prior selection of which part of each organ was selected for profiling, our results showed a very strong homogeneity in the pattern of gene expression for each organ type.
To establish the analysis of similarities and differences between the expression profiles for the 19 organs, we used hierarchical clustering. The dendrogram generated by this analysis reflected functional and morphological relationships between organs. Previously, Guo et al. (2003) showed by HC analysis that the expression pattern between testis and brain is similar. They interpreted this as a manifestation of the fact that both share the common specificity in the blood-barrier property (Sites et al. 1997). However, this is only partially compatible with our results because the stability analysis revealed that testis also neighbors with other hormone-predominant organs (ovary, prostate or adrenal) depending on which genes are used to perform the HC (Fig. 2B).
Our results show that gene expression profiling can distinguish each of the 19 organs. Furthermore, by an identification of overrepresented annotations using terms from the Gene Ontology, we demonstrated that the transcriptome reflects major organ functions. This result, although apparently not surprising, is nontrivial. Firstly, it provides experimental evidence that despite the importance of post-transcriptional regulation and signaling pathways, the regulation of transcript levels plays a major role in controlling normal cell functions. Secondly, our conclusions were based on an observation, which is unique to high-throughput measurements: collective phenomena. Rather than deriving conclusions from individual genes, we used ensembles of genes and identified overrepresented annotations in these ensembles. The fact that this procedure selected biologically meaningful annotations confirms that this type of information can be used to draw meaningful biological conclusions. The advantage of this “collective” approach is that it better reflects the underlying biology, which almost always involves multiple genes for specific biological processes and phenotypes. In addition, it avoids potential ambiguities that arise because individual genes often contribute to several different processes.
When we asked the question, how many of the genes in our experiment showed some form of differential expression, ANOVA analysis revealed that >90% of the 18,927 unique genes were differentially expressed at the 5% level of statistical significance after adjustment for multiple comparisons (data not shown). In order to exclude the possibility that this may have been caused by some unknown, technical bias not related to the function of the organ, we analyzed the stability of the HC structure. We found that randomly selected sets of genes of 100 or more could reproduce this HC structure, and the stability approached its maximum with 1000-2000 genes. Together with the observation that the HC dendrogram properly reflected known biological facts, this finding implies that small sets of arbitrarily selected genes contain sufficiently many differentially expressed genes to reconstruct the dendrogram and that these differential expressions are of biological origin, even though the absolute difference in transcript levels is often quite small.
Next, we used the reshuffling analysis and the Gene Ontology annotations to simultaneously compare the transcriptional patterns and biological processes shared by the 19 organs in our study. This method allows for a global analysis of the processes involved in each organ, and the interpretations of the results are based on the hypothesis that highly correlated or coexpressed genes may belong to the same functional pathway (Stuart et al. 2003). Our results corroborate this idea as we found, for instance, that the digestive, genitourinary, and respiratory organs showed a strong expression of extracellular matrix and immune-response genes in agreement with current knowledge (Sites et al. 1997). But, only stomach shows a lower expression of immune-related genes, which may be explained by the fact that it does not have Payer's patch (Sites et al. 1997). Also, cerebellum, cerebrum, testis, prostate, and pancreas show lower expression of immune genes and are known to contain a paucity of immunity-related cells (Abbas et al. 1997; Sites et al. 1997; Bernstorff et al. 2002).
Next, we identified the genes that are most highly expressed in one organ compared to all other organs. These identified genes can potentially be used as markers for the origin of metastatic tumors, where the primary location of the tumor is unknown. We demonstrated a strong relevance of these genes to the functions of the corresponding organs through GO analysis (Supplemental Table 4). Many of the high-ranking genes in each organ are associated with organogenesis and organ-specific functions. For example, the top-ranking cerebellum-specific gene, ZIC2, is known to control cerebellar development (Aruga et al. 2002), and the top-ranking cerebrum-specific gene, GABRA5, has been implicated in the pathogenesis of mood disorders (Papadimitriou et al. 2001; Supplemental Table 3). Additionally, we found GKN1 (CA11), which is known to be underexpressed in gastric cancer (Oien et al. 2004), uniquely overexpressed in stomach. The observation that colon, stomach, and ileum have the fewest number of uniquely expressed genes is probably due to a common pool of genes that are coexpressed in these organs since our pairwise t-test would have excluded these genes. Further analysis could be performed to identify these genes, but has not been done for the purpose of this study.
From these results we show that the global gene expression profiles for these 19 normal organs contain high-quality reliable data that reflect the biological function of each organ. We next showed one possible application of this database by the identification of potential targets for diagnosis and/or therapy in one cancer type. We chose neuroblastoma (NB), which is the most common extracranial pediatric solid tumor, and accounts for 7%-10% of all childhood cancers (Brodeur 2003). We identified 19 genes whose expression levels were significantly overexpressed when compared with all the normal samples that were associated with GO terms that would imply potential “druggable” targets, for example, apoptosis, growth, proliferation, and transcription. Since our comparison was between malignant and normal tissue, several of the genes identified (e.g., the cell cycle genes CCND1 and CDK6) are up-regulated in other cancers, but nevertheless represent legitimate targets for therapy. PHOX2B was the most highly expressed gene in NB that passed our selection process, and represents a NB-specific gene. It is a neurodevelopmental gene, expressed in both the central and the peripheral autonomic nervous system during human embryonic development (Amiel et al. 2003). It has recently been found to be mutated in familial neuroblastomas (Trochet et al. 2004). Other genes include MYCN, which is amplified in a subset of neuroblastomas and important in the etiology of these tumors, in that transgenic mice that express this gene develop NB with a high penetrance (Weiss et al. 1997). Indeed, in this mouse model the MYCN transgene is driven by the TH gene, which is also among these 19 genes. Other genes of note in this list include L1CAM,a membrane glycoprotein that belongs to a large class of immunoglobulin superfamily cell adhesion molecules that mediate cell-to-cell adhesion in the nervous system. Interestingly, this gene was originally named MIC5, because it was discovered through the use of monoclonal antibodies to be expressed in a NB cell line (Hope et al. 1982) and was subsequently shown to be a serum and immunohistochemical marker of NB as well as other embryonal cancers. All of these 19 genes represent potential biomarkers for diagnosis, targets for therapy as well for development of immune-based vaccine treatments.
In conclusion, we have developed a genome-wide database of mRNA expression across a large number of human organs. We demonstrate that the data are internally consistent and the global pattern of gene expression reflects the function of the organs. We have released the raw data for all 42,421 cDNA clones, which are open to the public at http://home.ccr.cancer.gov/oncology/oncogenomics/. This database allows investigators to make simple queries of the data to extract gene expression profiles based on IMAGE Clone ID, LocusLink number, Gene Ontology Terms, Gene Ontology ID, Gene Symbol, UniGene ID, Clone Title, Cytoband, and Chromosome. Additionally, investigators can identify correlated genes or download the entire or subsets of the data for their own analysis. We believe that our database will be of wide interest and utility to both basic and translational scientists.
Methods
Samples
A total of 158 tissue samples across 19 different organs were collected from the Brain and Tissue Banks for Developmental Disorders at the University of Maryland from 30 individual donors, consisting of 17 males and 13 females with a median age of 20 yr (range 3-469 mo). The donors came from three ethnic groups (Caucasian, African American, Pacific Ocean Islands) (see Supplemental Table 1). Soon after surgical removal (median postmortem hours [PMH] of 11 h), specimens were snap-frozen in liquid nitrogen and kept in a deep freezer (-80°C) until RNA extraction. The profiles from 100 primary NB samples from various tumors' banks consisting of equal numbers of stage 1-4 and stage 4 with MYCN amplification were obtained from a prior study (Krasnoselsky et al. 2004).
RNA extraction and construction of reference RNA
Total RNA extraction from tissue samples and seven human cancer cell lines (CHP212 RD, A204, RDES K562, CA46, and HeLa) was done by published protocols (Wei and Khan 2002). We used an Agilent BioAnalyzer 2100 (Agilent) to assess the integrity of total RNAs. Equal quantities of total RNA from seven cancer cell lines were pooled for RNA reference, which was used in all cDNA microarray experiments.
RNA amplification and labeling of cDNA probes
mRNA amplification was done by a modified Eberwine RNA amplification procedure (Sotiriou et al. 2002) followed by indirect fluorescent labeling of cDNA probes as described by Hegde et al. (2000). In brief, amplified RNA was converted into cDNA with incorporation of amynoallyl-dUTP (Sigma-Aldrich) by using Superscript II RT enzyme (Invitrogen). After purification and drying of cDNA, monoreactive-Cy3 for tissue samples or Cy5 (Amersham Pharmacia) for reference were conjugated with amynoallyl-dUTP on cDNA. Fluorescent-labeled probes were purified with QIAGEN PCR purification kits (QIAGEN) according to the manufacturer's instruction.
Fabrication of cDNA microarray, hybridization, image acquisition, and analysis
Sequence-verified cDNA libraries were purchased from Research Genetics, and a total of 42,421 cDNA clones, representing 25,933 UniGene clusters (13,606 known genes and 12,327 ESTs), were amplified and purified. Then cDNA microarrays were printed using a BioRobotics MicroGrid II spotter (Harvard Bioscience). After hybridization and washing of microarrays as described by Hegde et al. (2000), we acquired images by an Agilent DNA microarray scanner (Agilent), and analyzed them using the Microarray Suite program as described previously (Chen et al. 1997), coded in IPLab (Scanalytics).
Data normalization and principal component analysis
Fluorescence ratios were normalized for each microarray by setting the average log ratio for each subarray element to zero (commonly referred to as “pin-normalization”). The data were quality-filtered by removing those clones that had poor quality measurement (quality <0.5) for more than 20% of all the samples, modified from Chen et al. (1997). Out of the 42,421 clones on the chip, 36,153 passed this filter. For the clones that passed this filter, the ratio of low-quality spots was set to the average observation for the other samples of the same tissue. This procedure substituted one or more values per organ in 7% of the clones and more than three values in <1% of the clones. The clones were then assigned to UniGene Clusters (Unigene Build 166). For the UniGene clusters represented by multiple spots or clones, mean fluorescence ratios of those points are used. After these processes we had 18,927 unique UniGene clusters remaining from the initial 42,421 clones. Where mentioned in the text, expression levels averaged over organ samples were used. PCA was performed using the log2 gene expression ratios of all the samples of the 18, 927 genes using Matlab (The Mathworks).
Stability of hierarchical clustering
All HC was performed using the Pearson correlation coefficient distance metric. In order to analyze the stability of the results of hierarchical clustering, we introduced a measure of similarity between two trees based on a comparison of the geometry of the dendrograms. This approach was motivated by the observation that dendrograms that appear very different by visual inspection may actually encode identical relations. We therefore analyzed the distances between leaves of the HC structure and how those distances vary if different data sets were used for HC. The distance dAB between leaf A and B was defined in terms of the number N of internal nodes (branch points of the tree) passed to get from A to B: dAB = 2-N. The exponent reflects how strongly the average linkage algorithm of HC coupled the two leaves: at each node the contribution of the pattern of A and B is reduced by ½ because of averaging of both sides of the branch. The variability of dAB was defined as the variance of dAB when different sets of genes were used for the clustering. The variability index for whole HC structure was defined as the variability averaged over all pairs of leaves. These observables were estimated numerically by drawing 1000 random subsets of a given size from the genes on the microarray. In order to set a scale for this observable, we have also analyzed the variability index for trees generated from random data. Owing to the lack of information, this type of data does not generate a stable tree; therefore, the index obtained for these data set a scale for data containing no reproducible structure. These random data were generated from a Gaussian distribution. The decrease of variability for larger subsets, which was found even for the random data, is of a technical nature, because the random subsets have a considerable overlap if the size of the random subsets is not small compared to the whole data set.
Gene Ontology analysis
The Gene Ontology (GO) consortium (Ashburner et al. 2000) provides annotation for 47% of the clones in our experiment. From the directed acyclyic graph structure of the GO, each node is coupled to over- or underlying nodes via an “isa” or “part-of” relation. A clone mapped to any given annotation is therefore also associated with the parent nodes. The LocusLink database provides a link to GO terms. We therefore mapped each clone to a UniGene cluster and used the LocusLink identifier to associate the clones with GO terms.
Reshuffling
We first selected genes with a variance larger than 0.25 (n = 7020 genes). The reshuffling algorithm has been previously used by Cunliffe et al. (2003). A detailed description of the algorithm can be obtained from http://www.thep.lu.se/pub/Preprints/00/lu_tp_00_18.pdf. For this algorithm a Pearson correlation distance metric was used, and the end result of this procedure is to place the genes in a unique order where the most correlated pairs of genes are adjacent to each other. Using a window of size 160, we tested overrepresentation of GO terms in this subset of clones by estimating with the hypergeometric distribution the probability to find the same number or more clones in this GO term by chance alone. GO terms were selected as significantly enriched if at any location the P-value for a nonrandom density of genes was smaller than α = 0.0001. The P-values obtained in this way were visualized by a heat map, where higher intensities represent lower P-values [I = -log(P)].
Selection of genes
Differentially expressed genes were selected by a two-sided t-test. In the analysis of genes driving the bifurcation of the hierarchical clustering a threshold α =10-8 was used, which reflects a correction for multiple comparison. Genes were selected as “uniquely” up- or down-regulated in a tissue if by pairwise t-test with all the other tissue samples the gene was significantly up-regulated (down-regulated) with a threshold α = 0.01.
Identification of NB-specific genes
To select the genes that were highly overexpressed in NB compared to the normal samples, we used a set of highly stringent filters. We first identified differentially expressed clones using the t-test (P < 0.01) and Bonferroni adjustment for multiple comparisons between a set of 100 neuroblastoma tumors (unpubl.) of various clinical stages and a randomly selected set of 100 normal samples distributed evenly throughout all 19 organs. From this set of genes, we selected only those where the ratio of median gene expression in NB to normal samples was >3. Next, we selected those genes whose median ratio value in NB was twofold greater than the maximum median value of all 19 organs. Finally, we selected those genes that were associated with preselected GO terms that would suggest good targets (Fig. 3C).
Web-based database development
The database contains the raw data for all 42,421 cDNA clones, and is open to the public at http://home.ccr.cancer.gov/oncology/oncogenomics/. The backend of the database uses MySQL (http://www.mysql.com) to house the data. Perl scripts are used both to query the database (DBI) and generate the HTML (CGI) to display the query results. The publicly available databases GeneKeyDB (http://genereg.ornl.gov/gkdb/) and Gene Ontology (http://www.geneontology.org) form the backbone for producing rich annotation associated with each cDNA clone. Users will be able to query genes based on IMAGE Clone ID, LocusLink number, Gene Ontology Terms, Gene Ontology ID, Gene Symbol, UniGene ID, Clone Title, Cytoband, and Chromosome. Three normalization options exist for the user to choose from: log2, median-centered log2, and median z-scored log2. The results of the query can be viewed by either a heat map for multiple genes grouped by organ type or a bar chart for individual genes also grouped by organ type. The raw data are available and can be freely downloaded from http://www.genome.org or http://home.ccr.cancer.gov/oncology/oncogenomics/ to the local workstation for further individual analysis. Links to various external databases populate the details page when an individual clone is selected. In the details page, clones that correlate (Pearson |correlation| ≥0.5) with that individual clone can be extracted. Future database developments will focus on enhanced queries that will allow the user to search based on gene expression ratios for the various organ types.
Acknowledgments
This work was supported in part by the post-doctoral fellowship program of the Korea Science & Engineering Foundation (KOSEF).
Footnotes
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.3124505.
[Supplemental material is available online at www.genome.org and at http://home.ccr.cancer.gov/oncology/oncogenomics/.]
References
- Abbas, A.K., Lichtman, A.H., and Pober, J.S. 1997. Cellular and molecular immunology. W.B. Saunders, Philadelphia.
- Albert, B., Bray, D., Lewis, J., Raff, M., Roberts, K., and Watson, J.D. 1994. Molecular biology of the cell. Garland, New York.
- Amiel, J., Laudier, B., Attie-Bitach, T., Trang, H., de Pontual, L., Gener, B., Trochet, D., Etchevers, H., Ray, P., Simonneau, M., et al. 2003. Polyalanine expansion and frameshift mutations of the paired-like homeobox gene PHOX2B in congenital central hypoventilation syndrome. Nat. Genet. 33: 459-461. [DOI] [PubMed] [Google Scholar]
- Aruga, J., Inoue, T., Hoshino, J., and Mikoshiba, K. 2002. Zic2 controls cerebellar development in cooperation with Zic1. J. Neurosci. 22: 218-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner, M., Ball, C.A., Blake, J.A., Botstein, D., Butler, H., Cherry, J.M., Davis, A.P., Dolinski, K., Dwight, S.S., Eppig, J.T., et al. 2000. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 25: 25-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstorff, W.V., Glickman, J.N., Odze, R.D., Farraye, F.A., Joo, H.G., Goedegebuure, P.S., and Eberlein, T.J. 2002. Fas (CD95/APO-1) and Fas ligand expression in normal pancreas and pancreatic tumors. Implications for immune privilege and immune escape. Cancer 94: 2552-2560. [DOI] [PubMed] [Google Scholar]
- Brodeur, G.M. 2003. Neuroblastoma: Biological insights into a clinical enigma. Nat. Rev. Cancer 3: 203-216. [DOI] [PubMed] [Google Scholar]
- Chen, Y., Dougherty, E.R., and Bittner, M.L. 1997. Ratio-based decisions and the quantitative analysis of cDNA microarray image. Biomed. Optics 2: 364-374. [DOI] [PubMed] [Google Scholar]
- Cunliffe, H.E., Ringner, M., Bilke, S., Walker, R.L., Cheung, J.M., Chen, Y., and Meltzer, P.S. 2003. The gene expression response of breast cancer to growth regulators: Patterns and correlation with tumor expression profiles. Cancer Res. 63: 7158-7166. [PubMed] [Google Scholar]
- Guo, J., Zhu, P., Wu, C., Yu, L., Zhao, S., and Gu, X. 2003. In silico analysis indicates a similar gene expression pattern between human brain and testis. Cytogenet. Genome Res. 103: 58-62. [DOI] [PubMed] [Google Scholar]
- Haverty, P.M., Weng, Z., Best, N.L., Auerbach, K.R., Hsiao, L.L., Jensen, R.V., and Gullans, S.R. 2002. HugeIndex: A database with visualization tools for high-density oligonucleotide array data from normal human tissues. Nucleic Acids Res. 30: 214-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegde, P., Qi, R., Abernathy, K., Gay, C., Dharap, S., Gaspard, R., Hughes, J.E., Snesrud, E., Lee, N., and Quackenbush, J. 2000. A concise guide to cDNA microarray analysis. Biotechniques 29: 548-556. [DOI] [PubMed] [Google Scholar]
- Higgins, J.P., Wang, L., Kambham, N., Montgomery, K., Mason, V., Vogelmann, S.U., Lemley, K.V., Brown, P.O., Brooks, J.D., and van de Rijn, M. 2004. Gene expression in the normal adult human kidney assessed by complementary DNA microarray. Mol. Biol. Cell 15: 649-656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hope, R.M., Goodfellow, P.N., Solomon, E., and Bodmer, W.F. 1982. Identification of MIC5, a human X-linked gene controlling expression of a cell surface antigen: Definition by a monoclonal antibody raised against a human X mouse somatic cell hybrid. Cytogenet. Cell Genet. 33: 204-212. [DOI] [PubMed] [Google Scholar]
- Krasnoselsky, A.L., Whiteford, C.C., Wei, J.S., Bilke, S., Westermann, F., Chen, Q.R., and Khan, J. 2004. Altered expression of cell cycle genes distinguishes aggressive neuroblastoma. Oncogene [E-pub ahead of print, Dec. 13]. [DOI] [PubMed]
- Lander, E.S., Linton, L.M., Birren, B., Nusbaum, C., Zody, M.C., Baldwin, J., Devon, K., Dewar, K., Doyle, M., FitzHugh, W., et al. 2001. Initial sequencing and analysis of the human genome. Nature 409: 860-921. [DOI] [PubMed] [Google Scholar]
- Oien, K.A., McGregor, F., Butler, S., Ferrier, R.K., Downie, I., Bryce, S., Burns, S., and Keith, W.N. 2004. Gastrokine 1 is abundantly and specifically expressed in superficial gastric epithelium, down-regulated in gastric carcinoma, and shows high evolutionary conservation. J. Pathol. 203: 789-797. [DOI] [PubMed] [Google Scholar]
- Papadimitriou, G.N., Dikeos, D.G., Karadima, G., Avramopoulos, D., Daskalopoulou, E.G., and Stefanis, C.N. 2001. GABA-A receptor β3 and α5 subunit gene cluster on chromosome 15q11-q13 and bipolar disorder: A genetic association study. Am. J. Med. Genet. 105: 317-320. [DOI] [PubMed] [Google Scholar]
- Park, P.J., Cao, Y.A., Lee, S.Y., Kim, J.W., Chang, M.S., Hart, R., and Choi, S. 2004. Current issues for DNA microarrays: Platform comparison, double linear amplification, and universal RNA reference. J. Biotechnol. 112: 225-245. [DOI] [PubMed] [Google Scholar]
- Shmueli, O., Horn-Saban, S., Chalifa-Caspi, V., Shmoish, M., Ophir, R., Benjamin-Rodrig, H., Safran, M., Domany, E., and Lancet, D. 2003. GeneNote: Whole genome expression profiles in normal human tissues. C R Biol. 326: 1067-1072. [DOI] [PubMed] [Google Scholar]
- Sites, D.P., Terr, A.I., and Parslow, T.G. 1997. Medical immunology. Appleton & Lance, Stanford, CT.
- Sotiriou, C., Khanna, C., Jazaeri, A.A., Petersen, D., and Liu, E.T. 2002. Core biopsies can be used to distinguish differences in expression profiling by cDNA microarrays. J. Mol. Diagn. 4: 30-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart, J.M., Segal, E., Koller, D., and Kim, S.K. 2003. A gene-coexpression network for global discovery of conserved genetic modules. Science 302: 249-255. [DOI] [PubMed] [Google Scholar]
- Su, A.I., Cooke, M.P., Ching, K.A., Hakak, Y., Walker, J.R., Wiltshire, T., Orth, A.P., Vega, R.G., Sapinoso, L.M., Moqrich, A., et al. 2002. Large-scale analysis of the human and mouse transcriptomes. Proc. Natl. Acad. Sci. 99: 4465-4470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su, A.I., Wiltshire, T., Batalov, S., Lapp, H., Ching, K.A., Block, D., Zhang, J., Soden, R., Hayakawa, M., Kreiman, G., et al. 2004. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc. Natl. Acad. Sci. 101: 6062-6067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trochet, D., Bourdeaut, F., Janoueix-Lerosey, I., Deville, A., de Pontual, L., Schleiermacher, G., Coze, C., Philip, N., Frebourg, T., Munnich, A., et al. 2004. Germline mutations of the paired-like homeobox 2B (PHOX2B) gene in neuroblastoma. Am. J. Hum. Genet. 74: 761-764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venter, J., Adams, M.D., Myers, E.W., Li, P.W., Mural, R.J., Sutton, G.G., Smith, H.O., Yandell, M., Evans, C.A., Holt, R.A., et al. 2001. The sequence of the human genome. Science 291: 1304-1351. [DOI] [PubMed] [Google Scholar]
- Walker, J.R., Su, A.I., Self, D.W., Hogenesch, J.B., Lapp, H., Maier, R., Hoyer, D., and Bilbe, G. 2004. Applications of a rat multiple tissue gene expression data set. Genome Res. 14: 742-749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, J.S. and Khan, J. 2002. Purification of total RNA from mammalian cells and tissues. In DNA microarrays: A molecular cloning manual (eds. D. Bowtell and J. Sambrook), pp. 110-119. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [DOI] [PubMed]
- Weiss, W.A., Aldape, K., Mohapatra, G., Feuerstein, B.G., and Bishop, J.M. 1997. Targeted expression of MYCN causes neuroblastoma in transgenic mice. EMBO J. 16: 2985-2995. [DOI] [PMC free article] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- http://genereg.ornl.gov/gkdb/; GeneKeyDB database.
- http://home.ccr.cancer.gov/oncology/oncogenomics/; Authors' Web site with raw data for all 42,421 cDNA clones.
- http://medschool.umaryland.edu/btbank/; Brain and Tissue Banks for Developmental Disorders at the University of Maryland.
- http://www.geneontology.org; Gene Ontology database.
- http://www.mysql.com; MySQL.
- http://www.thep.lu.se/pub/Preprints/00/lu_tp_00_18.pdf; Lund technical report, reshuffling algorithm.