Abstract
WDFY3 is a master regulator of selective autophagy that we recently showed to interact with TRAF6 and augment RANKL-induced osteoclastogenesis in vitro and in vivo via the NF-κB pathway. Since the NF-κB pathway plays a major role in inflammation herein, we investigate the role of WDFY3 in an arthritis animal model. Our data show that WDFY3 conditional knockout mice (Wdfy3loxP/loxP-LysM-Cre+,) were protected in the K/BxN serum transfer-induced arthritis animal model. These effects were independent of alterations in starvation-induced autophagy as evidenced by Western blot analysis of the autophagy marker LC3, autophagosome formation in osteoclast precursors and lysosome formation in osteoclasts derived from WDFY3-cKO mice compared to controls. Moreover, we demonstrate by immunofluorescence and co-immunoprecipitation that WDFY3 interacts with SQSTM1 in macrophages and osteoclasts. Collectively, our data suggest that loss of WDFY3 in myeloid cells leads to reduced severity of inflammatory arthritis independently of WDFY3 function in starvation-induced autophagy.
Keywords: Autophagy, Autophagy-linked FYVE containing protein, ALFY, WDFY3, osteoclast, musculoskeletal diseases
1. Introduction
Rheumatoid arthritis (RA) is a chronic inflammatory disease that exhibits various clinical manifestations including synovial inflammation and bone loss [1]. Although the development of biologics such as anti-TNF is an effective treatment for the majority of RA patients, approximately 40% of patients do not respond to TNF inhibition, suggesting that RA disease mechanisms are only partly understood. Autophagy, a cellular process that degrades organelles and misfolded proteins and ensures cells survival at homeostatic and stress conditions was recently associated with autoimmunity in multiple studies [2]. Specifically, autophagy plays multiple roles immune functions in macrophages such as clearing intracellular pathogens [3], regulating inflammatory cytokines expression [4], and modulating (M1, M2) macrophage polarization [5]. Recent studies on osteoclasts, the cells responsible for bone and joint destruction in autoimmune diseases, showed that autophagy-related proteins 5 and 7 (ATG5 and ATG7) deficiency leads to defective ruffled border formation and osteoclast function in both in vitro and in vivo assays [6]. Moreover, mice deficient in autophagy-related protein ATG7 were protected from TNF-mediated joint destruction in experimental arthritis [7]. Mutations in autophagy-related proteins have been associated with autoimmune diseases such as systemic lupus erythematosus [8] and rheumatoid arthritis [9]. WDFY3 is a master regulator of selective autophagy, which can work in concert with adaptor protein SQSTM1 (p62) to recruit and degrade ubiquitinated protein aggregates [10].
During the process of ubiquitinated protein aggregates sequestration, SQSTM1 (p62) works in concert with WDFY3, which is tethered to autophagosomal membranes [11]. Upon the autophagosome formation, cytosolic LC3-I converts into membrane-bound LC3-II. Therefore, the LC3-II to LC3-I ratio has been used as a marker correlated to autophagosome production [12, 13]. WDFY3 can also form a complex with SQSTM1 and TNF receptor associated factor 6 (TRAF6) during midbody ring degradation by selective autophagy [14]. SQSTM1 has indispensable roles in osteoclast differentiation since SQSTM1 deficiency leads to defect osteoclast function in vitro and osteopetrosis phenotype in vivo [15]. Specifically, SQSTM1 acts as a bridge between receptor activator of NF-κB ligand (RANKL)/RANK/TRAF6 mediated NF-κB signaling [16]. Mutations of SQSTM1 at ubiquitin-associated domain lead to increased osteoclast differentiation and function that is linked to Paget’s disease of bone, a skeletal disorder characterized by focal increased bone remodeling and abnormal bone structure formation [17].
Although the WDFY3/SQSTM1 interaction is important in autophagy, WDFY3 and SQSTM1 have also been associated with synovial fibroblasts and osteoclasts, implicating new roles in rheumatoid arthritis pathology [18, 19]. The functional interaction between SQSTM1 and WDFY3 has been clearly documented in human osteoclasts [19], and we recently showed a novel role of WDFY3 in RANKL-induced osteoclastogenesis in the absence of inflammation [20]. To investigate the role of WDFY3 in arthritis we employed the K/BxN serum transfer-induced arthritis model, where anti-glucose-6-phosphate isomerase autoantibodies induce joint specific inflammation that closely resembles the rheumatoid arthritis pathologies in humans [21]. Our data show a new role of WDFY3 protein in the protection of autoimmune arthritis, which is independent of starvation-induced autophagy.
2. Methods
2.1 Antibodies and reagents
All cell incubations were performed in culture medium consisting of αMEM with 2mM L-glutamine, 10% heat-inactivated FBS, 100 IU/ml Penicillin and 100 IU/ml Streptomycin (Life Technologies, 10007D). Mouse soluble RANKL (R&D Systems, 462TR) and M-CSF ELISA (R&D Systems, DY416), were used for in vitro experiments. CMG14-12 (CMG) media was generated as described before [22]. Anti-Wdfy3 (Abnova, clone 2F12), anti-WDFY3 (Novus, NBP1-03332), anti-SQSTM1 (Progen, GP62-C), LC3 antibody (Novus, NB100-2220), anti-β-actin antibody (Cell Signaling, 4970), 800 or 680 secondary antibodies (Li-Cor) were used for in vitro experiments. Cyto-ID kit (Enzo biochem, ENZ-51031) was used for autophagosome staining. LysoTracker DND-99 was used for lysosome staining (Life Technologies, L-7528).
2.2 Mice and bone cell culture
Animal experiments were conducted in accordance with the protocol approved by the Institutional Animal Care and Use Committee of the University of California, Davis. Eight to twelve weeks old C57BL/6 (The Jackson Laboratory), Wdfy3 loxP/loxP-LysM-Cre+, Wdfy3 loxP/loxP animals [23] were sacrificed to extract bone marrow from both rear femurs and tibias bones. Bone marrow cells were cultured with 1:20 (v/v) CMG media [22] in absence or presence of 30 ng/mL RANKL. Media and cytokines were replenished every other day.
2.3 K/BxN serum-transfer arthritis model
Arthritis was induced in 8-week-old male and female mice by i.v. injection of 200 µl of pooled serum from K/BxN mice. Disease severity score was measured as previously described [21]. Briefly, mice were scored for paw swelling, 1 indicates mild swelling of the ankle insufficient to reverse the normal V shape of the foot; 2 indicates swelling sufficient to make the ankle and midfoot approximately equal in thickness to the forefoot; 3 indicates the reversal of the normal V shape of the foot. Every two days, disease severity score was recorded, and ankle thickness was measured using digital calipers [21].
2.4 Immunofluorescence staining
Osteoclasts grown on coverslip were imaged by Nikon C1 confocal microscopy. Cells were fixed in 4% paraformaldehyde for 10 minutes and were permeabilized in 0.2% Triton X-100 and blocked with 10% normal donkey serum buffer in PBS for 2 hours at room temperature. All antibodies were diluted in 5% normal donkey serum buffer in PBS. Primary antibodies were applied on samples overnight at 4°C. Secondary antibody AF-488 donkey and anti-guinea pig Ig or AF-594 donkey anti-rabbit Ig antibodies were added to samples for one hour incubation at room temperature. TRITC conjugated phalloidin was used for F-actin staining for 30 minutes incubation at room temperature. Coverslips were then mounted in mounting media with DAPI (Vector Laboratories, H1200) to stain nuclei.
2.5 Co-Immunoprecipitation
Osteoclast-like cells cultured from 8–12 weeks old wild type mice were lysed in lysis buffer consisting of 50 mM Tris-HCl pH 7.4, 150 mM NaCl, 1mM EDTA, 1%v/v Triton X-100 containing protease inhibitor cocktail tablet (Roche, 4693124001). The clear lysate was incubated with anti-Wdfy3 antibody overnight and was then co-immunoprecipitated by dynabeads protein A immunoprecipitation kit (Life technologies, 10006D).
2.6 Western Blotting
Bone marrow-derived macrophages and osteoclast-like cells starved in serum-free medium were stimulated with RANKL (100 ng/ml) or M-CSF (100 ng/ml) and lysed at indicated time points. Protein lysates obtained from cell cultures were run on a Nu-Page 3–8% Tris-Acetate gel or 12% Bis-Tris gels (Invitrogen). Proteins were transferred to PVDF membranes and blocked in Odyssey blocking buffer. Membranes were incubated with anti-Wdfy3 primary antibody diluted in Odyssey blocking buffer containing 0.1% Tween-20, overnight at 4°C. After washing, we incubated with secondary antibody (Li-Cor) in blocking buffer containing 0.1% Tween-20 and 0.02% SDS, washed, and imaged on the Li-Cor Odyssey scanner. The process was repeated with β-actin as loading control. Signal intensity relative to background was determined using Li-Cor Image Studio software.
2.7 Statistical analyses
Statistical significance was determined using Student’s t-test and p-values lower than 0.05 were considered significant.
3. Results
3.1 WDFY3-cKO mice show reduced disease severity in K/BxN serum-transfer induced arthritis
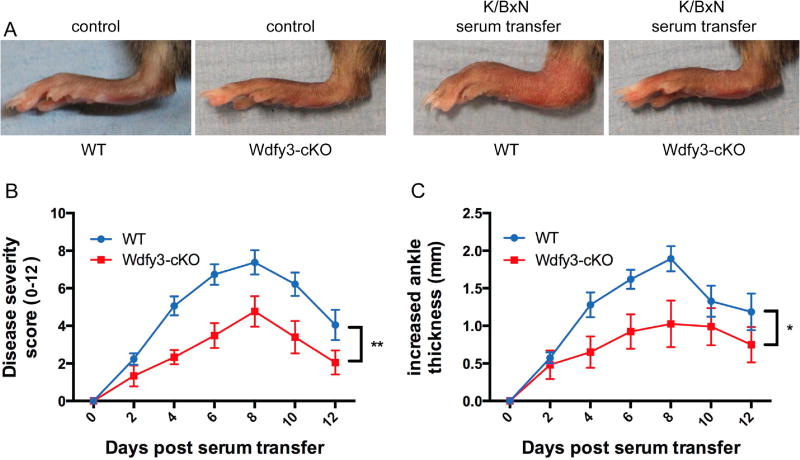
Since reduced WDFY3 expression has been associated with rheumatoid arthritis [18], we used the K/BxN serum-transfer model to determine whether loss of WDFY3 in myeloid cells affected the pathogenesis of inflammatory arthritis. We injected pooled K/BxN serum into Wdfy3-cKO and wild type littermates at day 0 and record disease severity scores and ankle thickness every two days throughout the disease course. We observed reduced ankle swelling in Wdfy3-cKO at day eight post-serum transfer compared to wild type littermates (Fig. 1A). Wdfy3-cKO mice displayed significantly reduced arthritis severity as evidence by disease severity score (Fig. 1B), and measurements of ankle swelling (Fig. 1C).
Figure 1.
Reduced disease severity in K/BxN serum-transfer arthritis in Wdfy3-cKO mice. Eight weeks old Wdfy3-cKO mice and wild type littermates were injected with 200 µl pooled K/BxN serum. (A) Representative pictures of swollen paws derived from wild type or Wdfy3-cKO mice at day 8 post serum transfer. (B) Diseases severity score and (C) increased ankle thickness of the K/BxN serum transferred animals (n=6 for each group). The results were pooled from two independent experiments. Error bars represent mean ± SEM. 2-way ANOVA is used for statistical analysis in B, C. *p ≤ 0.05, **p ≤ 0.01.
3.2 Macrophages derived from WDFY3-cKO mice exhibit normal autophagosome formation
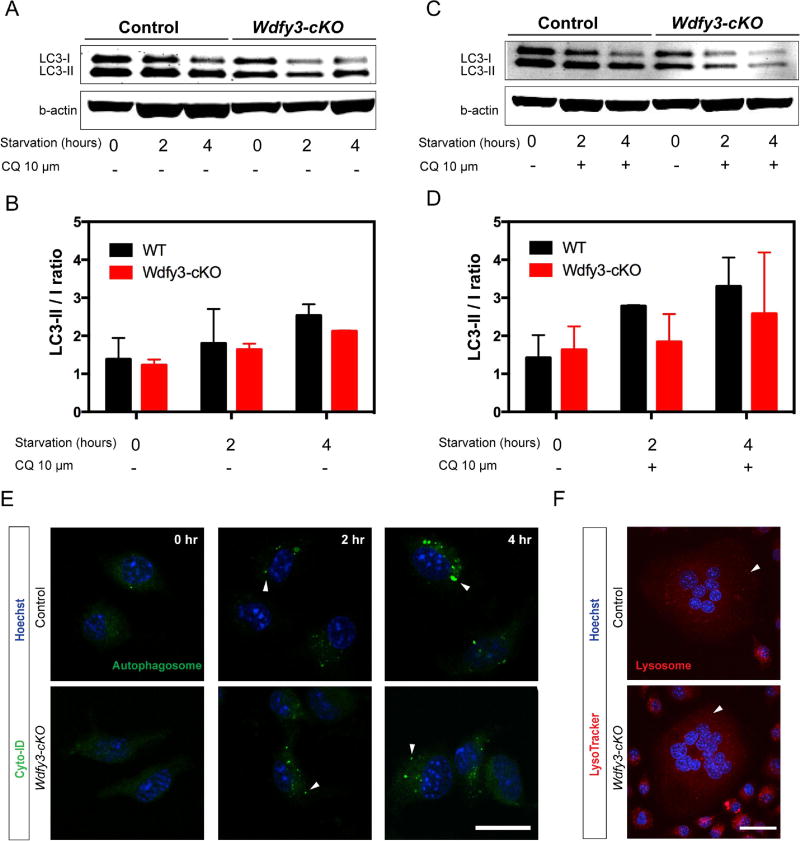
Since WDFY3 is involved in the autophagy pathway, we next investigated whether loss of WDFY3 affects starvation-induced autophagy. Western blot analysis of total cell lysates derived from wild type and WDFY3 deficient macrophages under starvation for 2 or 4 hours showed no significant differences of LC3-II to LC3-I ratio, an indicator of maturation of autophagosomes (Fig. 2A and B). Blockade of lysosomes and autophagosomes fusion with chloroquine again showed no significant difference in autophagy flux between wild type and WDFY3 deficient macrophages (Fig. 2C and 2D). We also used commercial available cationic amphiphilic tracer (CAT) dye to visualize autophagosomes in wild type and WDFY3 deficient macrophages with fluorescent microscopy and detected no differences in autophagosome formation (Fig. 2E). Furthermore, we observed no difference in lysosome formation using LysoTracker dye and fluorescent microscopy in both wild type and WDFY3 deficient macrophages, which suggested WDFY3 deficiency did not influence starvation-induced in macrophages, and lysosomes formation in RANKL-stimulated TRAP+ multinucleated giant cells (osteoclasts) (Fig. 2F).
Figure 2.
WDFY3 deficient macrophages demonstrate no difference in starvation-induced autophagy, lysosome and autophagosome formation. (A, B) Western blot analysis of total cell lysates from wild type and WDFY3 deficient bone marrow derived macrophages (pre-osteoclasts) starved for 0, 2 and 4 hours in the absence or (C, D) in the presence of 10µM chloroquine. (E) Immunofluorescent photomicrographs of wild type or WDFY3 deficient macrophages in serum free HBSS for 2 to 4 hours with 10 µM chloroquine treatment and stained with Cyto-ID kit to visualized autophagosomes (green) and nuclei (blue). (White arrowheads indicate autophagosomes). (F) Multinucleated giant cells cultured from wild type or Wdfy3-cKO mice were stained with LysoTracker DND-99 (red) and Hoechst (blue) (White arrowheads indicate lysosomes. Scale bars represent 20 µm in E and 30 µm in F.
3.3 WDFY3 interacts with SQSTM1 in macrophages and multinucleated giant cells
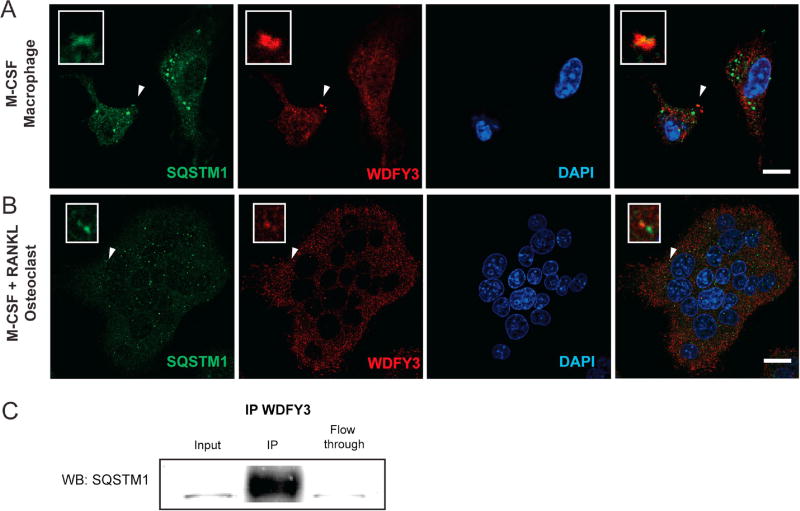
To examine the interaction between WDFY3 and SQSTM1, we performed immunostaining of WDFY3 and SQSTM1 in mouse bone marrow-derived macrophages and observed SQSTM1 puncta co-localized with WDFY3 (Fig. 3A). We then further differentiated the bone marrow-derived macrophages, in the presence of RANKL into TRAP+ multinucleated giant cells/osteoclast (80–100µm) and also observed the presence of SQSTM1 puncta co-localized with WDFY3 in osteoclasts (Fig. 3B). We further confirmed the interaction between WDFY3 and SQSTM1 by co-immunoprecipitation in total cell lysates derived from wild type osteoclasts using anti-WDFY3 antibody and immunoblotted with SQSTM1 (Fig. 3C).
Figure 3.
WDFY3 interacts with SQSTM1 in macrophages and multinucleated giant cells. (A) Immunofluorescent photomicrographs of macrophages (pre-osteoclasts) or (B) multinucleated giant cells (osteoclasts) were stained with anti-SQSTM1 (green), anti-WDFY3 (red), and DAPI (blue). (White arrowheads indicate WDFY3 and SQSTM1 co-localization). (C) Total cell lysates from osteoclast cultures co-immunoprecipitated with anti-WDFY3 antibodies and probed with SQSTM1 antibodies. Scale bars represent 10 µm in A, 20 µm in B.
3.4 Macrophages and osteoclasts derived from WDFY3-cKO mice show decreased level of SQSTM1
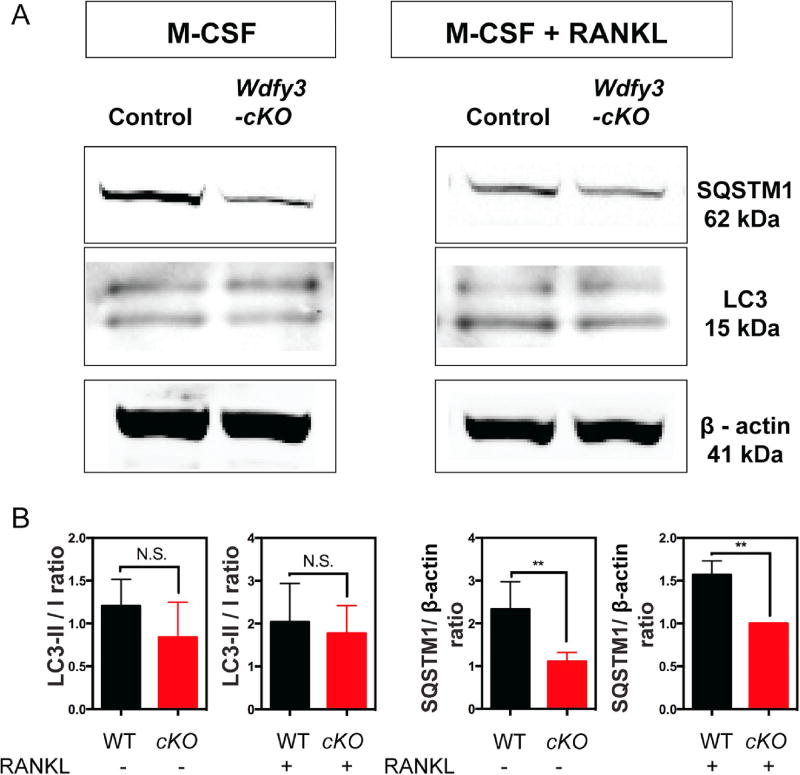
We then examined the adaptor protein SQSTM1, which plays multiple roles in both autophagy and RANKL signaling pathway in macrophages. Western blot analysis of total cell lysates from wild type and WDFY3 deficient macrophages treated in the presence or absence of RANKL showed no significant difference of autophagosome formation marker, LC3 but showed a reduced expression of SQSTM1 in WDFY3 deficient macrophages in the presence or absence of RANKL (Fig. 4A). We observed no difference when we quantified the LC3-II to LC3-I ratio and reduced SQSTM1 to β-actin ratio in WDFY3 deficient cells compared to wild type cells (Fig. 4B).
Figure 4.
Loss of WDFY3 leads to decrease level of SQSTM1 in macrophages. (A) Western blot analysis of total cell lysates from macrophages and multinucleated giant cells culture from wild type or Wdfy3-cKO mice. (B) Quantification of LC3-II/I ratio and SQSTM1 to β-actin ratio. Student t-test is used for statistical analysis in B. **p ≤ 0.01.
4. Discussion
WDFY3 together with WDFY1, WDFY2, WDFY4 belongs to the WDFY protein family. While all WDFY1, 2, 3 and 4 contain multiple WD40 repeats, their function is further diversified by the presence of an FYVE domain, in WDFY1, 2, 3, which is associated with vesicle trafficking. WDFY1 and WDFY2 are medium size proteins (around 400 amino acids) and have been associated with TLRs signaling and endocytosis respectively [24] and [25]. WDFY3 and WDFY4 are large proteins (>3000 amino acids) and contain PH and BEACH domains. WDFY4 has been associated with rheumatoid arthritis [26] and systemic lupus erythematosus [8] pathology in human genome-wide association studies (GWAS) studies, and WDFY3 has been detected in RA synovial fibroblasts [18] and osteoclasts [19]. Recently we showed that WDFY3 interacts with TRAF6 in RANKL-induced osteoclastogenesis in the absence of inflammation [20].
Herein, we investigated the role of WDFY3 in arthritis using the K/BxN serum transfer arthritis model. The deletion efficiency of WDFY3 in macrophages and osteoclasts derived from Wdfy3-cKO mice has been previously described [20]. We observed reduced joint swelling in Wdfy3-cKO mice compared to wild type littermates with serum transfer-induced arthritis suggesting that loss of WDFY3 in myeloid cells leads to a protective phenotype in arthritis. The protective effect from arthritis in Wdfy3-cKO mice was interesting since we previously observed increased NF-κB activation in RANKL induced osteoclastogenesis. We anticipated increased NF-κB activation under inflammatory conditions in the K/BxN serum transfer arthritis model. One plausible explanation for the protective phenotype in Wdfy3-cKO mice is that WDFY3 may play different roles in different types of myeloid cells such as neutrophils and mast cells where loss of WDFY3 in these myeloid cells may fail to initiate arthritis pathology due to inhibition of the secretory pathways. The protective effect from arthritis in Wdfy3-cKO mice was unlikely due to apoptosis of macrophages or osteoclasts as WDFY3 deletion in macrophages and osteoclasts has minimal effect on their viability [20]. Reduced expression of WDFY3 and the formation of SQSTM1-positive protein aggregates promote cell death in rheumatoid arthritis synovial fibroblasts (RASF) under severe ER stress [18]. Therefore enhanced apoptosis of inflammatory cells other than macrophages in K/BxN serum-transfer Wdfy3-cKO mice remains an additional possibility for the reduced pathology observed.
We then examined whether WDFY3 deficient macrophages had a defect in autophagy pathway and therefore lead to protective function in inflammatory arthritis in mice. In ATG5 and ATG7 deficient myeloid cells, autophagosome formation is impaired evidenced by the accumulation of LC3-I and absence of LC3-II [6]. However, WDFY3 deficiency in bone marrow-derived macrophages does not affect starvation-induced autophagy as evidenced by Western blot and immunofluorescence microscopy. These findings are in agreement with other studies that show WDFY3 is dispensable for starvation-induced autophagy [11]. WDFY3 targets specifically ubiquitinated protein aggregates in selective autophagy and has not been shown to play a role in other type of autophagy pathways such as LC3-associated phagocytosis [27].
WDFY3 interaction with SQSTM1 has been clearly documented in human multinucleated giant cells [19]. In agreement with the previous study, we also observed WDFY3 and SQSTM1 interaction in macrophages (pre-osteoclasts) and multinucleated giant cells by immunofluorescence microscopy and co-immunoprecipitation experiments. SQSTM1 has been shown to promote RANKL signaling, and complete ablation of SQSTM1 in vivo leads to dysfunctional osteoclastogenesis with increased bone mass [15]. Additionally, mutations of SQSTM1 that disrupt its C-terminal ubiquitin association (UBA) domain caused enhanced RANKL signaling, osteoclast differentiation, and bone resorption, which correlated to Paget’s disease of bone in human [28–30]. In this paper, we show that SQSTM1 interacts with WDFY3, a protein that we previously showed to modulate RANKL-induced osteoclastogenesis in vivo and in vitro [20]. Our new data show that although the interaction of SQSTM1 with WDFY3 is critical for bone remodeling, this interaction may play different roles under non-inflammatory and inflammatory condition as we observed increased osteoclastogenesis but reduced disease severity in inflammatory arthritis in our animal models. Accordingly, we observed a down-regulation of SQSTM1 in WDFY3 deficient macrophages, which may partly contribute to the protective phenotype in K/BxN serum-transfer Wdfy3-cKO mice. The reduced expression of SQSTM1 correlates with reduced inflammation as published observations have shown silencing SQSTM1 reduces inflammatory responses in HaCaT cells (keratinocyte cell line) [31]. Interestingly, we recently showed that Wdfy3-cKO mice are also protective in a mouse model of psoriasis and have reduced epidermal hyperplasia associated with Munro’s microabsecess; specifically, WDFY3 deficient neutrophils show reduced NETosis, neutrophil elastase, and ROS signaling but show no difference in neutrophil expansion, circulation and survival [32]. In conclusion, WDFY3 deficiency ameliorates disease severity in the serum transfer-induced arthritis animal model. Further experiments are needed to further address WDFY3 roles in the specific myeloid cell types to uncover the mechanisms of WDFY3 in arthritis pathologies.
Loss of WDFY3 ameliorates disease severity in an arthritis animal model.
WDFY3 is not essential in starvation-induced autophagy in myeloid cells.
Loss of WDFY3 leads to decreased level of SQSTM1 in myeloid cells.
Acknowledgments
This work was partly supported by NIH/NIAMS-R01AR062173 and SHC 85700 grants to IEA, and by the UC Davis, graduate group in immunology fellowship to DW. We thank Drs. Bottini and Stanford (LJI) for the gift of K/BxN serum.
Abbreviations
- ATG5 and ATG7
Autophagy-related proteins 5 and 7
- BEACH
Beige and Chediak-Higashi
- co-IP
co-immunoprecipitation
- F-actin
filamentous-actin
- M-CSF
Macrophage colony-stimulating factor
- MNCs
multinucleated cells
- PI3P
phosphatidylinositol 3-phosphate
- PH
Pleckstrin homology
- RANKL
receptor activator of NF-κB ligand
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest
None of the authors has any potential financial conflict of interest related to this manuscript.
References
- 1.Schett G, Gravallese E. Bone erosion in rheumatoid arthritis: mechanisms, diagnosis and treatment. Nature reviews. Rheumatology. 2012;8:656–664. doi: 10.1038/nrrheum.2012.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Deretic V, Saitoh T, Akira S. Autophagy in infection, inflammation and immunity. Nature reviews. Immunology. 2013;13:722–737. doi: 10.1038/nri3532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gutierrez MG, Master SS, Singh SB, Taylor GA, Colombo MI, Deretic V. Autophagy is a defense mechanism inhibiting BCG and Mycobacterium tuberculosis survival in infected macrophages. Cell. 2004;119:753–766. doi: 10.1016/j.cell.2004.11.038. [DOI] [PubMed] [Google Scholar]
- 4.Harris J, Hartman M, Roche C, Zeng SG, O'Shea A, Sharp FA, Lambe EM, Creagh EM, Golenbock DT, Tschopp J, Kornfeld H, Fitzgerald KA, Lavelle EC. Autophagy controls IL-1beta secretion by targeting pro-IL-1beta for degradation. The Journal of biological chemistry. 2011;286:9587–9597. doi: 10.1074/jbc.M110.202911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu K, Zhao E, Ilyas G, Lalazar G, Lin Y, Haseeb M, Tanaka KE, Czaja MJ. Impaired macrophage autophagy increases the immune response in obese mice by promoting proinflammatory macrophage polarization. Autophagy. 2015;11:271–284. doi: 10.1080/15548627.2015.1009787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.DeSelm CJ, Miller BC, Zou W, Beatty WL, van Meel E, Takahata Y, Klumperman J, Tooze SA, Teitelbaum SL, Virgin HW. Autophagy proteins regulate the secretory component of osteoclastic bone resorption. Developmental cell. 2011;21:966–974. doi: 10.1016/j.devcel.2011.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lin NY, Beyer C, Giessl A, Kireva T, Scholtysek C, Uderhardt S, Munoz LE, Dees C, Distler A, Wirtz S, Kronke G, Spencer B, Distler O, Schett G, Distler JH. Autophagy regulates TNFalpha-mediated joint destruction in experimental arthritis. Annals of the rheumatic diseases. 2012 doi: 10.1136/annrheumdis-2012-201671. [DOI] [PubMed] [Google Scholar]
- 8.Han JW, Zheng HF, Cui Y, Sun LD, Ye DQ, Hu Z, Xu JH, Cai ZM, Huang W, Zhao GP, Xie HF, Fang H, Lu QJ, Xu JH, Li XP, Pan YF, Deng DQ, Zeng FQ, Ye ZZ, Zhang XY, Wang QW, Hao F, Ma L, Zuo XB, Zhou FS, Du WH, Cheng YL, Yang JQ, Shen SK, Li J, Sheng YJ, Zuo XX, Zhu WF, Gao F, Zhang PL, Guo Q, Li B, Gao M, Xiao FL, Quan C, Zhang C, Zhang Z, Zhu KJ, Li Y, Hu DY, Lu WS, Huang JL, Liu SX, Li H, Ren YQ, Wang ZX, Yang CJ, Wang PG, Zhou WM, Lv YM, Zhang AP, Zhang SQ, Lin D, Li Y, Low HQ, Shen M, Zhai ZF, Wang Y, Zhang FY, Yang S, Liu JJ, Zhang XJ. Genome-wide association study in a Chinese Han population identifies nine new susceptibility loci for systemic lupus erythematosus. Nature genetics. 2009;41:1234–1237. doi: 10.1038/ng.472. [DOI] [PubMed] [Google Scholar]
- 9.Orozco G, Eyre S, Hinks A, Bowes J, Morgan AW, Wilson AG, Wordsworth P, Steer S, Hocking L, consortium U, Thomson W, Worthington J, Barton A. Study of the common genetic background for rheumatoid arthritis and systemic lupus erythematosus. Annals of the rheumatic diseases. 2011;70:463–468. doi: 10.1136/ard.2010.137174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Simonsen A, Birkeland HC, Gillooly DJ, Mizushima N, Kuma A, Yoshimori T, Slagsvold T, Brech A, Stenmark H. Alfy, a novel FYVE-domain-containing protein associated with protein granules and autophagic membranes. Journal of cell science. 2004;117:4239–4251. doi: 10.1242/jcs.01287. [DOI] [PubMed] [Google Scholar]
- 11.Filimonenko M, Isakson P, Finley KD, Anderson M, Jeong H, Melia TJ, Bartlett BJ, Myers KM, Birkeland HC, Lamark T, Krainc D, Brech A, Stenmark H, Simonsen A, Yamamoto A. The selective macroautophagic degradation of aggregated proteins requires the PI3P-binding protein Alfy. Molecular cell. 2010;38:265–279. doi: 10.1016/j.molcel.2010.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, Kominami E, Ohsumi Y, Yoshimori T. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. The EMBO journal. 2000;19:5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Klionsky DJ, Abdelmohsen K, Abe A, Abedin MJ, Abeliovich H, Acevedo Arozena A, Adachi H, Adams CM, Adams PD, Adeli K, Adhihetty PJ, Adler SG, Agam G, Agarwal R, Aghi MK, Agnello M, Agostinis P, Aguilar PV, Aguirre-Ghiso J, Airoldi EM, Ait-Si-Ali S, Akematsu T, Akporiaye ET, Al-Rubeai M, Albaiceta GM, Albanese C, Albani D, Albert ML, Aldudo J, Algul H, Alirezaei M, Alloza I, Almasan A, Almonte-Beceril M, Alnemri ES, Alonso C, Altan-Bonnet N, Altieri DC, Alvarez S, Alvarez-Erviti L, Alves S, Amadoro G, Amano A, Amantini C, Ambrosio S, Amelio I, Amer AO, Amessou M, Amon A, An Z, Anania FA, Andersen SU, Andley UP, Andreadi CK, Andrieu-Abadie N, Anel A, Ann DK, Anoopkumar-Dukie S, Antonioli M, Aoki H, Apostolova N, Aquila S, Aquilano K, Araki K, Arama E, Aranda A, Araya J, Arcaro A, Arias E, Arimoto H, Ariosa AR, Armstrong JL, Arnould T, Arsov I, Asanuma K, Askanas V, Asselin E, Atarashi R, Atherton SS, Atkin JD, Attardi LD, Auberger P, Auburger G, Aurelian L, Autelli R, Avagliano L, Avantaggiati ML, Avrahami L, Awale S, Azad N, Bachetti T, Backer JM, Bae DH, Bae JS, Bae ON, Bae SH, Baehrecke EH, Baek SH, Baghdiguian S, Bagniewska-Zadworna A, Bai H, Bai J, Bai XY, Bailly Y, Balaji KN, Balduini W, Ballabio A, Balzan R, Banerjee R, Banhegyi G, Bao H, Barbeau B, Barrachina MD, Barreiro E, Bartel B, Bartolome A, Bassham DC, Bassi MT, Bast RC, Jr, Basu A, Batista MT, Batoko H, Battino M, Bauckman K, Baumgarner BL, Bayer KU, Beale R, Beaulieu JF, Beck GR, Jr, Becker C, Beckham JD, Bedard PA, Bednarski PJ, Begley TJ, Behl C, Behrends C, Behrens GM, Behrns KE, Bejarano E, Belaid A, Belleudi F, Benard G, Berchem G, Bergamaschi D, Bergami M, Berkhout B, Berliocchi L, Bernard A, Bernard M, Bernassola F, Bertolotti A, Bess AS, Besteiro S, Bettuzzi S, Bhalla S, Bhattacharyya S, Bhutia SK, Biagosch C, Bianchi MW, Biard-Piechaczyk M, Billes V, Bincoletto C, Bingol B, Bird SW, Bitoun M, Bjedov I, Blackstone C, Blanc L, Blanco GA, Blomhoff HK, Boada-Romero E, Bockler S, Boes M, Boesze-Battaglia K, Boise LH, Bolino A, Boman A, Bonaldo P, Bordi M, Bosch J, Botana LM, Botti J, Bou G, Bouche M, Bouchecareilh M, Boucher MJ, Boulton ME, Bouret SG, Boya P, Boyer-Guittaut M, Bozhkov PV, Brady N, Braga VM, Brancolini C, Braus GH, Bravo-San Pedro JM, Brennan LA, Bresnick EH, Brest P, Bridges D, Bringer MA, Brini M, Brito GC, Brodin B, Brookes PS, Brown EJ, Brown K, Broxmeyer HE, Bruhat A, Brum PC, Brumell JH, Brunetti-Pierri N, Bryson-Richardson RJ, Buch S, Buchan AM, Budak H, Bulavin DV, Bultman SJ, Bultynck G, Bumbasirevic V, Burelle Y, Burke RE, Burmeister M, Butikofer P, Caberlotto L, Cadwell K, Cahova M, Cai D, Cai J, Cai Q, Calatayud S, Camougrand N, Campanella M, Campbell GR, Campbell M, Campello S, Candau R, Caniggia I, Cantoni L, Cao L, Caplan AB, Caraglia M, Cardinali C, Cardoso SM, Carew JS, Carleton LA, Carlin CR, Carloni S, Carlsson SR, Carmona-Gutierrez D, Carneiro LA, Carnevali O, Carra S, Carrier A, Carroll B, Casas C, Casas J, Cassinelli G, Castets P, Castro-Obregon S, Cavallini G, Ceccherini I, Cecconi F, Cederbaum AI, Cena V, Cenci S, Cerella C, Cervia D, Cetrullo S, Chaachouay H, Chae HJ, Chagin AS, Chai CY, Chakrabarti G, Chamilos G, Chan EY, Chan MT, Chandra D, Chandra P, Chang CP, Chang RC, Chang TY, Chatham JC, Chatterjee S, Chauhan S, Che Y, Cheetham ME, Cheluvappa R, Chen CJ, Chen G, Chen GC, Chen G, Chen H, Chen JW, Chen JK, Chen M, Chen M, Chen P, Chen Q, Chen Q, Chen SD, Chen S, Chen SS, Chen W, Chen WJ, Chen WQ, Chen W, Chen X, Chen YH, Chen YG, Chen Y, Chen Y, Chen Y, Chen YJ, Chen YQ, Chen Y, Chen Z, Chen Z, Cheng A, Cheng CH, Cheng H, Cheong H, Cherry S, Chesney J, Cheung CH, Chevet E, Chi HC, Chi SG, Chiacchiera F, Chiang HL, Chiarelli R, Chiariello M, Chieppa M, Chin LS, Chiong M, Chiu GN, Cho DH, Cho SG, Cho WC, Cho YY, Cho YS, Choi AM, Choi EJ, Choi EK, Choi J, Choi ME, Choi SI, Chou TF, Chouaib S, Choubey D, Choubey V, Chow KC, Chowdhury K, Chu CT, Chuang TH, Chun T, Chung H, Chung T, Chung YL, Chwae YJ, Cianfanelli V, Ciarcia R, Ciechomska IA, Ciriolo MR, Cirone M, Claerhout S, Clague MJ, Claria J, Clarke PG, Clarke R, Clementi E, Cleyrat C, Cnop M, Coccia EM, Cocco T, Codogno P, Coers J, Cohen EE, Colecchia D, Coletto L, Coll NS, Colucci-Guyon E, Comincini S, Condello M, Cook KL, Coombs GH, Cooper CD, Cooper JM, Coppens I, Corasaniti MT, Corazzari M, Corbalan R, Corcelle-Termeau E, Cordero MD, Corral-Ramos C, Corti O, Cossarizza A, Costelli P, Costes S, Cotman SL, Coto-Montes A, Cottet S, Couve E, Covey LR, Cowart LA, Cox JS, Coxon FP, Coyne CB, Cragg MS, Craven RJ, Crepaldi T, Crespo JL, Criollo A, Crippa V, Cruz MT, Cuervo AM, Cuezva JM, Cui T, Cutillas PR, Czaja MJ, Czyzyk-Krzeska MF, Dagda RK, Dahmen U, Dai C, Dai W, Dai Y, Dalby KN, Dalla Valle L, Dalmasso G, D'Amelio M, Damme M, Darfeuille-Michaud A, Dargemont C, Darley-Usmar VM, Dasarathy S, Dasgupta B, Dash S, Dass CR, Davey HM, Davids LM, Davila D, Davis RJ, Dawson TM, Dawson VL, Daza P, de Belleroche J, de Figueiredo P, de Figueiredo RC, de la Fuente J, De Martino L, De Matteis A, De Meyer GR, De Milito A, De Santi M, de Souza W, De Tata V, De Zio D, Debnath J, Dechant R, Decuypere JP, Deegan S, Dehay B, Del Bello B, Del Re DP, Delage-Mourroux R, Delbridge LM, Deldicque L, Delorme-Axford E, Deng Y, Dengjel J, Denizot M, Dent P, Der CJ, Deretic V, Derrien B, Deutsch E, Devarenne TP, Devenish RJ, Di Bartolomeo S, Di Daniele N, Di Domenico F, Di Nardo A, Di Paola S, Di Pietro A, Di Renzo L, DiAntonio A, Diaz-Araya G, Diaz-Laviada I, Diaz-Meco MT, Diaz-Nido J, Dickey CA, Dickson RC, Diederich M, Digard P, Dikic I, Dinesh-Kumar SP, Ding C, Ding WX, Ding Z, Dini L, Distler JH, Diwan A, Djavaheri-Mergny M, Dmytruk K, Dobson RC, Doetsch V, Dokladny K, Dokudovskaya S, Donadelli M, Dong XC, Dong X, Dong Z, Donohue TM, Jr, Doran KS, D'Orazi G, Dorn GW, 2nd, Dosenko V, Dridi S, Drucker L, Du J, Du LL, Du L, du Toit A, Dua P, Duan L, Duann P, Dubey VK, Duchen MR, Duchosal MA, Duez H, Dugail I, Dumit VI, Duncan MC, Dunlop EA, Dunn WA, Jr, Dupont N, Dupuis L, Duran RV, Durcan TM, Duvezin-Caubet S, Duvvuri U, Eapen V, Ebrahimi-Fakhari D, Echard A, Eckhart L, Edelstein CL, Edinger AL, Eichinger L, Eisenberg T, Eisenberg-Lerner A, Eissa NT, El-Deiry WS, El-Khoury V, Elazar Z, Eldar-Finkelman H, Elliott CJ, Emanuele E, Emmenegger U, Engedal N, Engelbrecht AM, Engelender S, Enserink JM, Erdmann R, Erenpreisa J, Eri R, Eriksen JL, Erman A, Escalante R, Eskelinen EL, Espert L, Esteban-Martinez L, Evans TJ, Fabri M, Fabrias G, Fabrizi C, Facchiano A, Faergeman NJ, Faggioni A, Fairlie WD, Fan C, Fan D, Fan J, Fang S, Fanto M, Fanzani A, Farkas T, Faure M, Favier FB, Fearnhead H, Federici M, Fei E, Felizardo TC, Feng H, Feng Y, Feng Y, Ferguson TA, Fernandez AF, Fernandez-Barrena MG, Fernandez-Checa JC, Fernandez-Lopez A, Fernandez-Zapico ME, Feron O, Ferraro E, Ferreira-Halder CV, Fesus L, Feuer R, Fiesel FC, Filippi-Chiela EC, Filomeni G, Fimia GM, Fingert JH, Finkbeiner S, Finkel T, Fiorito F, Fisher PB, Flajolet M, Flamigni F, Florey O, Florio S, Floto RA, Folini M, Follo C, Fon EA, Fornai F, Fortunato F, Fraldi A, Franco R, Francois A, Francois A, Frankel LB, Fraser ID, Frey N, Freyssenet DG, Frezza C, Friedman SL, Frigo DE, Fu D, Fuentes JM, Fueyo J, Fujitani Y, Fujiwara Y, Fujiya M, Fukuda M, Fulda S, Fusco C, Gabryel B, Gaestel M, Gailly P, Gajewska M, Galadari S, Galili G, Galindo I, Galindo MF, Galliciotti G, Galluzzi L, Galluzzi L, Galy V, Gammoh N, Gandy S, Ganesan AK, Ganesan S, Ganley IG, Gannage M, Gao FB, Gao F, Gao JX, Garcia Nannig L, Garcia Vescovi E, Garcia-Macia M, Garcia-Ruiz C, Garg AD, Garg PK, Gargini R, Gassen NC, Gatica D, Gatti E, Gavard J, Gavathiotis E, Ge L, Ge P, Ge S, Gean PW, Gelmetti V, Genazzani AA, Geng J, Genschik P, Gerner L, Gestwicki JE, Gewirtz DA, Ghavami S, Ghigo E, Ghosh D, Giammarioli AM, Giampieri F, Giampietri C, Giatromanolaki A, Gibbings DJ, Gibellini L, Gibson SB, Ginet V, Giordano A, Giorgini F, Giovannetti E, Girardin SE, Gispert S, Giuliano S, Gladson CL, Glavic A, Gleave M, Godefroy N, Gogal RM, Jr, Gokulan K, Goldman GH, Goletti D, Goligorsky MS, Gomes AV, Gomes LC, Gomez H, Gomez-Manzano C, Gomez-Sanchez R, Goncalves DA, Goncu E, Gong Q, Gongora C, Gonzalez CB, Gonzalez-Alegre P, Gonzalez-Cabo P, Gonzalez-Polo RA, Goping IS, Gorbea C, Gorbunov NV, Goring DR, Gorman AM, Gorski SM, Goruppi S, Goto-Yamada S, Gotor C, Gottlieb RA, Gozes I, Gozuacik D, Graba Y, Graef M, Granato GE, Grant GD, Grant S, Gravina GL, Green DR, Greenhough A, Greenwood MT, Grimaldi B, Gros F, Grose C, Groulx JF, Gruber F, Grumati P, Grune T, Guan JL, Guan KL, Guerra B, Guillen C, Gulshan K, Gunst J, Guo C, Guo L, Guo M, Guo W, Guo XG, Gust AA, Gustafsson AB, Gutierrez E, Gutierrez MG, Gwak HS, Haas A, Haber JE, Hadano S, Hagedorn M, Hahn DR, Halayko AJ, Hamacher-Brady A, Hamada K, Hamai A, Hamann A, Hamasaki M, Hamer I, Hamid Q, Hammond EM, Han F, Han W, Handa JT, Hanover JA, Hansen M, Harada M, Harhaji-Trajkovic L, Harper JW, Harrath AH, Harris AL, Harris J, Hasler U, Hasselblatt P, Hasui K, Hawley RG, Hawley TS, He C, He CY, He F, He G, He RR, He XH, He YW, He YY, Heath JK, Hebert MJ, Heinzen RA, Helgason GV, Hensel M, Henske EP, Her C, Herman PK, Hernandez A, Hernandez C, Hernandez-Tiedra S, Hetz C, Hiesinger PR, Higaki K, Hilfiker S, Hill BG, Hill JA, Hill WD, Hino K, Hofius D, Hofman P, Hoglinger GU, Hohfeld J, Holz MK, Hong Y, Hood DA, Hoozemans JJ, Hoppe T, Hsu C, Hsu CY, Hsu LC, Hu D, Hu G, Hu HM, Hu H, Hu MC, Hu YC, Hu ZW, Hua F, Hua Y, Huang C, Huang HL, Huang KH, Huang KY, Huang S, Huang S, Huang WP, Huang YR, Huang Y, Huang Y, Huber TB, Huebbe P, Huh WK, Hulmi JJ, Hur GM, Hurley JH, Husak Z, Hussain SN, Hussain S, Hwang JJ, Hwang S, Hwang TI, Ichihara A, Imai Y, Imbriano C, Inomata M, Into T, Iovane V, Iovanna JL, Iozzo RV, Ip NY, Irazoqui JE, Iribarren P, Isaka Y, Isakovic AJ, Ischiropoulos H, Isenberg JS, Ishaq M, Ishida H, Ishii I, Ishmael JE, Isidoro C, Isobe KI, Isono E, Issazadeh-Navikas S, Itahana K, Itakura E, Ivanov AI, Iyer AK, Izquierdo JM, Izumi Y, Izzo V, Jaattela M, Jaber N, Jackson DJ, Jackson WT, Jacob TG, Jacques TS, Jagannath C, Jain A, Jana NR, Jang BK, Jani A, Janji B, Jannig PR, Jansson PJ, Jean S, Jendrach M, Jeon JH, Jessen N, Jeung EB, Jia K, Jia L, Jiang H, Jiang H, Jiang L, Jiang T, Jiang X, Jiang X, Jiang X, Jiang Y, Jiang Y, Jimenez A, Jin C, Jin H, Jin L, Jin M, Jin S, Jinwal UK, Jo EK, Johansen T, Johnson DE, Johnson GV, Johnson JD, Jonasch E, Jones C, Joosten LA, Jordan J, Joseph AM, Joseph B, Joubert AM, Ju D, Ju J, Juan HF, Juenemann K, Juhasz G, Jung HS, Jung JU, Jung YK, Jungbluth H, Justice MJ, Jutten B, Kaakoush NO, Kaarniranta K, Kaasik A, Kabuta T, Kaeffer B, Kagedal K, Kahana A, Kajimura S, Kakhlon O, Kalia M, Kalvakolanu DV, Kamada Y, Kambas K, Kaminskyy VO, Kampinga HH, Kandouz M, Kang C, Kang R, Kang TC, Kanki T, Kanneganti TD, Kanno H, Kanthasamy AG, Kantorow M, Kaparakis-Liaskos M, Kapuy O, Karantza V, Karim MR, Karmakar P, Kaser A, Kaushik S, Kawula T, Kaynar AM, Ke PY, Ke ZJ, Kehrl JH, Keller KE, Kemper JK, Kenworthy AK, Kepp O, Kern A, Kesari S, Kessel D, Ketteler R, Kettelhut ID, Khambu B, Khan MM, Khandelwal VK, Khare S, Kiang JG, Kiger AA, Kihara A, Kim AL, Kim CH, Kim DR, Kim DH, Kim EK, Kim HY, Kim HR, Kim JS, Kim JH, Kim JC, Kim JH, Kim KW, Kim MD, Kim MM, Kim PK, Kim SW, Kim SY, Kim YS, Kim Y, Kimchi A, Kimmelman AC, Kimura T, King JS, Kirkegaard K, Kirkin V, Kirshenbaum LA, Kishi S, Kitajima Y, Kitamoto K, Kitaoka Y, Kitazato K, Kley RA, Klimecki WT, Klinkenberg M, Klucken J, Knaevelsrud H, Knecht E, Knuppertz L, Ko JL, Kobayashi S, Koch JC, Koechlin-Ramonatxo C, Koenig U, Koh YH, Kohler K, Kohlwein SD, Koike M, Komatsu M, Kominami E, Kong D, Kong HJ, Konstantakou EG, Kopp BT, Korcsmaros T, Korhonen L, Korolchuk VI, Koshkina NV, Kou Y, Koukourakis MI, Koumenis C, Kovacs AL, Kovacs T, Kovacs WJ, Koya D, Kraft C, Krainc D, Kramer H, Kravic-Stevovic T, Krek W, Kretz-Remy C, Krick R, Krishnamurthy M, Kriston-Vizi J, Kroemer G, Kruer MC, Kruger R, Ktistakis NT, Kuchitsu K, Kuhn C, Kumar AP, Kumar A, Kumar A, Kumar D, Kumar D, Kumar R, Kumar S, Kundu M, Kung HJ, Kuno A, Kuo SH, Kuret J, Kurz T, Kwok T, Kwon TK, Kwon YT, Kyrmizi I, La Spada AR, Lafont F, Lahm T, Lakkaraju A, Lam T, Lamark T, Lancel S, Landowski TH, Lane DJ, Lane JD, Lanzi C, Lapaquette P, Lapierre LR, Laporte J, Laukkarinen J, Laurie GW, Lavandero S, Lavie L, LaVoie MJ, Law BY, Law HK, Law KB, Layfield R, Lazo PA, Le Cam L, Le Roch KG, Le Stunff H, Leardkamolkarn V, Lecuit M, Lee BH, Lee CH, Lee EF, Lee GM, Lee HJ, Lee H, Lee JK, Lee J, Lee JH, Lee JH, Lee M, Lee MS, Lee PJ, Lee SW, Lee SJ, Lee SJ, Lee SY, Lee SH, Lee SS, Lee SJ, Lee S, Lee YR, Lee YJ, Lee YH, Leeuwenburgh C, Lefort S, Legouis R, Lei J, Lei QY, Leib DA, Leibowitz G, Lekli I, Lemaire SD, Lemasters JJ, Lemberg MK, Lemoine A, Leng S, Lenz G, Lenzi P, Lerman LO, Lettieri Barbato D, Leu JI, Leung HY, Levine B, Lewis PA, Lezoualc'h F, Li C, Li F, Li FJ, Li J, Li K, Li L, Li M, Li M, Li Q, Li R, Li S, Li W, Li W, Li X, Li Y, Lian J, Liang C, Liang Q, Liao Y, Liberal J, Liberski PP, Lie P, Lieberman AP, Lim HJ, Lim KL, Lim K, Lima RT, Lin CS, Lin CF, Lin F, Lin F, Lin FC, Lin K, Lin KH, Lin PH, Lin T, Lin WW, Lin YS, Lin Y, Linden R, Lindholm D, Lindqvist LM, Lingor P, Linkermann A, Liotta LA, Lipinski MM, Lira VA, Lisanti MP, Liton PB, Liu B, Liu C, Liu CF, Liu F, Liu HJ, Liu J, Liu JJ, Liu JL, Liu K, Liu L, Liu L, Liu Q, Liu RY, Liu S, Liu S, Liu W, Liu XD, Liu X, Liu XH, Liu X, Liu X, Liu X, Liu Y, Liu Y, Liu Z, Liu Z, Liuzzi JP, Lizard G, Ljujic M, Lodhi IJ, Logue SE, Lokeshwar BL, Long YC, Lonial S, Loos B, Lopez-Otin C, Lopez-Vicario C, Lorente M, Lorenzi PL, Lorincz P, Los M, Lotze MT, Lovat PE, Lu B, Lu B, Lu J, Lu Q, Lu SM, Lu S, Lu Y, Luciano F, Luckhart S, Lucocq JM, Ludovico P, Lugea A, Lukacs NW, Lum JJ, Lund AH, Luo H, Luo J, Luo S, Luparello C, Lyons T, Ma J, Ma Y, Ma Y, Ma Z, Machado J, Machado-Santelli GM, Macian F, MacIntosh GC, MacKeigan JP, Macleod KF, MacMicking JD, MacMillan-Crow LA, Madeo F, Madesh M, Madrigal-Matute J, Maeda A, Maeda T, Maegawa G, Maellaro E, Maes H, Magarinos M, Maiese K, Maiti TK, Maiuri L, Maiuri MC, Maki CG, Malli R, Malorni W, Maloyan A, Mami-Chouaib F, Man N, Mancias JD, Mandelkow EM, Mandell MA, Manfredi AA, Manie SN, Manzoni C, Mao K, Mao Z, Mao ZW, Marambaud P, Marconi AM, Marelja Z, Marfe G, Margeta M, Margittai E, Mari M, Mariani FV, Marin C, Marinelli S, Marino G, Markovic I, Marquez R, Martelli AM, Martens S, Martin KR, Martin SJ, Martin S, Martin-Acebes MA, Martin-Sanz P, Martinand-Mari C, Martinet W, Martinez J, Martinez-Lopez N, Martinez-Outschoorn U, Martinez-Velazquez M, Martinez-Vicente M, Martins WK, Mashima H, Mastrianni JA, Matarese G, Matarrese P, Mateo R, Matoba S, Matsumoto N, Matsushita T, Matsuura A, Matsuzawa T, Mattson MP, Matus S, Maugeri N, Mauvezin C, Mayer A, Maysinger D, Mazzolini GD, McBrayer MK, McCall K, McCormick C, McInerney GM, McIver SC, McKenna S, McMahon JJ, McNeish IA, Mechta-Grigoriou F, Medema JP, Medina DL, Megyeri K, Mehrpour M, Mehta JL, Mei Y, Meier UC, Meijer AJ, Melendez A, Melino G, Melino S, de Melo EJ, Mena MA, Meneghini MD, Menendez JA, Menezes R, Meng L, Meng LH, Meng S, Menghini R, Menko AS, Menna-Barreto RF, Menon MB, Meraz-Rios MA, Merla G, Merlini L, Merlot AM, Meryk A, Meschini S, Meyer JN, Mi MT, Miao CY, Micale L, Michaeli S, Michiels C, Migliaccio AR, Mihailidou AS, Mijaljica D, Mikoshiba K, Milan E, Miller-Fleming L, Mills GB, Mills IG, Minakaki G, Minassian BA, Ming XF, Minibayeva F, Minina EA, Mintern JD, Minucci S, Miranda-Vizuete A, Mitchell CH, Miyamoto S, Miyazawa K, Mizushima N, Mnich K, Mograbi B, Mohseni S, Moita LF, Molinari M, Molinari M, Moller AB, Mollereau B, Mollinedo F, Mongillo M, Monick MM, Montagnaro S, Montell C, Moore DJ, Moore MN, Mora-Rodriguez R, Moreira PI, Morel E, Morelli MB, Moreno S, Morgan MJ, Moris A, Moriyasu Y, Morrison JL, Morrison LA, Morselli E, Moscat J, Moseley PL, Mostowy S, Motori E, Mottet D, Mottram JC, Moussa CE, Mpakou VE, Mukhtar H, Mulcahy Levy JM, Muller S, Munoz-Moreno R, Munoz-Pinedo C, Munz C, Murphy ME, Murray JT, Murthy A, Mysorekar IU, Nabi IR, Nabissi M, Nader GA, Nagahara Y, Nagai Y, Nagata K, Nagelkerke A, Nagy P, Naidu SR, Nair S, Nakano H, Nakatogawa H, Nanjundan M, Napolitano G, Naqvi NI, Nardacci R, Narendra DP, Narita M, Nascimbeni AC, Natarajan R, Navegantes LC, Nawrocki ST, Nazarko TY, Nazarko VY, Neill T, Neri LM, Netea MG, Netea-Maier RT, Neves BM, Ney PA, Nezis IP, Nguyen HT, Nguyen HP, Nicot AS, Nilsen H, Nilsson P, Nishimura M, Nishino I, Niso-Santano M, Niu H, Nixon RA, Njar VC, Noda T, Noegel AA, Nolte EM, Norberg E, Norga KK, Noureini SK, Notomi S, Notterpek L, Nowikovsky K, Nukina N, Nurnberger T, O'Donnell VB, O'Donovan T, O'Dwyer PJ, Oehme I, Oeste CL, Ogawa M, Ogretmen B, Ogura Y, Oh YJ, Ohmuraya M, Ohshima T, Ojha R, Okamoto K, Okazaki T, Oliver FJ, Ollinger K, Olsson S, Orban DP, Ordonez P, Orhon I, Orosz L, O'Rourke EJ, Orozco H, Ortega AL, Ortona E, Osellame LD, Oshima J, Oshima S, Osiewacz HD, Otomo T, Otsu K, Ou JJ, Outeiro TF, Ouyang DY, Ouyang H, Overholtzer M, Ozbun MA, Ozdinler PH, Ozpolat B, Pacelli C, Paganetti P, Page G, Pages G, Pagnini U, Pajak B, Pak SC, Pakos-Zebrucka K, Pakpour N, Palkova Z, Palladino F, Pallauf K, Pallet N, Palmieri M, Paludan SR, Palumbo C, Palumbo S, Pampliega O, Pan H, Pan W, Panaretakis T, Pandey A, Pantazopoulou A, Papackova Z, Papademetrio DL, Papassideri I, Papini A, Parajuli N, Pardo J, Parekh VV, Parenti G, Park JI, Park J, Park OK, Parker R, Parlato R, Parys JB, Parzych KR, Pasquet JM, Pasquier B, Pasumarthi KB, Patschan D, Patterson C, Pattingre S, Pattison S, Pause A, Pavenstadt H, Pavone F, Pedrozo Z, Pena FJ, Penalva MA, Pende M, Peng J, Penna F, Penninger JM, Pensalfini A, Pepe S, Pereira GJ, Pereira PC, Perez-de la Cruz V, Perez-Perez ME, Perez-Rodriguez D, Perez-Sala D, Perier C, Perl A, Perlmutter DH, Perrotta I, Pervaiz S, Pesonen M, Pessin JE, Peters GJ, Petersen M, Petrache I, Petrof BJ, Petrovski G, Phang JM, Piacentini M, Pierdominici M, Pierre P, Pierrefite-Carle V, Pietrocola F, Pimentel-Muinos FX, Pinar M, Pineda B, Pinkas-Kramarski R, Pinti M, Pinton P, Piperdi B, Piret JM, Platanias LC, Platta HW, Plowey ED, Poggeler S, Poirot M, Polcic P, Poletti A, Poon AH, Popelka H, Popova B, Poprawa I, Poulose SM, Poulton J, Powers SK, Powers T, Pozuelo-Rubio M, Prak K, Prange R, Prescott M, Priault M, Prince S, Proia RL, Proikas-Cezanne T, Prokisch H, Promponas VJ, Przyklenk K, Puertollano R, Pugazhenthi S, Puglielli L, Pujol A, Puyal J, Pyeon D, Qi X, Qian WB, Qin ZH, Qiu Y, Qu Z, Quadrilatero J, Quinn F, Raben N, Rabinowich H, Radogna F, Ragusa MJ, Rahmani M, Raina K, Ramanadham S, Ramesh R, Rami A, Randall-Demllo S, Randow F, Rao H, Rao VA, Rasmussen BB, Rasse TM, Ratovitski EA, Rautou PE, Ray SK, Razani B, Reed BH, Reggiori F, Rehm M, Reichert AS, Rein T, Reiner DJ, Reits E, Ren J, Ren X, Renna M, Reusch JE, Revuelta JL, Reyes L, Rezaie AR, Richards RI, Richardson DR, Richetta C, Riehle MA, Rihn BH, Rikihisa Y, Riley BE, Rimbach G, Rippo MR, Ritis K, Rizzi F, Rizzo E, Roach PJ, Robbins J, Roberge M, Roca G, Roccheri MC, Rocha S, Rodrigues CM, Rodriguez CI, de Cordoba SR, Rodriguez-Muela N, Roelofs J, Rogov VV, Rohn TT, Rohrer B, Romanelli D, Romani L, Romano PS, Roncero MI, Rosa JL, Rosello A, Rosen KV, Rosenstiel P, Rost-Roszkowska M, Roth KA, Roue G, Rouis M, Rouschop KM, Ruan DT, Ruano D, Rubinsztein DC, Rucker EB, 3rd, Rudich A, Rudolf E, Rudolf R, Ruegg MA, Ruiz-Roldan C, Ruparelia AA, Rusmini P, Russ DW, Russo GL, Russo G, Russo R, Rusten TE, Ryabovol V, Ryan KM, Ryter SW, Sabatini DM, Sacher M, Sachse C, Sack MN, Sadoshima J, Saftig P, Sagi-Eisenberg R, Sahni S, Saikumar P, Saito T, Saitoh T, Sakakura K, Sakoh-Nakatogawa M, Sakuraba Y, Salazar-Roa M, Salomoni P, Saluja AK, Salvaterra PM, Salvioli R, Samali A, Sanchez AM, Sanchez-Alcazar JA, Sanchez-Prieto R, Sandri M, Sanjuan MA, Santaguida S, Santambrogio L, Santoni G, Dos Santos CN, Saran S, Sardiello M, Sargent G, Sarkar P, Sarkar S, Sarrias MR, Sarwal MM, Sasakawa C, Sasaki M, Sass M, Sato K, Sato M, Satriano J, Savaraj N, Saveljeva S, Schaefer L, Schaible UE, Scharl M, Schatzl HM, Schekman R, Scheper W, Schiavi A, Schipper HM, Schmeisser H, Schmidt J, Schmitz I, Schneider BE, Schneider EM, Schneider JL, Schon EA, Schonenberger MJ, Schonthal AH, Schorderet DF, Schroder B, Schuck S, Schulze RJ, Schwarten M, Schwarz TL, Sciarretta S, Scotto K, Scovassi AI, Screaton RA, Screen M, Seca H, Sedej S, Segatori L, Segev N, Seglen PO, Segui-Simarro JM, Segura-Aguilar J, Seki E, Seiliez I, Sell C, Semenkovich CF, Semenza GL, Sen U, Serra AL, Serrano-Puebla A, Sesaki H, Setoguchi T, Settembre C, Shacka JJ, Shajahan-Haq AN, Shapiro IM, Sharma S, She H, Shen CJ, Shen CC, Shen HM, Shen S, Shen W, Sheng R, Sheng X, Sheng ZH, Shepherd TG, Shi J, Shi Q, Shi Q, Shi Y, Shibutani S, Shibuya K, Shidoji Y, Shieh JJ, Shih CM, Shimada Y, Shimizu S, Shin DW, Shinohara ML, Shintani M, Shintani T, Shioi T, Shirabe K, Shiri-Sverdlov R, Shirihai O, Shore GC, Shu CW, Shukla D, Sibirny AA, Sica V, Sigurdson CJ, Sigurdsson EM, Sijwali PS, Sikorska B, Silveira WA, Silvente-Poirot S, Silverman GA, Simak J, Simmet T, Simon AK, Simon HU, Simone C, Simons M, Simonsen A, Singh R, Singh SV, Singh SK, Sinha D, Sinha S, Sinicrope FA, Sirko A, Sirohi K, Sishi BJ, Sittler A, Siu PM, Sivridis E, Skwarska A, Slack R, Slaninova I, Slavov N, Smaili SS, Smalley KS, Smith DR, Soenen SJ, Soleimanpour SA, Solhaug A, Somasundaram K, Son JH, Sonawane A, Song C, Song F, Song HK, Song JX, Song W, Soo KY, Sood AK, Soong TW, Soontornniyomkij V, Sorice M, Sotgia F, Soto-Pantoja DR, Sotthibundhu A, Sousa MJ, Spaink HP, Span PN, Spang A, Sparks JD, Speck PG, Spector SA, Spies CD, Springer W, Clair DS, Stacchiotti A, Staels B, Stang MT, Starczynowski DT, Starokadomskyy P, Steegborn C, Steele JW, Stefanis L, Steffan J, Stellrecht CM, Stenmark H, Stepkowski TM, Stern ST, Stevens C, Stockwell BR, Stoka V, Storchova Z, Stork B, Stratoulias V, Stravopodis DJ, Strnad P, Strohecker AM, Strom AL, Stromhaug P, Stulik J, Su YX, Su Z, Subauste CS, Subramaniam S, Sue CM, Suh SW, Sui X, Sukseree S, Sulzer D, Sun FL, Sun J, Sun J, Sun SY, Sun Y, Sun Y, Sun Y, Sundaramoorthy V, Sung J, Suzuki H, Suzuki K, Suzuki N, Suzuki T, Suzuki YJ, Swanson MS, Swanton C, Sward K, Swarup G, Sweeney ST, Sylvester PW, Szatmari Z, Szegezdi E, Szlosarek PW, Taegtmeyer H, Tafani M, Taillebourg E, Tait SW, Takacs-Vellai K, Takahashi Y, Takats S, Takemura G, Takigawa N, Talbot NJ, Tamagno E, Tamburini J, Tan CP, Tan L, Tan ML, Tan M, Tan YJ, Tanaka K, Tanaka M, Tang D, Tang D, Tang G, Tanida I, Tanji K, Tannous BA, Tapia JA, Tasset-Cuevas I, Tatar M, Tavassoly I, Tavernarakis N, Taylor A, Taylor GS, Taylor GA, Taylor JP, Taylor MJ, Tchetina EV, Tee AR, Teixeira-Clerc F, Telang S, Tencomnao T, Teng BB, Teng RJ, Terro F, Tettamanti G, Theiss AL, Theron AE, Thomas KJ, Thome MP, Thomes PG, Thorburn A, Thorner J, Thum T, Thumm M, Thurston TL, Tian L, Till A, Ting JP, Titorenko VI, Toker L, Toldo S, Tooze SA, Topisirovic I, Torgersen ML, Torosantucci L, Torriglia A, Torrisi MR, Tournier C, Towns R, Trajkovic V, Travassos LH, Triola G, Tripathi DN, Trisciuoglio D, Troncoso R, Trougakos IP, Truttmann AC, Tsai KJ, Tschan MP, Tseng YH, Tsukuba T, Tsung A, Tsvetkov AS, Tu S, Tuan HY, Tucci M, Tumbarello DA, Turk B, Turk V, Turner RF, Tveita AA, Tyagi SC, Ubukata M, Uchiyama Y, Udelnow A, Ueno T, Umekawa M, Umemiya-Shirafuji R, Underwood BR, Ungermann C, Ureshino RP, Ushioda R, Uversky VN, Uzcategui NL, Vaccari T, Vaccaro MI, Vachova L, Vakifahmetoglu-Norberg H, Valdor R, Valente EM, Vallette F, Valverde AM, Van den Berghe G, Van Den Bosch L, van den Brink GR, van der Goot FG, van der Klei IJ, van der Laan LJ, van Doorn WG, van Egmond M, van Golen KL, Van Kaer L, van Lookeren Campagne M, Vandenabeele P, Vandenberghe W, Vanhorebeek I, Varela-Nieto I, Vasconcelos MH, Vasko R, Vavvas DG, Vega-Naredo I, Velasco G, Velentzas AD, Velentzas PD, Vellai T, Vellenga E, Vendelbo MH, Venkatachalam K, Ventura N, Ventura S, Veras PS, Verdier M, Vertessy BG, Viale A, Vidal M, Vieira H, Vierstra RD, Vigneswaran N, Vij N, Vila M, Villar M, Villar VH, Villarroya J, Vindis C, Viola G, Viscomi MT, Vitale G, Vogl DT, Voitsekhovskaja OV, von Haefen C, von Schwarzenberg K, Voth DE, Vouret-Craviari V, Vuori K, Vyas JM, Waeber C, Walker CL, Walker MJ, Walter J, Wan L, Wan X, Wang B, Wang C, Wang CY, Wang C, Wang C, Wang C, Wang D, Wang F, Wang F, Wang G, Wang HJ, Wang H, Wang HG, Wang H, Wang HD, Wang J, Wang J, Wang M, Wang MQ, Wang PY, Wang P, Wang RC, Wang S, Wang TF, Wang X, Wang XJ, Wang XW, Wang X, Wang X, Wang Y, Wang Y, Wang Y, Wang YJ, Wang Y, Wang Y, Wang YT, Wang Y, Wang ZN, Wappner P, Ward C, Ward DM, Warnes G, Watada H, Watanabe Y, Watase K, Weaver TE, Weekes CD, Wei J, Weide T, Weihl CC, Weindl G, Weis SN, Wen L, Wen X, Wen Y, Westermann B, Weyand CM, White AR, White E, Whitton JL, Whitworth AJ, Wiels J, Wild F, Wildenberg ME, Wileman T, Wilkinson DS, Wilkinson S, Willbold D, Williams C, Williams K, Williamson PR, Winklhofer KF, Witkin SS, Wohlgemuth SE, Wollert T, Wolvetang EJ, Wong E, Wong GW, Wong RW, Wong VK, Woodcock EA, Wright KL, Wu C, Wu D, Wu GS, Wu J, Wu J, Wu M, Wu M, Wu S, Wu WK, Wu Y, Wu Z, Xavier CP, Xavier RJ, Xia GX, Xia T, Xia W, Xia Y, Xiao H, Xiao J, Xiao S, Xiao W, Xie CM, Xie Z, Xie Z, Xilouri M, Xiong Y, Xu C, Xu C, Xu F, Xu H, Xu H, Xu J, Xu J, Xu J, Xu L, Xu X, Xu Y, Xu Y, Xu ZX, Xu Z, Xue Y, Yamada T, Yamamoto A, Yamanaka K, Yamashina S, Yamashiro S, Yan B, Yan B, Yan X, Yan Z, Yanagi Y, Yang DS, Yang JM, Yang L, Yang M, Yang PM, Yang P, Yang Q, Yang W, Yang WY, Yang X, Yang Y, Yang Y, Yang Z, Yang Z, Yao MC, Yao PJ, Yao X, Yao Z, Yao Z, Yasui LS, Ye M, Yedvobnick B, Yeganeh B, Yeh ES, Yeyati PL, Yi F, Yi L, Yin XM, Yip CK, Yoo YM, Yoo YH, Yoon SY, Yoshida KI, Yoshimori T, Young KH, Yu H, Yu JJ, Yu JT, Yu J, Yu L, Yu WH, Yu XF, Yu Z, Yuan J, Yuan ZM, Yue BY, Yue J, Yue Z, Zacks DN, Zacksenhaus E, Zaffaroni N, Zaglia T, Zakeri Z, Zecchini V, Zeng J, Zeng M, Zeng Q, Zervos AS, Zhang DD, Zhang F, Zhang G, Zhang GC, Zhang H, Zhang H, Zhang H, Zhang H, Zhang J, Zhang J, Zhang J, Zhang J, Zhang JP, Zhang L, Zhang L, Zhang L, Zhang L, Zhang MY, Zhang X, Zhang XD, Zhang Y, Zhang Y, Zhang Y, Zhang Y, Zhang Y, Zhao M, Zhao WL, Zhao X, Zhao YG, Zhao Y, Zhao Y, Zhao YX, Zhao Z, Zhao ZJ, Zheng D, Zheng XL, Zheng X, Zhivotovsky B, Zhong Q, Zhou GZ, Zhou G, Zhou H, Zhou SF, Zhou XJ, Zhu H, Zhu H, Zhu WG, Zhu W, Zhu XF, Zhu Y, Zhuang SM, Zhuang X, Ziparo E, Zois CE, Zoladek T, Zong WX, Zorzano A, Zughaier SM. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition) Autophagy. 2016;12:1–222. doi: 10.1080/15548627.2015.1100356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Isakson P, Lystad AH, Breen K, Koster G, Stenmark H, Simonsen A. TRAF6 mediates ubiquitination of KIF23/MKLP1 and is required for midbody ring degradation by selective autophagy. Autophagy. 2013;9:1955–1964. doi: 10.4161/auto.26085. [DOI] [PubMed] [Google Scholar]
- 15.Duran A, Serrano M, Leitges M, Flores JM, Picard S, Brown JP, Moscat J, Diaz-Meco MT. The atypical PKC-interacting protein p62 is an important mediator of RANK-activated osteoclastogenesis. Developmental cell. 2004;6:303–309. doi: 10.1016/s1534-5807(03)00403-9. [DOI] [PubMed] [Google Scholar]
- 16.McManus S, Roux S. The adaptor protein p62/SQSTM1 in osteoclast signaling pathways. Journal of molecular signaling. 2012;7:1. doi: 10.1186/1750-2187-7-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morissette J, Laurin N, Brown JP. Sequestosome 1: mutation frequencies, haplotypes, and phenotypes in familial Paget's disease of bone. Journal of bone and mineral research : the official journal of the American Society for Bone and Mineral Research. 2006;21(Suppl 2):P38–44. doi: 10.1359/jbmr.06s207. [DOI] [PubMed] [Google Scholar]
- 18.Kato M, Ospelt C, Gay RE, Gay S, Klein K. Dual role of autophagy in stress-induced cell death in rheumatoid arthritis synovial fibroblasts. Arthritis and rheumatism. 2013 doi: 10.1002/art.38190. [DOI] [PubMed] [Google Scholar]
- 19.Hocking LJ, Mellis DJ, McCabe PS, Helfrich MH, Rogers MJ. Functional interaction between sequestosome-1/p62 and autophagy-linked FYVE-containing protein WDFY3 in human osteoclasts. Biochemical and biophysical research communications. 2010;402:543–548. doi: 10.1016/j.bbrc.2010.10.076. [DOI] [PubMed] [Google Scholar]
- 20.Wu DJ, Gu R, Sarin R, Zavodovskaya R, Chen CP, Christiansen BA, Adamopoulos IE. Autophagy-linked FYVE containing protein WDFY3 interacts with TRAF6 and modulates RANKL-induced osteoclastogenesis. J Autoimmun. 2016 doi: 10.1016/j.jaut.2016.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Monach PA, Mathis D, Benoist C. The K/BxN arthritis model. Current protocols in immunology / edited by John E. Coligan … [et al.], Chapter. 2008;15 doi: 10.1002/0471142735.im1522s81. Unit 15 22. [DOI] [PubMed] [Google Scholar]
- 22.Takeshita S, Kaji K, Kudo A. Identification and characterization of the new osteoclast progenitor with macrophage phenotypes being able to differentiate into mature osteoclasts. Journal of bone and mineral research : the official journal of the American Society for Bone and Mineral Research. 2000;15:1477–1488. doi: 10.1359/jbmr.2000.15.8.1477. [DOI] [PubMed] [Google Scholar]
- 23.Orosco LA, Ross AP, Cates SL, Scott SE, Wu D, Sohn J, Pleasure D, Pleasure SJ, Adamopoulos IE, Zarbalis KS. Loss of Wdfy3 in mice alters cerebral cortical neurogenesis reflecting aspects of the autism pathology. Nature communications. 2014;5:4692. doi: 10.1038/ncomms5692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hu YH, Zhang Y, Jiang LQ, Wang S, Lei CQ, Sun MS, Shu HB, Liu Y. WDFY1 mediates TLR3/4 signaling by recruiting TRIF. EMBO reports. 2015;16:447–455. doi: 10.15252/embr.201439637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hayakawa A, Leonard D, Murphy S, Hayes S, Soto M, Fogarty K, Standley C, Bellve K, Lambright D, Mello C, Corvera S. The WD40 and FYVE domain containing protein 2 defines a class of early endosomes necessary for endocytosis. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:11928–11933. doi: 10.1073/pnas.0508832103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Okada Y, Wu D, Trynka G, Raj T, Terao C, Ikari K, Kochi Y, Ohmura K, Suzuki A, Yoshida S, Graham RR, Manoharan A, Ortmann W, Bhangale T, Denny JC, Carroll RJ, Eyler AE, Greenberg JD, Kremer JM, Pappas DA, Jiang L, Yin J, Ye L, Su DF, Yang J, Xie G, Keystone E, Westra HJ, Esko T, Metspalu A, Zhou X, Gupta N, Mirel D, Stahl EA, Diogo D, Cui J, Liao K, Guo MH, Myouzen K, Kawaguchi T, Coenen MJ, van Riel PL, van de Laar MA, Guchelaar HJ, Huizinga TW, Dieude P, Mariette X, Bridges SL, Jr, Zhernakova A, Toes RE, Tak PP, Miceli-Richard C, Bang SY, Lee HS, Martin J, Gonzalez-Gay MA, Rodriguez-Rodriguez L, Rantapaa-Dahlqvist S, Arlestig L, Choi HK, Kamatani Y, Galan P, Lathrop M, consortium R, consortium G, Eyre S, Bowes J, Barton A, de Vries N, Moreland LW, Criswell LA, Karlson EW, Taniguchi A, Yamada R, Kubo M, Liu JS, Bae SC, Worthington J, Padyukov L, Klareskog L, Gregersen PK, Raychaudhuri S, Stranger BE, De Jager PL, Franke L, Visscher PM, Brown MA, Yamanaka H, Mimori T, Takahashi A, Xu H, Behrens TW, Siminovitch KA, Momohara S, Matsuda F, Yamamoto K, Plenge RM. Genetics of rheumatoid arthritis contributes to biology and drug discovery. Nature. 2014;506:376–381. doi: 10.1038/nature12873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu DJ, Adamopoulos IE. Autophagy and autoimmunity. Clin Immunol. 2017;176:55–62. doi: 10.1016/j.clim.2017.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rea SL, Walsh JP, Ward L, Yip K, Ward BK, Kent GN, Steer JH, Xu J, Ratajczak T. A novel mutation (K378X) in the sequestosome 1 gene associated with increased NF-kappaB signaling and Paget's disease of bone with a severe phenotype. Journal of bone and mineral research : the official journal of the American Society for Bone and Mineral Research. 2006;21:1136–1145. doi: 10.1359/jbmr.060405. [DOI] [PubMed] [Google Scholar]
- 29.Daroszewska A, van 't Hof RJ, Rojas JA, Layfield R, Landao-Basonga E, Rose L, Rose K, Ralston SH. A point mutation in the ubiquitin-associated domain of SQSMT1 is sufficient to cause a Paget's disease-like disorder in mice. Human molecular genetics. 2011;20:2734–2744. doi: 10.1093/hmg/ddr172. [DOI] [PubMed] [Google Scholar]
- 30.Layfield R, Cavey JR, Najat D, Long J, Sheppard PW, Ralston SH, Searle MS. p62 mutations, ubiquitin recognition and Paget's disease of bone. Biochemical Society transactions. 2006;34:735–737. doi: 10.1042/BST0340735. [DOI] [PubMed] [Google Scholar]
- 31.Chang CP, Su YC, Hu CW, Lei HY. TLR2-dependent selective autophagy regulates NF-kappaB lysosomal degradation in hepatoma-derived M2 macrophage differentiation. Cell death and differentiation. 2013;20:515–523. doi: 10.1038/cdd.2012.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Suzuki E, Maverakis E, Sarin R, Bouchareychas L, Kuchroo VK, Nestle FO, Adamopoulos IE. T Cell-Independent Mechanisms Associated with Neutrophil Extracellular Trap Formation and Selective Autophagy in IL-17A-Mediated Epidermal Hyperplasia. J Immunol. 2016;197:4403–4412. doi: 10.4049/jimmunol.1600383. [DOI] [PMC free article] [PubMed] [Google Scholar]