Abstract
We have combined genome-wide transcription factor binding and expression profiling to assemble a regulatory network controlling the myogenic differentiation program in mammalian cells. We identified a cadre of overlapping and distinct targets of the key myogenic regulatory factors (MRFs)—MyoD and myogenin—and Myocyte Enhancer Factor 2 (MEF2). We discovered that MRFs and MEF2 regulate a remarkably extensive array of transcription factor genes that propagate and amplify the signals initiated by MRFs. We found that MRFs play an unexpectedly wide-ranging role in directing the assembly and usage of the neuromuscular junction. Interestingly, these factors also prepare myoblasts to respond to diverse types of stress. Computational analyses identified novel combinations of factors that, depending on the differentiation state, might collaborate with MRFs. Our studies suggest unanticipated biological insights into muscle development and highlight new directions for further studies of genes involved in muscle repair and responses to stress and damage.
Keywords: ChIP-on-chip, myogenesis, transcriptional regulation network
Myogenic differentiation proceeds through irreversible cell cycle arrest of precursor cells (myoblasts), followed by a gradual increase in expression of muscle function genes, leading to fusion of myoblasts into multinucleate myofibers in the animal. This process can be recapitulated in vitro, wherein myoblasts can be converted to myotubes with high efficiency in well-established models. In adult skeletal muscle, monopotential precursor cells proliferate and differentiate in response to specific stimuli, such as injury or exercise.
Myogenesis is orchestrated through a series of transcriptional controls governed by the myogenic regulatory factors (MRFs). Ectopic expression of a single MRF, MyoD, is sufficient to force nonmuscle cells to complete the myogenic program (Tapscott et al. 1988). MyoD, a basic helix-loop-helix (bHLH) transcription factor that binds sequence elements termed E-boxes, is the founding member of the MRF family, which includes the closely related Myf5, myogenin, and MRF4 proteins (for review, see Buckingham 2001). These proteins cooperate with a second family of transcription factors, called Myocyte Enhancer Factor 2 (MEF2) (Molkentin and Olson 1996). MRFs are known to activate the expression of genes that specify muscle. The first steps in the regulatory cascade involve expression of MyoD and Myf5, which subsequently leads to expression of myogenin and MEF2, promoting conversion of myoblasts to myotubes. MyoD, which persists in myotubes, collaborates with myogenin to regulate the expression of genes necessary for terminal differentiation.
Beyond these first steps, our knowledge is somewhat fragmentary: Relatively few physiological targets of MRFs and MEF2 have been identified, and the number of genes known to be regulated by these factors is considerably smaller than the number of genes induced upon myogenic differentiation (Moran et al. 2002). In addition, the role of MyoD in myogenic differentiation appears to be considerably more complex, since recent studies point to a role for this factor in transcriptional repression as well as activation (Mal and Harter 2003). Furthermore, other critical, unresolved questions include the extent to which functional redundancy occurs within the MRF family and the functional impact of binding by individual family members to their targets. For these reasons, identification of direct transcriptional targets of MRFs and MEF2 and deconvolution of the transcriptional regulatory networks that operate in muscle cells represent a sine qua non to comprehensively understand not only how muscle differentiates but also how it responds to stress and damage, thereby allowing regeneration.
Recent attempts to identify MyoD-binding sites exploited gene expression profiling of cells that ectopically express MyoD (Bergstrom et al. 2002). While these experiments have certainly identified several bona fide, physiological targets, several complications arise using this approach. First, ectopic expression of a bHLH transcription factor can lead to promiscuous binding to E-boxes throughout the genome. The fact that (1) another E-box-binding factor, Myc, binds to an extensive portion of the genome (Fernandez et al. 2003) and (2) other MRFs (including Myf5 and myogenin) can similarly bind this sequence has hampered definitive conclusions regarding physiological occupancy by specific MRFs. In addition, secondary downstream effects resulting from the altered expression of transcription factor targets further complicate identification of direct targets. Thirdly, MyoD interacts with activator and repressor proteins, and ectopic expression could inappropriately titrate an inhibitory protein, such as Id, which blocks muscle differentiation by preventing DNA binding by MRFs (Benezra et al. 1990). Other approaches to identifying MRF targets have relied on mouse genetics. However, functional redundancy and compensation are likely to obscure the full impact of individual contributions of MRF family members on gene expression patterns. Recently, we and others have developed methods that circumvent these complications. This technology, termed ChIP-on-chip or location analysis, has been used to identify mammalian factor-binding sites in a genome-wide manner (Cam et al. 2004; Odom et al. 2004).
Here we describe a novel approach toward unraveling the regulatory mechanisms involved in mammalian muscle development. We constructed a mouse promoter DNA microarray and performed ChIP-on-chip analysis to identify targets of key muscle regulators MyoD, myogenin, and MEF2. In addition to confirming suspected targets of these transcription factors, we identified a cohort of genes involved in unanticipated pathways. Remarkably, these factors control stress response pathways and the specification and utilization of the neuromuscular synapse. Moreover, the regulatory power of MRFs and MEF2 extends beyond their immediate downstream targets, as they appear to control a vast array of transcription factors that propagate the signals initiated by MRFs. Using our genome-wide binding data, expression profiling, computational analyses, and previous observations regarding transcription factor function, we describe a complex, dynamic network that models the transcriptional activation cascades that govern skeletal myogenesis. We demonstrate that our approach is useful for uncovering unexplored aspects of muscle biology involved in the far-ranging processes of stress response, differentiation, and regeneration.
Results
ChIP-on-chip analysis of MRF and MEF2 binding
Conversion of mouse C2C12 myoblasts into myotubes represents a well-established and robust in vitro differentiation model. In this setting, proliferating myoblasts can be induced to differentiate into myotubes by growing them to confluence and switching them to reduced serum (differentiation medium, DM). Under these conditions, most cells fuse to form myotubes after 4 d. Furthermore, genome-wide comparison of gene expression in primary mouse myoblasts and C2C12 cells allows us to conclude that their overall transcriptional profiles are very similar (Supplementary Fig. S1; see below). In order to better understand the transcriptional regulatory networks that govern this process, we used a multifaceted approach that combines ChIP-on-chip, gene expression profiling, computational analyses, and gene ablation techniques (Fig. 1A). First, we performed ChIP on growing myoblasts and differentiated myotubes using antibodies that recognize MyoD, myogenin, and MEF2 and used PCR to verify enrichment of suspected or previously characterized targets. Each of these antibodies significantly enriched several E-box-containing promoters, including the Chrna1 and Mef2c genes, as expected (Fig. 1C; data not shown; see Table 1 for complete gene names; Piette et al. 1990; Ridgeway et al. 2000).
Figure 1.
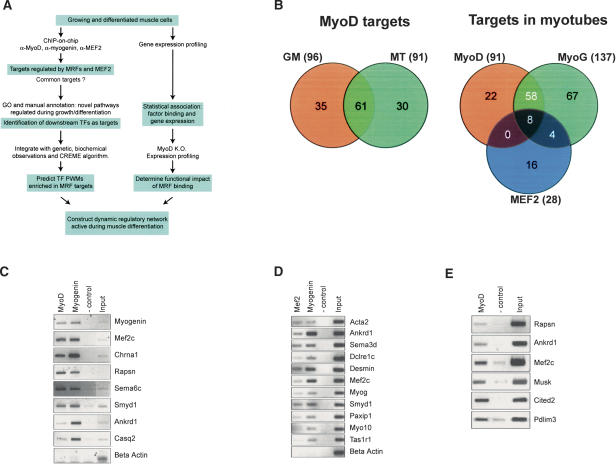
Identification of MRF and MEF2 targets. (A) Approach used to elucidate myogenic transcriptional networks. (MRFs) myogenic regulatory factors; (GO) Gene Ontology; (TFs) transcription factors; (PWMs) position-weight matrices. (B) Venn diagrams representing the overlap of MyoD targets in three different populations studied (left) and overlap of MyoD, myogenin, and MEF2 targets in myotubes (right). The total number of targets is indicated in parentheses. (C) ChIP assays in C2C12 myotubes with anti-MyoD and anti-myogenin antibodies, showing specific enrichment of selected target genes. (D) ChIP assays in C2C12 myotubes with anti-MEF2 and anti-myogenin antibodies. (E) Identification of MyoD targets in growing, primary human myoblasts using ChIP.
Table 1.
List of MRF and MEF2 target genes
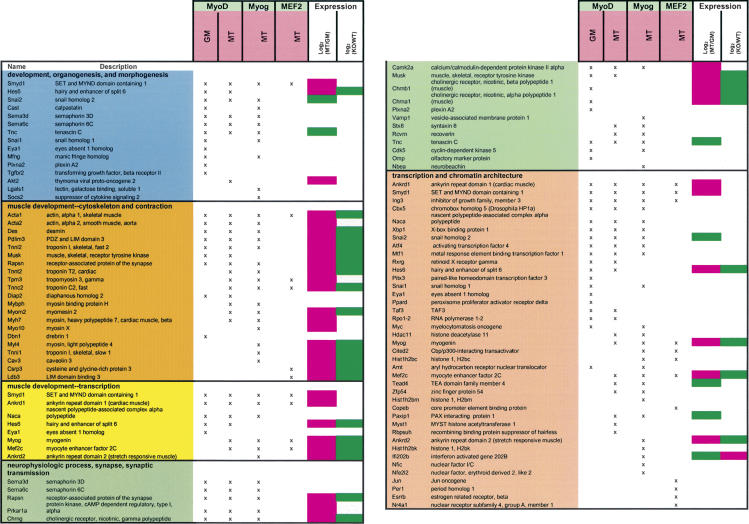
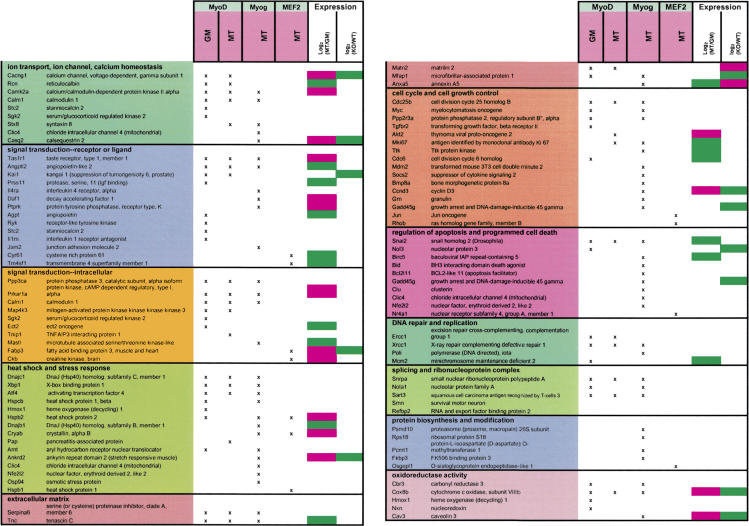
Target genes were classified according to Gene Ontology annotation. Some targets are represented in multiple categories. Uncharacterized genes are omitted here but are included in Supplementary Table S1. Myog indicates myogenin. Log2(MT/GM) indicates the change in gene expression that occurs when C2C12 myoblasts differentiate into myotubes. Log2(KO/WT) indicates the difference in gene expression between MyoD-/- and wild-type primary myoblasts. Green symbolizes values equal to or less than -1, whereas pink indicates values equal to or greater than 1.
To identify targets of MRFs in an unbiased, large-scale manner, we constructed a murine genomic DNA microarray (termed Mm4.7k) representing >4700 different loci and used ChIP-on-chip analysis to identify new targets of the muscle regulatory transcription factors MyoD, myogenin, and MEF2 in myoblasts and myotubes (see Supplemental Material). We isolated mature myotubes from differentiated cultures of C2C12 cells in order to separate them from reserve cells, a population of cells that does not fuse but remains quiescent after 4 d in differentiation medium (Carnac et al. 2000). Since Western blotting indicated that MyoD was expressed in proliferating and differentiated myotubes, we performed location analysis for this transcription factor in those two populations (growing cells, GM; and myotubes, MT) (Supplementary Fig. S1B). In contrast, myogenin and MEF2C are expressed exclusively in myotubes, and thus we restricted our analysis of these factors to myotubes. The lack of a high-quality, specific antibody against MYF5 prevented us from performing location analysis for this MRF.
We identified a total of 198 genes bound by MRFs or MEF2 (Fig. 1B; Table 1; Supplementary Table S1). Onehundred-twenty-six genes were bound by MyoD, although some targets were specifically bound by this factor in GM but not MT. In myotubes, myogenin and MEF2 occupied 137 and 28 promoters, respectively, whereas MyoD occupied 91 promoters. Some genes were bound by one factor but not the other two. These findings suggest that the MRFs recognize sets of distinct but overlapping targets, and that MyoD regulates a common set of genes in both growing and differentiated cells.
MRF binding in mouse and primary human cells
We confirmed binding by MyoD, myogenin, and MEF2 to a subset of targets that we identified using ChIP and semiquantitative PCR (Fig. 1C,D; data not shown). Importantly, we also performed ChIP assays using primary human skeletal myoblasts (Fig. 1E) and confirmed that MRF binding to a subset of targets is conserved in both mouse and human muscle cells. Moreover, we examined a larger cohort of genes identified by ChIP-on-chip with binding ratio values above or below our chosen threshold (see Supplemental Material; Supplementary Fig. S3; Supplementary Table S3) and determined that the falsepositive rate of our approach is similar to what we and others have observed using a similar human promoter microarray (Cam et al. 2004; Odom et al. 2004). Because not all mouse genes are represented on our microarray, and because our stringent criteria for identifying bound loci inevitably introduces false negatives, we may have overlooked several MRF and MEF2 targets. Although MRF-binding sites have been identified in enhancer regions several kilobases from their transcriptional start sites, the majority of E-boxes present in the MyoD and myogenin target promoters that we identified were located very close to the transcriptional start site (Supplementary Fig. S2E).
Identifying unexpected pathways that operate during myogenesis
We selectively enriched several known targets of MRFs and MEF2. However, a majority of the genes identified in our analysis have not been identified in previous studies and thus represent novel targets of these transcription factors. We clustered MRF and MEF2 target genes according to function using Gene Ontology (GO) annotation. Inspection of these clusters revealed that these factors bind to genes involved in a wide spectrum of pathways (Table 1; Fig. 2). We used an algorithm, EASE (Hosack et al. 2003), that allowed us to determine whether enrichment in a given category was statistically significant given the number of genes in each individual cluster represented on our microarray (Supplementary Table S4). Most strikingly, in growing myoblasts, MyoD bound a set of genes involved in synapse specification and utilization and neuromuscular function (Fig. 2B), whereas those bound by MyoD in MT play a role in muscle development and contraction. The repertoire of myogenin targets was also distinguished by genes involved in muscle development and contraction. MRFs also bound genes involved in the Notch pathway (Hes6, manic fringe homolog, Rbpsuh/RBP-J).
Figure 2.
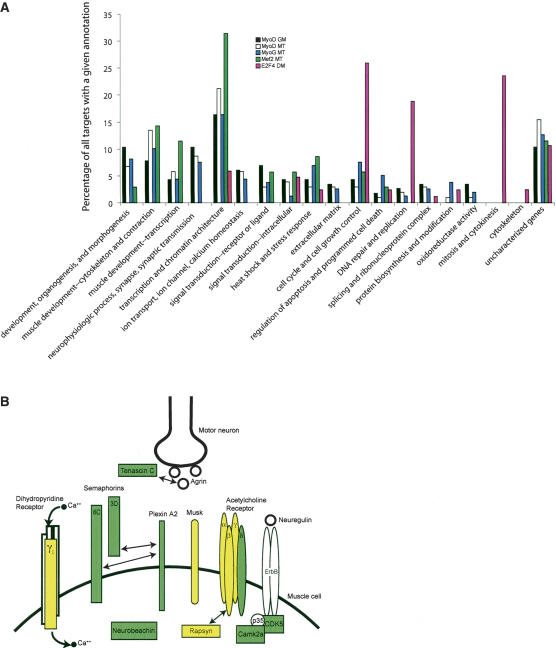
Distribution of Gene Ontology categories for biological function of MRF, MEF2, and E2F4 targets. (A) Histogram showing the distribution of GO categories for target genes bound by each factor in the indicated growth condition and the frequency of their occurrence. In some cases, genes appear in multiple categories. (B) Representation of components of the NMJ, showing MRF or MEF2 targets in green and yellow. Yellow indicates genes deregulated in MyoD-/- primary myoblasts. Protein interactions are indicated with bidirectional arrows.
We were also surprised by the relative lack of canonical cell cycle genes controlled by MRFs, since previous studies implicated MyoD in growth arrest prior to myogenic differentiation (Kitzmann and Fernandez 2001). We performed location analysis using antibodies against the E2F4 transcription factor and confirmed that genes involved in control of cell proliferation could be identified with the Mm4.7k microarray (Fig. 2; Supplementary Fig. S2D; Cam et al. 2004).
One remarkable feature common to all three factors investigated in both conditions is the large number of targets involved in transcription, the majority of which are sequence-specific regulators. The category of DNA-dependent transcription is the most prominent among MRF targets. Interestingly, of the 124 E2F4 targets we identified, only two are sequence-specific, DNA-binding transcription factors, in striking contrast to the number identified as targets of MyoD and myogenin (21 and 15, respectively). The repertoire of transcription factor targets expanded as cells progressed from a growing to differentiated state (Table 1), suggesting that the signal initiated by MyoD in myoblasts is amplified further through transcriptional cascades during the course of myogenesis.
Our data suggest several important conclusions. The MRFs appear to regulate a large cluster of transcription factor targets. This is consistent with models of gene activation during myogenesis, whereby genes are turned on sequentially, beginning with activation of myogenin and MEF2c by MyoD. Our data suggest that this cascade is more elaborate than expected, since we have discovered a significant number of novel targets of MRFs that are sequence-specific transcription factors, and several biochemical and genetic observations indicate that they are very likely to participate in the differentiation process (see Discussion).
A surprising role for stress-response pathways
One of the most unexpected results of our analysis was the identification of a cluster of MRF targets that play an established role in stress response and that are induced under conditions of stress in mice or humans (Table 2). Notably, 17 transcriptional regulators (including ATF4, XBP1, ARNT, Copeb/KLF6, NFE2l2/NRF2) cluster within this group, suggesting that the transcriptional regulatory program initiated by MRFs extends to pathways involved in response to, or protection from, stressful cues. These target genes take part in responding to diverse types of stress, including hypoxia, unfolded proteins (the unfolded protein response, UPR), heat shock, and oxidative damage. Moreover, another set of MRF targets (Ankrd1, Ankrd2, Csrp3 [MLP], and calcineurin subunits) appears to be involved in responding to muscle-specific stresses, such as stretch or sarcomeric dysfunction, by triggering fiber-type switching or by eliciting the so-called hypertrophic response, whereby an intracellular signaling cascade causes re-expression of fetal muscle genes, which leads to an increase in muscle fiber size.
Table 2.
MRF and MEF2 target genes involved in stress response
| Gene | Transcription factor activity | UPR or chaperonea | TM-inducedb | ATF4c | HIF1ad | Human stress responsee | Muscle-specificf |
|---|---|---|---|---|---|---|---|
| Xbp1 | x | x | x | ||||
| Atf4 | x | x | x | ||||
| Myc | x | x | |||||
| Rbpsuh | x | x | |||||
| Snai2 | x | x | |||||
| Arnt | x | x | |||||
| Nr4a1 | x | x | x | ||||
| Copeb | x | x | x | ||||
| Cited2 | x | x | x | ||||
| Ankrd1 | x | x | x | ||||
| Ankrd2 | x | x | |||||
| Csrp3 | x | x | |||||
| Tead4 | x | x | |||||
| Mtf1 | x | x | |||||
| Nfe212 | x | x | |||||
| Jun | x | x | |||||
| Snai1 | x | x | |||||
| Cyr61 | x | ||||||
| Dnajb1 | x | x | |||||
| Dnajc1 | x | ||||||
| Cryab | x | x | |||||
| Pfdn5 | x | x | |||||
| Hspb1 | x | ||||||
| Hspb2 | x | ||||||
| Hspcb | x | ||||||
| Osp94 | x | ||||||
| D19Ertd144e | x | ||||||
| 4632412E09Rik | x | ||||||
| Snrpa | x | ||||||
| Ifi202b | x | x | |||||
| Clic4 | x | ||||||
| Hmox1 | x | x | |||||
| Pgk1 | x | ||||||
| Sema6c | x | ||||||
| Stc2 | x | ||||||
| Bid | x | ||||||
| Sart3 | x | ||||||
| Tm4sf1 | x | ||||||
| Psmd10 | x | ||||||
| PPP3ca | x |
Stress-response genes expressed in muscle cells are shown. The annotations, indicated with footnotes, are based on previously described connections to specific processes (UPR and muscle stress), induction following specific treatments (tunicamycin and human stress response), regulation by a given transcription factor (HIF1α), or deregulation in the absence of a given transcription factor (ATF4).
Genes recognized as regulators of the unfolded protein response (Rutkowski and Kaufman 2004).
Genes shown to be induced by tunicamycin treatment in mouse (Harding et al. 2003; Lee et al. 2003).
Genes shown to be deregulated in Atf4-/- MEFs (Harding et al. 2003).
Genes recognized as interacting with HIF1α or regulated by HIF1α (Semenza 2002).
Genes for which the human homolog has been shown to be induced under conditions of stress (Murray et al. 2004).
Genes involved in the hypertrophic response or induced in responses to muscle stretch.
Linking expression with promoter occupancy by MRFs
To establish links between transcription factor binding and regulation of target genes during differentiation, we performed gene expression profiling using Affymetrix oligonucleotide arrays. We analyzed cells that were growing, in the process of differentiation (24 h in DM), or fully differentiated into myotubes. Globally, our results agree with data obtained previously (Moran et al. 2002). As expected, EASE analysis of genes induced during differentiation revealed large clusters involved in muscle development and contraction (self-organizing map clusters 1, 5, 9, and 13) (data not shown; see Supplementary Fig. S4; Supplementary Table S5). In contrast, other genes repressed in myotubes are related to cell cycle function (clusters 4, 8, and 12). One group of genes overlooked in previous studies comprises those that transiently decrease in expression at 24 h but then increase in myotubes (clusters 2 and 3). Strikingly, many genes in this group play a role in amino acid and tRNA metabolism (p = 7.4 × 10-8 and p = 6.3 × 10-7, respectively, Fisher exact test) and are induced in response to tunicamycin-induced endoplasmic reticulum (ER) stress in an ATF4-dependent manner (Harding et al. 2003), suggesting that the activity of ATF4 is modulated in a similar manner during the myogenic program.
Next, we linked factor binding with expression data for all genes on our Mm4.7k array by comparing the changes in transcript levels in myotubes versus growing cells (log2MT/GM) for genes that were bound by MRFs and MEF2 (Fig. 3, black dots). This analysis indicated a remarkable association between MRF and MEF2 binding and enhanced gene expression during myogenesis (Fig. 3, black dots, right portion of graph). A much smaller number of MRF and MEF2 target genes were down-regulated during myogenesis (Fig. 3, black dots, left portion of graph). Importantly, we verified this approach by analyzing our E2F4 ChIP-on-chip data obtained from differentiated muscle cells. In contrast with MRFs and MEF2, there was a pronounced association between E2F4 binding in myotubes and transcriptional repression, consistent with data implicating this factor as a dedicated repressor of genes in cells exiting the cell cycle (Cam et al. 2004). We used a statistical test of the difference of two proportions from binomial populations (Supplemental Material) to validate the observation that MRF and MEF2 binding is associated with induction of gene expression. In every case (with the notable exception of E2F4), the proportion of bound genes to total number of genes within the induced subset was significantly larger than for uninduced genes (p < 0.001). In contrast, E2F4 binding was strongly associated with repression. Our data suggest a pervasive role for MyoD, myogenin, and MEF2 in gene activation.
Figure 3.
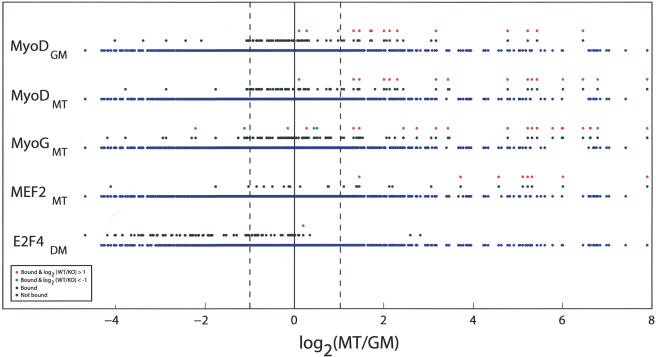
Linking binding of MRFs and MEF2 with gene expression during myogenesis. We plotted the distribution of gene expression values [log2(MT/GM)] along the horizontal axis for all genes present in our factor binding and expression data sets. (MT and GM) Normalized Affymetrix expression values for myotubes and growing cells, respectively. Blue and black dots represent genes that are not bound or bound by the indicated factors, respectively. Expression profiling results comparing genes bound by the indicated factor in primary wild-type and MyoD-/- mouse myoblasts are indicated with red and green dots that represent genes whose expression is up-regulated or down-regulated by greater than twofold, respectively, in wild-type compared with KO. Dotted lines indicate the zone within which no significant change in expression occurred. All genes are displayed in identical positions along the X-axis to compare binding of factors and conditions.
This approach also allowed us to analyze combinatorial contributions to gene expression. Strikingly, of the 12 genes bound by both MEF2 and either MRF, 10 are significantly induced during differentiation, suggesting that the combined occupancy by both factors has a greater impact than either factor alone. In comparison, 21 of the 83 genes bound in MT by MyoD but not MEF2, and 29 of the 130 genes bound by myogenin but not MEF2, were induced during differentiation. We also found a smaller number of genes bound by MyoD and myogenin that may be activated by MyoD in growing cells and repressed by MyoD and/or myogenin in myotubes (Fig. 3, left section of graph). Some of these genes are associated with cell cycle progression and proliferation.
Genetic evidence for the functional impact of MRF binding
In an effort to assess the functional impact of MyoD binding, we performed genome-wide expression profiling with primary myoblasts derived from MyoD-/- and matched wild-type mice (Supplementary Table S5; Sabourin et al. 1999). We observed that, in contrast with growing MyoD-/- cells, growing wild-type myoblasts activate a myogenic program, as evidenced by their elevated expression of muscle-specific marker genes such as desmin. However, 4 d after induction of differentiation, the gene expression profile of mutant cells was significantly closer to that of wild-type cells, and MyoD-/- cells formed myotubes (data not shown), confirming that null cells exhibit delayed activation of the myogenic program rather than complete impairment (Sabourin et al. 1999).
We compared the gene expression profiles of growing wild-type and MyoD-/- myoblasts and focused on those genes bound by MyoD in C2C12 cells (Fig. 3, red and green dots). We did not observe MyoD target genes that were up-regulated as a result of MyoD loss. Instead, we found a strong correlation between binding of MyoD to genes induced during differentiation of C2C12 myoblasts and diminished expression in cells that have lost this factor (Fig. 3, red dots). Interestingly, when we considered the functional annotation of those MyoD targets, we found genes involved in muscle development (desmin, troponin genes, myosin), transcriptional control (myogenin, MEF2c, Hes6), and synaptic function (Chrng, Chrna1, Musk, Rapsn) (Table 1; Fig. 2B). These experiments provide strong genetic support for our conclusion that we have identified a set of genes that are directly and physiologically activated by MRFs, indicate that both MRFs play a wider role in activation than repression, and suggest that MyoD is required for appropriate induction of these genes during myogenesis.
Computational analyses suggest combinatorial regulation of transcription by MRFs
Despite a strong association between MRF binding and gene expression (Fig. 3), promoter occupancy by MyoD in growing myoblasts does not ensure induction of gene expression. We reasoned that recruitment of additional factors, through combinatorial regulation, might underlie this observation. Indeed, combinatorial regulation of muscle-specific genes by MRFs and MEF2 has been documented (Molkentin and Olson 1996). Thus, we hypothesized that other transcription factors could also cooperate with MRFs to modulate transcriptional control. Our ChIP-on-chip data represent a rich source of information that allows us to test this hypothesis.
To analyze the myogenic transcriptional program in greater detail, we searched for position weight matrices (PWMs) of transcription factor-binding sites that are significantly enriched among the sets of genes bound by MRFs and MEF2. We used the CREME algorithm (Sharan et al. 2003b), based on 524 vertebrate PWMs from TRANSFAC (Matys et al. 2003). Importantly, we confirmed the validity of our approach by showing that the PWMs for MyoD, myogenin, and MEF2 were significantly enriched among the targets we identified, consistent with models of combinatorial regulation (Table 3; data not shown). Furthermore, we did not observe enrichment of any of these PWMs among our E2F4 targets (data not shown).
Table 3.
PWMs enriched among specific groups of targets
| Binding factors-motif name | p value | No. of hitsa |
|---|---|---|
| Total MRF and MEF2 targets | ||
| Group A. MyoD targets | ||
| E-box motif | 3.64E-21 to 5.01E-06 | 64 to 119 |
| MEF2 | 0.0018148 | 65 |
| SRF | 0.0021653 | 43 |
| TEAD/TEF family | 0.0033224 | 89 |
| ATF-2 and c-Jun | 0.005934 | 59 |
| Group B. Myogenin targets | ||
| E-box motif | 1.02E-34 to 2.41E-06 | 57 to 108 |
| SRF | 0.0004 | 40 |
| HNF2 | 0.0006 | 40 |
| MEF2 | 0.0007 | 62 |
| Nuclear hormone receptors (PR and GR) | 0.0024 | 41 |
| Group C. MEF2 targets | ||
| TEAD/TEF family | 4.77E-06 | 20 |
| MEF2 | 0.0002 | 16 |
| E-box motif | 0.0002 | 23 |
| Ikaros 2 | 0.0004 | 25 |
| CREB | 0.0080 | 6 |
| Zic and Gli factors | 0.0046 | 23 |
| Condition-specific binding sites | ||
| Group D. Targets of MyoD in GM but not in MT | ||
| NFE2L2 | 0.0004 | 29 |
| S8/Prrx2 | 0.0008 | 18 |
| ATF-2 and c-Jun | 0.0009 | 17 |
| Pax8 | 0.0028 | 29 |
| E-box motif | 0.0050 | 21 |
| Group E. Targets of MyoD in MT but not in GM | ||
| Ikaros 2 | 0.0008 | 21 |
| NF-κB | 0.0021 | 10 |
| Nuclear hormone receptors (PR and GR) | 0.0043 | 13 |
| Factor-specific binding sites | ||
| Group F. Bound in MT by MyoD but not by myogenin | ||
| Nuclear hormone receptors (PR and GR) | 8.92E-06 | 14 |
| Pou2f1/Octamer factor 1 | 0.0010 | 8 |
| Nkx2.1/Thyroid transcription factor 1 | 0.0027 | 18 |
| E4F1 | 0.0058 | 8 |
| NFE2L2 | 0.0080 | 19 |
| peroxisome proliferator activated receptor | 0.0094 | 8 |
| Group G. Bound in MT by myogenin but not by MyoD | ||
| E-box | 7.31E-18 | 54 |
| HNF-2 | 0.0003 | 24 |
| aryl hydrocarbon/dioxin receptor | 0.0004 | 23 |
| Zic and Gli factors | 0.0010 to 0.0014 | 47 to 53 |
| Expression-specific binding sites | ||
| Group H. Bound by MyoD and induced during differentiation | ||
| TEAD/TEF family | 0.0001 | 28 |
| SRF | 0.0003 | 16 |
| Retinoid and thyroid hormones receptors | 0.0030 | 19 |
| Zic and Gli factors | 0.0048 | 29 |
| MEF2 | 0.0052 | 22 |
| Group I. Bound by MyoD but not induced during differentiation | ||
| X-box binding protein 1 | 0.0038 | 63 |
| Myc | 0.0050 to 0.0074 | 7 to 26 |
| Pax8 | 0.0089 | 68 |
| Group J. Bound by myogenin and induced during differentiation | ||
| HNF-2 | 2.58E-06 | 17 |
| Nuclear hormone receptors (PR and GR) | 8.32E-05 | 37 |
| Smad proteins | 0.0001 to 0.0036 | 5 to 31 |
| TEAD/TEF family | 0.0005 | 29 |
| Zic and Gli factors | 0.0013 to 0.0069 | 33 to 37 |
| SRF | 0.0019 | 15 |
| MEF2 | 0.0022 to 0.0063 | 13 to 22 |
| COUP-TF1/NR2F1 | 0.0036 | 28 |
| Six1 homeobox transcription factor | 0.0093 | 35 |
| Group K. Bound by myogenin but not induced during differentiation | ||
| Tfcp2/Ubp1 | 5.43E-10 | 48 |
| Interferon Regulatory Factor 2 | 0.0007 | 7 |
| Olf1 (Olfactory Receptor) | 0.0044 | 10 |
| NF-KB | 0.0063 | 16 |
| Copeb, ZNF148, Sp6, KLF13 (CACCC motif) | 0.0070 | 6 |
Enrichment of transcription factor-binding site matrices in the promotor sequences (−1750 to 250 relative to transcription start site) of target genes was analyzed using the CREME algorithm (Sharan et al. 2003b). For the factor- and condition-specific PWMs, only those specific to the indicated sets that have a p-value <0.01 are shown. Not all redundant matrices (e.g., similar E-box motif PWMs) are shown. We indicate (in bold type) binding factors discovered in this study as MRF or MEF2 targets.
Number of different sequences with at least one occurrence of the motif.
Because some targets appear to be factor- or condition-specific, we looked for condition- or factor-specific PWMs. We found PWMs that appeared specific to MyoD (e.g., NFE2L2/NRF2, group F) and myogenin targets (e.g., Zic and Gli factors, group G). Certain PWMs were enriched among MyoD targets bound specifically in GM (e.g., ATF-2/c-Jun and Pax8, group D) or MT (Ikaros and NF-κB, group E). Assuming that recruitment of MRFs to their target genes can be altered by other DNA-binding proteins, these observations may explain why MyoD and myogenin have only partially overlapping sets of targets and how MyoD binds some promoters in one state but not in the other.
Remarkably, we also found that unique sets of PWMs enriched among MRF targets could distinguish whether or not expression of these targets is induced during C2C12 differentiation. In particular, binding sites for MEF2, Six1, SRF, nuclear receptors (for thyroid and retinoid hormones, and glucocorticoids and progestins), ZIC/Gli, and TEAD/TEF transcription factors were significantly enriched only in the induced subsets, suggesting that concerted binding of these proteins with MRFs enhances transactivation (Table 3, groups H and J). These findings contrast with the enrichment of other PWMs (including XBP-1, an MRF target) among the promoters bound by MyoD or myogenin but whose activity is not induced significantly during differentiation (Table 3, groups I and K). These results suggest that MRF recruitment is more likely to be associated with gene activation during myogenic differentiation if it is accompanied by binding of other specific factors.
Constructing a regulatory network controlling muscle differentiation
We used our expression profiling and ChIP-on-chip data to construct network diagrams representing an initial blueprint for myogenic differentiation (Fig. 4). We included additional layers of control by superimposing published genetic and biochemical observations on our experimental findings. We displayed factor binding data from both growing and differentiated cells together with our expression profiling results to indicate dynamic, temporal connections between both states. These networks reveal both anticipated and novel aspects of control by MRFs and MEF2.
Figure 4.
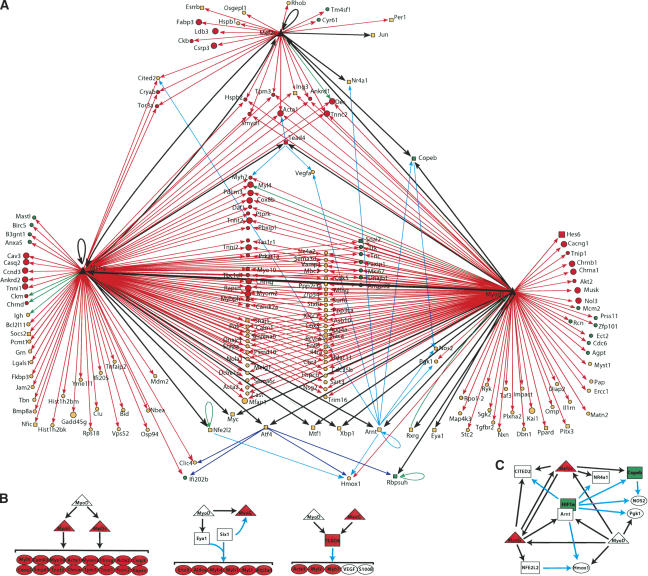
A transcriptional regulatory network in muscle cells MRFs and MEF2 (large black triangles) are connected by arrows to their target genes. The expression patterns of each target are color-coded to reflect changes in expression patterns during differentiation: red or green if their expression level is at least twofold higher or lower, respectively, in myotubes than in the GM state. Orange nodes represent genes with intermediate expression ratios. Transcription factors (TFs) are represented by squares and are connected with black arrows. Other genes (non-TFs) are represented as circles and are connected by red arrows. Literature mining was used to add additional layers of control as follows: Genes shown previously to be regulated by at least two TFs that are bound by MRFs in our experiments or shown to be regulated by at least one MRF, are represented as circles and are connected by green (binding relationship indicated in TRANSFAC), cyan (literature mining), and blue (genes deregulated in tunicamycin-treated Atf4-/- fibroblasts) (Harding et al. 2003) arrows. (B) MRFs regulate the expression of transcriptional regulators that induce the expression of muscle function genes. (C) Feed-forward loops and complex network motifs are illustrated.
Several features of this network are especially noteworthy (Fig. 4A). We observed several regulatory motifs, such as feedback and feed-forward loops, and multi-input and serial regulatory motifs. Feedback loops include regulation of MyoD by Fos and the retinoid receptor RXRγ, two targets of MyoD itself (Muscat et al. 1994; Pedraza-Alva et al. 1994), and the well-known regulation of myogenin and MEF2c by one another. We identified an indirect negative feedback loop, whereby MyoD regulates the expression of Rbpsuh/RBP-J, which mediates Notch-induced antagonism of MyoD expression (Kuroda et al. 1999).
Multi-input motifs were most prominent because many myogenin targets are also bound by MyoD (Fig. 1B, left), and both MRFs commonly bind MEF2 targets. The overall network was very rich in feed-forward loops owing to the large number of target genes that are themselves transcription factors. Some of the most interesting connections involve the transcription factors TEAD4/TEF-3, ARNT, Copeb/KLF6, NFE2l2/NRF2, and ATF4. Intriguingly, each of them exhibits multiple connections to the network, and each plays a role in stress response (Table 2).
Expression of many proteins that interact with one another appears to be coregulated. For example, MRFs and MEF2 target four acetylcholine receptor subunit genes, numerous myosin and troponin isoforms, Naca/SkNac and its transcriptional corepressor Smyd1, semaphorins, and one of their receptors, Plexin A2, as well as the MRF coactivator Csrp3/MLP (Fig. 2B). This network property suggests that efficiency in directing myogenesis could be achieved in part through usage of common paths coordinating the expression of interacting proteins.
When the topology of the network is considered along with the change in target gene expression levels from GM to MT, it becomes obvious that targets bound by MRFs and MEF2 are more likely to be induced than if they are bound by one MRF alone, in agreement with results presented in Figure 3 and our computational analyses described above (Table 3). The same is also apparent when we consider the ratio of induced to repressed genes bound by MyoD versus MyoD and myogenin: A greater proportion of MyoD target genes are induced when they are also targeted by myogenin.
We dissected this global regulatory network into subnetworks that displayed both low and high complexity (Fig. 4B,C). We depict in Figure 4B portions of the network where the repertoire of genes downstream of MRFs is amplified by transcriptional regulators that are MRF targets. These include examples of regulation of targets by MEF2 or myogenin, two genes downstream of MyoD. We also illustrate how TEAD4/Tef-3 and Eya1 and Six1 propagate the signaling cascade instigated by MyoD, by regulating the expression of additional muscle function genes. We show a portion of the network that exhibits extensive interconnectivity in Figure 4C.
We conclude that this network, which connects the MRFs and MEF2 to a large number of genes and considerably extends their repertoire of targets, explains how binding by these factors initiates and then sustains the cascade of events associated with myogenesis. Most importantly, the network we have constructed connects multiple genes without a previous connection to myogenesis. It implicates a functional role for transcription factors without known targets and for the first time provides a framework within which we can begin to explore their function.
Discussion
Here we present an initial attempt to define the transcriptional networks governing skeletal myogenesis using a multifaceted approach based on genome-wide location analysis, gene expression profiling, gene ablation techniques, and computational methods. To our knowledge, this is the first time that such an approach has been used to dissect a genetic program in mammalian cells. Although it is currently not technically possible to investigate transcription factor occupancy throughout the entire genome, our data suggest several fundamentally important biological conclusions regarding muscle differentiation.
Cell cycle control and MRFs
Our ChIP-on-chip studies identified a cadre of MRF and MEF2 target genes whose expression is expected to vary during myogenesis and that had been previously characterized as targets of these factors (Supplementary Table S2). More importantly, our approach also revealed several surprising pathways that operate during differentiation. The overall paucity of genes involved in cell cycle control was unexpected, given that one original function ascribed to MyoD was growth arrest and the observation here and elsewhere that cell cycle genes are down-regulated during the course of differentiation (Supplementary Fig. S4; Kitzmann and Fernandez 2001). Our findings suggest that this facet of the differentiation program may not be directly controlled by the MRFs. Rather, it may be indirectly controlled through the regulation of other transcription factors, and interestingly, we have shown that Myc, Fos, and Jun are targets of MRFs or MEF2 (data not shown).
A cohort of transcription factors amplifies signals initiated by MyoD
One of our most striking observations is that transcription factors represent the largest cluster of MRF targets. Although consistent with a cascade model of gene activation, the markedly high number of transcription factors regulated by MRFs and MEF2 suggests that the cascade may be more extensive than expected. Our analyses suggest the existence of new nodal points from which the transcriptional output of MyoD is relayed, greatly expanding the repertoire of indirect targets of MyoD. We contrast the role of MRFs in differentiation with E2F4, a repressor that plays a role in cell cycle exit (Cam et al. 2004): Only a handful of transcription factor genes are bound by E2F4, suggesting that gene regulatory programs involved in cell cycle control (and cell cycle exit) may be wired in fundamentally different ways from terminal differentiation.
We propose that transcriptional regulators (Eya1 and TEAD4/TEF-3) relay the differentiation signal initiated by MyoD (Fig. 4B). Several biochemical, computational, and genetic observations suggest that the Eya1/Six1 pathway is associated with MRF function. First, our ChIP-on-chip results indicate that Eya1 is a direct target of MyoD in growing myoblasts. Eya1 has the ability to switch the activity of Six1, a homeobox transcriptional regulator, from repressor to activator (Li et al. 2003). Second, the MEF3 PWM, a binding site for Six1, is specifically enriched among myogenin target genes that are induced during differentiation (Table 3, group J). Third, mice lacking Six1 display defects in embryonic myogenesis that are exacerbated when Eya1 function is also ablated (Li et al. 2003).
TEAD4 is closely related to TEAD1 (TEF-1), the founding member of a family of transcriptional regulators that bind M-CAT DNA elements (GGAATG) (Karasseva et al. 2003). By binding M-CAT sites, TEAD4 participates in muscle-fiber-type switching and mediates in part the transcriptional effects of hypoxia and α-adrenergic-stimulated muscular hypertrophy (Karasseva et al. 2003; Shie et al. 2004). Together with the observation that the M-CAT sequence is enriched among MRF targets, this suggests that, besides regulating additional genes during the muscle hypertrophic response (Ueyama et al. 2000), TEAD4 propagates the myogenic signal originating from MyoD and cooperates with MRFs to induce the expression of their targets.
Our studies identified other transcription factors likely to be involved in propagating gene expression cascades during myogenesis. These MRF targets include Naca (skNAC), a muscle-specific transcription factor involved in muscle repair (Munz et al. 1999), and Ankrd1 and Ankrd2, muscle-specific transcriptional modulators involved in myofibril-based hypertrophic response signaling. Identifying their as yet unknown targets will be essential to elucidate their role in response to activation signals originating from MyoD.
Myogenic regulatory factors and combinatorial control
We suggest that in some cases, binding of MRFs to target promoters marks them for subsequent activation upon exposure to appropriate cues rather than leading to instantaneous induction. The stimulus might be an inducer of differentiation (as presented here) or a specific condition likely to prevail later, such as innervation, stretch, or hypoxia (see below). This hypothesis is supported by the observation that two-thirds of MyoD targets bound in myotubes were also bound in undifferentiated myoblasts. Ultimately, chromatin remodeling or recruitment of additional factors may convert promoters from a “poised” to an active state.
As further support for this idea, it appears that several transcription factors collaborate with MRFs to regulate target gene expression. Although this concept is not novel per se, we believe that the circle of MRF collaborators is more expansive than anticipated. As noted above, MEF2 and SRF were identified as likely coregulators, in agreement with previous reports (Groisman et al. 1996; Molkentin and Olson 1996). Remarkably, our analyses also identified TEAD4, Six1, and Zic1 (a GLI-related transcription factor that appears to determine muscle lineage in ascidians) (Nishida and Sawada 2001; Imai et al. 2002) as additional, novel candidate coregulators (Table 3), reinforcing our factor binding studies and published observations (Fig. 4B).
A direct role for MRFs in synapse formation and synaptic transmission
Skeletal muscles express a large number of genes that allow transmission of signals from neurons through the neuromuscular synapse, and formation of this structure occurs through exchange of information between neuron and muscle (Burden 2002). Our genome-wide analysis strongly suggests that MRFs govern the expression of genes involved in different aspects of synapse formation and function (see Fig. 2B). Previous work demonstrated that development of synapses at the NMJ is regulated by contacts between the neuron and muscle cell and suggested that establishment of the NMJ involves muscleautonomous processes. Our data confirm that muscle cells have their own intrinsic transcriptional program for establishing synapses and that these networks are controlled at least in part by the MRFs, MyoD, and myogenin. We note that this program is likely to be conserved in human cells as several NMJ genes are bound by MRFs (Fig. 1E; data not shown).
MyoD-/- mice are subject to NMJ defects (Wang et al. 2003). Their aberrant neural branching phenotype led to the hypothesis that a myocyte-derived factor affecting axonal growth is expressed under the control of MyoD. Tenascin C, an extracellular matrix glycoprotein, was proposed as a candidate, because Tnc-/- mice display defective axonal growth following muscle denervation (Cifuentes-Diaz et al. 2002). Our results lend support to this hypothesis by demonstrating that the tenascin C gene is, indeed, regulated by MyoD and myogenin. Furthermore, we found that the plexin A2 gene and two members of the semaphorin family (sema3d, sema6c), which are essential for configuring appropriate nerve projections to peripheral muscles (Taniguchi et al. 1997; Winberg et al. 1998), are also under the control of MRFs (Table 1; Fig. 2B). Together, these observations strongly suggest that the regulation of axon guidance molecules by MRFs represents a novel muscle-intrinsic aspect of NMJ development, and they help explain the NMJ phenotype of MyoD-/- mice.
Stress pathway and response to damage and hypoxia
Another striking and novel outcome of our location analysis was the identification of a cohort of genes involved in the stress and UPR. Muscle contraction leads to increased mitochondrial function and production of free radicals that must be neutralized, as excessive oxidative conditions or failure of the cellular antioxidant system can lead to muscle damage (Rando 2002). That 17 of 40 genes identified (Table 2) as targets with a potential role in stress response are transcription factors further underscores how this response may be entrained by MRFs. It may also suggest that MRFs “delegate” tasks to “specialists.” For example, by regulating the expression of Arnt, the dimerization partner of HIF1α, MRFs may support an important role played by this transcription factor in the adaptive response of skeletal muscle to exercise-induced hypoxia and prevention of trauma (Mason et al. 2004).
Additional experiments will be required to determine whether genes involved in stress-response pathways are activated to enable future responses to high oxygen consumption, calcium fluxes, or tissue damage caused by mechanical overload or ischemia, or whether they underlie a general requirement for biogenesis of the sarcoplasmic reticulum (Kaufman 1999; Knoll et al. 2002; Harding et al. 2003; Blais et al. 2004; Iwawaki et al. 2004; Kojic et al. 2004; Schulz and Yutzey 2004). Regardless of the precise role of these pathways, our approach illustrates a powerful strategy for the identification of novel pathways involved in differentiation and suggests future studies of stress and damage-response pathways in myogenesis.
Properties of a gene regulatory network involved in muscle differentiation
We linked our genome-wide factor binding and expression data with previous genetic and biochemical observations to assemble an initial network diagram for muscle differentiation (Fig. 4). This network has several salient features, including the striking number of transcription factors that are targets of MRFs and MEF2. This feature has several important consequences for the network. First, many targets identified in our experiments were regulated by more than one transcription factor, and in many cases, these targets were connected by a feed-forward regulatory loop. As in other types of biological networks, this is likely to impart robustness to the network, making it less sensitive to perturbations (Alon 2003). The large number of transcription factors involved in stress-response pathways allows a further degree of adaptability. Moreover, the involvement of multiple transcription factors promotes the propagation and amplification of signals initiated by the MRFs.
We suggest that combining ChIP-on-chip analysis with PWM enrichment analysis, expression profiling, literature mining, and phylogenetic comparisons allows us to make powerful and testable hypotheses regarding gene regulatory programs. We surmise that such combined approaches will prove to be invaluable in elucidating the complex transcriptional networks that underlie mammalian development.
Materials and methods
Cell culture
The C2C12 murine myoblast cell line was obtained from the American Type Culture Collection (ATCC). Cells were grown in GM medium (DMEM supplemented with 10% FBS). To induce differentiation, cells approaching confluence were switched to differentiation medium (DM, DMEM supplemented with 2% horse serum). Cells were maintained in DM for 96 h, after which fully differentiated myotubes were separated from undifferentiated cells by mild trypsinization as described (Carnac et al. 2000). Since C2C12 myoblasts are asynchronous with respect to their differentiation kinetics, this prolonged differentiation period allowed us to obtain myotubes that were more homogeneous in their maturity level. Human primary myoblasts were obtained from Cambrex and grown according to the supplier's instructions. Primary mouse myoblasts from MyoD+/+ and MyoD-/- animals (gift of M. Rudnicki, Ottawa Health Research Institute, Ottawa, ON, Canada) were isolated and grown as described previously (Sabourin et al. 1999).
Chromatin immunoprecipitation assays and genome-wide location analysis
ChIP assays were performed as described previously (Rayman et al. 2002). Genome-wide location analysis was performed as described (Ren and Dynlacht 2004). Modifications to the ChIP and ChIP-on-chip protocols are indicated in Supplemental Material.
Functional classification of targets
We performed initial clustering of functional categories using GO. However, since a number of targets were classified in categories that were too general or poorly defined, we performed manual inspection using PubMed, and in some cases reclassified our targets in an attempt to achieve more precise biological categorization.
Gene expression profiling
Total RNA was isolated from cells using Trizol (Invitrogen), and mRNA was amplified from 8 μg of total RNA. Labeling of cRNA and hybridization to Affymetrix mouse 430A_v2 GeneChips were performed according to the manufacturer's instructions. Each experiment was performed in triplicate. Raw data were normalized and quantified using dCHIP (Li and Hung Wong 2001). Clustering into self-organizing maps was performed using Expander (Sharan et al. 2003a).
Promoter sequence analysis
Promoter sequences of MRF target genes were analyzed using the CREME algorithm (Sharan et al. 2003b) to search for enrichment of position weight matrices (PWMs) of transcription factor-binding sites in TRANSFAC (Matys et al. 2003). The background sequence set encompassed all sequences represented on the promoter microarray. Although our microarray contains sequences spanning from -750 to +250 bp relative to the transcription start site, we analyzed 2000 bp of promoter sequence (from -1750 to +250) in order to maximize the identification of relevant enriched PWMs. This is justified given the size of chromatin fragments generated in the ChIP assay and the possibility of productive hybridization between partially overlapping chromatin fragments and the spotted DNA on the microarray.
Acknowledgments
We are grateful to B. Mishra, G. Lerman, and J. McQuown for computational analysis of ChIP-on-chip results; to D. Xie and the NYU Cancer Institute Genomics Facility for helping us print our promoter microarrays; to V. Seale and M. Rudnicki for providing MyoD-/- and wild-type myoblasts; and to P.J. Houghton for the gift of anti-MyoD hybridoma. A.B. is supported by a post-doctoral training fellowship from the Fonds de la Recherche en Santé du Québec. R.S. was supported in part by NSF ITR grant CCR-0121555. B.D.D. is supported by NIH grant 1R01GM067132-01.
Supplemental material is available at http://www.genesdev.org.
Article published online ahead of print. Article and publication date are at http://www.genesdev.org/cgi/doi/10.1101/gad.1281105.
References
- Alon U. 2003. Biological networks: The tinkerer as an engineer. Science 301: 1866-1867. [DOI] [PubMed] [Google Scholar]
- Benezra R., Davis, R.L., Lockshon, D., Turner, D.L., and Weintraub, H. 1990. The protein Id: A negative regulator of helixloop-helix DNA binding proteins. Cell 61: 49-59. [DOI] [PubMed] [Google Scholar]
- Bergstrom D.A., Penn, B.H., Strand, A., Perry, R.L., Rudnicki, M.A., and Tapscott, S.J. 2002. Promoter-specific regulation of MyoD binding and signal transduction cooperate to pattern gene expression. Mol. Cell 9: 587-600. [DOI] [PubMed] [Google Scholar]
- Blais J.D., Filipenko, V., Bi, M., Harding, H.P., Ron, D., Koumenis, C., Wouters, B.G., and Bell, J.C. 2004. Activating transcription factor 4 is translationally regulated by hypoxic stress. Mol. Cell. Biol. 24: 7469-7482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckingham M. 2001. Skeletal muscle formation in vertebrates. Curr. Opin. Genet. Dev. 11: 440-448. [DOI] [PubMed] [Google Scholar]
- Burden S.J. 2002. Building the vertebrate neuromuscular synapse. J. Neurobiol. 53: 501-511. [DOI] [PubMed] [Google Scholar]
- Cam H., Balciunaite, E., Blais, A., Spektor, A., Scarpulla, R., Young, R.A., Kluger, Y., and Dynlacht, B.D. 2004. A common set of gene regulatory networks links metabolism and growth inhibition. Mol. Cell 16: 399-411. [DOI] [PubMed] [Google Scholar]
- Carnac G., Fajas, L., L'Honore, A., Sardet, C., Lamb, N.J., and Fernandez, A. 2000. The retinoblastoma-like protein p130 is involved in the determination of reserve cells in differentiating myoblasts. Curr. Biol. 10: 543-546. [DOI] [PubMed] [Google Scholar]
- Cifuentes-Diaz C., Faille, L., Goudou, D., Schachner, M., Rieger, F., and Angaut-Petit, D. 2002. Abnormal reinnervation of skeletal muscle in a tenascin-C-deficient mouse. J. Neurosci. Res. 67: 93-99. [DOI] [PubMed] [Google Scholar]
- Fernandez P.C., Frank, S.R., Wang, L., Schroeder, M., Liu, S., Greene, J., Cocito, A., and Amati, B. 2003. Genomic targets of the human c-Myc protein. Genes & Dev. 17: 1115-1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groisman R., Masutani, H., Leibovitch, M.P., Robin, P., Soudant, I., Trouche, D., and Harel-Bellan, A. 1996. Physical interaction between the mitogen-responsive serum response factor and myogenic basic-helix-loop-helix proteins. J. Biol. Chem. 271: 5258-5264. [DOI] [PubMed] [Google Scholar]
- Harding H.P., Zhang, Y., Zeng, H., Novoa, I., Lu, P.D., Calfon, M., Sadri, N., Yun, C., Popko, B., Paules, R., et al. 2003. An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol. Cell 11: 619-633. [DOI] [PubMed] [Google Scholar]
- Hosack D.A., Dennis Jr., G., Sherman, B.T., Lane, H.C., and Lempicki, R.A. 2003. Identifying biological themes within lists of genes with EASE. Genome Biol. 4: R70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai K.S., Satou, Y., and Satoh, N. 2002. Multiple functions of a Zic-like gene in the differentiation of notochord, central nervous system and muscle in Ciona savignyi embryos. Development 129: 2723-2732. [DOI] [PubMed] [Google Scholar]
- Iwawaki T., Akai, R., Kohno, K., and Miura, M. 2004. A transgenic mouse model for monitoring endoplasmic reticulum stress. Nat. Med. 10: 98-102. [DOI] [PubMed] [Google Scholar]
- Karasseva N., Tsika, G., Ji, J., Zhang, A., Mao, X., and Tsika, R. 2003. Transcription enhancer factor 1 binds multiple muscle MEF2 and A/T-rich elements during fast-to-slow skeletal muscle fiber type transitions. Mol. Cell. Biol. 23: 5143-5164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman R.J. 1999. Stress signaling from the lumen of the endoplasmic reticulum: Coordination of gene transcriptional and translational controls. Genes & Dev. 13: 1211-1233. [DOI] [PubMed] [Google Scholar]
- Kitzmann M. and Fernandez, A. 2001. Crosstalk between cell cycle regulators and the myogenic factor MyoD in skeletal myoblasts. Cell. Mol. Life Sci. 58: 571-579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoll R., Hoshijima, M., Hoffman, H.M., Person, V., Lorenzen-Schmidt, I., Bang, M.L., Hayashi, T., Shiga, N., Yasukawa, H., Schaper, W., et al. 2002. The cardiac mechanical stretch sensor machinery involves a Z disc complex that is defective in a subset of human dilated cardiomyopathy. Cell 111: 943-955. [DOI] [PubMed] [Google Scholar]
- Kojic S., Medeot, E., Guccione, E., Krmac, H., Zara, I., Martinelli, V., Valle, G., and Faulkner, G. 2004. The Ankrd2 protein, a link between the sarcomere and the nucleus in skeletal muscle. J. Mol. Biol. 339: 313-325. [DOI] [PubMed] [Google Scholar]
- Kuroda K., Tani, S., Tamura, K., Minoguchi, S., Kurooka, H., and Honjo, T. 1999. Delta-induced Notch signaling mediated by RBP-J inhibits MyoD expression and myogenesis. J. Biol. Chem. 274: 7238-7244. [DOI] [PubMed] [Google Scholar]
- Lee A.H., Iwakoshi, N.N., and Glimcher, L.H. 2003. XBP-1 regulates a subset of endoplasmic reticulum resident chaperone genes in the unfolded protein response. Mol. Cell. Biol. 23: 7448-7459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C. and Hung Wong, W. 2001. Model-based analysis of oligonucleotide arrays: Model validation, design issues and standard error application. Genome Biol. 2: RESEARCH0032. [DOI] [PMC free article] [PubMed]
- Li X., Oghi, K.A., Zhang, J., Krones, A., Bush, K.T., Glass, C.K., Nigam, S.K., Aggarwal, A.K., Maas, R., Rose, D.W., et al. 2003. Eya protein phosphatase activity regulates Six1-Dach-Eya transcriptional effects in mammalian organogenesis. Nature 426: 247-254. [DOI] [PubMed] [Google Scholar]
- Mal A. and Harter, M.L. 2003. MyoD is functionally linked to the silencing of a muscle-specific regulatory gene prior to skeletal myogenesis. Proc. Natl. Acad. Sci. 100: 1735-1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason S.D., Howlett, R.A., Kim, M.J., Olfert, I.M., Hogan, M.C., McNulty, W., Hickey, R.P., Wagner, P.D., Kahn, C.R., Giordano, F.J., et al. 2004. Loss of skeletal muscle HIF-1α results in altered exercise endurance. PLoS Biol. 2: E288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matys V., Fricke, E., Geffers, R., Gossling, E., Haubrock, M., Hehl, R., Hornischer, K., Karas, D., Kel, A.E., Kel-Margoulis, O.V., et al. 2003. TRANSFAC: Transcriptional regulation, from patterns to profiles. Nucleic Acids Res. 31: 374-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molkentin J.D. and Olson, E.N. 1996. Combinatorial control of muscle development by basic helix-loop-helix and MADS-box transcription factors. Proc. Natl. Acad. Sci. 93: 9366-9373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran J.L., Li, Y., Hill, A.A., Mounts, W.M., and Miller, C.P. 2002. Gene expression changes during mouse skeletal myoblast differentiation revealed by transcriptional profiling. Physiol. Genomics 10: 103-111. [DOI] [PubMed] [Google Scholar]
- Munz B., Wiedmann, M., Lochmuller, H., and Werner, S. 1999. Cloning of novel injury-regulated genes. Implications for an important role of the muscle-specific protein skNAC in muscle repair. J. Biol. Chem. 274: 13305-13310. [DOI] [PubMed] [Google Scholar]
- Murray J.I., Whitfield, M.L., Trinklein, N.D., Myers, R.M., Brown, P.O., and Botstein, D. 2004. Diverse and specific gene expression responses to stresses in cultured human cells. Mol. Biol. Cell 15: 2361-2374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muscat G.E., Mynett-Johnson, L., Dowhan, D., Downes, M., and Griggs, R. 1994. Activation of myoD gene transcription by 3,5,3′-triiodo-L-thyronine: A direct role for the thyroid hormone and retinoid X receptors. Nucleic Acids Res. 22: 583-591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida H. and Sawada, K. 2001. macho-1 encodes a localized mRNA in ascidian eggs that specifies muscle fate during embryogenesis. Nature 409: 724-729. [DOI] [PubMed] [Google Scholar]
- Odom D.T., Zizlsperger, N., Gordon, D.B., Bell, G.W., Rinaldi, N.J., Murray, H.L., Volkert, T.L., Schreiber, J., Rolfe, P.A., Gifford, D.K., et al. 2004. Control of pancreas and liver gene expression by HNF transcription factors. Science 303: 1378-1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedraza-Alva G., Zingg, J.M., and Jost, J.P. 1994. AP-1 binds to a putative cAMP response element of the MyoD1 promoter and negatively modulates MyoD1 expression in dividing myoblasts. J. Biol. Chem. 269: 6978-6985. [PubMed] [Google Scholar]
- Piette J., Bessereau, J.L., Huchet, M., and Changeux, J.P. 1990. Two adjacent MyoD1-binding sites regulate expression of the acetylcholine receptor α-subunit gene. Nature 345: 353-355. [DOI] [PubMed] [Google Scholar]
- Rando T.A. 2002. Oxidative stress and the pathogenesis of muscular dystrophies. Am. J. Phys. Med. Rehabil. 81: S175-S186. [DOI] [PubMed] [Google Scholar]
- Rayman J.B., Takahashi, Y., Indjeian, V.B., Dannenberg, J.H., Catchpole, S., Watson, R.J., te Riele, H., and Dynlacht, B.D. 2002. E2F mediates cell cycle-dependent transcriptional repression in vivo by recruitment of an HDAC1/mSin3B corepressor complex. Genes & Dev. 16: 933-947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren B. and Dynlacht, B.D. 2004. Use of chromatin immunoprecipitation assays in genome-wide location analysis of mammalian transcription factors. Methods Enzymol. 376: 304-315. [DOI] [PubMed] [Google Scholar]
- Ridgeway A.G., Wilton, S., and Skerjanc, I.S. 2000. Myocyte enhancer factor 2C and myogenin up-regulate each other's expression and induce the development of skeletal muscle in P19 cells. J. Biol. Chem. 275: 41-46. [DOI] [PubMed] [Google Scholar]
- Rutkowski D.T. and Kaufman, R.J. 2004. A trip to the ER: Coping with stress. Trends Cell Biol. 14: 20-28. [DOI] [PubMed] [Google Scholar]
- Sabourin L.A., Girgis-Gabardo, A., Seale, P., Asakura, A., and Rudnicki, M.A. 1999. Reduced differentiation potential of primary MyoD-/- myogenic cells derived from adult skeletal muscle. J. Cell Biol. 144: 631-643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz R.A. and Yutzey, K.E. 2004. Calcineurin signaling and NFAT activation in cardiovascular and skeletal muscle development. Dev. Biol. 266: 1-16. [DOI] [PubMed] [Google Scholar]
- Semenza G. 2002. Signal transduction to hypoxia-inducible factor 1. Biochem. Pharmacol. 64: 993-998. [DOI] [PubMed] [Google Scholar]
- Sharan R., Maron-Katz, A., and Shamir, R. 2003a. CLICK and EXPANDER: A system for clustering and visualizing gene expression data. Bioinformatics 19: 1787-1799. [DOI] [PubMed] [Google Scholar]
- Sharan R., Ovcharenko, I., Ben-Hur, A., and Karp, R.M. 2003b. CREME: A framework for identifying cis-regulatory modules in human-mouse conserved segments. Bioinformatics 19 Suppl 1: i283-i291. [DOI] [PubMed] [Google Scholar]
- Shie J.L., Wu, G., Wu, J., Liu, F.F., Laham, R.J., Oettgen, P., and Li, J. 2004. RTEF-1, a novel transcriptional stimulator of vascular endothelial growth factor in hypoxic endothelial cells. J. Biol. Chem. 279: 25010-25016. [DOI] [PubMed] [Google Scholar]
- Taniguchi M., Yuasa, S., Fujisawa, H., Naruse, I., Saga, S., Mishina, M., and Yagi, T. 1997. Disruption of semaphorin III/D gene causes severe abnormality in peripheral nerve projection. Neuron 19: 519-530. [DOI] [PubMed] [Google Scholar]
- Tapscott S.J., Davis, R.L., Thayer, M.J., Cheng, P.F., Weintraub, H., and Lassar, A.B. 1988. MyoD1: A nuclear phosphoprotein requiring a Myc homology region to convert fibroblasts to myoblasts. Science 242: 405-411. [DOI] [PubMed] [Google Scholar]
- Ueyama T., Zhu, C., Valenzuela, Y.M., Suzow, J.G., and Stewart, A.F. 2000. Identification of the functional domain in the transcription factor RTEF-1 that mediates α 1-adrenergic signaling in hypertrophied cardiac myocytes. J. Biol. Chem. 275: 17476-17480. [DOI] [PubMed] [Google Scholar]
- Wang Z.Z., Washabaugh, C.H., Yao, Y., Wang, J.M., Zhang, L., Ontell, M.P., Watkins, S.C., Rudnicki, M.A., and Ontell, M. 2003. Aberrant development of motor axons and neuromuscular synapses in MyoD-null mice. J. Neurosci. 23: 5161-5169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winberg M.L., Noordermeer, J.N., Tamagnone, L., Comoglio, P.M., Spriggs, M.K., Tessier-Lavigne, M., and Goodman, C.S. 1998. Plexin A is a neuronal semaphorin receptor that controls axon guidance. Cell 95: 903-916. [DOI] [PubMed] [Google Scholar]


