Supplemental Digital Content is available in the text
Keywords: biomarker, diagnosis, hepatocellular carcinoma, lncRNAs, meta-analysis
Abstract
Background:
Increasing evidences have shown that long noncoding RNAs (lncRNAs) are involved in cancer diagnosis and prognosis. However, the overall diagnostic accuracy of lncRNAs for hepatocellular carcinoma (HCC) remains unclear. Herein, we perform a meta-analysis to assess diagnostic value of lncRNAs for HCC.
Methods:
The online PubMed, Cochrane, Web of Science, and Embase database were searched for eligible studies published until October 5, 2016. Study quality was evaluated with the Quality Assessment for Studies of Diagnostic Accuracy (QUADAS). All statistical analyses were conducted with Stata 12.0 and Meta-Disc 1.4.
Results:
We included 19 studies from 10 articles with 1454 patients with HCC and 1300 controls. The pooled sensitivity, specificity, positive likelihood ratio (PLR), negative likelihood ratio (NLR), diagnostic odds ratio (DOR), and AUC for lncRNAs in the diagnosis of HCC were 0.83 (95% confidence interval [CI]: 0.76–0.88), 0.80 (95% CI: 0.73–0.86), 4.2 (95% CI: 3.00–5.80), 0.21 (95% CI: 0.15–0.31), 20 (95% CI: 11–34), and 0.88 (95% CI: 0.85–0.91), respectively. Additionally, the diagnostic value of lncRNAs varied based on sex ratio of cases and characteristics of methods (specimen type and reference gen).
Conclusion:
This meta-analysis suggests lncRNAs show a moderate diagnostic accuracy for HCC. However, prospective studies are required to confirm its diagnostic value.
1. Introduction
Hepatocellular carcinoma (HCC), accounting for 70% to 90% of all primary liver cancer, is one of the most common malignant cancers and the 2nd-leading cause of cancer-related death worldwide.[1,2] Surgical resection is a gold standard therapy for HCC[3]; however, HCC is often diagnosed at advanced stages due to inefficient screening, and many patients miss the chance of surgery, which leads to a very poor prognosis with the 5-year survival rate at 7%.[4,5] Therefore, early diagnosis of HCC is vital to improve patient's survival and facilitate cancer prevention.
Until now, serum biomarker detection[6,7] and imaging technology are commonly used for HCC screening.[8] However, the usefulness of serological markers is limited due to unsatisfied sensitivity and specificity. Alpha fetoprotein (AFP), the most widely used tumor marker for HCC, may remain normal in almost 40% of patients with early stage HCC, and even in 15% to 30% of advanced patients.[9,10] Furthermore, patients with chronic hepatitis B (HBV) and/or C may also be with increased AFP concentrations.[11] Ultrasonography is an ideal and cost-effective screening technique method to identify HCC patients in early screening, yet it fails to distinguish nodules of less than 3 cm.[12] CT and MRI have accredited sensitivity (55%–91%) and specificity (77%–96%) in diagnosis of the early stage of HCC.[13,14] However, owing to high expense and radiation exposure, it is unpractical for large-scale screening and routine surveillance. Therefore, there is an urgent need to identify a noninvasive, cost-effective, and sensitive diagnostic biomarkers to improve diagnosis and screening strategies of HCC.
Long noncoding RNAs (lncRNAs), a class of ncRNAs longer than 200 nucleotides, are disable to code proteins.[15] They have multiple functions, such as modulating protein and RNA activity, regulating transcription, protein trafficking and cell metabolism, and also acting as structural components.[15,16] Notably, many studies have evidently revealed the important roles of lncRNAs in the formation, progression, and prognosis of HCC.[17–19] Recently, some researchers have found that lncRNAs are stably detected, and mounting evidences indicate that these abnormal expressed lncRNAs may served as a diagnostic biomarker for multiple diseases.[20,21] However, considering the limits of single study, such as small sample size, heterogeneous populations, and differences in detection techniques, the diagnostic accuracy of lncRNAs for HCC is still unclear. Thus, this meta-analysis focused on assessing its overall diagnostic value for HCC.
2. Methods
2.1. Search strategy and selection criteria
We performed this meta-analysis in accordance with the PRISMA 2009 guidelines (Supplement S1).[22] The online PubMed, Cochrane, Web of Science, and Embase database were searched until October 5th 2016. The keywords for the search included: “liver cancer or neoplasm or carcinoma” AND “long non-coding RNAs or lncRNAs” AND “sensitivity or diagnosis or AUC or ROC or specificity.” References of eligible articles and relevant reviews were also manually searched to find out potential studies. As this meta-analysis was based on previous published studies, ethical approval and patient consent were not necessary.
The included studies must meet these criteria: about diagnostic performance of lncRNAs for HCC; HCC was diagnosed based on pathological examination; and published studies must provide sufficient data to construct the diagnostic 2-by-2 tables. The exclusion criteria were: duplicate articles; letters, reviews, meta-analyses, editorials, and case reports; and studies without sufficient diagnostic data. Any related articles were carefully assessed by 2 researchers (HQQ and CGY) independently. Disagreements were resolved by discussion.
2.2. Data extraction and quality assessment
We extracted information of studies as follows: details of studies (first author, published date, and country), clinical characteristics of subjects (number of participants, sex ratio, and sources of control), details of detection method (specimen type, reagents, cut-off value, reference gene, and lncRNAs profiles), and diagnostic performance (sensitivity, specificity, and data of 2-by-2 tables). If the article contained the overlapping data that evaluated the diagnostic accuracy of the same lncRNA, only the largest study was selected. If the study contains the training and validating cohorts, information from each cohort was all extracted and deemed as an individual study.
Study quality was evaluated with the Quality Assessment of Diagnostic Accuracy Studies (QUADAS) tool.[23] According to the 14-items scoring criteria, a score of 1 for “yes,” 0 for “unclear” and “no” (high risk) were given, respectively.[24]
Two reviewers (HQQ and CGY) performed data extraction and quality assessment independently. Any disagreements were resolved by consensus.
2.3. Statistical analysis
Statistical analysis was conducted with the STATA 12.0 and Meta-Disc 1.4. Pooled results were used to estimate sensitivity, specificity, diagnostic odds ratio (DOR), positive diagnostic likelihood ratio (positive likelihood ratio, PLR), and negative diagnostic likelihood ratio (negative likelihood ratio, NLR) with the bivariate analysis. The heterogeneity from the threshold and nonthreshold effects was assessed using the Spearman correlation analysis method, Cochran-Q, and inconsistency index (I2) tests, respectively. A P value (≤.05) and I2 value (≥50%) indicated significant heterogeneity existed across studies, then a random-effect model was conducted. Subgroup analysis and meta-regression were performed to explore the sources of heterogeneity. Country, sample size, sex ratio, source of control, specimen type, method, lncRNAs profiles, reference gene, and QUADAS scores were as covariates. Sensitivity and influence analysis were further performed to find the potential sources of heterogeneity.[24,25] At last, the publication bias was estimated with Deek funnel plot and a P value < .1 showed statistical significance.[26]
3. Results
3.1. Literature selection
As showed in Fig. 1, 287 articles from databases were initially identified; titles and abstracts were reviewed after 98 duplicated articles were excluded; due to letters, reviews, meta-analyses, or irrelevant research topic, further 166 articles were excluded, leaving 23 articles for full-text review; as a result, 13 articles were finally excluded due to unrelated to cancer diagnosis, insufficient data, or irrelevant to our topic. Finally, 10 articles[27–36] containing 19 studies were identified.
Figure 1.

Flow diagram of study selection process.
3.2. Study characteristics and quality assessment
Nineteen studies from 10 articles with 1454 HCC and 1300 matched controls were included. The patient demographics of each study were present in Supplement S2. The size of case and control groups ranged from 20 to 147 and 20 to 232, respectively. Nine studies were conducted in China, 1 in Japan, and 9 in Egypt; the patients with HCC were all confirmed by pathological examination; among these studies, lncRNAs levels were detected with the quantitative reverse transcription PCR (qRT-PCR) method, but the reference gene and specimen types were inconsistent; additionally, circulating lncRNAs were detected in 18 studies; 6 of 19 studies evaluated the diagnostic performance of a panel of lncRNAs for HCC, and the rest 13 assessed the diagnostic accuracy of single lncRNA (Table 1).
Table 1.
Main characteristics of 19 studies included in meta-analysis.
QUADAS-1 summary plot was presented in Fig. 2. According to the criteria, all the 19 studies achieved QUADAS scores equal or greater than 10 (Table 1), indicating moderate quality. The details of the quality assessment of each study were presented in Supplement S3.
Figure 2.

Study quality assessment using the Quality Assessment of Diagnostic Accuracy Studies (QUADAS) checklist.
3.3. Diagnostic accuracy and threshold analysis
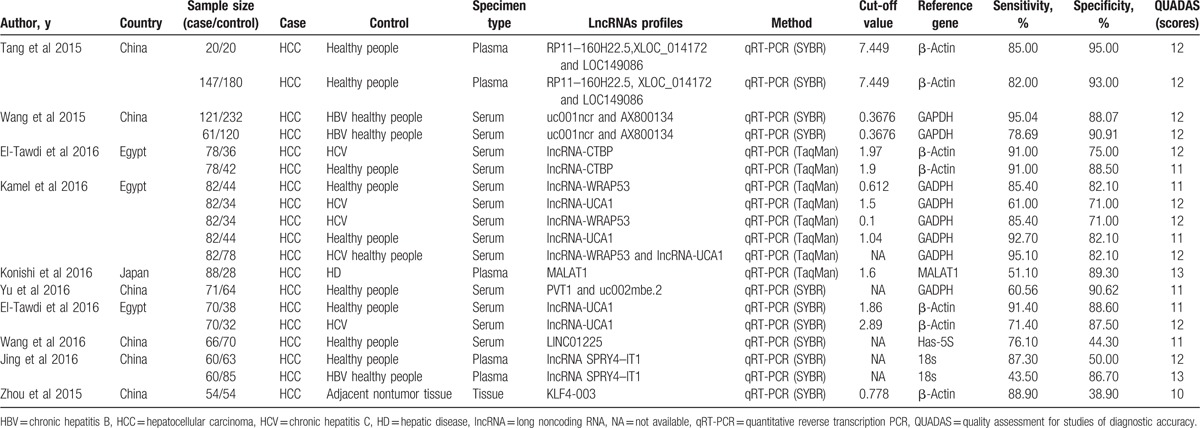
In our study, threshold effects, considered as one of the important reasons for heterogeneity, was assessed by Spearman correlation coefficient with Meta-Disc software. The Spearman correlation coefficient was 0.103 (P = .675), suggesting no obvious heterogeneity from threshold effect. Then heterogeneity from nonthreshold was evaluated by Cochran-Q and inconsistency index (I2) tests. There was substantial heterogeneity in pooled sensitivity (I2 = 91.58%, P < .01) and pooled specificity (I2 = 90.03%, P < .01), then, a random-effect model was conducted. The pooled sensitivity, specificity, PLR, NLR, and DOR of lncRNAs in HCC diagnosis were 0.83 (95% confidence interval [CI]: 0.76–0.88), 0.80 (95% CI: 0.73–0.86), 4.2 (95% CI: 3.00–5.80), 0.21 (95% CI: 0.15–0.31), and 20 (95% CI: 11–34), respectively (Fig. 3). The summary receiver operator characteristic curve was also plotted. As shown in Fig. 4, circulating lncRNAs achieved an AUC of 0.88 (95% CI: 0.85–0.91), which suggesting a moderate accuracy in HCC diagnosis.
Figure 3.

Forest plots of sensitivities and specificities of LncRNAs in HCC diagnosis for all included studies. HCC = hepatocellular carcinoma, lncRNA = long noncoding RNA.
Figure 4.
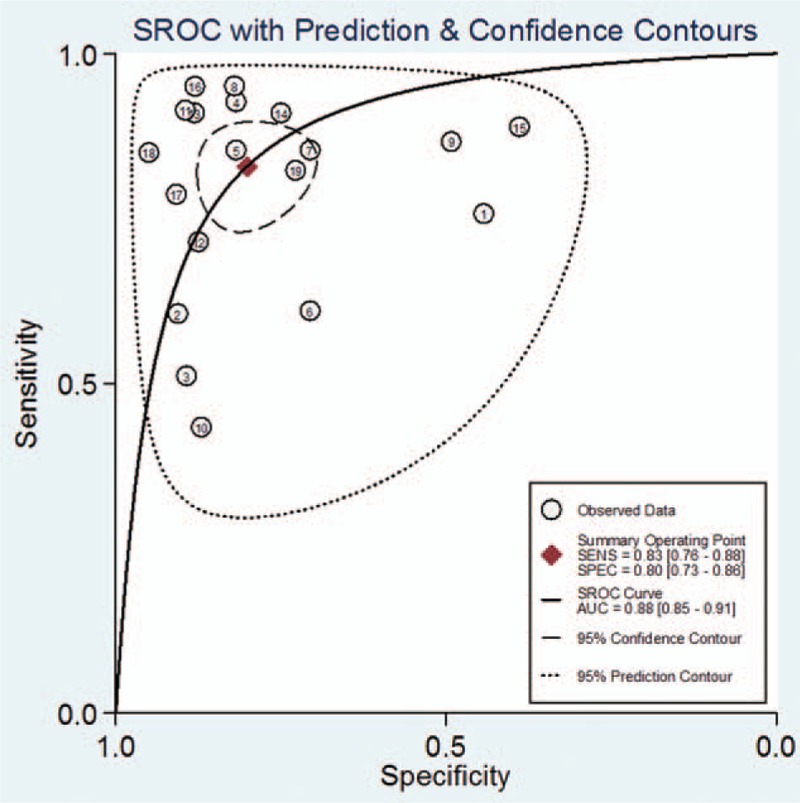
SROC curve for lncRNAs in the diagnosis of HCC. HCC = hepatocellular carcinoma, lncRNA = long noncoding RNA, SROC = summary receiver operator characteristic curve.
3.4. Subgroup analysis and meta-regression
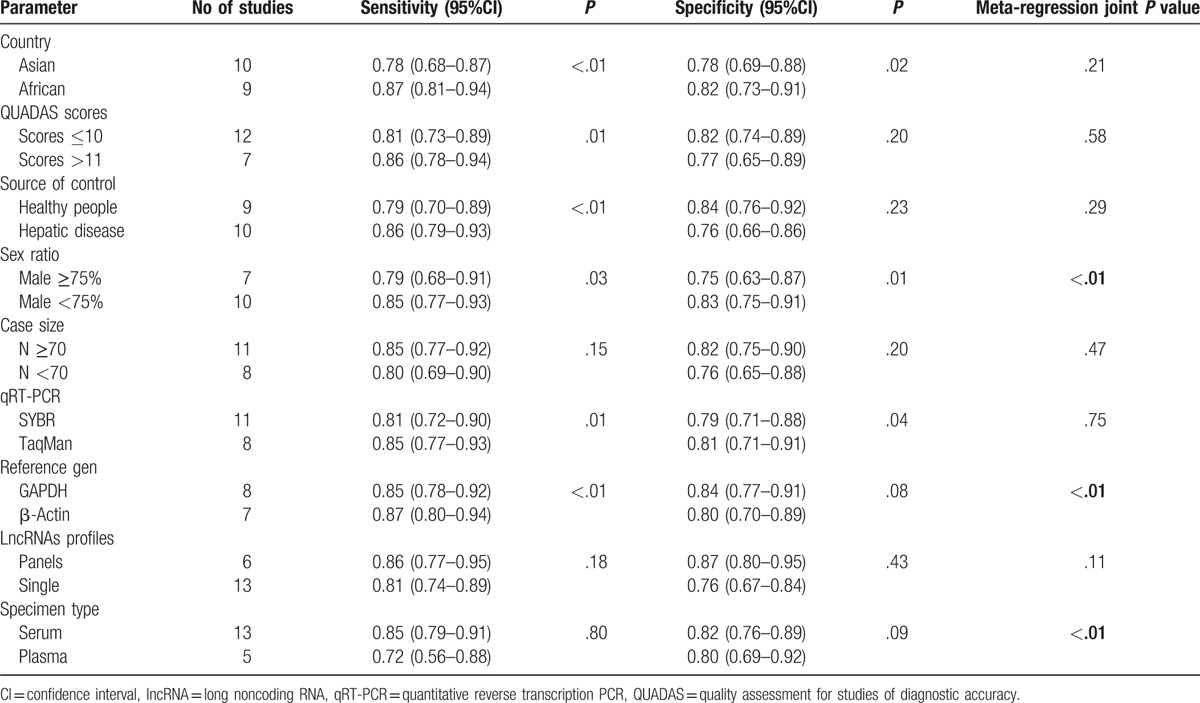
As displayed in Table 2, studies with male ≥75% or plasma as specimen tended to have significantly lower sensitivity and specificity than those with male <75% (joint P < .01) or serum as specimen (joint P < .01). Significantly lower sensitivity and higher specificity were reported in studies with GAPDH as reference gene than in those with β-actin as reference gene (joint P < .01). Additionally, studies with panels of lncRNAs as diagnostic biomarkers or qRT-PCR(TaqMan) as detection method revealed higher sensitivity and specificity than those with single lncRNAs or qRT-PCR(SYBR); however, the differences were not significant (joint P = .11, P = .75). And sensitivity and specificity did not change significantly, regardless of the country in which the studies were performed, QUADAS scores, size of cases, and the source of control.
Table 2.
Results of subgroup analyses and univariate meta-regression.
3.5. Sensitivity and influence analysis
We then performed the influence analysis to explore effects of each study. As indicated in Fig. 5, after individual study was separately omitted, 1 outlier study was identified, indicating that the Wang et al[33] study might be a source of heterogeneity. We then conducted sensitivity analyses. After the Wang et al study was excluded, the I2 and summary statistics altered minimally. Among 19 included studies, only Zhou et al[36] study assessed the diagnostic value of lncRNAs for HCC in tissue, and possessed the lowest quality simultaneously. After this study was excluded, the I2and summary statistics also did not alter significantly.
Figure 5.
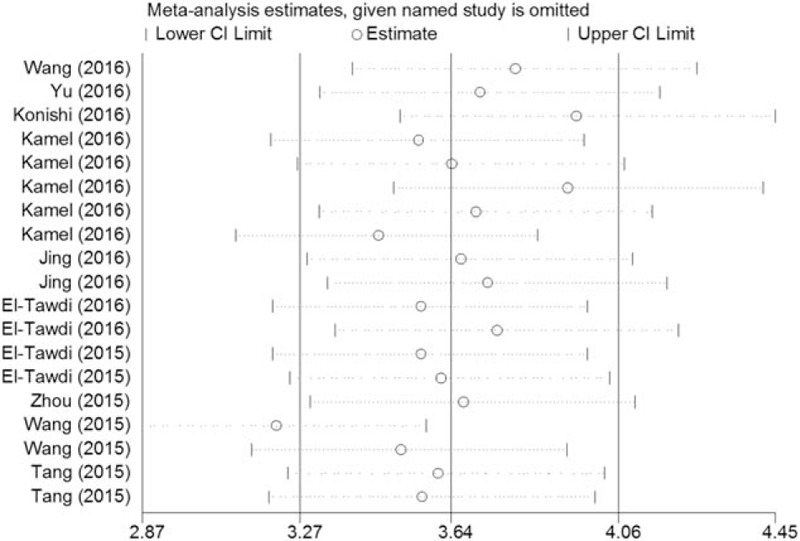
Influence analysis of the overall pooled study (outlier detection analysis). Influence analysis was performed with Stata 12.0.
3.6. Publication bias
To evaluate publication bias, Deeks funnel plot asymmetry test was performed. As displayed in Fig. 6, P = .42 suggested that the slope coefficient did not reveal obvious evidences of asymmetry, thus there was no potential publication bias among studies.
Figure 6.
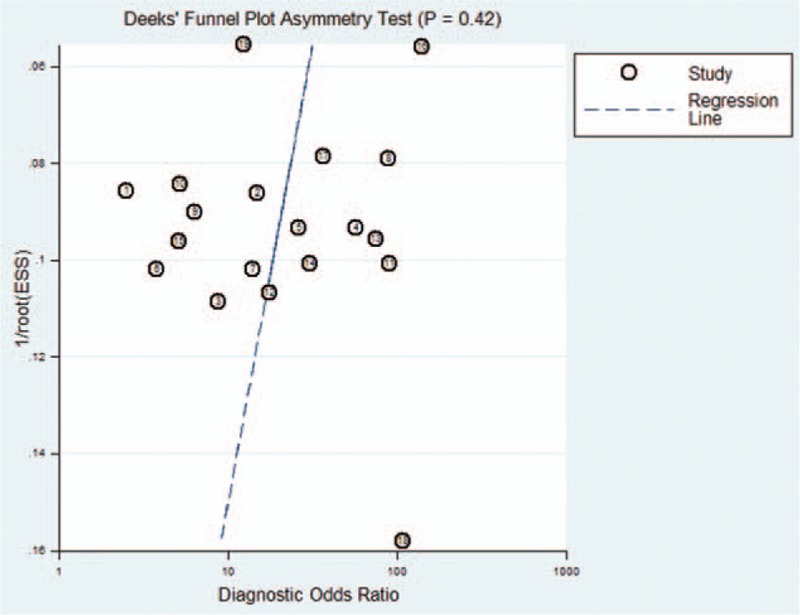
Deeks’ funnel plots for the overall studies included in the meta-analysis.
4. Discussion
HCC is the most common malignancy. Due to inefficient methods for screening and diagnosis, the prognosis is poor. Hence, identification of reliable diagnostic biomarkers for HCC is urgently needed.
In present study, the overall pooled sensitivity and specificity of lncRNAs for HCC detection were 0.83 and 0.80, with an AUC value of 0.88, indicating that the diagnostic accuracy of lncRNAs is moderate. DOR, another global index of diagnostic accuracy,[37] converts the strengths of sensitivity and specificity into a single indictor, and the larger the value of DOR is, the higher accuracy it indicates.[38] The overall pooled DOR of lncRNAs was 20 in this study, suggesting a moderate diagnostic accuracy. For a diagnostic test, a high PLR and a low NLR value present superior performance.[39] Given the moderate PLR and NLR in our study, these results are not sufficient to rule in or out the diagnosis of HCC.
However, there are still some points that support the potential clinical practice of lncRNAs as a diagnostic biomarker: First, lncRNAs are characterized with the relatively stable in body fluids and are detectable in tumor tissues or peripheral blood, which make them suitable noninvasive biomarkers.[40,41] Second, the diagnostic accuracy of single lncRNA may be relatively poor, a combination of multiple biomarkers therefore seems to be promising for HCC screening. As reported, lncRNAs could discriminate patients with HCC from healthy people with higher accuracy than the existing biomarkers (sensitivity: 91.4% vs 82.8%; specificity: 88.6% vs 82.8%), when combined with AFP, the sensitivity significant raises to 100%.[28] Likewise, a panel of lncRNAs (lncRNA-UCA1 and lncRNA-WRAP53) also achieved a higher accuracy than single lncRNA.[31] However, in contrast to these studies,[30,31,42] the difference in overall sensitivity and specificity was not statistical significant between panels of lncRNAs and single lncRNA in our study. This might be because of differences in the study quality, heterogeneous populations, and detection techniques.
Early-stage HCC, with 5-year survival rate from 50% to 75%, can be effectively treated. However, patients with advanced stage have a dismal prognosis (50% survival at 1 year). Even worse, the median survival is less than 3 months for patients with end stage.[43–46] Thus, it is vital to make an early diagnosis. As reported by Wang et al, HBV-positive HCC could be accurately diagnosed with a panel of lncRNAs, with AUC values of 0.9494 and 0.9491 for 2 cohorts, respectively. Excitingly, the diagnostic accuracy remained high at early Barcelona Clinic Liver Cancer stages (AUC values of 0.945 and 0.9564, respectively). Regrettably, due to incomplete clinical characteristics, we failed to estimate the diagnostic value of lncRNAs for early-stage HCC. Further researches are needed.
In Africa, Egypt has the highest prevalence of hepatitis C virus in the world.[30] And in Asia, HCC in China alone accounts for >50% of the cases worldwide due to the prevalence of HBV.[35] The diagnostic accuracy of lncRNAs may be affected by different virus infection of patients with HCC. However, according to subgroup analysis, the differences in diagnostic accuracy of lncRNAs between Asian and African county were not statistically significant. Thus, more studies are required to confirm this point in the future. Like miRNAs, the diagnostic value of lncRNAs varies based on differences of detection methods.[47,48] Therefore, to minimize protocol-based bias and make the results comparable, standardized protocol is needed to be established.[49] In our study, studies with GAPDH as reference gene had lower sensitivity and higher specificity than those with β-actin as reference gene. What is more, plasma-based lncRNA profile achieved lower accuracy than serum-based assay, indicating that matrix differences may influence the diagnostic accuracy of lncRNA and analysis using serum may be better.
Sex difference is one of risk factors for HCC, differences in lifestyle may be partly account for this.[50] Recently, sex hormones are found to play a vital role in the development of HCC. As reported by Naugler et al,[51] the gender disparity in HCC may be explained by estrogen-mediated inhibition of IL-6 production. Interestingly, our study demonstrated that sex differences also impact the diagnostic accuracy of lncRNAs for HCC, studies with male ≥75% tended to have lower diagnostic accuracy than those with male <75%.
In this study, substantial heterogeneity was found among overall studies. We found no evidence of heterogeneity from the threshold effect. Meta-regression and subgroup analysis were then performed. According to the results, the diagnostic value of lncRNAs differed depending on sex ratio of cases, and characteristics of methods (specimen type and reference gen). We also performed sensitivity and influence analysis, and found Wang et al study was an outlier. After this study and Zhou et al study with the lowest quality was excluded, respectively, the overall results did not alter significantly. There might be other potential sources of heterogeneity, such as, mean age, virus infection, tumor stage, status of smoking, and ethanol intake. Unfortunately, meta-regression based on these variables was failed to be done due to incomplete clinical data.
Finally, the following limitations merit consideration. First, it is vital for the diagnostic biomarkers that they could distinguish patients with HCC from not only healthy people but also patients with diseases, especially with similar symptom. However, the control sources of half of included studies were almost from healthy blood, which would lead to an overestimate of diagnostic value. Second, there was substantial heterogeneity among included studies. The results of subgroup analyses might not fully explain the observed heterogeneity. Due to limited clinical characteristics, we failed to find other sources of heterogeneity and estimate the values of lncRNAs as a diagnostic biomarker for HCC at early stages. Finally, only studies conducted in Asia and Africa were included, leading a population selection bias. It remains unknown whether these findings may be applied to other parts of the world.
In conclusion, the results of our study indicates that lncRNAs has moderate diagnostic accuracy for HCC. Nevertheless, because of substantial heterogeneity among the included studies, further large-scale, high-quality, and multicenter validation studies are required to confirm these findings.
Supplementary Material
Footnotes
Abbreviations: AFP = alpha fetoprotein, AUC = area under the SROC, BCLC = Barcelona Clinic Liver Cancer, CI = confidence interval, DOR = diagnostic odds ratio, HBV = chronic hepatitis B, HCC = hepatocellular carcinoma, lncRNA = long noncoding RNA, NLR = negative likelihood ratio, PLR = positive likelihood ratio, qRT-PCR = quantitative reverse transcription PCR, QUADAS = Quality Assessment of Diagnostic Accuracy Studies.
The authors have no funding and conflicts of interest to disclose.
Supplemental Digital Content is available for this article.
References
- [1].Shimakawa Y, Lemoine M, Njai HF, et al. Natural history of chronic HBV infection in West Africa: a longitudinal population-based study from The Gambia. Gut 2016;65:2007–16. [DOI] [PubMed] [Google Scholar]
- [2].Maluccio M, Covey A. Recent progress in understanding, diagnosing, and treating hepatocellular carcinoma. CA Cancer J Clin 2012;62:394–9. [DOI] [PubMed] [Google Scholar]
- [3].Ercolani G, Grazi GL, Ravaioli M, et al. Liver resection for hepatocellular carcinoma on cirrhosis: univariate and multivariate analysis of risk factors for intrahepatic recurrence. Ann Surg 2003;237:536–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Schwartz M, Roayaie S, Konstadoulakis M. Strategies for the management of hepatocellular carcinoma. Nat Clin Pract Oncol 2007;4:424–32. [DOI] [PubMed] [Google Scholar]
- [5].Ilikhan SU, Bilici M, Sahin H, et al. Assessment of the correlation between serum prolidase and alpha-fetoprotein levels in patients with hepatocellular carcinoma. World J Gastroenterol 2015;21:6999–7007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Marrero JA, Su GL, Wei W, et al. Des-gamma carboxyprothrombin can differentiate hepatocellular carcinoma from nonmalignant chronic liver disease in American patients. Hepatology (Baltimore, MD) 2003;37:1114–21. [DOI] [PubMed] [Google Scholar]
- [7].Lok AS, Seeff LB, Morgan TR, et al. Incidence of hepatocellular carcinoma and associated risk factors in hepatitis C-related advanced liver disease. Gastroenterology 2009;136:138–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].EASL-EORTC clinical practice guidelines: management of hepatocellular carcinoma. J Hepatol 2012;56:908–43. [DOI] [PubMed] [Google Scholar]
- [9].Zhang G, Ha SA, Kim HK, et al. Combined analysis of AFP and HCCR-1 as an useful serological marker for small hepatocellular carcinoma: a prospective cohort study. Dis Markers 2012;32:265–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Zhou L, Liu J, Luo F. Serum tumor markers for detection of hepatocellular carcinoma. World J Gastroenterol 2006;12:1175–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Corvalan AH. Early diagnosis of hepatocellular carcinoma by microRNAs: shining a light from the genome's “dark matter”. Digest Dis Sci 2012;57:2737–9. [DOI] [PubMed] [Google Scholar]
- [12].Sheu JC, Sung JL, Chen DS, et al. Early detection of hepatocellular carcinoma by real-time ultrasonography. A prospective study. Cancer 1985;56:660–6. [DOI] [PubMed] [Google Scholar]
- [13].Song ZZ. Diagnosis of hepatic nodules 20 mm or smaller in cirrhosis: prospective validation of the noninvasive diagnostic criteria for hepatocellular carcinoma. Hepatology (Baltimore, MD) 2008;47:2145–6. author reply 2146–7. [DOI] [PubMed] [Google Scholar]
- [14].Jain D. Tissue diagnosis of hepatocellular carcinoma. J Clin Exp Hepatol 2014;4(Suppl 3):S67–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell 2009;136:629–41. [DOI] [PubMed] [Google Scholar]
- [16].Shi X, Sun M, Liu H, et al. Long non-coding RNAs: a new frontier in the study of human diseases. Cancer Lett 2013;339:159–66. [DOI] [PubMed] [Google Scholar]
- [17].Yuan SX, Tao QF, Wang J, et al. Antisense long non-coding RNA PCNA-AS1 promotes tumor growth by regulating proliferating cell nuclear antigen in hepatocellular carcinoma. Cancer Lett 2014;349:87–94. [DOI] [PubMed] [Google Scholar]
- [18].Yuan JH, Yang F, Wang F, et al. A long noncoding RNA activated by TGF-beta promotes the invasion-metastasis cascade in hepatocellular carcinoma. Cancer Cell 2014;25:666–81. [DOI] [PubMed] [Google Scholar]
- [19].Zheng H, Yang S, Yang Y, et al. Epigenetically silenced long noncoding-SRHC promotes proliferation of hepatocellular carcinoma. J Cancer Res Clin Oncol 2015;141:1195–203. [DOI] [PubMed] [Google Scholar]
- [20].Lorenzen JM, Schauerte C, Kielstein JT, et al. Circulating long noncoding RNATapSaki is a predictor of mortality in critically ill patients with acute kidney injury. Clin Chem 2015;61:191–201. [DOI] [PubMed] [Google Scholar]
- [21].Toth K, Bartak BK, Tulassay Z, et al. Circulating cell-free nucleic acids as biomarkers in colorectal cancer screening and diagnosis. Expert Rev Mol Diagn 2016;16:239–52. [DOI] [PubMed] [Google Scholar]
- [22].Liberati A, Altman DG, Tetzlaff J, et al. The PRISMA statement for reporting systematic reviews and meta-analyses of studies that evaluate health care interventions: explanation and elaboration. Ann Intern Med 2009;151:W65–94. [DOI] [PubMed] [Google Scholar]
- [23].Whiting P, Rutjes AW, Reitsma JB, et al. The development of QUADAS: a tool for the quality assessment of studies of diagnostic accuracy included in systematic reviews. BMC Med Res Methodol 2003;3:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Cui Z, Chen Y, Xiao Z, et al. Long noncoding RNAs as auxiliary biomarkers for gastric cancer screening: a pooled analysis of individual studies. Oncotarget 2016;7:25791–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Zheng D, Guo Z, Schroder PM, et al. Accuracy of MR imaging and MR spectroscopy for detection and quantification of hepatic steatosis in living liver donors: a meta-analysis. Radiology 2016;152571. [DOI] [PubMed] [Google Scholar]
- [26].Wei D, Wan Q, Li L, et al. MicroRNAs as potential biomarkers for diagnosing cancers of central nervous system: a meta-analysis. Mol Neurobiol 2015;51:1452–61. [DOI] [PubMed] [Google Scholar]
- [27].El-Tawdi AH, Matboli M, Shehata HH, et al. Evaluation of circulatory RNA-based biomarker panel in hepatocellular carcinoma. Mol Diagn Ther 2016;20:265–77. [DOI] [PubMed] [Google Scholar]
- [28].El-Tawdi AHF, Matboli M, El-Nakeep S, et al. Association of long noncoding RNA and c-JUN expression in hepatocellular carcinoma. Expert Rev Gastroenterol Hepatol 2016;10:869–77. [DOI] [PubMed] [Google Scholar]
- [29].Jing W, Gao SS, Zhu M, et al. Potential diagnostic value of lncRNA SPRY4-IT1 in hepatocellular carcinoma. Oncol Rep 2016;36:1085–92. [DOI] [PubMed] [Google Scholar]
- [30].Kamel MM, Matboli M, Sallam M, et al. Investigation of long noncoding RNAs expression profile as potential serum biomarkers in patients with hepatocellular carcinoma. Transl Res 2016;168:134–45. [DOI] [PubMed] [Google Scholar]
- [31].Konishi H, Ichikawa D, Yamamoto Y, et al. Plasma level of metastasis-associated lung adenocarcinoma transcript 1 is associated with liver damage and predicts development of hepatocellular carcinoma. Cancer Sci 2016;107:149–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Tang J, Jiang R, Deng L, et al. Circulation long non-coding RNAs act as biomarkers for predicting tumorigenesis and metastasis in hepatocellular carcinoma. Oncotarget 2015;6:4505–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Wang K, Guo WX, Li N, et al. Serum LncRNAs profiles serve as novel potential biomarkers for the diagnosis of HBV-positive hepatocellular carcinoma. PloS One 2015;10:e0144934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Wang X, Zhang W, Tang J, et al. LINC01225 promotes occurrence and metastasis of hepatocellular carcinoma in an epidermal growth factor receptor-dependent pathway. Cell Death Dis 2016;7:e2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Yu J, Han J, Zhang J, et al. The long noncoding RNAs PVT1 and uc002mbe.2 in sera provide a new supplementary method for hepatocellular carcinoma diagnosis. Medicine 2016;95:e4436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Zhou JW, Lai PBS, Tsui SKW. Identification of a non-coding KLF4 transcript generated from intron retention and downregulated in human hepatocellular carcinoma. Int J Oncol 2015;47:1554–62. [DOI] [PubMed] [Google Scholar]
- [37].Dai M, Chen X, Liu X, et al. Diagnostic value of the combination of golgi protein 73 and alpha-fetoprotein in hepatocellular carcinoma: a meta-analysis. PloS One 2015;10:e0140067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Duval S, Tweedie R. Trim and fill: a simple funnel-plot-based method of testing and adjusting for publication bias in meta-analysis. Biometrics 2000;56:455–63. [DOI] [PubMed] [Google Scholar]
- [39].Cai Q, Wu Y, Guo Z, et al. Urine BLCA-4 exerts potential role in detecting patients with bladder cancers: a pooled analysis of individual studies. Oncotarget 2015;6:37500–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Maruyama R, Suzuki H. Long noncoding RNA involvement in cancer. BMB Rep 2012;45:604–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Tong YS, Wang XW, Zhou XL, et al. Identification of the long non-coding RNA POU3F3 in plasma as a novel biomarker for diagnosis of esophageal squamous cell carcinoma. Mol Cancer 2015;14:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Li J, Wang X, Tang J, et al. HULC and Linc00152 act as novel biomarkers in predicting diagnosis of hepatocellular carcinoma. Cell Physiol Biochem 2015;37:687–96. [DOI] [PubMed] [Google Scholar]
- [43].Bruix J, Sherman M. American association for the study of liver D. Management of hepatocellular carcinoma: an update. Hepatology (Baltimore, MD) 2011;53:1020–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Llovet JM, Burroughs A, Bruix J. Hepatocellular carcinoma. Lancet 2003;362:1907–17. [DOI] [PubMed] [Google Scholar]
- [45].Llovet JM, Di Bisceglie AM, Bruix J, et al. Design and endpoints of clinical trials in hepatocellular carcinoma. J Natl Cancer Inst 2008;100:698–711. [DOI] [PubMed] [Google Scholar]
- [46].Zhang BH, Yang BH, Tang ZY. Randomized controlled trial of screening for hepatocellular carcinoma. J Cancer Res Clin Oncol 2004;130:417–22. [DOI] [PubMed] [Google Scholar]
- [47].Dong L, Qi P, Xu MD, et al. Circulating CUDR, LSINCT-5 and PTENP1 long noncoding RNAs in sera distinguish patients with gastric cancer from healthy controls. Int J Cancer 2015;137:1128–35. [DOI] [PubMed] [Google Scholar]
- [48].Baraniskin A, Kuhnhenn J, Schlegel U, et al. Identification of microRNAs in the cerebrospinal fluid as marker for primary diffuse large B-cell lymphoma of the central nervous system. Blood 2011;117:3140–6. [DOI] [PubMed] [Google Scholar]
- [49].Fan YH, Ye MH, Wu L, et al. BRAF-activated lncRNA predicts gastrointestinal cancer patient prognosis: a meta-analysis. Oncotarget 2017;8:6295–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Nakagawa H, Maeda S, Yoshida H, et al. Serum IL-6 levels and the risk for hepatocarcinogenesis in chronic hepatitis C patients: an analysis based on gender differences. Int J Cancer 2009;125:2264–9. [DOI] [PubMed] [Google Scholar]
- [51].Naugler WE, Sakurai T, Kim S, et al. Gender disparity in liver cancer due to sex differences in MyD88-dependent IL-6 production. Science (New York, NY) 2007;317:121–4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


