Abstract
Aminoacyl-tRNA (aa-tRNA) formation, an essential process in protein biosynthesis, is generally achieved by direct attachment of an amino acid to tRNA by the aa-tRNA synthetases. An exception is Gln-tRNA synthesis, which in eukaryotes is catalyzed by glutaminyl-tRNA synthetase (GlnRS), while most bacteria, archaea, and chloroplasts employ the transamidation pathway, in which a tRNA-dependent glutamate modification generates Gln-tRNA. Mitochondrial protein synthesis is carried out normally by mitochondrial enzymes and organelle-encoded tRNAs that are different from their cytoplasmic counterparts. Early work suggested that mitochondria use the transamidation pathway for Gln-tRNA formation. We found no biochemical support for this in Saccharomyces cerevisiae mitochondria, but demonstrated the presence of the cytoplasmic GlnRS in the organelle and its involvement in mitochondrial Gln-tRNA synthesis. In addition, we showed in vivo localization of cytoplasmic tRNAGln in mitochondria and demonstrated its role in mitochondrial translation. We furthermore reconstituted in vitro cytoplasmic tRNAGln import into mitochondria by a novel mechanism. This tRNA import mechanism expands our knowledge of RNA trafficking in the eukaryotic cell. These findings change our view of the evolution of organellar protein synthesis.
Keywords: Saccharomyces cerevisiae, aminoacyl-tRNA, glutaminyl-tRNA synthetase, mitochondrion, protein synthesis, tRNA import
Gln-tRNA formation is the least conserved route of aminoacyl-tRNA (aa-tRNA) synthesis found in nature, and is generated by kingdom-specific pathways that guarantee proper glutamine incorporation during protein synthesis (Ibba and Söll 2004). In the eukaryotic cytoplasm and in some bacteria, glutaminyl-tRNA synthetase (GlnRS) attaches glutamine directly to tRNA (Fig. 1). A different pathway, found in most bacteria and all archaea, employs a pretranslational modification to generate Gln-tRNAGln (Fig. 1). tRNAGln is first misaminoacylated with glutamate by a nondiscriminating glutamyl-tRNA synthetase (GluRS), which is able to synthesize both Glu-tRNAGlu and Glu-tRNAGln (Lapointe et al. 1986; Sekine et al. 2001). The resulting mischarged tRNA is then converted by glutamyl-tRNAGln amidotransferase into Gln-tRNAGln (Feng et al. 2004). Mitochondria and chloroplasts, in line with their bacterial ancestry, are thought to share the same routes of aa-tRNA synthesis with bacteria and archaea (Yang et al. 1985; Schön et al. 1988; Woese et al. 2000). This is underscored by the fact that most chloroplast and mitochondrial genomes encode a complete set of tRNAs necessary for organellar protein synthesis (Barrell et al. 1980; Bonitz et al. 1980; Heckman et al. 1980). In line with this expectation, Gln-tRNA formation in barley chloroplasts was shown to occur by transamidation, and this led to the suggestion that the same route of Gln-tRNA synthesis is also prevalent in mitochondria (Schön et al. 1988).
Figure 1.
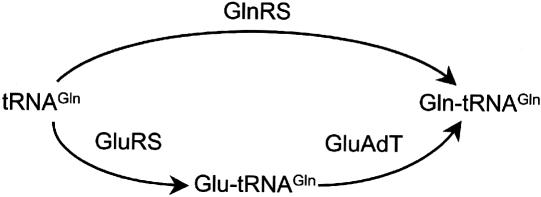
Pathways to Gln-tRNA formation. The direct aminoacylation pathway (top) and the transamidation pathway (bottom) are both abundant in nature. GluAdT refers to the tRNA-dependent amidotransferase of the transamidation pathway.
Proteins orthologous to bacterial amidotransferases are frequently found in the nuclear genomes of most eukaryotes. Bacterial amidotransferases are heterotrimeric enzymes consisting of A, B, and C subunits. Orthologs to the A and B subunits have been annotated in the Saccharomyces cerevisiae genome (PET112 and YMR293c) and are essential to mitochondrial function (Mulero et al. 1994; Hughes et al. 2000). Most eukaryotes also contain mitochondrial GluRSs that are distinct from their cytoplasmic counterparts and could potentially participate in the transamidation pathway. Despite these observations, a mitochondrial amidotransferase has not been characterized and the ability of a mitochondrial GluRS (mGluRS) to form mischarged Glu-tRNAGln has not been rigorously established.
To elucidate the mechanism of Gln-tRNA formation in mitochondria we have focused our investigations on S. cerevisiae. Our results support a new general pathway for mitochondrial Gln-tRNA formation and highlight a common feature relevant to mitochondrial protein synthesis.
Results
Direct formation of mitochondrial Gln-tRNA by the cytoplasmic GlnRS
Analysis of the S. cerevisiae genome reveals three open reading frames possibly involved in formation of Glu-tRNA and Gln-tRNA: GluRS, mGluRS, and GlnRS. The mGluRS and the GlnRS from S. cerevisiae are essential for mitochondrial protein synthesis and cytosolic Gln-tRNA formation, respectively; however, their relevance to mitochondrial Gln-tRNA formation has not been examined (Ludmerer and Schimmel 1987; Tzagoloff and Shtanko 1995). We therefore investigated the specificity of the S. cerevisiae mGluRS and cytoplasmic GlnRS. We purified both recombinant enzymes (expressed in yeast or Escherichia coli) and examined their enzymatic activities with purified, mature mitochondrial tRNAs (tRNAm) from S. cerevisiae (Fig. 2A). As expected, the mGluRS formed Glu-tRNAmGlu, but surprisingly, it could not generate Glu-tRNAmGln, the required intermediate for the transamidation pathway. This finding was consistent with our previous unsuccessful attempts at identifying a mitochondrial amidotransferase activity, suggesting that the transamidation pathway may not be functioning in yeast. Similar data were obtained with purified native tRNAnGln and are consistent with previous reports of yeast GlnRS activity (Ludmerer et al. 1993). The nature of the [14C]-labeled amino acid attached to the different tRNA species was confirmed by thin-layer chromatography (TLC) of in vitro generated aa-tRNA (Fig. 2B). Cytoplasmic GlnRS attaches glutamine to both mitochondrial tRNAmGln and cytoplasmic tRNAnGln. Fundamentally, these two types of tRNA are separated by the location of their genes in the nuclear and mitochondrial genomes. However, because the GlnRS can utilize both tRNAs as substrates, we examined the possibility that the mitochondrion may contain tRNAGln outside of its own genomic context. To this end, we designed specific 32P-labeled oligonucleotide probes for tRNAnGln and tRNAmGln (Fig. 2C). The positive identification of each tRNA was aided by the fact that they have distinct electrophoretic mobilities (Fig. 2C). Utilizing these probes and acid urea gel electrophoresis, we consistently observed a second set of aminoacylated tRNAGln in total mitochondrial tRNA that hybridized to probe specific for tRNAnGln (Fig. 2D, lane 1). Acid urea gel electrophoresis allowed us to further confirm the identity of this tRNA species by comparing the aa-tRNAs extracted in vivo with the products of in vitro aminoacylation with GlnRS (Varshney et al. 1991). The signals for aa-tRNAs, which are distinct from the tRNAmGln species encoded in the mitochondrial genome, are consistent with the Gln-tRNAGln in the in vivo aminoacylated tRNA fraction extracted from mitochondria (Fig. 2D, lanes 1,3). Furthermore, neither tRNAnGln in the total mitochondrial fraction is a substrate for the S. cerevisiae mGluRS (data not shown). Taken together, these results indicate that mitochondrial Gln-tRNAGln is formed by direct charging of a tRNAGln species not encoded in the mitochondrial genome.
Figure 2.
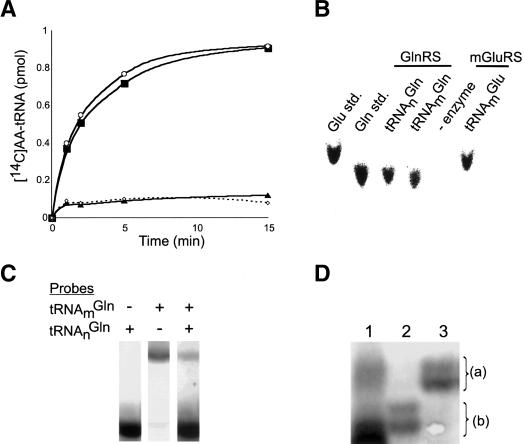
Aminoacylation of S. cerevisiae mitochondrial tRNAs. (A) Pure tRNAmGlu (○) and tRNAmGln (▴) in the presence of S. cerevisiae mGluRS and [14C]-glutamate compared to tRNAmGln (▪) in the presence of S. cerevisiae GlnRS and [14C]-glutamine; relative to minus enzyme control (⋄). (B) TLC analysis of [14C]-aminoacyl-tRNAs (aa-tRNAs) formed by the enzymes as indicated. [14C]-amino acids are compared to standards (std). (C) Total unfractionated yeast tRNAs were probed with [32P]-labeled oligonucleotides specific for tRNAmGln and tRNAnGln as indicated. The intensity of the two tRNA bands reflects the very different abundances of the cytoplasmic and mitochondrial tRNAGln.(D) Acid urea gel analysis of aa-tRNAs extracted from total mitochondria (lane 1) and from in vitro aminoacylation reactions (lanes 2,3). Total mitochondrial tRNA was deacylated under basic condition (lane 2) and reaminoacylated with glutamine in vitro by GlnRS (lane 3). Aminoacylated mitochondrial tRNA Gln-tRNAnGln (a) and deacylated tRNAnGln (b) was visualized by Northern blot.
Mitochondrial tRNAGln is of nuclear origin
The presence of aminoacylated tRNAnGln in the mitochondrion raised the question as to how these nucleus-encoded tRNAs became localized in the organelle. To investigate this further, we first probed total RNA from purified, micrococcal nuclease-treated mitochondria by Northern hybridization using radioactive oligonucleotides specific for either organelle- or nucleus-encoded tRNAGln. The data (Fig. 3A) show the presence of tRNAnGln, implying import of a fraction of cytosolic tRNAnGln into mitochondria. Probing of the same mitochondrial RNA fraction with the nuclear/cytosolic marker U6 snRNA showed this fraction to be devoid of any significant nuclear or cytosolic contamination.
Figure 3.
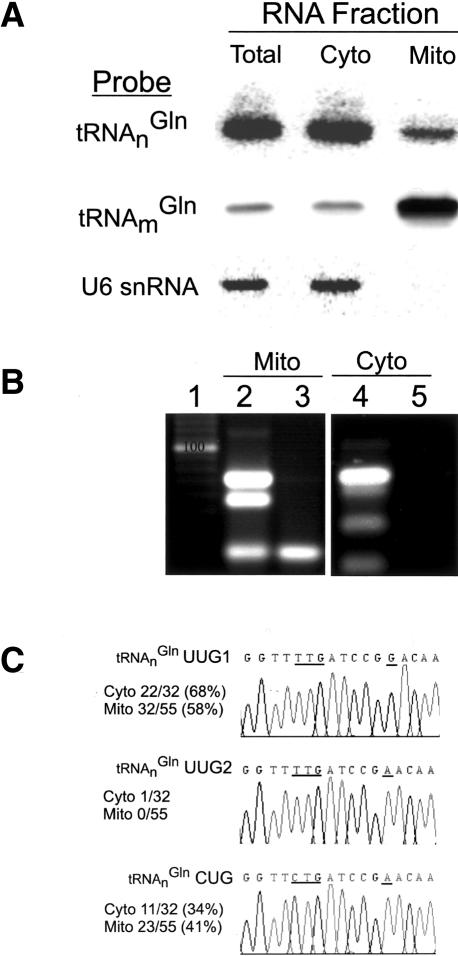
Localization of tRNAnGln to yeast mitochondria. (A) Northern analysis of total, cytosolic, and mitochondrial RNA fractions from wild-type yeast W303. RNAs were visualized with probes specific for tRNAnGln, tRNAmGln, and U6 snRNA. (B) RT-PCR analysis of total mitochondrial (Mito) and cytosolic (Cyto) RNA. Reactions with DNase-treated, total mitochondrial RNA (lane 2) or cytosolic RNA (lane 4), or in the absence of reverse transcriptase (RT) to control for contaminating DNA (lanes 3,5), were compared to a standard (lane 1). (C) Sequencing results of cloned RT-PCR products from lanes 2 and 4. Fiftyfive mitochondrial clones and 32 cytoplasmic clones were examined. The total number of each tRNAGln clones identified in the respective RNA pools is indicated. Representative chromatograms of RT-PCR clones are shown for tRNAnGlnUUG1, tRNAnGlnUUG2, and tRNAnGlnCUG. The anticodon and position 42 are underlined.
RT-PCR with oligonucleotides that allow amplification of all four isoacceptors permitted determination of the population of tRNAnGln isoacceptors present in mitochondrial and cytoplasmic fractions (Fig. 3B). The S. cerevisiae nuclear genome encodes a single tRNAnGlnCUG and three tRNAnGlnUUG genes differing at two or three nucleotide positions apart from the anticodon (Fig. 4). There was only tRNAnGlnUUG1 and tRNAnGlnCUG in the mitochondrial fraction; 23 out of 55 clones examined were tRNAnGlnCUG and 32 clones corresponded to tRNAnGlnUUG1 with a G at position 42 (Fig. 3C). We detected one tRNAnGlnUUG2 containing A42 in the cytoplasm, whereas this tRNA was not observed in the 55 clones examined from the mitochondrial fraction. This result further establishes that tRNAnGlnUUG and tRNAnGlnCUG are imported into the mitochondria from the cytoplasm and suggests a difference in the distribution of tRNA species between the two compartments.
Figure 4.

S. cerevisiae glutamine tRNAs. tRNA sequences are aligned under the predicted secondary structure (top line) where dots indicate unpaired bases and paired bases are indicated with a < or > (Lowe and Eddy 1997). Nuclear tRNAnGln sequences, their chromosomal locations and anticodon designations are indicated (left), are compared to the single tRNAmGln encoded in the mitochondrial genome. Conserved bases are shaded in gray. The anti-codon and position 42 are in bold with unique base changes shaded black to highlight differences between the tRNAs.
In vitro import of cytoplasmic tRNAGln into mitochondria
In light of the above observations, we tested the in vitro import of radioactive tRNAnGln purified from total cytosolic RNA fractions (isolated as above). Incubation of this tRNA with isolated yeast mitochondria led to 4% import of the labeled tRNA in vitro (Fig. 5A, lane 1). No import was observed in the absence of exogenous ATP, indicating that tRNAGln import is ATP dependent (Fig. 5A, lane 4). Import of tRNALys, previously the only imported tRNA known in S. cerevisiae, requires the addition of both the cytosolic and mitochondrial aa-tRNA synthetases during import (Tarassov et al. 1995; Kolesnikova et al. 2000). The mode of tRNAnGln import into yeast mitochondria is more similar to the mechanism of tRNA import described previously in trypanosomatids and plants (Rubio et al. 2000; Delage et al. 2003). This was demonstrated in a head-to-head comparison in which the tRNAnLys was not imported in vitro (Fig. 5B). The specificity of the tRNA import was further demonstrated by including the Leishmania spliced-leader RNA as a nonimported RNA control (Fig. 5B). Thus, the system described here is certainly different, as no exogenous cytosolic factors are required for in vitro import of tRNAnGln. Therefore, S. cerevisiae has more than one mechanism for the import of nucleus-encoded tRNAs into mitochondria. The direct demonstration of tRNAnGln import into isolated mitochondria also corroborates the cytosolic origin of the tRNA substrates used for the Gln-tRNA synthesis in S. cerevisiae mitochondria.
Figure 5.
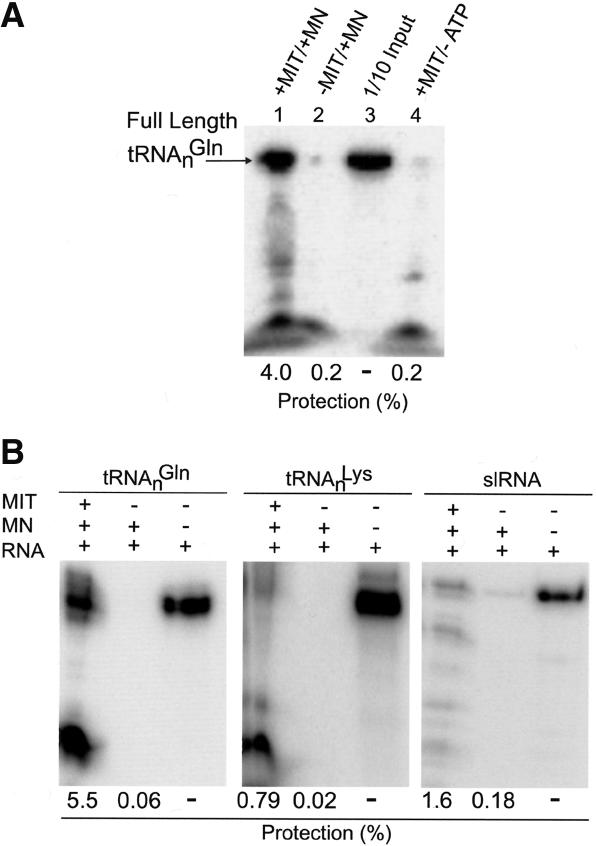
In vitro import of native tRNAnGln into isolated yeast mitochondria. (A) Nuclease protection of native tRNAnGln incubated with isolated yeast mitochondria. Native tRNAnGln radioactively labeled at the 5′ end with γ32P-ATP (1 × 105 cpm) was incubated with isolated yeast mitochondria in the presence (lane 1) and absence (lane 4) of ATP and subsequently digested with micrococcal nuclease (MN). The arrow depicts the migration of the full-length RNA protected from MN digestion. (Lane 2) As a control, input RNA was digested with MN in the absence of mitochondria. Lane 3 represents one-tenth of the input RNA, without MN digestion and in the absence of mitochondria, used for quantification. (B) In vitro import of native S. cerevisiae tRNAnGln compared to S. cerevisiae tRNAnLys and the Leishmania spliced leader RNA (slRNA) into isolated yeast mitochondria. Import experiments are presented as in A with the third lane in each panel representing an RNA-only control (one-tenth of the input RNA) used for quantification.
The S. cerevisiae GlnRS is also localized to the mitochondrion
The cytoplasmic S. cerevisiae GlnRS is an essential enzyme required for cytoplasmic Gln-tRNA synthesis (Ludmerer and Schimmel 1987). We examined the intracellular localization of the GlnRS protein to determine whether it also could play a role in mitochondrial Gln-tRNA formation. The gene for an N-terminal GlnRS-GFP fusion protein was transformed into S. cerevisiae; this protein became localized in the cytoplasm and the mitochondrion, as demonstrated by coincident GFP fluorescence and DAPI staining (Fig. 6A). A second experiment utilizing a C-terminally V5-tagged GlnRS was carried out in an S. cerevisiae GlnRS deletion strain. Plasmid exchange experiments with the GlnRS-V5 and GlnRS-GFP showed that both fusion proteins could rescue growth of a GlnRS deletion strain, suggesting that both proteins were fully functional (data not shown). Coexpression of the GlnRS-V5 and of a COXIV-GFP fusion protein targeted separately to mitochondria showed colocalization of both proteins in mitochondrial structures with the GlnRS-V5 showing additional staining of the cytoplasm (Fig. 6B). These results suggest that the native GlnRS enzyme may also be localized in the mitochondrion in vivo.
Figure 6.
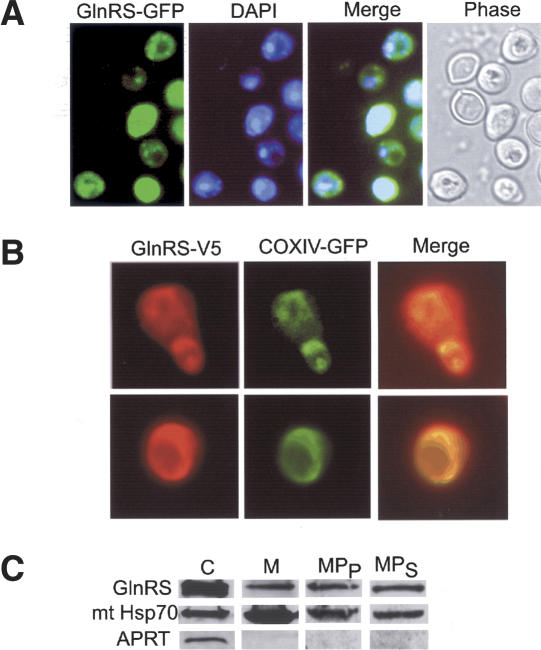
Localization of yeast GlnRS to the mitochondrion. (A) GFP fluorescence in living cells shows GlnRS-GFP is colocalized in the cytoplasm and the mitochondria. DAPI staining of mitochondria can be overlapped with the mitochondrial GlnRS-GFP fluorescence. (B) Immunofluorescence shows a V5-GlnRS fusion protein is also colocalized to the cytosol and the mitochondria of a GlnRS deletion strain. Mitochondrial structures are visualized by coexpression of a COXIV-GFP fusion protein, which is localized only to the mitochondrion (Sesaki and Jensen 1999). (C) Western blot analysis of fractions from wild-type yeast W303: (C) 40 μg total cytosolic proteins; (M) 40 μg mitochondria; (MPP) 80 μg mitoplast pellet; (MPS) 40 μg mitoplast supernatant. GlnRS was visualized by an anti-GlnRS antibody and compared with marker antibodies for APRT (cytoplasm) and mtHsp70 (mitochondrial matrix).
To investigate this further we examined GlnRS localization by Western blot analysis using pure mitochondria from wild-type S. cerevisiae. A clear GlnRS signal was evident in total mitochondrial and mitoplast fractions (Fig. 6C). All mitochondrial fractions were negative for the cytoplasmic marker protein adenine phosphoribosyltransferase (APRT), indicating that the signal observed for GlnRS is not due to cytosolic contamination of the mitochondrial fraction. Additionally, mitoplasts were prepared under conditions rigorous enough to release some of the mitochondrial matrix into the supernatant fraction. Even under these conditions, we observed colocalization of GlnRS and the mitochondrial marker mtHsp70, indicating that GlnRS is localized in the mitochondrial matrix of wild-type cells. However, the mitochondrial localization signal is not obvious. The first N-terminal 127 amino acids of the GlnRS are dispensable and do not affect the respiratory efficiency of wild-type cells and the remainder of the open reading frame contains no predictable targeting sequence (Ludmerer and Schimmel 1987). Furthermore, the fact that both the GFP and V5-tagged GlnRS proteins are localized in the mitochondrion suggests that the GlnRS does not contain a traditional cleavable targeting signal at its N or C terminus.
A single nuclear tRNAGln supports translation in both the cytoplasm and the mitochondrion
The collective in vitro evidence supports a new model of yeast mitochondrial Gln-tRNA function, in which tRNAnGln is partitioned between the cytoplasm and mitochondrion to participate in translation in both compartments. To demonstrate this in vivo we introduced a point mutation in a plasmid-borne copy of a yeast tRNAnGlnCUG gene to create an amber suppressor tRNAnGlnCUA. We utilized the tRNAnGlnCUA to suppress in frame amber stop codons in both a cytoplasmic and mitochondrial gene (Fig. 7). A yeast strain containing a mutant trp1-1UAG allele could grow in the absence of tryptophan only when transformed with the tRNAnGlnCUA (Fig. 7A). This showed that the tRNAnGlnCUA was a substrate for translation in the cytoplasm. The yeast strain HM4, which contains a cox2-114UAG mutation, was used to demonstrate the import of tRNAnGlnCUA and its subsequent role in mitochondrial translation (Fig. 7B). The HM4 lacked any detectable Cox2p whereas there was a clear signal when the same strain contained the plasmid-borne tRNAnGlnCUA gene. The HM4 strain is respiration-deficient and tRNAnGlnCUA expression did not rescue this phenotype (data not shown). In contrast to the Trp1p, the Cox2p may not be functional with a glutamine inserted at the site of the amber stop codon. BLAST searches revealed a conserved alanine at position 114 in most yeast and plant Cox2. Although the protein is not functional, tRNAnGlnCUA-mediated suppression clearly produces immunologically detectable Cox2p. Taken together, these data clearly show that yeast partitions a functional tRNAnGln between the cytoplasm and mitochondria and uses this imported tRNA in mitochondrial translation.
Figure 7.
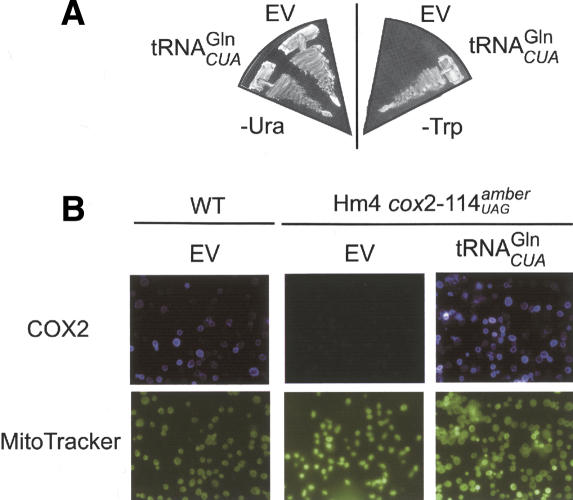
A single suppressor tRNA is active in both cytoplasmic and mitochondrial translation. (A) The yeast strain W303 (ura3-52 and trp1-1UAG) containing an empty p316(URA3+) vector (EV) or p316 with an amber suppressor mutant tRNAnGlnCUG gene (tRNAnGlnCUA) was grown on media lacking uracil (-URA) or tryptophan (-Trp). tRNAnGlnCUA-dependent suppression of the trp1-1UAG mutation is shown by the ability of W303 to grow in the absence of tryptophan. (B) Cox2p expression in wild-type yeast (WT) or cox2-114UAG (HM4) (Kolesnikova et al. 2000) visualized via immunofluorescence with an anti-Cox2p antibody. tRNAnGlnCUA-dependent suppression of the cox2-114UAG mutation is shown by the increased levels of Cox2p staining in HM4 cells containing the suppressor tRNA. Cells were costained with Mito Tracker.
Discussion
Here we describe a newly discovered strategy used by S. cerevisiae mitochondria to activate glutamine for protein synthesis. Following import into mitochondria, the cytosolic GlnRS directly attaches glutamine to tRNAnGln. In addition, a fraction of the tRNAnGln present in mitochondria is a consequence of import from the cytoplasm. This pathway contrasts with the mechanism operative in chloroplasts, which, in line with their bacterial ancestry, rely on the transamidation of mischarged Glu-tRNAGln (Schön et al. 1988). This indirect biosynthesis pathway requires two enzyme activities, a GluRS forming Glu-tRNAGln and an amidotransferase able to convert this mischarged tRNA species to the correctly charged Gln-tRNAGln (Curnow et al. 1997). While paralogs of the amidotransferase genes are found in the yeast genome, the lack of a mitochondrial activity capable of synthesizing Glu-tRNAGln, the required amidotransferase precursor, may explain why the transamidation route does not function in mitochondria. As switching of tRNA specificity between discriminating and nondiscriminating synthetases has been shown to be quite feasible (Charron et al. 2003; Feng et al. 2003), one may hypothesize that during mitochondrial evolution the organellar GluRS lost its tRNAGln specificity and with it the ability to form Glu-tRNAGln. This event may have prompted the organism to import the total pathway of Gln-tRNA formation from the cytoplasm. The tRNA import may have been advantageous, since the cytoplasmic tRNAs had been evolutionarily adapted to the cytoplasmic GlnRS, and are probably better substrates than the mitochondrial tRNAGln still present. A similar mechanism, by which amidotransferase is replaced, is established for the introduction of GlnRS, by lateral gene transfer, into some bacterial lineages, especially γ-proteobacteria (Fig. 8; Lamour et al. 1994; Brown and Doolittle 1999; Handy and Doolittle 1999). However, mitochondria only needed a horizontal gene “product” transfer, and perhaps this feature increased the chances of the simultaneous acquisition of tRNAGln and GlnRS from the cytoplasm.
Figure 8.
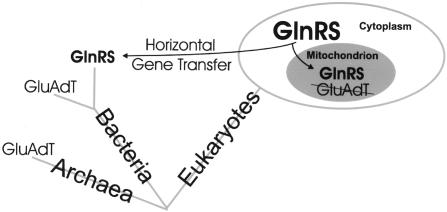
Origins of GlnRS from the eukaryotic cytoplasm. GlnRS, acquired by horizontal gene transfer, has replaced Glu-tRNA amidotransferase (GluAdT) in some bacteria and is of eukaryotic origin. In the mitochondrion, the GlnRS has replaced the bacterial-type GluAdT, which is part of a more ancient pathway.
Mitochondrial translation requires two tRNAGln isoacceptors
The observed import of a set of glutamine tRNAs explains how Gln-tRNA is synthesized in yeast mitochondria, but raises new and important questions. What roles are played by the imported tRNAnGlnCUG and the tRNAnGlnUUG? The yeast mitochondrial genome encodes a tRNAmGln with anticodon UUG. Two mitochondrial tRNA species, tRNAmGlnUUG and tRNAmLysUUU, were shown to contain 5-carboxymethylaminomethyl-2-thiouridine (mcm5s2U) in position 34, the first anticodon nucleotide (Nakai et al. 2004). Wobble base pairing would be restricted by mcm5s2U at position 34; thus tRNAmGlnUUG would only decode CAA codons (for review, see Yokoyama and Nishimura 1995). This should necessitate the import of a tRNAnGlnCUG isoacceptor to decode the CAG codons in mitochondrial mRNA. Similarly, mcm5s2U34 in tRNAmLysUUU would also explain the need for the imported tRNAnLysCUU (Kolesnikova et al. 2000). The function of the imported tRNAnGlnUUG is less clear. The S. cerevisiae nuclear genome encodes three different tRNAnGlnUUG species (Fig. 4), and the redundancy in this tRNA population may have some important and as of yet unknown function common to both the cytoplasm and the mitochondria. The selectivity of tRNAGln import and what features of the import machinery control intracellular tRNA distribution will be the subject of future investigations.
The S. cerevisiae mitochondrion has two tRNA import mechanisms
Prior to this report, yeast mitochondria were believed to encode and synthesize all of the necessary tRNAs for protein synthesis. Thus, the notion of mitochondria-imported tRNAs was not seriously considered for yeast and the import of a single functionally redundant tRNALys was an interesting exception (Kolesnikova et al. 2000). The ability to import uncharged tRNAGln in vitro in the absence of added cytosolic factors differs radically from the mechanism of tRNALys import. In the latter mechanism, an aminoacylated tRNALys and a precursor mitochondrial lysyl-tRNA synthetase are both required for import. The function of the imported tRNALys is unclear given that the mitochondrial lysyl-tRNA synthetase does not aminoacylate it (Tarassov et al. 1995). We observe localization of both cytoplasmic tRNAGln and GlnRS to the mitochondrion where they are most likely required for protein synthesis. Future studies will focus on the essential nature of each mitochondrial tRNAGln, of both nuclear and mitochondrial origin, in protein synthesis.
Is mitochondrial tRNA import, at least of tRNAnGln, more general and also occurring in higher eukaryotes? The ability to import both the cytoplasmic synthetase together with its cognate tRNA might not be unique to yeast mitochondria. This idea arises from the observation that a tRNAGln with anticodon CUG is not encoded in any of the >500 mitochondrial genomes sequenced to date (NCBI Organelle Genomes, http://www.ncbi.nlm.nih.gov/genomes/static/euk_o.html). The only exception is the Ascobolus immersus tRNAmGlnCUG “pseudogene” (with an unusual DNA sequence insertion) that is probably not functional (Goyon et al. 1996). Recent studies showed the presence of a nucleus-encoded GlnRS and the direct synthesis of Gln-tRNA in the mitochondria of kinetoplastid protozoans (Leishmania tarentolae and Trypanosoma brucei) (Nabholz et al. 1997; Rinehart et al. 2004). The concept of import of both GlnRS and tRNAGln is more obvious in these organisms because their mitochondrial genomes do not contain tRNA genes. Indeed tRNA import was first described in these and other organisms, but largely unexplored in higher eukaryotes (for reviews, see Schneider and Marechal-Drouard 2000; Hopper and Phizicky 2003). If our prediction of the essentiality of cytoplasmic tRNAnGln import into mitochondria were true, then GlnRS-catalyzed Gln-tRNA synthesis should be widespread in the mitochondria of most organisms.
Materials and methods
Yeast strains
The wild-type S. cerevisiae strain W303 mat a ade2-1 his3 leu2-3, 112 trp1-1 ura3-52 was used for tRNA preparation and protein expression. The mutant trp1-1 allele in W303 is the result of an amber nonsense codon; therefore, this strain was also utilized in suppression experiments. The strain HM4 mat a leu2-3, 112 lys2 ura3-52 arg8::hisG [p + cox2-114amber] (Kolesnikova et al. 2000) was used in mitochondrial suppression studies. INVSc1 mat a/α his3Δ1/his3Δ1 leu2/leu2 trp1-289/trp1-289 ura3-52/ura3-52 (Invitrogen) was used for immunofluorescence localization. The heterozygous GlnRS deletion strain, Y22424 BY4743 mat a/α his3Δ1/his3Δ1 leu2Δ0/leu2Δ0 lys2Δ0/LYS2 MET15/ met15Δ0 ura3Δ0/ura3Δ0 YOR168w::kanMX4/YOR168w (Research Genetics), was used to generate strains for immunofluorescence localization, as well as complementation via plasmid exchange (5-FOA selection). The GlnRS was subcloned (described below) into a p416GPD (CEN/ARS, URA+) vector and used to create a viable GlnRS chromosomal deletion strain (exchange strain). Tetrad dissection and other genetic manipulations were carried out as before (Rosas-Sandoval et al. 2002). All media were prepared as described (Guthrie et al. 1991).
Isolation of mitochondria and Western blot
Yeast W303 was grown in rich media containing 2% galactose, and mitochondria were isolated as described (Daum et al. 1982; Celis 1994; Glick and Pon 1995). Spheroplasts were prepared by Zymolyase-20T (ICN) treatment and broken by three passes through a EmulsiFlex-C5 (AVESTIN) in 0.6 M sorbitol (Fluka), 10 mM Tris-HCL (pH 7.4), and 1 mM phenylmethylsulfonyl fluoride. Mitochondria were purified by centrifugation and extensive washing in the same buffer as described (Daum et al. 1982). The cytosolic fraction obtained during this procedure was clarified by ultracentrifugation at 100,000 × g and used for protein localization experiments and to prepare total cytoplasmic tRNA. Highly purified organelles were obtained by centrifugation at 100,000 × g (Beckman SW41) in a Percoll step gradient (Amersham Biosciences) (40% Percoll in 0.6 M sorbitol [Fluka], 10 mM Tris-HCL at pH 7.4 overlaid with 20% Percoll in the same buffer). Percoll pure mitochondria (44 mg wet weight) were resuspended in 33% glycerol, 0.6 M sorbitol, and 10 mM Tris-HCL (pH 7.4) to a final concentration of 2 mg/mL and stored at -80°C for in vitro import experiments. A fraction of the Percoll pure mitochondria were treated with Proteinase K (Boehringer) (as in Hartl et al. 1987), for protein localization experiments, or with 100 U of micrococcal nuclease (Roche) for RNA localization. Mitoplasts were prepared from Proteinase-K-treated mitochondria by osmotic shock as described (Hartl et al. 1987). The supernatant was TCA precipitated for Western blot analysis and the pellet (mitoplast) fraction was lysed with 2% Triton-X and clarified by ultracentrifugation at 100,000 × g. Proteins were separated by SDS-PAGE and electroblotted to nitrocellulose membranes (0.2 μm; Schleicher & Schuell). Membranes were blocked, washed, and stripped according to the manufacturer's directions. Rabbit anti-GlnRS was used at 1: 40,000, rabbit anti-mtHsp70 was used as a mitochondrial marker at 1:20,000, and rabbit anti-APRT was used as a cytoplasmic marker at 1:20,000. Detection was carried out with HRP-conjugated secondary antibodies and the ECL system (Amersham Biosciences) with autoradiography.
Cloning and purification of GlnRS and mGluRS
Oligonucleotides were synthesized and DNAs were sequenced by the Keck Foundation Biotechnology Resource Laboratory at Yale University. PCR primers were designed for the S. cerevisiae GlnRS (GLN4/YOR168W) and mGluRS (MSE1/YOL033W) according to the sequences published in the Saccharomyces Genome Database (http://www.yeastgenome.org). The genes were cloned by PCR from a yeast genomic DNA library (ATCC, #77164) using the Expand High Fidelity PCR System (Roche). Primers GCATATGATCATGTTGAGAATACCGAC and CCTCGAGTGCCTTTTTCTCTCTTTTGC were used to clone the mGluRS in frame with the C-terminal His6 tag of the pET20b vector (Novagen) by utilizing NdeI and XhoI restriction sites. The recombinant mGluRS was expressed and purified from E. coli essentially as described (Rinehart et al. 2004). The mGluRS was purified first over Ni-NTA resin (Qiagen) followed by ion-exchange chromatography on Mono S HR 5/5 (Amersham). Primers GACTAGTATGTCTTCTGTAGAAGAATTGACTCAG and CCGCTCGAGTCACTTGGAAGTTGCGTCCTTCAAGCTAAC were used to clone the GlnRS in the HIS-CEN vector p413GPD between the SpeI and XhoI restriction sites, which allowed the overexpression of the native protein in yeast (Mumberg et al. 1995). Wild-type yeast W303 was transformed with the GlnRS-p413GPD construct and cells were cultured in semi-synthetic, 2% glucose media lacking histidine. Native GlnRS expression and purification was performed essentially as described (Ludmerer et al. 1993). Ion-exchange chromatography was carried out, in succession, over S-sepharose, Heparin HP, and Mono S HR (Amersham) columns in 50 mM Tris-HCL (pH 7.0-7.5), 5% glycerol, 5 mM β-mercaptoethanol (β-ME), protease inhibitor cocktail for fungal and yeast cells (Sigma), and the appropriate counter-ion. Purity was judged by Coomassie-stained SDS-polyacrylamide gel electrophoresis and protein concentrations were determined by the Quantigold (Diversified Biotech) colloidal gold protein assay (Stoscheck 1987).
tRNA localization with Northern blot and RT-PCR sequencing
RNA was isolated from total, micrococcal nuclease-treated mitochondrial fractions and/or cytosolic yeast fractions by the guanidinium thiocyanate/phenol/chloroform extraction method (Chomczynski and Sacchi 1987). RNA fractions were separated on denaturing 8% polyacrylamide gels with 8 M urea and electroblotted to a Zeta-probe (Bio-Rad) membrane. Membranes were probed with [32P] 5′ end-labled olionucleotides complementary to tRNAmGln (TACCTATTAGTCTACGACTCA) tRNAnGln (ATACCACTACACTATAGGACC) and U6 snRNA (CGAAGGGTTACTTCGCGAAC). Hybridization, washing, and stripping of the probes were carried out according to the manufacturer's directions (Bio-Rad). Images were taken with a Storm PhosphorImager and analyzed with ImageQuant (Molecular Dynamics). RNA from micrococcal nuclease-treated mitochondrial fractions was treated with RQ1 RNase free DNAse (Promega). Primers AGGTCCTACCCGGATTCGAACCGGG-RT and PCR forward primer and GGTCCTATAGTGTAGTGGTTATCAC-PCR reverse primer were used for RT PCR reactions. The RT primer was annealed to 5 μg of total mitochondrial RNA or 5 μg of total cytoplasmic tRNA at 50°C, followed by the addition of reverse transcriptase. The RT reaction was carried out as described in the SuperScript II reverse transcriptase first strand synthesis protocol (Invitrogen). Following RT, 1 μL of the 20 μL RT reaction was used as a template in a PCR reaction with the forward and reverse primers (above). PCR reactions were performed using Taq polymerase following manufacturer's instructions and included a 30-sec 95°C denaturation step, 55°C annealing for 30 sec, and 30 sec of 72°C elongation for a total of 30 cycles. Controls included a mock reaction where the RT was left out of the reaction and used as a negative control to test for DNA contamination in the RNA samples. RT PCR products were cloned with the TOPO method according to the manufacturer's instructions (Invitrogen).
tRNA purification
Wild-type yeast (W303) was cultured in rich media supplemented with 2% galactose to increase the mitochondrial content of the cells. Unfractionated tRNA and total cytoplasmic tRNA was prepared with a QIAGEN-tip 100 column (QIAGEN) as described (Rinehart et al. 2004). Following established procedures, individual tRNAs were purified from unfractionated tRNA with 5′ biotinylated DNA oligonucleotides, complementary to the target tRNA immobilized on streptavidin agarose beads (Pierce) (Kaneko et al. 2003). Biotinylated oli gos (150 μg), XTTGGTAACTTACCTATTAGTCTACGACTCA for tRNAmGln (X = 5′ biotin), XGTGATGTCGTAACCATTAGACGATAAGGTC for tRNAmGlu, and XCGAAAGTGATAACCACTACACTATAGGACC for tRNAnGln, were bound to the streptavidin beads in 10 mM Tris-HCl (pH 7.5). The oligo-beads were equilibrated in 6× NTE solution (20× NTE solution is 4 M NaCl, 0.1 M Tris-HCl at pH 7.5, 50 mM EDTA, 5 mM 2-ME) and 3 mg of unfractionated yeast tRNA (10 mg/mL in 6× NTE) was then added and incubated at 65°C for 30 min. After incubation, the temperature of the mixture was decreased by natural heat dissipation until it reached 30°C and washed, in succession, with 3× NTE three times, 1× NTE twice, and a final 0.1× NTE wash until the absorbance at 260 nm of the wash was zero. tRNAs retained on the beads were eluted with 0.1× NTE at 65°C and ethanol precipitated. Pure tRNAs were examined with Northern blot by hybridization with a 5′ 32P-labeled oligodeoxyribonucleotide probe specific to each tRNA (described above).
Aminoacylation assays: filter binding, thin-layer chromatography, and acid urea gel electrophoresis
Filter binding assays were performed essentially as described (Ahel et al. 2002). Aminoacylation reactions were performed at 30°C in 100 mM HEPES-Na (pH 7.2), 30 mM KCl, 12 mM MgCl2, 10 mM ATP, 10 mM DTT, 1 μM pure tRNA, and 50 μM [14C]Gln (50 μCi/mL, 242 mCi/mmol; Amersham) or 50 μM [14C]Glu (50 μCi/mL, 254 mCi/mmol; Amersham) using 0.1 μM GlnRS or mGluRS. The identity of the [14C] amino acid was confirmed by thin layer chromatography. In vitro aminoacylation reactions were stopped after 30 min with 1 volume of 0.6 M sodium acetate (pH 4.5) and the aa-tRNA was extracted with acidic-phenol. Samples were ethanol precipitated, and the aa-tRNA (∼1 pmol) was treated with 0.5 M borate (pH 9.0) to remove the [14C] amino acid, dried in a speed vac, spotted in water on 20 × 20 cellulose plates (Sigma), and resolved in isopropanol/formic acid/water (80/20/4, vol/vol/vol). TLC plates were dried and visualized with the Storm phosphorimageing system described above. Acid urea gel electrophoresis of tRNA and aa-tRNA was carried out essentially as described (Varshney et al. 1991). Total aa-tRNAs from pure mitochondria were extracted under acidic conditions (equilibrated with 0.3 M sodium acetate at pH 4.5 and 10 mM EDTA), ethanol precipitated, and resuspended in 10 mM sodium acetate (pH 4.5) and 1 mM EDTA. A fraction of the total aa-tRNA was stored at -80°C to be used for in vivo aa-tRNA controls. The remaining tRNA was deacylated by incubation in 100 mM Tris-OAc (pH 8.5) at 37°C for 30 min, ethanol precipitated, and used for in vitro aminoacylation assays. In vitro aminoacylation reactions were carried out as described above except with unlabeled amino acids and 10 μg of total deacylated tRNA from mitochondria. The reactions were stopped at 30 min with 1 volume of 0.6 M sodium acetate (pH 4.5). Parallel aminoacylation reactions with radiolabeled amino acids were performed under the same conditions to monitor aminoacylation. In vitro aminoacylation reactions were analyzed on acid urea gels essentially as described (Polycarpo et al. 2003). After acidic-phenol extraction and ethanol precipitation, the aa-tRNAs were dissolved in 1.5 μL of 10 mM sodium acetate (pH 4.5) and 1 mM EDTA. Once resuspended, 1.5 μL of loading buffer (7 M urea, 0.1 M sodium acetate at pH 4.5, 0.1% bromophenol blue, 0.1% xylene cyanol) was added to each sample. Samples were loaded on a 9.5% polyacrylamide gel (50 × 20 cm, 0.4 mm thick) containing 7 M urea, 0.1 M sodium acetate (pH 5.0). The gel was run at 4°C, at 600 V in 0.1 M sodium acetate (pH 5.0) for 38 h. The 0.1 M sodium acetate (pH 5.0) was changed after 24 h. tRNAs were subsequently electroblotted onto a Hybond N+ nylon membrane (Amersham Biosciences) and visualized via Northern blot with the appropriate 5′ 32P-labeled probe (described above).
Immunofluorescence analysis
V5-tagged GlnRS was cloned into a URA-2 μm pYES-V5 TOPO vector (Invitrogen) utilizing primers similar to those described above except lacking a stop codon and XhoI restriction site in the reverse primer. The resulting constructs allowed stable expression of each V5 fusion protein in the yeast strain INVSc1 as verified by Western blot with an HRP-conjugated anti-V5 antibody (Invitrogen). The GlnRS-V5 plasmid construct was transformed into the yeast strain Y22424, which was then used to generate a GlnRS deletion strain (as described above). Each strain was cotransformed with a HIS-CEN plasmid that carries an ADH1-COX4-GFP and directs GFP to the mitochondrion (Sesaki and Jensen 1999). Cells were cultured to early log phase in semi-synthetic, 2% galactose media lacking histidine and uracil, fixed with 4% paraformaldehyde, permeabilized with zymolyase, and prepared with organic solvents for immunostaining as described (Harlow and Lane 1999). The cells were incubated with mouse anti-V5 (Invitrogen) primary antibody (1:60) followed by Cy3-conjugated goat anti-mouse (Jackson Immuno Research) secondary antibody (1:200) and mounted with Pro-Long (Molecular Probes) mounting media. Cox2p immunofluorescence was carried out in a similar fashion with mouse anti-Cox2p (Molecular Probes) primary antibody (1:200) and Cy5-conjugated anti-mouse (Jackson Immuno Research) secondary antibody (1:500) followed by staining with 20 nM of MitoTraker Green FM (Molecular Probes) according to the manufacturer's instructions. An N-terminal GlnRS-GFP was constructed by inserting GFP into the GlnRS-p413GPD. Utilizing PCR primers GACTAGTATGAGTAAAGGAGAAGAACTTTTCACTG and GACTAGTTTTGTATAGTTCATCCATG, the stop codon was removed and 5′ and 3′ SpeI restriction sites were added to the GFP gene. The gene was then inserted at the SpeI site in GlnRS-p413GPD and the proper orientation of the GlnRS-GFP fusion gene was checked by MscI/XhoI restriction analysis. W303 yeast expressing GlnRS-GFP were cultured to early log phase in semi-synthetic, 2% galactose media lacking histidine, and stained with DAPI (Molecular Probes). Yeast strains expressing both GFP- and V5-tagged proteins were examined on an Axioscope (Carl Zeiss MicroImaging) fitted with an Axiocam, a ChromaGFP filter set (model C2909), DAPI and TR filters (Chroma Technology), and a Uniblitz Shutter assembly. Images were processed with OpenLab version 3.1.5.
In vivo suppression
Primers GGACTAGTCCGTTAAAATAGGGACCATCTTTC and GGACTAGTCCGATGAGCGATTGTATCTGT with flanking SpeI sites were used to clone the native yeast tRNAGlnCUG gene into the low copy yeast vector p316(URA3+). The tRNAGlnCUA gene was then generated from this clone via PCR using the primers TTGTTCGGATTAGAACCGAAAGT and ACTTTCGGTTCTAATCCGAACAA. Yeast strain W303 was transformed with the tRNAGlnCUA-p316 construct and cells were cultured in semi-synthetic, 2% glucose media lacking uracil. Supression was scored by growth on semi-synthetic, 2% glucose media lacking tryptophan. Yeast strain HM4 was transformed with the tRNAGlnCUA-p316 construct and cells were cultured in semi-synthetic, 2% glucose media lacking uracil. Cells were cultured to early log phase and prepared for immunofluorescence as described above.
In vitro tRNA import assays
Native tRNAGln (purified as above) were first treated with calf intestinal Alkaline Phosphatase (Invitrogen) followed by phenol extraction and ethanol precipitation. The sequence for the imported yeast tRNA1Lys was as described (Kolesnikova et al. 2000). In vitro transcript of tRNAnLys and slRNA were prepared by T7 transcription as described (Rubio et al. 2000; Ahel et al. 2002). The tRNA was then 5′-end-labeled with [γ32P]ATP and T4 polynucleotide kinase (GIBCO-BRL) followed by gel purification on a 7 M urea/8% polyacrylamide gel. In vitro RNA import assays were performed in a 20 μL reaction volume containing 100,000 cpm of 5′-end-labeled tRNA, 1 μg of mitochondria, 0.6 M sorbitol, 20 mM Tris-HCl (pH 8.0), 5 mM ATP, 2 mM DTT, 20 mM MgCl2, and 2 mM EDTA. After incubation at 27°C for 10 min, 100 U of micrococcal nuclease (MN) (Roche) and 5 mM CaCl2 were added and the reaction was incubated an additional 30 min to digest the tRNAs that were not imported into the mitochondria. MN was then inhibited by the addition of 10 mM EGTA (pH 8.0). To isolate protected tRNAs, the mitochondria were washed with 0.6 M sorbitol/20 mM Tris-HCl (pH 8.0), pelleted, resuspended in 90 μL of 10 mM Tris-HCl (pH 8.0), 1 mM EDTA, and 0.1% SDS, and extracted with 100 μL of water-saturated phenol (pH 4.5) followed by ethanol precipitation. The radioactively labeled tRNAs were separated by electrophoresis through a 7 M urea/6% acrylamide gel. After electrophoresis, the gels were dried onto Whatman 3MM chromatography paper and visualized with the Storm PhosphorImager imaging system as described above.
Acknowledgments
We thank Michael Ibba, Hubert Becker, and Jeffrey Sabina for valuable discussions and critical review of the manuscript. We thank Janet Shaw (University of Utah) for anti-mtHsp70 antibody, Franco Fasiolo (Strasbourg) for anti-GlnRS antibody, and Milton Taylor (Indiana University) for anti-APRT antibody. We also thank Mark Hochstrasser (Yale University) for the use of microscopy equipment and Robert Jensen (Johns Hopkins University) for providing the COX4-GFP plasmid construct. Many members of the Hochstrasser laboratory, especially Oliver Kerscher and David Schwartz, were extremely helpful. We also thank Thomas Fox (Cornell University) for providing the HM4 strain. J.R. was a F.W. and Elsie L. Heyl Predoctoral Fellow. This work was supported by grants from the National Institute of General Medical Sciences (D.S.), the Department of Energy (D.S.), and the American Heart Association (J.D.A.).
Article published online ahead of print. Article and publication date are at http://www.genesdev.org/cgi/doi/10.1101/gad.1269305.
References
- Ahel I., Stathopoulos, C., Ambrogelly, A., Sauerwald, A., Toogood, H., Hartsch, T., and Söll, D. 2002. Cysteine activation is an inherent in vitro property of prolyl-tRNA synthetases. J. Biol. Chem. 277: 34743-34748. [DOI] [PubMed] [Google Scholar]
- Barrell B.G., Anderson, S., Bankier, A.T., de Bruijn, M.H., Chen, E., Coulson, A.R., Drouin, J., Eperon, I.C., Nierlich, D.P., Roe, B.A., et al. 1980. Different pattern of codon recognition by mammalian mitochondrial tRNAs. Proc. Natl. Acad. Sci. 77: 3164-3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonitz S.G., Berlani, R., Coruzzi, G., Li, M., Macino, G., Nobrega, F.G., Nobrega, M.P., Thalenfeld, B.E., and Tzagoloff, A. 1980. Codon recognition rules in yeast mitochondria. Proc. Natl. Acad. Sci. 77: 3167-3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown J.R. and Doolittle, W.F. 1999. Gene descent, duplication, and horizontal transfer in the evolution of glutamyl- and glutaminyl-tRNA synthetases. J. Mol. Evol. 49: 485-495. [DOI] [PubMed] [Google Scholar]
- Celis J.E. 1994. Cell biology: A laboratory handbook. Academic Press, San Diego, CA.
- Charron C., Roy, H., Blaise, M., Giege, R., and Kern, D. 2003. Non-discriminating and discriminating aspartyl-tRNA synthetases differ in the anticodon-binding domain. EMBO J. 22: 1632-1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomczynski P. and Sacchi, N. 1987. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenolchloroform extraction. Anal. Biochem. 162: 156-159. [DOI] [PubMed] [Google Scholar]
- Curnow A.W., Hong, K., Yuan, R., Kim, S., Martins, O., Winkler, W., Henkin, T.M., and Söll, D. 1997. Glu-tRNAGln amidotransferase: A novel heterotrimeric enzyme required for correct decoding of glutamine codons during translation. Proc. Natl. Acad. Sci. 94: 11819-11826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daum G., Bohni, P.C., and Schatz, G. 1982. Import of proteins into mitochondria. Cytochrome b2 and cytochrome c peroxidase are located in the intermembrane space of yeast mitochondria. J. Biol. Chem. 257: 13028-13033. [PubMed] [Google Scholar]
- Delage L., Dietrich, A., Cosset, A., and Marechal-Drouard, L. 2003. In vitro import of a nuclearly encoded tRNA into mitochondria of Solanum tuberosum. Mol. Cell. Biol. 23: 4000-4012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng L., Tumbula-Hansen, D., Toogood, H., and Söll, D. 2003. Expanding tRNA recognition of a tRNA synthetase by a single amino acid change. Proc. Natl. Acad. Sci. 100: 5676-5681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng L., Tumbula-Hansen, D., Min, B., Namgoong, S., Salazar, J., Orellana, O., and Söll, D. 2004. Transfer RNA-dependent amidotransferases: Key enzymes for Asn-tRNA and Gln-tRNA synthesis in nature. In The aminoacyl-tRNA synthetases (eds. M. Ibba et al.), pp. 314-319. Landes Biosciences, Georgetown, TX.
- Glick B.S. and Pon, L.A. 1995. Isolation of highly purified mitochondria from Saccharomyces cerevisiae. Methods Enzymol. 260: 213-223. [DOI] [PubMed] [Google Scholar]
- Goyon C., Rossignol, J.L., and Faugeron, G. 1996. Native DNA repeats and methylation in Ascobolus. Nucleic Acids Res. 24: 3348-3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guthrie C., Fink, G., Abelson, J., and Simon, M. 1991. Guide to yeast genetics & molecular biology: Methods in enzymology, Vol. 194. Academic Press, San Diego, CA. [PubMed]
- Handy J. and Doolittle, R.F. 1999. An attempt to pinpoint the phylogenetic introduction of glutaminyl-tRNA synthetase among bacteria. J. Mol. Evol. 49: 709-715. [DOI] [PubMed] [Google Scholar]
- Harlow E. and Lane, D. 1999. Using antibodies: A laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Hartl F.U., Ostermann, J., Guiard, B., and Neupert, W. 1987. Successive translocation into and out of the mitochondrial matrix: Targeting of proteins to the intermembrane space by a bipartite signal peptide. Cell 51: 1027-1037. [DOI] [PubMed] [Google Scholar]
- Heckman J.E., Sarnoff, J., Alzner-DeWeerd, B., Yin, S., and RajBhandary, U.L. 1980. Novel features in the genetic code and codon reading patterns in Neurospora crassa mitochondria based on sequences of six mitochondrial tRNAs. Proc. Natl. Acad. Sci. 77: 3159-3163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopper A.K. and Phizicky, E.M. 2003. tRNA transfers to the limelight. Genes & Dev. 17: 162-180. [DOI] [PubMed] [Google Scholar]
- Hughes T.R., Marton, M.J., Jones, A.R., Roberts, C.J., Stoughton, R., Armour, C.D., Bennett, H.A., Coffey, E., Dai, H., He, Y.D., et al. 2000. Functional discovery via a compendium of expression profiles. Cell 102: 109-126. [DOI] [PubMed] [Google Scholar]
- Ibba M. and Söll, D. 2004. Aminoacyl-tRNAs: Setting the limits of the genetic code. Genes & Dev. 18: 731-738. [DOI] [PubMed] [Google Scholar]
- Kaneko T., Suzuki, T., Kapushoc, S.T., Rubio, M.A., Ghazvini, J., Watanabe, K., and Simpson, L. 2003. Wobble modification differences and subcellular localization of tRNAs in Leishmania tarentolae: Implication for tRNA sorting mechanism. EMBO J. 22: 657-667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolesnikova O.A., Entelis, N.S., Mireau, H., Fox, T.D., Martin, R.P., and Tarassov, I.A. 2000. Suppression of mutations in mitochondrial DNA by tRNAs imported from the cytoplasm. Science 289: 1931-1933. [DOI] [PubMed] [Google Scholar]
- Lamour V., Quevillon, S., Diriong, S., N'Guyen, V.C., Lipinski, M., and Mirande, M. 1994. Evolution of the Glx-tRNA synthetase family: The glutaminyl enzyme as a case of horizontal gene transfer. Proc. Natl. Acad. Sci. 91: 8670-8674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapointe J., Duplain, L., and Proulx, M. 1986. A single glutamyl-tRNA synthetase aminoacylates tRNAGlu and tRNAGln in Bacillus subtilis and efficiently misacylates Escherichia coli tRNAGln1 in vitro. J. Bacteriol. 165: 88-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe T.M. and Eddy, S.R. 1997. tRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25: 955-964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludmerer S.W. and Schimmel, P. 1987. Construction and analysis of deletions in the amino-terminal extension of glutamine tRNA synthetase of Saccharomyces cerevisiae. J. Biol. Chem. 262: 10807-10813. [PubMed] [Google Scholar]
- Ludmerer S.W., Wright, D.J., and Schimmel, P. 1993. Purification of glutamine tRNA synthetase from Saccharomyces cerevisiae. A monomeric aminoacyl-tRNA synthetase with a large and dispensable NH2-terminal domain. J. Biol. Chem. 268: 5519-5523. [PubMed] [Google Scholar]
- Mulero J.J., Rosenthal, J.K., and Fox, T.D. 1994. PET112, a Saccharomyces cerevisiae nuclear gene required to maintain rho+ mitochondrial DNA. Curr. Genet. 25: 299-304. [DOI] [PubMed] [Google Scholar]
- Mumberg D., Muller, R., and Funk, M. 1995. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene 156: 119-122. [DOI] [PubMed] [Google Scholar]
- Nabholz C.E., Hauser, R., and Schneider, A. 1997. Leishmania tarentolae contains distinct cytosolic and mitochondrial glutaminyl-tRNA synthetase activities. Proc. Natl. Acad. Sci. 4: 7903-7908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai Y., Umeda, N., Suzuki, T., Nakai, M., Hayashi, H., Watanabe, K., and Kagamiyama, H. 2004. Yeast Nfs1p is involved in thio-modification of both mitochondrial and cytoplasmic tRNAs. J. Biol. Chem. 279: 12363-12368. [DOI] [PubMed] [Google Scholar]
- Polycarpo C., Ambrogelly, A., Ruan, B., Tumbula-Hansen, D., Ataide, S.F., Ishitani, R., Yokoyama, S., Nureki, O., Ibba, M., and Söll, D. 2003. Activation of the pyrrolysine suppressor tRNA requires formation of a ternary complex with class I and class II lysyl-tRNA synthetases. Mol. Cell. 12: 287-294. [DOI] [PubMed] [Google Scholar]
- Rinehart J., Horn, E.K., Wei, D., Söll, D., and Schneider, A. 2004. Non-canonical eukaryotic glutaminyl- and glutamyl-tRNA synthetases form mitochondrial aminoacyl-tRNA in Trypanosoma brucei. J. Biol. Chem. 279: 1161-1166. [DOI] [PubMed] [Google Scholar]
- Rosas-Sandoval G., Ambrogelly, A., Rinehart, J., Wei, D., CruzVera, L.R., Graham, D.E., Stetter, K.O., Guarneros, G., and Söll, D. 2002. Orthologs of a novel archaeal and of the bacterial peptidyl-tRNA hydrolase are nonessential in yeast. Proc. Natl. Acad. Sci. 99: 16707-16712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio M.A., Liu, X., Yuzawa, H., Alfonzo, J.D., and Simpson, L. 2000. Selective importation of RNA into isolated mitochondria from Leishmania tarentolae. RNA 6: 988-1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider A. and Marechal-Drouard, L. 2000. Mitochondrial tRNA import: Are there distinct mechanisms? Trends. Cell Biol. 10: 509-513. [DOI] [PubMed] [Google Scholar]
- Schön A., Kannangara, C.G., Gough, S., and Söll, D. 1988. Protein biosynthesis in organelles requires misaminoacylation of tRNA. Nature 331: 187-190. [DOI] [PubMed] [Google Scholar]
- Sekine S., Nureki, O., Shimada, A., Vassylyev, D.G., and Yokoyama, S. 2001. Structural basis for anticodon recognition by discriminating glutamyl-tRNA synthetase. Nat. Struct. Biol. 8: 203-206. [DOI] [PubMed] [Google Scholar]
- Sesaki H. and Jensen, R.E. 1999. Division versus fusion: Dnm1p and Fzo1p antagonistically regulate mitochondrial shape. J. Cell. Biol. 147: 699-706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoscheck C.M. 1987. Protein assay sensitive at nanogram levels. Anal. Biochem. 160: 301-305. [DOI] [PubMed] [Google Scholar]
- Tarassov I., Entelis, N., and Martin, R.P. 1995. Mitochondrial import of a cytoplasmic lysine-tRNA in yeast is mediated by cooperation of cytoplasmic and mitochondrial lysyl-tRNA synthetases. EMBO J. 14: 3461-3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzagoloff A. and Shtanko, A. 1995. Mitochondrial and cytoplasmic isoleucyl-, glutamyl- and arginyl-tRNA synthetases of yeast are encoded by separate genes. Eur. J. Biochem. 230: 582-586. [DOI] [PubMed] [Google Scholar]
- Varshney U., Lee, C.P., and RajBhandary, U.L. 1991. Direct analysis of aminoacylation levels of tRNAs in vivo. Application to studying recognition of Escherichia coli initiator tRNA mutants by glutaminyl-tRNA synthetase. J. Biol. Chem. 266: 24712-24718. [PubMed] [Google Scholar]
- Woese C.R., Olsen, G.J., Ibba, M., and Söll, D. 2000. Aminoacyl-tRNA synthetases, the genetic code, and the evolutionary process. Microbiol. Mol. Biol. Rev. 64: 202-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D., Oyaizu, Y., Oyaizu, H., Olsen, G.J., and Woese, C.R. 1985. Mitochondrial origins. Proc. Natl. Acad. Sci. 82: 4443-4447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama S. and Nishimura, S. 1995. Modified nucleosides and codon recognition. In tRNA: Structure, biosynthesis and function (eds. U. RajBhandary and D. Söll), pp. 207-233. American Society for Microbiology Press, Washington, DC.


