Abstract
Macrophomina phaseolina is the most devastating pathogen which causes charcoal rot and root rot diseases in various economically important crops. Three strains M. phaseolina 1156, M. phaseolina 1160, and M. phaseolina PCMC/F1 were tested for their virulence on sunflower (Helianthus annuus L.) and chickpea (Cicer arietinum L.). The strains showed high virulence on both hosts with a disease score of 2 on chickpea and sunflower. The strains also increased the hydrogen per oxide (H2O2) content by 1.4- to 1.6-fold in root as well as shoot of chickpea and sunflower. A significant increase in antioxidant enzymes was observed in fungal infected plants which indicated prevalence of oxidative stress during pathogen propagation. The M. phaseolina strains also produced hydrolytic enzymes such as lipase, amylase, and protease with solubilization zone of 5–43 mm, 5–45 mm, and 12–35 mm, respectively. The M. phaseolina strains were identified by 18S rRNA and analyzed for genetic diversity by using random amplified polymorphic DNA (RAPD) markers. The findings based on RAPD markers and 18S rRNA sequence analysis clearly indicate genetic variation among the strains collected from different hosts. The genetically diverse strains were found to be pathogenic to sunflower and chickpea.
Keywords: Macrophomina phaseolina, diversity, pathogenicity, RAPD
Introduction
Macrophomina phaseolina is a soil borne phytopathogenic fungus having a wide host range of about 500 cultivated and wild plant species worldwide (Khan, 2007). Important diseases caused by M. phaseolina include color rot, damping off, charcoal rot, stem rot, root rot, and seedling blight in economically important crops (Babu et al., 2007). The plants affected with fungus show necrotic lesion on different parts such as branches, peduncles, and stems. A higher temperature and low moisture favors the disease development (Aegerter et al., 2000). The microsclerotia of the pathogen can survive on infected plant debris and soil for a long period, i.e., 2–15 years depending on the environmental conditions (Baird et al., 2003; Vasebi et al., 2013). Microsclerotia are usually spherical, black, and oblong. However, there is a great variation in their shape and size depending on substrate, isolates, and temperature (Khan, 2007).
Macrophomina phaseolina affects the plant by secreting an array of cell wall degrading enzymes which depolymerize the cell wall components such as cellulose, xylan, pectin, polygalacturonic acid, and other proteins (Javaid and Saddique, 2012). The most significant enzymes secreted by M. phaseolina are pectinases, xylanases, cellulases, and proteases (Tonukari, 2003). Pectinases break the pectin of the host cell and use carbon as a source of pathogenesis whereas exo- and endopolygalacturonase are produced in early pathogenesis and colonization of host tissues (Murad and Azzaz, 2011). Lipases are also produced by M. phaseolina and cause hydrolysis of the fats, mono and diglycerides into free fatty acids and glycerol (Kakde and Chavan, 2011). It also produces certain toxins such as phaseolinone and botryodiplodin which facilitate the infection (Ramezani, 2008; Bressano et al., 2010). The production of hydrolytic enzymes have been reported to play a crucial role in the development of disease (Kaur et al., 2012).
Any pathogen’s attack stimulates the plant defense mechanism through hypersensitive response (HR). As a result of HR, reactive oxygen species (ROS) such as hydroxyl radicals, superoxide radicals, and hydrogen peroxide (H2O2) are produced. The unbalanced production of ROS could damage the plants severely. Hence, the production of ROS stimulates the deployment of antioxidant enzymes which scavenge ROS to maintain a balance. Thus, activity of antioxidant enzymes could serve as good marker for estimating oxidative stress in certain plant caused by pathogen (Anthony et al., 2017).
Morphological identification of M. phaseolina is very difficult and often not reliable because of the variation among isolates (Babu et al., 2010b; Saleh et al., 2010). Biochemical and serological techniques are being employed to identify the fungus but their specificity is limited (Srivastava and Arora, 1997). Advances in molecular techniques have provided alternative techniques for the reliable identification of fungi. Internal transcribed spacers (ITS) and 18S rRNA have been one of the most conserved genes used to identify the fungus (Babu et al., 2010b).
Genetic techniques such as random amplified polymorphic DNA (RAPD), restriction fragment length polymorphism (RFLP), and amplified fragment length polymorphism (AFLP) have helped researchers to understand more about the genetic variation in M. phaseolina (Mayék-Pérez et al., 2001; Su et al., 2001; Purkayastha et al., 2006; Reyes-Franco et al., 2006). Variation in pathogenicity, physiology, morphology, and genotype of M. phaseolina have been reported widely (Jana et al., 2003; Edraki and Banihashemi, 2010). Genetic and pathogenic variation in M. phaseolina strains have been assessed in numerous isolates but so far, it is hard to discriminate of M. phaseolina isolates from exact hosts or geographic locations due to heterogeneous nature. Lack of a strong association between the geographical origin and genotype propose high diversity among M. phaseolina strains (Jana et al., 2005).
The housekeeping gene 18S rRNA sequence is an ideal marker to identify the fungi at genus level but has some limitations to discriminate the intra species of fungi. The sequence of hyper-variable regions like V2, V4, V7, and V9 play an important role in identification of fungi. The region V7 has been utilized to discriminate various strains by phylogenetic analysis (Wu et al., 2015).
Random amplified polymorphic DNA are useful markers to measure genetic relatedness and variation within and among various fungi thus facilitating the understanding of their ecology (Kini et al., 2002; Cramer et al., 2003). The present study aims to identify the virulent strains of M. phaseolina showing cross host pathogenicity and characterize their genetic diversity by inferring their phylogenetic lineage and RAPD markers.
Materials and Methods
Strain and Culture Conditions
Three strains of M. phaseolina strain 1156, 1161, and PCMC-F1 isolated from sesame, cowpea, and cotton were obtained from first fungal bank, University of Punjab, Lahore, Pakistan. The strains were routinely grown on potato dextrose agar (PDA) (oxoid) at 28 ± 2°C and preserved in slants containing 2% PDA.
Detection of Hydrolytic Enzymes
Production of various hydrolytic enzymes such as protease, amylase, and lipase was detected on specific medium as described by Schinke and Germani (2012). For protease activity, the fungal strains were grown on skim milk agar. Amylase activity was detected on Pontecorvo’s minimal medium amended with 1% w/v soluble starch, respectively. Solubilization zone of amylase was detected by spreading the 1% lugol solution on the plates, respectively. Productions of lipases were detected with sterilized rhodamine B agar amended with 1% (v/v) filter sterilized olive oil. The inoculated plates were incubated at 28 ± 2°C for 4–5 days. The production zones of protease on gelatin and amylase on soluble starch were detected by pouring saturated solution of ammonium sulfate and 1% lugol solution, respectively. The zone of lipase production was observed on the basis of yellow–orange color appeared under UV light while that of protease on skim milk agar was observed as transparent area appeared after casein degradation. The experiments were repeated thrice.
Pathogenicity of M. phaseolina Strains under Net House (Pot Experiment)
Pathogenicity of three strains M. phaseolina 1156, M. phaseolina 1161, and M. phaseolina PCMC-F1 was tested on its two hosts, i.e., chick pea (Cicer arietinum) and sunflower (Helianthus annuus L.) under net house conditions.
Chick Pea (Cicer arietinum)
Pathogenicity test was conducted on the chickpea variety Sheenghar. Seeds were obtained from Agriculture Research Station Ahmad Wala, Karak, Khyber Pakhtunkhwa Khah (KPK), Pakistan and sown in pots in net house conditions. The experiment was laid out as complete randomized design (CRD). There were three replication per treatment and four plants per replication. A 9 mm disk of each strain of M. phaseolina was grown on Richard’s liquid medium at 28 ± 2°C for 14 days. The fungus was drenched near plant roots by using disposable syringes at 30th day of sowing. The disease was scored by using the scale (0–2) at 40th day post inoculation by measuring the damaged root (Bhattacharyya et al., 1985). Growth retardation was observed by measuring different growth parameters such as shoot (SL) and root lengths (RL), shoot dry weight (SDW) and root dry weight (RDW).
Sunflower (Helianthus annuus L.)
Sunflower seeds of Variety SMH0917 were obtained from National Agriculture Research Centre (NARC), Islamabad, Pakistan and sown in pots under net house conditions. The experiment was laid out as described in Section “Chick Pea (Cicer arietinum).” The M. phaseolina strains were inoculated as described in Section “Pathogenicity of M. phaseolina Strains under Field (Field Experiment).” However, a syringe was used instead of toothpick. The growth retardation of sunflower plants infected with M. phaseolina strains was observed by measuring root length (RL), shoot length (SL), fresh weight and dry weight of shoots and roots. The disease was scored (0–2) on the basis of root and shoot damage.
In Planta Pathogenicity Mechanism
In planta pathogenicity mechanism of Macrophomina phaseolina was explored by quantifying the ROS and their scavengers. Oxidative stress related metabolites such as H2O2 and ROS scavenging enzymes like peroxidase (POD), ascorbate peroxidase (APX), and catalase (CAT) were quantified from the roots and shoots of chickpea and sunflower. Root and shoot of 5- to 6-week-old plants were sampled and weighed. A 0.5 g of each part was grinded to fine powder in mortar pestle with liquid nitrogen. The grinded powder was dissolved in 10 mL of 100 mM phosphate buffer (pH 7.0) and centrifuged at 10,000 rpm for 20 min. The supernatant was used as crude extract.
Hydrogen Peroxide (H2O2)
Amount of H2O2 was quantitatively estimated as described by Jana and Choudhuri (1981). A 3 mL of crude extract was mixed with 1 mL of solution containing titanium sulfate (0.1%) and sulfuric acid (20%). The mixture was centrifuged at 10,000 rpm for 15 min. A yellow color was developed in solution whose intensity was measured at 410 nm (Singh and Jha, 2016).
Ascorbate Peroxidase (APX)
Activity of APX enzyme was assayed as described by Sarkar et al. (2014). The reaction mixture was prepared by mixing potassium phosphate buffer (50 mM, pH 7.0), H2O2 (0.1 mM), and ascorbate (0.5 mM). The crude extract was added to the mixture to initiate the reaction and H2O2 -dependent oxidation of ascorbate was measured at 290 nm.
Catalase (CAT)
Catalase activity was determined by following the method of Sarkar et al. (2014). Briefly, the crude extract was mixed with potassium phosphate buffer (50mM, pH 7.5) and H2O2 (0.1 mM). The absorbance was measured at 240 nm. CAT activity was calculated on the basis of H2O2 utilization (extinction coefficient = 43.6 M-1cm-1) (Aebi, 1984).
Peroxidase (POD)
Peroxidase activity assay was conducted as described by Singh and Jha (2016). Briefly, the phosphate buffer (0.1 M, pH 7.0), pyrogallol (0.1 mM), and H2O2 (5 mM) were mixed with 100 μL of crude extract. The mixture was incubated at 25°C for 5 min. A 1.0 ml of 2.5 N H2SO4 was used to stop the reaction. The absorbance was read at 420 nm.
Pathogenicity of M. phaseolina Strains under Field (Field Experiment)
Pathogenicity test of three fungal strains was conducted on the sunflower hybrids hysun 33 and SMH 0917 at the experimental fields of National Agriculture Research Center (NARC), Islamabad, Pakistan. The experiment was laid out in randomized completely block design (RCBD). The optimum temperature for sunflower was 70 to 78°F. Plants were sown with row to row distance of 75 cm and seeds were embedded in the ridges 1 to 2 cm with plant to plant distance of 12 cm. Each fungal strain was considered as single treatment with three replications and eight to ten plants per replication. The fungus was inoculated by tooth pick method (Simonetti et al., 2015). Briefly, the tooth picks (bamboo) were boiled for 30 min in a glass beaker and then dried on sterilized blotting paper. The dried tooth picks were transferred to three glass jars containing potato dextrose broth. Tooth picks were placed in jars vertically in such a way that half of the parts were dipped in broth. The pure culture of three pathogenic strains of M. phaseolina were poured into three jars with the help of cork borer and incubated at 28 ± 2°C for 14 days. After 14 days, tooth picks were ready to use.
At the flowering stage of sunflower plants, a hole was made in stem with a needle one foot above the soil surface. The tooth picks inoculated with respective fungus were incorporated into the holes. The inoculated plants were tagged with ribbons of different colors.
At physiological maturity, the disease was assessed by cutting the plant stem into two parts and measuring the symptoms up and down of the inoculated tooth picks. The infected part of the stem where microsclerotia developed was measured and scored on 0–6 scale, i.e., 1- up to 5 cm; 2- up to 10 cm; 3- up to 20 cm; 4- up to 30 cm; 5- up to 40 cm; 6- over 50 cm or completely wilted plant.
Molecular Identification
The fungal strains were identified by 18S rRNA gene sequencing. The genomic DNA of the fungal pathogens was extracted by cetyltrimethyl ammonium bromide (CTAB) method (Brandfass and Karlovsky, 2008). Part of 18S rRNA region was amplified by using universal primers nu-SSU-0817-5′5′TTAGCATGGAATAATRRAATAGGA3′ and nu-SSU-1536-3′5′ATTGCAATGCYCTATCCCCA3′ (Borneman and Hartin, 2000). PCR reaction mixture consisted of 5 ng genomic DNA, 1.5 mM MgCl2, 10 μM of each primer, 1XTaq buffer (Fermentas) and 180–200 μM of each dNTPs. The reaction mixture was amplified in thermo cycler (Eppendorf) with the following amplifying conditions; Initial denaturation at 95°C for 5 min followed by 30 cycles of denaturation at 95°C for 45 s, annealing at 49°C for 45 s, extension at 72°C for 60 s and final extension at 72°C for 10 min. The amplified PCR product was analyzed on 1.2% agarose gel, purified by PCR purification kit (Fermentas) and sequenced by Macrogen Inc., Korea. Sequences were annotated and analyzed at BLAST to search the closest homolog.
Random Amplified Polymorphism DNA (RAPD)
Random Amplified Polymorphic DNA analysis was used to detect the variations among the isolates of M. phaseolina. A total of six random primers of OPA series were used in this study (Prabhu et al., 2012). The sequences of primers used in the study are shown in the Supplementary Table S1.
Phylogenetic Analysis
Full length Sequences of 18S rRNA were retreived from silva database. The sequences were aligned in Mega 6 with MUSCLE and clustered by using UPGMB method (Edgar, 2004). All sequences were trimed mannualy to remove a fair comparison with our own strains and realligned. The tree was constructed by neighbor joining to infer the evolutionary history (Saitou and Nei, 1987). The phylogeny was tested by applying bootstrap method (Felsenstein, 1985). The evolutionary analyses were conducted in MEGA6 which draw the tree to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree and compute the by using the Maximum Composite Likelihood method (Tamura et al., 2004, 2013).
Statistical Analysis
The data was analyzed with analysis of variance (ANOVA) using the statistical software MSTAT-C. The mean values were separated on the basis of Fisher’s least significant difference (LSD) test.
Results
Virulence of M. phaseolina Strains on Chick Pea (Cicer arietinum)
All the M. phaseolina strains caused high virulence on chickpea (Cicer arietinum). A significant reduction in root length (RL) (44–49%), shoot length (SL) (5–16%), and fresh weight (55–63%) was observed in plants treated with pathogenic fungi as compared to that of untreated control (Figure 1). The dry weight of infected plants was found to be higher (26–38%) than that of healthy plants, i.e., (control). All the strains showed high virulence with a disease score 2 (Figure 1).
FIGURE 1.
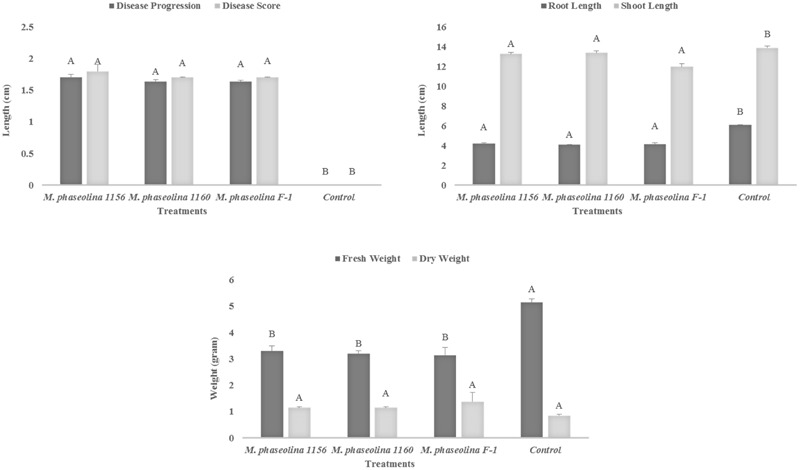
Growth retardation and disease progression in chickpea caused by M. phaseolina. Mean values followed by different letters are significantly different (P < 0.05).
Virulence of M. phaseolina Strains on Sunflower
The M. phaseolina strains caused disease score of 2 on sunflower plants in net house conditions (Figure 2) while a disease score of 5.8–6.0 under field conditions (Table 1). A significant reduction in RL (53–58%), SL (22–31%), and fresh weight (41–52%) was observed in sunflower plants inoculated with fungi as compared to that of control (Figure 2). The virulence of strains on sunflower hybrids under field conditions was highly significant (Table 1).
FIGURE 2.
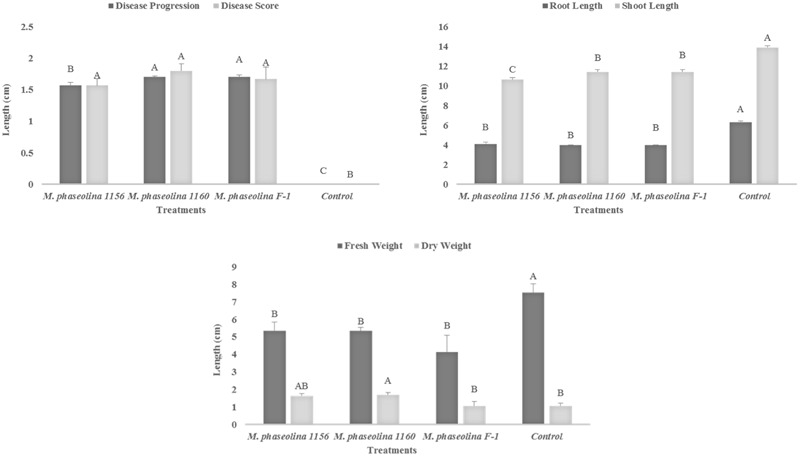
Growth retardation and disease progression in sunflower caused by M. phaseolina. Mean values followed by different letters are significantly different (P < 0.05).
Table 1.
Pathogenicity of Macrophomina phaseolina strains on sunflower under field conditions.
| Fungal strains | Disease progression (DP) | Disease score (DS) |
|---|---|---|
| M. phaseolina strain PCMC/F1 | 81 A | 6 A |
| M. phaseolina strain 1161 | 82 A | 6 A |
| M. phaseolina strain 1156 | 81 A | 6 A |
| No fungus (control) | 0 B | 0 B |
| Hybrids | ||
| H1 | 82 A | 6 A |
| H2 | 80 A | 6 A |
Values are mean of three replications and separated by different letters in the same column are statistically different from each other.
H1 = Sunflower hybrid Hysun 33; Sunflower hybrid SMH-0917.
In Planta Pathogenicity Mechanism of M. phaseolina
Determinants of in planta pathogenicity and metabolic markers of oxidative stress viz H2O2 and other antioxidant enzymes were produced in both crops upon fungal infection.
Chick Pea (Cicer arietinum)
A significant impact of M. phaseolina strains was observed on the H2O2 content and activity of ROS scavenging enzymes in chickpea. The H2O2 content was increased 1.4- to 1.6-fold in shoot of the plants treated with virulent strains of M. phaseolina over the untreated plants (Figure 3). Similarly the activities of ROS scavenging enzymes POD, APX, and CAT were increased 1.2- to 1.6-fold in the chickpea plants treated with M. phaseolina over untreated plants (control).
FIGURE 3.
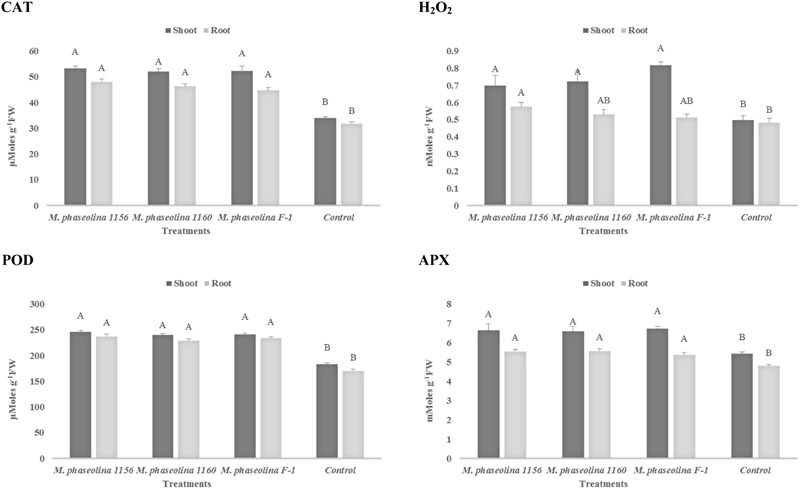
Activity of H2O2 and antioxidant enzymes in chickpea during M. phaseolina infection. Mean values followed by different letters are significantly different (P < 0.05). H2O2, hydrogen peroxide; CAT, catalase; POD, peroxidase; APX, ascorbate peroxidase.
Sunflower (Helianthus annuus L)
In sunflower plants, M. phaseolina strains altered the concentration of H2O2 and the other enzymes in a similar trend as observed in chickpea. The H2O2 content was increased 1.2- to 1.3-fold in roots and 1.3- to 1.4-fold in shoots of the fungal treated plants over the untreated ones (control). The activities of ROS scavenging enzymes POD, APX, and CAT were also increased by 1.3- to 1.7-fold upon fungal infection (Figure 4). The phenotypic effect of M. phaseolina strains on sunflower and chick pea is shown in Figures 5A,B.
FIGURE 4.
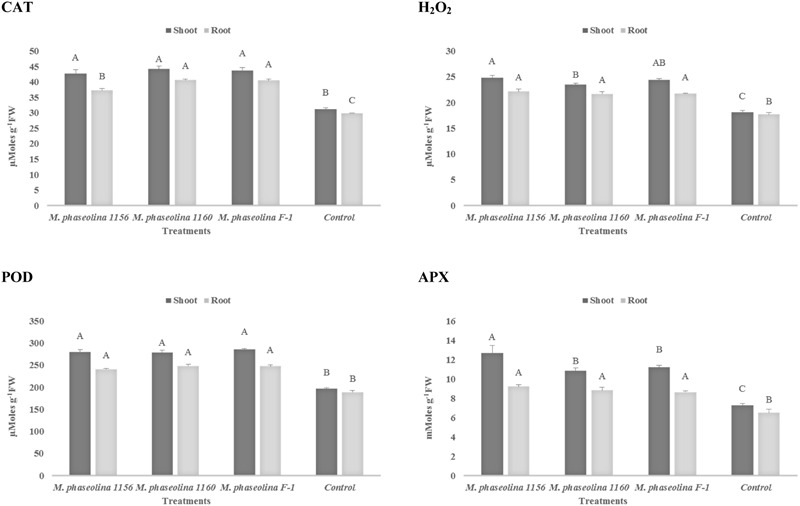
Activity of H2O2 and antioxidant enzymes in sunflower during M. phaseolina infection. Mean values followed by different letters are significantly different (P < 0.05). H2O2, hydrogen peroxide; CAT, catalase; POD, peroxidase; APX, ascorbate peroxidase.
FIGURE 5.

(A) Disease symptoms appeared on sunflower plants after M. phaseolina inoculation. (B) Disease symptoms appeared on chickpea plants after M. phaseolina inoculation.
Production of Hydrolytic Enzymes
Macrophomina phaseolina strains produced different hydrolytic enzymes. PCMC/F1 produced amylase and lipase with solubilization zone of 41mm and 43 mm, respectively (Figure 6). M. phaseolina strain 1156 also produced highest amylase with the solubilization zone of 45 mm but least lipase with the solubilization zone of 5 mm. The strain 1160 showed minimal production of amylase and lipase with a solubilization zone of 5 and 8 mm, respectively. Production of protease enzymes showed substrate specificity. The effect of substrates ‘starch’ and ‘gelatin’ was highly significant on the production of protease (Table 2). M. phaseolina strain 1160 showed maximum protease production with a solubilization zone of 35 mm followed by the strain PCMC/F1 and 1156 with a solubilization zone of 14 and 12 mm, respectively. Growth of fungus on respective substrate is shown in Supplementary Figure S3.
FIGURE 6.
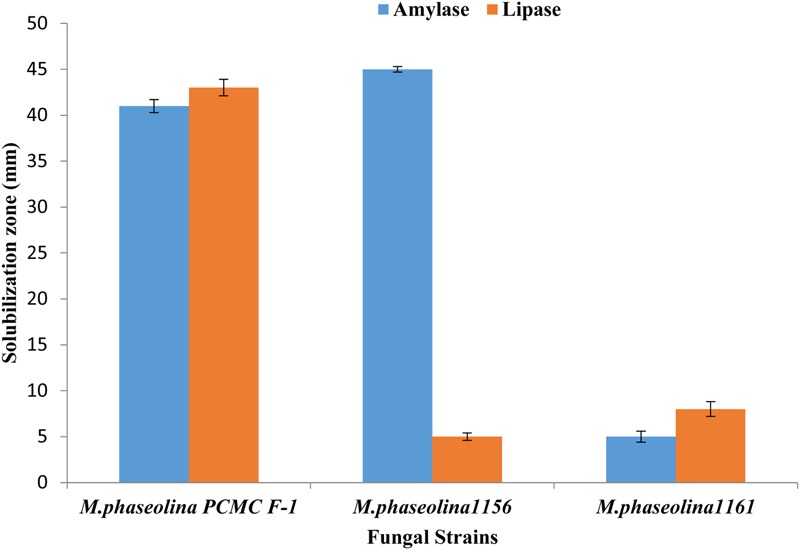
Production of hydrolytic enzymes by the M. phaseolina strains. Values are mean of three replications. Bars represent standard error.
Table 2.
Production of protease on different substrates by M. phaseolina strains.
| Treatment | Solubilization zone (mm) |
|---|---|
| Strain | |
| M. phaseolina strain PCMC/F1 | 14.3 B |
| M. phaseolina strain 1160 | 35.3 A |
| M. phaseolina strain 1156 | 11.7 C |
| LSD strains | 2.4 |
| Substrate | |
| Gelatin | 10.6 E |
| Starch | 30.3 D |
| LSD substrate | 2.0 |
Values are mean of three replications and separated by different letters in the same column are statistically different from each other.
18S rRNA Amplification and Accession No.
A 762 bp of 18S rRNA gene was amplified as shown in Supplementary Figure S1. Sequences of 18S rRNA gene have been deposited in Gene Bank under the accession numbers KC422671, KP174124, and KP174125. Phylogenetic tree of M. phaseolina strains is shown in Figure 7. The strains revealed its relatedness with M. phaseolina. The strain PCMC was found to be different from other strains.
FIGURE 7.
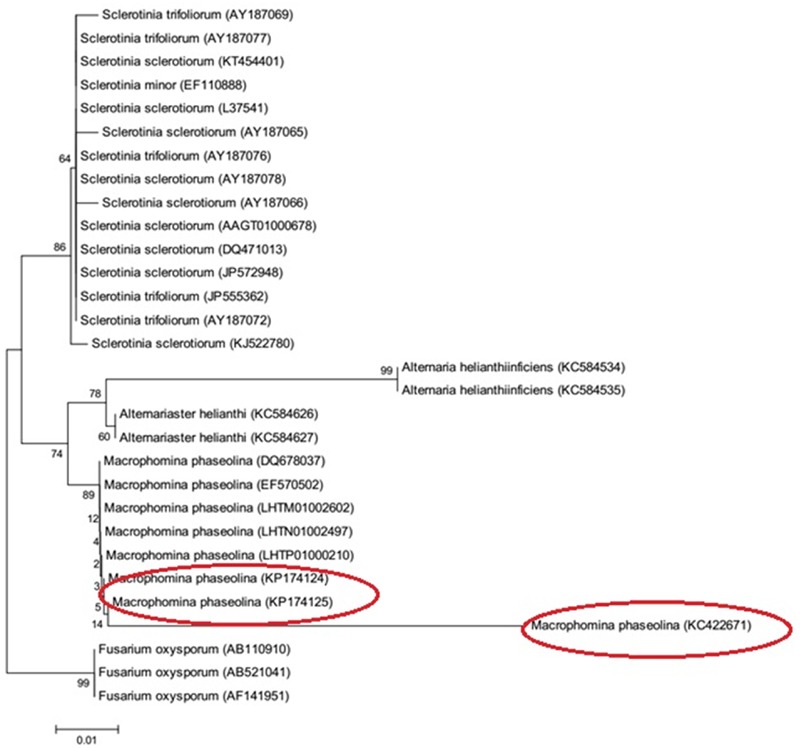
Phylogenetic tree of M. phaseolina strains based on the 18S rRNA gene.
RAPD Result (Dendrogram)
Random Amplified Polymorphic DNA analysis showed genetic variations among the M. phaseolina strains. Out of the six primers used for amplification, OPA01 and OPA04 showed more than 80 percent polymorphism among isolates followed by OPA02 and OPA03 which showed 60 and 44% while OPA7 and OPA10 53 and 70% polymorphism. The Dendrogram constructed on the basis of band patterns using Jaccard’s coefficient in PAST software is shown in Figure 8. Different band patterns on agarose gel are shown in the Supplementary Figure S2.
FIGURE 8.
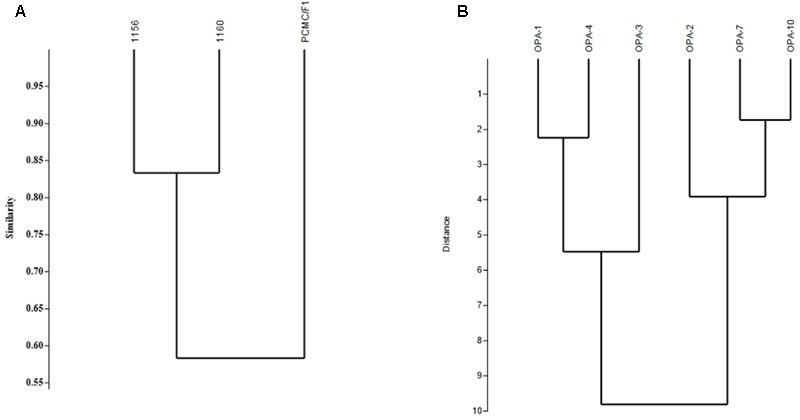
Genetic diversity of M. phaseolina strains based on the RAPD analysis. (A) Similarity index between M. phaseolina strains. (B) Similarity index between OPA primers.
Discussion
Macrophomina phaseolina is one of the most devastating pathogens that infect more than 500 plant species throughout the world. It can grow swiftly in infected plants and afterward produces a large quantity of sclerotia that plugs the vessels, resulting in wilting of the plant. In present investigation, native strains of M. phaseolina isolated from sesame (Sesamum indicum), cotton (Gossypium hirsutum), and cowpea (Vigna unguiculata) showed highly virulence on chickpea and sunflower irrespective of their origin.
Macrophomina phaseolina, being a necrotrophic pathogen, kills the cells of host and reduces growth rate. In chick pea plant, we focused on root rot symptoms because pathogen enters through the roots leading to the appearance of symptoms firstly on the roots. While in sunflower, basal node is the entry point of this pathogen. The M. phaseolina strains reduced the growth of root and shoot of both crops without any significant difference among their virulence.
Host adaptability of M. phaseolina strains has been found to be dependent on many factors. The adaptation of soybean associated strains was highly impacted by crop rotation while other strains showed specificity with corn (Su et al., 2001; Almeida et al., 2008). These findings depict the diversity in different M. phaseolina strains. In the present era of intensive cultivation and irrational use of fertilizers/pesticides, may tend toward the emergence of new races of the strains.
Macrophomina phaseolina strains significantly enhanced the quantities of metabolites/enzymes causing oxidative stress in cell. As the necrotrophic pathogens prefer to feed on cell remain, they kill the host cells through creation of oxidative stress (Laluk and Mengiste, 2010). Thus, measurement of oxidative stress determinants could be ideal to estimate in planta pathogenicity mechanism of phytopathogens.
A higher quantity of H2O2 was observed during the M. phaseolina infection on both hosts. H2O2 is an important component of ROS and found to be associated with virulence of numerous pathogens (Yu et al., 2017).
In oxidative stress, ROS scavengers and antioxidant enzymes are highly activated to neutralize the negative effect. The fungal inoculation also enhanced the activity of ROS scavenging enzymes CAT, POD, and APX which advocated the presence of high oxidative stress during fungal infection. Similar findings on other hosts under different pathogens stress have been reported (Anthony et al., 2017).
In our study, a consistency between the phenotypic symptoms and biochemical changes was observed depicting that M. phaseolina cause virulence by creating strong oxidative stress in the host cells.
The strains also showed variable potential to secret hydrolytic enzymes viz pectinase, amylase, and protease. Hydrolytic enzymes are secreted by virulent strains of pathogens which they utilize to overcome the physical barriers by degrading various cell wall components of plants like pectin, lipid, cellulose, and proteins (Wheeler, 1975). In present study, all the M. phaseolina strains produced amylases. This is in contrast to the earlier report in which Macrophomina sp. was unable to produce amylase (Sohail et al., 2009). However, the strains produce variable amount of protease which were consistent to the earlier findings (Ahmad et al., 2006).
Production hydrolytic enzymes by the pathogenic fungi have been reported in determining their pathogenic potential (Ghannoum, 2000; Lawrence et al., 2000; López-Otín and Overall, 2002). These enzymes not only facilitate the fungal penetration by dissolving different plant structures like peptide bonds, phospholipids but also act as signaling molecules for the induction of HR (Sarkar et al., 2014). The proteolytic processing mediated by protease enzyme, cause change in certain protein function, and used as common mechanism for controlling the biological processes at cellular level (López-Otín and Overall, 2002).
Sequence of 18S rRNA of all the strains showed closest homology, i.e., 99% similarity with the reference sequences deposited in Gene Bank and confirmed their identification as M. phaseolina. The clustering of 18S rRNA sequences of test isolates with that of “Gene Bank reference sequences” further validated the identification results. A greater similarity in a 490–491 bp of 18S rRNA gene validated its conserved nature among the M. phaseolina strains. The Identification of fungi based on 18S rDNA is authenticated and well reported (Romanelli et al., 2014).
Phylogenetic analysis delineated the strains 1156 (KP174124) and 1160 (KP174125) were into same clades while strain PCMC F1 (KC422671) into different clade. These findings showed that M. phaseolina is not confined to either geographical location and/or a specific host. The distribution of M. phaseolina groups have been found to be independent of sampling location and host in numerous studies except a few reports in which M. phaseolina strains exhibited host specificity (Jana et al., 2003; Purkayastha et al., 2006; Bashasab and Kuruvinashetti, 2007; Rayatpanah et al., 2009; Baird et al., 2010; Saleh et al., 2010).
DNA markers such as RAPDs using polymerase chain reactions (PCR) have been widely used for detecting genetic diversity among the microorganisms (Sharma et al., 2013). In present study, RAPD analysis clearly indicated high polymorphism among the M. phaseolina strains depicting its effectiveness in evaluation of genetic diversity in M. phaseolina.
The genetic diversity within strains of M. phaseolina have been widely studied using the RAPD and rDNA sequencing (Jana et al., 2003; Purkayastha et al., 2006; Bashasab and Kuruvinashetti, 2007; Babu et al., 2010a; Saleh et al., 2010; Sundravadana et al., 2011; Mudalige et al., 2012).
These reports on the basis of RAPD markers and 18S rRNA sequence analysis clearly indicate genetic variation among the strains collected from different hosts and shore up the possibility of emergence of various pathotypes in M. phaseolina independent of host specificity. Variation in the pathogenicity may be associated with their ability to produce hydrolytic enzymes and genetic diversity. The broad host range enables M. phaseolina to survive in soil for a long time and cause great damage to all the crops. Therefore, there is need to design the new control strategies for the management of emerging genetically diverse pathotypes of M. phaseolina.
Author Contributions
AK performed the RAPD PCR analysis and involved in write up. FS analyzed the RAPD data and was involved in writing the manuscript, KM conducted pathogenicity test on sunflower plants. ZH helped in setting up the PCR experiments. MK monitored the field experiments. FH edited the manuscript and co supervised the experiments. MH designed the study, edited and revised the manuscript and supervised the experiments conducted at his laboratory.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are thankful to International Foundation for Science (IFS) for providing funds, Gram Research Institute Ahmad Wala, Khyber Pakhtunkhwa Khah (KPK), Pakistan for providing seeds of chickpea varieties. A special thanks to Dr. Arshan Nasir (Department of Biosciences, CIIT, Islamabad) for analyzing the DNA sequences.
Footnotes
Funding. The funds were provided by the International foundation for science under project entitled “Biocontrol of Macrophomina phaseolina causing charcol rot in oil seed crops by using Plant growth promoting rhizobacteria (PGPR) Grant No: C/4977-1.”
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2017.01309/full#supplementary-material
References
- Aebi H. (1984). Catalase in vitro. Methods Enzymol. 105 121–126. 10.1016/S0076-6879(84)05016-3 [DOI] [PubMed] [Google Scholar]
- Aegerter B., Gordon T., Davis R. (2000). Occurrence and pathogenicity of fungi associated with melon root rot and vine decline in California. Plant Dis. 84 224–230. 10.1094/PDIS.2000.84.3.224 [DOI] [PubMed] [Google Scholar]
- Ahmad Y., Hameed A., Ghaffar A. (2006). Enzymatic activity of fungal pathogens in corn. Pak. J. Bot. 38:1305. [Google Scholar]
- Almeida Á. M., Sosa-Gomez D. R., Binneck E., Marin S. R., Zucchi M. I., Abdelnoor R. V., et al. (2008). Effect of crop rotation on specialization and genetic diversity of Macrophomina phaseolina. Trop. Plant Pathol. 33 257–264. [Google Scholar]
- Anthony K. K., George D. S., Baldev Singh H. K., Fung S. M., Santhirasegaram V., Razali Z., et al. (2017). Reactive oxygen species activity and antioxidant properties of Fusarium infected bananas. J. Phytopathol. 165 213–222. 10.1111/jph.12552 [DOI] [Google Scholar]
- Babu B. K., Reddy S., Yadav M. K., Sukumar M., Mishra V., Saxena A., et al. (2010a). Genetic diversity of Macrophomina phaseolina isolates from certain agro-climatic regions of India by using RAPD markers. Indian J. Microbiol. 50 199–204. 10.1007/s12088-010-0033-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babu B. K., Saikia R., Arora D. K. (2010b). “Molecular characterization and diagnosis of Macrophomina phaseolina: a charcoal rot fungus,” in Molecular Identification of Fungi eds Gherbawy Y., Voigt K. (New York, NY: Springer; ) 179–193. [Google Scholar]
- Babu B. K., Saxena A. K., Srivastava A. K., Arora D. K. (2007). Identification and detection of Macrophomina phaseolina by using species-specific oligonucleotide primers and probe. Mycologia 99 797–803. 10.1080/15572536.2007.11832511 [DOI] [PubMed] [Google Scholar]
- Baird R. E., Wadl P. A., Allen T., McNeill D., Wang X., Moulton J. K., et al. (2010). Variability of United States isolates of Macrophomina phaseolina based on simple sequence repeats and cross genus transferability to related genera within Botryosphaeriaceae. Mycopathologia 170 169–180. 10.1007/s11046-010-9308-3 [DOI] [PubMed] [Google Scholar]
- Baird R. E., Watson C. E., Scruggs M. (2003). Relative longevity of Macrophomina phaseolina and associated mycobiota on residual soybean roots in soil. Plant Dis. 87 563–566. 10.1094/PDIS.2003.87.5.563 [DOI] [PubMed] [Google Scholar]
- Bashasab R., Kuruvinashetti M. S. (2007). Genetic variability of sorghum charcoal rot pathogen (Macrophomina phaseolina) assessed by random DNA markers. Plant Pathol. J. 23 45–50. 10.5423/PPJ.2007.23.2.045 [DOI] [Google Scholar]
- Bhattacharyya D., Basu S., Chattapadhyay J., Bose S. (1985). Biocontrol of Macrophomina root-rot disease of jute by an antagonistic organism, Aspergillus versicolor. Plant Soil 87 435–438. 10.1007/BF02181910 [DOI] [Google Scholar]
- Borneman J., Hartin R. J. (2000). PCR primers that amplify fungal rRNA genes from environmental samples. Appl. Environ. Microbiol. 66 4356–4360. 10.1128/AEM.66.10.4356-4360.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandfass C., Karlovsky P. (2008). Upscaled CTAB-based DNA extraction and real-time PCR assays for Fusarium culmorum and F. graminearum DNA in plant material with reduced sampling error. Int. J. Mol. Sci. 9 2306–2321. 10.3390/ijms9112306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bressano M., Giachero M. L., Luna C. M., Ducasse D. A. (2010). An in vitro method for examining infection of soybean roots by Macrophomina phaseolina. Physiol. Mol. Pathol. 74 201–204. 10.1016/j.pmpp.2009.12.003 [DOI] [Google Scholar]
- Cramer R. A., Byrne P. F., Brick M. A., Panella L., Wickliffe E., Schwartz H. F. (2003). Characterization of Fusarium oxysporum isolates from common bean and sugar beet using pathogenicity assays and random-amplified polymorphic DNA markers. J. Phytopathol. 151 352–360. 10.1046/j.1439-0434.2003.00731.x [DOI] [Google Scholar]
- Edgar R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32 1792–1797. 10.1093/nar/gkh340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edraki V., Banihashemi Z. (2010). Phenotypic diversity among isolates of Macrophomina phaseolina and its relation to pathogenicity. Iran. J. Plant Pathol. 46 93–100. [Google Scholar]
- Felsenstein J. (1985). Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39 783–791. 10.1111/j.1558-5646.1985.tb00420.x [DOI] [PubMed] [Google Scholar]
- Ghannoum M. A. (2000). Potential role of phospholipases in virulence and fungal pathogenesis. Clin. Microbiol. Rev. 13 122–143. 10.1128/CMR.13.1.122-143.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jana S., Choudhuri M. A. (1981). Glycolate metabolism of three submersed aquatic angiosperms during ageing. Aquatic Bot. 12 345–354. 10.1016/0304-3770(82)90026-2 [DOI] [Google Scholar]
- Jana T., Sharma T. R., Prasad R. D., Arora D. K. (2003). Molecular characterization of Macrophomina phaseolina and Fusarium species by a single primer RAPD technique. Microbiol. Res. 158 249–257. 10.1078/0944-5013-00198 [DOI] [PubMed] [Google Scholar]
- Jana T., Singh N., Koundal K., Sharma T. (2005). Genetic differentiation of charcoal rot pathogen, Macrophomina phaseolina, into specific groups using URP-PCR. Can. J. Microbiol. 51 159–164. 10.1139/w04-122 [DOI] [PubMed] [Google Scholar]
- Javaid A., Saddique A. (2012). Control of charcoal rot fungus Macrophomina phaseolina by extracts of Datura metel. Nat. Prod. Res. 26 1715–1720. 10.1080/14786419.2011.605363 [DOI] [PubMed] [Google Scholar]
- Kakde R. B., Chavan A. M. (2011). Extracellular lipase enzyme production by seed-borne fungi under the influence of physical factors. Int. J. Biol. 3:94. [Google Scholar]
- Kaur S., Dhillon G. S., Brar S. K., Chauhan V. B. (2012). Carbohydrate degrading enzyme production by plant pathogenic mycelia and microsclerotia isolates of Macrophomina phaseolina through koji fermentation. Ind. Crops Prod. 36 140–148. 10.1016/j.indcrop.2011.08.020 [DOI] [Google Scholar]
- Khan S. N. (2007). Macrophomina phaseolina as causal agent for charcoal rot of sunflower. Mycopath 5 111–118. [Google Scholar]
- Kini K. R., Leth V., Mathur S. B. (2002). Genetic variation in Fusarium moniliforme isolated from seeds of different host species from Burkina Faso based on random amplified polymorphic DNA analysis. J. Phytopathol. 150 209–212. 10.1046/j.1439-0434.2002.00739.x [DOI] [Google Scholar]
- Laluk K., Mengiste T. (2010). Necrotroph attacks on plants: wanton destruction or covert extortion? Arabidopsis Book 8:e0136 10.1199/tab.0136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence C. B., Singh N. P., Qiu J., Gardner R. G., Tuzun S. (2000). Constitutive hydrolytic enzymes are associated with polygenic resistance of tomato to Alternaria solani and may function as an elicitor release mechanism. Physiol. Mol. Plant Pathol. 57 211–220. 10.1006/pmpp.2000.0298 [DOI] [Google Scholar]
- López-Otín C., Overall C. M. (2002). Protease degradomics: a new challenge for proteomics. Nat. Rev. Mol. Cell Biol. 3 509–519. 10.1038/nrm858 [DOI] [PubMed] [Google Scholar]
- Mayék-Pérez N., López-Castañeda C., González-Chavira M., Garcia-Espinosa R., Acosta-Gallegos J., de la Vega O. M. N., et al. (2001). Variability of Mexican isolates of Macrophomina phaseolina based on pathogenesis and AFLP genotype. Physiol. Mol. Plant Pathol. 59 257–264. 10.1006/pmpp.2001.0361 [DOI] [Google Scholar]
- Mudalige C., Girisha S., Raghavendra V., Niranjan M., Ravikumar K., Sumantha M. (2012). Molecular characterization of Macrophomina phaseolina, the incitant of Coleus forskohlii revealed by RAPD markers. Int. J. Life Sci. 5 44–50. 10.3126/ijls.v5i1.5598 [DOI] [Google Scholar]
- Murad H., Azzaz H. (2011). Microbial pectinases and ruminant nutrition. Res. J. Microbiol. 6 246–269. 10.3923/jm.2011.246.269 [DOI] [Google Scholar]
- Prabhu H. V., Adiver S., Bhat R., Narayana Y., Jahagirdar S., Parameshwarappa K. (2012). Genetic variability in Macrophomina phaseolina (Tassi.) Goid., causal agent of charcoal rot of sorghum. Karnataka J. Agric. Sci. 25 72–76. [Google Scholar]
- Purkayastha S., Kaur B., Dilbaghi N., Chaudhury A. (2006). Characterization of Macrophomina phaseolina, the charcoal rot pathogen of cluster bean, using conventional techniques and PCR-based molecular markers. Plant Pathol. 55 106–116. 10.1111/j.1365-3059.2005.01317.x [DOI] [Google Scholar]
- Ramezani H. (2008). Biological control of root-rot of eggplant caused by Macrophomina phaseolina. Am. Eurasian J. Agric. Environ. Sci. 4 218–220. [Google Scholar]
- Rayatpanah S., Nanagulyan S., Alavi S., Yasari E., Prof A., Carvil O., et al. (2009). Phenotypic variations of isolates of Macrophomina phaseolina from different hosts in Northern Iran. Austr. J. Basic Appl. Sci. 3 2908–2913. [Google Scholar]
- Reyes-Franco M., Hernández-Delgado S., Beas-Fernández R., Medina-Fernández M., Simpson J., Mayek-Pérez N. (2006). Pathogenic and genetic variability within Macrophomina phaseolina from Mexico and other countries. J. Phytopathol. 154 447–453. 10.1111/j.1439-0434.2006.01127.x [DOI] [Google Scholar]
- Romanelli A. M., Fu J., Herrera M., Wickes B. (2014). A universal DNA extraction and PCR amplification method for fungal rDNA sequence-based identification. Mycoses 57 612–622. 10.1111/myc.12208 [DOI] [PubMed] [Google Scholar]
- Saitou N., Nei M. (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4 406–425. [DOI] [PubMed] [Google Scholar]
- Saleh A., Ahmed H., Todd T., Travers S., Zeller K., Leslie J., et al. (2010). Relatedness of Macrophomina phaseolina isolates from tallgrass prairie, maize, soybean and sorghum. Mol. Ecol. 19 79–91. 10.1111/j.1365-294X.2009.04433.x [DOI] [PubMed] [Google Scholar]
- Sarkar T. S., Biswas P., Ghosh S. K., Ghosh S. (2014). Nitric oxide production by necrotrophic pathogen Macrophomina phaseolina and the host plant in charcoal rot disease of jute: complexity of the interplay between necrotroph–host plant interactions. PLoS ONE 9:e107348 10.1371/journal.pone.0107348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schinke C., Germani J. C. (2012). Screening Brazilian Macrophomina phaseolina isolates for alkaline lipases and other extracellular hydrolases. Int. Microbiol. 15 1–7. [DOI] [PubMed] [Google Scholar]
- Sharma G., Verma H., Sharma R. (2013). RAPD Analysis to study metagenome diversity in soil microbial community of arid zone plants. Proc. Natl. Acad. Sci. India Sect. B Biol. Sci. 83 135–139. 10.1007/s40011-012-0041-1 [DOI] [Google Scholar]
- Simonetti E., Viso N. P., Montecchia M., Zilli C., Balestrasse K., Carmona M. (2015). Evaluation of native bacteria and manganese phosphite for alternative control of charcoal root rot of soybean. Microbiol. Res. 180 40–48. 10.1016/j.micres.2015.07.004 [DOI] [PubMed] [Google Scholar]
- Singh R. P., Jha P. N. (2016). The multifarious PGPR Serratia marcescens CDP-13 augments induced systemic resistance and enhanced salinity tolerance of wheat (Triticum aestivum L.). PLoS ONE 11:e0155026 10.1371/journal.pone.0155026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohail M., Naseeb S., Sherwani S. K., Sultana S., Aftab S., Shahzad S., et al. (2009). Distribution of hydrolytic enzymes among native fungi: Aspergillus the pre-dominant genus of hydrolase producer. Pak. J. Bot. 41 2567–2582. [Google Scholar]
- Srivastava A., Arora D. (1997). Evaluation of a polyclonal antibody immunoassay for detection and quantification of Macrophomina phaseolina. Plant Pathol. 46 785–794. 10.1046/j.1365-3059.1997.d01-64.x [DOI] [Google Scholar]
- Su G., Suh S.-O., Schneider R., Russin J. (2001). Host specialization in the charcoal rot fungus, Macrophomina phaseolina. Phytopathology 91 120–126. 10.1094/PHYTO.2001.91.2.120 [DOI] [PubMed] [Google Scholar]
- Sundravadana S., Thirumurugan S., Alice D. (2011). Exploration of molecular variability in Rhizoctonia bataticola, the incitant of root rot disease of pulse crops. J. Plant Prot. Res. 51 184–189. 10.2478/v10045-011-0032-x [DOI] [Google Scholar]
- Tamura K., Nei M., Kumar S. (2004). Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc. Natl. Acad. Sci. U.S.A. 101 11030–11035. 10.1073/pnas.0404206101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30 2725–2729. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonukari N. (2003). Enzymes and fungal virulence. J. Appl. Sci. Environ. Manage. 7 5–8. 10.4314/jasem.v7i1.17158 [DOI] [Google Scholar]
- Vasebi Y., Safaie N., Alizadeh A. (2013). Biological control of soybean charcoal root rot disease using bacterial and fungal antagonists In Vitro and greenhouse condition. J. Crop Prot. 2 139–150. [Google Scholar]
- Wheeler H. (ed.) (1975). “Mechanisms of pathogenesis,” in Plant Pathogenesis (New York, NY: Springer; ) 5–32. 10.1007/978-3-642-66184-6_2 [DOI] [Google Scholar]
- Wu S., Xiong J., Yu Y. (2015). Taxonomic resolutions based on 18S rRNA genes: a case study of subclass copepoda. PLoS ONE 10:e0131498 10.1371/journal.pone.0131498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu S.-X., Feng Q.-N., Xie H.-T., Li S., Zhang Y. (2017). Reactive oxygen species mediate tapetal programmed cell death in tobacco and tomato. BMC Plant Biol. 17:76 10.1186/s12870-017-1025-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


