Abstract
Tomato (Solanum lycopersicum) fruit weight (FW), soluble solid content (SSC), fruit shape and fruit color are crucial for yield, quality and consumer acceptability. In this study, a 192 accessions tomato association panel comprising a mixture of wild species, cherry tomato, landraces, and modern varieties collected worldwide was genotyped with 547 InDel markers evenly distributed on 12 chromosomes and scored for FW, SSC, fruit shape index (FSI), and color parameters over 2 years with three replications each year. The association panel was sorted into two subpopulations. Linkage disequilibrium ranged from 3.0 to 47.2 Mb across 12 chromosomes. A set of 102 markers significantly (p < 1.19–1.30 × 10−4) associated with SSC, FW, fruit shape, and fruit color was identified on 11 of the 12 chromosomes using a mixed linear model. The associations were compared with the known gene/QTLs for the same traits. Genetic analysis using F2 populations detected 14 and 4 markers significantly (p < 0.05) associated with SSC and FW, respectively. Some loci were commonly detected by both association and linkage analysis. Particularly, one novel locus for FW on chromosome 4 detected by association analysis was also identified in F2 populations. The results demonstrated that association mapping using limited number of InDel markers and a relatively small population could not only complement and enhance previous QTL information, but also identify novel loci for marker-assisted selection of fruit traits in tomato.
Keywords: Solanum lycopersicum, fruit traits, association analysis, linkage disequilibrium, InDel markers
Introduction
Tomato (Solanum lycopersicum) is one of the most consumed vegetables and ranks second in production among the solanaceous crops worldwide (http://www.fao.org/statistics/en/). It is also an excellent plant genetic analysis system, particularly for investigating the mechanisms of fruit development, color formation, and nutrient accumulation. Furthermore, clinical nutrition studies have suggested that increased consumption of tomato products can improve human health and reduce the risk of developing certain cancers (Giovannucci et al., 2002; Etminan et al., 2004; Burton-Freeman and Sesso, 2014; Perveen et al., 2015; Pourahmadi et al., 2015; Raiola et al., 2015). The combination of the economic importance and the potential health benefits make tomato fruits an important target for increasing the nutritional properties.
It has been well-known that wild tomato species bear small, round, red, or green fruits (Alpert et al., 1995). These fruits usually contain high concentration of nutritional properties such as lycopene (Hyman et al., 2004; Kinkade and Foolad, 2013) and soluble solid content (SSC; Rick, 1974; Osborn et al., 1987). Domestication and breeding alternating fruit characters result in a wide diversity of fruit color, weight, and shape (Grandillo et al., 1999; Bai and Lindhout, 2007). Unfortunately, the nutritional properties are usually lower in cultivated varieties than in wild species due to the increase of fruit size (Markovic et al., 1997; Hyman et al., 2004). Many studies have been carried out to identify genes/QTLs for fruit traits using structural populations (e.g., F2 population, backcross population, advanced backcross population) derived from crosses between cultivated varieties and wild species. This approach maximizes the marker polymorphisms and has led to the discovery of new genes. To date, many loci for weight (FW), shape, color, and SSC of tomato fruits have been reported from various species using this classical genetic analysis approach, and some of them have been validated using different mapping populations (Chen et al., 1999; Grandillo et al., 1999; Liu et al., 2003; Ashrafi et al., 2012; Zhou et al., 2016). However, only two loci for fruit weight (fw2.2 and fw3.2), four loci for fruit shape or size (ovate, sun, lc, and fas), three loci for SSC (LIN5, sucr, and Agp-L1), and several loci for color (e.g., r, gf, ogc, t, y, hp1, hp2, hp3, and del) have been well-characterized (Chetelat et al., 1995; Liu et al., 2003; Fridman et al., 2004; Petreikov et al., 2009; The Tomato Genome Consortium, 2012; van der Knaap et al., 2014).
Association analysis represents an alternative to bi-parental linkage mapping for the determination of the genetic basis of traits by allowing evaluation of a large number of alleles in diverse populations, which provides high mapping resolution and reduction in time to develop a mapping population (Rafalski, 2010; Cericola et al., 2014; Pace et al., 2015). Several attempts have been performed to identify loci for fruit traits using association analysis in tomato. These attempts can be classified into three strategies based on the techniques used for genotyping. The first strategy is re-sequencing the whole genomes of certain number of tomato accessions to conduct genome-wide association analysis, resulting in identification of the well-characterized fas gene for fruit size (Shirasawa et al., 2013), fw2.2 locus for fruit weight and y gene for pink fruit (Lin et al., 2014), and 251 association signals for flavor chemicals in tomato fruit (Tieman et al., 2017). The second strategy is to run large-scale genotyping of various collections of tomato accessions using the tomato array platform of Solanaceae Coordinated Agricultural Project (SolCAP) or Centre of BioSystems Genomics (CBSG) for association analysis. This approach has identified phenotype/genotype associations for over 20 fruit traits including fruit color, FW, SSC, and fruit shape (Ruggieri et al., 2014; Sauvage et al., 2014; Sacco et al., 2015; Bauchet et al., 2017). The third strategy is to genotype a collection of tomato accessions with a relatively small amount of markers for association, which has detected hundreds of genotype/phenotype associations for fruit traits (Xu et al., 2013; Zhang et al., 2015, 2016; Zhao et al., 2016). All these efforts suggest that association mapping can be used to identify loci conferring agricultural traits in tomato. However, except for the re-sequencing approach, only few loci have been connected to known loci in other association studies. There is no published report of using marker-trait associations discovered through association mapping in fruit quality improvement programs for the purpose of marker-assisted selection in tomato.
Due to the abundance and wide distribution of single nucleotide polymorphisms (SNPs) in the whole genome and the availability of automatic large-scale genotyping platform, SNPs have been popularly used in association analysis in tomato (Robbins et al., 2011; Shirasawa et al., 2013; Lin et al., 2014; Ruggieri et al., 2014; Sauvage et al., 2014; Sacco et al., 2015; Sim et al., 2015; Bauchet et al., 2017). However, as the second abundant form of genetic variation in the whole genome (Yang et al., 2014), InDel markers have not been widely used in genetic study. Particularly, the use of limited number of InDel markers along with a relatively small population has not been tested in association analysis. The present study used a strategy of combining association mapping and classical genetic analysis to identify loci for four fruit traits including FW, SSC, fruit shape, and color. The cost-effective InDel markers were used to genotype a diverse collection of 192 tomato accessions consisted of S. lycopersicum, S. lycopersicum var. cerasiforme, and Solanum pimpinellifolium. The experience gained here will help refine strategies for genome-wide identification of quantitative loci conferring traits with economic importance in tomato and other species at an affordable level.
Materials and methods
Plant materials and experimental design
The association mapping panel consisted of 192 tomato accessions including 10 of S. pimpinellifolium, 18 of S. lycopersicum var. cerasiforme or cherry tomato, and 164 of S. lycopersicum, which were obtained from various sources (Table S1). The 164 S. lycopersicum accessions included 23 vintage varieties, 20 Latin American cultivars, 54 fresh-market lines, 59 processing lines, and 8 lines with unknown type. The association panel was grown in a randomized complete block design with three blocks containing each accession in two independent experiments conducted in 2013 and 2014. Plots of each accession consisted of at least four plants.
To identify loci for SSC and FW using the structural population mapping approach, two F2 populations were developed by crossing processing tomato varieties OH88119 and OH9242, respectively, to a cherry tomato line Black cherry. Both OH88119 and OH9242 have medium-sized fruit (average FW 48.3 g for OH88119 and 73.7 g for OH9242 in 2013 and 2014) with relatively low SSC (average 4.1% for OH88119 and 4.6% for OH9242 in 2013 and 2014), while Black cherry is a small-fruited tomato line (average FW 18.6 g in 2013 and 2014) with a relatively high SSC (average 6.3% in 2013 and 2014). A sub-population of 503 individuals from the F2 population of OH88119 × Black cherry were grown in the fall season of 2012, and another sub-population consisting of 752 individuals from the same cross as well as 276 individuals from the F2 population of OH 9242 × Black cherry were grown in the spring season of 2013.
Tomato seeds for all studies were sown in 288 cell flats filled with a mixture of peat and vermiculite (3:1) in a protected greenhouse. Seedlings were transplanted ~50 days after germination to field. All experiments were conducted at Shangzhuang Research Station of China Agricultural University in Beijing, China. Production practices, plant spacing, and row spacing were as recommended for commercial growers (Gao et al., 2010).
Phenotypic data collection and analysis
Phenotypic data for association analysis were collected on a plot basis. Five to ten ripe fruits were harvested from each plot and subjected to SSC, FW, fruit height, fruit width, and color measurements.
Total weight of fruits collected in each plot was obtained by weighing all fruits using a pair of balances. Mean was obtained by dividing the total FW by the number of fruits and used as FW for individual fruit in each plot. The maximum height and width of a fruit were measured using a vernier caliper (Hangzhou Tool and Measuring Tool Company, Hangzhou, China). The fruit shape index (FSI) was calculated as the ratio of maximum height to maximum width (Brewer et al., 2006). Numeric descriptions of the red, green, yellow, and blue components of tomato color were obtained using the software Tomato Analyzer 3.0 (Brewer et al., 2006) following the description in Darrigues et al. (2008). The software generated a set of L*, a*, b*, hue, and chroma values representing absolute color for each fruit. SSC was measured using a WAY-2S digital ABBE refractometer (Shanghai Precision Scientific Instrument Company, Shanghai, China). Plot means for FSI, values of color parameters and SSC were calculated based on measurements of all fruits in each plot. Pearson correlation coefficients for each trait between 2 years and among traits were obtained using SAS v9.4 (SAS Institute, Cary NC, USA). Analysis of variance was conducted using PROC GLM in SAS with the model show to best fit the data: Xijb = μ + Gi + Yj + Rb/j + Mij+ εijb, where Xijb is the trait value of the bth replication of the ith genotype in the jth year, Gi is the random effect of the ith genotype, Yj represents the fixed effect of the jth year, Rb/j is the fixed effect of the bth replication in the jth year, Mij the random effect of the genotype by year interaction and εijb is the residual. Broad sense heritability (H2) for each trait was calculated based on the plot level using the equation H2 = σG2/(σG2 +σGY2 +σε2) according to the description in Nyquist and Baker (1991), where σG2 is genotypic variance, σGY2 is the variance due to interaction between genotype and year, andσε2 is the error variance.
Marker analysis
A total of 547 InDel markers (Table S2) evenly distributed across the tomato genome were used to genotype the association panel. These InDel markers were chosen from our previous study and were polymorphic within 10 accessions of S. lycopersicum (Yang et al., 2014). Genomic DNA was isolated from fresh-collected young leaves of each accession using the modified CTAB method (Kabelka et al., 2002). PCR and genotypic data collection were conducted according to the description in Yang et al. (2014).
Nei's genetic distance (Nei, 1972) was calculated for each pair of accessions and marker allele frequency was obtained using the software PowerMarker V3.25 (Liu and Muse, 2005). Polymorphism information content (PIC) was calculated using the formula (Weir, 1990) of PIC = 1-Σpi2, where pi is the frequency of ith allele for each marker locus. Markers with a minor allele frequency below 5% were removed from the marker data set to calculate population structure, kinship, and to perform association analysis.
Linkage disequilibrium analysis
Marker genotypes were used to measure the extent of linkage disequilibrium (LD) within the 164 accessions of S. lycopersicum and 18 accessions of S. lycopersicum var. cerasiforme. The TASSEL 2.1 (Bradbury et al., 2007) software was used to calculate pair-wise r2-values for markers polymorphic within the 164 accessions. P-values for each r2 estimate were calculated using 1,000 permutations in TASSEL.
LD decay was calculated by plotting pair-wise r2-values onto genetic distance in base pairs on the same WGS chromosome (SL2.40) of tomato variety Heinz1706 (The Tomato Genome Consortium, 2012). All markers with <25% missing data and a minor allele frequency >5% were used to calculate LD decay. Critical values of r2 as an evidence of linkage were derived from the parametric 95th percentile of the distribution of the unlinked markers (Breseghello and Sorrells, 2006).
Population structure and association analysis
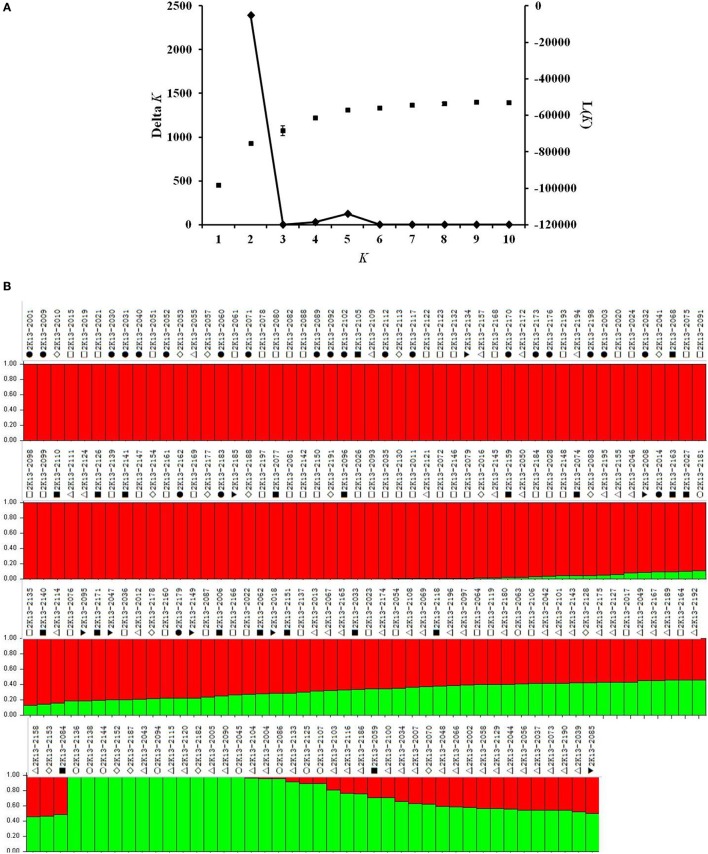
Population structure (Q matrix) was estimated using Structure 2.3.4 software (Pritchard et al., 2000; Falush et al., 2003, 2007). Number of populations (K) was determined following the instruction in Pritchard et al. (2000) with a burn-in period of 100,000 iterations and Markov Chain Monte Carlo of 100,000. Twenty independent runs were done for K varying from 1 to 10. The most probable K-value was defined according to the method proposed by Evanno et al. (2005).
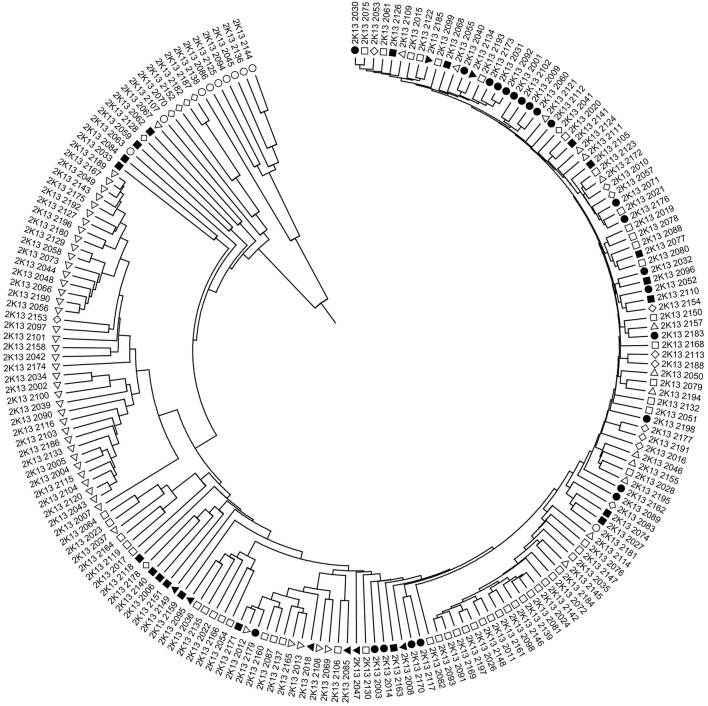
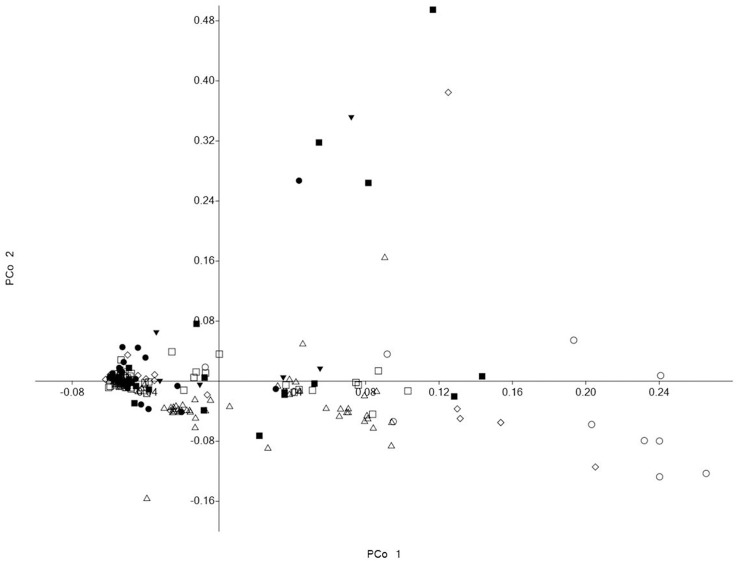
Unweighted Pair Group Method with Arithmetic Mean (UPGMA) cluster analysis was performed to develop a phylogenetic tree using the software PowerMarker V3.25 and the tree was viewed in MEGA5 (Tamura et al., 2011). Principal coordinate analysis (PCoA) was conducted using the Past 3.13 software (Hammer et al., 2001). The Loiselle kinship coefficients between tomato lines (K matrix) were calculated using the software SpAGeDi (Hardy and Vekemans, 2002).
The software program TASSEL 2.1 was used to conduct association analysis. A mixed linear model (MLM) taking into account both population structure (Q matrix) and the kinship matrix (K matrix), and a general linear model (GLM) using population structure (Q matrix) as a fixed factor were used for association identification of loci conferring fruit traits. Significance of marker-trait association was determined based on p-value at a level of 5% after Bonferroni (1936) multiple test correction. Since it has been popularly proved that MLM+Q+K model is a more effective approach than other models for detecting loci (Yu et al., 2006; Malosetti et al., 2007; Cericola et al., 2014; Pace et al., 2015; Sim et al., 2015), only the data of the MLM+Q+K model was presented in the current study. The phenotypic variation explained by each marker was the R2-value obtained from GLM model.
Linkage analysis of loci for FW and SSC using F2 populations
Since most markers significantly associated with loci for FW or SSC were not polymorphic between Black cherry and OH88119 or OH9242, we selected InDel markers that showing polymorphisms between the parents in Yang et al. (2014). Our goal was to identify two markers per chromosome arm in order to survey the whole genome. Thus, a total of 56 additional markers (Table S3) distributed on 12 chromosomes were used for initial identification of loci conferring FW and SSC in the F2 population of OH 88119 × Black cherry. ANOVA using SAS v9.4 with a general linear model Xi = μ + Mi+ εi (Yang et al., 2005) was performed to determine an association between trait and marker genotype of 126 individuals randomly picked from the F2 population of OH88119 × Black cherry. Once a marker was identified to be significantly (P < 0.05) associated with a trait, the marker was used to genotype the whole F2 population of OH88119 × Black cherry and the F2 population of OH9242 × Black cherry for validation.
Results
Marker polymorphisms
The 547 markers generated 1295 alleles in the 192 tomato accession with a range of two to nine alleles (average 2.4 alleles) for individual markers (Table S2). Among the polymorphic markers, ~93% had two or three alleles with the dominance of bi-allele markers (Figure 1). As expected, all markers were polymorphic in the 192 accessions. However, polymorphisms within species was decreased from wild species S. pimpinellifolium to S. lycopersicum var. cerasiforme and then to S. lycopersicum, the cultivated tomato. The proportion of polymorphic markers were 97.4% in 10 accessions of S. pimpinellifolium, 85.2% in 18 accessions of S. lycopersicum var. cerasiforme, 64.7% in 23 vintage accessions, 78.4% in 20 Latin American Cultivars, 67.3% in 54 fresh-market lines, and 73.3% in 59 processing lines (Table S2). Subsequently, the average polymorphic information content (PIC) also decreased from wild species S. pimpinellifolium to S. lycopersicum var. cerasiforme and then to S. lycopersicum (Table S2).
Figure 1.
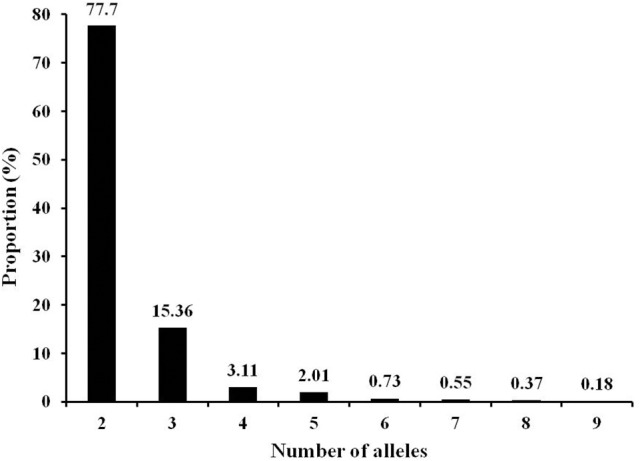
Frequency distribution of alleles in 192 tomato accessions generated by 547 InDel markers.
Phenotypic variation
Significant difference for each trait was observed among genotypes, and FSI was the only trait that showed no significant difference between 2 years or three replications (Table S4). However, the phenotypic data for each trait in 2 years were highly correlated with the correlation coefficient (r) of 0.85 for SSC, 0.91 for FW, 0.96 for FSI, 0.86 for L*, 0.92 for Hue, and 0.83 for Chroma. Thus, the mean of the 2-year data for each trait was used for the following analysis.
A wide range of variation was observed for SSC, FW, FSI, L*, Hue, and Chroma in the 192 tomato accessions (Table 1, Figure S1). The average of SSC was higher in S. pimpinellifolium than in S. lycopersicum, while the mean of FW was smaller in S. pimpinellifolium than in S. lycopersicum, and the means of SSC and FW in S. lycopersicum var. cerasiforme were between those in S. pimpinellifolium and S. lycopersicum. Significant negative correlation was observed between SSC and FW (r = −0.43, p < 0.0001). No differences of FSI were observed among three species. The means for fruit color parameters L*, Hue, and Chroma were lower in S. pimpinellifolium than in S. lycopersicum var. cerasiforme and S. lycopersicum. Broad-sense heritability for each trait was obviously different. Chorma was the least heritable trait with heritability of 0.69, while FSI was the most heritable trait with heritability of 0.91 (Table 1).
Table 1.
Range, mean, standard deviations (SD), coefficients of variation (CV) and broad-sense heritabilities (H2) collected for all traits in 192 tomato accessions.
| Trait | Code | Species | Minimum | Maximum | Mean | SD | CV | H2 |
|---|---|---|---|---|---|---|---|---|
| Soluble solid content (%) | SSC | Combined | 2.84 | 7.67 | 4.43 | 0.83 | 0.19 | 0.71 |
| Solanum lycopersicum | 2.84 | 5.55 | 4.22 | 0.48 | 0.11 | |||
| Solanum lycopersicum var. cerasiforme | 3.55 | 6.73 | 5.3 | 1.01 | 0.19 | |||
| Solanum pimpinellifolium | 4.93 | 7.67 | 6.90 | 0.90 | 0.13 | |||
| Fruit weight (g) | FW | Combined | 0.78 | 311.16 | 91.98 | 56.57 | 0.62 | 0.80 |
| Solanum lycopersicum | 15.57 | 311.16 | 105.38 | 49.85 | 0.47 | |||
| Solanum lycopersicum var. cerasiforme | 2.04 | 41.10 | 17.92 | 11.67 | 0.65 | |||
| Solanum pimpinellifolium | 0.78 | 2.60 | 1.31 | 0.57 | 0.43 | |||
| Fruit shape index | FSI | Combined | 0.64 | 2.19 | 1.02 | 0.27 | 0.26 | 0.91 |
| Solanum lycopersicum | 0.64 | 2.19 | 1.02 | 0.27 | 0.27 | |||
| Solanum lycopersicum var. cerasiforme | 0.86 | 1.71 | 1.14 | 0.26 | 0.23 | |||
| Solanum pimpinellifolium | 0.95 | 1.10 | 1.00 | 0.05 | 0.05 | |||
| Color—darkness or lightness | L* | Combined | 26.12 | 68.23 | 48.43 | 6.82 | 0.14 | 0.75 |
| Solanum lycopersicum | 35.66 | 68.23 | 49.11 | 5.60 | 0.11 | |||
| Solanum lycopersicum var. cerasiforme | 26.12 | 67.65 | 47.66 | 11.49 | 0.24 | |||
| Solanum pimpinellifolium | 33.89 | 58.97 | 38.86 | 8.26 | 0.21 | |||
| Color—basic tint | Hue | Combined | 37.69 | 99.87 | 55.66 | 14.13 | 0.26 | 0.84 |
| Solanum lycopersicum | 40.12 | 99.87 | 53.51 | 11.69 | 0.22 | |||
| Solanum lycopersicum var. cerasiforme | 43.58 | 95.42 | 68.81 | 22.95 | 0.33 | |||
| Solanum pimpinellifolium | 37.69 | 94.62 | 49.51 | 18.86 | 0.38 | |||
| Color—saturation or vividness | Chroma | Combined | 25.99 | 55.68 | 45.46 | 4.54 | 0.10 | 0.69 |
| Solanum lycopersicum | 33.55 | 55.68 | 46.03 | 4.31 | 0.09 | |||
| Solanum lycopersicum var. cerasiforme | 25.99 | 50.09 | 42.25 | 5.49 | 0.13 | |||
| Solanum pimpinellifolium | 37.95 | 44.09 | 41.61 | 2.08 | 0.05 |
Population structure
Although, the 192 tomato accessions were from three species, model without prior population information was used to assign individual accession to a subpopulation using the software package of STRUCTURE2.3.4. In order to define the number of subpopulations within the 192 accessions, a series of independent runs of the data were run at a range of K-values from 1 to 10. The summary plot of membership coefficients (Q) and ΔK analysis (Figure 2) from STRUCTURE software, and the genetic relationships revealed by the phylogenetic analysis (Figure 3) and the PCoA (Figure 4), suggested that the mapping population was sorted into two subpopulations (K = 2). The larger subpopulation was composed of 134 accessions including all fresh-market, vintage, and unknown type accessions. One accession of S. pimpinellifolium LA2183, two-thirds of S. lycopersicum var. cerasiforme or cherry tomato, one-third of processing accessions, and four-fifths of Latin American Cultivars were also assigned into the larger subpopulation. Although, there was no certain trend of clustering in the larger subpopulation, the fresh-market accessions from Florida, USA formed one independent cluster. The smaller subpopulation consisted of 58 accessions including nine accessions of S. pimpinellifolium, two-thirds of processing, one-third of S. lycopersicum var. cerasiforme or cherry tomato, and four Latin American Cultivars. Within the smaller subpopulation, 38 processing accessions were clustered together, while the seven accessions of S. pimpinellifolium formed one cluster.
Figure 2.
(A) L(K) and ΔK plots derived from the InDel data. ΔK is calculated as the mean of the absolute values of L”(K) averaged over 20 runs divided by the standard deviation of L(K). ΔK = m(|L”(K)|)/s[L(K)], which expands to ΔK = m(|L(K+1)-2L(K)+L(K-1)|)/s[L(K)]. L(K) is the Pr(X|K) referred as “Ln P(D)” in the output of STRUCTURE software. (B) Population structure estimates based on 547 InDel markers distributed across the tomato genome. The area of two different colors (Red and Green) illustrates the proportion of each subpopulation based on these InDel markers. Symbols represent accessions from different species or market class of tomato: ○ Solanum pimpinellifolium, ◊ S. lycopersicum var. cerasiforme or cherry tomato, ■ Latin American Cultivar, □ Fresh-market, Δ Processing, • Vintage, ▼ Unknown type. The accession of each plot number can be found in Table S1.
Figure 3.
UPGMA dendrogram of 192 tomato accessions based on 547 InDel marker data. Symbols represent accessions from different species or market class of tomato: ○ Solanum pimpinellifolium, ◊ S. lycopersicum var. cerasiforme or cherry tomato, ■ Latin American Cultivar, □ Fresh-market, Δ Processing, • Vintage, ▼ Unknown type. The accession of each plot number can be found in Table S1.
Figure 4.
PCoA visualization of the genetic relationships between members of the association panel. Symbols represent accessions from different species or market class of tomato: ◊ Solanum pimpinellifolium, ○ S. lycopersicum var. cerasiforme or cherry tomato, ■ Latin American Cultivar, □ Fresh-market, Δ Processing, • Vintage, ▼ Unknown type.
Linkage disequilibrium evaluation
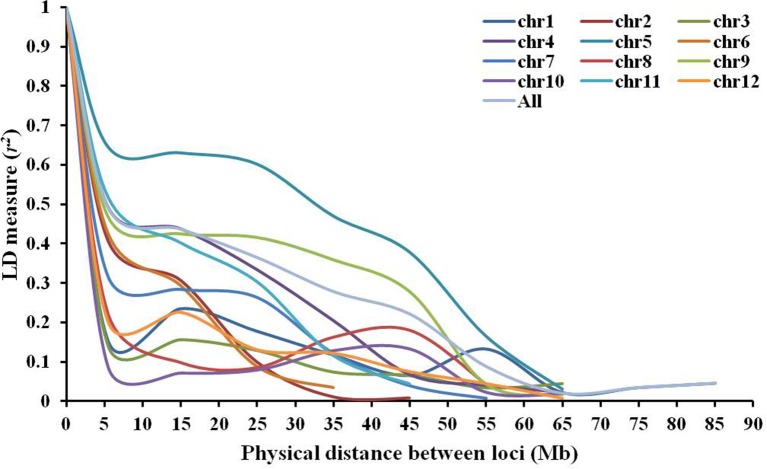
A mean r2 of 0.393 was observed between all pairs of linked loci throughout the genome, with a mean maximum r2-value of 0.571 on chromosome 5 and a mean minimum r2-value of 0.086 on chromosome 10 (Table 2). The rate of LD decay was different across chromosomes (Figure 5). The baseline r2-values varied from 0.283 (chromosome 8) to 0.381 (chromosome 10) estimated by the 95th percentile method. Based on LOESS curves, the baseline r2-values corresponded to physical distances varying from 3.0 to 47.2 Mb. LD decayed within 6.5 Mb on chromosomes 1, 2, 3, 7, 8, 10, and 12, while chromosome 5 showed the lowest decay with a baseline r2-value of 0.335 reached ~47.2 Mb.
Table 2.
Summary of linkage disequilibrium (LD) analysis.
| Chromosome | No. marker pairs | r2 estimates | LD decay (Mb) | |||
|---|---|---|---|---|---|---|
| Median | Mean | St. Dev. | 95% percentile | |||
| 1 | 12,866 | 0.008 | 0.156 | 0.081 | 0.359 | 3.2 |
| 2 | 21,501 | 0.010 | 0.277 | 0.098 | 0.368 | 6.5 |
| 3 | 13,450 | 0.008 | 0.112 | 0.062 | 0.359 | 3.2 |
| 4 | 37,357 | 0.020 | 0.357 | 0.141 | 0.337 | 24.5 |
| 5 | 43,986 | 0.011 | 0.571 | 0.205 | 0.335 | 47.2 |
| 6 | 29,774 | 0.012 | 0.308 | 0.105 | 0.331 | 12.5 |
| 7 | 10,039 | 0.009 | 0.224 | 0.067 | 0.353 | 4.8 |
| 8 | 8,838 | 0.005 | 0.144 | 0.055 | 0.283 | 4.5 |
| 9 | 27,333 | 0.012 | 0.384 | 0.118 | 0.313 | 42.5 |
| 10 | 9,973 | 0.010 | 0.086 | 0.060 | 0.381 | 3.0 |
| 11 | 20,063 | 0.010 | 0.370 | 0.116 | 0.314 | 24.0 |
| 12 | 13,451 | 0.014 | 0.167 | 0.061 | 0.319 | 3.9 |
| All | 131,381 | 0.012 | 0.393 | 0.169 | 0.333 | 28.5 |
Figure 5.
Linkage disequilibrium decay across all 12 chromosomes within the association mapping panel of 164 Solanum lycopersicum accessions revealed by plots of linkage disequilibrium (LD) values (r2) against physical genetic distance (Mb) between pairs of InDel markers.
Association identification of loci conferring tomato fruit traits
Using the MLM K + Q model and Bonferroni correction for p-values, a total of 102 genotype/phenotype associations with phenotypic variation explained >2.0% were detected under the threshold p-values of 1.30 × 10−4 for SSC, 1.19 × 10−4 for FW and FSI, 1.22 × 10−4 for L* and Chroma, and 1.24 × 10−4 for Hue. The numbers of associations per trait ranged from three for Hue to 27 for Chroma. Regions carrying the presumed genes/QTLs were identified on 11 of the 12 chromosomes, but none were detected on chromosome 10 (Figure S2, Table 3). In order to match the associations with previously identified QTL, loci linked to one another within 3 Mb, the lowest LD, on the same chromosome were considered as a unit.
Table 3.
Markers significantly associated with soluble solid content, fruit weight, fruit shape index, and fruit color parameters (L*, Hue, and Chroma) identified by association analysis.
| Trait | Marker name | Chromosome | Physical position (SL2.40) | p-value | Phenotypic variation explained (%) |
|---|---|---|---|---|---|
| Soluble solid content | sli2868 | 1 | 70500607 | 1.27E-11 | 14.5 |
| sli2875 | 1 | 72511190 | 8.11E-07 | 20.6 | |
| sli2916 | 1 | 81603104 | 6.53E-06 | 25.5 | |
| sli311 | 2 | 13572187 | 7.59E-05 | 13.9 | |
| sli2686 | 2 | 25824814 | 4.55E-07 | 8.9 | |
| sli2221 | 4 | 9883293 | 8.35E-08 | 5.6 | |
| sli3316 | 4 | 54162751 | 3.29E-11 | 13.8 | |
| sli493 | 5 | 3635186 | 2.36E-05 | 2.6 | |
| sli408 | 5 | 14801010 | 4.11E-06 | 4.5 | |
| sli500 | 5 | 37859546 | 9.52E-05 | 3.7 | |
| sli508 | 5 | 39790245 | 3.00E-06 | 3.6 | |
| sli562 | 5 | 54572647 | 3.65E-06 | 5.4 | |
| sli571 | 5 | 57656536 | 4.35E-07 | 10.7 | |
| sli589 | 5 | 61241179 | 7.27E-05 | 12.2 | |
| sli634 | 6 | 7801283 | 1.60E-07 | 6.2 | |
| sli660 | 6 | 13668472 | 1.47E-06 | 9.8 | |
| sli665 | 6 | 14558967 | 4.14E-06 | 6.3 | |
| sli761 | 6 | 37324771 | 1.37E-14 | 22.3 | |
| sli919 | 7 | 38991134 | 1.71E-07 | 15.3 | |
| sli1147 | 8 | 31663632 | 8.68E-06 | 24.9 | |
| sli4003 | 9 | 13623231 | 4.85E-05 | 16.5 | |
| sli1324 | 9 | 14404856 | 2.21E-05 | 4.6 | |
| sli1490 | 9 | 15826796 | 1.95E-08 | 10.4 | |
| sli1368 | 9 | 31114697 | 1.12E-06 | 18.4 | |
| Fruit weight | sli201 | 1 | 29372090 | 1.63E-05 | 5.2 |
| sli2887 | 1 | 75604440 | 2.95E-08 | 20.5 | |
| sli2438 | 3 | 2044312 | 1.45E-05 | 17.9 | |
| sli2221 | 4 | 9883293 | 3.26E-12 | 4.5 | |
| sli3003 | 4 | 16716471 | 2.56E-05 | 2.1 | |
| sli2307 | 4 | 33951126 | 3.36E-05 | 2.8 | |
| sli2329 | 4 | 41110769 | 3.36E-05 | 2.8 | |
| sli2388 | 4 | 55504498 | 4.73E-05 | 6.5 | |
| sli580 | 5 | 59621517 | 8.94E-05 | 2.9 | |
| sli1146 | 8 | 31471959 | 7.76E-14 | 22.9 | |
| sli1147 | 8 | 31663632 | 3.15E-07 | 12.6 | |
| sli1497 | 9 | 20876005 | 2.61E-15 | 3.4 | |
| sli1498 | 9 | 22109103 | 7.22E-06 | 10.5 | |
| sli1516 | 9 | 39903394 | 1.25E-08 | 5.8 | |
| sli1464 | 9 | 65041065 | 1.28E-09 | 18.9 | |
| sli1873 | 11 | 33962309 | 5.13E-06 | 9.2 | |
| sli1886 | 11 | 37929907 | 6.86E-21 | 7.9 | |
| Fruit shape index | sli2653 | 3 | 60117959 | 2.17E-05 | 3.5 |
| sli2655 | 3 | 60656932 | 2.31E-06 | 2.0 | |
| sli2216 | 4 | 7766862 | 2.61E-05 | 7.4 | |
| sli2347 | 4 | 46429902 | 4.04E-08 | 2.0 | |
| sli2378 | 4 | 53631402 | 2.68E-07 | 2.5 | |
| sli2384 | 4 | 54772171 | 1.23E-08 | 5.2 | |
| sli2419 | 4 | 62034593 | 1.03E-04 | 2.9 | |
| sli45 | 5 | 8155946 | 1.07E-05 | 5.4 | |
| sli451 | 5 | 23416656 | 1.92E-08 | 5.1 | |
| sli470 | 5 | 29201547 | 8.39E-05 | 5.6 | |
| sli565 | 5 | 55625013 | 2.63E-05 | 5.0 | |
| sli571 | 5 | 57656536 | 9.59E-05 | 8.1 | |
| sli665 | 6 | 14558967 | 4.63E-05 | 3.3 | |
| sli855 | 7 | 21689257 | 4.96E-05 | 4.2 | |
| sli1414 | 9 | 53917837 | 2.81E-13 | 2.4 | |
| L* | sli2799 | 1 | 43606482 | 2.71E-09 | 5.9 |
| sli2940 | 1 | 87328257 | 4.05E-11 | 23.4 | |
| sli2556 | 3 | 33299542 | 8.81E-05 | 5.1 | |
| sli2215 | 4 | 7234645 | 4.12E-05 | 5.1 | |
| sli2272 | 4 | 25200675 | 9.71E-06 | 2.0 | |
| sli2299 | 4 | 32139951 | 1.03E-06 | 2.6 | |
| sli2302 | 4 | 32770761 | 1.03E-06 | 2.6 | |
| sli2323 | 4 | 39113326 | 8.79E-07 | 3.6 | |
| sli2333 | 4 | 42413320 | 1.03E-06 | 2.6 | |
| sli477 | 5 | 30794762 | 2.76E-14 | 3.0 | |
| sli495 | 5 | 36780476 | 2.40E-09 | 2.9 | |
| sli508 | 5 | 39790245 | 2.18E-06 | 3.1 | |
| sli571 | 5 | 57656536 | 4.10E-07 | 6.1 | |
| sli685 | 6 | 20250580 | 4.80E-14 | 14.7 | |
| sli1346 | 9 | 23125992 | 1.42E-08 | 3.7 | |
| sli1970 | 12 | 10754442 | 2.34E-06 | 9.2 | |
| Hue | sli2792 | 2 | 48225893 | 7.92E-14 | 3.2 |
| sli2309 | 4 | 34621455 | 7.81E-05 | 2.2 | |
| sli685 | 6 | 20250580 | 7.90E-13 | 4.2 | |
| Chroma | sli2799 | 1 | 43606482 | 8.80E-07 | 9.2 |
| sli2940 | 1 | 87328257 | 5.00E-09 | 10.3 | |
| sli296 | 2 | 9872987 | 2.89E-10 | 11.0 | |
| sli325 | 2 | 16913369 | 2.89E-10 | 11.0 | |
| sli2792 | 2 | 48225893 | 1.40E-14 | 5.9 | |
| sli3313 | 4 | 53694137 | 1.19E-05 | 14.3 | |
| sli3316 | 4 | 54162751 | 1.52E-11 | 14.6 | |
| sli45 | 5 | 8155946 | 1.17E-25 | 4.8 | |
| sli418 | 5 | 16965721 | 4.47E-08 | 3.5 | |
| sli477 | 5 | 30794762 | 8.51E-05 | 8.0 | |
| sli508 | 5 | 39790245 | 1.77E-05 | 8.2 | |
| sli522 | 5 | 43463720 | 1.33E-08 | 7.9 | |
| sli556 | 5 | 52455728 | 7.35E-06 | 8.5 | |
| sli568 | 5 | 56274851 | 1.33E-05 | 5.9 | |
| sli571 | 5 | 57656536 | 6.72E-19 | 17.0 | |
| sli588 | 5 | 61020978 | 5.16E-14 | 26.3 | |
| sli652 | 6 | 12252957 | 1.08E-06 | 14.3 | |
| sli685 | 6 | 20250580 | 1.80E-08 | 16.6 | |
| sli28 | 6 | 39518119 | 1.08E-06 | 7.5 | |
| sli1322 | 9 | 14058288 | 2.82E-06 | 9.8 | |
| sli1324 | 9 | 14404856 | 1.64E-06 | 6.7 | |
| sli1346 | 9 | 23125992 | 9.95E-21 | 8.7 | |
| sli1520 | 9 | 41701481 | 1.51E-09 | 11.6 | |
| sli1423 | 9 | 56541008 | 9.77E-13 | 6.7 | |
| sli1864 | 11 | 30139161 | 3.77E-05 | 7.6 | |
| sli1970 | 12 | 10754442 | 2.02E-05 | 11.5 | |
| sli2056 | 12 | 38855927 | 7.97E-09 | 9.7 |
Of the 24 genotype/phenotype associations identified for SSC, the phenotypic variation explained by each marker varied from 2.6 to 25.5% (Table 3). Based on the physical distance between markers, at least 19 loci were detected on eight chromosomes and 13 of them had the phenotypic contribution >10%. The 17 markers associated with FW could be assigned to 15 chromosomal regions on seven chromosomes and explained 2.1–22.9% of the phenotypic variation. Five loci on four chromosomes had the phenotypic contribution >10%. Of the 15 associations between FSI and markers detected at 12 chromosomal regions on six chromosomes, the phenotypic variation explained by each marker ranged from 2.0 to 8.1%.
A total of 46 genotype/phenotype associations were detected for three parameters of fruit color, of which 16, 3, and 27 were for L*, Hue, and Chroma, respectively (Table 3). These associations were on nine of the 12 chromosomes except chromosomes 7, 8, and 10. The phenotypic variation explained by these markers ranged from 2.0 to 23.4% for L*, 2.2–4.2% for Hue, and 3.5–26.3% for Chroma. One marker Sli685 on chromosome 6 was common for all three parameters, seven markers (Sli2799, Sli2940, Sli477, Sli508, Sli571, Sli1346, and Sli1970) were common for L* and Chroma, and one marker Sli2792 on chromosome 2 was common for Hue and Chroma.
Co-localization of QTLs was also observed (Table 3). Seven phenotype/genotype associations for SSC also contributed to FW, fruit shape or fruit color. One QTL on chromosome 5 for fruit shape conferred to Chroma as well. For the three parameters for fruit color, it was not surprised that the QTLs for Hue and Chroma were co-localized because the value of Hue was derived from Chroma (Yang et al., 2004). However, eight QTLs for L* also had impact to Chroma, which was consisted with that the increase of chromaticity makes a color becomes more intense (Yang et al., 2014).
Identification of loci for SSC and FW in F2 populations
To identify loci for FW and SSC in the F2 populations, the F2 population of OH88119 × Black cherry grown in the spring season 2013 was subjected to initial test using 56 InDel markers. Four were significantly (p < 0.05) associated with FW (Table 4). The alleles from Black cherry contributed to small fruit. Two markers Sli2788 and Sli2772 on chromosome 2 contributed 24.2 and 17.9% of total phenotypic variation for FW, respectively. One marker Sli2377 on chromosome 4 explained 6.1% phenotypic variation of FW was close to the marker Sli2388 explaining 6.5% phenotypic variation in the 192 association panel. These four marker-trait associations were validated in the F2 population of OH88119 × Black cherry grown in fall 2012. However, only the two markers on chromosome 2 could be validated in the F2 population of OH9242 × Black cherry (Table 4).
Table 4.
Markers significantly (P < 0.05) associated with fruit weight (FW) and soluble solid content (SSC) identified in F2 populations.
| Trait | F2 population | Season | Marker name | Chromosome | Physical position (SL2.40) | p | R2 | Effect of Black cherry allele |
|---|---|---|---|---|---|---|---|---|
| Fruit weight | OH88119 × Black cherry | Spring 2013 | Sli2788 | 2 | 47404370 | < 0.0001 | 0.242 | − |
| Sli2772 | 2 | 44259569 | < 0.0001 | 0.179 | − | |||
| Sli2377 | 4 | 53360768 | < 0.0001 | 0.061 | − | |||
| Sli1926 | 11 | 53303453 | 0.0002 | 0.022 | − | |||
| OH88119 × Black cherry | Fall 2012 | Sli2788 | 2 | 47404370 | < 0.0001 | 0.142 | − | |
| Sli2772 | 2 | 44259569 | < 0.0001 | 0.156 | − | |||
| Sli2377 | 4 | 53360768 | 0.0034 | 0.023 | − | |||
| Sli1926 | 11 | 53303453 | 0.0003 | 0.033 | − | |||
| OH9242 × Black cherry | Spring 2013 | Sli2788 | 2 | 47404370 | < 0.0001 | 0.079 | − | |
| Sli2772 | 2 | 44259569 | < 0.0001 | 0.077 | − | |||
| Sli2377 | 4 | 53360768 | 0.055 | 0.021 | − | |||
| Sli1926 | 11 | 53303453 | 0.4005 | 0.007 | − | |||
| Soluble solid content | OH88119 × Black cherry | Spring 2013 | Sli2184 | 4 | 808145 | 0.0366 | 0.055 | + |
| Sli2416 | 4 | 61406781 | 0.0315 | 0.058 | + | |||
| Sli742 | 6 | 33534324 | 0.0002 | 0.139 | + | |||
| Sli743 | 6 | 33733158 | < 0.0001 | 0.149 | + | |||
| Sli744 | 6 | 34024097 | 0.0001 | 0.139 | + | |||
| Sli745 | 6 | 34110927 | 0.001 | 0.118 | + | |||
| Sli762 | 6 | 37511917 | 0.0003 | 0.131 | − | |||
| Sli775 | 6 | 40230850 | 0.0011 | 0.112 | + | |||
| Sli1003 | 7 | 58815972 | 0.0039 | 0.092 | + | |||
| Sli1290 | 9 | 3428268 | 0.0047 | 0.087 | + | |||
| Sli1926 | 11 | 53303453 | 0.0106 | 0.079 | − | |||
| Sli1958 | 12 | 6425452 | 0.0233 | 0.062 | + | |||
| Sli2009 | 12 | 20925959 | 0.0401 | 0.053 | + | |||
| Sli2112 | 12 | 55874351 | 0.0389 | 0.054 | + |
A total of 14 markers were identified to be significantly (p < 0.05) associated with SSC in the F2 population of OH88119 × Black cherry grown in the spring season 2013 (Table 4). These markers were from 10 regions on six chromosomes and contributed 5.3–14.9% of total phenotypic variation. Alleles of most markers from Black cherry contributed to high SSC. However, alleles of the marker Sli762 on chromosome 6 and the marker Sli1926 on chromosome 11 from Black cherry contributed to low SSC. Six markers spanning ~6.7 Mb region on chromosome 6 explained the highest phenotypic variation (11.2–14.9%) was in the same region of the marker Sli761 detecting high association with SSC in the 192 association panel.
Discussion
The LD in cultivated tomatoes has been investigated using various molecular markers and different collections of germplasm. Analyzing a set of 94 cultivars for commercial greenhouse production in Europe with 887 AFLP markers indicates that the LD decay is 15–20 cM (van Berloo et al., 2008), while analyzing 24 fresh market varieties and 39 processing varieties using 434 PCR-based markers shows the LD decay is 6–14 cM within processing cultivars and 3–16 cM within fresh-market cultivars (Robbins et al., 2011). Furthermore, the rate of LD decay depends on chromosomes and tomato types. Processing varieties have greater LD on chromosomes 1, 2, and 5, while fresh-market cultivars have higher LD on chromosomes 6 and 9 (Robbins et al., 2011). Re-sequencing genomes of 360 accessions reveals that the LD decay with physical distance between SNPs occurred at 8.8 kb in S. pimpinellifolium, 256.8 kb in S. lycopersicum var. cerasiforme, and 865.7 kb in S. lycopersicum accessions (Lin et al., 2014). A recent study analyzing 300 tomato accessions with 11000 SNPs suggests that the LD decay ranges from 0.2 cM (73 kb) to 49 cM (47 Mb) at chromosomal level (Bauchet et al., 2017). In this study, the LD decay range from 3.0 to 47.2 Mb on 12 chromosomes with an overall of 28.5 Mb in the whole genome, which is larger than previous reports. Molecular marker types, calculation methods and types of tomato accessions could contribute to this difference. SNPs marker data (Lin et al., 2014) provides a smaller LD decay than PCR-based marker data (van Berloo et al., 2008; Robbins et al., 2011), which suggests that high density markers might provide more accurate calculation of LD decay. In addition, previous studies calculated LD decay separately for each market type or species of tomato, while we combined tomato accessions from S. lycopersicum var. cerasiforme and S. lycopersicum to calculate LD decay for each chromosome. The rates of LD decay on seven chromosomes were <6.5 Mb, ~8.7 cM based on the estimation of 1 cM equals ~750 kb (Tanksley et al., 1992), which is consistent with previous data (van Berloo et al., 2008; Robbins et al., 2011). The remaining five chromosomes 4, 5, 6, 9, and 11 with relatively large LD could be biased by chromosome fragments that have been introgressed from wild species in the past decades (van Berloo et al., 2008; Bauchet et al., 2017), e.g., fragments containing resistance genes on chromosomes 5, 9, and 11. However, all studies suggest that the LD is strong in tomato than in other species and association mapping is theoretically feasible with a small number of markers (van Berloo et al., 2008; Robbins et al., 2011; Lin et al., 2014).
Hundreds of genes/QTLs for fruit traits have been detected in tomato using both classical genetic analysis and association mapping. However, due to the lack of direct comparison between genes/QTLs identified by association mapping and classical genetic analysis, only known genes (fas, y) or /QTL (fw2.2) can be validated through association analysis approach (Shirasawa et al., 2013; Lin et al., 2014). In the present study, a total of 102 phenotype/genotype associations for FW, SSC, fruit shape, and fruit color were detected in the association mapping panel consisted of 192 tomato accessions. Two markers were located within the known gene regions. The marker Sli28 associated with Chroma was at the ogc region on chromosome 6 and the marker Sli2799 associated with L* and Chroma was at the hp2 region of chromosome 1. The ogc gene can increase the accumulation of lycopene while the hp2 gene is responsible for more deep pigment in tomato fruits (Mustilli et al., 1999; Ronen et al., 2000). The marker Sli3313 associated with Chroma was at the previously identified QTL region on chromosome 4 (Yang et al., 2004). In their study, the marker LEOH37 explains 21.6% of phenotypic variation of Chroma in the F2 population of OH8245 × OH2349 consisted of 160 individuals, while the marker Sli3313 identified in the current study contributed to 14.3% of phenotypic variation in the association mapping panel. No other cloned genes for FW, fruit shape or size, and color were detected. This could be due to several reasons. First, the lack of detection of ovate, sun, gf, t, and del genes was due to only few accessions for each gene were included in the association mapping panel. Second, since the phenotypes of yellow and pink colors were out of our objectives, we did not record these traits. Thus, it was reasonable for not detecting phenotype/genotype associations for yellow and pink colors conditioned by the r and y genes, respectively. Third, we used a relatively high stringency for association analysis in this study. The p-values of 1.19–1.30 × 10−4 used here were much lower than 0.005 used in other studies (Xu et al., 2013; Zhang et al., 2016; Bauchet et al., 2017), which could decrease the power of association analysis. Fourth, it has been reported that the genome of cherry tomato accessions is a mosaic composed of polymorphisms of S. pimpinellifolium and S. lycopersicum (Blanca et al., 2015), which might also cause the failure of association mapping.
Among the 102 phenotype/genotype associations for FW, SSC, fruit shape, and fruit color, 22 with the p < 1E-10 were considered as the strongest associations. By searching the tomato genome annotation database (ITAG-cDNA 3.1, http://solgenomics.net), the numbers of predicted genes within 2 Mb regions corresponding to the 22 markers varied from 19 to 286, of which 615 for SSC, 102 for FW, 41 for FSI, 430 for L*, 1,000 for Chroma, and 342 for Hue (Table S5). A lot of predicted genes can be considered as candidate genes for each trait based on their predicted roles in biological process, cellular component, and molecular function. For examples, Solyc04g018020, Solyc04g018030, and Solyc09g031560 are putative serine/threonine-protein phosphatase genes. Solyc04g018063 is a putative cytochrome P450 gene. Solyc04g018147 is a putative DnaJ heat shock amino-terminal domain protein gene. Solyc09g030420 is a putative auxin response factor gene. Solyc11g04555 is a putative Myb domain protein gene. All these kinds of genes are considered as candidates for FW (Huang and van der Knaap, 2011). The use of structural populations for genetic analysis results in identification many loci conferring fruit traits in tomato. However, only 28 loci for FW and 11 loci for fruit shape can be detected in at least two independent studies (Grandillo et al., 1999). To identify and validate real QTLs conferring FW, we made crosses between the small fruited tomato accession Black cherry and two medium-sized tomato accessions OH88119 and OH9242 to develop F2 populations. The OH88119 × Black cherry F2 was divided into two sub-populations grown in two seasons. Three QTLs for FW were identified from the F2 sub-population of OH88119 × Black cherry grown in the spring season of 2013 and validated in the F2 sub-population of the same cross, but two of them could be detected in the F2 population of OH9242 × Black cherry. This suggests that the QTL for FW is affected by genetic background. Two markers Sli2788 and Sli2772 on chromosome 2 contributed 24.2 and 17.9% of total phenotypic variation, respectively, were at each side of fw2.2 region (Frary et al., 2000). The marker Sli1926 on chromosome 11 contributing 2.2% phenotypic variation located at the known locus fw11.3 region (Huang and van der Knaap, 2011). This result suggests that both fw2.2 and fw1.3 contribute small fruit in Black cherry, which is not in our interest. The marker Sli2377 was significantly associated with FW in both F2 sub-populations of OH88119 × Black cherry and marginally (p = 0.055) associated with FW in the F2 population of OH9242 × Black cherry. The physical distance between Sli2377 and Sli2388, a marker detected significant association with FW in the association mapping panel but not polymorphic among OH88119, OH9242 and Black cherry, was 2.14 Mb that was smaller than the LD on chromosome 4. Thus, this region could be one unit conferring FW. Comparing the physical positions of markers (Grandillo et al., 1999) linked to known loci for FW on chromosome 4, there is no known loci between markers Sli2377 and Sli2388. Therefore, this locus could be a novel one for FW.
In conclusion, association mapping using InDel marker data was applied to uncover the genomic regions harboring genes underlying FW, SSC, shape, and color in tomato fruits followed by confirmation with F2 populations in this study. The results demonstrated that the use of limited number of InDel markers and a relatively small number of accessions was effective in validating known genes/QTLs and identifying novel genotype/phenotype associations for marker-assisted selection of fruit traits in tomato.
Author contributions
Conceived and designed the experiments: HS, WY. Performed the experiments: XL, XG, HZ. Analyzed the data: XL, XG, WY. Contributed reagents/materials/analysis tools: HS, WY. Wrote the paper: XL, XG, WY.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to thank all persons and institutes listed in Table S1 for providing the seeds of tomato lines. The work was partly supported by the National Natural Science Foundation of China (31171973), National Program on Key Basic Research Projects (The 973 Program: 2012CB113900), the 111 Project (B17043) and Beijing Innovation Consortium of Agriculture Research System (Grant No. BAIC01-2016).
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2017.01269/full#supplementary-material
Distribution of phenotypic data in 192 tomato accessions.
Physical map of InDels and QTLs for fruit weight (FW), soluble solid content (SSC), fruit shape index (FSI), and color parameters (L*, Hue, and Chroma) on tomato chromosomes. The relative map position of each InDel can be found in Table S2.
Description of 192 tomato accessions used in this study.
Chromosomal position, primer sequence, number of alleles, and polymorphic information content (PIC) for markers used in this study.
Information of the 56 InDel markers used for genotyping the F2 populations.
Analysis of variance for soluble solid content (SSC), fruit weight (FW), sruit shape index (FSI), fruit color parameters (L, Hue, and Chroma) in 192 tomato accessions grown in 2013 and 2014.
Annotated genes in 2 cM region around the markers associated with traits with e < 1E−10.
References
- Alpert K. B., Grandillo S., Tanksley S. D. (1995). fw2.2: a major QTL controlling fruit weight is common to both red- and green-fruited tomato species. Theor. Appl. Genet. 91, 994–1000. [DOI] [PubMed] [Google Scholar]
- Ashrafi H., Kinkade M. P., Merk H. L., Foolad M. R. (2012). Identification of novel quantitative trait loci for increased lycopene content and other fruit quality traits in a tomato recombinant inbred line population. Mol. Breed. 30, 549–567. 10.1007/s11032-011-9643-1 [DOI] [Google Scholar]
- Bai Y., Lindhout P. (2007). Domestication and breeding of tomatoes: what have we gained and what can we gain in the future? Ann. Bot. 100, 1085–1094. 10.1093/aob/mcm150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauchet G., Grenier S., Samson N., Nonnet J., Grivet L., Causse M. (2017). Use of modern tomato breeding germplasm for deciphering the genetic control of agronomical traits by Genome Wide Association study. Theor. Appl. Genet. 130, 875–889. 10.1007/s00122-017-2857-9 [DOI] [PubMed] [Google Scholar]
- Blanca J., Montero-Pau J., Sauvage C., Bauchet G., Illa Berengur E., Díez M. J., et al. (2015). Genomic variation in tomato, from wild ancestors to contemporary breeding accessions. BMC Genomics 16:257. 10.1186/s12864-015-1444-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonferroni C. E. (1936). Teoria Statistica delle Classi e Calcolo delle Probabilità. Pubblicazioni del Regio Istituto Superiore di Scienze Economiche e Commerciali di Firenze 8, 3–62. [Google Scholar]
- Bradbury P. J., Zhang Z. W., Kroon D. E., Casstevens T. M., Ramdoss Y., Buckler E. S. (2007). TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics 23, 2633–2635. 10.1093/bioinformatics/btm308 [DOI] [PubMed] [Google Scholar]
- Breseghello F., Sorrells M. E. (2006). Association mapping of kernel size and milling quality in wheat (Triticum aestivum L.) cultivars. Genetics 172, 1165–1177. 10.1534/genetics.105.044586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer M. T., Lang L. X., Fujimura K., Dujmovic N., Gray S., van der Knaap E. (2006). Development of a controlled vocabulary and software application to analyze fruit shape variation in tomato and other plant species. Plant Physiol. 141, 15–25. 10.1104/pp.106.077867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton-Freeman B. M., Sesso H. D. (2014). Whole food versus supplement: comparing the clinical evidence of tomato intake and lycopene supplementation on cardiovascular risk factors. Adv. Nutr. 5, 457–485. 10.3945/an.114.005231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cericola F., Portis E., Lanteri S., Toppino L., Barchi L., Acciarri N., et al. (2014). Linkage disequilibrium and genome-wide association analysis for anthocyanin pigmentation and fruit color in eggplant. BMC Genomics 15:896. 10.1186/1471-2164-15-896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F. Q., Foolad M. R., Hyman J., St Clair D. A., Beeleman R. B. (1999). Mapping of QTLs for lycopene and other fruit traits in a Lycopersicon esculentum × L. pimpinellifolium cross and comparison of QTLs across tomato species. Mol. Breed. 5, 283–299. 10.1023/A:1009656910457 [DOI] [Google Scholar]
- Chetelat R. T., Deverna J. W., Bennett A. B. (1995). Effects of the Lycopersicon chmielewskii sucrose accumulator gene (sucr) on fruit yield and quality parameters following introgression into tomato. Theor. Appl. Genet. 91, 334–339. 10.1007/bf00220896 [DOI] [PubMed] [Google Scholar]
- Darrigues A., Hall J., van der Knaap E., Francis D. M., Dujmovic N., Gray S. (2008). Tomato analyzer-color test: a new tool for efficient digital phenotyping. J. Am. Soc. Hortic. Sci. 133, 579–586. [Google Scholar]
- Etminan M., Takkouche B., Caamano-Isorna F. (2004). The role of tomato products and lycopene in the prevention of prostate cancer: A meta-analysis of observational studies. Cancer Epidem. Biomar. 3, 340–345. [PubMed] [Google Scholar]
- Evanno G., Regnaut S., Goudet J. (2005). Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol. Ecol. 14, 2611–2620. 10.1111/j.1365-294X.2005.02553.x [DOI] [PubMed] [Google Scholar]
- Falush D., Stephens M., Pritchard J. K. (2003). Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164, 1567–1587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falush D., Stephens M., Pritchard J. K. (2007). Inference of population structure using multilocus genotype data: dominant markers and null alleles. Mol. Ecol. Notes 7, 574–578. 10.1111/j.1471-8286.2007.01758.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frary A., Nesbitt T. C., Grandillo S., Knaap E., Cong B., Liu J., et al. (2000). fw2.2: a quantitative trait locus key to the evolution of tomato fruit size. Science 289, 85–88. 10.1126/science.289.5476.85 [DOI] [PubMed] [Google Scholar]
- Fridman E., Carrari F., Liu Y. S., Fernie A. R., Zamir D. (2004). Zooming in on a quantitative trait for tomato yield using interspecific introgressions. Science 305, 1786–1789. 10.1126/science.1101666 [DOI] [PubMed] [Google Scholar]
- Gao L. H., Qu M., Ren H. Z., Sui X. L., Chen Q. Y., Zhang Z. X. (2010). Structure, function, application and ecological benefit of single slope energy efficient solar greenhouse in China. Horttechnology 20, 626–631 [Google Scholar]
- Giovannucci E., Rimm E. B., Liu Y., Stampfer M. J., Willet W. C. (2002). A prospective study of tomato products, lycopene, and prostate cancer risk. J. Natl. Cancer Inst. 94, 391–398. 10.1093/jnci/94.5.391 [DOI] [PubMed] [Google Scholar]
- Grandillo S., Ku H. M., Tanksley S. D. (1999). Identifying the loci responsible for natural variation in fruit size and shape in tomato. Theor. Appl. Genet. 99, 978–987. 10.1007/s001220051405 [DOI] [Google Scholar]
- Hammer Ø., Harper D. A. T., Ryan P. D. (2001). Past: paleontological statistics software package for education and data analysis. Palaeontol. Electron. 4, 1–9. [Google Scholar]
- Hardy O. J., Vekemans X. (2002). SPAGeDi: a versatile computer program to analyse spatial genetic structure at the individual or population levels. Mol. Ecol. Notes 2, 618–620. 10.1046/j.1471-8286.2002.00305.x [DOI] [Google Scholar]
- Huang Z. J., van der Knaap E. (2011). Tomato fruit weight 11.3 maps close to fasciated on the bottom of chromosome 11. Theor. Appl. Genet. 123, 465–474. 10.1007/s00122-011-1599-3 [DOI] [PubMed] [Google Scholar]
- Hyman J. R., Gaus J., Foolad M. R. (2004). A rapid and accurate method for estimating tomato lycopene content by measuring chromaticity values of fruit puree. J. Am. Soc. Hortic. Sci. 129, 717–723. [Google Scholar]
- Kabelka E., Franchino B., Francis D. M. (2002). Two loci from Lycopersicon hirsutum LA407 confer resistance to strains of Clavibacter michiganensis subsp. michiganensis. Phytopathology 92, 504–510. 10.1094/PHYTO.2002.92.5.504 [DOI] [PubMed] [Google Scholar]
- Kinkade M. P., Foolad M. R. (2013). Validation and fine mapping of lyc12.1, a QTL for increased tomato fruit lycopene content. Theor. Appl. Genet. 126, 2163–2175. 10.1007/s00122-013-2126-5 [DOI] [PubMed] [Google Scholar]
- Lin T., Zhu G. T., Zhang J. H., Xu X. Y., Yu Q. H., Zheng Z., et al. (2014). Genomic analyses provide insights into the history of tomato breeding. Nat. Genet. 46, 1220–1226. 10.1038/ng.3117 [DOI] [PubMed] [Google Scholar]
- Liu K. J., Muse S. V. (2005). PowerMarker: an integrated analysis environment for genetic marker analysis. Bioinformatics 21, 2128–2129. 10.1093/bioinformatics/bti282 [DOI] [PubMed] [Google Scholar]
- Liu Y. S., Gur A., Ronen G., Causse M., Damidaux R., Buret M., et al. (2003). There is more to tomato fruit colour than candidate carotenoid genes. Plant Biotechnol. J. 1, 195–207. 10.1046/j.1467-7652.2003.00018.x [DOI] [PubMed] [Google Scholar]
- Malosetti M., van der Linden C. G., Vosman B., van Eeuwijk F. A. (2007). A mixed-model approach to association mapping using pedigree information with an illustration of resistance to Phytophthora infestans in potato. Genetics 175, 879–889. 10.1534/genetics.105.054932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markovic Z., Zdravkovic J., Damjanovic M. (1997). Correlation between the morphological characteristics and the biochemical components of tomato fruit quality. Acta Hortic. 462, 151–156. 10.17660/ActaHortic.1997.462.19 [DOI] [Google Scholar]
- Mustilli A. C., Fenzi F., Ciliento R., Alfano F., Bowler C. (1999). Phenotype of the tomato high pigment-2 mutant is caused by a mutation in the tomato homolog of DEETIOLATED1. Plant Cell 11, 145–157. 10.1105/tpc.11.2.145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M. (1972). Genetic distance between populations. Am. Nat. 106, 283–292. 10.1086/282771 [DOI] [Google Scholar]
- Nyquist W. E., Baker R. J. (1991). Estimation of heritability and prediction of selection response in plant populations. Crit. Rev. Plant Sci. 10, 235–322. 10.1080/07352689109382313 [DOI] [Google Scholar]
- Osborn T. C., Alexander D. C., Fobes J. F. (1987). Identification of restriction fragment length polymorphisms linked to genes controlling soluble solids content in tomato fruit. Theor. Appl. Genet. 73, 350–356. 10.1007/BF00262500 [DOI] [PubMed] [Google Scholar]
- Pace J., Gardner C., Romay C., Ganapathysubramanian B., Lübberstedt T. (2015). Genome-wide association analysis of seedling root development in maize (Zea mays L.). BMC Genomics 16:47. 10.1186/s12864-015-1226-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perveen R., Suleria H. A., Anjum F. M., Butt M. S., Pasha I., Ahmad S. (2015). Tomato (Solanum lycopersicum) carotenoids and lycopenes chemistry; metabolism, absorption, nutrition, and allied health claims-a comprehensive review. Crit. Rev. Food Sci. 55, 919–929. 10.1080/10408398.2012.657809 [DOI] [PubMed] [Google Scholar]
- Petreikov M., Yeselson L., Shen S., Levin I., Schaffer A. A. (2009). Carbohydrate balance and accumulation during development of near-isogenic tomato lines differing in the AGPase-L1 allele. J. Am. Soc. Hort. Sci. 134, 134–140. [Google Scholar]
- Pourahmadi Z., Mahboob S., Saedisomeolia A., Reykandeh M. T. (2015). The effect of tomato juice consumption on antioxidant status in overweight and obese females. Women Health 55, 795–804. 10.1080/03630242.2015.1050546 [DOI] [PubMed] [Google Scholar]
- Pritchard J. K., Stephens M., Donnelly P. (2000). Inference of population structure using multilocus genotype data. Genetics 155, 945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rafalski J. A. (2010). Association genetics in crop improvement. Curr. Opin. Plant Biol. 13, 174–180. 10.1016/j.pbi.2009.12.004 [DOI] [PubMed] [Google Scholar]
- Raiola A., Tenore G. C., Barone A., Frusciante L., Rigano M. M. (2015). Vitamin E content and composition in tomato fruits: beneficial roles and bio-fortification. Int. J. Mol. Sci. 16, 29250–29264. 10.3390/ijms161226163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rick C. M. (1974). High soluble-solids content in large-fruited tomato lines derived from a wild green-fruited species. Hildardia 42, 493– 510. 10.3733/hilg.v42n15p493 [DOI] [Google Scholar]
- Robbins M. D., Sim S. C., Yang W. C., Van Deynze A., van der Knaap E., et al. (2011). Mapping and linkage disequilibrium analysis with a genome-wide collection of SNPs that detect polymorphism in cultivated tomato. J. Exp. Bot. 62, 1831–1845. 10.1093/jxb/erq367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronen G., Carmel-Goren L., Zamir D., Hirschberg J. (2000). An alternative pathway to β-carotene formation in plant chromoplasts discovered by map-based cloning of Beta and old-gold color mutations in tomato. Proc. Natl. Acad. Sci. U.S.A. 97, 11102–11107. 10.1073/pnas.190177497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruggieri V., Francese G., Sacco A., D'Alessandro A., Rigano M. M., Parisi M., et al. (2014). An association mapping approach to identify favourable alleles for tomato fruit quality breeding. BMC Plant Biol. 14:337. 10.1186/s12870-014-0337-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacco A., Ruggieri V., Parisi M., Festa G., Rigano M. M., Picarella M. E., et al. (2015). Exploring a tomato landraces collection for fruit-related traits by the aid of a high-throughput genomic platform. PLoS ONE 10:e0137139. 10.1371/journal.pone.0137139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauvage C., Segura V., Bauchet G., Stevens R., Do P. T., Nikoloski Z., et al. (2014). Genome-wide association in tomato reveals 44 candidate loci for fruit metabolic traits. Plant Physiol. 165, 1120–1132. 10.1104/pp.114.241521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirasawa K., Fukuoka H., Matsunaga H., Kobayashi Y., Kobayashi I., Hirakawa H., et al. (2013). Genome-wide association studies using single nucleotide polymorphism markers developed by re-sequencing of the genomes of cultivated tomato. DNA Res. 20, 593–603. 10.1093/dnares/dst033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sim S. C., Robbins M. D., Wijeratne S., Wang H., Yang W. C., Francis D. M. (2015). Association analysis for bacterial spot resistance in a directionally selected complex breeding population of tomato. Phytopathology 105, 1437–1445. 10.1094/PHYTO-02-15-0051-R [DOI] [PubMed] [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739. 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanksley S. D., Ganal M. W., Prince J. P., de Vicente M. C., Bonierbale M. W., Broun P., et al. (1992). High density molecular linkage maps of the tomato and potato genomes. Genetics 132, 1141–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Tomato Genome Consortium (2012). The tomato genome sequence provides insights into fleshy fruit evolution. Nature 485, 635–641. 10.1038/nature11119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tieman D., Zhu G., Resende M. F., Jr., Lin T., Nguyen C., Bies D., et al. (2017). A chemical genetic roadmap to improved tomato flavor. Science 355, 391–394. 10.1126/science.aal1556 [DOI] [PubMed] [Google Scholar]
- van Berloo R., Zhu A., Ursem R., Verbakel H., Gort G., van Eeuwijk F. A. (2008). Diversity and linkage disequilibrium analysis within a selected set of cultivated tomatoes. Theor. Appl. Genet. 117, 89–101. 10.1007/s00122-008-0755-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Knaap E., Chakrabarti M., Chu Y., Clevenger J., Illa-Berenguer E., Huang Z., et al. (2014). What lies beyond the eye: the molecular mechanisms regulating tomato fruit weight and shape. Front. Plant Sci. 5:227. 10.3389/fpls.2014.00227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir B. S. (1990). Genetic Data Analysis-Methods for Discrete Population Genetic Data. Sinauer Associates, Massachusett. [Google Scholar]
- Xu J. X., Ranc N., Mu-os S., Rolland S., Bouchet J. P., Desplat N., et al. (2013). Phenotypic diversity and association mapping for fruit quality traits in cultivated tomato and related species. Theor. Appl. Genet. 126, 567–581. 10.1007/s00122-012-2002-8 [DOI] [PubMed] [Google Scholar]
- Yang J. J., Wang Y. Y., Shen H. L., Yang W. C. (2014). In silico identification and experimental validation of insertion-deletion polymorphisms in tomato genome. DNA Res. 21, 429–438. 10.1093/dnares/dsu008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W. C., Bai X. D., Kabelka E., Eaton C., Kamoun S., Van der Knaap E., et al. (2004). Discovery of single nucleotide polymorphisms in Lycopersicon esculentum by computer aided analysis of expressed sequence tags. Mol. Breed. 14, 21–34. 10.1023/B:MOLB.0000037992.03731.a5 [DOI] [Google Scholar]
- Yang W. C., Sacks E. J., Ivey M. L. L., Miller S. A., Francis D. M. (2005). Resistance in Lycopersicon esculentum intraspecific crosses to race T1 strains of Xanthomonas campestris pv. vesicatoria causing bacterial spot of tomato. Phytopathology 95, 519–527. 10.1094/PHYTO-95-0519 [DOI] [PubMed] [Google Scholar]
- Yu J. M., Pressoir G., Briggs W. H., Vroh Bi I., Yamasaki M., Doebley J. F., et al. (2006). A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nat. Genet. 38, 203–208. 10.1038/ng1702 [DOI] [PubMed] [Google Scholar]
- Zhang J., Zhao J. T., Yao X., Liang J., Chang P. P., Yan F., et al. (2015). Genome-wide association mapping for tomato volatiles positively contributing to tomato flavor. Front. Plant Sci. 6:1042. 10.3389/fpls.2015.01042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Zhao J., Laing Y., Zou Z. (2016). Genome-wide association-mapping for fruit quality traits in tomato. Euphytica 207, 439–451. 10.1007/s10681-015-1567-0 [DOI] [Google Scholar]
- Zhao J. T., Xu Y., Ding Q., Huang X. L., Zhang Y. T., Zou Z. R., et al. (2016). Association mapping of main tomato fruit sugars and organic acids. Front. Plant Sci. 7:1286. 10.3389/fpls.2016.01286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H., Wang X., Huang Z., Gao J., Guo Y., Du Y., et al. (2016). Identification of quantitative trait loci for fruit weight, soluble solids content, and plant morphology using an introgression line population of Solanum pennellii in a fresh market tomato inbred line. Hortic. Plant J. 2, 26–34. 10.1016/j.hpj.2016.02.007 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Distribution of phenotypic data in 192 tomato accessions.
Physical map of InDels and QTLs for fruit weight (FW), soluble solid content (SSC), fruit shape index (FSI), and color parameters (L*, Hue, and Chroma) on tomato chromosomes. The relative map position of each InDel can be found in Table S2.
Description of 192 tomato accessions used in this study.
Chromosomal position, primer sequence, number of alleles, and polymorphic information content (PIC) for markers used in this study.
Information of the 56 InDel markers used for genotyping the F2 populations.
Analysis of variance for soluble solid content (SSC), fruit weight (FW), sruit shape index (FSI), fruit color parameters (L, Hue, and Chroma) in 192 tomato accessions grown in 2013 and 2014.
Annotated genes in 2 cM region around the markers associated with traits with e < 1E−10.