Abstract
Diatoms are one of the most successful marine eukaryotic algal groups, responsible for up to 20% of the annual global CO2 fixation. The evolution of a CO2-concentrating mechanism (CCM) allowed diatoms to overcome a number of serious constraints on photosynthesis in the marine environment, particularly low [CO2]aq in seawater relative to concentrations required by the CO2 fixing enzyme, ribulose-1,5-bisphosphate carboxylase/oxygenase (RubisCO), which is partly due to the slow diffusion rate of CO2 in water and a limited CO2 formation rate from 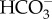 in seawater. Diatoms use two alternative strategies to take up dissolved inorganic carbon (DIC) from the environment: one primarily relies on the direct uptake of
in seawater. Diatoms use two alternative strategies to take up dissolved inorganic carbon (DIC) from the environment: one primarily relies on the direct uptake of 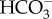 through plasma-membrane type solute carrier (SLC) 4 family
through plasma-membrane type solute carrier (SLC) 4 family 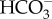 transporters and the other is more reliant on passive diffusion of CO2 formed by an external carbonic anhydrase (CA). Bicarbonate taken up into the cytoplasm is most likely then actively transported into the chloroplast stroma by SLC4-type transporters on the chloroplast membrane system. Bicarbonate in the stroma is converted into CO2 only in close proximity to RubisCO preventing unnecessary CO2 leakage. CAs play significant roles in mobilizing DIC as it is progressively moved towards the site of fixation. However, the evolutionary types and subcellular locations of CAs are not conserved between different diatoms, strongly suggesting that this DIC mobilization strategy likely evolved multiple times with different origins. By contrast, the recent discovery of the thylakoid luminal θ-CA indicates that the strategy to supply CO2 to RubisCO in the pyrenoid may be very similar to that of green algae, and strongly suggests convergent coevolution in CCM function of the thylakoid lumen not only among diatoms but among eukaryotic algae in general. In this review, both experimental and corresponding theoretical models of the diatom CCMs are discussed.
transporters and the other is more reliant on passive diffusion of CO2 formed by an external carbonic anhydrase (CA). Bicarbonate taken up into the cytoplasm is most likely then actively transported into the chloroplast stroma by SLC4-type transporters on the chloroplast membrane system. Bicarbonate in the stroma is converted into CO2 only in close proximity to RubisCO preventing unnecessary CO2 leakage. CAs play significant roles in mobilizing DIC as it is progressively moved towards the site of fixation. However, the evolutionary types and subcellular locations of CAs are not conserved between different diatoms, strongly suggesting that this DIC mobilization strategy likely evolved multiple times with different origins. By contrast, the recent discovery of the thylakoid luminal θ-CA indicates that the strategy to supply CO2 to RubisCO in the pyrenoid may be very similar to that of green algae, and strongly suggests convergent coevolution in CCM function of the thylakoid lumen not only among diatoms but among eukaryotic algae in general. In this review, both experimental and corresponding theoretical models of the diatom CCMs are discussed.
This article is part of the themed issue ‘The peculiar carbon metabolism in diatoms’.
Keywords: marine diatom, CO2-concentrating mechanism, HCO3− transport, photosynthesis, carbonic anhydrase
1. Introduction
The carbon-dioxide-concentrating mechanism (CCM) is a major evolutionary innovation for aquatic photosynthetic organisms that helps them to overcome constraints on acquisition of the photosynthetic substrate, CO2. These systems are of particular relevance in the marine environment, where CO2 availability is consistently low because of the aqueous chemistry of seawater, most notably its alkaline pH, high pH buffer capacity and high salinity. The solubility of CO2 rarely exceeds 25 µM and the rate of dehydration of 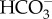 to CO2 is slow. Carbon dioxide as high as 25 µM is generally below the concentration needed to support fixation by ribulose-1,5-bisphosphate carboxylase/oxygenase (RubisCO) in most marine microalgae [1–4].
to CO2 is slow. Carbon dioxide as high as 25 µM is generally below the concentration needed to support fixation by ribulose-1,5-bisphosphate carboxylase/oxygenase (RubisCO) in most marine microalgae [1–4].
Marine diatoms are major primary producers in the oceans and are responsible for up to 20% of annual global primary production [5,6]. A long history of physiological experiments showed that diatoms take up both CO2 and 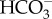 and concentrate dissolved inorganic carbon (DIC) in their cells [7–12], strongly suggesting the occurrence of a CCM. However, only a select number of molecular components of diatom CCMs had been elucidated until very recently. The first breakthrough in identifying the molecular components of diatom CCMs was the isolation and characterization of two pyrenoidal β-carbonic anhydrases (CAs), PtCA1 and PtCA2, in the marine pennate diatom Phaeodactylum tricornutum [13–17]. Studies on CA in the context of the CCM had also revealed interesting new subsets of CAs in the marine centric species Thalassiosira weissflogii and Thalassiosira pseudonana, reinforcing the notion that CAs have evolved many times through convergent evolution and leading to new marine type CA classes denoted δ-CAs, ζ-CAs and θ-CAs [18–22]. In the mid to late 2000s, the sequencing of diatom genomes and development of molecular tools for diatoms including genetic transformation systems enabled more rapid identification of CCM components. CAs have been identified and localized in the model diatoms P. tricornutum and T. pseudonana, revealing a considerable diversity of origins and subcellular localizations of these CAs [15,23]. In contrast with such divergent features of diatom CCMs, convergent aspects in the function of the pyrenoid have recently begun to be recognized [22,24].
and concentrate dissolved inorganic carbon (DIC) in their cells [7–12], strongly suggesting the occurrence of a CCM. However, only a select number of molecular components of diatom CCMs had been elucidated until very recently. The first breakthrough in identifying the molecular components of diatom CCMs was the isolation and characterization of two pyrenoidal β-carbonic anhydrases (CAs), PtCA1 and PtCA2, in the marine pennate diatom Phaeodactylum tricornutum [13–17]. Studies on CA in the context of the CCM had also revealed interesting new subsets of CAs in the marine centric species Thalassiosira weissflogii and Thalassiosira pseudonana, reinforcing the notion that CAs have evolved many times through convergent evolution and leading to new marine type CA classes denoted δ-CAs, ζ-CAs and θ-CAs [18–22]. In the mid to late 2000s, the sequencing of diatom genomes and development of molecular tools for diatoms including genetic transformation systems enabled more rapid identification of CCM components. CAs have been identified and localized in the model diatoms P. tricornutum and T. pseudonana, revealing a considerable diversity of origins and subcellular localizations of these CAs [15,23]. In contrast with such divergent features of diatom CCMs, convergent aspects in the function of the pyrenoid have recently begun to be recognized [22,24].
While molecular-level analysis of diatom CAs advanced, diatom DIC transport mechanisms were far less understood until recently. Nakajima et al. [25] reported the first discovery of a diatom DIC transport mechanism when they characterized a plasma membrane solute carrier (SLC) 4 type transporter, PtSLC4-2 in P. tricornutum which was shown to be a Na+-dependent 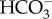 transporter allowing this species to access the abundant extracellular
transporter allowing this species to access the abundant extracellular 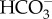 in seawater. Orthologous SLC4-type transporters are found in many diatom genomes and may also be located in chloroplast membranes and probably function as DIC transporters.
in seawater. Orthologous SLC4-type transporters are found in many diatom genomes and may also be located in chloroplast membranes and probably function as DIC transporters.
CCMs are highly responsive to environmental conditions, especially CO2 levels, and the regulatory mechanisms that mediate these responses are of great interest. In P. tricornutum, transcriptional level regulation of the nuclear-encoded pyrenoidal β-CA genes, Ptca1 and Ptca2, have been investigated extensively as representative genes of the diatom CCM. The role of the promoter regions of these genes (PPtca1 and PPtca2) in responding to changes in CO2 levels has been studied using the β-glucuronidase (GUS) reporter assay. These studies showed that transcriptional regulation of PtCA1 and PtCA2 are governed by the second messenger cAMP via interactions of a basic-ZIP (bZIP) type transcription factor, PtbZIP11, with its counterpart cis-element in the promoter regions, which were denoted CO2–cAMP-responsive elements (CCREs) [26,27]. Through transcriptomic analysis, CCRE-bZIP was implicated in CO2-based regulation of the CCM and photorespiration in T. pseudonana [28], strongly suggesting CCRE-bZIP CO2 response systems are a general feature in marine diatoms. Recent work on the CCRE-bZIP-based regulation system revealed that light intensity signals are also integrated into this signalling pathway [29,30], indicating a pivotal function of the cAMP-mediated CO2 signal transduction system as a point of cross-talk between CO2 and light signals.
Taking this molecular information on CCM components and their regulation into account, mathematical modelling of the diatom CCM and its dynamics are also being refined [16,31,32]. However, the functions of the four-layered chloroplast envelopes of diatoms and the pyrenoid are still largely unknown at the molecular level, knowledge which is essential for further refinement of diatom CCM models.
In this review, recent progress in understanding the diatom CCM is updated and perspectives on future directions of the field are discussed. Finally, it should be noted that some diatom species are reported to incorporate a C4-like biochemical CCM system. For brevity, however, this review focuses on the biophysical CCM.
2. Membrane dissolved inorganic carbon transport systems
(a). Plasma membrane 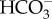 transport systems
transport systems
The uptake of DIC across the plasma membrane represents the critical first step in supplying DIC for photosynthesis, and is especially challenging because environmental conditions are variable and in some cases unpredictable. Diatoms have been shown to actively take up 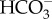 and/or CO2, which has been demonstrated using a range of physiological approaches [7–12,33–35]. However, until recently, the molecular mechanisms responsible for DIC uptake and internal transport, and the regulatory mechanisms controlling these fluxes were not known.
and/or CO2, which has been demonstrated using a range of physiological approaches [7–12,33–35]. However, until recently, the molecular mechanisms responsible for DIC uptake and internal transport, and the regulatory mechanisms controlling these fluxes were not known.
In a recent study, it was demonstrated that a plasma membrane-localized transporter in the marine diatom P. tricornutum homologous to the mammalian SLC4 family, PtSLC4-2, functions as a major uptake mechanism for the acquisition of extracellular DIC under low-CO2 conditions [25] (figure 1). PtSLC4-2 specifically transports 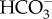 in the presence of a high concentration of sodium ions with a saturation level of about 100 mM Na+. The
in the presence of a high concentration of sodium ions with a saturation level of about 100 mM Na+. The 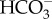 uptake rate of this protein reached its maximum level at pH 8.2, which is the typical pH of seawater [25]. Like PtSLC4-2, two other closely related putative
uptake rate of this protein reached its maximum level at pH 8.2, which is the typical pH of seawater [25]. Like PtSLC4-2, two other closely related putative 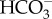 transporters, PtSLC4-1 and PtSLC4-4, are induced specifically under low CO2 conditions, suggesting that these transporters also make a significant contribution to
transporters, PtSLC4-1 and PtSLC4-4, are induced specifically under low CO2 conditions, suggesting that these transporters also make a significant contribution to 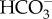 influx into the cell in low CO2 environments like seawater [25,31], although their localizations and functional details have yet to be determined.
influx into the cell in low CO2 environments like seawater [25,31], although their localizations and functional details have yet to be determined.
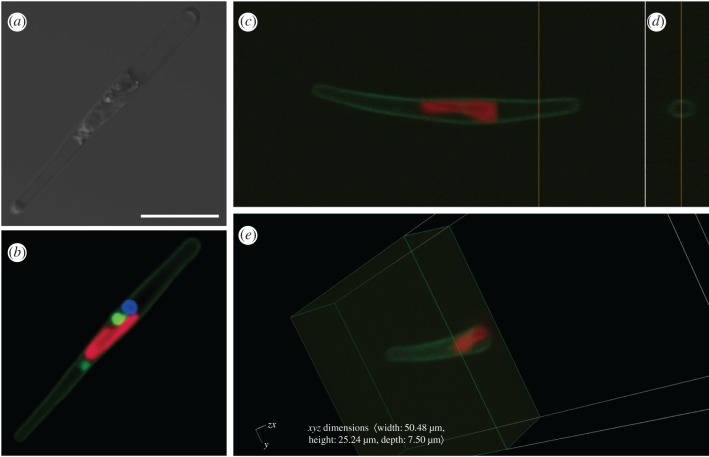
Figure 1.
Localization of exogenously introduced PtSLC4-2:green fluorescent protein fusion in diatom cells. (a,b) Localization of PtSLC4-2:GFP fusion in P. tricornutum: (a) light image; (b) merged image of PtSLC4-2:GFP fusion (green), Hoechst-stained nucleus (blue) and auto-fluorescence of the chloroplast (red). (c–e) Z-stacked images with GFP signals (green) and chlorophyll auto-fluorescence. (d) The cross-section at the orange line in (c). (e) The cross-section of tiled image c at the middle part of the cell. Scale bar,10 µm.
Interestingly, SLC4-type transporters in diatoms form a diatom-specific cluster and this group is phylogenetically close to members of the human SLC4 family with high bootstrap support [25]. In addition, 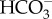 transporters already identified in cyanobacteria and the green alga Chlamydomonas reinhardtii do not share homology with the SLC4 transporters identified in diatoms. This strongly indicates that eukaryotic algae acquired
transporters already identified in cyanobacteria and the green alga Chlamydomonas reinhardtii do not share homology with the SLC4 transporters identified in diatoms. This strongly indicates that eukaryotic algae acquired 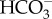 transporters independently from various ancestral eukaryotic hosts. Indeed, it has been suggested that diatom transporters share a common origin with those in human cells [25]. However, more detailed investigation is needed to substantiate this claim because several scenarios have been postulated for the evolution of the Heterokontphyta [36] and consequently, the origin of diatom SLCs may be complex. There has been no molecular work on plasma-membrane-type
transporters independently from various ancestral eukaryotic hosts. Indeed, it has been suggested that diatom transporters share a common origin with those in human cells [25]. However, more detailed investigation is needed to substantiate this claim because several scenarios have been postulated for the evolution of the Heterokontphyta [36] and consequently, the origin of diatom SLCs may be complex. There has been no molecular work on plasma-membrane-type 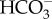 transporters in freshwater diatoms. Physiological studies on the freshwater diatom Navicula pelliculosa have revealed that this species is able to take up
transporters in freshwater diatoms. Physiological studies on the freshwater diatom Navicula pelliculosa have revealed that this species is able to take up 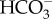 without external CA activity [37], suggesting it uses a specific
without external CA activity [37], suggesting it uses a specific 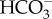 transporter. However, the freshwater environment does not fulfil the Na+ requirement needed by the SLC4 studied in the marine diatom P. tricornutum, strongly suggesting the occurrence of a different type of plasma membrane
transporter. However, the freshwater environment does not fulfil the Na+ requirement needed by the SLC4 studied in the marine diatom P. tricornutum, strongly suggesting the occurrence of a different type of plasma membrane 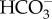 transporter.
transporter.
(b). Bicarbonate transport in the plastidic membrane system
The four-layered chloroplast membranes represent a series of barriers that prevent DIC imported into the cytosol from making its way to the chloroplast for fixation. Therefore, it has been suggested that 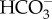 transporters are located at each four-layered chloroplast membrane [31] and that these transporters, in conjunction with CAs densely packed in the spaces between chloroplast membranes, control the permeation of DIC [15,23]. However, chloroplast-membrane-type
transporters are located at each four-layered chloroplast membrane [31] and that these transporters, in conjunction with CAs densely packed in the spaces between chloroplast membranes, control the permeation of DIC [15,23]. However, chloroplast-membrane-type 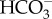 transporters have not yet been identified in diatoms.
transporters have not yet been identified in diatoms.
There are seven PtSLC4 genes and three PtSLC26 genes in the genome of P. tricornutum [25]. A sequence alignment of seven PtSLC4s is given in the electronic supplementary material, figure S1, in comparison with human SLC4s, hsSLC4A1 and hsSLC4A4, and SLC4s in T. pseudonana, TpSLC4-1, TpSLC4-2 and TpSLC4-3. Eleven to 12 membrane-spanning helices of human SLC4A1 were highly conserved from PtSLC4-1 to PtSLC4-5 with the exception of the first transmembrane domain unique to PtSLC4-2 (electronic supplementary material, figure S1, TM2–TM13 of PtSLC4-1–PtSLC4-5). By contrast, TM2, 4, 5, 8 and 9 were relatively well conserved throughout SLCs as compared in the electronic supplementary material, figure S1, but there were significant variations in TM 6, 7 and 10–14 in PtSLC4-6, PtSLC4-7 and three TpSLC4s. Three of the PtSLC4 genes, PtSLC4-1, PtSLC4-2 and PtSLC4-4, are CO2 responsive as mentioned earlier. Two other PtSLC4 genes, PtSLC4-6 and PtSLC4-7, encode proteins previously predicted to localize in the four-layered chloroplast membrane systems based on the presence of targeting sequences [38]. In addition, the N-terminal transit peptide sequence in PtSLC4-6 (GSA-FTS), PtSLC4-7 (SAA-FHT), TpSLC4-2 (SFS-FAP) and TpSLC4-3 (VNA-FPT) includes both an endoplasmic reticulum (ER) signal and a plastid-transit sequence at the predicted cleavage site of ER signal, corresponding to one of the variants of the ASA-FAP motif [39] (the upper N-terminal sequence including these transit sequences is not shown in the electronic supplementary material, figure S1). Interestingly, PtSLC4-6 and PtSLC4-7 cluster phylogentically with heterokont genes which are related to the human/diatom cluster, as mentioned above. The expression levels of PtSLC4-6 and PtSLC4-7 genes were constitutive under high CO2 and low CO2 conditions [25], suggesting that PtSLC4-6 and PtSLC4-7 perhaps constantly regulate DIC flow from the cytosol to the plastid regardless of ambient CO2 concentrations. Currently, very little is known about the intracellular localizations and functions of either PtSLC4-6 and PtSLC4-7 as DIC transporters. The driving forces for transporters within the four-layered chloroplast membranes are also not known. Most probably, these transporters would work with pH and/or ionic gradients across these membranes. In fact, there are some studies reporting the periplastidal compartment (PPC) to be an acidified compartment [40,41]. However, pH and ion regulation systems within the four-layered chloroplast membranes in secondary endosymbionts have not been identified. These are clearly enticing targets for future research.
(c). Energy to drive 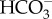 transport
transport
Bicarbonate uptake is an energy-dependent active transport process, but how energy consumption is coupled with 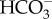 transport remains elusive. In P. tricornutum,
transport remains elusive. In P. tricornutum, 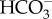 uptake across the plasma membrane is mediated by PtSLC4-2, and this activity is dependent on the Na+ concentration. Most likely PtSLC4-2 is a secondary active transporter, co-transporting Na+ and
uptake across the plasma membrane is mediated by PtSLC4-2, and this activity is dependent on the Na+ concentration. Most likely PtSLC4-2 is a secondary active transporter, co-transporting Na+ and 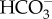 , making use of a transmembrane Na+ gradient [25], which can be made quite large due to the high salinity of seawater. As Na+ continuously flows into the cell with
, making use of a transmembrane Na+ gradient [25], which can be made quite large due to the high salinity of seawater. As Na+ continuously flows into the cell with 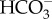 , an efflux of Na+ back out of the cell is required to maintain a [Na+] gradient across the plasma membrane. This would require an ATP-dependent primary transporter to export Na+. While the molecular identity of this putative Na+ efflux pump has not been identified, there is some suggestive evidence for the occurrence of Na+/K+-ATPase and involvement of K+ in DIC acquisition in some diatoms [42–44]. However, orthologous genes encoding known Na+/Ka+-ATPases were not found in diatom genomes. Alternatively, there is a possibility that a secondary Na+/H+ antiporter maintains the Na+ gradient as suggested in the cyanobacterial CCM [45]. Further study is required to reveal the molecular mechanism of Na+-dependent
, an efflux of Na+ back out of the cell is required to maintain a [Na+] gradient across the plasma membrane. This would require an ATP-dependent primary transporter to export Na+. While the molecular identity of this putative Na+ efflux pump has not been identified, there is some suggestive evidence for the occurrence of Na+/K+-ATPase and involvement of K+ in DIC acquisition in some diatoms [42–44]. However, orthologous genes encoding known Na+/Ka+-ATPases were not found in diatom genomes. Alternatively, there is a possibility that a secondary Na+/H+ antiporter maintains the Na+ gradient as suggested in the cyanobacterial CCM [45]. Further study is required to reveal the molecular mechanism of Na+-dependent 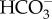 uptake. There are several possible mechanisms to generate the ATP ultimately required for
uptake. There are several possible mechanisms to generate the ATP ultimately required for 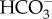 uptake including photophosphorylation, cyclic electron flow (CEF) around photosystem I (PSI) and respiration. In the case of cyanobacterial and green algal CCMs, the involvement of CEF in active DIC uptake has been suggested [46,47]. By contrast, in diatoms, the rate of CEF is reported to be negligibly low relative to total electron transport activity [48], making CEF an unlikely ATP source. Most probably, ATP is generated by linear electron transport through the photosynthetic electron transport chain or respiration, although further study is needed to distinguish between the two.
uptake including photophosphorylation, cyclic electron flow (CEF) around photosystem I (PSI) and respiration. In the case of cyanobacterial and green algal CCMs, the involvement of CEF in active DIC uptake has been suggested [46,47]. By contrast, in diatoms, the rate of CEF is reported to be negligibly low relative to total electron transport activity [48], making CEF an unlikely ATP source. Most probably, ATP is generated by linear electron transport through the photosynthetic electron transport chain or respiration, although further study is needed to distinguish between the two.
(d). Diffusive CO2 uptake
In addition to the 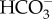 uptake facilitated by plasma membrane SLC4, diatoms take up CO2 from the external environment to support photosynthesis [7,10,11]. Because lipid bilayer membranes are permeable to CO2 [49], CO2 uptake cannot proceed through a typical membrane-embedded transporter mechanism. Instead, organisms take up CO2 through a diffusive mechanism by generating a CO2 deficit inside the cell, which then draws CO2 in from the external environment. In cyanobacteria, this deficit is generated by active conversion of CO2 to
uptake facilitated by plasma membrane SLC4, diatoms take up CO2 from the external environment to support photosynthesis [7,10,11]. Because lipid bilayer membranes are permeable to CO2 [49], CO2 uptake cannot proceed through a typical membrane-embedded transporter mechanism. Instead, organisms take up CO2 through a diffusive mechanism by generating a CO2 deficit inside the cell, which then draws CO2 in from the external environment. In cyanobacteria, this deficit is generated by active conversion of CO2 to 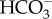 in the cytoplasm through the action of NADPH dehydrogenase (NDH-1) complexes [50] with ferredoxin likely acting as the electron donor to NDH-1 [51,52]. In diatoms, this deficit was instead proposed to be generated by the active transport of
in the cytoplasm through the action of NADPH dehydrogenase (NDH-1) complexes [50] with ferredoxin likely acting as the electron donor to NDH-1 [51,52]. In diatoms, this deficit was instead proposed to be generated by the active transport of 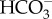 out of the cytoplasm and into the chloroplast resulting in a low
out of the cytoplasm and into the chloroplast resulting in a low 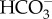 concentration in the cytoplasm [16,31]. The cytoplasmic CO2 concentration is then lowered through the action of a cytoplasmic CA, which when the
concentration in the cytoplasm [16,31]. The cytoplasmic CO2 concentration is then lowered through the action of a cytoplasmic CA, which when the 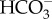 concentration is below equilibrium with CO2 will drive a net hydration of CO2 to
concentration is below equilibrium with CO2 will drive a net hydration of CO2 to 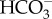 . The CO2 gradient passively draws CO2 into the cell across the plasma membrane, and continued export of
. The CO2 gradient passively draws CO2 into the cell across the plasma membrane, and continued export of 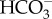 from the cytoplasm maintains a constant cytoplasmic CO2 deficit resulting in sustained CO2 uptake. The activity of the transporter exporting
from the cytoplasm maintains a constant cytoplasmic CO2 deficit resulting in sustained CO2 uptake. The activity of the transporter exporting 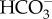 from the cytoplasm must match or exceed the rates of CO2 and
from the cytoplasm must match or exceed the rates of CO2 and 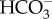 influx in order to maintain the inward CO2 gradient.
influx in order to maintain the inward CO2 gradient.
New evidence suggests this model for CO2 uptake needs to be modified because there is no known CA localized at the cytoplasm in P. tricornutum [15,23] and cytoplasmic CA localized in T. pseudonana is a γ type [23], which is confirmed to be a CA enzyme only in bacteria and archaea [53,54] but not in eukaryotes [55]. Even under alkaline pH of diatom cytoplasm at around 7.6 [8,56], it would not be possible without CA activity to maintain an inward CO2 gradient between the cytoplasm and the external environment, because the uncatalysed hydration rate of CO2 is slow. Instead, the CO2 deficit may be generated in one of the membrane-bound compartments surrounding the chloroplast (the chloroplastic ER or the periplastidal space) where CAs are definitely present rather than the cytoplasm. Modelling studies of this modified CO2 uptake mechanism indicate that it would be functionally equivalent to the older mechanism involving the cytoplasm [31].
An additional emerging complexity in the CO2 uptake pathway involves the possible role that aquaporins play in enhancing membrane permeability to CO2. While aquaporins were originally identified as water channels, they are also believed to facilitate CO2 diffusion in cyanobacteria [47,57], higher plant mesophyll cells [57,58] and red blood cells [59,60] and are found in diatom genomes. While lipid bilayers are inherently permeable to CO2, they do present some resistance to diffusion that could be reduced by the presence of aquaporins. CO2 permeation through diatom membranes is very rapid [16], and this high permeability may be in part due to the presence of channels such as aquaporins.
3. Carbonic anhydrase as a mobilizer and an insulator for dissolved inorganic carbon movement
(a). Carbonic anhydrases and their locations in diatom cells
CA is typically an extremely fast enzyme and catalyses CO2 hydration and 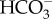 dehydration moving the CO2/
dehydration moving the CO2/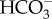 system towards equilibrium [61]. As CA interconverts CO2 and
system towards equilibrium [61]. As CA interconverts CO2 and 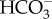 , and these inorganic carbon species have contrasting membrane permeability, CA must have significant roles in controlling the direction and magnitude of DIC fluxes, as will be described in the next section. In the marine diatom P. tricornutum, at least 10 putative CA genes from four families have been identified in its genome. The most prominent CA family is the α-CA family with five members, but there are also two β-CAs, two γ-CAs and one CA from the newly discovered θ family [15,22]. These 10 CAs have all been localized and interestingly, they display specific localizations based upon subtype: α-CAs are located at the four-layered chloroplastic membrane, β-CAs in the pyrenoid, γ-CAs in the mitochondria and the θ-CA in the thylakoid lumen [15,22]. No cytosolic CAs have been identified so far in P. tricornutum. Of these 10 CAs, CA activity has only been confirmed with two β-CAs and one θ-CA, but the Zn binding sites in most of the other CAs are intact, suggesting they are functional [17,22]. Most of the CAs are not regulated transcriptionally by CO2, but the two pyrenoidal β-CAs, PtCA1 and PtCA2, are highly CO2 responsive at the transcriptional level via signal transduction involving a second messenger cAMP [26,27,29]. Transcription of Ptca1 and Ptca2 is also controlled by light using the same signal transduction pathway as that used to respond to CO2 [29], illustrating that there is a cross-talk between light and CO2 signals, two factors that have a major effect on the need to generate CO2 for photosynthesis [29]. Interestingly, these two CAs are also regulated at the post-translational level by the redox state of the chloroplast through the counteraction of thioredoxins (Trxs) and molecular oxygen [17]. The reduced forms of PtCA1 and PtCA2 showed higher activity than oxidized forms [17]. Oxygen from PSII and reduced Trxs would competitively modulate CA activity, suggesting the occurrence of a system to fine tune pyrenoidal CA activities during photosynthesis. In P. tricornutum, there are no external, cytosolic nor free stromal CAs.
, and these inorganic carbon species have contrasting membrane permeability, CA must have significant roles in controlling the direction and magnitude of DIC fluxes, as will be described in the next section. In the marine diatom P. tricornutum, at least 10 putative CA genes from four families have been identified in its genome. The most prominent CA family is the α-CA family with five members, but there are also two β-CAs, two γ-CAs and one CA from the newly discovered θ family [15,22]. These 10 CAs have all been localized and interestingly, they display specific localizations based upon subtype: α-CAs are located at the four-layered chloroplastic membrane, β-CAs in the pyrenoid, γ-CAs in the mitochondria and the θ-CA in the thylakoid lumen [15,22]. No cytosolic CAs have been identified so far in P. tricornutum. Of these 10 CAs, CA activity has only been confirmed with two β-CAs and one θ-CA, but the Zn binding sites in most of the other CAs are intact, suggesting they are functional [17,22]. Most of the CAs are not regulated transcriptionally by CO2, but the two pyrenoidal β-CAs, PtCA1 and PtCA2, are highly CO2 responsive at the transcriptional level via signal transduction involving a second messenger cAMP [26,27,29]. Transcription of Ptca1 and Ptca2 is also controlled by light using the same signal transduction pathway as that used to respond to CO2 [29], illustrating that there is a cross-talk between light and CO2 signals, two factors that have a major effect on the need to generate CO2 for photosynthesis [29]. Interestingly, these two CAs are also regulated at the post-translational level by the redox state of the chloroplast through the counteraction of thioredoxins (Trxs) and molecular oxygen [17]. The reduced forms of PtCA1 and PtCA2 showed higher activity than oxidized forms [17]. Oxygen from PSII and reduced Trxs would competitively modulate CA activity, suggesting the occurrence of a system to fine tune pyrenoidal CA activities during photosynthesis. In P. tricornutum, there are no external, cytosolic nor free stromal CAs.
In T. pseudonana, at least 13 putative CA genes have been identified [23]. In sharp contrast with the case of P. tricornutum, subcellular locations of T. pseudonana CAs are not related to the CA subtypes [15,23]. Also in contrast with P. tricornutum, there is no identified pyrenoidal CA, but there is a stromal α-CA, a cytosolic γ-CA, and two external CAs, one δ-CA and one ζ-CA [23]. There are also three γ-CAs and one δ-CA in the mitochondria, and one δ-CA in the four-layered chloroplast membrane system [23]. Of these CAs, the activity of δ-CA is confirmed [62,63] and also ζ-CA activity has been confirmed in T. weissflogii [21]. Among these putative CAs in T. pseudonana, transcript levels of two external CAs (Tpδ-CA1 and Tpζ-CA1) and a CA in the PPC (Tpδ-CA1) were greatly increased in air-grown cells relative to those in high CO2-grown cell [23]. The result strongly suggests contributions of these putative CAs to DIC acquisition and/or recapturing leaked CO2 under CO2-limited conditions.
Interestingly, in addition to these 13 potential CAs, the occurrence of the new subtype θ-CA family with chloroplast and thylakoid targeting motifs at its N-terminus was discovered recently in the T. pseudonana genome [20], suggesting the generality of occurrence of this type of CA in the lumen of diatom thylakoid.
The localization of CAs in these two model diatoms are updated in figure 2. Even within diatoms, the subtype of CAs found in each species and their localizations are extremely diverse, strongly suggesting that CA genes were acquired by diatoms from diverse origins after they had undergone substantial diversification and that these CAs may be involved in diverse strategies for DIC flux control depending upon the diatom species.
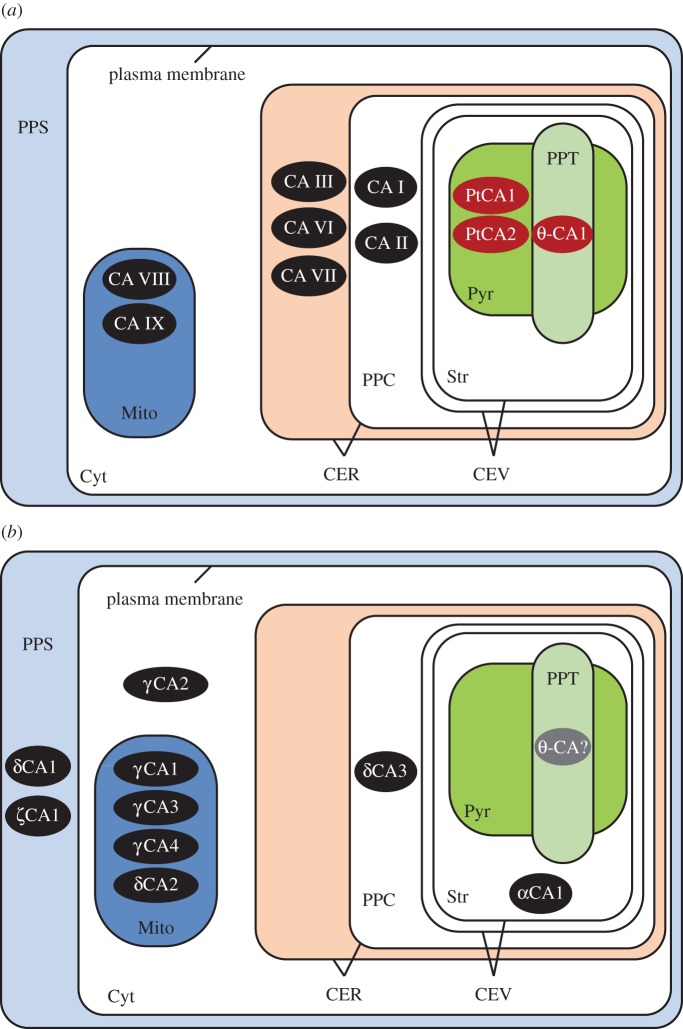
Figure 2.
Subtypes and localization of CAs in P. tricornutum (a) and T. pseudonana (b). Black colouring indicates that the CA has been localized, but activity has not been confirmed, whereas red colouring indicates the CA has been both localized and confirmed to have CA activity. CAs shown in grey have been identified in the genome, but have not been localized or tested for CA activity. The compartmental abbreviations are: periplasmic space (PPS), cytoplasm (Cyt), chloroplastic endoplasmic reticulum (CER), mitochondria (Mito), periplastidal compartment (PPC), chloroplast envelope (CEV), stroma (Str), pyrenoid (Pyr) and pyrenoid-penetrating thylakoid (PPT).
(b). Function of carbonic anhydrases in diatom CO2-concentrating mechanisms
CAs may have multiple critical functions in diatom CCMs, including roles in CO2 uptake, generation of CO2 in the chloroplast for fixation and recovery of CO2 leaking out of the chloroplast. As discussed above, CO2 uptake proceeds by the generation of an internal CO2 deficit, proximally generated by the CA-catalysed hydration of CO2 to HCO−3 in the cytoplasm or other compartment [16]. External CAs, located in the periplasmic space [15,23], facilitate CO2 uptake by generating CO2 from 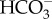 at the cell surface [64]. These external CAs are apparently very important for diatom CCMs as they occur in a number of diatoms species and are expressed at very high levels, in contrast with other microalgae where they are less commonly found [11,65,66].
at the cell surface [64]. These external CAs are apparently very important for diatom CCMs as they occur in a number of diatoms species and are expressed at very high levels, in contrast with other microalgae where they are less commonly found [11,65,66].
Bicarbonate imported into the cell must be converted to CO2 for fixation by RubisCO and CAs are necessary to catalyse this conversion, because the intrinsic dehydration rate of bicarbonate is slow. In P. tricornutum, a newly discovered route to produce CO2 involves import of 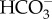 into the pyrenoid-penetrating thylakoid lumen where the low pH and action of a θ-CA rapidly converts
into the pyrenoid-penetrating thylakoid lumen where the low pH and action of a θ-CA rapidly converts 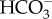 to CO2 for RubisCO [22]. β-CAs inside the pyrenoid also convert bicarbonate that diffuses in the stroma into CO2, elevating the CO2 concentration around RubisCO [14,15]. A portion of the CO2 supplied to RubisCO is fixed, but the CCM is not perfectly efficient, and a significant fraction of the CO2 leaks out of the chloroplast [16]. This leaked CO2 is recovered by CA-catalysed conversion to
to CO2 for RubisCO [22]. β-CAs inside the pyrenoid also convert bicarbonate that diffuses in the stroma into CO2, elevating the CO2 concentration around RubisCO [14,15]. A portion of the CO2 supplied to RubisCO is fixed, but the CCM is not perfectly efficient, and a significant fraction of the CO2 leaks out of the chloroplast [16]. This leaked CO2 is recovered by CA-catalysed conversion to 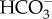 back in the cytoplasm or other outlying compartment, and the regenerated
back in the cytoplasm or other outlying compartment, and the regenerated 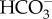 can be transported back into the chloroplast. Like many other aspects of the diatom CCM, such CA-based CO2 recovery systems seem to be highly diverse. In T. pseudonana, either the cytoplasmic, chloroplast envelope or stromal CAs could function in CO2 recovery. In sharp contrast, P. tricornutum has numerous chloroplast envelope CAs but lacks cytosolic and stromal CAs [15,23], strongly suggesting that the main recovery points are in the four-layered chloroplastic envelope. It should also be noted that pyrenoidal β-CAs in P. tricornutum may be a part of such a recovery system of leaking CO2, which will be discussed in the next section. The detailed molecular role of these CAs in diatoms however has yet to be determined and await further reverse genetics approaches to be confirmed.
can be transported back into the chloroplast. Like many other aspects of the diatom CCM, such CA-based CO2 recovery systems seem to be highly diverse. In T. pseudonana, either the cytoplasmic, chloroplast envelope or stromal CAs could function in CO2 recovery. In sharp contrast, P. tricornutum has numerous chloroplast envelope CAs but lacks cytosolic and stromal CAs [15,23], strongly suggesting that the main recovery points are in the four-layered chloroplastic envelope. It should also be noted that pyrenoidal β-CAs in P. tricornutum may be a part of such a recovery system of leaking CO2, which will be discussed in the next section. The detailed molecular role of these CAs in diatoms however has yet to be determined and await further reverse genetics approaches to be confirmed.
(c). Pyrenoid functions with carbonic anhydrase
Apart from the significant divergence in the origins of chloroplast-based CAs, examples of convergence can also be found in the chloroplast and pyrenoid. The recent discovery of θ-CA (Pt43233) in the lumen of the thylakoid membrane suggests dramatic convergent coevolution of CCM function in eukaryotes. In the green alga C. reinhardtii, it has long been known that the final conversion of 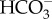 to CO2 occurs within a portion of the thylakoid that penetrates the pyrenoid.
to CO2 occurs within a portion of the thylakoid that penetrates the pyrenoid. 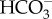 is imported into the thylakoid lumen where the low pH and action of CA convert it to CO2, which then diffuses out into the pyrenoid [67]. The newly discovered θ-CA in P. tricornutum is also localized specifically to the region of the thylakoid that passes through the pyrenoid and it is critical for CCM function, suggesting this final CO2 generation step works quite similarly in the disparate diatom and green algal groups. Notably, the θ-CA sequence is not related to that of the C. reinhardtii thylakoid CA (an α-CA).
is imported into the thylakoid lumen where the low pH and action of CA convert it to CO2, which then diffuses out into the pyrenoid [67]. The newly discovered θ-CA in P. tricornutum is also localized specifically to the region of the thylakoid that passes through the pyrenoid and it is critical for CCM function, suggesting this final CO2 generation step works quite similarly in the disparate diatom and green algal groups. Notably, the θ-CA sequence is not related to that of the C. reinhardtii thylakoid CA (an α-CA).
The P. tricornutum θ-CA possesses a Cys-Gly-His rich (CGHR) domain with an N-terminal chloroplast-thylakoid targeting motif and similar putative thylakoid targeted CGHR family proteins also exist in T. pseudonana, strongly suggesting this new class θ-CA occurs throughout the diatoms [22]. The RNAi suppression of this protein in P. tricornutum resulted in inhibition of growth and reduced photosynthetic DIC affinity [22], indicating the pivotal function of the thylakoid luminal CA for the diatom CCM. Interestingly, CGHR-containing proteins also occur in C. reinhardtii (denoted as LCIB, C, D and E). An LCIB/C hexamer complex was localized at the peripheral pyrenoid and is critical for CCM function [64,65]. CA activity of the LCIB/C complex in C. reinhardtii has not yet been tested, but the occurrence of proteins homologous to diatom luminal θ-CA as a pyrenoid-localized component critical to the CCM in the distant green algae [68,69] indicates that this CA class is commonly involved in CCM functions associated with the pyrenoid.
It is hypothesized that the LCIB/C complex in C. reinhardtii functions as a part of the system for recapturing CO2 leaking out of the pyrenoid by hydrating it to 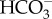 [69–71]. If C. reinhardtii LCIB/C serves as CA, CO2 hydration occurs via LCIB/C itself in the pyrenoid, which potentially competes with the reaction of RubisCO for CO2. This consideration also applies to the case of P. tricornutum pyrenoid, which possesses pyrenoidal PtCAs outside the thylakoid membrane and θ-CA at the lumen [22]. It is noteworthy that, in C. reinhardtii and P. tricornutum, the LCIB/C complex and PtCAs revealed a clumped distribution to highly localized parts in the pyrenoid, while RubisCO disperses over the pyrenoid [15,70,72], suggesting that pyrenoidal CAs localize differentially from RubisCO. Detailed localization of these components is needed to clarify their functions. Recently, a novel pyrenoidal protein EPYC1/LCI5 was identified in the green alga C. reinharditii. EPYC1/LCI5 is essential to the formation of a dense aggregation of RubisCO in the pyrenoid, suggesting EPYC1/LCI5 is an essential structural component for pyrenoid formation [24]. A putative structural analogue of EPYC1/LCI5 occurs in the diatom genome [24], suggesting the involvement of such structural proteins in the arrangement of diatom pyrenoidal proteins.
[69–71]. If C. reinhardtii LCIB/C serves as CA, CO2 hydration occurs via LCIB/C itself in the pyrenoid, which potentially competes with the reaction of RubisCO for CO2. This consideration also applies to the case of P. tricornutum pyrenoid, which possesses pyrenoidal PtCAs outside the thylakoid membrane and θ-CA at the lumen [22]. It is noteworthy that, in C. reinhardtii and P. tricornutum, the LCIB/C complex and PtCAs revealed a clumped distribution to highly localized parts in the pyrenoid, while RubisCO disperses over the pyrenoid [15,70,72], suggesting that pyrenoidal CAs localize differentially from RubisCO. Detailed localization of these components is needed to clarify their functions. Recently, a novel pyrenoidal protein EPYC1/LCI5 was identified in the green alga C. reinharditii. EPYC1/LCI5 is essential to the formation of a dense aggregation of RubisCO in the pyrenoid, suggesting EPYC1/LCI5 is an essential structural component for pyrenoid formation [24]. A putative structural analogue of EPYC1/LCI5 occurs in the diatom genome [24], suggesting the involvement of such structural proteins in the arrangement of diatom pyrenoidal proteins.
4. CO2 sensing at the ocean surface
(a). CO2/light response
Like most algal CCMs, diatom CCMs are highly responsive to the ambient CO2 concentration [73]. In general, components are more highly expressed at low CO2 when the CCM is most needed to maintain high CO2 concentrations around RubisCO [15,23,25,26,74,75]. Changes in the partial CO2 pressure in the atmosphere result in a suite of changes in the concentrations of aqueous DIC species, including increased CO2 and 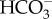 and decreased concentrations of
and decreased concentrations of 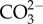 in the range of pH relevant to marine systems. Typically, it is unclear which DIC species are eliciting the observed modifications of the algal CCM. However, a few quantitative investigations have been carried out on this topic. In cyanobacteria, the critical factor governing CCM expression is known to be the total DIC concentration, and photorespiratory metabolism is involved in the CO2 signal transduction process [76–78]. By contrast, in freshwater green algae, Chlorella and Chlamydomonas, dissolved CO2 is known to be a critical DIC species controlling CCM expression [79–81], strongly suggesting the involvement of some direct sensing systems in this process in eukaryotic algae. Similarly in diatoms, several physiological studies have shown that CO2 is the critical determinant of the extent of CCM expression [8,10,11,34,73].
in the range of pH relevant to marine systems. Typically, it is unclear which DIC species are eliciting the observed modifications of the algal CCM. However, a few quantitative investigations have been carried out on this topic. In cyanobacteria, the critical factor governing CCM expression is known to be the total DIC concentration, and photorespiratory metabolism is involved in the CO2 signal transduction process [76–78]. By contrast, in freshwater green algae, Chlorella and Chlamydomonas, dissolved CO2 is known to be a critical DIC species controlling CCM expression [79–81], strongly suggesting the involvement of some direct sensing systems in this process in eukaryotic algae. Similarly in diatoms, several physiological studies have shown that CO2 is the critical determinant of the extent of CCM expression [8,10,11,34,73].
The molecular mechanisms mediating responses to CO2 in diatoms are most well studied with regard to the transcriptional control system of two pyrenoidal β-CAs in P. tricornutum. It has been clearly demonstrated that transcription of both Ptca1 and Ptca2 are CO2 and light responsive; i.e. the expression of these genes are stimulated by low (at least atmospheric level) CO2 and largely repressed in enriched (1–5%) CO2, and this expressional control requires light [26,74,75]. Interestingly, such stimulation (or de-repression) of these CA genes under low CO2 conditions are efficiently suppressed by cAMP analogues and/or cAMP phosphodiesterase inhibitors [26,27,29], indicating an involvement of the cAMP second messenger system downstream of the CO2 sensing mechanism (figure 3). A detailed GUS reporter assay targeting the region 1.3 kbp upstream of the transcription-start sites of both Ptca1 and Ptca2 revealed new cis-elements critical for the CO2 response, termed CO2/cAMP-responsive elements (CCREs: ACGTCA/G) [27,29]. Three CCRE sequences in the Ptca1 promoter region reside within −90 bp relative to the transcription-start site and these sequences run in opposite directions to each other with 15–18 bp intervals [27]. Gel shift assays showed that these CCREs are a target of a group 4 basic-zipper (bZIP) transcription factor, PtbZIP11 in P. tricornutum [27,83]. The promoter region of the Ptca2 gene also possesses three CCRE sequences with a different arrangement from that of the Ptca1 gene promoter, and these sequences were also shown to be critical cis-elements mediating the CO2 response of this gene [29]. A further transcriptomics study using T. pseudonana demonstrated that the transcriptional response of the CCM and photorespiratory gene clusters are responsive to CO2 using CCRE sequences for their regulation [28], indicating a common mechanism for CO2 signalling in diatoms.
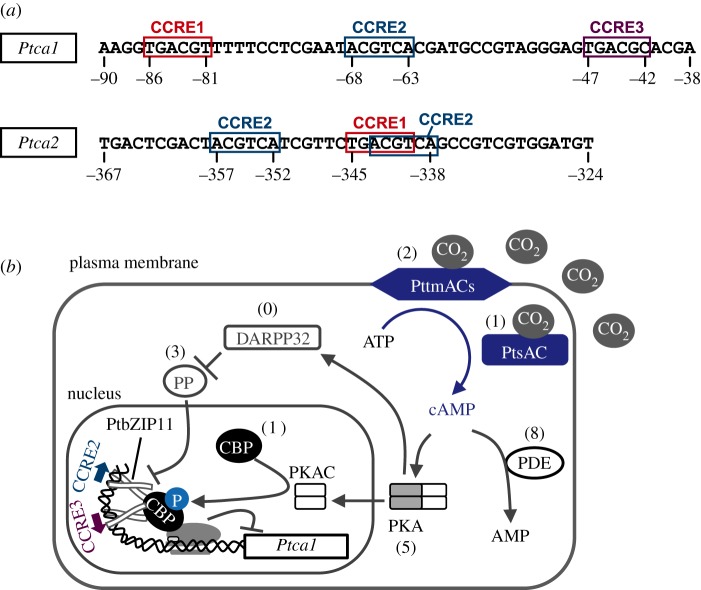
Figure 3.
The CO2-responsive elements in the Ptca1 and Ptca2 promoters and the putative CO2 signalling pathway. (a) Structures of the core-regulatory region of the Ptca1 and the Ptca2 promoter. (b) A suggested model of the cAMP-mediated CO2 signalling pathway based upon the mammalian cAMP signalling cascade. (Redrawn from Matsuda & Kroth [82]). This model describes the signalling route under high CO2 conditions. PDE, cAMP phosphodiesterase; PKA, protein kinase A; PKAC, C subunit of PKA; PP, protein phosphatase; CBP, CREB (cAMP binding protein) binding protein; DARPP, dopamine- and cAMP-regulated phosphoprotein. Parentheses indicate the number of candidate genes in the P. tricornutum genome. No functional analogue of DARPP has been identified so far.
A recent study further demonstrated that Ptca1 and Ptca2 genes are also light responsive and this response is governed by the same set of CCREs that are involved in the CO2 response [29] (figure 3), strongly suggesting that the light signal is integrated with the CO2/cAMP signal by some type of cross-talk mechanism [48]. Interestingly, a very weak dose of 2,6-dichlorophenol-indophenol, which oxidizes the acceptor side of PSI, efficiently suppressed the transcriptions of Ptca1 and Ptca2 under low-CO2 and illuminated conditions [29]. This strongly suggests that light may generate a retrograde signal at the acceptor side of PSI as a part of an electron sorting system from ferredoxin, which is one of the first examples of an involvement of PSI in a CO2/light retrograde signal which ultimately manipulates nuclear gene expression.
(b). CO2 with diel light and Fe signals
The micronutrient iron has a major role in diatom metabolism, particularly in the light reaction of photosynthesis and in nitrate assimilation. For both of these metabolic pathways, light levels greatly influence the regulation, and thus potentially Fe requirements. A recently published study examined the global transcriptional response of P. tricornutum to diel light cycles at three different Fe concentrations [84], and we have examined the expression of the CCM components detailed in this review. As part of that study, individual gene expression profiles were determined to be statistically responsive to either light or iron levels using multiple methods.
Of the various CCM components, PtCA2 is by far the most highly expressed component, with PtCA1 being expressed below statistical significance. PtCA2 had a statistically similar expression profile to PtSLC4-1, PtSLC4-5, the θ-CA and PtbZIP11. Each of these components are upregulated in light conditions relative to dark conditions and are downregulated at lower Fe concentrations. PtbZIP11 is induced by both light conditions but also by low Fe concentrations. Curiously, both PtSLC4-3 and PtSLC4-52bd are upregulated in the dark relative to the light, suggesting different functional roles to the other putative outermembrane-localized PtSLC4. PtSLC4-7 shows constitutive expression, while PtSLC4-6 is mildly upregulated in light conditions. The names of the CCM components in P. tricornutum referred to in this paper are listed with their corresponding Protein ID and updated annotations in table 1.
Table 1.
CCM-associated proteins encoded in the genome of P. tricornutum and T. pseudonana, and role of each protein shown in this review. PM and CM indicate plasma membrane and chloroplast membrane, respectively.
| annotation name | Protein ID | role |
|---|---|---|
| PtbZIP11 | 49205a | regulating |
| PtCA1 | 45433a | pyrenoidal carbonic anhydrase |
| PtCA2 | 51305a | pyrenoidal carbonic anhydrase |
| θ-CA | 43233a | pyrenoid-penetrating thylakoidal lumen carbonic anhydrase |
| PtSLC4-1 | 1677a | PM-type bicarbonate transporter? |
| PtSLC4-2 | bd1806b | PM-type bicarbonate transporter |
| PtSLC4-3 | bd1743b | PM-type bicarbonate transporter? |
| PtSLC4-4 | bd714b | PM-type bicarbonate transporter? |
| PtSLC4-5 | 54405a | PM-type bicarbonate transporter? |
| PtSLC4-52bd | bd52b | PM-type bicarbonate transporter? |
| PtSLC4-6 | 43194a | CM-type bicarbonate transporter? |
| PtSLC4-7 | 45656a | CM-type bicarbonate transporter? |
| PtSLC26-1 | 42555a | PM-type bicarbonate transporter? |
| PtSLC26-2 | 42556a | PM-type bicarbonate transporter? |
| PtSLC26-3 | 50075a | PM-type bicarbonate transporter? |
| TpSLC4-1 | 266801c | PM-type bicarbonate transporter? |
| TpSLC4-2 | 1403c | CM-type bicarbonate transporter? |
| TpSLC4-3 | 267979c | CM-type bicarbonate transporter? |
aProtein IDs refer to JGI genome database (http://genome.jgi.doe.gov/Phatr2/Phatr2.home.html).
bProtein IDs refer to JGI genome database (http://genome.jgi.doe.gov/Phatr2_bd/Phatr2_bd.home.html).
cProtein IDs refer to JGI genome database (http://genome.jgi.doe.gov/Thaps3/Thaps3.home.html).
5. Models of dissolved inorganic carbon flux and its dynamic control
The basic components used to create CCMs are few in number—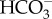 transporters, CAs, RubisCO—and it is the careful spatial arrangement of these components and manipulation of CO2/
transporters, CAs, RubisCO—and it is the careful spatial arrangement of these components and manipulation of CO2/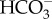 equilibrium and gradients that combine to form efficient CCMs. Consequently, numerical models of CCMs have been especially helpful in establishing the essential functional characteristics of CCMs in diverse microalgae [1,85,86]. In the case of diatoms, the most extensive numerical modelling of the CCM has involved the diatom P. tricornutum. Here, model-data comparisons were used to infer quantitative rates of inorganic carbon fluxes into the cell and within the major cellular compartments [16,31]. These models suggested that a high rate of active
equilibrium and gradients that combine to form efficient CCMs. Consequently, numerical models of CCMs have been especially helpful in establishing the essential functional characteristics of CCMs in diverse microalgae [1,85,86]. In the case of diatoms, the most extensive numerical modelling of the CCM has involved the diatom P. tricornutum. Here, model-data comparisons were used to infer quantitative rates of inorganic carbon fluxes into the cell and within the major cellular compartments [16,31]. These models suggested that a high rate of active 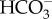 transport out of the cytoplasm and into the chloroplast (the ‘chloroplast pump’) was the major driver of the CCM in P. tricornutum leading to
transport out of the cytoplasm and into the chloroplast (the ‘chloroplast pump’) was the major driver of the CCM in P. tricornutum leading to 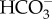 accumulation in the chloroplast stroma and the CO2 deficit in the cytoplasm. Furthermore, according to the transcriptional analysis of some CCM components in response to CO2 (and partially to light), acclimation responses of the CCM to CO2 concentration seemed to take place primarily in the DIC acquisition system from the bulk medium, while by contrast, putative components of a ‘chloroplast pump’ moving DIC from the cytoplasm to RubisCO were not affected by CO2 availability [15,22,23,25]. The latest iteration of these P. tricornutum CCM models [31] emphasized the coherence between molecular and physiological data when incorporated into a modelling framework, and suggested that the system was largely understood. However, the recent discovery of the role of the thylakoid and its associated θ-CA in CO2 supply require a reassessment of this conclusion. There is also the need to develop CCM models for other diatoms, and this should be feasible for additional species such as T. pseudonana as more molecular and physiological data on these species become available.
accumulation in the chloroplast stroma and the CO2 deficit in the cytoplasm. Furthermore, according to the transcriptional analysis of some CCM components in response to CO2 (and partially to light), acclimation responses of the CCM to CO2 concentration seemed to take place primarily in the DIC acquisition system from the bulk medium, while by contrast, putative components of a ‘chloroplast pump’ moving DIC from the cytoplasm to RubisCO were not affected by CO2 availability [15,22,23,25]. The latest iteration of these P. tricornutum CCM models [31] emphasized the coherence between molecular and physiological data when incorporated into a modelling framework, and suggested that the system was largely understood. However, the recent discovery of the role of the thylakoid and its associated θ-CA in CO2 supply require a reassessment of this conclusion. There is also the need to develop CCM models for other diatoms, and this should be feasible for additional species such as T. pseudonana as more molecular and physiological data on these species become available.
6. Conclusion
The molecular components, their origin, their localization and some of their putative functions in the CCM described in this review illustrate the diversity of diatom CCMs. In particular, putative DIC flux control systems between the external medium and the cytosol seem to be divided into at least two low-CO2 inducible strategies: one uses active 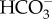 pumps across plasma membranes and the other relies more significantly on passive CO2 entry aided by external CAs, and these likely use contrasting diffusion barrier systems against CO2 leakage (figure 4). However, in both cases, as a convergent aspect, we assume a strong constitutive
pumps across plasma membranes and the other relies more significantly on passive CO2 entry aided by external CAs, and these likely use contrasting diffusion barrier systems against CO2 leakage (figure 4). However, in both cases, as a convergent aspect, we assume a strong constitutive 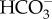 pumping action at the chloroplast envelope, which allows an efficient removal of CO2 from the cytosol and an efficient accumulation of
pumping action at the chloroplast envelope, which allows an efficient removal of CO2 from the cytosol and an efficient accumulation of 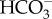 in the stroma. Furthermore, both systems most likely use the pyrenoid as a hub to generate ample CO2 flux from accumulated
in the stroma. Furthermore, both systems most likely use the pyrenoid as a hub to generate ample CO2 flux from accumulated 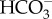 aided by thylakoid luminal θ-CA to support fixation by RubisCO. In this process, pyrenoidal CA activity may function as a critical part of the CCM in concert with luminal CA, which appears to be a general feature of algal CCM, but details of this mechanism in diatoms requires further fine structural analysis of the pyrenoid.
aided by thylakoid luminal θ-CA to support fixation by RubisCO. In this process, pyrenoidal CA activity may function as a critical part of the CCM in concert with luminal CA, which appears to be a general feature of algal CCM, but details of this mechanism in diatoms requires further fine structural analysis of the pyrenoid.
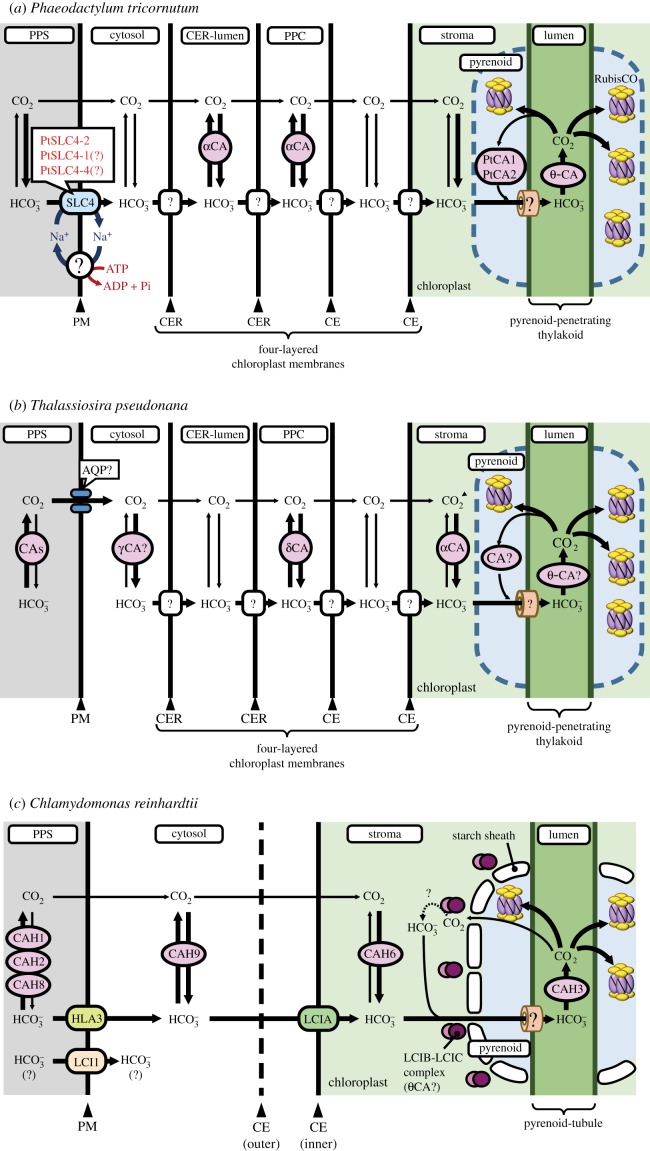
Figure 4.
Models of DIC transport in marine diatoms. (a) In P. tricornutum, plasma-membrane-located-SLC4s (PtSLC4-2, possibly PtSLC4-1 and PtSLC4-4) transport 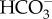 from seawater in a Na+-dependent manner, most probably Na+–
from seawater in a Na+-dependent manner, most probably Na+–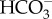 cotransport. This would require an unidentified ATP-dependent primary Na+ pump or a proton-dependent secondary pump to maintain a [Na+] gradient across the plasma membrane. The mechanism of DIC transport from the cytosol to the stroma is unknown. Unidentified transporters on CER and CE might be involved in this process in combination with CAs in intermembrane spaces. In the pyrenoid, CO2 is generated by luminal θ-CA in the pyrenoid-penetrating thylakoid (PPT). CO2 released from the lumen is fixed by RubisCO, or otherwise recaptured at specialized pyrenoidal loci by β-type CAs, PtCA1 and/or PtCA2 by which
cotransport. This would require an unidentified ATP-dependent primary Na+ pump or a proton-dependent secondary pump to maintain a [Na+] gradient across the plasma membrane. The mechanism of DIC transport from the cytosol to the stroma is unknown. Unidentified transporters on CER and CE might be involved in this process in combination with CAs in intermembrane spaces. In the pyrenoid, CO2 is generated by luminal θ-CA in the pyrenoid-penetrating thylakoid (PPT). CO2 released from the lumen is fixed by RubisCO, or otherwise recaptured at specialized pyrenoidal loci by β-type CAs, PtCA1 and/or PtCA2 by which 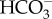 is regenerated to be transported back to the thylakoid lumen. (b) In T. pseudonana, extracellular CA accelerates the dehydration of
is regenerated to be transported back to the thylakoid lumen. (b) In T. pseudonana, extracellular CA accelerates the dehydration of 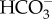 , resulting in high diffusive permeation of CO2. Active
, resulting in high diffusive permeation of CO2. Active 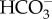 transport into the chloroplast would lower cytoplasmic [
transport into the chloroplast would lower cytoplasmic [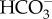 ], and then CO2 that diffuses into the cell is readily converted into
], and then CO2 that diffuses into the cell is readily converted into 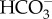 by cytosolic CA (alternatively this could occur in the PPC). As in P. tricornutum, transporters on CER and CE have not been identified. In T. pseudonana, a putative θ-CA is predicted in the PPT, suggesting a role for θ-CA in CO2 generation as proposed in P. tricornutum. There is no known CA localized in the pyrenoid. (c) In C. reinharditii,
by cytosolic CA (alternatively this could occur in the PPC). As in P. tricornutum, transporters on CER and CE have not been identified. In T. pseudonana, a putative θ-CA is predicted in the PPT, suggesting a role for θ-CA in CO2 generation as proposed in P. tricornutum. There is no known CA localized in the pyrenoid. (c) In C. reinharditii, 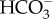 is delivered into the stroma by plasma-membrane-located HLA3 and chloroplast-envelope-located LCIA. LCIA is shown to be on the chloroplast envelope, but its exact localization, whether inner or outer membrane, is unknown. Here, we assumed that LCIA is on the inner envelope because the outer membrane is generally permeable to low molecular weight compounds which most probably allows spontaneous
is delivered into the stroma by plasma-membrane-located HLA3 and chloroplast-envelope-located LCIA. LCIA is shown to be on the chloroplast envelope, but its exact localization, whether inner or outer membrane, is unknown. Here, we assumed that LCIA is on the inner envelope because the outer membrane is generally permeable to low molecular weight compounds which most probably allows spontaneous 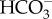 permeation. LCI1 on plasma membrane was strongly suggested to be a DIC transporter, but the inorganic carbon species transported by LCI1 have not been determined. CAH3, a luminal CA in the thylakoid tubule, generates CO2 from
permeation. LCI1 on plasma membrane was strongly suggested to be a DIC transporter, but the inorganic carbon species transported by LCI1 have not been determined. CAH3, a luminal CA in the thylakoid tubule, generates CO2 from 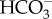 . Unlike P. tricornutum, the luminal CAH3 is α-type CA, suggesting convergent evolution of a mechanism to supply CO2 by using luminal acidification. Similarly, the LCIB-C complex is a putative θ-CA, another example of convergent evolution of pyrenoidal CA which may play a role in recapturing leaking out CO2. CE, chloroplast envelopes; CER, chloroplast ER; PM, plasma membrane; PPC, periplastidal compartment; PPS, periplasmic space.
. Unlike P. tricornutum, the luminal CAH3 is α-type CA, suggesting convergent evolution of a mechanism to supply CO2 by using luminal acidification. Similarly, the LCIB-C complex is a putative θ-CA, another example of convergent evolution of pyrenoidal CA which may play a role in recapturing leaking out CO2. CE, chloroplast envelopes; CER, chloroplast ER; PM, plasma membrane; PPC, periplastidal compartment; PPS, periplasmic space.
Supplementary Material
Data accessibility
This article has no data.
Competing interests
We declare we have no competing interests.
Funding
This work was supported by Grant-in-Aid for Scientific Research B (grant no. 24310015 to Y.M.), by Grant-in-Aid for Scientific Research on Innovative Areas (grant no. 16H06557 to Y.M.), by Grant-in-Aid for Young Scientists B (grant no. 26870750 to K.N., and grant nos 15K16156 and 17K15326 to Y.T.) from Japan Society for the Promotion of Science (JSPS), by MEXT-Supported program for the Strategic Research Foundation for the Advancement of Environmental Protection Technology and for Development of Intelligent Self-Organized Biomaterials and by the Science Research Promotion Fund of the Promotion and Mutual Aid Corporation for Private Schools of Japan (to Y.M.). B.M.H. was supported by the US National Science Foundation (EF 1041023, MCB 1129326) and the Alfred P. Sloan Foundation (BR2014-049).
References
- 1.Badger MR, Andrews TJ, Whitney SM, Ludwig M, Yellowlees DC, Leggat W, Price GD. 1998. The diversity and coevolution of Rubisco, plastids, pyrenoids, and chloroplast-based CO2-concentrating mechanisms in algae. Can. J. Bot. 76, 1052–1071. ( 10.1139/b98-074) [DOI] [Google Scholar]
- 2.Losh JL, Young JN, Morel FMM. 2013. Rubisco is a small fraction of total protein in marine phytoplankton. New Phytol. 198, 52–58. ( 10.1111/nph.12143) [DOI] [PubMed] [Google Scholar]
- 3.Young JN, Goldman JAL, Kranz SA, Tortell PD, Morel FMM. 2015. Slow carboxylation of Rubisco constrains the rate of carbon fixation during Antarctic phytoplankton blooms. New Phytol. 205, 172–181. ( 10.1111/nph.13021) [DOI] [PubMed] [Google Scholar]
- 4.Young JN, Heureux AMC, Sharwood RE, Rickaby REM, Morel FMM, Whitney SM. 2016. Large variation in the Rubisco kinetics of diatoms reveals diversity among their carbon-concentrating mechanisms. J. Exp. Bot. 67, 3445–3456. ( 10.1093/jxb/erw163) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tréguer P, Nelson DM, Van Bennekom AJ, Demaster DJ, Leynaert A, Quéguiner B. 1995. The silica balance in the world ocean: a reestimate. Science 268, 375–379. ( 10.1126/science.268.5209.375) [DOI] [PubMed] [Google Scholar]
- 6.Falkowski P, et al. 2000. The global carbon cycle: a test of our knowledge of earth as a system. Science 290, 291–296. ( 10.1126/science.290.5490.291) [DOI] [PubMed] [Google Scholar]
- 7.Rotatore C, Colman B, Kuzma M. 1995. The active uptake of carbon dioxide by the marine diatoms Phaeodactylum ticornutum and Cyclotella sp. Plant Cell Environ. 18, 913–918. ( 10.1111/j.1365-3040.1995.tb00600.x) [DOI] [Google Scholar]
- 8.Colman B, Rotatore C. 1995. Photosynthetic inorganic carbon uptake and accumulation in two marine diatoms. Plant Cell Environ. 18, 919–924. ( 10.1111/j.1365-3040.1995.tb00601.x) [DOI] [Google Scholar]
- 9.Korb RE, Saville PJ, Johnston AM, Raven JA. 1997. Sources of inorganic carbon for photosynthesis by three species of marine diatom. J. Phycol. 33, 433–440. ( 10.1111/j.0022-3646.1997.00433.x) [DOI] [Google Scholar]
- 10.Burkhardt S, Amoroso G, Riebesell U. 2001. CO2 and HCO3− uptake in marine diatoms acclimated to different CO2 concentrations. Limnol. Oceanogr. 46, 1378–1391. ( 10.4319/lo.2001.46.6.1378) [DOI] [Google Scholar]
- 11.Rost B, Burkhardt S, Sültemeyer D, Riebesell U, Burkhardt S, Sültemeyer D. 2003. Carbon acquisition of bloom-forming marine phytoplankton. Limnol. Oceanogr. 48, 55–67. ( 10.4319/lo.2003.48.1.0055) [DOI] [Google Scholar]
- 12.Trimborn S, Lundholm N, Thoms S, Richter KU, Krock B, Hansen PJ, Rost B. 2008. Inorganic carbon acquisition in potentially toxic and non-toxic diatoms: the effect of pH-induced changes in seawater carbonate chemistry. Physiol. Plant. 133, 92–105. ( 10.1111/j.1399-3054.2007.01038.x) [DOI] [PubMed] [Google Scholar]
- 13.Satoh D, Hiraoka Y, Colman B, Matsuda Y. 2001. Physiological and molecular biological characterization of intracellular carbonic anhydrase from the marine diatom Phaeodactylum tricornutum. Plant Physiol. 126, 1459–1470. ( 10.1104/pp.126.4.1459) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tanaka Y, Nakatsuma D, Harada H, Ishida M, Matsuda Y. 2005. Localization of soluble β-carbonic anhydrase in the marine diatom Phaeodactylum tricornutum. Sorting to the chloroplast and cluster formation on the girdle lamellae. Plant Physiol. 138, 207–217. ( 10.1104/pp.104.058982) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tachibana M, Allen AE, Kikutani S, Endo Y, Bowler C, Matsuda Y. 2011. Localization of putative carbonic anhydrases in two marine diatoms, Phaeodactylum tricornutum and Thalassiosira pseudonana. Photosynth. Res. 109, 205–221. ( 10.1007/s11120-011-9634-4) [DOI] [PubMed] [Google Scholar]
- 16.Hopkinson BM, Dupont CL, Allen AE, Morel FMM. 2011. Efficiency of the CO2-concentrating mechanism of diatoms. Proc. Natl Acad. Sci. USA 108, 3830–3837. ( 10.1073/pnas.1018062108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kikutani S, Tanaka R, Yamazaki Y, Hara S, Hisabori T, Kroth PG, Matsuda Y. 2012. Redox regulation of carbonic anhydrases via thioredoxin in chloroplast of the marine diatom Phaeodactylum tricornutum. J. Biol. Chem. 287, 20 689–20 700. ( 10.1074/jbc.M111.322743) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roberts SB, Lane TW, Morel FMM. 1997. Carbonic anhydrase in the marine diatom Thalassiosira weissflogii (Bacillariophyceae). J. Phycol. 33, 845–850. ( 10.1111/j.0022-3646.1997.00845.x) [DOI] [Google Scholar]
- 19.Lane TW, Morel FM. 2000. A biological function for cadmium in marine diatoms. Proc. Natl Acad. Sci. USA 97, 4627–4631. ( 10.1073/pnas.090091397) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lane TW, Saito MA, George GN, Pickering IJ, Prince RC, Morel FMM. 2005. Biochemistry: a cadmium enzyme from a marine diatom. Nature 435, 42 ( 10.1038/435042a) [DOI] [PubMed] [Google Scholar]
- 21.Xu Y, Feng L, Jeffrey PD, Shi Y, Morel FMM. 2008. Structure and metal exchange in the cadmium carbonic anhydrase of marine diatoms. Nature 452, 56–61. ( 10.1038/nature06636) [DOI] [PubMed] [Google Scholar]
- 22.Kikutani S, Nakajima K, Nagasato C, Tsuji Y, Miyatake A, Matsuda Y. 2016. Thylakoid luminal θ-carbonic anhydrase critical for growth and photosynthesis in the marine diatom Phaeodactylum tricornutum. Proc. Natl Acad. Sci. USA 113, 9828–9833. ( 10.1073/pnas.1603112113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Samukawa M, Shen C, Hopkinson BM, Matsuda Y. 2014. Localization of putative carbonic anhydrases in the marine diatom, Thalassiosira pseudonana. Photosynth. Res. 121, 235–249. ( 10.1007/s11120-014-9967-x) [DOI] [PubMed] [Google Scholar]
- 24.Mackinder LCM, et al. 2016. A repeat protein links Rubisco to form the eukaryotic carbon-concentrating organelle. Proc. Natl Acad. Sci. USA 113, 5958–5963. ( 10.1073/pnas.1522866113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nakajima K, Tanaka A, Matsuda Y. 2013. SLC4 family transporters in a marine diatom directly pump bicarbonate from seawater. Proc. Natl Acad. Sci. USA 110, 1767–1772. ( 10.1073/pnas.1216234110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harada H, Nakajima K, Sakaue K, Matsuda Y. 2006. CO2 sensing at ocean surface mediated by cAMP in a marine diatom. Plant Physiol. 142, 1318–1328. ( 10.1104/pp.106.086561) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ohno N, Inoue T, Yamashiki R, Nakajima K, Kitahara Y, Ishibashi M, Matsuda Y. 2012. CO2-cAMP-responsive cis-elements targeted by a transcription factor with CREB/ATF-like basic zipper domain in the marine diatom Phaeodactylum tricornutum. Plant Physiol. 158, 499–513. ( 10.1104/pp.111.190249) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hennon GMM, Ashworth J, Groussman RD, Berthiaume C, Morales RL, Baliga NS, Orellana MV, Armbrust EV. 2015. Diatom acclimation to elevated CO2 via cAMP signalling and coordinated gene expression. Nat. Clim. Chang. 5, 761–765. ( 10.1038/nclimate2683) [DOI] [Google Scholar]
- 29.Tanaka A, Ohno N, Nakajima K, Matsuda Y. 2016. Light and CO2/cAMP signal cross talk on the promoter elements of chloroplastic β-carbonic anhydrase genes in the marine diatom Phaeodactylum tricornutum. Plant Physiol. 170, 1105–1116. ( 10.1104/pp.15.01738) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huysman MJJ, et al. 2013. AUREOCHROME1a-mediated induction of the diatom-specific cyclin dsCYC2 controls the onset of cell division in diatoms (Phaeodactylum tricornutum). Plant Cell 25, 215–228. ( 10.1105/tpc.112.106377) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hopkinson BM. 2014. A chloroplast pump model for the CO2 concentrating mechanism in the diatom Phaeodactylum tricornutum. Photosynth. Res. 121, 223–233. ( 10.1007/s11120-013-9954-7) [DOI] [PubMed] [Google Scholar]
- 32.Hopkinson BM, Dupont CL, Matsuda Y. 2016. The physiology and genetics of CO2 concentrating mechanisms in model diatoms. Curr. Opin. Plant Biol. 31, 51–57. ( 10.1016/j.pbi.2016.03.013) [DOI] [PubMed] [Google Scholar]
- 33.Patel BN, Merrett MJ. 1986. Inorganic-carbon uptake by the marine diatom Phaeodactylum tricornutum. Planta 169, 222–227. ( 10.1007/BF00392318) [DOI] [PubMed] [Google Scholar]
- 34.Johnston AM, Raven JA. 1996. Inorganic carbon accumulation by the marine diatom Phaeodactylum tricornutum. Eur. J. Phycol. 31, 285–290. ( 10.1080/09670269600651491) [DOI] [Google Scholar]
- 35.Mitchell C, Beardall J. 1996. Inorganic carbon uptake by an Antarctic sea-ice diatom, Nitzschia frigida. Polar Biol. 16, 95–99. ( 10.1007/BF02390429) [DOI] [Google Scholar]
- 36.Keeling PJ. 2013. The number, speed, and impact of plastid endosymbioses in eukaryotic evolution. Annu. Rev. Plant Biol. 64, 583–607. ( 10.1146/annurev-arplant-050312-120144) [DOI] [PubMed] [Google Scholar]
- 37.Rotatore C, Colman B. 1992. Active uptake of CO2 by the diatom Navicula pelliculosa. J. Exp. Bot. 43, 571–576. ( 10.1093/jxb/43.4.571) [DOI] [Google Scholar]
- 38.Kroth PG, et al. 2008. A model for carbohydrate metabolism in the diatom Phaeodactylum tricornutum deduced from comparative whole genome analysis. PLoS ONE 3, e1426 ( 10.1371/journal.pone.0001426) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gruber A, Vugrinec S, Hempel F, Gould SB, Maier UG, Kroth PG. 2007. Protein targeting into complex diatom plastids: functional characterisation of a specific targeting motif. Plant Mol. Biol. 64, 519–530. ( 10.1007/s11103-007-9171-x) [DOI] [PubMed] [Google Scholar]
- 40.Lee RE, Kugrens P. 1998. Hypothesis: the ecological advantage of chloroplast ER—the ability to outcompete at low dissolved CO2 concentrations. Protist 149, 341–345. ( 10.1016/S1434-4610(98)70040-9) [DOI] [PubMed] [Google Scholar]
- 41.Lee RE, Kugrens P. 2000. Ancient atmospheric CO2 and the timing of evolution of secondary endosymbioses. Phycologia 39, 167–172. ( 10.2216/i0031-8884-39-2-167.1) [DOI] [Google Scholar]
- 42.Sullivan CW, Volcani BE. 1974. Synergistically stimulated (Na+,K+)-adenosine triphosphatase from plasma membrane of a marine diatom. Proc. Natl Acad. Sci. USA 71, 4376–4380. ( 10.1073/pnas.71.11.4376) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rees TAV. 1984. Sodium dependent photosynthetic oxygen evolution in a marine diatom. J. Exp. Bot. 35, 332–337. ( 10.1093/jxb/35.3.332) [DOI] [Google Scholar]
- 44.Chen X, Qiu CE, Shao JZ. 2006. Evidence for K+-dependent HCO3− utilization in the marine diatom Phaeodactylum tricornutum. Plant Physiol. 141, 731–736. ( 10.1104/pp.106.079616) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kaplan A, Scherer S, Lerner M. 1989. Nature of the light-induced H+ efflux and Na+ uptake in cyanobacteria. Plant Physiol. 89, 1220–1225. ( 10.1104/PP.89.4.1220) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sültemeyer DF, Fock HP, Canvin DT. 1991. Active uptake of inorganic carbon by Chlamydomonas reinhardtii: evidence for simultaneous transport of HCO3− and CO2 and characterization of active CO2 transport. Can. J. Bot. 69, 995–1002. ( 10.1139/b91-128) [DOI] [Google Scholar]
- 47.Tchernov D, Helman Y, Keren N, Luz B, Ohad I, Reinhold L, Ogawa T, Kaplan A. 2001. Passive entry of CO2 and its energy-dependent intracellular conversion to HCO3− in cyanobacteria are driven by a photosystem I-generated ΔμH+. J. Biol. Chem. 276, 23 450–23 455. ( 10.1074/jbc.M101973200) [DOI] [PubMed] [Google Scholar]
- 48.Bailleul B, et al. 2015. Energetic coupling between plastids and mitochondria drives CO2 assimilation in diatoms. Nature 524, 366–369. ( 10.1038/nature14599) [DOI] [PubMed] [Google Scholar]
- 49.Gutknecht J, Bisson MA, Tosteson FC. 1977. Diffusion of carbon dioxide through lipid bilayer membranes: effects of carbonic anhydrase, bicarbonate, and unstirred layers. J. Gen. Physiol. 69, 779–794. ( 10.1085/jgp.69.6.779) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ogawa T, Mi H. 2007. Cyanobacterial NADPH dehydrogenase complexes. Photosynth. Res. 93, 69–77. ( 10.1007/s11120-006-9128-y) [DOI] [PubMed] [Google Scholar]
- 51.Mi H, Endo T, Ogawa T, Asada K. 1995. Thylakoid membrane-bound, NADPH-specific pyridine nucleotide dehydrogenase complex mediates cyclic electron transport in the cyanobacterium Synechocystis sp. PCC 6803. Plant Cell Physiol. 36, 661–668. [Google Scholar]
- 52.He Z, Zheng F, Wu Y, Li Q, Lv J, Fu P, Mi H. 2015. NDH-1 L interacts with ferredoxin via the subunit NdhS in Thermosynechococcus elongatus. Photosynth. Res. 126, 341–349. ( 10.1007/s11120-015-0090-4) [DOI] [PubMed] [Google Scholar]
- 53.Peña KL, Castel SE, de Araujo C, Espie GS, Kimber MS. 2001. Structural basis of the oxidative activation of the carboxysomal γ-carbonic anhydrase, CcmM. Proc. Natl Acad. Sci. USA 107, 2455–2460. ( 10.1073/pnas.0910866107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Alber BE, Ferry JG. 1994. A carbonic anhydrase from the archaeon Methanosarcina thermophila. Proc. Natl Acad. Sci. USA 91, 6909–6913. ( 10.1073/pnas.91.15.6909) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.DiMario RJ, Clayton H, Mukherjee A, Ludwig M, Moroney JV. 2016. Plant carbonic anhydrases: structures, locations, evolution, and physiological roles. Mol. Plant 10, 30–46. ( 10.1016/j.molp.2016.09.001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Burns BD, Beardall J. 1987. Utilization of inorganic carbon by marine microalgae. J. Exp. Mar. Biol. Ecol. 107, 75–86. ( 10.1016/0022-0981(87)90125-0) [DOI] [Google Scholar]
- 57.Kaldenhoff R, Kai L, Uehlein N. 2014. Aquaporins and membrane diffusion of CO2 in living organisms. Biochim. Biophys. Acta 1840, 1592–1595. ( 10.1016/j.bbagen.2013.09.037) [DOI] [PubMed] [Google Scholar]
- 58.Uehlein N, Lovisolo C, Siefritz F, Kaldenhoff R. 2003. The tobacco aquaporin NtAQP1 is a membrane CO2 pore with physiological functions. Nature 425, 734–737. ( 10.1038/nature02027) [DOI] [PubMed] [Google Scholar]
- 59.Tu C, Wynns GC, McMurray RE, Silverman DN. 1978. CO2 kinetics in red cell suspensions measured by 18O exchange. J. Biol. Chem. 253, 8178–8184. [PubMed] [Google Scholar]
- 60.Endeward V, et al. 2006. Evidence that aquaporin 1 is a major pathway for CO2 transport across the human erythrocyte membrane. FASEB J. 20, 1974–1981. ( 10.1096/fj.04-3300com) [DOI] [PubMed] [Google Scholar]
- 61.Chegwidden WR, Carter ND. 2000. Introduction to the carbonic anhydrases. In The carbonic anhydrases: new horizons, vol. 9 (eds WR Chegwidden, ND Carter, YH Edwards), pp. 13–28. Basel, Switzerland: Birkhaeuser. ( 10.1007/978-3-0348-8446-4_2) [DOI] [PubMed] [Google Scholar]
- 62.Lee RBY, Smith JAC, Rickaby REM. 2013. Cloning, expression and characterization of the δ-carbonic anhydrase of Thalassiosira weissflogii (Bacillariophyceae). J. Phycol. 49, 170–177. ( 10.1111/j.1529-8817.2012.01226.x) [DOI] [PubMed] [Google Scholar]
- 63.Del Prete S, Vullo D, Scozzafava A, Capasso C, Supuran CT. 2014. Cloning, characterization and anion inhibition study of the δ-class carbonic anhydrase (TweCA) from the marine diatom Thalassiosira weissflogii. Bioorg. Med. Chem. 22, 531–537. ( 10.1016/j.bmc.2013.10.045) [DOI] [PubMed] [Google Scholar]
- 64.Hopkinson BM, Meile C, Shen C. 2013. Quantification of extracellular carbonic anhydrase activity in two marine diatoms and investigation of its role. Plant Physiol. 162, 1142–1152. ( 10.1104/pp.113.217737) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nimer NA, Iglesias-Rodriguez MD, Merrett MJ. 1997. Bicarbonate utilization by marine phytoplankton species. J. Phycol. 33, 625–631. ( 10.1111/j.0022-3646.1997.00625.x) [DOI] [Google Scholar]
- 66.Shen C, Hopkinson BM. 2015. Size scaling of extracellular carbonic anhydrase activity in centric marine diatoms. J. Phycol. 51, 255–263. ( 10.1111/jpy.12269) [DOI] [PubMed] [Google Scholar]
- 67.Moroney JV, Ma Y, Frey WD, Fusilier KA, Pham TT, Simms TA, DiMario RJ, Yang J, Mukherjee B. 2011. The carbonic anhydrase isoforms of Chlamydomonas reinhardtii: intracellular location, expression, and physiological roles. Photosynth. Res. 109, 133–149. ( 10.1007/s11120-011-9635-3) [DOI] [PubMed] [Google Scholar]
- 68.Wang Y, Spalding MH. 2006. An inorganic carbon transport system responsible for acclimation specific to air levels of CO2 in Chlamydomonas reinhardtii. Proc. Natl Acad. Sci. USA 103, 10 110–10 115. ( 10.1073/pnas.0603402103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mitra M, Lato SM, Ynalvez RA, Xiao Y, Moroney JV. 2004. Identification of a new chloroplast carbonic anhydrase in Chlamydomonas reinhardtii. Plant Physiol. 135, 173–182. ( 10.1104/pp.103.037283) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yamano T, Tsujikawa T, Hatano K, Ozawa S, Takahashi Y, Fukuzawa H. 2010. Light and low-CO2-dependent LCIB-LCIC complex localization in the chloroplast supports the carbon-concentrating mechanism in Chlamydomonas reinhardtii. Plant Cell Physiol. 51, 1453–1468. ( 10.1093/pcp/pcq105) [DOI] [PubMed] [Google Scholar]
- 71.Wang Y, Spalding MH. 2014. Acclimation to very low CO2: contribution of limiting CO2 inducible proteins, LCIB and LCIA, to inorganic carbon uptake in Chlamydomonas reinhardtii. Plant Physiol. 166, 2040–2050. ( 10.1104/pp.114.248294) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jenks A, Gibbs SP. 2000. Immunolocalization and distribution of form II Rubisco in the pyrenoid and chloroplast stroma of Amphidinium carterae and form I Rubisco in the symbiont-derived plastids of Peridinium foliaceum (Dinophyceae). J. Phycol. 36, 127–138. ( 10.1046/j.1529-8817.2000.99114.x) [DOI] [Google Scholar]
- 73.Matsuda Y, Hara T, Colman B. 2001. Regulation of the induction of bicarbonate uptake by dissolved CO2 in the marine diatom, Phaeodactylum tricornutum. Plant Cell Environ. 24, 611–620. ( 10.1046/j.1365-3040.2001.00702.x) [DOI] [Google Scholar]
- 74.Harada H, Matsuda Y. 2005. Identification and characterization of a new carbonic anhydrase in the marine diatom Phaeodactylum tricornutum. Can. J. Bot. 83, 909–916. ( 10.1139/b05-078) [DOI] [Google Scholar]
- 75.Harada H, Nakatsuma D, Ishida M, Matsuda Y. 2005. Regulation of the expression of intracellular β-carbonic anhydrase in response to CO2 and light in the marine diatom Phaeodactylum tricornutum. Plant Physiol. 139, 1041–1050. ( 10.1104/pp.105.065185) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mayo WP, Williams TG, Birch DG, Turpin DH. 1986. Photosynthetic adaptation by Synechococcus leopoliensis in response to exogenous dissolved inorganic carbon. Plant Physiol. 80, 1038–1040. ( 10.1104/PP.80.4.1038) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Marcus Y, Harel E, Kaplan A. 1983. Adaptation of the cyanobacterium Anabaena variabilis to low CO2 concentration in their environment. Plant Physiol. 71, 208–210. ( 10.1104/PP.71.1.208) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Nishimura T, Takahashi Y, Yamaguchi O, Suzuki H, Maeda S, Omata T. 2008. Mechanism of low CO2-induced activation of the cmp bicarbonate transporter operon by a LysR family protein in the cyanobacterium Synechococcus elongatus strain PCC 7942. Mol. Microbiol. 68, 98–109. ( 10.1111/j.1365-2958.2008.06137.x) [DOI] [PubMed] [Google Scholar]
- 79.Shiraiwa Y, Satoh H, Hirokawa T. 1988. Factors controlling induction of carbonic anhydrase in Chlorella vulgaris 11h: effects of CO2 and O2. Plant Cell Physiol. 29, 731–734. ( 10.1093/oxfordjournals.pcp.a077554) [DOI] [Google Scholar]
- 80.Matsuda Y, Colman B. 1995. Induction of CO2 and bicarbonate transport in the green alga Chlorella ellipsoidea (II. Evidence for induction in response to external CO2 concentration). Plant Physiol. 108, 253–260. ( 10.1104/PP.108.1.253) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Vance P, Spalding MH. 2005. Growth, photosynthesis, and gene expression in Chlamydomonas over a range of CO2 concentrations and CO2/O2 ratios: CO2 regulates multiple acclimation states. Can. J. Bot. 83, 796–809. ( 10.1139/b05-064) [DOI] [Google Scholar]
- 82.Matsuda Y, Kroth PG. 2014. Carbon fixation in diatoms. In The structural basis of biological energy generation, vol. 39 (ed. MF Hohmann-Marriott), pp. 335–362. Dordrecht, The Netherlands: Springer; ( 10.1007/978-94-017-8742-0_18) [DOI] [Google Scholar]
- 83.Bowler C, et al. 2008. The Phaeodactylum genome reveals the evolutionary history of diatom genomes. Nature 456, 239–244. ( 10.1038/nature07410) [DOI] [PubMed] [Google Scholar]
- 84.Smith SR, et al. 2016. Transcriptional orchestration of the global cellular response of a model pennate diatom to diel light cycling under iron limitation. PLoS Genet. 12, e1006490 ( 10.1371/journal.pgen.1006490) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Aizawa K, Miyachi S. 1986. Carbonic anhydrase and CO2 concentrating mechanisms in microalgae and cyanobacteria. FEMS Microbiol. Lett. 39, 215–233. ( 10.1016/0378-1097(86)90447-7) [DOI] [Google Scholar]
- 86.Reinhold L, Zviman M, Kaplan A. 1987. Inorganic carbon fluxes and photosynthesis in cyanobacteria—a quantitative model. In Progress in photosynthesis research, vol. 4 (ed. J Biggins), pp. 289–296. Dordrecht, The Netherlands: Springer; ( 10.1007/978-94-017-0519-6_62) [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This article has no data.