Abstract
Diatoms accumulate triacylglycerols in spherical organelles called oil bodies when exposed to nutrient deprivation conditions. Oil body biology in diatoms has attracted significant attention due to the complexity of the intracellular organelles and the unique combination of genes generated by the evolutionary history of secondary endosymbiosis. The demand for biofuel production has further increased the interest in and importance of a better understanding of oil body biology in diatoms, because it could provide targets for genetic engineering to further enhance their promising lipid accumulation. This review describes recent progress in studies of the structure and properties of diatom oil bodies. Firstly, the general features of diatom oil bodies are described, in particular, their number, size and morphology, as well as the quantity and quality of lipids they contain. Subsequently, the diatom oil body-associated proteins, which were recently discovered through oil body proteomics, are introduced. Then, the metabolic pathways responsible for the biogenesis and degradation of diatom oil bodies are summarized. During biogenesis and degradation, oil bodies interact with other organelles, including chloroplasts, the endoplasmic reticulum and mitochondria, suggesting their dynamic nature in response to environmental changes. Finally, the functions of oil bodies in diatoms are discussed.
This article is part of the themed issue ‘The peculiar carbon metabolism in diatoms’.
Keywords: diatom, oil body, triacylglycerol, oil body-associated protein, lipid metabolism, biofuels
1. Introduction
Diatoms are photosynthetic microalgae that are believed to be responsible for 20% of the primary production on the Earth [1,2]. Primary production occurs principally through CO2 fixation via photosynthesis, in which inorganic carbon is converted into organic biomacromolecules that are stored as reserve materials or used to construct cellular components. In diatoms, the main carbon storage compounds are triacylglycerols (TAGs, which are neutral lipids) and chrysolaminarin (a carbohydrate). Diatoms tend to produce TAGs rather than chrysolaminarin when exposed to nutrient (e.g. nitrogen, phosphorus and silicon) deprivation, and the massively produced TAGs are accumulated in the specific lipid-storage droplets. The terminology to represent the lipid-storage droplets in diatoms is not unified, and they are called oil bodies [3,4], lipid droplets [5,6], oil droplets [7] or lipid bodies [8]. In this article, we use the term ‘oil bodies’ because the lipid-storage droplets are likely to be the organelle which is structurally composed of lipids and other biomacromolecules including proteins and is functionally dynamic as described hereinafter, rather than inert droplets of lipid molecules. Although TAG accumulation in oil bodies occurs in a wide range of organisms, including animals, plants and microalgae, oil body biology in diatoms has attracted broad interest because of their high species diversity [9], the evolutionary history of secondary endosymbiosis and the resulting complex intracellular structures [10,11], and the unique combination of genes involved, including not only plant-like and diatom-specific genes but also a substantial number of animal-like [11] and bacterial genes [12]. Nonetheless, although there are a number of reviews describing oil body biology in green algae (such as Chlamydomonas reinhardtii), including their chemical composition, TAG biosynthetic pathways, contributing organelles, variation of intracellular structures during TAG accumulation, and function [13–15], there are few reviews of oil body biology in diatoms [16].
Lipid production in diatoms has also drawn attention for its potential use in biofuel production. As we face the exhaustion of fossil fuel reserves and their negative impact on climate, energy production without the release of massive amounts of CO2 is critical for sustainable development of our society. Biodiesel fuels derived from microalgal lipids have been recognized as promising resources to meet this demand; thus, a number of oleaginous microalgae have been intensively studied as candidate lipid producers (e.g. Nannochloropsis (Eustigmatophyceae) [17,18], Chlorella vulgaris (Chlorophyceae) [19], Fistulifera solaris (Bacillariophyceae) [20] and Cyclotella cryptica (Bacillariophyceae) [21,22]). Diatoms have high potential for biofuel production due to their high lipid productivity, ease of mass cultivation, availability of omics data and genetic manipulation methods, and simultaneous production of value-added products [16]. However, additional efforts (e.g. genetic engineering) are needed to make biofuels economically viable. Towards this goal, a better understanding of oil body biology at the molecular level in diatoms is essential.
In this review, we summarize a wide range of recent studies on oil body biology in diatoms. Firstly (§2), the general features of diatom oil bodies are presented, including their number, size and morphology, as well as their fatty acid composition and neutral lipid content (mainly TAGs), which are the major components of diatom oil bodies. Subsequently (§3), oil body-associated proteins, another important component, are described. Recent progress on oil body proteomes in diatoms is reviewed, and compared to similar studies in diverse microalgae (§3c). Finally (§4), the metabolic pathways for oil body biogenesis and the functional roles of the oil body are described and discussed. Recent studies on the biogenesis and degradation of oil bodies in diatoms have suggested that oil bodies are not simply inert lipid droplets that served as an irreversible storage of energy and carbon, but rather dynamic organelles. By reading this review, readers will obtain comprehensive information on oil body biology in diatoms.
2. General features of oil bodies in diatoms
(a). Size, number and morphology of diatom oil bodies
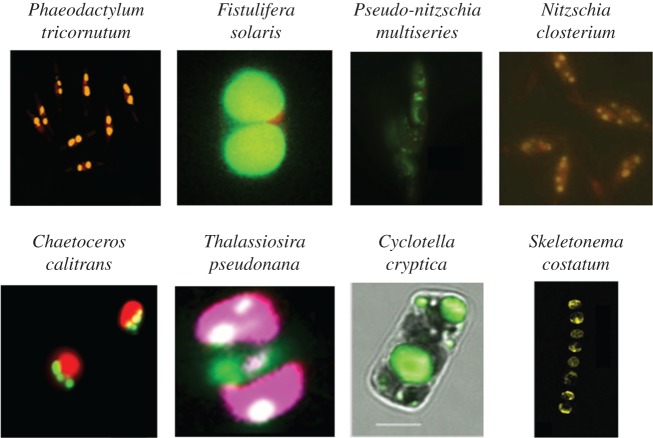
Microalgae accumulate neutral lipids under various stress conditions, and the accumulated lipids are stored in specific organelles called oil bodies [13–15]. Diatoms also accumulate neutral lipids in oil bodies in response to nutrient deprivation (figure 1) [20,23–29], most commonly studied under nitrogen deprivation [4], but also under phosphorus [30] and silicon [5,31,32] deprivation. Oil bodies are readily observed by staining with lipophilic fluorescence dyes such as Nile Red or BODIPY 505/515 [26,33,34]. These staining methods allow us to measure the size and number of oil bodies in the cells. The size and number of oil bodies in several diatoms are summarized in table 1 [20,22–29,32], although these features vary according to lipid accumulation phase. It is important to know these variation ranges because, from this information, we can estimate whether the observed cells are going to accumulate oil, or have already accumulated extensive oil. The localization of oil bodies is species-dependent (figure 1). For example, the oleaginous diatom Fistulifera solaris always has two oil bodies at the polar regions of the cell. This suggests that there are diverse mechanisms that control oil body localization in the cells of diatoms. Oil bodies are generally spherical. However, in some oleaginous diatoms, morphological changes were observed in the late oil accumulation phase [35]. These changes may be due to spatial constraints caused by the shape of the cells and other intracellular organelles, suggesting the elastic nature of the oil body surface.
Figure 1.
Morphology of the oil bodies in various diatom species [20,23–29]. Oil bodies are stained with Nile Red (yellow) or BODIPY 505/515 (green). The red fluorescence is derived from the chloroplasts, except in Thalassiosira pseudonana in which fluorescent protein-labelled chloroplasts are shown in magenta. The figures are reproduced from previous studies [20,23–29], with some modifications. Pseudo-nitzschia multiseries is reproduced from [24] with permission. Copyright © Elsevier Masson SAS.
Table 1.
| order | species | size (µm) | number | references |
|---|---|---|---|---|
| Pennales | Phaeodactylum tricornutum | 0.4–2.6 | 1–5 | [23] |
| Fistulifera solaris | 1.3–5.6 | 2 | [20] | |
| Pseudo-nitzschia multiseries | 0.7–1.4 | 6–15 | [24] | |
| Nitzschia closterium | 0.8–1.3 | 3–4 | [25] | |
| Centrales | Chaetoceros calitrans | 1.0–2.0 | 3–4 | [26] |
| Thalassiosira pseudonana | 0.3–1.5 | 3–5 | [27,32] | |
| Cyclotella cryptica | 1.1–3.9 | 1–10 | [22,28] | |
| Skeletonema costatum | 1.1–2.3 | 3–5 | [29] |
(b). Lipid content and productivity in diatoms
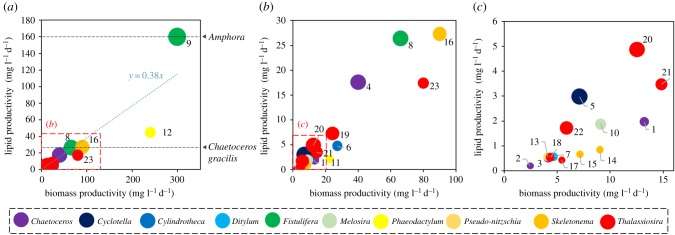
The microscale phenotypes of the size and number of oil bodies in the cells are directly correlated to the lipid productivity of each microalga which is cultured in large-scale bioreactors. For efficient biofuel production, it is important to investigate the lipid productivity of microalgae in order to select promising candidates of biofuel producers. These values vary widely among diatom species (table 2 and figure 2) [17,36–41]. In addition, even in the same species, biomass and lipid productivity can change drastically under different culture conditions (for example, compare nos. 8 and 9 for F. solaris and nos. 11 and 12 for Phaeodactylum tricornutum in table 2 and figure 2). It should be noted that the values in table 2 and figure 2 may not represent the upper limits of biomass and lipid productivity for each diatom species, because the culture conditions used in these studies may not be the optimal conditions for lipid production. Among the listed diatom species, F. solaris has one of the highest rates of lipid production (biomass productivity, 299 mg l−1 d−1; lipid productivity, 160 mg l−1 d−1; lipid content, 53.5% [38]). In addition, three strains of Amphora (AMPHO27, AMPHO45 and AMPHO46) were reported to show high lipid productivity (345, 63 and 71 mg l−1 d−1, respectively; 160 mg l−1 d−1 on average) [39,40]. However, no further investigations were performed. Although the lipid productivity of Chaetoceros gracilis was reported to be 2210 mg l−1 d−1 [42], a recent study using the same species determined that TAG productivity under optimal conditions was 27.2 mg l−1 d−1 [41]. This difference might be attributed to culture conditions and strains used in each study. Because the biomass productivity of Amphora and C. gracilis was not reported in the same units (mg l−1 d−1), only the lipid productivity is presented in figure 2. The slope of the fitted curve in the linear approximation is 0.38, suggesting a general lipid content of 38%, which is comparable to that of other microalgal classes [43].
Table 2.
| species | no. in figure 2 | biomass productivity (mg l−1 day−1) | lipid productivity (mg l−1 day−1) | lipid content (% dry mass) | culture conditions (gas supply, lighta, temperature) | references |
|---|---|---|---|---|---|---|
| Amphora spp. (AMPHO27, AMPHO45, and AMPHO46) | — | — | av. 160 (345, 63, and 71) | — | air, 200 µmol m−2 s−1 (12 L : 12 D cycle), 30°C | [39,40] |
| Chaetoceros curvisetus CCMP 3260 | 1 | 13.22 | 1.97 | 14.86 | air, 200 µmol m−2 s−1 (14 L : 10 D cycle), 20°C | [36] |
| Chaetoceros socialis CCMP 3263 | 2 | 2.47 | 0.19 | 7.63 | air, 200 µmol m−2 s−1 (14 L : 10 D cycle), 20°C | [36] |
| Chaetoceros affinis CCMP 3259 | 3 | 4.13 | 0.5 | 12.11 | air, 200 µmol m−2 s−1 (14 L : 10 D cycle), 20°C | [36] |
| Chaetoceros calcitrans CS 178 | 4 | 40 | 17.6 | 39.8 | air, 200 µmol m−2 s−1 (14 L : 10 D cycle), 20°C | [17] |
| Chaetoceros gracilis Schutt UTEX LB 2658 | — | — | 27.2 | — | air, 50 µmol m−2 s−1, 20°C, 1% NaCl | [41] |
| Cyclotella cryptica CCMP 331 | 5 | 7.11 | 2.98 | 41.97 | air, 200 µmol m−2 s−1 (14 L : 10 D cycle), 20°C | [36] |
| Cylindrotheca fusiformis CCMP 343 | 6 | 27.27 | 4.78 | 17.51 | air, 200 µmol m−2 s−1 (14 L : 10 D cycle), 20°C | [36] |
| Ditylum brightwelli CCMP 358 | 7 | 4.66 | 0.57 | 12.2 | air, 200 µmol m−2 s−1 (14 L : 10 D cycle), 20°C | [36] |
| Fistulifera solaris JPCC DA0580 | 8 | 66 | 26.4 | 40 | air, 140 µmol m−2 s−1 (24 h) | [37] |
| Fistulifera solaris JPCC DA0580 | 9 | 299 | 160 | 53.5 | 2% CO2, 200 µmol m−2 s−1 (24 h), 25°C, nutrient deprivation | [38] |
| Melosira octogona CCMP 483 | 10 | 9.1 | 1.88 | 20.6 | air, 200 µmol m−2 s−1 (14 L : 10 D cycle), 20°C | [36] |
| Phaeodactylum tricornutum CCMP 632 | 11 | 22.44 | 2.09 | 9.32 | air, 200 µmol m−2 s−1 (14 L : 10 D cycle), 20°C | [36] |
| Phaeodactylum tricornutum F&M-M40 | 12 | 240 | 44.8 | 18.7 | 5% CO2, 100 µmol m−2 s−1 (24 h), 25°C | [17] |
| Pseudo-nitzschia pseudodelicatissima B317 | 13 | 4.12 | 0.56 | 13.5 | air, 200 µmol m−2 s−1 (14 L : 10 D cycle), 20°C | [36] |
| Skeletonema marinoi CCMP 2092 | 14 | 9.02 | 0.85 | 9.38 | air, 200 µmol m−2 s−1 (14 L : 10 D cycle), 20°C | [36] |
| Skeletonema marinoi CCMP 2052 | 15 | 7.17 | 0.66 | 9.14 | air, 200 µmol m−2 s−1 (14 L : 10 D cycle), 20°C | [36] |
| Skeletonema sp. CS 252 | 16 | 90 | 27.3 | 31.8 | 5% CO2, 100 µmol m−2 s−1 (24 h), 25°C | [17] |
| Thalassiosira rotula CCMP 1647 | 17 | 5.44 | 0.43 | 7.95 | air, 200 µmol m−2 s−1 (14 L : 10 D cycle), 20°C | [36] |
| Thalassiosira rotula CCMP 3264 | 18 | 4.35 | 0.54 | 12.42 | air, 200 µmol m−2 s−1 (14 L : 10 D cycle), 20°C | [36] |
| Thalassiosira weissflogii P09 | 19 | 24.29 | 7.27 | 29.94 | air, 200 µmol m−2 s−1 (14 L : 10 D cycle), 20°C | [36] |
| Thalassiosira weissflogii CCMP 1010 | 20 | 12.53 | 4.87 | 38.84 | air, 200 µmol m−2 s−1 (14 L : 10 D cycle), 20°C | [36] |
| Thalassiosira weissflogii CCMP 1336 | 21 | 14.84 | 3.48 | 23.48 | air, 200 µmol m−2 s−1 (14 L : 10 D cycle), 20°C | [36] |
| Thalassiosira pseudonana CCMP 1335 | 22 | 5.87 | 1.72 | 29.33 | air, 200 µmol m−2 s−1 (14 L : 10 D cycle), 20°C | [36] |
| Thalassiosira pseudonana CS 173 | 23 | 80 | 17.4 | 20.6 | 5% CO2, 100 µmol m−2 s−1 (24 h), 25°C | [17] |
a(12 L : 12 D) stands for 12 h light and 12 h dark conditions.
Figure 2.
The biomass and lipid productivity of diatoms [17,36–41]. For Amphora (the average value of three strains [39,40]) and Chaetoceros gracilis [41], only lipid productivity is shown. (b) and (c) Magnifications of the insets in (a) and (b), respectively. Circle size represents lipid content (%). The numbers correspond to those shown in table 2.
(c). Fatty acid composition
As is the case with lipid quantity, lipid quality, which is mainly dictated by fatty acid composition, is greatly affected by physiological stage and culture conditions. The variations in fatty acid composition between the different oil accumulation phases were investigated for several diatom species, including P. tricornutum, Thalassiosira pseudonana [31,32,44], F. solaris [38], Nitzschia closterium [45] and C. gracilis [41], and these studies provided insights into lipid metabolism during adaptation to environmental changes. Table 3 shows the fatty acid composition of the oil bodies produced in these diatoms under oil accumulation conditions. In general, the major fatty acids are C14 : 0, C16 : 0 and C16 : 1, and the major polyunsaturated fatty acid (PUFA) is eicosapentaenoic acid (EPA, C20 : 5); these are much different from those of green algae, which contain substantial amounts of C18 fatty acids [43]. Fatty acid composition is a critical factor for evaluating the potential of using microalgae for biodiesel application, because it directly determines biodiesel quality [46]. There are some standards for evaluating the quality of biodiesel (e.g. American Society for Testing and Materials (ASTM) D6751 in the United States and EN 14214 in Europe). The automotive fuels-fatty acid methyl ester as a blend stock specification, and the Japanese Industrial Standard K 2390 : 2008, has also been established with reference to the EN14214 standard. To meet these standards, the PUFA content (in particular, the number of PUFAs containing at least three double bonds) should be low. Therefore, the fatty acid composition of diatoms, which is predominantly C16 : 0 and C16 : 1, would be advantageous for biofuel applications.
Table 3.
Fatty acid composition of the oil bodies in diatoms [38,41,44,45] ((%mol), n.d., not detected; —, no data).
| species | C14 : 0 | C16 : 0 | C16 : 1 | C16 : 2 | C16 : 3 | C18 : 0 | C18 : 1 | C18 : 2 | C18 : 3 | C20 : 5 | C22 : 6 | references |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Phaeodactylum tricornutum | 6.7 | 30.59 | 51.91 | — | — | n.d. | 3.61 | — | — | 5.38 | n.d. | [44] |
| Thalassiosira pseudonana | 5.98 | 29.43 | 28.77 | — | — | 0.36 | 2 | — | — | 7.51 | 0.69 | [44] |
| Fistulifera solaris | 3.7 | 40.6 | 45.2 | 0.2 | 0.2 | 0.5 | 1.5 | 0.4 | 0.7 | 5.8 | — | [38] |
| Nitzschia closterium | 5.4 | 21.82 | 47.54 | 2.24 | 2.4 | 0.9 | 7.12 | 0.31 | 0.25 | 10.43 | 0.22 | [45] |
| Chaetoceros gracillis | 6.7 | 42.1 | 40.8 | 2.7 | 1.5 | 1.1 | — | — | — | 3.9 | — | [41] |
3. Diatom oil body-associated proteins
It has been widely accepted that the oil bodies in animals [47] and higher plants [48] are functional organelles which are composed of neutral lipid droplets, phospholipids surrounding the droplets and several types of proteins which are associated with these lipid molecules [49,50]. These oil body-associated proteins likely play important roles in oil body biology, and some of such proteins make (or help) the oil body's functionally active organelles. In animals, perilipin family proteins were identified as major proteins on the surface of oil bodies [51,52]. In plants, several oil body-associated proteins were identified, such as oleosin, caleosin and steroleosin [53–55]. Compared to animals and plants, investigation of algal oil body-associated proteins has been hampered, mainly due to a lack of genomic information. However, the genomes of several microalgae have recently been analysed, which has facilitated proteomic studies of microalgal oil bodies. Consequently, a number of unique oil body-associated proteins were isolated and identified from a wide range of microalgal taxonomies [56–65], including diatoms [3,6]. These findings support the notion that oil bodies in microalgae, including diatoms, are also functional organelles, although it might still be a matter of debate. As there are in depth reviews of a variety of microalgal oil body-associated proteins identified in diverse microalgae [35] and proteomic approaches for microalgae towards biofuel production [66], the structural and functional features of microalgal oil body-associated proteins are only very briefly described in this review. Hereinafter, we focus more on the unique studies of diatom oil body-associated proteins by comparing these results to those in other microalgae.
(a). Methodologies for investigation of algal oil body-associated proteins
The general approach for identifying microalgal oil body-associated proteins is divided into four steps: (i) disruption of the cells, (ii) isolation of oil bodies, (iii) extraction of the proteins from the oil bodies and (iv) identification of the proteins. The major difficulty in proteomic studies of oil body-associated proteins is isolating intact, pure oil bodies. If this is not achieved, the oil body fraction can be contaminated with proteins from other fractions (e.g. the cytoplasm, chloroplasts, nucleus and mitochondria). Therefore, (i) cell disruption and (ii) oil body isolation are the key steps for successful identification.
Table 4 summarizes the methodologies used for proteomic studies of microalgal oil body-associated proteins. For the cell wall-less mutant of the green alga C. reinhardtii strain cw15 [56] and the natively cell wall-less green alga Dunaliella strains (e.g. D. salina [57] and D. bardawil [65]), osmotic stress was frequently employed to disrupt the cells, because it was a relatively gentle method to keep the intracellular organelles intact and minimize contamination of the oil body fraction by other proteins. In some cases, other cell disruption methods, such as homogenization [63] and French press [62], were also applied. Disruption of other cell wall-containing microalgal cells was performed using harsher methods. The green alga Haematococcus pluvialis was disrupted by bead beating [58], Chlorella sp. was ground with a mortar and pestle in liquid nitrogen [64] and French press was used for Nannochloropsis oceanica (Eustigmatophyceae) and Tisochrysis lutea (Haptophyceae). Diatoms (Bacillariophyceae) present a unique methodological challenge due to the presence of silica cell walls called frustules. When studying the oleaginous diatom F. solaris [3], we used bead beating which was harsh enough to break the cells. Yoneda et al. [6] used a French press for P. tricornutum. Despite being a model diatom, P. tricornutum does not always have a silica cell wall. Thus, it might be relatively easy to disrupt these cells.
Table 4.
| taxonomic group | microalgal strain | cell disruption | oil body isolation | protein extraction | references |
|---|---|---|---|---|---|
| Chlorophyceae | Chlamydomonas reinhardtii (cell wall-less strain) | osomtic shock | centrifugation in scrose-including buffer | as is | [56] |
| Chlamydomonas reinhardtii (cell wall-less strain) | homoginator | centrifugation in scrose-including buffer | — | [63] | |
| Chlamydomonas reinhardtii (cell wall-less strain) | French press (1500 psi) | wash with detergent, salt, urea/discontinuous sucrose gradient centrifugation | as is | [62] | |
| Chlamydomonas reinhardtii | homogenator | Percoll gradient centrifugation | — | [60] | |
| Chlorella sp. | ground in liquid nitrogen by pestle and mortar | discontinuous sucrose gradient centrifugation/wash with detergent | as is | [64] | |
| Haematococcus pluvialis | beads beating | discontinuous sucrose gradient centrifugation | acetone precipitation | [58] | |
|
Dunaliella salina Dunaliella bardawill Dunaliella parva (native cell wall-less microalgae) |
osomtic shock | discontinuous sucrose gradient centrifugation | acetone precipitation | [57,65] | |
| Eustigmatophyceae | Nannochloropsis oceanica | French press (20 kpsi) | discontinuous sucrose gradient centrifugation/wash with detergent | — | [59] |
| Haptophyceae | Tisochrysis lutea (non-calcareous) | French press (0.8 kpsi) | discontinuous sucrose gradient centrifugation | acetone precipitation | [61] |
| Bacillariophyceae | Fistulifera solaris | beads beating | discontinuous sucrose gradient centrifugation | acetone precipitation | [3] |
| Phaeodactylum tricornutum | French press (1 kpsi) | discontinuous sucrose gradient centrifugation/wash with detergent | acetone precipitation | [6] |
After the cell disruption step, oil bodies are isolated from the cell debris and other intracellular fractions by centrifugation, using a discontinuous sucrose gradient for better fractionation. The oil body fraction floats to the top surface of the centrifugation sample, and forms a solidified oil phase, or the so-called lipid pad (figure 3). Therefore, it is easy to collect the oil body fraction. The oil body fraction was washed to remove proteins that are non-specifically associated with the oil body. Subsequently, the oil body-associated proteins are extracted and condensed by acetone precipitation, and subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and subsequent liquid chromatography-mass spectrometry (MS) for identification.
Figure 3.
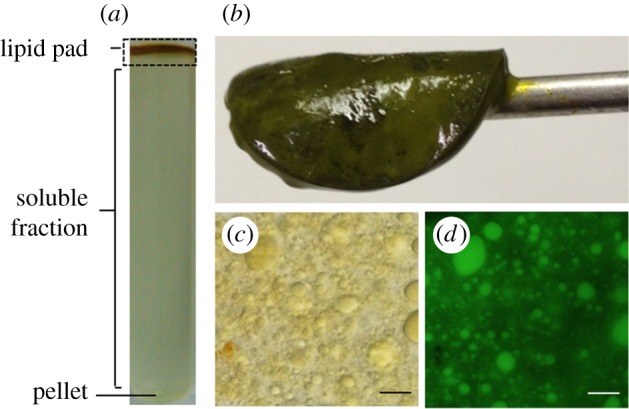
Isolation of oil bodies from the oleaginous diatom Fistulifera solaris. (a) Sucrose gradient ultracentrifugation sample to separate the lipid pad. (b) The lipid pad (solidified phase) collected from the centrifuged sample. The separated lipid pad contains the oil bodies (c, differential interference contrast observation; d, BODIPY 505/515 staining). Scale bar, 10 µm. The figure is reproduced from a previous study [3], with some modifications.
(b). Diversity of the major algal oil body-associated proteins
Major lipid droplet protein (MLDP, 27 kDa) was identified as the most abundant protein in the oil body fraction isolated from C. reinhardtii [63]. MLDP might function as a regulator of oil body size because repression of mldp gene expression causes an increase in oil body size. The mldp orthologues were identified in the genome sequences of other green algae, including Chlorella sp. NC64A, Chlorella vulgaris, H. pluvialis, D. salina, D. bardawil, D. parva and Volvox carteri [13,57,63]. In addition, MLDPs were detected in the oil bodies of D. salina (30 kDa) [57] and H. pluvialis (Haematococcus oil globule protein (HOGP), 33 kDa) [58].
Caleosin is the second most abundant protein on the lipid droplet surface in higher plants, following oleosin. Microalgal oil bodies in the green alga Auxenochlorella protothecoides (formerly Chlorella sp.) in the class Trebouxiophyceae [64,67] were shown to possess a 28 kDa caleosin-like protein showing cross-reactivity with an anti-caleosin antibody [68]. A major oil body-associated protein identified in an endosymbiotic alga, Symbiodinium sp. (Symbiodinium lipid droplet protein (SLDP), 20 kDa) showed the same cross-reactivity.
The Eustigmatophyceae N. oceanica contains lipid droplet surface protein (LDSP, 17 kDa) as a predominant oil body-associated protein. This protein is Nannochloropsis species-specific and has structural and functional properties that are similar to those of oleosin in plants and MLDP in green algae. The Haptophyceae comprise both calcifying species, called coccolithophores, and non-calcifying species, and produce unique lipid molecules called alkenones. Proteomic analysis of the proteins associated with the oil bodies that include alkenones, called alkenone bodies, was determined in the non-calcifying species T. lutea [61]. Proton pumps (V-ATPases) were found to be abundant in the alkenone bodies and might be assembled in the endoplasmic reticulum (ER). This result raises the hypothesis that neutral lipids containing alkenones might firstly accumulate in the internal space of the ER membrane and then bud from ER to form alkenone bodies in T. lutea.
(c). Proteomic analysis of diatom oil body-associated proteins
We confirmed that the homologues of plant oil body-associated proteins (oleosin, caleosin and steroleosin) and microalgal oil body-associated proteins (MLDP, HOGP, SLDP and LDSP) were not encoded in the genomes of F. solaris, P. tricornutum and T. pseudonana (unpublished data). Therefore, diatoms could have unique oil body-associated proteins. Proteomic analysis of diatom oil body-associated proteins was performed by two groups using different diatom species, an oleaginous diatom F. solaris [3] and the model diatom P. tricornutum [6], both of which belong to the pennate diatoms. In both studies, contamination issue was carefully taken into account, and different approaches were employed to identify the unique oil body-associated proteins in diatoms.
Our group published the first study of oil body proteomics in F. solaris [3]. After cell disruption, the oil body fraction was collected and analysed by SDS-PAGE and nanoLC-MS. In the SDS-PAGE, there was no abundant specific band in the oil body fraction, suggesting that major oil body-associated proteins such as MLDP in green algae might not exist in the oil bodies of F. solaris. In addition, protein contamination from the soluble fraction was detected in the oil body fraction in the SDS-PAGE. Therefore, we compared the proteomic data derived from the oil body and soluble fractions and estimated the proteins specific to the oil body fraction by subtracting the proteins found in both fractions from the proteins found in the oil body fraction. This subtraction strategy successfully identified 14 candidate proteins specific to the oil body fraction, of which, five were predicted to have at least one transmembrane region potentially associated with lipid droplets or a phospholipid monolayer membrane (table 5). Using a GFP–gene fusion technique, we conducted a localization study, and confirmed the localization of two proteins, G4301 and G6574, in the oil bodies. Based on Protein BLAST, G6574 was predicted to be a potassium channel, whereas the function of G4301 was unknown. A domain search based on InterProScan found an alcohol dehydrogenase-like superfamily domain, which was also detected in the oil body proteome of the plant Camelina sativa [69]. G4301 also has an ER-targeting signal sequence [70]. The presence of oil body-associated proteins in both the oil body and ER fractions was also reported in T. lutea, which belongs to the Haptophyceae [61], a secondary symbiotic microalgal group in which diatoms also belong. These results suggest that in secondary symbiotic microalgae, oil bodies are closely related to the ER.
Table 5.
| gene/protein ID | DDBJ/NCBI reference no. | annotationa | size (kDa) | transmembrane regionb | GRAVY scorec | references |
|---|---|---|---|---|---|---|
| g4796 | AB836658 | transmembrane protein | 85 | 6 | −0.11 | [3] |
| g6705 | AB836659 | ABC transporter transmembrane region | 149 | 6 | −0.33 | |
| g6574 | AB836660 | potassium channel, NKT2-like protein | 58 | 1 | −0.16 | |
| g4301 | AB836661 | hypothetical protein (diatom oil body-associated protein, DOAP1) | 53 | 1 | −0.03 | |
| g5708 | AB836662 | hypothetical protein | 49 | 1 | −0.27 | |
| g10552 | — | hypothetical protein | 88 | 0 | −0.71 | |
| g19744 | — | hypothetical protein | 80 | 0 | −0.45 | |
| g1204 | — | hypothetical protein | 43 | 0 | −0.77 | |
| g5858 | — | hypothetical protein | 45 | 0 | −0.18 | |
| g11870 | — | hypothetical protein | 27 | 0 | −0.35 | |
| g17204 | — | hypothetical protein | 105 | 0 | −0.4 | |
| g2223 | — | no hit | 88 | 0 | −0.45 | |
| g12717 | — | hypothetical protein | 40 | 0 | −0.18 | |
| g2311 | — | sec14-like cytosolic factor | 109 | 0 | −0.076 | |
| PHATRDRAFT_48859 | XP_002183367 | hypothetical protein (stramenopile-type lipid droplet protein, StLDP) | 49 | 4 | 0.26 | [6] |
| PHATRDRAFT_48778 | XP_002183443 | acyl-CoA-binding protein | 38 | 4 | 0.22 | |
| PHATRDRAFT_54019 | XP_002177351 | heat shock protein Hsp70 | 71 | 0 | −0.42 | |
| PHATRDRAFT_45894 | XP_002180271 | hypothetical protein | 38 | 0 | −0.62 | |
| PHATRDRAFT_49981 | XP_002184813 | hypothetical protein | 61 | 0 | −0.64 |
aAnnotated by BlastP (e-value < 10−10).
bPredicted by SOSUI program.
cCalculated by GRAVY calculator program.
Yoneda et al. [6] established a protocol of sucrose density gradient centrifugation which enabled them to isolate the oil bodies from the model diatom P. tricornutum with minimal contamination. The purity of the isolated oil bodies was assessed by measurement of the absorption at 663 nm (A663), corresponding to the light absorbance of chlorophyll a, and the detection of ribulose bisphosphate carboxylase/oxygenase (Rubisco) in the oil body fraction. Both the A663 and Rubisco signal were low; thus, chloroplast contamination in the oil body fraction was minimal. Subsequent SDS-PAGE exhibited major protein bands that were specific to the oil body fraction, and five proteins were identified by MS analysis (table 5). The most abundant protein was Phatr48859. Highly homologous genes were also detected in the genome sequences of several other diatoms, including F. solaris and two other heterokonts, the Eustigmatophyceae N. gaditana and the brown alga Ectocarpus siliculosus; therefore, Phatr48859 was renamed stramenopile-type lipid droplet protein (StLDP). In addition to StLDP, an acyl-CoA-binding protein (Phatr48778), which may be related to fatty acid metabolism, a heat shock protein 70 (Hsp70), which might function as a molecular chaperone on the oil body, and two other proteins, Phatr45894 and Phatr49981, which might undergo redox reactions, were identified.
Interestingly, there were no common oil body-associated proteins identified in the two proteomic studies in diatoms described above. This result suggests that, although both F. solaris and P. tricornutum belong to the pennate diatoms, their oil bodies could have different sets of oil body-associated proteins. In particular, StLDP was found to be a major oil body-associated protein in P. tricornutum, but was not detected in F. solaris. Although the function of StLDP is unknown, its sequence is similar to that of oleosin, implying that this protein may regulate lipid droplet size by preventing lipid droplet fusion [35]. When F. solaris was exposed to stress conditions, large oil bodies almost completely filled the cells (figure 1). Such giant oil bodies have also been observed in avocado and olive, which do not possess oleosin on their surfaces [71]. These facts suggest that the oil bodies of F. solaris lack oleosin-like proteins (including StLDP) on the surface, resulting in giant lipid droplets. It should be noted that StLDP was discovered through a sequence similarity search of the F. solaris genome [20].
4. Biogenesis and dynamics of oil bodies in diatoms
(a). Triacylglycerol synthesis in diatoms
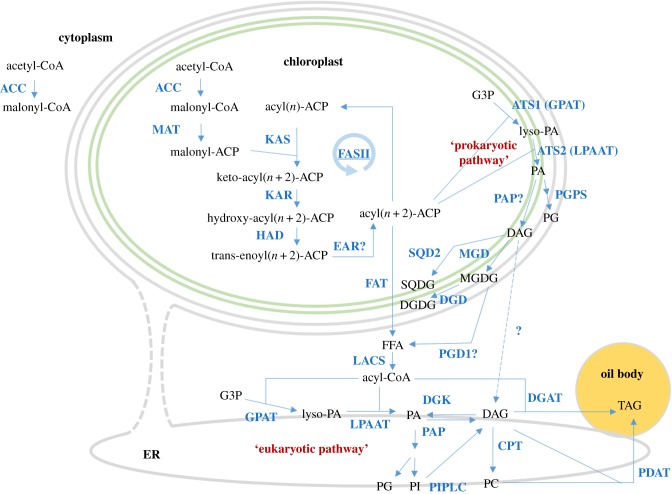
The de novo synthesis of TAGs begins with the esterification of glycerol-3-phosphate (G3P) and acyl-CoA (or acyl ACP) catalysed by glycerol-3-phosphate acyltransferase (GPAT). A second esterification step of the resulting lysophosphatidic acid (lyso-PA) and acyl-CoA (or acyl ACP) generates phosphatidic acid (PA), which is catalysed by lysophosphatidic acid acyltransferase (LPAAT). The remaining phosphate group is dephosphorylated by phosphatidic acid phosphatase (PAP), generating diacylglycerol (DAG). There are two main routes to form TAG from DAG. One is catalysed by diacylglycerol acyltransferase (DGAT), in which acyl-CoA provides the third acyl group [8,72]. The other is catalysed by phospholipid:diacylglycerol acyltransferase (PDAT), in which phospholipid provides the acyl group.
Chloroplasts and the ER are responsible for de novo synthesis of TAGs in higher plants [73] and some microalgae [74]. The chloroplast pathway is called the prokaryotic pathway, and the ER pathway is called the eukaryotic pathway (figure 4). The TAGs assembled via the prokaryotic pathway have a C16 fatty acid at the sn-2 position, while those assembled via the eukaryotic pathway have a C18 fatty acid at this position. In diatoms, fatty acid moieties at the sn-2 position are mainly C16 [38,75], suggesting that the prokaryotic pathway in the chloroplast could play a central role in TAG assembly in diatoms, as is the case in the green alga C. reinhardtii [74]. The enzymes which are involved in the TAG assembly are predicted to localize in the chloroplast or ER (figure 4) according to the bioinformatics pipeline which we established previously [20,76]. Nonetheless, we noticed that no DGAT and PDAT proteins identified in diatoms [8,20,72] are predicted to localize in the chloroplast due to the lack of the chloroplast-targeting signal motif at the N-termini, which transports the proteins encoded in the nuclear genome into the complex chloroplasts in diatoms [77–79]. Therefore, the final step in TAG synthesis in the prokaryotic pathway in diatoms remains to be determined.
Figure 4.
Fatty acids and TAG biosynthetic pathways in diatoms. Abbreviations for the enzymes (in light blue) are as follows: ACC, acetyl-CoA carboxylase; MAT, malonyl-CoA ACP transacylase; FASII, fatty acid synthase II; KAS, beta-ketoacyl-ACP synthase; KAR, beta-ketoacyl-ACP reductase; HAD, beta-hydroxyacyl-ACP dehydrase; EAR, enoyl-ACP reductase; FAT, acyl-ACP thioesterases; LACS, long-chain acyl-CoA synthetase; GPAT, glycerol-3-phosphate acyltransferase; LPAAT, lysophosphatidic acid acyltransferase; PAP, phosphatidate phosphatase; PIPLC, phosphoinositide phospholipase C; CPT, choline phosphotranseferase; DGAT diacylglycerol acyltransferase; PDAT, phospholipid:diacylglycerol acyltransferase; PGPS, phosphatidylglycerol phosphate synthase; SQD2, sulfoquinovosyltransferase; MGD, monogalactosyldiacylglycerol synthase; DGD, digalactosyldiacylglycerol synthase; and PGD1, galactoglycerolipid lipase. Abbreviations for the substrates (in black) are as follows: ACP, acyl carrier protein; G3P, glycerol 3-phosphate; lyso-PA, lysophosphatidic acid; PA, phosphatidic acid; PG, phosphatidylglycerol; PI, phosphatidyl inositol; PC, phosphatidyl choline; DAG, diacylglycerol; TAG, triacylglycerol; SQDG, sulfoquinovosyldiacylglycerol; MGDG, monogalactosyldiacylglycerol; and DGDG, digalactosyldiacylglycerol. The figure is reproduced from a previous study [20], with some modifications.
In addition to de novo synthesis, remodelling of chloroplast glycolipids (e.g. monogalactosyldiacylglycerol (MGDG) and digalactosyldiacylglycerol (DGDG)) can be considered as an alternative TAG synthesis pathway (described in §4b). The pgd1 gene, encoding galactoglycerolipid lipase, which is responsible for glycolipid turnover, was identified in the green alga C. reinhardtii [80]. Nonetheless, no homologues have been found in diatom genomes [20].
(b). Variations in subcellular ultrastructure during oil body biogenesis
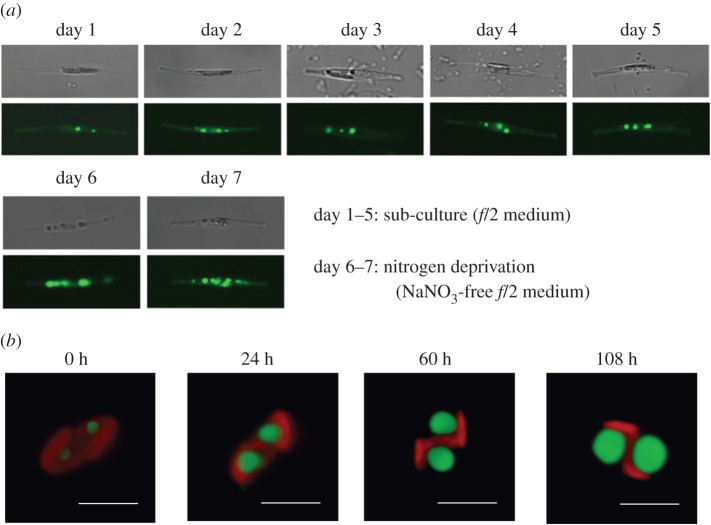
When exposed to nutrient deprivation (e.g. nitrogen deprivation), the subcellular ultrastructures of the diatoms change drastically. Time course monitoring of the cells during oil body growth was studied in P. tricornutum [4] and F. solaris [81] by confocal laser scanning microscopy (figure 5).
Figure 5.
Time course monitoring of oil accumulation in diatoms. (a) Phaeodactylum tricornutum was subcultured in f/2 medium for 5 days, and then subjected to nitrogen deprivation conditions in NaNO3-free f/2 medium for 2 days [4]. Oil bodies were stained with Nile Red (shown in green). (b) Fistulifera solaris was incubated in artificial seawater free of nutrient salts (scale bar, 5 µm) [81]. Oil bodies were stained with BODIPY 505/515 and are shown in green. The red fluorescence is derived from the chloroplast. The figures are reproduced from previous studies [4,81], with some modifications.
In P. tricornutum, under nitrogen deprivation, the number of oil bodies increased, but their size did not significantly change (most were 0.1–0.3 µm in diameter, although a few were much larger, approx. 2 µm). Meanwhile, the membrane systems surrounding the chloroplast and thylakoid membrane tended to be dispersed and poorly organized compared to those of cells cultivated in nutrient-rich conditions. In F. solaris, the size of subcellular organelles drastically changed during oil accumulation [81]. In this diatom, two large oil bodies are always observed, and their volume increased more than eightfold in the artificial seawater free from nutrient supplements (approx. 3 µm in diameter). By contrast, chloroplast volume decreased by half during oil accumulation. Given these results, oil accumulation appears to be species-dependent, typically either increasing the number of oil bodies (e.g. P. tricornutum) or increasing the size of the oil bodies (e.g. F. solaris). As described above (§3c), oleosin or oleosin-like proteins are responsible for regulating the size of oil bodies in higher plants and microalgae. Although P. tricornutum expresses the oleosin-like protein StLDP on oil bodies, the oil bodies in F. solaris lack a similar protein. These oil body-associated proteins could be related to the way in which oil accumulates, and their relationship requires further investigation.
The concomitant degradation of chloroplast membrane systems along with oil accumulation [4,81] indicates that remodelling of chloroplast membrane lipids, at least in part, provides the substrates for TAG synthesis. Indeed, in P. tricornutum, MGDG and DGDG, which are the major chloroplast membrane glycolipids, decreased by 50% under nitrogen deprivation conditions, while TAG content increased twofold [4]. The increase in the levels of C20 : 5 (EPA) in TAG corresponded to the decrease in MGDG in P. tricornutum under nitrogen deprivation, further supporting the transfer of fatty acid moieties (e.g. EPA, C20 : 5) from MGDG to TAG [30]. Although the enzyme involved in this lipid remodelling step in diatoms has not been identified, it has been identified in the green alga C. reinhardtii [80].
However, intracellular lipid turnover cannot solely account for the massive TAG accumulation. An isotope-labelling experiment using 13C-bicarbonate revealed that 60% of the lipids were de novo synthesized through photosynthetic CO2 fixation under nitrogen deprivation in P. tricornutum [82]. This has been demonstrated in another diatom species, F. solaris [81]. In this oleaginous diatom, the increase in oil body volume and decrease in chloroplast volume were nearly equal for the first 72 h of the oil accumulation phase, suggesting that remodelling of chloroplast membranes largely accounts for the TAG accumulation. However, after 72 h, the oil bodies kept expanding but the chloroplast no longer shrank. In this late oil accumulation phase, de novo synthesis of TAG through photosynthetic CO2 fixation could be a major means of TAG accumulation. This notion is supported by the fact that, even after the chloroplast membrane tended to be disordered [4] or its volume decreased [81], the photosynthetic capability did not reach zero. Therefore, oil accumulation in diatoms seems likely to be driven by the coordinated action of membrane lipid remodelling and de novo TAG synthesis.
From the viewpoint of efficient biofuel production, oil accumulation driven by de novo synthesis is particularly promising. Vinayak et al. [83] proposed a conceptual approach called ‘diatom milking’ for the extraction and purification of microalgal oils. In this approach, culture medium containing the cells is exposed to organic solvents to directly extract the neutral lipids without harvesting and killing the cells. After releasing the intracellular neutral lipids, the cells will grow again and accumulate more oil. This milking process avoids the high energy-consuming processes in biofuel production (i.e. cell harvesting and dewatering) [84,85]. Because this approach promises sustainable cultivation of intact cells, de novo synthesis would be preferable rather than breakdown of intracellular components. Nonetheless, at present, the balance between lipid remodelling and de novo synthesis is uncontrollable because the regulation mechanisms are poorly understood.
(c). Oil bodies acting as an electron sink
Why microalgae accumulate TAGs when exposed to nitrogen deprivation is a long-standing question. Although there is no clear answer, including detailed underlying mechanisms, one possibility is based on the electron sink function of TAG synthesis. Under nitrogen deprivation, cell growth is arrested due to a lack of the building blocks of biomacromolecules, including proteins and nucleic acids. At the same time, subcellular structures change drastically (e.g. chloroplasts shrink), while photosynthesis does not completely shut down. As a result, chloroplasts keep generating electrons from water molecules, and excess electrons accumulate in the photosynthetic electron transport chain, leading to the generation of reactive oxygen species. To attenuate this photo-oxidative stress, fatty acid synthesis and lipid assembly are likely to be induced. The de novo synthesis of TAGs effectively consumes electrons. Hu et al. [43] estimated that the formation of a C18 fatty acid consumes approximately 24 NADPH molecules as a reducing equivalent derived from the photosynthetic electron transport chain, which is twice that required for the synthesis of the same mass of carbohydrate or protein.
This notion leads to another technologically important question: whether it is possible to enhance algal lipid productivity by artificially providing excess reducing equivalents. This question was addressed in P. tricornutum [86] and F. solaris [87]. Because reinforcement of NADPH production through photosynthesis is difficult due to the complexity of photosynthesis systems (for example, it was shown that simple overexpression of ferredoxin-NADP+ reductase (FNR), which transfers an electron to NADP+ at the final step of the photosynthesis, did not significantly improve NADPH production in tobacco plants [88]), in these studies, alternative pathways for NADPH production were induced. Xue et al. [86] overexpressed a malic enzyme (ME) that catalyses oxidative decarboxylation of malate to pyruvate and produces NADPH. Overexpression of ME has also been attempted in numerous non-photosynthetic organisms, such as the fungi Mucor circinelloides and Rhodotorula glutinis [89,90] and Escherichia coli [91], and this overexpression led to increased lipid production. In P. tricornutum, it was reported that the genetically engineered diatom overexpressing ME showed greater numbers of oil bodies with increased size; consequently, oil accumulation increased 2.5-fold [86]. Our group employed a different strategy to artificially provide excess NADPH in F. solaris, through improvement of the pentose phosphate pathway (PPP) [87]. A recent study suggested that the NADPH-producing enzymes in PPP, i.e. glucose-6-phosphate dehydrogenase (G6PD) and phosphogluconate dehydrogenase (PGD), may play a more important role in lipid metabolism than previously thought [92]. We overexpressed G6PD or PGD in F. solaris, and both enhanced lipid productivity by accelerating oil accumulation. G6PD overexpression showed greater enhancement, and it elevated lipid productivity 1.5-fold. These studies show that providing excess reducing equivalents is a promising strategy for enhancement of lipid productivity. It should be noted that, for both ME and G6PD overexpression, their predicted subcellular localization is the mitochondrion and cytoplasm, respectively. In diatoms, the chloroplast is responsible for fatty acid synthesis; thus, reducing equivalents produced by ME or G6PD should be somehow transported into the chloroplast. Recently, energetic coupling between the mitochondrion and chloroplast in diatoms was proposed [93]. These organelles accommodate ATP and reducing equivalents, and several molecular mechanisms (e.g. malate shuttle [93], glutamine-ornithine shunt [94], and mitochondrial glycolysis [95]) are proposed to be involved in this transportation. These molecular mechanisms might be related to the transportation of the reducing equivalents produced by ME or G6PD into the chloroplast, and assist lipid production.
(d). Degradation of the storage lipids in oil bodies
Zienkiewicz et al. [15] recently summarized the differences between microalgal oil bodies and plant oil bodies, and reported that the most striking difference is the dynamic nature of microalgal oil bodies. The lipids in plant oil bodies mainly function as long-term carbon and energy stores, and are irreversibly mobilized in particular structures (e.g. seed or pollen germination). By contrast, microalgal oil bodies appear to function more as transient reservoirs, as the storage lipids are degraded in quick response to environmental changes. Degradation of the storage lipids (mainly TAGs) induced by re-supplementation of starved cells with nitrate has been well documented in green algae [96–99]. TAG degradation produces ATP through β-oxidation and provides the carbon required for reproduction of cell components, such as proteins, primary pigments and chloroplast membranes. Thus, microalgal oil bodies could serve as an energy and carbon source during recovery when nutrient conditions are favourable. Mobilization of the storage lipids in oil bodies of green microalgae after re-supplementation was investigated in detail by lipidomic analysis and radiolabelling experiments for Parietochloris incisa [96], biomass composition analysis for Chromochloris zofingiensis [99], and morphological analysis using fluorescence and transmission electron microscopy for Chlorella vulgaris [98]. A proteomic analysis of Chlamydomonas oil bodies identified several lipase-like proteins that could be involved in lipid degradation [62].
Compared to green algae, our understanding of lipid catabolism in diatoms is still in its infancy. After P. tricornutum accumulated neutral lipids under stress conditions, re-supplementation with nitrate and/or phosphate rapidly induced neutral lipid degradation [100]. However, cell growth and chlorophyll a content only recovered when nitrate was re-supplied, suggesting that nitrate supplementation triggered the metabolic shift from lipid accumulation to cellular growth, along with active photosynthesis. Nearly, all the storage lipids in the oil bodies were used, and no oil bodies were observed in the re-supplemented cells. Elucidation of the molecular mechanisms underlying the lipid catabolism in diatoms, including lipid degradation, energy generation and reconstruction of cellular components, has just been launched, and some TAG lipases which might be involved in the lipid degradation were identified as described below.
Trentacoste et al. [5] performed a microarray experiment to examine lipase expression in T. pseudonana under silicon-limited conditions. The stress conditions arrested diatom cell growth and induced oil accumulation. They focused on lipases that were downregulated during oil accumulation, and Thaps3_264297 was identified as the lipase gene exhibiting the greatest decrease in transcript abundance. The Thaps3_264297 protein shares sequence similarity with the human comparative gene identification 58 (CGI-58) protein (29% identity), which does not have lipase activity because the serine residue in its catalytic centre is replaced by an asparagine; however, it activates other lipases to break down storage lipids in adipose cells [101]. By contrast, Thaps3_264297 has an active catalytic centre. Indeed, in vitro assays using a recombinant Thaps3_264297 protein showed TAG lipase activity as well as phospholipase and LPAAT activities. Involvement of Thaps3_264297 in the lipid degradation was verified by a knockdown experiment, which showed significantly increased TAG storage. Thaps3_264297 knockdown also resulted in an increase in polar lipids, suggesting the role of this enzyme in membrane lipid turnover and lipid homeostasis. Barka et al. [102] focused on another lipase, sugar-dependent 1 (SDP1)-patatin like lipase. In Arabidopsis thaliana, a major TAG lipase, SDP1, is associated with the oil body of seeds, and it initiates storage lipid breakdown during germination [103]. Among the 49 genes with predicted lipase functions in the P. tricornutum genome, Phatrdraft_1971 was identified as the only candidate that is similar to a SDP1-patatin like lipase. Like Thaps3_264297, an in vitro assay using a recombinant Phatrdraft_1971 protein showed TAG lipase activity, and knockdown of Phatrdraft_1971 resulted in increased TAGs. However, mutation of Phatrdraft_1971 decreased total polar lipid content to 80–85%, suggesting a different role in lipid turnover.
Interestingly, although knockdown of Thaps3_264297 and Phatrdraft_1971 led to significant increases in TAG accumulation, cell growth was not significantly decreased. This suggests that although these enzymes could be the major TAG lipases in each diatom species, they are likely not the only lipases involved in lipid degradation. Other lipases that cooperate with Thaps3_264297 and Phatrdraft_1971 remain to be determined. From the viewpoint of biofuel production, knockdown of Thaps3_264297 or Phatrdraft_1971 can enhance lipid productivity without decreasing biomass productivity; thus they are attractive targets for better biofuel yield.
5. Conclusion
During lipid accumulation in diatoms, the size and/or number of oil bodies increases, and the fatty acid composition of the oil bodies varies. These variations are highly species-dependent; thus, it is difficult to generalize about diverse diatom species, although it is well known that nitrogen deprivation commonly triggers the metabolic shift from cellular growth to lipid accumulation. Some oleaginous diatoms accumulate high levels of TAGs of appropriate quality for biofuel applications, and their lipid productivities are comparative to those of other oleaginous microalgae.
In addition to the TAGs, oil body-associated proteins are another important component of oil bodies. Although, thus far only the oil bodies of two diatom species, P. tricornutum and F. solaris, have been subjected to proteomic analysis, these studies successfully identified unique proteins that are potentially involved in the formation and stabilization of oil bodies. In addition, some TAG lipases that may be related to TAG degradation in diatoms were also identified. The temporal and spatial distribution of these proteins could be elucidated in future studies, which could provide more insights into the mechanisms governing oil body dynamics in diatoms.
The de novo synthesis of TAGs is catalysed by a variety of enzymes, including DGAT and PDAT, which use acyl-CoA and polar lipids as acyl chain donors, respectively. Both the chloroplast and ER contribute to this de novo synthesis, while the chloroplast pathway (termed the prokaryotic pathway) plays a more central role than the ER pathway (termed the eukaryotic pathway). In addition to de novo synthesis, remodelling of chloroplast membrane lipids (mainly the glycolipids) appears also to be involved in TAG synthesis. However, the enzymes involved in these TAG synthetic pathways have not yet been identified.
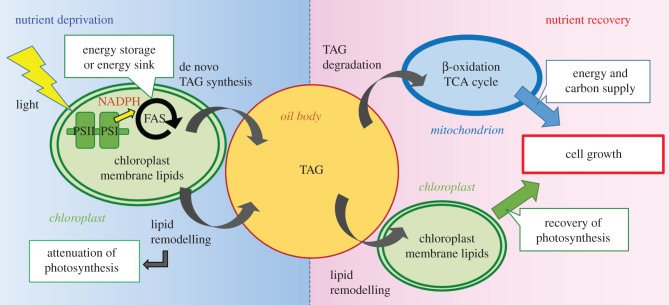
Remodelling of the chloroplast membrane lipids into TAGs leads to a decrease in photosynthetic capacity in the oil-accumulating cells. However, photosynthesis is not completely inhibited, and excess reducing power continues to be generated, which is potentially toxic for the cells. Fatty acid synthesis efficiently consumes NADPH, which cancels out the excess reducing power. Therefore, de novo TAG synthesis with fatty acid synthesis works as an electron sink when the cells are exposed to nutrient deprivation. When nutrient conditions are more favourable, degradation of TAGs is observed. TAG degradation supplies the energy and carbon needed for cell growth by passing through β-oxidation and the TCA cycle in mitochondria. An increase in chloroplast volume and recovery of photosynthetic capacity were confirmed during oil body degradation, suggesting that remodelling of TAGs into chloroplast membrane lipids could also contribute to cell growth after nutrient recovery.
The dynamics and functions of the oil bodies in diatoms, which were estimated by recent studies, are summarized in figure 6. We expect that further investigations of oil body biology in diatoms will be performed, and such studies will provide a better understanding of lipid metabolism in diatoms, and provide efficient strategies for improved biofuel production.
Figure 6.
Schematic of TAG synthesis and degradation in diatoms responding to nutrient deprivation and recovery.
Data accessibility
This article has no additional data.
Authors' contributions
T.T. designed the study. Y.M., D.N., T.Y. and T.T. analysed the data and prepared the manuscript.
Competing interests
We have no competing interests.
Funding
This study was supported in part through a project commissioned by the New Energy and Industrial Technology Development Organization (NEDO).
References
- 1.Field CB, Behrenfeld MJ, Randerson JT, Falkowski P. 1998. Primary production of the biosphere: integrating terrestrial and oceanic components. Science 281, 237–240. ( 10.1126/science.281.5374.237) [DOI] [PubMed] [Google Scholar]
- 2.Falkowski PG, Barber RT, Smetacek VV. 1998. Biogeochemical controls and feedbacks on ocean primary production. Science 281, 200–207. ( 10.1126/science.281.5374.200) [DOI] [PubMed] [Google Scholar]
- 3.Nojima D, Yoshino T, Maeda Y, Tanaka M, Nemoto M, Tanaka T. 2013. Proteomics analysis of oil body-associated proteins in the oleaginous diatom. J. Proteome Res. 12, 5293–5301. ( 10.1021/pr4004085) [DOI] [PubMed] [Google Scholar]
- 4.Yang ZK Niu YF, Ma YH, Xue J, Zhang MH, Yang WD, Liu JS, Lu SH, Guan Y, Li HY. 2013. Molecular and cellular mechanisms of neutral lipid accumulation in diatom following nitrogen deprivation. Biotechnol. Biofuels 6, 67 ( 10.1186/1754-6834-6-67) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Trentacoste EM, Shrestha RP, Smith SR, Gle C, Hartmann AC, Hildebrand M, Gerwick WH. 2013. Metabolic engineering of lipid catabolism increases microalgal lipid accumulation without compromising growth. Proc. Natl Acad. Sci. USA 110, 19 748–19 753. ( 10.1073/pnas.1309299110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yoneda K, Yoshida M, Suzuki I, Watanabe MM. 2016. Identification of a major lipid droplet protein in a marine diatom Phaeodactylum tricornutum. Plant Cell Physiol. 57, 397–406. ( 10.1093/pcp/pcv204) [DOI] [PubMed] [Google Scholar]
- 7.Ge F, Huang W, Chen Z, Zhang C, Xiong Q, Bowler C, Yang J, Xu J, Hu H. 2014. Methylcrotonyl-CoA carboxylase regulates triacylglycerol accumulation in the model diatom Phaeodactylum tricornutum. Plant Cell 26, 1681–1697. ( 10.1105/tpc.114.124982) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guiheneuf F, Leu S, Zarka A, Khozin-Goldberg I, Khalilov I, Boussiba S. 2011. Cloning and molecular characterization of a novel acyl-CoA:diacylglycerol acyltransferase 1-like gene (PtDGAT1) from the diatom Phaeodactylum tricornutum. FEBS J. 278, 3651–3666. ( 10.1111/j.1742-4658.2011.08284.x) [DOI] [PubMed] [Google Scholar]
- 9.Armbrust EV. 2009. The life of diatoms in the world's oceans. Nature 459, 185–192. ( 10.1038/nature08057) [DOI] [PubMed] [Google Scholar]
- 10.Keeling PJ. 2010. The endosymbiotic origin, diversification and fate of plastids. Phil. Trans. R. Soc. B 365, 729–748. ( 10.1098/rstb.2009.0103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Armbrust EV, et al. 2004. The genome of the diatom Thalassiosira pseudonana: ecology, evolution, and metabolism. Science 306, 79–86. ( 10.1126/science.1101156) [DOI] [PubMed] [Google Scholar]
- 12.Bowler C, et al. 2008. The Phaeodactylum genome reveals the evolutionary history of diatom genomes. Nature 456, 239–244. ( 10.1038/nature07410) [DOI] [PubMed] [Google Scholar]
- 13.Goold H, Beisson F, Peltier G, Li-Beisson Y. 2015. Microalgal lipid droplets: composition, diversity, biogenesis and functions. Plant Cell Rep. 34, 545–555. ( 10.1007/s00299-014-1711-7) [DOI] [PubMed] [Google Scholar]
- 14.Liu B, Benning C. 2013. Lipid metabolism in microalgae distinguishes itself. Curr. Opin. Biotechnol. 24, 300–309. ( 10.1016/j.copbio.2012.08.008) [DOI] [PubMed] [Google Scholar]
- 15.Zienkiewicz K, Du ZY, Ma W, Vollheyde K, Benning C. 2016. Stress-induced neutral lipid biosynthesis in microalgae: molecular, cellular and physiological insights. Biochim. Biophys. Acta 1861, 1269–1281. ( 10.1016/j.bbalip.2016.02.008) [DOI] [PubMed] [Google Scholar]
- 16.Hildebrand M, Davis AK, Smith SR, Traller JC, Abbriano R. 2012. The place of diatoms in the biofuels industry. Biofuels 3, 221–240. ( 10.4155/bfs.11.157) [DOI] [Google Scholar]
- 17.Rodolfi L, Chini Zittelli G, Bassi N, Padovani G, Biondi N, Bonini G, Tredici MR. 2009. Microalgae for oil: strain selection, induction of lipid synthesis and outdoor mass cultivation in a low-cost photobioreactor. Biotechnol. Bioeng. 102, 100–112. ( 10.1002/bit.22033) [DOI] [PubMed] [Google Scholar]
- 18.Radakovits R, Jinkerson RE, Fuerstenberg SI, Tae H, Settlage RE, Boore JL, Posewitz MC. 2012. Draft genome sequence and genetic transformation of the oleaginous alga Nannochloropis gaditana. Nat. Commun. 3, 686 ( 10.1038/ncomms1688) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Converti A, Casazza AA, Ortiz EY, Perego P, Del Borghi M. 2009. Effect of temperature and nitrogen concentration on the growth and lipid content of Nannochloropsis oculata and Chlorella vulgaris for biodiesel production. Chem. Eng. Process 48, 1146–1151. ( 10.1016/j.cep.2009.03.006) [DOI] [Google Scholar]
- 20.Tanaka T Maeda Y, Veluchamy A, Tanaka M, Abida H, Maréchal E, Bowler C, Muto M, Sunaga Y, Tanaka M. 2015. Oil accumulation by the oleaginous diatom Fistulifera solaris as revealed by the genome and transcriptome. Plant Cell 27, 162–176. ( 10.1105/tpc.114.135194) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roessler PG. 1988. Effects of silicon deficiency on lipid composition and metabolism in the diatom Cyclotella cryptica. J. Phycol. 24, 394–400. ( 10.1111/j.1529-8817.1988.tb00189.x) [DOI] [Google Scholar]
- 22.Traller JC, Hildebrand M. 2013. High throughput imaging to the diatom Cyclotella cryptica demonstrates substantial cell-to-cell variability in the rate and extent of triacylglycerol accumulation. Algal Res. 2, 244–252. ( 10.1016/j.algal.2013.03.003) [DOI] [Google Scholar]
- 23.Wu S, Zhang B, Huang A, Huan L, He L, Lin A, Niu J, Wang G. 2014. Detection of intracellular neutral lipid content in the marine microalgae Prorocentrum micans and Phaeodactylum tricornutum using Nile red and BODIPY 505/515. J. Appl. Phycol. 26, 1659–1668. ( 10.1007/s10811-013-0223-0) [DOI] [Google Scholar]
- 24.Lelong A, Hegaret H, Soudant P. 2011. Cell-based measurements to assess physiological status of Pseudo-nitzschia multiseries, a toxic diatom. Res. Microbiol. 162, 969–981. ( 10.1016/j.resmic.2011.06.005) [DOI] [PubMed] [Google Scholar]
- 25.Liu W, Huang Z, Li P, Xia J, Chen B. 2012. Formation of triacylglycerol in Nitzschia closterium f. minutissima under nitrogen limitation and possible physiological and biochemical mechanisms. J. Exp. Mar. Biol. Ecol. 418, 24–29. ( 10.1016/j.jembe.2012.03.005) [DOI] [Google Scholar]
- 26.Govender T, Ramanna L, Rawat I, Bux F. 2012. BODIPY staining, an alternative to the Nile Red fluorescence method for the evaluation of intracellular lipids in microalgae. Bioresour. Technol. 114, 507–511. ( 10.1016/j.biortech.2012.03.024) [DOI] [PubMed] [Google Scholar]
- 27.Manandhar-Shrestha K, Hildebrand M. 2015. Characterization and manipulation of a DGAT2 from the diatom Thalassiosira pseudonana: improved TAG accumulation without detriment to growth, and implications for chloroplast TAG accumulation. Algal Res. 12, 239–248. ( 10.1016/j.algal.2015.09.004) [DOI] [Google Scholar]
- 28.Wong DM, Franz AK. 2013. A comparison of lipid storage in Phaeodactylum tricornutum and Tetraselmis suecica using laser scanning confocal microscopy. J. Microbiol. Methods 95, 122–128. ( 10.1016/j.mimet.2013.07.026) [DOI] [PubMed] [Google Scholar]
- 29.Popovich CA, Damiani C, Constenla D, Leonardi PI. 2012. Lipid quality of the diatoms Skeletonema costatum and Navicula gregaria from the South Atlantic Coast (Argentina): evaluation of its suitability as biodiesel feedstock. J. Appl. Phycol. 24, 1–10. ( 10.1007/s10811-010-9639-y) [DOI] [Google Scholar]
- 30.Abida H, et al. 2015. Membrane glycerolipid remodeling triggered by nitrogen and phosphorus starvation in Phaeodactylum tricornutum. Plant Physiol. 167, 118–136. ( 10.1104/pp.114.252395) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eizadora TY, Zendejas FJ, Lane PD, Gaucher S, Simmons BA, Lane TW. 2009. Triacylglycerol accumulation and profiling in the model diatoms Thalassiosira pseudonana and Phaeodactylum tricornutum (Baccilariophyceae) during starvation. J. Appl. Phycol. 21, 669–681. ( 10.1007/s10811-008-9400-y) [DOI] [Google Scholar]
- 32.Smith SR, Gle C, Abbriano RM, Traller JC, Davis A, Trentacoste E, Vernet M, Allen AE, Hildebrand M. 2016. Transcript level coordination of carbon pathways during silicon starvation-induced lipid accumulation in the diatom Thalassiosira pseudonana. New Phytol. 210, 890–904. ( 10.1111/nph.13843) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cooksey KE, Guckert JB, Williams SA, Callis PR. 1987. Fluorometric determination of the neutral lipid content of microalgal cells using Nile Red. J. Microbiol. Methods 6, 333–345. ( 10.1016/0167-7012(87)90019-4) [DOI] [Google Scholar]
- 34.Rumin J, Bonnefond H, Saint-Jean B, Rouxel C, Sciandra A, Bernard O, Cadoret JP, Bougaran G. 2015. The use of fluorescent Nile red and BODIPY for lipid measurement in microalgae. Biotechnol. Biofuels 8, 42 ( 10.1186/s13068-015-0220-4) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nojima D, Nonoyama T, Yoshino T, Tanaka T. In press Lipid droplet dynamics in microalgae revealed by proteomic analysis. Perspect. Phycol. 4, 25–32. ( 10.1127/pip/2017/0069) [DOI] [Google Scholar]
- 36.d'Ippolito G, Sardo A, Paris D, Vella FM, Adelfi MG, Botte P, Gallo C, Fontana A. 2015. Potential of lipid metabolism in marine diatoms for biofuel production. Biotechnol. Biofuels 8, 28 ( 10.1186/s13068-015-0212-4) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Matsumoto M, Sugiyama H, Maeda Y, Sato R, Tanaka T, Matsunaga T. 2010. Marine diatom, Navicula sp. strain JPCC DA0580 and marine green alga, Chlorella sp. strain NKG400014 as potential sources for biodiesel production. Appl. Biochem. Biotechnol. 161, 483–490. ( 10.1007/s12010-009-8766-x) [DOI] [PubMed] [Google Scholar]
- 38.Liang Y, Maeda Y, Yoshino T, Matsumoto M, Tanaka T. 2014. Profiling of fatty acid methyl esters from the oleaginous diatom Fistulifera sp. strain JPCC DA0580 under nutrition-sufficient and -deficient conditions. J. Appl. Phycol. 26, 2295–2302. ( 10.1007/s10811-014-0265-y) [DOI] [Google Scholar]
- 39.Griffiths MJ, Harrison ST. 2009. Lipid productivity as a key characteristic for choosing algal species for biodiesel production. J. Appl. Phycol. 21, 493–507. ( 10.1007/s10811-008-9392-7) [DOI] [Google Scholar]
- 40.Sheehan J, Dunahay T, Benemann J, Roessler P, Weissman JC. 1998 A look back at the US Department of Energy's aquatic species program: biodiesel from algae. National Renewable Energy Laboratory Technical Report NREL/TP-580-24190. Golden, CO: NREL. See http://www.nrel.gov/docs/legosti/fy98/24190.pdf . [Google Scholar]
- 41.Tokushima H, Inoue-Kashino N, Nakazato Y, Masuda A, Ifuku K, Kashino Y. 2016. Advantageous characteristics of the diatom Chaetoceros gracilis as a sustainable biofuel producer. Biotechnol. Biofuels 9, 235 ( 10.1186/s13068-016-0649-0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Araujo GS, Matos LJ, Goncalves LR, Fernandes FA, Farias WR. 2011. Bioprospecting for oil producing microalgal strains: evaluation of oil and biomass production for ten microalgal strains. Bioresour. Technol. 102, 5248–5250. ( 10.1016/j.biortech.2011.01.089) [DOI] [PubMed] [Google Scholar]
- 43.Hu Q, Sommerfeld M, Jarvis E, Ghirardi M, Posewitz M, Seibert M, Darzins A. 2008. Microalgal triacylglycerols as feedstocks for biofuel production: perspectives and advances. Plant J. 54, 621–639. ( 10.1111/j.1365-313X.2008.03492.x) [DOI] [PubMed] [Google Scholar]
- 44.Tonon T, Harvey D, Larson TR, Graham IA. 2002. Long chain polyunsaturated fatty acid production and partitioning to triacylglycerols in four microalgae. Phytochemistry 61, 15–24. ( 10.1016/S0031-9422(02)00201-7) [DOI] [PubMed] [Google Scholar]
- 45.Su X, Xu J, Yan X, Zhao P, Chen J, Zhou C, Zhao F, Li S. 2013. Lipidomic changes during different growth stages of Nitzschia closterium f. minutissima. Metabolomics 9, 300–310. ( 10.1007/s11306-012-0445-1) [DOI] [Google Scholar]
- 46.Knothe G. 2005. Dependence of biodiesel fuel properties on the structure of fatty acid alkyl esters. Fuel Process Technol. 86, 1059–1070. ( 10.1016/j.fuproc.2004.11.002) [DOI] [Google Scholar]
- 47.Christiansen K, Jensen PK. 1972. Membrane-bound lipid particles from beef heart chemical composition and structure. Biochim. Biophys. Acta 260, 449–459. ( 10.1016/0005-2760(72)90060-4) [DOI] [PubMed] [Google Scholar]
- 48.Tzen J, Cao Y, Laurent P, Ratnayake C, Huang A. 1993. Lipids, proteins, and structure of seed oil bodies from diverse species. Plant Physiol. 101, 267–276. ( 10.1104/pp.101.1.267) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Farese RV, Walther TC. 2009. Lipid droplets finally get a little RESPECT. Cell 139, 855–860. ( 10.1016/j.cell.2009.11.005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Martin S, Parton RG. 2006. Lipid droplets: a unified view of a dynamic organelle. Nat. Rev. Mol. Cell Biol. 7, 373–378. ( 10.1038/nrm1912) [DOI] [PubMed] [Google Scholar]
- 51.Kimmel AR, Brasaemle DL, McAndrews-Hill M, Sztalryd C, Londos C. 2010. Adoption of PERILIPIN as a unifying nomenclature for the mammalian PAT-family of intracellular lipid storage droplet proteins. J. Lipid Res. 51, 468–471. ( 10.1194/jlr.R000034) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lu X, Gruia-Gray J, Copeland NG, Gilbert DJ, Jenkins NA, Londos C, Kimmel AR. 2001. The murine perilipin gene: the lipid droplet-associated perilipins derive from tissue-specific, mRNA splice variants and define a gene family of ancient origin. Mamm. Genome 12, 741–749. ( 10.1007/s00335-01-2055-5) [DOI] [PubMed] [Google Scholar]
- 53.Huang A. 1996. Oleosins and oil bodies in seeds and other organs. Plant Physiol. 110, 1055 ( 10.1104/pp.110.4.1055) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lin LJ, Tai SS, Peng CC, Tzen JT. 2002. Steroleosin, a sterol-binding dehydrogenase in seed oil bodies. Plant Physiol. 128, 1200–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Næsted H, Frandsen GI, Jauh GY, Hernandez-Pinzon I, Nielsen HB, Murphy DJ, Rogers JC, Mundy J. 2000. Caleosins: Ca2+-binding proteins associated with lipid bodies. Plant Mol. Biol. 44, 463–476. ( 10.1023/A:1026564411918) [DOI] [PubMed] [Google Scholar]
- 56.Wang ZT, Ullrich N, Joo S, Waffenschmidt S, Goodenough U. 2009. Algal lipid bodies: stress induction, purification, and biochemical characterization in wild-type and starchless Chlamydomonas reinhardtii. Eukaryot. Cell 8, 1856–1868. ( 10.1128/EC.00272-09) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Davidi L, Katz A, Pick U. 2012. Characterization of major lipid droplet proteins from Dunaliella. Planta 236, 19–33. ( 10.1007/s00425-011-1585-7) [DOI] [PubMed] [Google Scholar]
- 58.Peled E, Leu S, Zarka A, Weiss M, Pick U, Khozin-Goldberg I, Boussiba S. 2011. Isolation of a novel oil globule protein from the green alga Haematococcus pluvialis (Chlorophyceae). Lipids 46, 851–861. ( 10.1007/s11745-011-3579-4) [DOI] [PubMed] [Google Scholar]
- 59.Vieler A, Brubaker SB, Vick B, Benning C. 2012. A lipid droplet protein of Nannochloropsis with functions partially analogous to plant oleosins. Plant Physiol. 158, 1562–1569. ( 10.1104/pp.111.193029) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Huang NL, Huang MD, Chen TL, Huang AH. 2013. Oleosin of subcellular lipid droplets evolved in green algae. Plant Physiol. 161, 1862–1874. ( 10.1104/pp.112.212514) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shi Q, Araie H, Bakku RK, Fukao Y, Rakwal R, Suzuki I, Shiraiwa Y. 2015. Proteomic analysis of lipid body from the alkenone-producing marine haptophyte alga Tisochrysis lutea. Proteomics 15, 4145–4158. ( 10.1002/pmic.201500010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nguyen HM, et al. 2011. Proteomic profiling of oil bodies isolated from the unicellular green microalga Chlamydomonas reinhardtii: with focus on proteins involved in lipid metabolism. Proteomics 11, 4266–4273. ( 10.1002/pmic.201100114) [DOI] [PubMed] [Google Scholar]
- 63.Moellering ER, Benning C. 2010. RNA interference silencing of a major lipid droplet protein affects lipid droplet size in Chlamydomonas reinhardtii. Eukaryot. Cell 9, 97–106. ( 10.1128/EC.00203-09) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lin IP, Jiang PL, Chen CS, Tzen JT. 2012. A unique caleosin serving as the major integral protein in oil bodies isolated from Chlorella sp. cells cultured with limited nitrogen. Plant Physiol. Biochem. 61, 80–87. ( 10.1016/j.plaphy.2012.09.008) [DOI] [PubMed] [Google Scholar]
- 65.Davidi L, Levin Y, Ben-Dor S, Pick U. 2015. Proteome analysis of cytoplasmatic and plastidic beta-carotene lipid droplets in Dunaliella bardawil. Plant Physiol. 167, 60–79. ( 10.1104/pp.114.248450) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ndimba BK, Ndimba RJ, Johnson TS, Waditee-Sirisattha R, Baba M, Sirisattha S, Shiraiwa Y, Agrawal GK, Rakwal R. 2013. Biofuels as a sustainable energy source: an update of the applications of proteomics in bioenergy crops and algae. J. Proteomics 93, 234–244. ( 10.1016/j.jprot.2013.05.041) [DOI] [PubMed] [Google Scholar]
- 67.Gao C, Wang Y, Shen Y, Yan D, He X, Dai J, Wu Q. 2014. Oil accumulation mechanisms of the oleaginous microalga Chlorella protothecoides revealed through its genome, transcriptomes, and proteomes. BMC Genomics 15, 1 ( 10.1186/1471-2164-15-582) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pasaribu B, Lin IP, Chen CS, Lu CY, Jiang PL. 2014. Nutrient limitation in Auxenochlorella protothecoides induces qualitative changes of fatty acid and expression of caleosin as a membrane protein associated with oil bodies. Biotechnol. Lett. 36, 175–180. ( 10.1007/s10529-013-1332-1) [DOI] [PubMed] [Google Scholar]
- 69.Jolivet P, Acevedo F, Boulard C, d'Andrea S, Faure JD, Kohli A, Nesi N, Valot B, Chardot T. 2013. Crop seed oil bodies: from challenges in protein identification to an emerging picture of the oil body proteome. Proteomics 13, 1836–1849. ( 10.1002/pmic.201200431) [DOI] [PubMed] [Google Scholar]
- 70.Maeda Y, Sunaga Y, Yoshino T, Tanaka T. 2014. Oleosome-associated protein of the oleaginous diatom Fistulifera solaris contains an endoplasmic reticulum-targeting signal sequence. Mar. Drugs 12, 3892–3903. ( 10.3390/md12073892) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ross JH, Sanchez J, Millan F, Murphy DJ. 1993. Differential presence of oleosins in oleogenic seed and mesocarp tissues in olive (Olea europaea) and avocado (Persea americana). Plant Sci. 93, 203–210. ( 10.1016/0168-9452(93)90050-A) [DOI] [Google Scholar]
- 72.Gong Y, Zhang J, Guo X, Wan X, Liang Z, Hu CJ, Jiang M. 2013. Identification and characterization of PtDGAT2B, an acyltransferase of the DGAT2 acyl-coenzyme A: diacylglycerol acyltransferase family in the diatom Phaeodactylum tricornutum. FEBS Lett. 587, 481–487. ( 10.1016/j.febslet.2013.01.015) [DOI] [PubMed] [Google Scholar]
- 73.Somerville C, Browse J. 1991. Plant lipids: metabolism, mutants, and membranes. Science 252, 80–87. ( 10.1126/science.252.5002.80) [DOI] [PubMed] [Google Scholar]
- 74.Fan J, Andre C, Xu C. 2011. A chloroplast pathway for the de novo biosynthesis of triacylglycerol in Chlamydomonas reinhardtii. FEBS Lett. 585, 1985–1991. ( 10.1016/j.febslet.2011.05.018) [DOI] [PubMed] [Google Scholar]
- 75.Yongmanitchai W, Ward OP. 1993. Molecular species of triacylglycerols from the freshwater diatom, Phaeodactylum tricornutum. Phytochemistry 32, 1137–1139. ( 10.1016/S0031-9422(00)95078-7) [DOI] [Google Scholar]
- 76.Sunaga Y, Maeda Y, Yabuuchi T, Muto M, Yoshino T, Tanaka T. 2015. Chloroplast-targeting protein expression in the oleaginous diatom Fistulifera solaris JPCC DA0580 toward metabolic engineering. J. Biosci. Bioeng. 119, 28–34. ( 10.1016/j.jbiosc.2014.06.008) [DOI] [PubMed] [Google Scholar]
- 77.Kilian O, Kroth PG. 2005. Identification and characterization of a new conserved motif within the presequence of proteins targeted into complex diatom plastids. Plant J. 41, 175–183. ( 10.1111/j.1365-313X.2004.02294.x) [DOI] [PubMed] [Google Scholar]
- 78.Apt KE, Zaslavkaia L, Lippmeier JC, Lang M, Kilian O, Wetherbee R, Grossman AR, Kroth PG. 2002. In vivo characterization of diatom multipartite plastid targeting signals. J. Cell Sci. 115, 4061–4069. ( 10.1242/jcs.00092) [DOI] [PubMed] [Google Scholar]
- 79.Gruber A, Vugrinec S, Hempel F, Gould SB, Maier UG, Kroth PG. 2007. Protein targeting into complex diatom plastids: functional characterisation of a specific targeting motif. Plant Mol. Biol. 64, 519–530. ( 10.1007/s11103-007-9171-x) [DOI] [PubMed] [Google Scholar]
- 80.Li X, Moellering ER, Liu B, Johnny C, Fedewa M, Sears BB, Kuo MH, Benning C. 2012. A galactoglycerolipid lipase is required for triacylglycerol accumulation and survival following nitrogen deprivation in Chlamydomonas reinhardtii. Plant Cell 24, 4670–4686. ( 10.1105/tpc.112.105106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Liang Y, Osada K, Sunaga Y, Yoshino T, Bowler C, Tanaka T. 2015. Dynamic oil body generation in the marine oleaginous diatom Fistulifera solaris in response to nutrient limitation as revealed by morphological and lipidomic analysis. Algal Res. 12, 359–367. ( 10.1016/j.algal.2015.09.017) [DOI] [Google Scholar]
- 82.Burrows EH, Bennette NB, Carrieri D, Dixon JL, Brinker A, Frada M, Baldassano SN, Falkowski PG, Dismukes GC. 2012. Dynamics of lipid biosynthesis and redistribution in the marine diatom Phaeodactylum tricornutum under nitrate deprivation. Bioenergy Res. 5, 876–885. ( 10.1007/s12155-012-9201-7) [DOI] [Google Scholar]
- 83.Vinayak V, Manoylov KM, Gateau H, Blanckaert V, Herault J, Pencreac'h G, Marchand J, Gordon R, Schoefs B. 2015. Diatom milking: a review and new approaches. Mar. Drugs 13, 2629–2665. ( 10.3390/md13052629) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Uduman N, Qi Y, Danquah MK, Forde GM, Hoadley A. 2010. Dewatering of microalgal cultures: a major bottleneck to algae-based fuels. J. Renew. Sustain. Energy 2, 012701 ( 10.1063/1.3294480) [DOI] [Google Scholar]
- 85.Sander K, Murthy GS. 2010. Life cycle analysis of algae biodiesel. Int. J. Life Cycle Assess. 15, 704–714. ( 10.1007/s11367-010-0194-1) [DOI] [Google Scholar]
- 86.Xue J, Niu YF, Huang T, Yang WD, Liu JS, Li HY. 2015. Genetic improvement of the microalga Phaeodactylum tricornutum for boosting neutral lipid accumulation. Metab. Eng. 27, 1–9. ( 10.1016/j.ymben.2014.10.002) [DOI] [PubMed] [Google Scholar]
- 87.Osada K, Maeda Y, Yoshino T, Nojima D, Bowler C, Tanaka T. 2017. Enhanced NADPH production in the pentose phosphate pathway accelerates lipid accumulation in the oleaginous diatom Fistulifera solaris. Algal Res. 23, 126–134. ( 10.1016/j.algal.2017.01.015) [DOI] [Google Scholar]
- 88.Rodriguez RE, et al. 2007. Transgenic tobacco plants overexpressing chloroplastic ferredoxin-NADP(H) reductase display normal rates of photosynthesis and increased tolerance to oxidative stress. Plant Physiol. 143, 639–649. ( 10.1104/pp.106.090449) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Li Z, Sun HX, Mo XM, Li XY, Xu B, Tian P. 2013. Overexpression of malic enzyme (ME) of Mucor circinelloides improved lipid accumulation in engineered Rhodotorula glutinis. Appl. Microbiol. Biotechnol. 97, 4927–4936. ( 10.1007/s00253-012-4571-5) [DOI] [PubMed] [Google Scholar]
- 90.Zhang Y, Adams IP, Ratledge C. 2007. Malic enzyme: the controlling activity for lipid production? Overexpression of malic enzyme in Mucor circinelloides leads to a 2.5-fold increase in lipid accumulation. Microbiology 153, 2013–2025. ( 10.1099/mic.0.2006/002683-0) [DOI] [PubMed] [Google Scholar]
- 91.Meng X, Yang J, Cao Y, Li L, Jiang X, Xu X, Liu W, Xian M, Zhang Y. 2011. Increasing fatty acid production in E. coli by simulating the lipid accumulation of oleaginous microorganisms. J. Ind. Microbiol. Biotechnol. 38, 919–925. ( 10.1007/s10295-010-0861-z) [DOI] [PubMed] [Google Scholar]
- 92.Chen HQ, Hao GF, Wang L, Wang HC, Gu Z, Liu LM, Zhang H, Chen W, Chen YQ. 2015. Identification of a critical determinant that enables efficient fatty acid synthesis in oleaginous fungi. Sci. Rep. 5, 11247 ( 10.1038/srep11247) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Bailleul B, et al. 2015. Energetic coupling between plastids and mitochondria drives CO2 assimilation in diatoms. Nature 524, 366–369. ( 10.1038/nature14599) [DOI] [PubMed] [Google Scholar]
- 94.Levering J Broddrick J, Dupont CL, Peers G, Beeri K, Mayers J, Gallina AA, Allen AE, Palsson BO, Zengler K. 2016. Genome-scale model reveals metabolic basis of biomass partitioning in a model diatom. PLoS ONE 11, e0155038 ( 10.1371/journal.pone.0155038) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kim J, Fabris M, Baart G, Kim MK, Goossens A, Vyverman W, Falkowski PG, Lun DS. 2016. Flux balance analysis of primary metabolism in the diatom Phaeodactylum tricornutum. Plant J. 85, 161–176. ( 10.1111/tpj.13081) [DOI] [PubMed] [Google Scholar]
- 96.Khozin-Goldberg I, Shrestha P, Cohen Z. 2005. Mobilization of arachidonyl moieties from triacylglycerols into chloroplastic lipids following recovery from nitrogen starvation of the microalga Parietochloris incisa. Biochim. Biophys. Acta 1738, 63–71. ( 10.1016/j.bbalip.2005.09.005) [DOI] [PubMed] [Google Scholar]
- 97.Siaut M, et al. 2011. Oil accumulation in the model green alga Chlamydomonas reinhardtii: characterization, variability between common laboratory strains and relationship with starch reserves. BMC Biotechnol. 11, 7 ( 10.1186/1472-6750-11-7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Přibyl P, Cepák V, Zachleder V. 2013. Production of lipids and formation and mobilization of lipid bodies in Chlorella vulgaris. J. Appl. Phycol. 25, 545–553. ( 10.1007/s10811-012-9889-y) [DOI] [Google Scholar]
- 99.Mulders KJ, Lamers PP, Wijffels RH, Martens DE. 2015. Dynamics of biomass composition and growth during recovery of nitrogen-starved Chromochloris zofingiensis. Appl. Microbiol. Biotechnol. 99, 1873–1884. ( 10.1007/s00253-014-6181-x) [DOI] [PubMed] [Google Scholar]
- 100.Valenzuela J, Carlson RP, Gerlach R, Cooksey K, Peyton BM, Bothner B, Fields MW. 2013. Nutrient resupplementation arrests bio-oil accumulation in Phaeodactylum tricornutum. Appl. Microbiol. Biotechnol. 97, 7049–7059. ( 10.1007/s00253-013-5010-y) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Yamaguchi T, Osumi T. 2009. Chanarin-Dorfman syndrome: deficiency in CGI-58, a lipid droplet-bound coactivator of lipase. Biochim. Biophys. Acta 1791, 519–523. ( 10.1016/j.bbalip.2008.10.012) [DOI] [PubMed] [Google Scholar]
- 102.Barka F, Angstenberger M, Ahrendt T, Lorenzen W, Bode HB, Buchel C. 2016. Identification of a triacylglycerol lipase in the diatom Phaeodactylum tricornutum. Biochim. Biophys. Acta 1861, 239–248. ( 10.1016/j.bbalip.2015.12.023) [DOI] [PubMed] [Google Scholar]
- 103.Eastmond PJ. 2006. SUGAR-DEPENDENT1 encodes a patatin domain triacylglycerol lipase that initiates storage oil breakdown in germinating Arabidopsis seeds. Plant Cell 18, 665–675. ( 10.1105/tpc.105.040543) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This article has no additional data.