Abstract
In microalgae, the photosynthesis-driven CO2 assimilation delivers cell building blocks that are used in different biosynthetic pathways. Little is known about how the cell regulates the subsequent carbon allocation to, for example, cell growth or for storage. However, knowledge about these regulatory mechanisms is of high biotechnological and ecological importance. In diatoms, the situation becomes even more complex because, as a consequence of their secondary endosymbiotic origin, the compartmentation of the pathways for the primary metabolic routes is different from green algae. Therefore, the mechanisms to manipulate the carbon allocation pattern cannot be adopted from the green lineage. This review describes the general pathways of cellular energy distribution from light absorption towards the final allocation of carbon into macromolecules and summarizes the current knowledge of diatom-specific allocation patterns. We further describe the (limited) knowledge of regulatory mechanisms of carbon partitioning between lipids, carbohydrates and proteins in diatoms. We present solutions to overcome the problems that hinder the identification of regulatory elements of carbon metabolism.
This article is part of the themed issue ‘The peculiar carbon metabolism in diatoms’.
Keywords: carbon allocation, photosynthesis, energy balance
1. Introduction
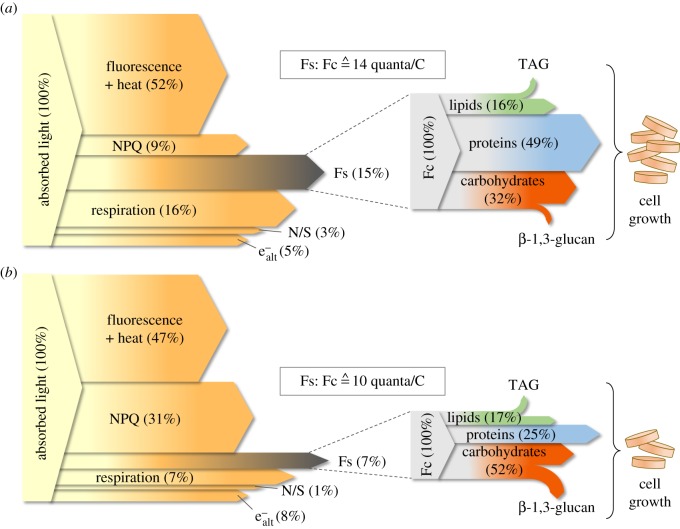
Carbon allocation in diatoms is defined as the partitioning of photosynthetically assimilated carbon into the pools of different macromolecules, i.e. proteins, lipids, carbohydrates and nucleic acids. Under optimal conditions of light, nutrient availability and temperature, the carbon flux for the synthesis of cell macromolecules (figure 1; Fc) is in equilibrium with the energy flux provided by photosynthesis (Fs). However, changing environmental conditions can result in a loss of equilibrium between these two fluxes. Therefore, microalgae require efficient regulatory responses to promptly adjust them and meet their needs for growth. To illustrate the energetic pathways in diatom cells from absorbed light to carbon allocation to sustain cell growth, figure 1 depicts two different scenarios based on results of the study of Jakob et al. [1]. Scenario 1 (figure 1a) represents diatom cells cultivated under nutrient-rich conditions in combination with moderate light. The carbon allocation pattern revealed a high proportion of proteins, which is necessary to support a high growth rate. As a consequence, there was a high demand for electrons per assimilated carbon. In this case, Fs is in equilibrium with Fc and the necessity for the cells to dissipate excessively absorbed light or to store excessive carbon is low (see below). By contrast, scenario 2 summarizes the energy and carbon fluxes of cells grown under nitrogen-limited conditions in combination with high light (figure 1b). Nitrogen limitation drastically reduced the growth rate and the carbon flux (Fc) and, more importantly, changed the carbon allocation pattern to a biomass with a significant lower proportion of proteins and an increased fraction of carbohydrates. As the synthesis of carbohydrates requires fewer electrons per carbon than the synthesis of proteins, the cells were forced to strongly downregulate the photosynthetic energy flux (Fs) by the increase in energy dissipation measured as non-photochemical quenching (NPQ) and alternative electron pathways (e−alt).
Figure 1.
Schematic of cellular pathways of photosynthetic energy and carbon allocation in diatoms based on two different scenarios with either (a) optimal growth conditions or (b) nitrogen-limited conditions according to the results of Jakob et al. [1]. The fractions of the different pathways of photosynthetic energy were calculated based on the amount of absorbed radiation. At PSII, photosynthetically absorbed photons are converted into electrons that are fed to the electron transport chain. Here, different options (energy sinks) exist to dissipate excess energy (see text for details). At PSII, energy can be dissipated unregulated as fluorescence and heat. In addition, cells can actively regulate energy dissipation via the mechanism of non-photochemical quenching (NPQ). Additional electron sinks are represented by alternative electron pathways (e−alt) and the reduction in nitrate and sulphate (N/S). Basic metabolic energy demand is covered by respiration. Finally, the remaining net energy flux (denoted as ‘Fs’) delivers the reductants and the energy equivalents to reduce CO2 in the Calvin cycle to synthesize building blocks for the subsequent biochemical pathways. In this respect, ‘Fc’ denotes the cellular carbon flux that is designated to afford cell growth/cell division and storage. Excess carbon could be stored as neutral lipids (triacylglyceroles, TAG) and polysaccharides (chrysolaminarin, β-1,3-glucan). The calculation of Fc was based on total amount of carbon in the cell biomass, whereas the carbon allocation pattern was derived from the relative fractions of lipids, proteins and carbohydrates in the biomass. The scaling of Fc in relation to Fs was based on the ratio Fs : Fc which was calculated from the measured net fluxes of photosynthetic energy and carbon production, respectively [1].
Diatoms are species particularly successful in highly dynamic habitats [2]. As a consequence, diatoms are forced to constantly re-adjust the equilibrium between Fs and Fc under these environmental conditions. Whereas regulatory elements of the photosynthetic flux are sufficiently characterized in diatoms [3], there is only limited knowledge about the way carbon allocation is regulated. This is an important issue because carbon allocation does not only influence Fc but also Fs (see above). The circumstance is further complicated by the fact that diatoms differ from green algae due to their distinct metabolic compartmentation [4,5] and to the presence of additional metabolic pathways (e.g. urea cycle, see below) [6]. Therefore, the identification of the regulatory elements in carbon allocation is of paramount importance not only in the context of predicting the changes in global element cycling under climate change conditions but also for biotechnological applications. The modern techniques of genetic transformation by means of gene editing open new perspectives of metabolic engineering to use algal cells as factories for the production of highly valuable products [7]. However, a sound engineering strategy by gene manipulation must be based on knowledge about the sensors involved, the signal transduction pathways and the target genes of regulatory elements.
From these observations, it is obvious that photosynthetic energy and carbon allocation cannot be regulated independently of each other. Therefore, this review presents the current knowledge about the regulation of metabolic pathways in balancing the energy and carbon fluxes in diatoms, starting from light absorption to end with the partitioning of carbon within the cellular macromolecule pools. Furthermore, we highlight the current restrictions to identifying the regulatory elements of carbon allocation and present possible solutions to overcome these problems.
2. Photosynthetic energy allocation and metabolic costs
The quantum efficiency of biomass production is determined not only by the yield of photosynthetic light reactions but also by the overall energetic costs of cellular metabolism. Thus, metabolic costs depend on, for example, energy requirements for nutrient acquisition, defence mechanisms, the biomass composition, the cell cycle activity and stress-induced repair activities (reviewed in [8]). An understanding of the ecological success of algal groups or even species under natural conditions (e.g. in the presence of competition or nutrient limitation) requires knowledge of the quantum efficiency of specific metabolic processes and the partitioning of photosynthetically acquired carbon. Moreover, to use algae/diatoms for biotechnological applications, knowledge of metabolic pathways, their quantum efficiency and regulation is essential, either to search for suitable strains or to apply metabolic engineering to generate cells that preferentially incorporate carbon into desired products with an optimized efficiency.
A suitable approach to reveal the quantum efficiency of different levels of carbon allocation in algae under specific growth conditions is the measurement of energy balances [1,9]. The principle of these measurements is the differentiation between certain levels of cellular energy/carbon allocation by the application of a set of methods, e.g. measurement of cellular light absorption (Qphar), active Chl a-fluorescence, O2/CO2 gas exchange and the analysis of biomass composition. In this way, at least five levels of cellular metabolism can be distinguished and analysed: (i) the efficiency of the primary photosynthetic reaction at photosystem II (PSII), (ii) the dissipation of absorbed light energy by regulated/non-regulated non-photochemical processes and energy dissipation by fluorescence, (iii) the activity of alternative electron sinks, (iv) mitochondrial respiratory losses, and (v) the degree of reduction in cellular biomass as a function of its macromolecular composition. In the following sections, it is intended to describe the relevance and the quantitative impact of these metabolic levels on the energy balance in diatoms.
(a). Light absorption
A prerequisite to calculating energy balances of the carbon that is allocated into different metabolic sinks (growth, storage or excretion) is the knowledge of the amount of absorbed light energy (Qphar; [10]). Qphar can be estimated by measuring the in vivo-absorption spectra of cell suspensions and by calculating the Chl a-absorption coefficient (a*phy). This coefficient can be defined as the effective absorption area per Chl and depends on the packaging of Chl molecules within the chloroplast membrane. It was shown that a*phy varies drastically depending on the size and the nutrient and light acclimation status of microalgae [1,11,12]. Besides the requirement of Qphar for the calculation of the quantum efficiency of photosynthesis and carbon allocation, this information is also useful to estimate, for example, the maximum cell loading for specific reactor designs/dimensions, or to estimate the potential of biomass production of metabolically engineered algae [13].
(b). Quantum yield of photosystem II and dissipation of excess energy
For an energy balance, the measurement of the effective PSII quantum yield under illumination is also required. This effective quantum yield is influenced by the absorption efficiency of the cell (see above) in relation to the capacity of the electron transport chain (ETC). In the case of a sudden increase in illumination, the capacity of the ETC may not be sufficient, thus an increasing excitation pressure at PSII requires pathways that can safely dissipate excessive absorbed energy via regulated NPQ. Therefore, the effective quantum yield of PSII and the extent of NPQ are indirectly correlated [14].
The advantage to diatoms of using NPQ as an energy dissipation mechanism is the very high capacity in combination with fast activation kinetics (i.e. in the order of a few minutes [15]). In particular, diatoms profit from the NPQ mechanism because their habitats are zones of high turbidity with strong light gradients [16]. Diatoms generally possess a higher regulated NPQ capacity than green algae, and therefore, the ecological importance of NPQ in diatoms has been intensively investigated during the last 10 years [17]. However, it was also shown that diatoms can be divided into two types with either higher or lower NPQ capacity, indicating that excessive energy may be dissipated by other pathways in the low NPQ type [18–20].
A high NPQ capacity has the disadvantage that specific pigments, proteins and enzymes are required. Moreover, the dissipated energy is inevitably lost as heat, which is in contrast with the energy dissipation via alternative electron pathways (see below). On the other hand, the regulated NPQ prevents the formation of harmful oxygen species and its capacity exceeds the fluxes of alternative electron pathways by several factors.
(c). Alternative electron pathways
In the linear electron transport pathway, electrons released at PSII will be used to reduce NADP+ to NADPH. This process takes place at photosystem I (PSI) and requires a second electron excitation step induced by the absorption of an additional photon. PSI is also the branching point for electrons consumed in the so-called alternative electron pathways. Alternative electrons will not be used for NADPH production but for the reduction in nitrate, sulfate, oxygen (Mehler reaction) and for electron cycling around PSI. These alternative electron pathways have in common that they do not produce reducing equivalents, but they contribute to the proton gradient across the thylakoid membrane and, thus, to ATP production. Therefore, the energy of alternative electrons is not lost per se, but it is used to adjust the NADPH/ATP ratio in the chloroplast under unbalanced cell conditions.
The activity of cyclic electron transfer around PSI appears to be low in diatoms and thus, is not involved in the regulation of the ATP/NADPH ratio. Instead, the interaction and energetic exchange between chloroplast and mitochondrium plays an important role in the optimization of the cellular demand for ATP and NADPH in diatoms, a situation very different from cells of the green lineage [21].
Owing to methodological constraints, only limited quantitative data for electron consumption by Mehler reaction or the PSI cycle in diatoms are available. Nevertheless, light-induced oxygen uptake by Mehler reaction was estimated to be an important alternative electron sink (20–50% of gross oxygen production at PSII) under unbalanced growth conditions [22–24]. The electron requirement for the reduction in nitrate and sulfate to meet the cellular demand for synthesis of new biomass is rather low. Under nitrogen-replete conditions (low C : N ratio), it could be estimated to yield a maximum of 15% of electrons released at PSII (calculated on the basis of published data [1,20]). However, the study of Lomas & Glibert [25] shows that diatoms can store significant amounts of nitrate in excess with respect to the biosynthetic demand. They can further excrete significant amounts of reduced nitrogen into the surrounding medium, particularly under conditions of changing water temperature. In this way, reduction in nitrate may represent an important electron sink in diatoms. This assumption is further supported by a recent study showing that the nitrogen metabolism in the diatom Phaeodactylum tricornutum is redox-regulated [26]. These characteristics enable the nitrate reduction pathway and subsequent reactions, such as the urea cycle and photorespiration [6], to be a fast-responding electron sink under changing environmental conditions.
There are three additional alternative electron transport pathways that transfer electrons back to oxygen but do not take place at PSI: photorespiration, chlororespiration and cyclic electron transport around PSII. Owing to the presence of carbon-concentrating mechanisms (CCMs) in diatoms, the activity of photorespiration can be assumed to be relatively weak [3,27]. On the other hand, based on the analysis of transcript levels of photorespiratory enzymes, there is evidence for increased photorespiratory activity in a diatom under conditions of high light and cool temperatures [28]. This is a very interesting finding, with respect to the excessive reduction in nitrate in diatoms under comparable growth conditions (see above) and it implies a role of photorespiration by the incorporation of reduced N (ammonium) into amino acids (see also [29,30]). However, there are only very limited quantitative data about the impact of photorespiration as a sink for alternative electrons. In Skeletonema costatum [31], it was shown that photorespiration consumes only a few per cent of the total photosynthetic electron transport.
The chlororespiratory reduction in oxygen by stromal reductants can be important in diatoms under certain environmental conditions such as prolonged darkness [32]. However, it is assumed that chlororespiration is not active under actinic illumination [33] and will not be considered in detail in this review.
The cyclic electron transport around PSII re-donates electrons to the reaction centre and can be detected as a deficit of oxygen evolution under illumination with single-turnover flashes [34]. In diatoms, a strong activity of the PSII cycle was observed under high light stress [18]. Recently, it was shown that nitrogen limitation in combination with dynamic light conditions significantly enhances PSII-cycle activity in a diatom [35]. From the correlation of the measured oxygen deficit and the extent of alternative electron flow, it was further concluded that the PSII cycle could play a significant role in the dissipation of excessive photosynthetic energy under unbalanced growth conditions. However, a precise quantification of PSII cyclic electron transport is not possible due to methodological constraints.
(d). Respiratory losses and degree of reduction in biomass
Mitochondrial respiration (including the citric acid cycle and oxidative phosphorylation) is not easy to integrate in quantitative terms into energy/carbon balances of microalgae [36]. On the one hand, the final reduction in molecular oxygen to water is coupled to a release of CO2, thereby influencing the amount and the quantum efficiency of carbon fixed into the cellular biomass. On the other hand, respiration delivers NADH and ATP for the synthesis of cellular metabolites and thus, it is not a loss process with respect to cellular energy. However, the latter could be biased by alternative respiration, which dissipates excess reductants by the mitochondrial alternative oxidase (AOX) in combination with strongly reduced ATP production [37]. In diatoms, alternative respiration can be an important valve to dissipate cellular energy because it contributes to 50% of mitochondrial respiration [21].
The impact of respiratory activity on the energy balance of microalgae depends on their metabolic activity. Thus, respiration is different in slow- or fast-growing cells [36] and in response to changed environmental conditions, like temperature [38], nutrient status [1] or day length [39,40]. The metabolic activity of microalgae is also influenced by their macromolecular stoichiometry and the degree of reduction in cellular macromolecules. This is explained by the fact that the synthesis of carbohydrates requires four electrons per carbon, whereas the synthesis of lipids and protein strongly increases the electron requirement to a value of greater than 6 [41]. The metabolic energy for biosynthesis of macromolecules is delivered by both respiration and photosynthesis. Therefore, depending on the relative fractions of carbohydrates, lipids and proteins in the biomass, the photosynthetic quotient (net O2 evolution rate/net CO2 consumption rate) may vary in diatoms between 1.1 and 1.8 ([1]; W Su et al. 2012, unpublished data for S. costatum and Cyclotella meneghiniana).
3. The biochemical phenotype of diatoms
(a). Biomass composition under optimal growth conditions
From previous studies, it is evident that the macromolecular composition of microalgae depends on environmental/experimental conditions and on the resulting growth rate. It is, therefore, not surprising that the relative cellular proportions of carbohydrates, proteins and lipids vary tremendously in the comparison of different studies (see §3b). However, a recent work allows a general description of the biochemical phenotype of diatoms in comparison to other microalgae groups [42]. Accordingly, under nutrient-replete conditions, the mean lipid content of diatoms accounts for 19% of dry weight (DW) which is higher than the median values over all microalgae groups (17% lipids of DW), whereas the mean carbohydrate content (12% DW) and protein content (27% DW) of diatoms is lower (15% carbohydrates, 32% proteins; mean of all species). Obviously, diatoms possess a significantly lower protein : lipid ratio (1.7) and carbohydrate : lipid ratio (0.6) compared with the microalgae group-independent mean values of 2.2 and 0.9, respectively [42]. An additional peculiarity of diatoms is the presence of a silica shell that can make up to 10% of cellular DW [43]. This increases the cellular ash content in diatoms to 27% compared with a median value over all microalgae groups of 17% ash content [42]. It has to be pointed out that the diatom P. tricornutum is an important exception that contains only a marginal amount of silica as this species does not develop a typical silica shell. Nevertheless, this diatom species is used as a model organism for physiological studies and is one of the few diatom species with a fully sequenced genome [44].
(b). Biochemical spillover and biomass composition under unbalanced conditions
Under optimum growth conditions (high growth rate), photosynthetically fixed carbon is initially guided into carbohydrates during the light period and is detectable as diurnal variation of the cellular carbohydrate content [35,36,45]. Apart from these diurnal changes of macromolecular composition, changes in growth conditions can force microalgae into unbalanced conditions of fluxes (changed ratio of ratio of Fs : Fc; figure 1) and will drastically change the carbon allocation pattern.
The equilibrium between Fs and Fc can be easily disturbed when low light acclimated cells are transferred to supersaturating light conditions (high Fs : Fc). Besides the induction of photoprotective mechanisms (e.g. NPQ), a typical reaction to high light stress is the reorganization of the proteome. As a consequence, metabolic carbon fluxes (Fc) are adjusted by increasing cell growth and carbohydrate storage, whereas Fs is decreased by antenna size reduction [46]. Especially in diatoms, this could also result in a higher proportion of lipids at the expense of proteins and carbohydrates [47–49].
Not only light but also nutrient availability disturbs the balance between Fs and Fc, leading to decreased growth rates and to drastic changes in the allocation of carbon to the different macromolecular pools [36,50]. In particular, N limitation was intensively studied in diatoms, because the shift from replete to starved conditions is a common process in the natural environment and strongly influences the function of diatoms as a CO2 sink in the ocean. During a transition from replete to N-limited conditions, the cells must adjust their carbon allocation pattern to the N availability by reducing the N-containing components such as amino acids, proteins and RNAs [48]. This reduction leads to an altered allocation of the photosynthetically produced building blocks into other pathways, most probably towards storage compound synthesis (metabolic spillover) [51]. In addition, N-limited cells must reduce the amount of energy provided by photochemistry in a feedback regulatory mechanism. Thus, phenomenological, N-limited cells are characterized by less chlorophyll, less thylakoids and a higher content of storage compounds. In diatoms, the latter can be neutral lipids [52,53], chrysolaminarin [52,53] or even both components [54]. Consequently, the response of the cells to N shortage requires a complex reorganization of the metabolic fluxes into the different pathways of protein, carbohydrate and lipid synthesis [55].
Besides nitrogen, diatoms are known to be frequently limited by the availability of iron, silicon and phosphate. Iron-limited cells in the open ocean as well as in laboratory-controlled experiments are characterized by a decreased size of the photosynthetic apparatus mainly due to a lower amount of PSI, of the cytochrome complex and to a minor extent to a lower amount of PSII [56]. This reorganization of the ETC causes lower photosynthetic performance [57,58]. Although there are species-specific differences in the response to iron limitation [57,59], generally, diatom cells tend to have lower growth rates, lower iron per carbon amounts, but increased specific cell size [37,56,58]. It was further observed that the release of cells from iron starvation induced changes in transcription levels of components of the metabolic network [60] and resulted in a considerable reorganization of the proteome [61]. Thus, changes in carbon allocation pattern may also be expected. Unfortunately, detailed analyses of diatoms' macromolecular composition in response to iron limitation are scant as yet [62].
Further studies have been performed on freshwater diatoms in response to phosphorus and silicon limitation [43,63–65]. Both nutrients play an important role in freshwater systems, because phosphorus can be the main limiting factor of microalgae growth and is a key factor in controlling water quality. In summary, almost all of the studies describing diatoms' response to phosphate and/or silicon limitation show a similar pattern: the growth rate decreases, the pigment as well as protein content are downregulated and storage products tend to accumulate [43,66]. Interestingly, in diatoms, the preference to store either lipids or carbohydrates is species-dependent [43,67,68].
From these data, one can conclude that changes of environmental conditions force microalgae into an unbalanced state. Typically, under these settings, the ratio of energy flux from photosynthesis (Fs) to the flux of carbon (Fc) is much higher and cells must react to avoid futile carbon waste or damage. The response includes changes at the level of the photosynthetic electron pathways that downregulate Fs, but also changes at the level of carbon allocation to keep Fc as high as possible. In the latter case, it is obvious that even within the group of diatoms, there is not only one strategy to handle the excess of metabolic energy (see above). Unfortunately, our knowledge about the regulation of the different pathways of metabolic spillover is very scarce (see below).
4. Regulatory elements determining the biochemical phenotype
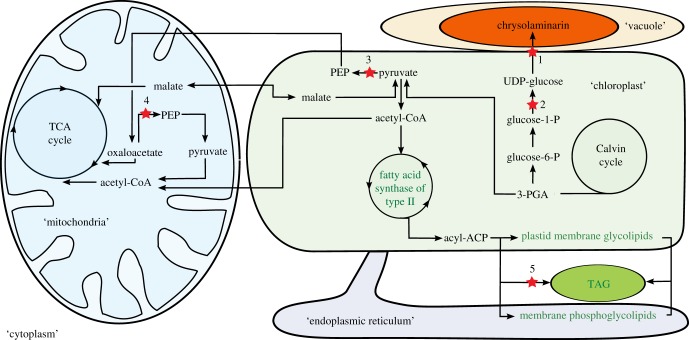
Today, a variety of newly developed methods can be applied for the identification and characterization of regulatory factors of cell metabolism. Genome sequence data are available and genetic transformation has been made possible in diatoms [69]. With the recent application of the so-called ‘omics’ approaches, huge amounts of metabolic data, as well as transcriptomes, became available and the outcome of those data will further increase in the future [70–72]. Furthermore, complex metabolic models can illustrate processes related to macromolecular metabolism [73–75]. Based on these recent studies, a couple of metabolic key processes and possible regulators in diatoms have been described and will be briefly discussed here (figure 2).
Figure 2.
Schematic map of a diatom's carbon metabolism and the metabolic compartmentalization of carbon allocation. Lipid metabolism (green pathways) ends up with TAG (triacylglycerol) as storage compound, whereas carbohydrate storage (orange) as chrysolaminarin is located in the vacuole. Red stars indicate genetically modified genes altering the carbon allocation pattern described within the text: 1, chrysolaminarin synthase [76]; 2, UDP-glucose pyrophosphorylase (UGPase) [77]; 3, PPDK [78]; 4, PEP carboxykinase (PEPCK) [79]; 5, diacylglycerol acyltransferase [80].
A specific biochemical feature of diatoms is the storage of carbohydrates outside of the chloroplast as chrysolaminarin, a β-1,3-glucan. Unfortunately, until now, it was not possible to uncover the entire biochemistry of the chrysolaminarin synthetic pathway. However, whole-genome analysis could identify key enzymes and their subcellular distribution [73]. Genetic transformation of these identified genes can be further used to investigate their regulatory function. The silencing of the chrysolaminarin synthase, for example, resulted in a re-distribution of assimilated carbon into lipids [76]. A similar carbon allocation pattern was observed upon silencing the chloroplast-located UDP-glucose pyrophosphorylase [77,81]. Thus, it could be hypothesized that the branching and regulatory point for the storage of excess carbon as carbohydrates or as lipids is localized in the chloroplast (figure 2).
This assumption is supported by investigations on diatoms impaired in specific C4-cycle enzymes. Originally, the C4 cycle in diatoms was suggested to operate as a CCM [82]. However, more recent studies suggest a role in dissipation of excess energy and in pH homeostasis [78]. Genome analysis could identify the proteins required for the C4 pathway and their putative subcellular localization in a diatom [73]. The pyruvate-orthophosphate dikinase (PPDK), for example, catalyses the phosphorylation of pyruvate into phosphoenolpyruvate (PEP) (figure 2). Interestingly, the knockdown of PPDK induced an accumulation of lipids and carbohydrates in P. tricornutum [78]. From this result, it was concluded that the decreased consumption of pyruvate by PPDK can induce a metabolic imbalance in the chloroplast. It should be noted that the growth rate of wild-type and mutant strains of P. tricornutum was similar in the study of Haimovich-Dayan et al. [78]. This is a strong indication that silencing of PPDK did not change the cellular metabolic carbon flux in quantitative terms, which is in contrast with, for example, experiments under nutrient limitation. Therefore, the evaluation of an energy balance and the quantitative knowledge of the macromolecular composition would allow the analysis of the energy/carbon flux into different pools of mutant strains under, for example, different intensity of illumination and a thereby induced metabolic imbalance.
The investigation of C4 pathway enzymes in diatoms revealed another possible site of carbon allocation regulation in the mitochondrial metabolism. The PEP carboxykinase (PEPCK) decarboxylates oxaloacetate to PEP, thereby draining carbon skeletons from the tricarboxylic acid (TCA) cycle (figure 2). In the study of Yang et al. [79], it was shown that the downregulation of PEPCK in P. tricornutum was accompanied by a slight increase in the content of neutral lipids. Moreover, in the PEPCK-silenced cells, the transcription level of the fumarate hydratase increased significantly, indicating that intermediates of the TCA cycle may have accumulated. From the results, it could be concluded that the PEPCK is involved in lipid metabolism in diatoms. However, the regulatory mechanism remains unknown.
As lipid production is in the research focus of biotechnical applications, there is strong interest in uncovering the whole pathway in diatoms. In very early studies, the evaluation started with the phenotypical selection of lipid-producing strains and it became clear that high lipid production is always coupled to conditions that severely decrease growth rates. To increase lipid productivity, it is attempted to understand lipid metabolism and its regulation in order to generate high-lipid-containing cell lines by specific genetic modifications. The study of Tanaka et al. [83] demonstrates that genome and transcriptome data of diatoms could be used to select promising candidate species. Moreover, such an approach provides valuable information at least on targets of regulation (e.g. fatty acid biosynthesis versus β-oxidation) but not yet on the regulatory mechanism itself. In a more specific approach, the overexpression of diacylglycerol acyltransferase, for example, increased lipid concentration and the number of lipid bodies [80] (figure 2) which points to a potential regulatory step.
Another approach to reveal metabolic pathways and even metabolic fluxes is genome-scale modelling [75,84]. Such a modelling allows in silico prediction of carbon allocation under different growth conditions and the prediction of metabolic effects of genetic modifications [75]. However, these models are based on assumptions about the pathway regulatory elements which are not yet proved for diatoms.
5. Experimental set-up for the identification of regulatory elements of carbon allocation
As outlined above, previous approaches to identify regulatory mechanisms of cellular metabolism (e.g. nutrient limitation) mostly employed conditions where the cellular source/sink ratio (e.g. capacity of primary photosynthetic reaction versus carbon fixed in the biomass) was changed drastically. This resulted in changes of the excitation pressure at photosystems but also in shifting the degree of reduction in the ETC or even of the cellular redox balance. Therefore, under these conditions, it is difficult or even impossible to distinguish between primary and secondary regulatory effects on cellular metabolism. Therefore, it does not matter which approach is used, either nutrient limitation or genetic transformation. Blocking one or more key enzymes as a possible regulatory step does not explain regulation by itself, but results in a changed source/sink ratio of the metabolites, thereby inducing secondary reactions from the regulatory network.
An alternative approach to investigate changes in cell metabolism and to identify targets of metabolic regulation in diatoms is the carbon flux analysis of wild-type cells grown under different light quality but equal amounts of absorbed photosynthetically active quanta. In this case, the primary reactions of photosynthesis are not influenced by the light climate, thus changes in the metabolic phenotype are only due to different metabolic regulations. In diatoms, light quality is sensed by photoreceptors of the cryptochrome and aureochrome family [85]. It was shown that the growth of P. tricornutum under red light (RL) or blue light (BL) illumination with the same amount of absorbed quanta resulted in similar steady-state growth rates but in a very different photoacclimation status of the cells [86]. This means that these growth conditions did not change the source/sink ratio but obviously influenced the regulatory network of the cells. Moreover, a recent study revealed that a quantum-controlled shift RL to BL illumination or vice versa induced severe changes in the metabolome and in the macromolecular composition of P. tricornutum within a time frame of minutes to hours [87]. Accordingly, the shift from RL to BL induced a significant increase in metabolites of glycolysis, TCA cycle and amino acids within 15–30 min followed by a significant increase in carbon partitioning into proteins within 2 h after the shift. By contrast, the metabolite profile revealed a decreased abundance of amino acids, an increase in metabolites of the pentose phosphate pathway and a significant increase in the carbohydrate pool during the shift from BL to RL illumination. As the BL receptor aureochrome is a light-regulated transcription factor, these changes can be interpreted both as a result of regulated enzyme activity and of changes on the transcriptional level. This assumption is in agreement with the study of Rosenwasser et al. [26] where the redox regulation of enzymes of glycolysis, the nitrate reduction pathway and amino acid synthesis was proven. In addition to redox regulation, the analysis of changes in the transcriptome of P. tricornutum WT cells in comparison to Aureochrome-1a knockout mutants revealed that aureochromes play a crucial role in the regulation of cellular processes on the transcriptomic level (M Mann et al. 2017, unpublished results). Currently, it is not known whether aureochromes are also involved in short-term regulation (e.g. as a master regulator) of cellular metabolism in diatoms.
Importantly, shifts in light quality on a short-term scale do not change the availability of nutrients or the absorption efficiency of the cells. Therefore, it could be concluded that the source/sink ratio is not changed during light quality shift experiments and any changes in the energy/carbon allocation profile of cells should be induced by primary regulatory mechanisms. This opens the possibility to study targets of regulation of cellular metabolism by means of light quality shift experiments.
6. Conclusion
In diatoms, the metabolic pathways from trioses to the macromolecules forming new cells differ from the green algal lineage due to a different compartmentalization of enzymes and to peculiar mechanisms of regulation. Owing to the silica shell, the cell wall has only a minor demand in carbohydrates with the consequence that the diatom biomass is enriched in lipids and proteins compared with other algal taxa. All together these facts result in a different C-allocation regulation in diatoms, whose mechanisms are not yet understood. Recent advances in transgenic cell lines and the discovery of aureochromes as genetic and metabolic regulators open the future perspective to understand the regulatory network in more detail. This progress would help to generate diatom cells engineered for bio-production of valuable product on a knowledge-based strategy.
Data accessibility
This article has no additional data.
Authors' contributions
H.W., T.J. and C.W. conducted conception and design; H.W., A.F. and T.J. performed research and analysed data; A.F., H.W., T.J. and C.W. wrote the paper and finally approved the version to be published.
Competing interests
We declare we have no competing interests.
Funding
We would like to thank the Bundesanstalt für Gewässerkunde (BfG) for financial support as well as the Deutsche Forschungsgemeinschaft (DFG) (grant nos Wi 764/10, Wi 764/14 and Wi 764/19).
References
- 1.Jakob T, Wagner H, Stehfest K, Wilhelm C. 2007. A complete energy balance from photons to new biomass reveals a light- and nutrient-dependent variability in the metabolic costs of carbon assimilation. J. Exp. Bot. 58, 2101–2112. ( 10.1093/jxb/erm084) [DOI] [PubMed] [Google Scholar]
- 2.Denman KL, Gargett AE. 1983. Time and space scales of vertical mixing and advection of phytoplankton in the upper ocean. Limnol. Oceanogr. 28, 801–815. ( 10.4319/lo.1983.28.5.0801) [DOI] [Google Scholar]
- 3.Wilhelm C, et al. 2006. The regulation of carbon and nutrient assimilation in diatoms is significantly different from green algae. Protist 157, 91–124. ( 10.1016/j.protis.2006.02.003) [DOI] [PubMed] [Google Scholar]
- 4.Smith SR, Abbriano RM, Hildebrand M. 2012. Comparative analysis of diatom genomes reveals substantial differences in the organization of carbon partitioning pathways. Algal Res. 1, 2–16. ( 10.1016/j.algal.2012.04.003) [DOI] [Google Scholar]
- 5.Hockin NL, Mock T, Mulholland F, Kopriva S, Malin G. 2012. The response of diatom central carbon metabolism to nitrogen starvation is different from that of green algae and higher plants. Plant Physiol. 158, 299–312. ( 10.1104/pp.111.184333) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Armbrust EV, et al. 2004. The genome of the diatom Thalassiosira pseudonana: ecology, evolution, and metabolism. Science 306, 79–86. ( 10.1126/science.1101156) [DOI] [PubMed] [Google Scholar]
- 7.Ho S-H, Ye X, Hasunuma T, Chang J-S, Kondo A. 2014. Perspectives on engineering strategies for improving biofuel production from microalgae—a critical review. Biotechnol. Adv. 32, 1448–1459. ( 10.1016/j.biotechadv.2014.09.002) [DOI] [PubMed] [Google Scholar]
- 8.Wilhelm C, Jakob T. 2011. From photons to biomass and biofuels: evaluation of different strategies for the improvement of algal biotechnology based on comparative energy balances. Appl. Microbiol. Biotechnol. 92, 909–919. ( 10.1007/s00253-011-3627-2) [DOI] [PubMed] [Google Scholar]
- 9.Wagner H, Jakob T, Wilhelm C. 2006. Balancing the energy flow from captured light to biomass under fluctuating light conditions. New Phytol. 169, 95–108. ( 10.1111/j.1469-8137.2005.01550.x) [DOI] [PubMed] [Google Scholar]
- 10.Gilbert M, Wilhelm C, Richter M. 2000. Bio-optical modelling of oxygen evolution using in vivo fluorescence: comparison of measured and calculated photosynthesis/irradiance (P–I) curves in four representative phytoplankton species. J. Plant Physiol. 157, 307–314. ( 10.1016/S0176-1617(00)80052-8) [DOI] [Google Scholar]
- 11.Kirk JTO. 1994. Light and photosynthesis in aquatic ecosystems. Cambridge, UK: Cambridge University Press. [Google Scholar]
- 12.Fujiki T, Taguchi S. 2002. Variability in chlorophyll a specific absorption coefficient in marine phytoplankton as a function of cell size and irradiance. J. Plankton Res. 24, 859–874. ( 10.1093/plankt/24.9.859) [DOI] [Google Scholar]
- 13.Schramm A, Jakob T, Wilhelm C. 2016. The impact of the optical properties and respiration of algal cells with truncated antennae on biomass production under simulated outdoor conditions. Curr. Biotechnol. 5, 142–153. ( 10.2174/2211550105666151222175121) [DOI] [Google Scholar]
- 14.Schreiber U, Schliwa U, Bilger W. 1986. Continuous recording of photochemical and nonphotochemical chlorophyll fluorescence quenching with a new type of modulation fluorometer. Photosynth. Res. 10, 51–62. ( 10.1007/BF00024185) [DOI] [PubMed] [Google Scholar]
- 15.Goss R, Jakob T. 2010. Regulation and function of xanthophyll cycle-dependent photoprotection in algae. Photosynth. Res. 106, 103–122. ( 10.1007/s11120-010-9536-x) [DOI] [PubMed] [Google Scholar]
- 16.Tozzi S, Schofield O, Falkowski P. 2004. Historical climate change and ocean turbulence as selective agents for two key phytoplankton functional groups. Mar. Ecol. Prog. Ser. 274, 123–132. ( 10.3354/meps274123) [DOI] [Google Scholar]
- 17.Brunet C, Lavaud J. 2010. Can the xanthophyll cycle help extract the essence of the microalgal functional response to a variable light environment? J. Plankton Res. 32, 1609–1617. ( 10.1093/plankt/fbq104) [DOI] [Google Scholar]
- 18.Lavaud J, Strzepek RF, Kroth PG. 2007. Photoprotection capacity differs among diatoms: possible consequences on the spatial distribution of diatoms related to fluctuations in the underwater light climate. Limnol. Oceanogr. 52, 1188–1194. ( 10.4319/lo.2007.52.3.1188) [DOI] [Google Scholar]
- 19.Dimier C, Corato F, Tramontano F, Brunet C. 2007. Photoprotection and xanthophyll-cycle activity in three marine diatoms. J. Phycol. 43, 937–947. ( 10.1111/j.1529-8817.2007.00381.x) [DOI] [Google Scholar]
- 20.Su W, Jakob T, Wilhelm C. 2012. The impact of nonphotochemical quenching of fluorescence on the photon balance in diatoms under dynamic light conditions. J. Phycol. 48, 336–346. ( 10.1111/j.1529-8817.2012.01128.x) [DOI] [PubMed] [Google Scholar]
- 21.Bailleul B, et al. 2015. Energetic coupling between plastids and mitochondria drives CO2 assimilation in diatoms. Nature 524, 366–369. ( 10.1038/nature14599) [DOI] [PubMed] [Google Scholar]
- 22.Claquin P, Kromkamp JC, Martin-Jezequel V. 2004. Relationship between photosynthetic metabolism and cell cycle in a synchronized culture of the marine alga Cylindrotheca fusiformis (Bacillariophyceae). Eur. J. Phycol. 39, 33–41. ( 10.1080/0967026032000157165) [DOI] [Google Scholar]
- 23.Eisenstadt D, Barkan E, Luz B, Kaplan A. 2010. Enrichment of oxygen heavy isotopes during photosynthesis in phytoplankton. Photosynth. Res. 103, 97–103. ( 10.1007/s11120-009-9518-z) [DOI] [PubMed] [Google Scholar]
- 24.Waring J, Klenell M, Bechtold U, Underwood GJC, Baker NR. 2010. Light-induced responses of oxygen photoreduction, reactive oxygen species production and scavenging in two diatom species. J. Phycol. 46, 1206–1217. ( 10.1111/j.1529-8817.2010.00919.x) [DOI] [Google Scholar]
- 25.Lomas MW, Glibert PM. 2000. Comparisons of nitrate uptake, storage, and reduction in marine diatoms and flagellates. J. Phycol. 36, 903–913. ( 10.1046/j.1529-8817.2000.99029.x) [DOI] [Google Scholar]
- 26.Rosenwasser S, et al. 2014. Mapping the diatom redox-sensitive proteome provides insight into response to nitrogen stress in the marine environment. Proc. Natl Acad. Sci. USA 111, 2740–2745. ( 10.1073/pnas.1319773111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Allen AE, Vardi A, Bowler C. 2006. An ecological and evolutionary context for integrated nitrogen metabolism and related signaling pathways in marine diatoms. Curr. Opin. Plant Biol. 9, 264–273. ( 10.1016/j.pbi.2006.03.013) [DOI] [PubMed] [Google Scholar]
- 28.Parker MS, Armbrust EV. 2005. Synergistic effects of light, temperature, and nitrogen source on transcription of genes for carbon and nitrogen metabolism in the centric diatom Thalassiosira pseudonana (Bacillariophyceae). J. Phycol. 41, 1142–1153. ( 10.1111/j.1529-8817.2005.00139.x) [DOI] [Google Scholar]
- 29.Allen AE. 2005. Defining the molecular basis for energy balance in marine diatoms under fluctuating environmental conditions. J. Phycol. 41, 1073–1076. ( 10.1111/j.1529-8817.2005.00156.x) [DOI] [Google Scholar]
- 30.Bender SJ, Parker MS, Armbrust EV. 2012. Coupled effects of light and nitrogen source on the urea cycle and nitrogen metabolism over a diel cycle in the marine diatom Thalassiosira pseudonana. Protist 163, 232–251. ( 10.1016/j.protis.2011.07.008) [DOI] [PubMed] [Google Scholar]
- 31.Mortain-Bertrand A, Descolas-Gros C, Jupin H. 1988. Growth, photosynthesis and carbon metabolism in the temperate marine diatom Skeletonema costatum adapted to low temperature and low photon-flux density. Mar. Biol. 100, 135–141. ( 10.1007/BF00392963) [DOI] [Google Scholar]
- 32.Jakob T, Goss R, Wilhelm C. 2001. Unusual pH-dependence of diadinoxanthin de-epoxidase activation causes chlororespiratory induced accumulation of diatoxanthin in the diatom Phaeodactylum tricornutum. J. Plant Physiol. 158, 383–390. ( 10.1078/0176-1617-00288) [DOI] [Google Scholar]
- 33.Beardall J, Quigg A, Raven JA. 2003. Oxygen consumption: photorespiration and chlororespiration. In Photosynthesis in algae (eds Larkum AWD, Douglas SE, Raven JA), pp. 157–181. Dordrecht, The Netherlands: Springer. [Google Scholar]
- 34.Lavaud L, van Gorkom HJ, Etienne AL. 2002. Photosystem II electron transfer cycle and chlororespiration in planktonic diatoms. Photosynth. Res. 74, 51–59. ( 10.1023/A:1020890625141) [DOI] [PubMed] [Google Scholar]
- 35.Wagner H, Jakob T, Lavaud J, Wilhelm C. 2016. Photosystem II cycle activity and alternative electron transport in the diatom Phaeodactylum tricornutum. Photosynth. Res. 128, 151–161. ( 10.1007/s11120-015-0209-7) [DOI] [PubMed] [Google Scholar]
- 36.Halsey KH, O'Malley RT, Graff JR, Milligan AJ, Behrenfeld MJ. 2013. A common partitioning strategy for photosynthetic products in evolutionarily distinct phytoplankton species. New Phytol. 198, 1030–1038. ( 10.1111/nph.12209) [DOI] [PubMed] [Google Scholar]
- 37.Allen AE, LaRoche J, Maheswari U, Lommer M, Schauer N, Lopez PJ, Finazzi G, Fernie AR, Bowler C. 2008. Whole-cell response of the pennate diatom Phaeodactylum tricornutum to iron starvation. Proc. Natl Acad. Sci. USA 105, 10 438–10 443. ( 10.1073/pnas.0711370105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fanesi A, Wagner H, Wilhelm C. 2016. Temperature affects the partitioning of absorbed light energy in freshwater phytoplankton. Freshw. Biol. 61, 1365–1378. ( 10.1111/fwb.12777) [DOI] [Google Scholar]
- 39.Tilzer MM, Dubinsky Z. 1987. Effects of temperature and day length on the mass balance of Antarctic phytoplankton. Polar Biol. 7, 35–42. ( 10.1007/BF00286822) [DOI] [Google Scholar]
- 40.Gilstad M, Johnsen G, Sakshaug E. 1993. Photosynthetic parameters, pigment composition and respiration rates of the marine diatom Skeletonema costatum grown in continuous light and a 12:12 h light–dark cycle. J. Plankton Res. 15, 939–951. ( 10.1093/plankt/15.8.939) [DOI] [Google Scholar]
- 41.Kroon BMA, Thoms S. 2006. From electron to biomass: a mechanistic model to describe phytoplankton photosynthesis and steady-state growth rates. J. Phycol. 42, 593–609. ( 10.1111/j.1529-8817.2006.00221.x) [DOI] [Google Scholar]
- 42.Finkel ZV, Follows MJ, Liefer JD, Brown CM, Benner I, Irwin AJ. 2016. Phylogenetic diversity in the macromolecular composition of microalgae. PLoS ONE 11, e0155977 ( 10.1371/journal.pone.0155977) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jungandreas A, Wagner H, Wilhelm C. 2012. Simultaneous measurement of the silicon content and physiological parameters by FTIR spectroscopy in diatoms with siliceous cell walls. Plant Cell Physiol. 53, 2153–2162. ( 10.1093/pcp/pcs144) [DOI] [PubMed] [Google Scholar]
- 44.Bowler C, et al. 2008. The Phaeodactylum genome reveals the evolutionary history of diatom genomes. Nature 456, 239–244. ( 10.1038/nature07410) [DOI] [PubMed] [Google Scholar]
- 45.Langner U, Jakob T, Stehfest K, Wilhelm C. 2009. An energy balance from absorbed photons to new biomass for Chlamydomonas reinhardtii and Chlamydomonas acidophila under neutral and extremely acidic growth conditions. Plant Cell Environ. 32, 250–258. ( 10.1111/j.1365-3040.2008.01917.x) [DOI] [PubMed] [Google Scholar]
- 46.Wilhelm C, Jungandreas A, Jakob T, Goss R. 2014. Light acclimation in diatoms: from phenomenology to mechanisms. Mar. Genomics 6, 5–15. ( 10.1016/j.margen.2013.12.003) [DOI] [PubMed] [Google Scholar]
- 47.Norici A, Bazzoni AM, Pugnetti A, Raven JA, Giordano M. 2011. Impact of irradiance on the C allocation in the coastal marine diatom Skeletonema marinoi Sarno and Zingone. Plant Cell Environ. 34, 1666–1677. ( 10.1111/j.1365-3040.2011.02362.x) [DOI] [PubMed] [Google Scholar]
- 48.Xia S, Wan L, Li A, Sang M, Zhang C. 2013. Effects of nutrients and light intensity on the growth and biochemical composition of a marine microalga Odontella aurita. Chin. J. Oceanol. Limnol. 31, 1163–1173. ( 10.1007/s00343-013-2092-4) [DOI] [Google Scholar]
- 49.Fimbres-Olivarria D, Lopez-Elias JA, Martinez-Cordova LR, Carvajal-Millan E, Enriquez-Ocana F, Valdez-Holguin E, Miranda-Baeza A. 2015. Growth and biochemical composition of Navicula sp. cultivated at two light intensities and three wavelengths. Isr. J. Aquac. Bamidgeh 67, 1–7. (doi:hdl.handle.net/10524/49183) [Google Scholar]
- 50.Shifrin NS, Chisholm SW. 1981. Phytoplankton lipids: interspecific differences and effects of nitrate, silicate and light–dark cycles. J. Phycol. 17, 374–384. ( 10.1111/j.1529-8817.1981.tb00865.x) [DOI] [Google Scholar]
- 51.Guerra LT, Levitan O, Frada MJ, Sun JS, Falkowski PG, Dismukes GC. 2013. Regulatory branch points affecting protein and lipid biosynthesis in the diatom Phaeodactylum tricornutum. Biomass Bioenergy 59, 306–315. ( 10.1016/j.biombioe.2013.10.007) [DOI] [Google Scholar]
- 52.Alipanah L, Rohloff J, Winge P, Bones AM, Brembu T. 2015. Whole-cell response to nitrogen deprivation in the diatom Phaeodactylum tricornutum. J. Exp. Bot. 66, 6281–6296. ( 10.1093/jxb/erv340) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yu ET, Zendejas FJ, Lane PD, Gaucher S, Simmons BA, Lane TW. 2009. Triacylglycerol accumulation and profiling in the model diatoms Thalassiosira pseudonana and Phaeodactylum tricornutum (Baccilariophyceae) during starvation. J. Appl. Phycol. 21, 669 ( 10.1007/s10811-008-9400-y) [DOI] [Google Scholar]
- 54.Bertozzini E, Galluzzi L, Ricci F, Penna A, Magnani M. 2013. Neutral lipid content and biomass production in Skeletonema marinoi (Bacillariophyceae) culture in response to nitrate limitation. Appl. Biochem. Biotechnol. 170, 1624–1636. ( 10.1007/s12010-013-0290-3) [DOI] [PubMed] [Google Scholar]
- 55.Bittar TB, Lin Y, Sassano LR, Wheeler BJ, Brown SL, Cochlan WP, Johnson ZI. 2013. Carbon allocation under light and nitrogen resource gradients in two model marine phytoplankton. J. Phycol. 49, 523–535. ( 10.1111/jpy.12060) [DOI] [PubMed] [Google Scholar]
- 56.Strzepek RF, Harrison PJ. 2004. Photosynthetic architecture differs in coastal and oceanic diatoms. Nature 431, 689–692. ( 10.1038/nature02954) [DOI] [PubMed] [Google Scholar]
- 57.Marchetti A, Varela DE, Lance VP, Johnson Z, Palmucci M, Giordano M, Armbrust EV. 2010. Iron and silicic acid effects on phytoplankton productivity, diversity, and chemical composition in the central equatorial Pacific Ocean. Limnol. Oceanogr. 55, 11–29. ( 10.4319/lo.2010.55.1.0011) [DOI] [Google Scholar]
- 58.Strzepek RF, Hunter KA, Frew RD, Harrison PJ, Boyd PW. 2012. Iron-light interactions differ in Southern Ocean phytoplankton. Limnol. Oceanogr. 57, 1182–1200. ( 10.4319/lo.2012.57.4.1182) [DOI] [Google Scholar]
- 59.de Baar HJW, et al. 2005. Synthesis of iron fertilization experiments: from the Iron Age in the Age of Enlightenment. J. Geophys. Res. Oceans 110, C09S16 ( 10.1029/2004JC002601) [DOI] [Google Scholar]
- 60.Smith SR, et al. 2016. Transcriptional orchestration of the global cellular response of a model pennate diatom to diel light cycling under iron limitation. PLoS Genet. 12, e1006490 ( 10.1371/journal.pgen.1006490) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nunn BL, Faux JF, Hippmann AA, Maldonado MT, Harvey HR, Goodlett DR, Boyd PW, Strzepek RF. 2013. Diatom proteomics reveals unique acclimation strategies to mitigate Fe limitation. PLoS ONE 8, e75653 ( 10.1371/journal.pone.0075653) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sackett O, et al. 2014. Taxon-specific responses of Southern Ocean diatoms to Fe enrichment revealed by synchrotron radiation FTIR microspectroscopy. Biogeosciences 11, 5795–5808. ( 10.5194/bg-11-5795-2014) [DOI] [Google Scholar]
- 63.Bothwell ML. 1988. Growth rate responses of lotic periphytic diatoms to experimental phosphorus enrichment: the influence of temperature and light. Can. J. Fish. Aquat. Sci. 45, 261–270. ( 10.1139/f88-031) [DOI] [Google Scholar]
- 64.Cruz de Carvalho MH, Sun H-X, Bowler C, Chua N-H. 2016. Noncoding and coding transcriptome responses of a marine diatom to phosphate fluctuations. New Phytol. 210, 497–510. ( 10.1111/nph.13787) [DOI] [PubMed] [Google Scholar]
- 65.Litchman E, Steiner D, Bossard P. 2003. Photosynthetic and growth responses of three freshwater algae to phosphorus limitation and daylength. Freshw. Biol. 48, 2141–2148. ( 10.1046/j.1365-2427.2003.01157.x) [DOI] [Google Scholar]
- 66.Suroy M, Panagiotopoulos C, Boutorh J, Goutx M, Moriceau B. 2015. Degradation of diatom carbohydrates: a case study with N- and Si-stressed Thalassiosira weissflogii. J. Exp. Mar. Biol. Ecol. 470, 1–11. ( 10.1016/j.jembe.2015.04.018) [DOI] [Google Scholar]
- 67.Soler C, Claquin P, Goutx M, Ragueneau O, Moriceau B. 2010. Impact of nutrient starvation on the biochemical composition of the marine diatom Thalassiosira weissflogii: from the whole cell to the frustule fraction. Biogeosci. Discuss. 7, 5953–5995. ( 10.5194/bgd-7-5953-2010) [DOI] [Google Scholar]
- 68.Sackett O, Petrou K, Reedy B, De Grazia A, Hill R, Doblin M, Beardall J, Ralph P, Heraud P. 2013. Phenotypic plasticity of Southern Ocean diatoms: key to success in the sea ice habitat? PLoS ONE 8, e81185 ( 10.1371/journal.pone.0081185) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Qin S, Lin H, Jiang P. 2012. Advances in genetic engineering of marine algae. Biotechnol. Adv. 30, 1602–1613. ( 10.1016/j.biotechadv.2012.05.004) [DOI] [PubMed] [Google Scholar]
- 70.Fernie AR, Obata T, Allen AE, Araújo WL, Bowler C. 2012. Leveraging metabolomics for functional investigations in sequenced marine diatoms. Trends Plant Sci. 17, 395–403. ( 10.1016/j.tplants.2012.02.005) [DOI] [PubMed] [Google Scholar]
- 71.Muhseen ZT, Xiong Q, Chen Z, Ge F. 2015. Proteomics studies on stress responses in diatoms. Proteomics 15, 3943–3953. ( 10.1002/pmic.201500165) [DOI] [PubMed] [Google Scholar]
- 72.Palenik B. 2015. Molecular mechanisms by which marine phytoplankton respond to their dynamic chemical environment. Annu. Rev. Mar. Sci. 7, 325–340. ( 10.1146/annurev-marine-010814-015639) [DOI] [PubMed] [Google Scholar]
- 73.Kroth PG, et al. 2008. A model for carbohydrate metabolism in the diatom Phaeodactylum tricornutum deduced from comparative whole genome analysis. PLoS ONE 3, e1426 ( 10.1371/journal.pone.0001426) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Fabris M, Matthijs M, Rombauts S, Vyverman W, Goossens A, Baart GJ. E. 2012. The metabolic blueprint of Phaeodactylum tricornutum reveals a eukaryotic Entner–Doudoroff glycolytic pathway. Plant J. 70, 1004–1014. ( 10.1111/j.1365-313X.2012.04941.x) [DOI] [PubMed] [Google Scholar]
- 75.Levering J, et al. 2016. Genome-scale model reveals metabolic basis of biomass partitioning in a model diatom. PLoS ONE 11, e0155038 ( 10.1371/journal.pone.0155038) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hildebrand M, Manandhar-Shrestha K, Abbriano R. 2017. Effects of chrysolaminarin synthase knockdown in the diatom Thalassiosira pseudonana: implications of reduced carbohydrate storage relative to green algae. Algal Res. 23, 66–77. ( 10.1016/j.algal.2017.01.010) [DOI] [Google Scholar]
- 77.Daboussi F, et al. 2014. Genome engineering empowers the diatom Phaeodactylum tricornutum for biotechnology. Nat. Commun. 5, 3831 ( 10.1038/ncomms4831) [DOI] [PubMed] [Google Scholar]
- 78.Haimovich-Dayan M, Garfinkel N, Ewe D, Marcus Y, Gruber A, Wagner H, Kroth PG, Kaplan A. 2013. The role of C4 metabolism in the marine diatom Phaeodactylum tricornutum. New Phytol. 197, 177–185. ( 10.1111/j.1469-8137.2012.04375.x) [DOI] [PubMed] [Google Scholar]
- 79.Yang J, Pan Y, Bowler C, Zhang L, Hu H. 2016. Knockdown of phosphoenolpyruvate carboxykinase increases carbon flux to lipid synthesis in Phaeodactylum tricornutum. Algal Res. 15, 50–58. ( 10.1016/j.algal.2016.02.004) [DOI] [Google Scholar]
- 80.Niu Y-F, Zhang M-H, Li D-W, Yang W-D, Liu J-S, Bai W-B, Li H-Y. 2013. Improvement of neutral lipid and polyunsaturated fatty acid biosynthesis by overexpressing a Type 2 diacylglycerol acyltransferase in marine diatom Phaeodactylum tricornutum. Mar. Drugs 11, 4558–4569. ( 10.3390/md11114558) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhu B-H, Shi H-P, Yang G-P, Lv N-N, Yang M, Pan K-H. 2016. Silencing UDP-glucose pyrophosphorylase gene in Phaeodactylum tricornutum affects carbon allocation. New Biotechnol. 33, 237–244. ( 10.1016/j.nbt.2015.06.003) [DOI] [PubMed] [Google Scholar]
- 82.Reinfelder JR. 2011. Carbon concentrating mechanisms in eukaryotic marine phytoplankton. Annu. Rev. Mar. Sci. 3, 291–315. ( 10.1146/annurev-marine-120709-142720) [DOI] [PubMed] [Google Scholar]
- 83.Tanaka T, et al. 2015. Oil accumulation by the oleaginous diatom Fistulifera solaris as revealed by the genome and transcriptome. Plant Cell 27, 162–176. ( 10.1105/tpc.114.135194) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kim J, Fabris M, Baart G, Kim MK, Goossens A, Vyverman W, Falkowski PG, Lun DS. 2016. Flux balance analysis of primary metabolism in the diatom Phaeodactylum tricornutum. Plant J. 85, 161–176. ( 10.1111/tpj.13081) [DOI] [PubMed] [Google Scholar]
- 85.Lepetit B, Dietzel L. 2015. Light signaling in photosynthetic eukaryotes with ‘green’ and ‘red’ chloroplasts. Environ. Exp. Bot. 114, 30–47. ( 10.1016/j.envexpbot.2014.07.007) [DOI] [Google Scholar]
- 86.Schellenberger BS, Jungandreas A, Jakob T, Weisheit W, Mittag M, Wilhelm C. 2013. Blue light is essential for high light acclimation and photoprotection in the diatom Phaeodactylum tricornutum. J. Exp. Bot. 64, 483–493. ( 10.1093/jxb/ers340) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Jungandreas A, Schellenberger Costa B, Jakob T, von Bergen M, Baumann S, Wilhelm C. 2014. The acclimation of Phaeodactylum tricornutum to blue and red light does not influence the photosynthetic light reaction but strongly disturbs the carbon allocation pattern. PLoS ONE 9, e99727 ( 10.1371/journal.pone.0099727) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This article has no additional data.